Summary
Recent advances and accomplishments of artificial intelligence (AI) and deep generative models have established their usefulness in medicinal applications, especially in drug discovery and development. To correctly apply AI, the developer and user face questions such as which protocols to consider, which factors to scrutinize, and how the deep generative models can integrate the relevant disciplines. This review summarizes classical and newly developed AI approaches, providing an updated and accessible guide to the broad computational drug discovery and development community. We introduce deep generative models from different standpoints and describe the theoretical frameworks for representing chemical and biological structures and their applications. We discuss the data and technical challenges and highlight future directions of multimodal deep generative models for accelerating drug discovery.
Deep generative models hold great promise as powerful approaches for drug design. Zeng et al. review the current deep generative models and their applications in discovering small molecules and macromolecules, discuss the data and technical challenges, and highlight future directions in improving deep generative models for drug discovery communities.
Introduction: Deep generative models in drug discovery
A recent study estimates that pharmaceutical companies spent $2.6 billion in 2015 for the development of new, US Food and Drug Administration-approved drugs, up from $802 million in 2003.1 Although more direct costs are incurred during clinical trials, since the preclinical investment comes earlier the capitalized costs of the two stages are roughly equal. Recent advances in computational sciences and technologies capture the requisites and urgencies and provide a set of potentially promising approaches. Among these, the developers can select the right artificial intelligence (AI) to target the problem at hand, in particular deep generative models, appropriate protocol, and factors. Collectively, they map paths that integrate biology, chemistry, computational science, pharmacology, and disease treatments.
The rapid growth in computing power, amount of data, and advanced algorithms has led to breakthroughs in AI for drug discovery,2 especially in the application of deep generative models.3,4,5 The models have emerged as high potential tools to transform the design, optimization, and synthesis of small molecules, and macromolecules (Figure 1). Applications of deep generative models have already delivered new partially optimized candidate leads, in some cases in less time typically required by conventional sequential approaches.6,7,8,9,10 If applied on a large scale, deep generative modeling has the potential of boosting the development (R&D) process.
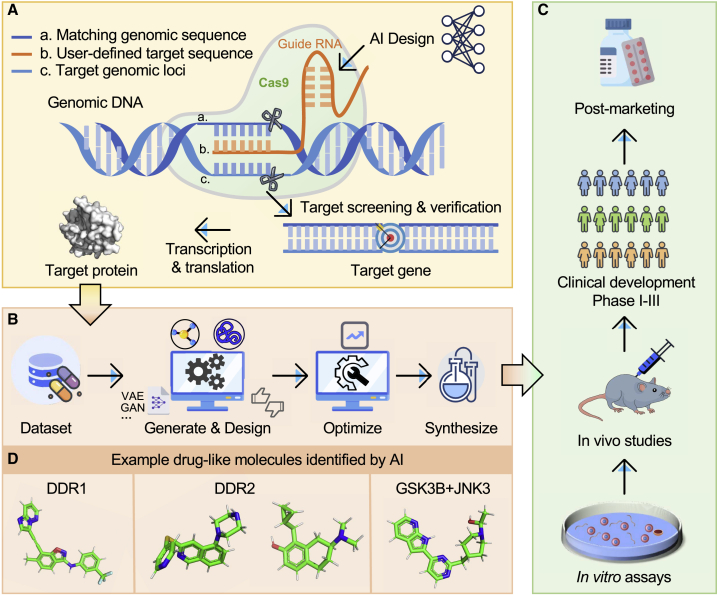
Figure 1.
AI and deep generative model applications in the drug discovery pipeline
Several successful applications of AI and deep generative models in various stage of the drug development pipeline: (A) AI-assistant target selection and validation, (B) molecular design, lead optimization, and chemical synthesis, (C) biological evaluation (in vitro and in vivo), clinical development, and post marketing surveillance, and (D) several successful preclinical and clinical molecules identified by AI and deep generative models. DDR1, discoidin domain receptor 1; DDR2, discoidin domain receptor tyrosine kinase 2; GSK3B, glycogen synthase kinase 3 beta; JNK3, c-Jun N-terminal kinase 3.
Deep generative models correspond to a theoretical framework for generating novel chemical and biological structures with desired properties using data structures, such as graphs and fingerprints, and operations, such as the flow of functional or experimental information. Creative deep generative models can significantly promote algorithm development and application in drug discovery. In this “big data” era, deep generative models would offer a cutting-edge technology that could revolutionize an informatics view of biology, disease, and therapeutics. In this review, we describe classical and state-of-the-art deep generative models and their applications (Figure 1) in computational drug discovery and discuss limitations and challenges. Our aim is to provide an overview of current tools and techniques (the toolbox) of deep generative models in multiple applications on small-molecule and macromolecular systems.
The toolboxes for deep generative models
Designing a novel drug is a complex undertaking that needs to satisfy pre-defined criteria for on-target potency, specificity relative to off-targets, physical properties, and other chemistry and biology measures. Traditional methods, which require chemists to select and validate candidate molecules experimentally from a vast chemical space, are ineffective. Deep generative models have become popular because they can automatically generate new bioactive and synthesizable molecules in a time- and cost-effective way.
Big biomedical datasets for drug discovery
We begin with a brief overview of several commonly used chemical and bioinformatics databases, which provide both labeled and unlabeled data to train, validate, and test deep generative models for the drug discovery community. Pharmaceutical companies have their in-house proprietary collections on the order of 2–3M compounds with associated data from past drug discovery quests. In the public domain, the ZINC database collected nearly 2 billion purchasable, commercially available, “drug-like” compounds for in silico screening.11 Its massive size makes it also useful for learning molecular patterns for pre-training generative models. Bioactive molecules, such as those in the manually curated ChEMBL database, which approaches 1.5M of real bioactive molecules with every molecule having at least one experimental bioactivity measurement,12 are of particular interest. They can be used for training models to generate molecules with certain properties. The GDB-17 database13 enumerates most organic molecules (166.4 billion) of up to 17 heavy atoms of C, N, O, S, and halogens. This includes many of the lower-molecular-weight small-molecule drugs as well as the smaller typical lead compounds. Ultra-large chemical databases,14 such as Enamine (https://enamine.net) and REALdb,15 contain billions of synthesizable compounds identified by chemoinformatics approaches and expert-system type rules. These ultra-large databases offer an opportunity to train models with broadened applicability. In addition to small-molecule resources, several macromolecular databases offer enriched data for generative model training in macromolecule design, such as the PDB.16
Representation of compounds/molecules
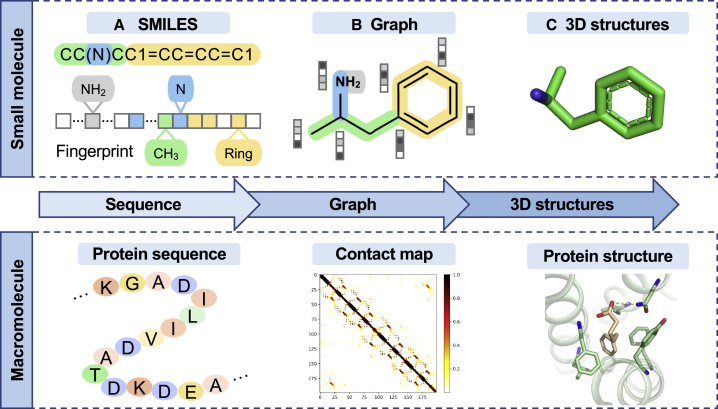
The representation of molecules is important for generative models. There are three types of representations: (1) sequence based, (2) graph based, and (3) images (Figure 2). The unprecedented success of natural language processing (NLP) inspired the idea to describe molecules in symbols in a way analogous to human language. Semantics and grammars in biological structures bear a resemblance to human language; hence, molecules can be represented as sequences of characters. De novo small-molecule designs generally use simplified molecular input line entry systems (SMILES).17 The sequence-based structure is generated by following the SMILES grammar rules encoded into vectors (Figure 2A). A more direct method to represent molecules is graph based.18 In the graph representation, the atoms of a small molecule form a set of nodes and the bonds are regarded as edges (Figure 2B). For macromolecules, a contact map19 is a graph that denotes the distance between any two amino acid residue pairs. Training graph-based models on a large number of nodes is expensive because the space complexity increases with the square of their number.20 Compared with sequence-based approaches, graph-based representations are easy to implement as graph convolutional layers, and bond weights can be optimized in message-passing networks. Sequence-based representations are in general compact, memory-efficient, and easily searchable. However, both sequence-based and graph-based approaches cannot capture the 3D information of ligands or proteins in biologically meaningful ligand-protein interactions. The 3D conformation of a molecule captures the relative orientation of atoms21,22,23,24 (Figure 2C). Several latest 3D representations were presented as well.25,26,27 DEVELOP incorporate an existing graph-based deep generative model, De-Linker, along with a convolutional neural network to utilize 3D representations of molecules and target pharmacophores.28 DeepLigBuilder is a graph-based generative model that utilizes 3D structural representation of ligand-receptor interactions for the end-to-end design of chemically and conformationally valid 3D molecules with drug-likeness properties.29 Traditional image or 3D representation of proteins requires accurate 3D structural data from cryoelectron microscopy and crystallography, which is challenging to obtain. Recent AI approaches, such as AlphaFold2, can provide massive protein 3D data to address these challenges.30
Figure 2.
