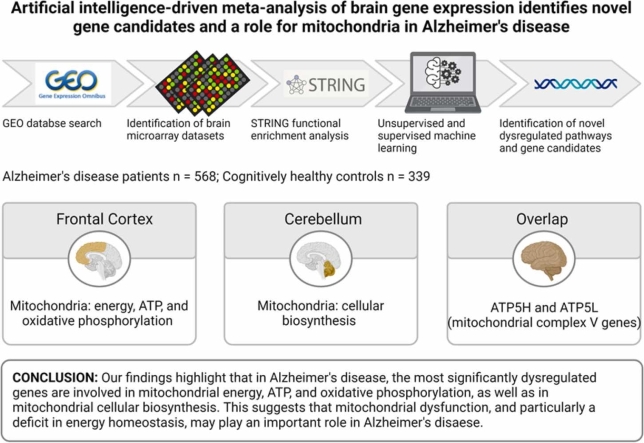
Abstract
Alzheimer’s disease (AD) is the most common form of dementia. There is no treatment and AD models have focused on a small subset of genes identified in familial AD. Microarray studies have identified thousands of dysregulated genes in the brains of patients with AD yet identifying the best gene candidates to both model and treat AD remains a challenge. We performed a meta-analysis of microarray data from the frontal cortex (n = 697) and cerebellum (n = 230) of AD patients and healthy controls. A two-stage artificial intelligence approach, with both unsupervised and supervised machine learning, combined with a functional network analysis was used to identify functionally connected and biologically relevant novel gene candidates in AD. We found that in the frontal cortex, genes involved in mitochondrial energy, ATP, and oxidative phosphorylation, were the most significant dysregulated genes. In the cerebellum, dysregulated genes were involved in mitochondrial cellular biosynthesis (mitochondrial ribosomes). Although there was little overlap between dysregulated genes between the frontal cortex and cerebellum, machine learning models comprised of this overlap. A further functional network analysis of these genes identified that two downregulated genes, ATP5L and ATP5H, which both encode subunits of ATP synthase (mitochondrial complex V) may play a role in AD. Combined, our results suggest that mitochondrial dysfunction, particularly a deficit in energy homeostasis, may play an important role in AD.
Keywords: Machine learning, Alzheimer’s disease, Microarray, Gene expression, Mitochondria
Graphical Abstract
1. Introduction
Alzheimer’s disease (AD) is the most common form of dementia with 50 million suffering from it globally and almost 10 million people developing it yearly [1]. There are currently no effective treatments for AD and over 99.6% of clinical trials of AD have failed so far [2]. Further, most of what is currently known about AD has been established using familial AD (FAD) models, characterized by mutations in amyloid precursor protein (APP), presenilin 1 (PSEN1) and presenilin 2 (PSEN2), which account for less than 1% of AD cases [3], [4], [5], [6].
The recent advent of high-throughput genetic analysis technologies have allowed us to delve into the dysregulated genetic landscape of sporadic, or late onset, AD. Gene expression profiling via microarray analysis allows quantitation of a large number of mRNA transcripts and their variation in a relatively unbiased approach [7], but have previously been confined to relatively small sample numbers, making statistical analysis challenging [8]. More specifically, the dataset is pruned using arbitrary fold changes as well as p-values that require multiple corrections, both of which potentially overlook genes that may be important. Additionally, there is a tendency to use microarray data to confirm a priori hypotheses, perhaps biasing reported outcomes rather than using microarray as a purely exploratory method.
Recent meta-analyses of publicly available microarray data have proven to be a rich vein for identifying novel genes contributing to several diseases, including influenza [9], atherosclerosis [10], chronic pain [11], cancer [12], [13], and Parkinson’s disease [14]. To date, however, few studies have used this approach to investigate possible genetic contribution to AD. One such study took a comparative approach to find overlapping gene expression profiles in neurodegenerative disorders including AD, Lewy body disease, amyotrophic lateral sclerosis and frontotemporal dementia [15]. Another study took a similar approach to compare cross-species transcriptional overlap between mouse models of AD and humans [16]. These meta-analyses, however, are limited in two ways. First, human transcriptomic data is high-dimensional (i.e. high ratio of samples to genes) and complex, making it difficult to identify disease associated patterns in the datasets [17]. Second, as discussed above, these studies identified thousands of dysregulated genes without identifying which of these may be the best target(s) for developing new mouse and cell models of AD to interrogate and identify novel therapeutic approaches.
