Abstract
Advancements in AI enable personalizing healthcare, for example by investigating disease origins at the genetic or molecular level, understanding intraindividual drug effects, and fusing multi-modal personal physiological, behavioral, laboratory, and clinical data to uncover new aspects of pathophysiology. Future efforts should address equity, fairness, explainability, and generalizability of AI models.
The incorporation of artificial intelligence (AI) into medicine has revolutionized and continues to dramatically change the landscape of healthcare. In this commentary, Shandhi and Dunn highlight the potential and challenges of AI in personalized and precision medicine with specific examples of its current deployment and anticipated future implementations.
Main text
Introduction
Personalized medicine is a novel approach to understanding health, disease, and treatment outcomes based on personal data, including medical diagnoses, clinical phenotype, laboratory studies, imaging, environmental, demographic, and lifestyle factors. Precision medicine often overlaps with this concept and also includes utilizing genomic data to tailor a plan of treatment or prevention of a particular disease. Personalized and precision medicine promise a better understanding of health and medicine, improvements in the early detection of diseases, and better long-term health and chronic disease management. The multi-modal information originating from multiple domains can be collected from individuals because of recent advancements in sensing, cloud infrastructure, medicine, genetics, metabolomics, and imaging technologies, among others. However, with such innovations in sensing and diagnostic testing capabilities, an incredible amount of personal data can now be generated for each patient. Appropriately storing and analyzing this voluminous personal data can be a challenging and daunting task. Thankfully, advances are also being made in these directions to increase the capabilities and efficiency with which we can digitize, store, and analyze these large volumes of personal and population-level data. Ultimately, the combined advancements in biomolecular, imaging, and sensing technologies, along with hardware, software, and data science, and the ability of the medical community to leverage these technologies effectively, have together enabled personalized/precision medicine.
Personalized/precision medicine
The human genome project (https://www.genome.gov/human-genome-project) spurred an entirely new way of thinking about the role of biology in healthcare. Completed in 2003, the fully sequenced human genome was anticipated to address a large proportion of open issues in our understanding of health and disease. However, two decades later, it had decidedly not delivered on that promise.1 This is because biomolecular pathways are more complex than were appreciated at the time, and the many layers of interactions between the genome, phenome, and environment mean that the one gene, one disease (i.e., Mendelian disease) framework is surprisingly infrequent and that more complex relationships within the genome itself (e.g., non-coding regions) and between the genome and other biomolecular species remain to be uncovered.
The advent of new technologies for high-throughput biomolecular measurements over the past two decades has increased our knowledge of the biomolecular milieu, which is a snapshot of the entirety of biomolecules existing in any one compartment at any one point in time. One such technology, next-generation sequencing (NGS), enables transcriptomic analyses through RNA-seq, as well as 16S, meta-genomic, and metatranscriptomic analyses to characterize the microbial milieu and its activities. Further, proteomic and metabolomic profiling spawned from mass spectrometry technologies enable profiling of the biomolecules that are the machines and byproducts of life-sustaining processes. New knowledge generated by these methods has enabled a move from expensive and dirty “shotgun” approaches that capture a broad but poorly annotated set of biomolecules to fast, low-cost, and targeted panel profiling approaches (e.g., genotyping arrays). Further, new laboratory methods to profile various configurations of biomolecules have emerged (e.g., protein post-translational modifications; epigenomic histone and DNA methylation alterations; chromatin conformations, etc.). These methods have uncovered new knowledge about the importance of not only the biomolecules themselves, but also the role of their spatial arrangements and modifications.
Through these emerging technologies and integration of the data generated by them, the promise of a truly “multi-omics” approach to research and medicine is increasingly being realized.2 Examples of the multi-omics approach in practice include: (1) gut microbiome relationships to health and disease, as mediated and characterized by the human metabolome and microbial meta-omes (i.e., metagenome, metatranscriptome, and meta-metabolome), (2) applications of machine learning to large multi-modal multi-omic datasets to uncover new relationships, and (3) variations on sequencing-based technologies to uncover higher-order information, including epigenetics and chromatin configuration (e.g., ATAC-Seq, RRBS, Hi-C). Further, a new appreciation for the key role of drug-metabolizing genes has come to light. Specifically, a host of genes in the Cyp (Cytochrome P450) family have been found to control the metabolism of a vast array of drugs, which in turn enables a precision approach to drug dosing on an individualized level. Applying these approaches complementarily will be key to a more comprehensive assessment of the biomolecular milieu.