A diagram illustrating three molecular representation approaches
Three molecular representation approaches include: (A) one-dimensional (1D) sequence-based representation; (B) graph-based representation; and (C) 3D representation for both small molecules and macromolecules (i.e., proteins). The value of contact map matrix is 1 if the distance is greater than a predetermined threshold, otherwise it is 0.
Recurrent neural networks
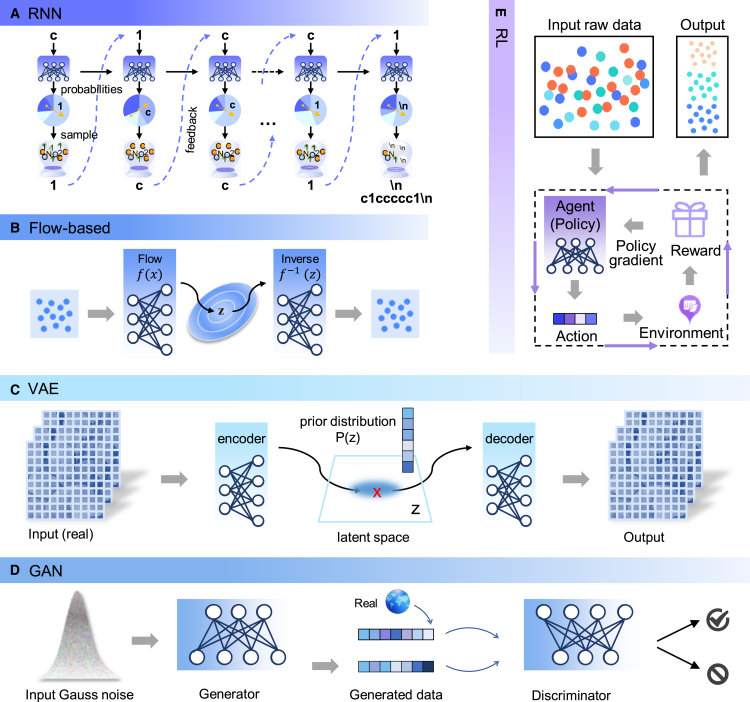
Recurrent neural networks (RNNs) are fundamental components of generative neural networks in processing human language. They are useful for modeling systems that have a sequential or time component and have been powerful in NLP automated computer code generation31 and musical composition.32 The language of molecules, such as SMILES, is similar to human language. Thus, it is natural to use RNNs for generating molecules based on sequential representation. As depicted in Figure 3A, SMILES (i.e., “c1cc … c1”) can be generated by RNNs in the following way. RNNs receive the first character “c” and assign different probabilities to possible next characters: character “1” would receive a high probability and may be sampled as the next one. “1” is feedback input to RNNs. This process is repeated until the end token “\n” is generated. Long short-term memory (LSTM)33 and gated recurrent unit (GRU)34 introduce a gate mechanism to remember valuable input information for a long series of steps, lacking in traditional RNNs. Whether LSTM or GRU is preferable may depend on the specific application. LSTM cell can hold much longer history than GRU. However, additional parameters in LSTM may increase the risk of overfitting. RNNs with LSTM or GRU are among the most promising for the generation of de novo small molecules under the representation of SMILES.35
Figure 3.
A diagram illustrating the theory framework of five deep generative models (A–E) in the drug discovery applications
RNN, recurrent neural networks; VAE, variational autoencoder; GAN, generative adversarial networks; RL, reinforcement learning.
Variational autoencoder
An autoencoder (AE) is constructed of two networks: (1) one (the encoder) is trained to map the input into a low-dimensional latent vector, and (2) the other (the decoder) to map the latent vector into the inputted data. The original AE creates a latent space by reproducing the input. To avoid overfitting and discontinuities in the original AE, variational AE (VAE) regularizes the latent space by replacing latent space points with distributions. In a pioneering work, VAE was employed for molecule generation, ushering in a new strategy in de novo drug design.10 As shown in Figure 3C, the encoder is trained to map the molecules (e.g., SMILES) into a low-dimensional latent vector that is assumed to be sampled from a Gaussian distribution, and the decoder to map the latent vector into the inputted molecules (e.g., SMILES). The latent vectors are constrained to follow a probability distribution (usually Gaussian distribution) so that a molecule is represented as an explicit probability distribution over latent space. When the encoder and decoder are trained jointly, the output must reconstruct the training samples’ probability distribution. Recently, learning disentangled representations for VAE has attracted increasing attention, where the main goal is to make each latent variable of the latent vector encode an independent property or factor of data.36 If disentangled VAE is successfully introduced for molecular generation, a molecular property can be edited without changing other properties, by editing the latent variables associated with that property.
Generative adversarial networks
The invention of generative adversarial networks (GANs)37 started a flurry of generative models. Unlike VAE, GANs do not work with an explicit probability density function (Figure 3D), but provide an adversarial training framework composed of a generator and a discriminator. The discriminator trains a classification model aiming at maximizing the error rate of synthetic molecules from the generator, which resemble the real data. The generator and the discriminator are trained together in an adversarial, zero-sum game, until the discriminator model is fooled, meaning the generator network is generating plausible (i.e., realistic fake) molecules.
Flow-based models
VAE and GAN do not explicitly model the real probability density function. VAE implicitly optimizes the log likelihood of the data by maximizing a lower bound on a likelihood function, whereas GAN avoids modeling the distribution but learns in an adversarial way to measure the difference between “valid molecules” and “synthetic molecules.” Deep flow-based models resolve the intractability issue of explicit density estimation by leveraging normalizing flow.38 A normalizing flow is an invertible deterministic transformation between the raw data space and latent space (Figure 3B). For example, a recent method called MoFlow learns a chain of transformation to map valid molecules to their latent representations, and the reverse chain of transformation to map the latent representations to valid molecules.39 One major limitation for the flow-based models is that they are time consuming due to the complex hyperparameter tuning processes. To take full advantage of the flow-based models, the molecular graphs must be transformed into continuous data by incorporating real-value noise into the molecular generation flow.
Reinforcement learning
Deep RL has emerged as one of the most prominent toolboxes for optimizing an objective, especially with recent breakthroughs, such as AlphaGo.40 The immensity of the chemical space is similar to Go’s enormous possible solution space; hence, RL is a potential method for exploring the chemical space by a dynamic decision process.41 As depicted in Figure 3E, RL—consisting of an agent, a reward function, and environment—aims to optimize toward a user-directed target. The agent chooses the next action, and the reward function evaluates the quality of the actions according to the environment (domain-specific rules) and provides feedback to the agent. After the generative model is trained on a large and general set of molecules to learn the SMILES grammar, RL can be applied as a technique for fine-tuning of target properties, such as synthetic accessibility42 and quantitative estimate of druglikeness,43 which assesses physical properties. For example, policy gradient for forward synthesis (PGFS) (more below) was proposed to generate synthetically accessible molecules using RL.44 For this, (1) the agent is a neural network; (2) the policy actions are chemical transformations executed by modifying a molecule by adding or removing atoms and bonds; and (3) the reward is synthetic accessibility.44
Applications in small-molecule drug design
Conventional exploration, such as virtual screening,45,46 needs to navigate a vast chemical space, posing time and cost challenges. De novo design, a technique of automatically generating molecules with desired properties from scratch, has benefitted from advances in deep generative models.47 Here, we describe their applications toward various design purposes.