The use of artificial intelligence (AI) is a way to overcome the above-mentioned limitations. Importantly, not only is AI able to unravel patterns within complex data in an unbiased way [17], it also has the ability to reveal which gene target(s) should be investigated further. An example of this is that machine learning models have successfully differentiated Parkinson’s disease patients from healthy controls based on their gene expression profiles [18]. Further, a recent study using a similar approach, in inflammatory bowel disease (IBD), performed a meta-analysis of human gene expression data, followed by machine learning, to identify novel disease causing genes [19]. Their role in disease was confirmed in mice, representing the development of a novel model of IBD. Using these recently developed tools, patient-derived IBD organoids were successfully ‘treated’, identifying novel therapeutic targets and therapies for IBD [19]. In AD, as far as we are aware, a meta-analysis combined with machine learning approach has been used once. However the identified genes had little to no functional interactions in STRING pathway analysis, suggesting that they are unlikely to be biologically relevant [20]. In the context of AD, these previous data establish a clear need to for methods that identify functionally connected and biologically relevant candidate genes as has been done in IBD [19]. Hopefully, this approach will lead to the development of novel animal models of AD as well as new treatments.
To identify the most biologically relevant genes in AD that inform disease pathophysiology, we performed a meta-analysis of an unprecedented number of microarray datasets from the frontal cortex and cerebellum of patients with AD compared to healthy controls. We then used a combination of unsupervised and supervised machine learning along with functional network analyses in STRING to determine genes with clearly established interaction networks indicating that may be biologically relevant to AD.
2. Methods
2.1. Systematic review of publicly available data repositories
To identify publicly available transcriptomic datasets, a systematic review of the Gene Expression Omnibus (GEO) database was performed. The key search terms used included “Alzheimer’s disease” and “homo sapiens”. Datasets were screened based on the following inclusion criteria: (a) gene expression data generated using microarray platforms, (b) gene expression specific to the amygdala, hippocampus, entorhinal cortex, frontal cortex, temporal cortex or cerebellum, (c) clinically confirmed Alzheimer’s disease patients and (d) inclusion of cognitively normal healthy controls. Datasets were excluded for the following: (a) use of other high throughput gene expression assays (e.g. RNA-sequencing), (b) gene expression in the periphery (e.g. blood), (c) not specifying confirmed AD diagnosis, and (d) not including cognitively healthy controls (e.g. use of controls with mild cognitive impairment). It is important to note that we excluded RNA-sequencing datasets because of their small sample sizes of around 25 total donors and inability to be merged with microarray data, because of their differing sensitivity of gene detection. Additionally, exclusive analysis of available RNAseq data using our methods, runs the risk of developing overfitted models, limiting model predictive efficacy.
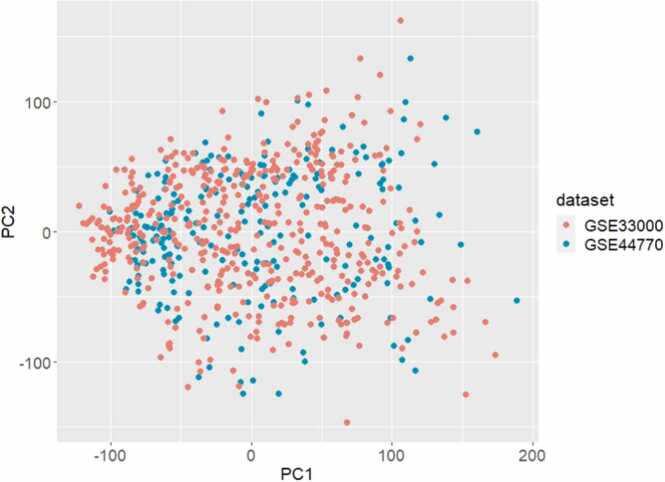
After identifying the respective datasets for the frontal cortex and cerebellum, we first confirmed that the data uploaded to the GEO database in each respective dataset was pre-normalized and therefore did not require any additional normalization procedures. We did not perform any log transformations of the data as our analyses did not use traditional statistical analyses and fold change measures. Each dataset was then processed independently. Duplicated entries for genes were removed and a list of common genes for all datasets was created. For all datasets, genes that were not present in the common list were discarded. We then converted gene expression to a z-score in each dataset independently. Data processing and merging was done in R Studio v1.2.5033 (R v3.6.3) with packages GEOquery, and dplyr. After merging the datasets, we performed a principal component analysis (PCA) to visualize whether any batch effects existed between the frontal cortex datasets [21] (Fig. 1). PCA was performed using PCA in R Studio v1.2.5033 (R v3.6.3) and visualized using ggplot.