Collection and computational processing of these samples remains expensive and sometimes challenging. As costs decreases, we can obtain improved temporal resolution of the biomolecular dynamics, for example by dense sampling in time, which will enable us to better understand how this symphony evolves over the lifespan and during health or disease. As these new data become available, new complementary time series methods development will be needed. Further, methods for multi-modal data fusion remain relatively rudimentary and can be advanced through new AI methods that will support more data-driven rather than hypothesis-driven approaches. A contributor to the bottleneck on this front is the lack of samples on such high-dimensional data, which will be solved by decreasing cost and increasing accessibility of technologies, as well as continued pushes toward open science including the deposition of new data into public repositories.
Artificial intelligence and machine learning
Artificial intelligence (AI) and related technologies (machine learning [ML]) are becoming increasingly prevalent across diverse sectors including business, society, and healthcare. These technologies have the potential to transform many aspects of patient care, as well as administrative processes within provider, payer, and pharmaceutical organizations. Wearable sensor development and the wide adoption of these devices in people’s day-to-day lives provides a unique opportunity to monitor the physiology and behavior of an individual and to generate an unprecedented amount of personalized data in real-world settings. This opportunity is further enabled by complementary technological advancement in analyzing this enormous amount of data (a.k.a. “big data”) using AI/ML algorithms.
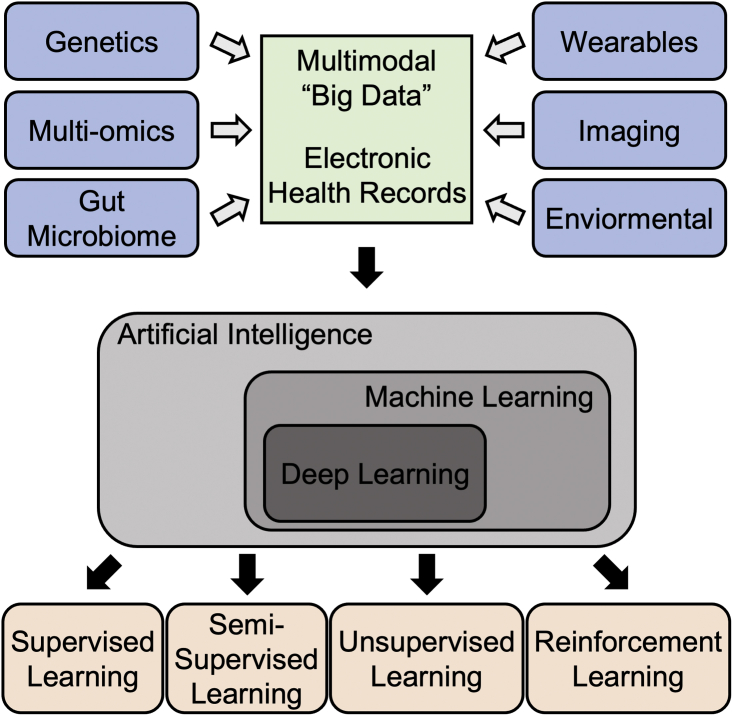
Machine learning (ML), a branch of AI, has gained popularity in the past few decades due to advances in computing machinery which make previously inaccessible methods within practical reach. ML is a method of data analysis that automates analytical model building, which evolved as a way to recognize patterns in data without explicitly programming for that particular task. Although many of today’s top-performing ML algorithms were invented more than half a century ago, the computational power and resources required were unavailable to make this process practical and feasible. Advancements in the semiconductor industry, for example, has reduced computation times, lessened power requirements, and improved the cost effectiveness of computing, all of which has rejuvenated the field of ML in the last two to three decades. New ML applications have ranged from social networking to financial services, transportation, healthcare, and more. The advantages of ML are adaptability, scalability, automation, and the capability of leveraging multi-dimensional and multivariate data to learn new aspects of systems. ML can be divided into four overarching types of learning: supervised, semi-supervised, unsupervised, and reinforcement (Figure 1). Supervised learning is most common because of its utility for prediction. In supervised learning, the prediction target value (i.e., outcome variable, or label) is known, and the algorithm attempts to learn the relationship between it and other, often more easily measured, variables. For example, one may attempt to predict the size of a tumor after a drug treatment based on genetic characteristics of the patient. Supervised learning can be further classified for the intended tasks of either regression (where the target variable is continuous, e.g., heart rate) or classification (where the target variable is discrete, e.g., the presence or absence of arrhythmias). Its methods range from simple linear regression to random forests to neural nets and beyond. Often, simpler methods are preferred because they are