Generating valid small molecules
As deep generative models for de novo small-molecule design were emerging, research initially focused on how to generate molecules with high validity, with a particular emphasis on the grammar and semantics of small molecules. In 2016, Gómez-Bombarelli et al. pioneered a data-driven method that generates molecules by mapping discrete high-dimensional chemical space to and from continuous latent space.10 The model showed that training VAE jointly with a molecular property prediction task and optimizing via a Gaussian process were promising. This paradigm promoted the development of de novo small-molecule design, even if the output included invalid molecules. Subsequently, inspired by the compiler theory where the syntax and semantics check is done via syntax-directed translation (SDT), Dai et al. incorporated SDT into VAE for constraining the decoder.48 The proposed model (SD-VAE) can generate both syntactically and semantically valid molecules.48
Previous works achieved high validity by incorporating extra constraints. Inspired by fragment-based drug discovery, Jin et al. proposed junction tree variational encoder (JT-VAE).49 JT-VAE considers chemically valid substructures, such as aromatic rings as nodes in the graph structure. A molecular graph assembled by these nodes can maintain chemical validity without implementing additional chemical rules. JT-VAE reached 100% validity due to obeying the ground truth in chemistry by generating bioactive molecules from fragments. A new AE, the Wasserstein autoencoder character (cWAE),50 incorporates adversarial training and has shown improved model accuracy. When applied to molecular design and trained on 1.6 billion compounds, compared with JT-VAE, cWAE produces an accurate generative model (the compound reconstruction error is reduced by over 80%).51 MoFlow39 generates a molecular graph in a one-shot manner that generates bonds and atoms by a flow-based model and then assembles them into a molecular graph. Instead, MolGrow52 generates a molecular graph in an iterative manner, termed a hierarchical normalizing flow model via generating molecular graphs from a single-node graph by recursively splitting every node into two. Experimental results show that both MoFlow and MolGrow can generate 100% valid molecules.
Generating molecules with drug-like properties
With the gradual maturity of generative models, molecular generative models have been aiming to find molecules with specific properties, not only focusing on their validity. Drug-like properties, such as biological activity and synthetic accessibility, are critical for the success of drug candidates. In 2020, a molecular GAN model53 conditioned on gene expression signatures was shown to generate molecules with a high probability to induce a desired transcriptomic profile.
Generative tensorial reinforcement learning (GENTRL)54 was designed to generate novel molecules that can inhibit DDR1 (discoidin domain receptor 1) by designing a reward function. The generated molecules were evaluated using in vitro and in vivo mouse assays to verify the binding affinity on DDR1 and the preclinical and pharmacokinetic properties. With a time frame of 46 days from target selection to partially validated molecule, GENTRL validated a promising outlook for accelerating drug discovery (Figure 1D). Notably, GENTRL leveraged a set of relevant information which is frequently available, such as crystal structure data and information related to active compounds. This model is not generalizable to cases where target-specific activity data are unavailable, and a model requiring less information could be more practical in such cases.
PGFS44 was designed to generate molecules that can be feasibly synthesized. PGFS treats the molecular generation problem as a sequential decision process of selecting reactant molecules and reaction transformation in a linear synthetic sequence, where the choice of reactants is considered an action and synthetic accessibility a reward. PGFS has been validated in an in-silico proof-of-concept associated with three HIV targets.44
Generating molecules with multi-objective drug-like properties
Generative models for de novo molecular generation are able to design molecules with multiple design constraints such as potency, safety, and desired metabolic profile. Molecules with such constraints will better meet the requirements of drug discovery. RationaleRL55 trained a graph-based RL model to complete a pre-selected molecular subgraph into an integral molecule with several desired co-existing properties, such as bioactivities toward multiple targets (e.g., GSK3β and JNK3; Figure 1D), quantitative estimate of drug-likeness, and synthetic accessibility. As part of multi-objective optimization, the predictiveness to drug-likeness has been significantly improved by combining individual classifiers and calculating their Bayesian errors. The difficulty lies in how to define and characterize non-drug-like molecules.56
Generating better bioavailable molecules with optimization
Molecular optimization aims toward desired properties for a given starting molecule. This process is analogous to image-to-image translation (e.g., turn horses into zebras) in computer vision or style transfer in NLP. Jin et al. presented an optimization method inspired by style transfer.57 Molecular optimization can be formulated as graph-to-graph translation via converting one molecular graph to another with better properties using the paired training sets.
Inspired by the image-to-image translation approach that CycleGAN58 learned to translate an image from a source domain X to a target domain Y in the absence of paired examples, Mol-CycleGAN59 was proposed and trained on two datasets with and without a desired property. The training framework consists of two GANs forming a cycle: (1) the first GAN is used to generate molecules with the desired property when the input is not equipped with the target property, and (2) the second network has the opposite input/output order. The objective of the model is to minimize the distance between the original molecules and the generated molecules of the second network.
Capturing 3D information of ligand-protein interactions
In an attempt to bring 3D protein structure information directly into generative molecule creation rather than by post-generation docking, a high-quality target family sequence alignment was leveraged to identify binding site residues across the kinase family and train 1D string representation of the PaccMann model.60 The quantitative structure-activity relationship (QSAR) model built with this reduced dataset outperformed the QSAR model built with the conventional full-sequence approach, and the molecules created with the generative model were likewise encouraging in terms of their similarity to validated kinase inhibitors.61
Applications in macromolecular drug design
In addition to designing small molecules, the application of AI has been extended to the design of medicinal macromolecules, such as designing antimicrobial peptides (AMPs), therapeutic proteins, and CRISPR-Cas9 systems design and optimization, as detailed below.
AMP generation
The emergence of antibiotic-resistant bacteria led to nearly 1 million deaths worldwide each year from bacterial infections that cannot be treated with ordinary antibiotics.62 AMPs increase the repertoire and deep generative models are a promising way of designing them. Das et al. augmented a variant of VAE (Wasserstein Autoencoder)63 with molecular dynamics information to generate AMPs with broad-spectrum potency and low toxicity.64 For a controlled sequence generation, linear binary classifiers conditional latent (attribute) space sampling (CLaSS) for attribute prediction was trained on the latent space, and then rejected sampling was utilized for screening the molecules of interest. CLaSS can be trained for binary classification of antimicrobial function, broad-spectrum efficacy, presence of secondary structures, and toxicity at the same time. Within 48 days, two new antimicrobial peptides with high potency against Gram+ and Gram− bacteria were synthesized and tested in vitro and in mice. Both resulted in low resistance in Escherichia coli and low toxicity. Another example of antibiotic discovery emerged from combining the message-passing approach and experimental assays to predict the growth inhibition of E. coli followed by screening an existing compound library to identify molecules with antimicrobial activity and different structures from known antibiotics.9 In the message-passing approach, the processors execute a task independently and communicate data between them by exchanging messages.
Therapeutic protein generation
De novo protein design plays important roles in protein therapies. For instance, a de novo design strategy was proposed to produce rapidly and accurately decoy proteins by replicating the protein interface of human angiotensin I-converting enzyme 2 (hACE2) for a potential treatment of coronavirus disease 2019 (COVID-19).65 Deep generative models can also be used to design protein therapies by modeling the spatial properties of the amino acid sequence. ProteinGAN,66 which incorporated a self-attention mechanism into GAN and learned the evolutionary relationships of protein sequences, was a generalizable framework to generate protein sequences with specific functions. About 24% of the generated sequences were soluble and showed activity comparable with the wild types, including some highly mutated sequences. The generated sequences include 119 novel structural sequence motifs, not present in the training dataset, showcasing de novo generation of functional proteins for therapeutic development.