Fig. 1.
Principal component analysis (PCA) demonstrating no batch effects between the two frontal cortex GEO datasets (GSE33000 and GSE44770) prior to merging.
2.2. Machine Learning and STRING Network Analysis
The conventional approach for identifying differentially expressed genes typically uses fold change, p-values, or a combination of the two. These methods, however, are limited and are unlikely to provide the information needed to make strong conclusions about dysregulated genes that may be important for AD. First, using standard fold change cutoffs to select differentially expressed genes is inherently problematic. Genes with either low, or high, absolute expression are more likely to either easily meet or miss the fold change threshold, respectively, regardless of whether or not the gene is truly differentially expressed [22], [23]. Further, the p-value is well-known to not only provide limited information about the data at hand, but to also be easily misinterpreted, thus likely contributing to the replication crisis [24], [25], [26]. Specifically, calculating statistical significance using a p-value does not account for the degree to which the genes are, or are not, involved in differences between groups (in this case, between AD patients and healthy controls). One way to circumvent the inherent limitations of conventional methods is to approach the problem of identifying novel disease-driving genes through the lens of artificial intelligence, such as machine learning. In other words, we treat the problem as a machine learning problem: we would like to determine which genes best predict the classification of a given sample as being from either an AD patient or healthy control.
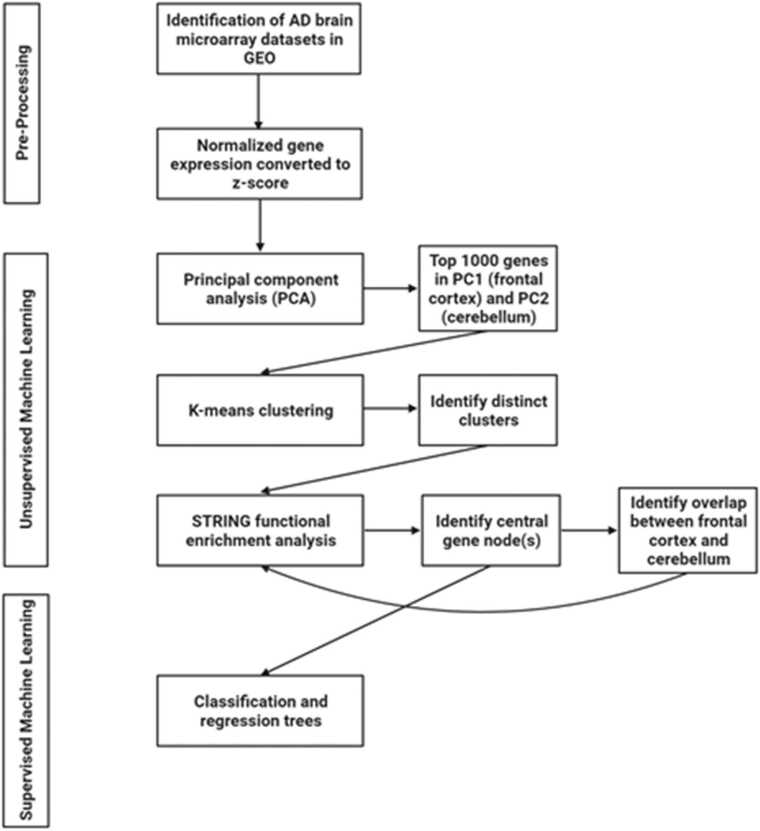
In the present study, we used a two-stage machine learning approach (Fig. 1). In the first stage, we used unsupervised machine learning to perform an initial feature selection: identifying the genes that are likely to be important candidates for distinguishing an AD patient from a healthy control. Feature selection is an important step to reduce the number of features and thus avoid the curse of dimensionality for the final machine learning algorithm [27]. Unsupervised machine learning is unique in the sense that it does not predefine any sample as being either an AD or healthy control sample. Instead it identifies similarities between the various samples that exist, irrespective of their group membership [28]. Samples with high similarity will cluster together and will inform us how the data is grouped and what the drivers of this differentiation are [28]. If true differences exist between AD patients and healthy controls, the unique clusters will be representative of this, and we will identify which genes are driving these differences. The unsupervised machine learning approach used here was a PCA, an important technique that reduces dimensionality within a dataset while simultaneously minimizing any information loss [29]. PCA has an established use in the analysis of high throughput datasets, such as microarray, to reveal hidden patterns within the thousands of identified genes [30], [31], [32]. All the genes (>15,000 genes per dataset) that were identified were analyzed using PCA in R Studio v1.2.5033 (R v3.6.3) and visualized using ggplot (Fig. 2). The top 1000 genes that correlated with the principal components (PCs) were identified and selected as potential gene candidates for AD.