understandable and generalize more readily. Unsupervised learning is used to uncover groups or patterns in data where there is no obvious outcome of interest or where the label is not known. Such methods include clustering and dimension reduction. Often, unsupervised learning can be used as a precursor to supervised learning to generate labels for prediction targets. Semi-supervised learning is used in places where labels are known for only a part of the data. Reinforcement learning algorithms learn from trial and error, where the model learns from past experiences and adapts its approaches to achieve the lowest possible prediction error.
Figure 1.
Pipeline of data analysis in healthcare using artificial intelligence
Multi-modal personal “big data” are curated from diversified sources and integrated with electronic health records that are analyzed using artificial intelligence, more specifically machine learning or deep learning algorithms (e.g., supervised, semi-supervised, unsupervised, and reinforcement learning).
In recent years, deep learning (DL) models, a branch of ML, have become more and more popular, achieving great performance in biomedical applications, including diagnoses of tumors from radiology images.3 DL models are in general more complex in nature and structure of the algorithm and generally consist of many artificial neural network and recurrent neural network, which is relatively opaque and hard to interpret. In addition to that, DL models require a lot more data for training purposes compared to traditional ML algorithms (e.g., support vector machine, random forest) and tend to overfit.
There are already a number of research studies suggesting that ML, and more specifically DL models, can perform as well as or better than humans at key healthcare tasks, such as diagnosing diseases and detecting malignant tumors from radiology images.3 While some algorithms can outperform humans, for example in disease detection (e.g., discovering arrhythmias like atrial fibrillation from longitudinal electrocardiogram data) and guiding researchers in how to best construct cohorts for costly clinical trials, AI technologies are most effective for augmenting rather than replacing humans. For example, AI can reduce human errors, augment knowledge capacity, and free up time consumed by menial tasks. However, it is important that we consider both the potential of AI/ML as well as the challenges and concerns in general and in areas that are specific to health care before applying these tools and models in clinical decision support system.
AI and ML in personalized/precision medicine
AI/ML models have been successfully applied to a wide variety of genomics data, particularly in cases with high dimensional and complex data that are challenging to process using traditional statistical methods. Genome-wide association studies, which involve rapidly scanning markers across the genomes of thousands or more people to discover genetic variants associated with a particular disease, have benefitted substantially from ML. One example is in Type 1 diabetes, where there is an improved risk assessment using ML algorithms that can account for interactions between a large collection of biomarkers.4
Using treatment outcomes data generated from previous patients treated for a disease, ML models can identify which future patients may benefit from a specific treatment based on their characteristics. An example of this is genetically informed therapeutic planning (using support vector machine-based anti-cancer drug sensitivity prediction method using genomic data5) for patients with pharmacogenomically actionable variants, where titrated prescription and dosing are critical. This advancement can avoid unnecessary treatments in non-responders and support titrated prescription and dosing to maximize the anti-cancer effect in responders.