CRISPR-Cas9 systems design and optimization
The CRISPR-Cas9 system, consisting of a Cas9 nuclease and a guide RNA (gRNA), is a technology for genome editing and a tool to identify targets in drug discovery (Figure 1A). Based on the principle of complementary base pairing, gRNA guides Cas protein localization to the genome and CRISPR KO (knockout). CRISPRi (interference) and CRISPRa (activation) technologies then determine whether the candidate genes are the key to disease and thus a therapeutic target. The selection of gRNA sequences affects knockout efficacy and is essential for target identification. Recent studies have demonstrated the power of deep learning algorithms, such as CNNs and RNNs, to design and optimize CRISPR-Cas9 systems. Recently, Chuai et al. proposed a design tool called DeepCRISPR for gRNA with high sensitivity and specificity, which adopts a combination of unsupervised and supervised CNNs to learn the representations of gRNAs.67 DeepCRISPR can predict on-target knockout efficacy and off-target profile in the same framework. In addition, it automatically detects important features of optimized gRNAs to promote effective CRISPR design. SpCas9 genome editing tools68 can address the off-target issue. A DeepHF model, which combined RNNs with the secondary structure, GC content, and thermodynamics features was developed, but could not be automatically obtained by RNNs.69 Although deep learning models have conveniently facilitated CRISPR-Cas9 systems design, these data-driven approaches are subject to the problems of data heterogeneity, sparsity, and imbalance.67 CRISPR-Cas9 systems design can be further optimized using advanced algorithms with higher-quality data.
Outstanding questions, perspective, and future direction
Despite the enthusiasm for AI-enabled drug discovery, questions and challenges abound. For decades, translational science has been facing the challenge of how to translate research findings into a novel, more effective medicine.70 In fact, the “ultimate goal of the translational challenge is to eliminate the Valley of Death, through scientific understanding and innovation.”71 Most machine learning models in the drug discovery pipeline require large volumes of data for training and validation, particularly deep learning models.72 The lack of adequate quality and robust data-sharing practices remain critical barriers for machine learning models to positively impact drug discovery.73 Inadequate data quality can lead to models that have poor generalizability. Data harmonization, which improves the data quality and utilization via domain knowledge and machine learning techniques, plays a crucial role in the development and application of drug discovery.74 Here, we briefly discuss several challenges and potential future directions as follows.
Interpretable generative models
While generative models and other deep learning-based approaches offer great potential, they are often essentially “black boxes” that require objective algorithmic interpretation of the predictions to provide confidence and actionability. Drug discovery is a highly complex process involving interactions between compounds and targets and interconnected biological systems. Current deep generative models are limited to capturing shallow statistical correlations of the data, which cannot explain mechanisms and results, possibly misleading decisions. Thus, model users must understand how the algorithms are constructed, which data they rely on, and to what extent the models are reliable. It is also important for AI scientists to involve biologists and clinicians in experimental design and data interpretation.
Models should be made interpretable.75 One way is to perturb the input or parameters in the model and observe how the results change. For example, controllable molecular generation can be achieved by disentanglement, which decomposes the latent space into interpretable and independent factors that correspond to each property,76 such as bioactivity and synthesizability. In this way, molecules with desired properties can be generated. Another solution can be displaying more semantic information from the algorithm to explain the causality of the results. The reasoning of relationships between molecular structures and drug-like properties may guide the construction of causal graphs followed by molecule generation. Models can also be made transparent. Algorithms rationalize their prediction processes in a way that a human can understand. A hierarchical generative model may better trace each step back to previous levels, allowing for human-computer interaction to achieve targeted optimization.77
Few-shot generative models
Current AI techniques rely on learning from large amounts of data. However, the available data are often quantitatively imbalanced due to, e.g., privacy, security, ethics,78 or a small number of patients suffering from rare diseases, leading to little clinical data about the toxicity and poor bioactivity. Such situations could be alleviated by machines that learn from few samples. Combined with past knowledge, they can achieve good performance. Here, we highlight strategies to address insufficient data.
Starting from the source is the intuitive way to solve problems. Increasing the sample size can be achieved through data augmentation. Some approaches change the starting atom and the branching order in SMILES to enrich the data, taking advantage of the non-uniqueness of SMILES sequences for a structure.79 Graph-based data can be varied by adding or removing edges using appropriate strategies,80 such as 3D conformations.81 This can be compounded by information at different granularity (e.g., atomic, pharmacophore, and toxicophore levels).
Insufficient training data of specific targets is inevitable in de novo molecular generation, especially for peptide or protein design. Transfer learning aims to transfer knowledge learned from one domain to a target domain related to the source domain, as solving data scarcity of the target domain.82 Transfer learning drives molecule generation toward desired properties commonly in a fine-tuning manner from a pre-trained model.83 The parameters obtained from the pre-trained model serve as the initialization of the specific task.
If no bioactive molecules are available, zero-shot learning, where a model can learn to recognize effects, or conditions, that were not observed, can be employed. Zero-shot learning requires more knowledge and alleviates the dependence on data. In rare diseases or orphan targets, learning compound-target interactions from big datasets, such as ChEMBL,12 and designing molecules through disease-related targets instead of fitting molecular distributions, builds on “understanding the drug-target interactions.”
Considering that AlphaFold has uncovered 98.5% of human protein structures,84 the target-based molecule generation can be converted into a classical image captioning problem. For example, image is the distance map (or 3D image) for a protein and captioning is the molecular SMILES code to be generated. In this configuration, target-based molecule generation can generally be handled with pipelines composed of a target visual encoder and a language model for SMILES generation.
Multimodal generative models
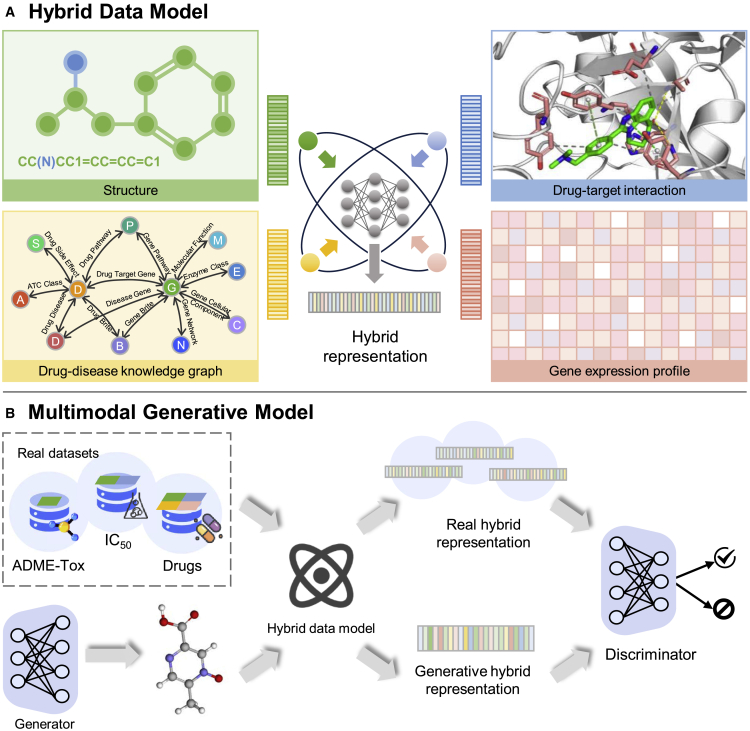
The promise of successful drug discovery lies in the diversity of multiple data modalities that offer complementary perspectives and enable triangulating the evidence for discovery.85 Deep generative models using multimodal data may have significant advantages over unimodal counterparts since the multimodal data contain complementary insights.77 Current studies usually focus on the molecular structural data, and do not fully use other data modalities, such as drug-target interactions, drug-disease knowledge, and relevant gene expression in specific cells following drug treatment (Figure 4A). Therefore, how to make full use of diverse and heterogeneous biological data is a matter worth discussing. There are multiple possible solutions to this challenge. First is “modality alignment,” which means connecting all modalities with an intermediate modality. Because establishing relationships with molecular structures is easier, the structure modality is chosen as the intermediary to other modalities, such as drug-induced gene expression. We then connect the structure modality with other modalities and finally align all modalities in the middle space. “Modality fusion,” which drops the median modality converter, is another possibility. All modalities are directly mapped to a common latent space and indicated by a hybrid representation (Figure 4A). Different modalities describing the same molecules should be closer in the modality-shared space, while the same modalities reflecting diverse molecules should be farther apart.