Fig. 2.
AI workflow used in the current study to identify new AD related genes.
To further narrow down the list of gene candidates, we entered each list of the top 1000 contributing genes from the PCs into STRING v11 [33]. Importantly, STRING allows for the identification of interaction networks and gene-enrichment analysis [33]. We then identified possible distinct network clusters using k-means clustering in STRING (Fig. 2). K-means clustering is an unsupervised machine learning approach which, here, is a way to identify genes in the network that have similar interconnections and overlapping pathways [34]. Subsequently, each distinct k-means cluster for the two brains regions were separately entered into STRING and a network analysis was performed using the following active interaction sources: experiments, databases, co-expression, neighborhood, and gene fusion and a minimum required interaction score of 0.7 (high confidence) (Fig. 2). Importantly, using STRING, and the wealth of experimental data that it relies on, to identify pathways and interaction networks increases the likelihood of identifying strong, biologically relevant gene candidates for AD. More specifically, this approach allows for the identification of biologically relevant pathways rather than stand-alone genes that may not have any evidence of interactions thus increasing the chance of network effects and identifying therapeutic candidates AD. In STRING, each k-means cluster were characterized using biological processes identified by Gene Ontology [35]. We then selected the central node(s) in each k-means cluster based on their connections with other genes in the network, with identified central node(s) having the highest number of connections (Fig. 2). In other words, those with the most connections are preferentially chosen. Selecting highly interconnected gene nodes increases the likelihood of identifying candidates that are fundamentally important biologically relevant targets in AD.
After identifying the central nodes in each k-means cluster for the frontal cortex and cerebellum, respectively, we then undertook the second stage of our two-stage machine learning approach: supervised machine learning using decision trees (classification and regression trees (CART)) [36] (Fig. 2). Here, the CART identifies which genes are best able to separate AD patients from healthy controls within the machine learning model. The use of CART has been well-established in large clinical and public health projects characterized by high-dimensional, heterogeneous data [36], [37]. In the current study, the central gene nodes for each k-means cluster were used as features in the CART. All datasets were each split into training (75%) and testing (25%) datasets. Training, tuning, and validating the model was done on the training dataset. Here, a 5-fold cross-validation was repeated three times to improve the accuracy estimates of the models [38]. Cross-validation was used also as a tool for identifying the top performing gene predictors. The final evaluation of the CART model performance was done on the previously withheld testing dataset. Performance indicators used included optimal sensitivity (correctly identifies AD patients), specificity (correctly identifies healthy controls), accuracy (correct number of classifications (AD / healthy control)) and AUC-ROC curve (capability of the model to distinguish between AD patients and healthy controls). The optimal performing CART was selected based on a high specificity and sensitivity as well as AUC. CART was performed in R Studio v1.2.5033 (R v3.6.3) with libraries rpart, caret, and pROC.
2.3. Comparison of central node genes between brain regions
In parallel with our supervised machine learning analysis, we also sought to identify if there were any common central node genes between the included brain regions. Identifying overlapping functional nodes is important to establish any common mechanisms underlying AD pathology. After identifying the overlap, we then performed a functional network analysis of these genes in STRING v11 to determine enriched pathways and biological processes that are common. We were also able to use the STRING analysis to confirm if any of the overlapping central node genes were themselves central nodes in the overlap functional network (Fig. 2).
3. Results
3.1. Included datasets and characteristics
A total number of 1010 datasets in GEO were screened. Based on the inclusion and exclusion criteria, 14 datasets were identified as being eligible for inclusion in the meta-analysis. The meta-data of each dataset was analyzed to determine if there was a sufficient sample size to undertake further analyses and machine learning. Of these datasets, 11 were excluded due to an insufficient sample size and were unable to be combined to create a new dataset of sufficient size (e.g. sample size of only 10). Consequently, we were unable to include the following brain regions in the analyses: the amygdala, hippocampus, entorhinal cortex, and temporal cortex. In the three identified data sets, we included two brains regions, the frontal cortex and cerebellum. Data from the frontal cortex originated from two GEO datasets: GSE44770 and GSE33000. Data for the cerebellum originated from the GEO dataset GSE44768. Samples from the studies consisted of AD patients, with confirmed antemortem clinical diagnosis and postmortem neuropathological assessment, and normal, non-demented, healthy controls (Table 1) and all donors were Caucasian [39], [40].