ML algorithms have also demonstrated remarkable performance in identifying novel biomarkers that can support early disease detection, prediction of treatment response, and provide indicators of disease progression. A recent comprehensive integrative molecular analysis of a complete set of tumors in The Cancer Genome Atlas (TCGA) employed unsupervised clustering to learn similarities and differences in tumors across 33 different cancer types.6 Further exploration into the resulting tumor subgroups has improved our understanding of, for example, how cancers mutate and the various factors that modulate these tumors.6
Similar to its applications in genomics, the application of ML algorithms and in particular DL to radiology and histopathology have improved the accuracy of image analysis while also reducing the time required. Currently, radiologists visually assess radiological images to detect, characterize, and monitor diseases—this work is manual, tedious, subjective, and prone to human error. Deep learning models, including convolutional neural networks and variational autoencoders, have automated such tasks via quantitative assessment of radiographic characteristics, e.g., cancer diagnosis, staging, and segmentation of tumors from neighboring healthy tissue.3 These methods fall under radiomics, a data-centric, radiology-based research field. Radiogenomics is a new and complimentary field in precision medicine that uses ML/DL to combine radiology images (e.g., cancer imaging) with gene expression signatures to stratify patients’ risk, guide therapeutic strategies, and perform prognoses.7 Similar to radiologic images, ML/DL models have been used to examine histopathology images which can contain billions of pixels and are difficult to process without computer-aided diagnosis.8 The utility of AI-assisted precision medicine has been demonstrated in other areas of precision medicine, including cardiovascular and neurological disease. Here, it has been used to identify novel genotypes and phenotypes in heterogeneous cardiovascular diseases and to improve genetic diagnostics in neurodevelopment disorders. For example, recent work has demonstrated methods to predict heart failure and other serious cardiovascular diseases in asymptomatic individuals,9 which can be acted upon with personalized prevention plans to delay disease onset and reduce negative health outcomes.
Overall, the current value and future potential of AI in personalized health care, early detection of disease, tracking disease progression, and as a clinical decision support tool have been amply demonstrated (Figure 2). AI-assisted precision medicine has demonstrated promise not only for personalization of therapies for existing diseases, but also for individual risk prediction and personalized prevention plans.
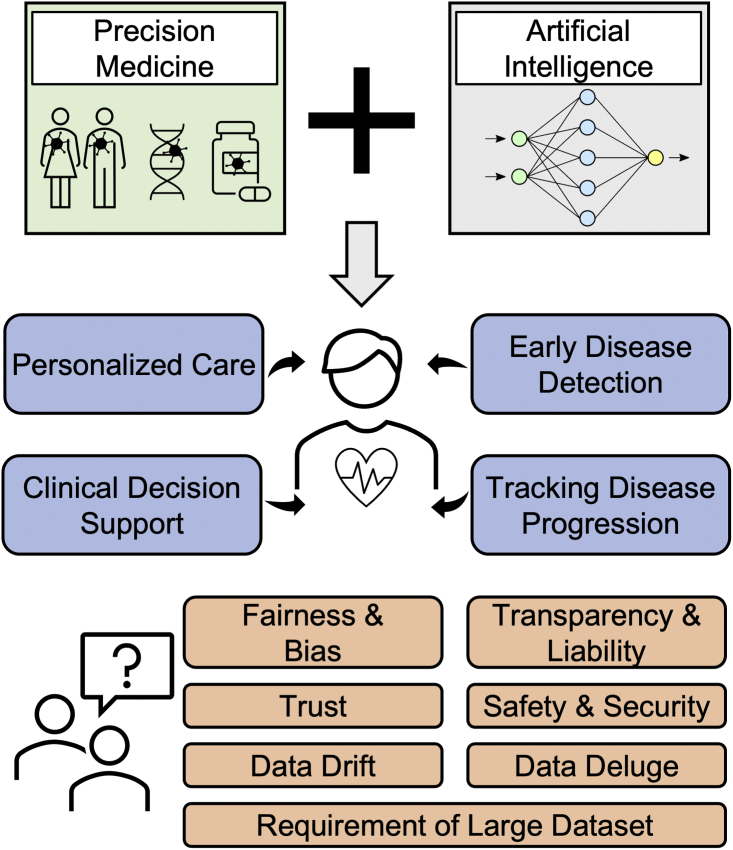
Figure 2.