Figure 4.
A proposed multimodal generative model in the drug discovery applications
(A) A hybrid data model can fully capture diverse information during drug design, including chemical, drug-target interactions, drug-disease knowledge, and disease-relevant expression of target (protein/gene).
(B) A multimodal generative model can consider various drug discovery pipeline components to increase likelihood of success of clinical trials. ADME-Tox, absorption, distribution, metabolism, and excretion-toxicity; IC50, half-maximal inhibitory concentration.
The above discussion is based on training data with sufficient and complete modalities, but the reality often does not satisfy such assumptions. To further exploit these partial data, we need to consider how to complement the missing modality. One possible way is to generate synthetic modalities through established relationships between modalities covering biological activities and pharmacokinetics and pharmacodynamics properties of molecules (Figure 4B). There is an urgent need to seek ways to integrate multimodal information that can generate molecules meaningfully to speed up the process of drug discovery.
Generative models from data consumer to data producer
Unprecedented provision of data is pivotal to boosting data-driven drug discovery, in addition to the emergence of deep-learning algorithms and advances in high-performance computations based on the graphics processing unit. Pharmaceutical companies possess vast amounts of labeled data associated with their ∼2–3M proprietary molecules and generated from the assays routinely run to support lead optimization. In addition, unlabeled data can be used for training as can computationally generated data such as from docking or molecular dynamics trajectories.86
The quantity of high-quality data87 alone does not guarantee actionable decisions in drug discovery.88 For example, leveraging a deep learning algorithm, AlphaFold predicts the 3D structure of proteins from their amino acid sequences and multi-sequence alignments with superior performance.30 Yet critical details of the sites of molecular recognition, the active site for ligand binding or quaternary structure for protein-protein interaction, both vital for structure-based therapeutics design, remain unresolved. The affinity of the drug to the protein versus that of the substrate (or cofactor) determines its effectiveness. Yet, thermodynamic and dynamic properties are even farther from being routinely deployed in deep-learning models for drug design, despite their recognized importance. Free energy calculations are frequently applied in lead optimization with a manageable size (>∼100 s) of molecules, and, recently, protein-ligand binding kinetics have attracted attention in medicinal chemistry. However, the protein-ligand binding/unbinding dynamics is impractical to observe even in a long trajectory (∼ms) from conventional molecular dynamics due to transition states separated by high energy barriers, thus locking the system in configuration around its initial state, lacking conformational sampling.89
In this regard, a considerable effort employing deep-learning methods has been focused on enhanced samplings for extracting the free energy surface and kinetics, computing thermodynamics variables, constructing coarse-grained models, and generative modeling for molecular structure sampling.90 For example, a VAE-based generative network was employed to learn low-dimensional, non-linear embeddings by reconstructing time-lagged conformations, revealing the slow dynamics from the stochastic protein motions.91 With a modified VAE in another example, weighted reaction coordinates optimized by maximizing a predictive information bottleneck framework can efficiently guide a biased simulation for capturing rare events in a short trajectory as well as calculating free energy and kinetics.92
Generative networks combined with molecular simulations solidly rooted in physics, could provide not only meaningful insights but also an invaluable framework for producing statistically reliable protein dynamics data for drug discovery, including COVID-19.93 Still, in its infancy, it poses open questions, including some related to applications of generative modeling, e.g., accurate and efficient force field parameterization, enhanced sampling for kinetic modeling, and scalable generative modeling for a biological system. While current drug discovery is primarily devoted to small-molecule systems due to the data of proteins is severely limited, once the protein conformational dynamics data become more feasible, drug design would be driven toward enhanced safety and effectivity.
Conclusions and outlook
Drug discovery platforms are becoming increasingly industrialized with the ability to both consume and generate big data using AI to drive new molecule design.94 Ageing,95,96 Alzheimer’s disease,97,98 COVID-19,6,65,93 antimicrobial resistance,9 and developments assisting the diagnosis and therapeutics of the COVID-19 pandemic6,99,100,101 provide examples. These successes encourage us to embrace the challenges in further optimization and validation of AI approaches in medical applications. Increased enterprise architecture and infrastructure, including exascale computing,102 quantum computers,103,104 hardware, and connectivity, are a priority in drug discovery data strategies in industries, academia, and governments. Strong data stewardship practices enable the realization of interoperability and adherence to standards. Three rules have been highly recommended:
-
1.
Data stewardship must ensure that data ownership rights (which lays the groundwork for data-sharing models) are operationalized and considered for data acquisition, use, and distribution practices.
-
2.
Representative data (including diverse chemical and target coverage) is critical to ensuring the absence of data biases to allow deep learning models to cover a wide range of applications.
-
3.
Big data’s volume, variety, velocity, and veracity (4Vs) require automated and rigorous data harmonization and validation.
Data harmonization and validation from diverse biological endpoints and different assays can ensure data quality (completeness, consistency, integrity, fairness, and transparency) and data accuracy. In addition, advanced data-sharing and model-learning strategies, such as swarm learning105,106 and federated learning,74,107,108 will accelerate data sharing among industries, academics, governments, and health care systems for drug development. For example, a recent platform called collaborative Profile-QSAR74 developed collaborative models from previously reported biological assays to broaden the domain of applicability without sharing any of the training data, offering a way to address data scarcity.
In summary, recent advances triggered by the rapidly growing deep generative molecular design have brought new momentum for drug discovery, including the production and optimization of small molecules and macromolecules. However, the bottlenecks of AI technologies, such as lack of or limited interpretability of the model, inaccessibility, and lack of availability of high-quality data, currently restrict their application and affect their performance. There is a critical need to further develop and evaluate intelligent generative models in realistic real-world drug discovery contexts in order for deep learning to reach its full potential. Under such developments, the intelligent generative model paradigms will have the potential to transform from theoretical research to practical generation of therapeutics and provide easy-to-use toolkits for chemists and chemistry modelers in their daily work.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261201500003I (to R.N.). This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, and Center for Cancer Research to R.N. This project was supported by the IBM-Cleveland Clinic Accelerator Initiative to F.C. and W.C.
Author contributions
F.C. conceived the manuscript. X.Z., F.C., F.W., J.T., F.C.L., S.K., W.C., and E.F.F. contributed to critical discussion. X.Z. drafted the manuscript. X.Z., F.C., Y.L., S.K, W.C., and R.N. critically revised the manuscript.
Declaration of interests
E.F.F. has a CRADA arrangement with ChromaDex (USA) and is consultant to Aladdin Healthcare Technologies (UK and Germany), the Vancouver Dementia Prevention Centre (Canada), Intellectual Labs (Norway), and MindRank AI (China). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. S.K. and W.C. are employees of IBM TJ Watson Research Center. The other authors declare no competing interests.
References
- 1.Avorn J. The $2.6 billion pill–methodologic and policy considerations. N. Engl. J. Med. 2015;372:1877–1879. doi: 10.1056/NEJMp1500848. [DOI] [PubMed] [Google Scholar]
- 2.Fleming N. How artificial intelligence is changing drug discovery. Nature. 2018;557:S55–S57. doi: 10.1038/d41586-018-05267-x. [DOI] [PubMed] [Google Scholar]
- 3.Schütt K.T., Gastegger M., Tkatchenko A., Müller K.R., Maurer R.J. Unifying machine learning and quantum chemistry with a deep neural network for molecular wavefunctions. Nat. Commun. 2019;10:5024. doi: 10.1038/s41467-019-12875-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng X., Zhu S., Lu W., Liu Z., Huang J., Zhou Y., Fang J., Huang Y., Guo H., Li L., et al. Target identification among known drugs by deep learning from heterogeneous networks. Chem. Sci. 2020;11:1775–1797. doi: 10.1039/c9sc04336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hie B., Zhong E.D., Berger B., Bryson B. Learning the language of viral evolution and escape. Science. 2021;371:284–288. doi: 10.1126/science.abd7331. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y., Wang F., Tang J., Nussinov R., Cheng F. Artificial intelligence in COVID-19 drug repurposing. Lancet. Digit. Health. 2020;2:e667–e676. doi: 10.1016/S2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider P., Walters W.P., Plowright A.T., Sieroka N., Listgarten J., Goodnow R.A., Jr., Fisher J., Jansen J.M., Duca J.S., Rush T.S., et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 2020;19:353–364. doi: 10.1038/s41573-019-0050-3. [DOI] [PubMed] [Google Scholar]
- 8.Riesselman A.J., Ingraham J.B., Marks D.S. Deep generative models of genetic variation capture the effects of mutations. Nat. Methods. 2018;15:816–822. doi: 10.1038/s41592-018-0138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokes J.M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N.M., MacNair C.R., French S., Carfrae L.A., Bloom-Ackermann Z., et al. A deep learning approach to antibiotic discovery. Cell. 2020;181:475–483. doi: 10.1016/j.cell.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Gómez-Bombarelli R., Wei J.N., Duvenaud D., Hernández-Lobato J.M., Sánchez-Lengeling B., Sheberla D., Aguilera-Iparraguirre J., Hirzel T.D., Adams R.P., Aspuru-Guzik A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 2018;4:268–276. doi: 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irwin J.J., Tang K.G., Young J., Dandarchuluun C., Wong B.R., Khurelbaatar M., Moroz Y.S., Mayfield J., Sayle R.A. ZINC20-A free ultralarge-scale chemical database for ligand discovery. J. Chem. Inf. Model. 2020;60:6065–6073. doi: 10.1021/acs.jcim.0c00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., Overington J.P. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruddigkeit L., van Deursen R., Blum L.C., Reymond J.L. Enumeration of 166 billion organic small molecules in the chemical universe database GDB-17. J. Chem. Inf. Model. 2012;52:2864–2875. doi: 10.1021/ci300415d. [DOI] [PubMed] [Google Scholar]
- 14.Patel H., Ihlenfeldt W.D., Judson P.N., Moroz Y.S., Pevzner Y., Peach M.L., Delannée V., Tarasova N.I., Nicklaus M.C. SAVI, in silico generation of billions of easily synthesizable compounds through expert-system type rules. Sci. Data. 2020;7:384. doi: 10.1038/s41597-020-00727-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann T., Gastreich M. The next level in chemical space navigation: going far beyond enumerable compound libraries. Drug Discov. Today. 2019;24:1148–1156. doi: 10.1016/j.drudis.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weininger D. A chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988;28:31–36. [Google Scholar]
- 18.Schwalbe-Koda D., Gómez-Bombarelli R. Machine Learning Meets Quantum Physics. Springer; 2020. Generative models for automatic chemical design; pp. 445–467. [Google Scholar]
- 19.Gupta N., Mangal N., Biswas S. Evolution and similarity evaluation of protein structures in contact map space. Proteins. 2005;59:196–204. doi: 10.1002/prot.20415. [DOI] [PubMed] [Google Scholar]
- 20.David L., Thakkar A., Mercado R., Engkvist O. Molecular representations in AI-driven drug discovery: a review and practical guide. J. Cheminform. 2020;12:56. doi: 10.1186/s13321-020-00460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gainza P., Sverrisson F., Monti F., Rodolà E., Boscaini D., Bronstein M.M., Correia B.E. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods. 2020;17:184–192. doi: 10.1038/s41592-019-0666-6. [DOI] [PubMed] [Google Scholar]
- 22.Wójcikowski M., Kukiełka M., Stepniewska-Dziubinska M.M., Siedlecki P. Development of a protein–ligand extended connectivity (PLEC) fingerprint and its application for binding affinity predictions. Bioinformatics. 2019;35:1334–1341. doi: 10.1093/bioinformatics/bty757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahmoud A.H., Masters M.R., Yang Y., Lill M.A. Elucidating the multiple roles of hydration for accurate protein-ligand binding prediction via deep learning. Commun. Chem. 2020;3:19. doi: 10.1038/s42004-020-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones D., Kim H., Zhang X., Zemla A., Stevenson G., Bennett W.F.D., Kirshner D., Wong S.E., Lightstone F.C., Allen J.E. Improved protein–ligand binding affinity prediction with structure-based deep fusion inference. J. Chem. Inf. Model. 2021;61:1583–1592. doi: 10.1021/acs.jcim.0c01306. [DOI] [PubMed] [Google Scholar]
- 25.Xu M., Wang W., Luo S., Shi C., Bengio Y., Gomez-Bombarelli R., Tang J. International Conference on Machine Learning. PMLR; 2021. An end-to-end framework for molecular conformation generation via bilevel programming; pp. 11537–11547. [Google Scholar]
- 26.Shi C., Luo S., Xu M., Tang J. International Conference on Machine Learning. PMLR; 2021. Learning gradient fields for molecular conformation generation; pp. 9558–9568. [Google Scholar]
- 27.Axelrod S., Gómez-Bombarelli R. GEOM, energy-annotated molecular conformations for property prediction and molecular generation. Sci. Data. 2022;9:185–214. doi: 10.1038/s41597-022-01288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imrie F., Hadfield T.E., Bradley A.R., Deane C.M. Deep generative design with 3D pharmacophoric constraints. Chem. Sci. 2021;12:14577–14589. doi: 10.1039/d1sc02436a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Pei J., Lai L. Structure-based de novo drug design using 3D deep generative models. Chem. Sci. 2021;12:13664–13675. doi: 10.1039/d1sc04444c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Z., Zhu Q., Mou L., Xiong Y., Li G., Zhang L. A grammar-based structural cnn decoder for code generation. Proc. AAAI Conf. Artif. Intell. 2019;33:7055–7062. [Google Scholar]
- 32.Hadjeres G., Nielsen F. Enforcing unary constraints in sequence generation, with application to interactive music generation. Neural Comput. Appl. 2020;32:995–1005. [Google Scholar]
- 33.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 34.Cho K., Merrienboer B.V., Gulcehre C., Bahdanau D., Bougares F., Schwenk H., Bengio Y. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, EMNLP 2014, October 25-29, 2014, Doha, Qatar. ACL; 2014. Learning phrase representations using RNN encoder-decoder for statistical machine translation. A meeting of SIGDAT, a special interest Group of the ACL 1724-1734. [Google Scholar]
- 35.Brown N., Fiscato M., Segler M.H.S., Vaucher A.C. GuacaMol: benchmarking models for de novo molecular design. J. Chem. Inf. Model. 2019;59:1096–1108. doi: 10.1021/acs.jcim.8b00839. [DOI] [PubMed] [Google Scholar]
- 36.Mita G., Filippone M., Michiardi P. International Conference on Machine Learning. PMLR; 2021. An identifiable double VAE for disentangled representations; pp. 7769–7779. [Google Scholar]
- 37.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., Courville A., Bengio Y. Generative adversarial networks. Commun. ACM. 2020;63:139–144. [Google Scholar]
- 38.Rezende D., Mohamed S. International Conference on Machine Learning. PMLR; 2015. Variational inference with normalizing flows; pp. 1530–1538. [Google Scholar]
- 39.Zang C., Wang F. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2020. MoFlow: an invertible flow model for generating molecular graphs; pp. 617–626. [Google Scholar]