Overview of incorporation of artificial intelligence into precision medicine
The incorporation of artificial intelligence into precision medicine has demonstrated tremendous potential and progress in personalized care, clinical decision support systems, early disease detection, and tracking disease progression. However, there are technical and ethical challenges (e.g., fairness and bias, transparency and liability, trust, safety, and security) that may hinder the progress and reliability of the field and delay clinical implementation.
Challenges and considerations
Although tremendous progress has been made using AI techniques and high-volume data, there is substantial room for improvement. While the potential of AI/ML in personalized/precision medicine is clear, there are many remaining concerns and challenges, both technical and ethical, that may hinder progress and reliability of the field and delay clinical implementation. Some examples of this are:
-
(1)
Fairness and bias: Data and algorithms can reflect, reinforce, and perpetuate biases. When the data utilized to train AI is either incomplete (e.g., lack of representation from underserved and underrepresented communities) or inherently biased (e.g., collected in settings where existing stereotypes affect the data itself), the models built will be problematic and can serve to further exacerbate disparities and biases.
-
(2)
Limited data availability: In recent years, DL models have become increasingly popular, achieving great performance in many areas of biomedical applications. However, due to their data-hungry nature, DL models cannot easily learn from small datasets, and available datasets are often insufficient in size to train deep learning models.
-
(3)
Data deluge: As a society we are generating ever more data both in and out of clinical settings, with healthcare data storage projected to exceed 2,000 exabytes by 2020.10 Collecting such multi-modal data on a large scale opens new data storage and organization challenges and costs.
-
(4)
Transparency and liability: DL and other high complexity ML models may demonstrate greater accuracy than simpler models, but are opaque and do not provide users with insights as to how the algorithm arrives at its conclusions. This “black box” nature of the technology is particularly concerning in healthcare where lives are on the line. Additionally, where the responsibility for AI-assisted medical decisions lies is not always clear.
-
(5)
Data drift: A common assumption in AI (particularly ML) algorithms is that data from the past (used for the model training) is a representation of the data from the future (for which the model will eventually be deployed). Unfortunately, this is rarely the case in real-world settings and so changes in the data may affect the model’s behavior and accuracy in a real-world deployment.
-
(6)
Data safety and privacy: AI algorithms use significant amounts of personal data for their decision making. However, software and corresponding hardware may have security flaws which can lead to theft of personal and health information.
-
(7)
Trust: Similar to other technologies, early mistakes and failures in technologies can lead to general mistrust which can reduce adoption and utilization of the technology.
Current advances to overcome the challenges
Recent advances in sensing and measurement technologies and novel data science and AI methods have the potential to mitigate many of the challenges described above. The wide adoption of wearables in the general population (1 in 4 Americans has a wearable) enables continuous collection of large volumes of personal physiological and behavioral data in addition to environmental information (e.g., location, ambient condition) in real-world settings at minimal cost. This has been aided by advancements in miniaturization and improvements in hardware including better signal-to-noise ratios, battery life, compute power, and more. Investing in ways to intelligently reduce the sampling rates and developing computationally efficient data compression tools to store a large volume of data without losing key information can reduce the costs of data storage as well as computation. Another recent advancement in AI, federated learning,11 introduces a decentralized data usage infrastructure for training the algorithms, which can mitigate challenges associated with data safety and privacy. In traditional ML methodology, datasets from multiple sources would be centrally combined and then used to train an algorithm, whereas in federated learning, an algorithm is trained locally on edge devices or local servers without sharing that data and later only the model parameters are combined centrally and optimized iteratively. As only model parameters are shared, not the actual data, data safety and privacy remain intact in federated learning.
Recent initiatives from multiple stakeholders including researchers, policymakers, clinicians, and ethicists emphasize inclusivity and inference-centered explainability of AI algorithms in health care12 as opposed to the traditional prediction-centered black box approach of the past. These new tools and frameworks can enable dissection and interpretation of predictions made by an algorithm and may even improve model performance further by allowing for the injection of domain knowledge into their design. These new tools can help detect and resolve bias, drift, and other gaps in data and models.