- 40.Silver D., Schrittwieser J., Simonyan K., Antonoglou I., Huang A., Guez A., Hubert T., Baker L., Lai M., Bolton A., et al. Mastering the game of go without human knowledge. nature. 2017;550:354–359. doi: 10.1038/nature24270. [DOI] [PubMed] [Google Scholar]
- 41.Popova M., Isayev O., Tropsha A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018;4:eaap7885. doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ertl P., Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform. 2009;1:8. doi: 10.1186/1758-2946-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J., Xu P., Hao Y., Yu T., Liu L., Song Y., Li Y. Multi-constraint molecular generation based on conditional transformer, knowledge distillation and reinforcement learning. BMC Cancer. 2021;21:914–922. [Google Scholar]
- 44.Gottipati S.K., Sattarov B., Niu S., Pathak Y., Wei H., Liu S., Thomas K.M.J., Blackburn S., Coley C.W., Tang J., et al. International Conference on Machine Learning. PMLR; 2020. Learning to navigate the synthetically accessible chemical space using reinforcement learning; pp. 3668–3679. [Google Scholar]
- 45.Kitchen D.B., Decornez H., Furr J.R., Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 46.Bleicher K.H., Böhm H.J., Müller K., Alanine A.I. Hit and lead generation: beyond high-throughput screening. Nat. Rev. Drug Discov. 2003;2:369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 47.Chen H., Engkvist O., Wang Y., Olivecrona M., Blaschke T. The rise of deep learning in drug discovery. Drug Discov. Today. 2018;23:1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 48.Dai H., Tian Y., Dai B., Skiena S., Song L. Proceedings of the International Conference on Learning Representations. 2018. Syntax-directed variational autoencoder for molecule generation. [Google Scholar]
- 49.Jin W., Barzilay R., Jaakkola T. International Conference on Machine Learning. PMLR; 2018. Junction tree variational autoencoder for molecular graph generation; pp. 2323–2332. [Google Scholar]
- 50.Tolstikhin I., Bousquet O., Gelly S., Schölkopf B. 6th International Conference on Learning Representations (ICLR) 2018. Wasserstein auto-encoders. [Google Scholar]
- 51.Jacobs S.A., Moon T., McLoughlin K., Jones D., Hysom D., Ahn D.H., Gyllenhaal J., Watson P., Lightstone F.C., Allen J.E., et al. Enabling rapid COVID-19 small molecule drug design through scalable deep learning of generative models. Int. J. High Perform. Comput. Appl. 2021;35:469–482. [Google Scholar]
- 52.Kuznetsov M., Polykovskiy D. MolGrow: a graph normalizing flow for hierarchical molecular generation. Proc. AAAI Conf. Artif. Intell. 2021;35:8226–8234. [Google Scholar]
- 53.Méndez-Lucio O., Baillif B., Clevert D.-A., Rouquié D., Wichard J. De novo generation of hit-like molecules from gene expression signatures using artificial intelligence. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-019-13807-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhavoronkov A., Ivanenkov Y.A., Aliper A., Veselov M.S., Aladinskiy V.A., Aladinskaya A.V., Terentiev V.A., Polykovskiy D.A., Kuznetsov M.D., Asadulaev A., et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019;37:1038–1040. doi: 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- 55.Jin W., Barzilay R., Jaakkola T. International Conference on Machine Learning. PMLR; 2020. Multi-objective molecule generation using interpretable substructures; pp. 4849–4859. [Google Scholar]
- 56.Beker W., Wołos A., Szymkuć S., Grzybowski B.A. Minimal-uncertainty prediction of general drug-likeness based on Bayesian neural networks. Nat. Mach. Intell. 2020;2:457–465. [Google Scholar]
- 57.Jin W., Yang K., Barzilay R., Jaakkola T.S. 7th International Conference on Learning Representations, ICLR 2019, New Orleans, LA, USA, May 6-9, 2019. OpenReview.net; 2019. Learning multimodal graph-to-graph translation for molecule optimization. [Google Scholar]
- 58.Zhu J.-Y., Park T., Isola P., Efros A.A. Proceedings of the IEEE International Conference on Computer Vision. 2017. Unpaired image-to-image translation using cycle-consistent adversarial networks; pp. 2223–2232. [Google Scholar]
- 59.Maziarka Ł., Pocha A., Kaczmarczyk J., Rataj K., Danel T., Warchoł M. Mol-CycleGAN: a generative model for molecular optimization. J. Cheminform. 2020;12:2–18. doi: 10.1186/s13321-019-0404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cadow J., Born J., Manica M., Oskooei A., Rodríguez Martínez M. A web service for interpretable anticancer compound sensitivity prediction. Nucleic Acids Res. 2020;48:W502–W508. doi: 10.1093/nar/gkaa327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Born J., Huynh T., Stroobants A., Cornell W.D., Manica M. Active site sequence representations of human kinases outperform full sequence representations for affinity prediction and inhibitor generation: 3D effects in a 1D model. J. Chem. Inf. Model. 2021;62:240–257. doi: 10.1021/acs.jcim.1c00889. [DOI] [PubMed] [Google Scholar]
- 62.Ghosh D., Veeraraghavan B., Elangovan R., Vivekanandan P. Antibiotic resistance and epigenetics: more to it than meets the eye. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.02225-19. 022255-e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arjovsky M., Chintala S., Bottou L. International Conference on Machine Learning. PMLR; 2017. Wasserstein generative adversarial networks; pp. 214–223. [Google Scholar]
- 64.Das P., Sercu T., Wadhawan K., Padhi I., Gehrmann S., Cipcigan F., Chenthamarakshan V., Strobelt H., Dos Santos C., Chen P.Y., et al. Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations. Nat. Biomed. Eng. 2021;5:613–623. doi: 10.1038/s41551-021-00689-x. [DOI] [PubMed] [Google Scholar]
- 65.Linsky T.W., Vergara R., Codina N., Nelson J.W., Walker M.J., Su W., Barnes C.O., Hsiang T.Y., Esser-Nobis K., Yu K., et al. De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2. Science. 2020;370:1208–1214. doi: 10.1126/science.abe0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Repecka D., Jauniskis V., Karpus L., Rembeza E., Rokaitis I., Zrimec J., Poviloniene S., Laurynenas A., Viknander S., Abuajwa W., et al. Expanding functional protein sequence spaces using generative adversarial networks. Nat. Mach. Intell. 2021;3:324–333. [Google Scholar]
- 67.Chuai G., Ma H., Yan J., Chen M., Hong N., Xue D., Zhou C., Zhu C., Chen K., Duan B., et al. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19:80. doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Casini A., Olivieri M., Petris G., Montagna C., Reginato G., Maule G., Lorenzin F., Prandi D., Romanel A., Demichelis F., et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018;36:265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang D., Zhang C., Wang B., Li B., Wang Q., Liu D., Wang H., Zhou Y., Shi L., Lan F., Wang Y. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat. Commun. 2019;10:4284–4314. doi: 10.1038/s41467-019-12281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gelijns A.C. National Academies Press; 1989. Institute of Medicine Committee on Technological Innovation in, M. Technological Innovation: Comparing Development of Drugs, Devices, and Procedures in Medicine. [PubMed] [Google Scholar]
- 71.Austin C.P. Opportunities and challenges in translational science. Clin. Transl. Sci. 2021;14:1629–1647. doi: 10.1111/cts.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.AlQuraishi M., Sorger P.K. Differentiable biology: using deep learning for biophysics-based and data-driven modeling of molecular mechanisms. Nat. Methods. 2021;18:1169–1180. doi: 10.1038/s41592-021-01283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bender A., Cortes-Ciriano I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 2: a discussion of chemical and biological data. Drug Discov. Today. 2021;26:1040–1052. doi: 10.1016/j.drudis.2020.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin E.J., Zhu X.W. Collaborative profile-QSAR: a natural platform for building collaborative models among competing companies. J. Chem. Inf. Model. 2021;61:1603–1616. doi: 10.1021/acs.jcim.0c01342. [DOI] [PubMed] [Google Scholar]
- 75.Weber J.K., Morrone J.A., Bagchi S., Pabon J.D.E., Kang S.G., Zhang L., Cornell W.D. Simplified, interpretable graph convolutional neural networks for small molecule activity prediction. J. Comput. Aided Mol. Des. 2022;36:391–404. doi: 10.1007/s10822-021-00421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Higgins I., Matthey L., Pal A., Burgess C., Glorot X., Botvinick M., Mohamed S., Lerchner A. 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, April 24-26, 2017, Conference Track Proceedings. OpenReview.net; 2017. Beta-VAE: learning basic visual concepts with a constrained variational framework. [Google Scholar]
- 77.Manica M., Oskooei A., Born J., Subramanian V., Sáez-Rodríguez J., Rodríguez Martínez M. Toward explainable anticancer compound sensitivity prediction via multimodal attention-based convolutional encoders. Mol. Pharm. 2019;16:4797–4806. doi: 10.1021/acs.molpharmaceut.9b00520. [DOI] [PubMed] [Google Scholar]
- 78.Wang Y., Yao Q., Kwok J.T., Ni L.M. Generalizing from a few examples: a survey on few-shot learning. ACM Comput. Surv. 2020;53:1–34. [Google Scholar]
- 79.Arús-Pous J., Johansson S.V., Prykhodko O., Bjerrum E.J., Tyrchan C., Reymond J.L., Chen H., Engkvist O. Randomized SMILES strings improve the quality of molecular generative models. J. Cheminform. 2019;11:71. doi: 10.1186/s13321-019-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao T., Liu Y., Neves L., Woodford O., Jiang M., Shah N. Data augmentation for graph neural networks. Proc. AAAI Conf. Artif. Intell. 2021;35:11015–11023. [Google Scholar]
- 81.Hemmerich J., Asilar E., Ecker G.F. COVER: conformational oversampling as data augmentation for molecules. J. Cheminform. 2020;12:18. doi: 10.1186/s13321-020-00420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhuang F., Qi Z., Duan K., Xi D., Zhu Y., Zhu H., Xiong H., He Q. A comprehensive survey on transfer learning. Proc. IEEE. 2021;109:43–76. [Google Scholar]
- 83.Segler M.H.S., Kogej T., Tyrchan C., Waller M.P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 2018;4:120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., Bridgland A., Cowie A., Meyer C., Laydon A., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo Y., Eran A., Palmer N., Avillach P., Levy-Moonshine A., Szolovits P., Kohane I.S. A multidimensional precision medicine approach identifies an autism subtype characterized by dyslipidemia. Nat. Med. 2020;26:1375–1379. doi: 10.1038/s41591-020-1007-0. [DOI] [PubMed] [Google Scholar]
- 86.Bayarri G., Hospital A., Orozco M. 3dRS, a web-based tool to share interactive representations of 3D biomolecular structures and molecular dynamics trajectories. Front. Mol. Biosci. 2021;8:726232. doi: 10.3389/fmolb.2021.726232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nigam A., Pollice R., Hurley M.F.D., Hickman R.J., Aldeghi M., Yoshikawa N., Chithrananda S., Voelz V.A., Aspuru-Guzik A. Assigning confidence to molecular property prediction. Expert Opin. Drug Discov. 2021;16:1009–1023. doi: 10.1080/17460441.2021.1925247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bender A., Cortés-Ciriano I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 1: ways to make an impact, and why we are not there yet. Drug Discov. Today. 2021;26:511–524. doi: 10.1016/j.drudis.2020.12.009. [DOI] [PubMed] [Google Scholar]
- 89.Allison J.R. Computational methods for exploring protein conformations. Biochem. Soc. Trans. 2020;48:1707–1724. doi: 10.1042/BST20200193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Noé F., Tkatchenko A., Müller K.R., Clementi C. Machine learning for molecular simulation. Annu. Rev. Phys. Chem. 2020;71:361–390. doi: 10.1146/annurev-physchem-042018-052331. [DOI] [PubMed] [Google Scholar]
- 91.Wehmeyer C., Noé F. Time-lagged autoencoders: deep learning of slow collective variables for molecular kinetics. J. Chem. Phys. 2018;148:241703. doi: 10.1063/1.5011399. [DOI] [PubMed] [Google Scholar]
- 92.Wang Y., Ribeiro J.M.L., Tiwary P. Past-future information bottleneck for sampling molecular reaction coordinate simultaneously with thermodynamics and kinetics. Nat. Commun. 2019;10:3573. doi: 10.1038/s41467-019-11405-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sztain T., Ahn S.H., Bogetti A.T., Casalino L., Goldsmith J.A., Seitz E., McCool R.S., Kearns F.L., Acosta-Reyes F., Maji S., et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 2021;13:963–968. doi: 10.1038/s41557-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sadybekov A.A., Sadybekov A.V., Liu Y., Iliopoulos-Tsoutsouvas C., Huang X.P., Pickett J., Houser B., Patel N., Tran N.K., Tong F., et al. Synthon-based ligand discovery in virtual libraries of over 11 billion compounds. Nature. 2022;601:452–459. doi: 10.1038/s41586-021-04220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aman Y., Frank J., Lautrup S.H., Matysek A., Niu Z., Yang G., Shi L., Bergersen L.H., Storm-Mathisen J., Rasmussen L.J., et al. The NAD(+)-mitophagy axis in healthy longevity and in artificial intelligence-based clinical applications. Mech. Ageing Dev. 2020;185:111194. doi: 10.1016/j.mad.2019.111194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mkrtchyan G.V., Abdelmohsen K., Andreux P., Bagdonaite I., Barzilai N., Brunak S., Cabreiro F., de Cabo R., Campisi J., Cuervo A.M., et al. Ardd 2020: from aging mechanisms to interventions. Aging (Albany NY) 2020;12:24484–24503. doi: 10.18632/aging.202454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fang J., Zhang P., Zhou Y., Chiang C.W., Tan J., Hou Y., Stauffer S., Li L., Pieper A.A., Cummings J., Cheng F. Endophenotype-based in-silico network medicine discovery combined with insurance records data mining identifies sildenafil as a candidate drug for Alzheimer’s disease. Nat. Aging. 2021;1:1175–1188. doi: 10.1038/s43587-021-00138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taubes A., Nova P., Zalocusky K.A., Kosti I., Bicak M., Zilberter M.Y., Hao Y., Yoon S.Y., Oskotsky t., Pineda S., et al. Experimental and real-world evidence supporting the computational repurposing of bumetanide for APOE4-related Alzheimer’s disease. Nat. Aging. 2021;1:932–947. doi: 10.1038/s43587-021-00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou Y., Hou Y., Shen J., Mehra R., Kallianpur A., Culver D.A., Gack M.U., Farha S., Zein J., Comhair S., et al. A network medicine approach to prediction and population-based validation of disease manifestations and drug repurposing for COVID-19. PLoS Biol. 2020;18:e3000970. doi: 10.1371/journal.pbio.3000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Galindez G., Matschinske J., Rose T.D., Sadegh S., Salgado-Albarrán M., Späth J., Baumbach J., Pauling J.K. Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies. Nat. Comput. Sci. 2021;1:33–41. doi: 10.1038/s43588-020-00007-6. [DOI] [PubMed] [Google Scholar]
- 102.Nussinov R., Jang H., Nir G., Tsai C.J., Cheng F. A new precision medicine initiative at the dawn of exascale computing. Signal Transduct. Target. Ther. 2021;6:3. doi: 10.1038/s41392-020-00420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abbott A. Quantum computers to explore precision oncology. Nat. Biotechnol. 2021;39:1324–1325. doi: 10.1038/s41587-021-01116-x. [DOI] [PubMed] [Google Scholar]
- 104.Satzinger K.J., Liu Y.J., Smith A., Knapp C., Newman M., Jones C., Chen Z., Quintana C., Mi X., Dunsworth A., et al. Realizing topologically ordered states on a quantum processor. Science. 2021;374:1237–1241. doi: 10.1126/science.abi8378. [DOI] [PubMed] [Google Scholar]
- 105.Warnat-Herresthal S., Schultze H., Shastry K.L., Manamohan S., Mukherjee S., Garg V., Sarveswara R., Händler K., Pickkers P., Aziz N.A., et al. Swarm Learning for decentralized and confidential clinical machine learning. Nature. 2021;594:265–270. doi: 10.1038/s41586-021-03583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ferrer E.C., Hardjono T., Pentland A., Dorigo M. Secure and secret cooperation in robot swarms. Sci. Robot. 2021;6:eabf1538. doi: 10.1126/scirobotics.abf1538. [DOI] [PubMed] [Google Scholar]
- 107.Chen S., Xue D., Chuai G., Yang Q., Liu Q. A federated learning-based QSAR prototype for collaborative drug discovery. Bioinformatics. 2021;36:5492–5498. doi: 10.1093/bioinformatics/btaa1006. [DOI] [PubMed] [Google Scholar]
- 108.Rieke N., Hancox J., Li W., Milletarì F., Roth H.R., Albarqouni S., Bakas S., Galtier M.N., Landman B.A., Maier-Hein K., et al. The future of digital health with federated learning. NPJ Digit. Med. 2020;3:119. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]