Conclusions
Technological advancements in sensor hardware and AI algorithms equipped with personalized/precision medicine have already resulted in an unprecedented acceleration of personalized therapy, early disease detection, and personalized disease prevention strategies. Overall, the synergy between AI and personalized/precision medicine could ultimately decrease the disease burden for the public at large, and, therefore, the cost of preventable health care for all. However, we should remain vigilant with an ethics and equity lens to ensure that these advancements are not increasing health-related disparities and exacerbating existing inequities or creating new divides in care or health-related outcomes.
Acknowledgments
Declaration of interests
J.P.D. is on the scientific advisory board of Human Engineering Health Oy and is a consultant for ACI Gold Track.
References
- 1.Shendure J., Findlay G.M., Snyder M.W. Genomic medicine–progress, Pitfalls, and promise. Cell. 2019;177:45–57. doi: 10.1016/j.cell.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kellogg R.A., Dunn J., Snyder M.P. Personal omics for precision health. Circ. Res. 2018;122:1169–1171. doi: 10.1161/circresaha.117.310909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosny A., Parmar C., Quackenbush J., Schwartz L.H., Aerts H.J.W.L. Artificial intelligence in radiology. Nat. Rev. Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei Z., Wang K., Qu H.-Q., Zhang H., Bradfield J., Kim C., Frackleton E., Hou C., Glessner J.T., Chiavacci R., et al. From disease association to risk assessment: an Optimistic View from genome-wide association studies on type 1 diabetes. PLoS Genet. 2009;5:e1000678. doi: 10.1371/journal.pgen.1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong Z., Zhang N., Li C., Wang H., Fang Y., Wang J., Zheng X. Anticancer drug sensitivity prediction in cell lines from baseline gene expression through recursive feature selection. BMC Cancer. 2015;15:489. doi: 10.1186/s12885-015-1492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoadley K.A., Yau C., Hinoue T., Wolf D.M., Lazar A.J., Drill E., Shen R., Taylor A.M., Cherniack A.D., Thorsson V., Akbani R., Bowlby R., Wong C.K., Wiznerowicz M., Sanchez-Vega F., Robertson A.G., Schneider B.G., Lawrence M.S., Noushmehr H., Malta T.M., Cancer Genome Atlas Network. Stuart J.M., Benz C.C., Laird P.W. Cell-of-Origin patterns Dominate the molecular classification of 10, 000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodalal Z., Trebeschi S., Nguyen-Kim T.D.L., Schats W., Beets-Tan R. Radiogenomics: bridging imaging and genomics. Abdom. Radiol. 2019;44:1960–1984. doi: 10.1007/s00261-019-02028-w. [DOI] [PubMed] [Google Scholar]
- 8.Komura D., Ishikawa S. Machine learning methods for Histopathological image analysis. J. 2018;16:34–42. doi: 10.1016/j.csbj.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambale-Venkatesh B., Yang X., Wu C.O., Liu K., Hundley W.G., McClelland R., Gomes A.S., Folsom A.R., Shea S., Guallar E., et al. Cardiovascular event prediction by machine learning. Circ. Res. 2017;121:1092–1101. doi: 10.1161/circresaha.117.311312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanford Medicine Health Trends Report. 2017. Harnessing the Power of Data in Health https://repository.gheli.harvard.edu/repository/collection/resource-pack-big-data-and-health/resource/11902.
- 11.Li T., Sahu A.K., Talwalkar A., Smith V. Federated learning: challenges, methods, and future directions. Mag. 2020;37:50–60. doi: 10.1109/msp.2020.2975749. [DOI] [Google Scholar]
- 12.Lauritsen S.M., Kristensen M., Olsen M.V., Larsen M.S., Lauritsen K.M., Jørgensen M.J., Lange J., Thiesson B. Explainable artificial intelligence model to predict acute critical illness from electronic health records. Nat. Commun. 2020;11:3852. doi: 10.1038/s41467-020-17431-. [DOI] [PMC free article] [PubMed] [Google Scholar]