Table 1.
Characteristics of the included GEO datasets.
| Frontal Cortex |
Cerebellum |
Hippocampus |
||||||
|---|---|---|---|---|---|---|---|---|
|
GSE44770 |
GSE33000 |
GSE44768 |
GSE36890 |
|||||
| AD | Control | AD | Control | AD | Control | AD | Control | |
| Sample Size | 129 | 101 | 310 | 157 | 129 | 101 | 5 | 10 |
| Age (Average +/- SEM) | 80 + 0.8 | 62 + 1.1 | 80 + 0.5 | 64 + 0.8 | 80 + 0.8 | 62 + 1.1 | 94 + 2.4 | 77 + 2.8 |
| Sex (M:F) | 62:67 | 82:19 | 135:175 | 123:34 | 62:67 | 82:19 | 2:3 | 5:5 |
| AD Diagnostic Criteria | Pathological assessment (Braak stage, general and regional atrophy, grey and white matter atrophy, ventricular enlargement) | Neurohistopathology traits (Braak stge, specific regional atrophy on gross and microscopic scale, ventricular enlargement) | Pathological assessment (Braak stage, general and regional atrophy, grey and white matter atrophy, ventricular enlargement) | Neuropathological changes (Consortium to Establish a Registry for Alzheimer’s disease (CREAD)) guidelines and Braak stage | ||||
| RNA Quality (RIN average +/- SEM) | 7.1 + 0.05 | 7.3 + 0.05 | Not specified | 6.7 + 0.04 | 6.6 + 0.04 | > 6.9 | ||
For the frontal cortex only, genes common to both datasets were selected and then the two datasets were merged into one. The final, unified frontal cortex dataset had a total sample size of n = 697 (AD n = 439, healthy control n = 238). The cerebellum dataset had a total sample size of n = 230 (AD n = 129, healthy control n = 101). We also included a tertiary dataset of hippocampal gene expression data to further test our identified central gene nodes and models. This dataset had a total sample of n = 15 (AD n = , healthy control n = ) and all donors were Japanese [41].
The quality of RNA (RIN) was included in the meta-data for two datasets (GSE44770 and GSE44768) and all samples were of high quality (Table 1). One dataset GSE33000 did not include any RIN details but stated in their publication that RINs were of high quality [40]. The remaining dataset GSE36890 also did not include RINs in their meta-data but did specify in their publication that all RINs were> 6.9 [41].
3.2. Meta-analysis and unsupervised machine learning identifies novel dysregulated pathways and genes in AD
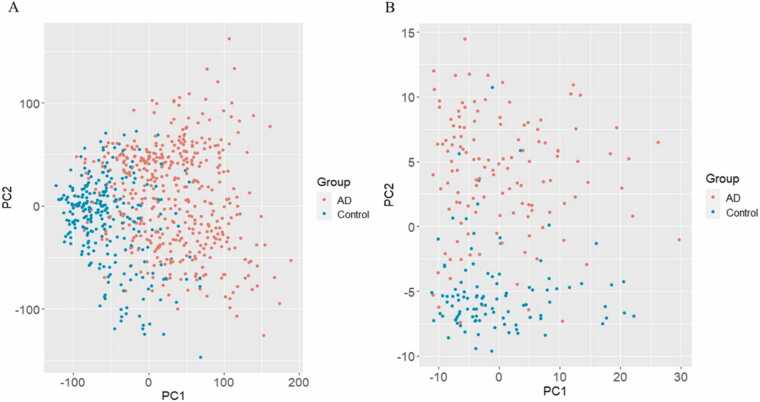
Unsupervised machine learning using principal component analysis (PCA) of the frontal cortex and cerebellum datasets demonstrated a clear clustering between the AD patients and healthy control groups along principal component (PC) 1 for the frontal cortex and PC2 for the cerebellum (Fig. 3a and b).
Fig. 3.
Principal component analysis of genes identified in the microarray meta-analysis reveals a clear between-group separation between the AD patients (red) and healthy controls (blue). (a) Frontal cortex. (b) Cerebellum.
Based on this finding, the top 1000 genes from frontal cortex PC1 and the top 1000 genes from cerebellum PC2 were identified as the most dysregulated genes in AD as they contributed the most to between-group variance (Supplementary Table 1). To determine the number of common pathways represented in these genes, a second unsupervised machine learning approach was performed using k-means clustering in STRING to identify genes in the network that have similar interconnections and overlapping pathways [34]. Three k-means clusters were identified in both the frontal cortex and cerebellum (Fig. 4). We then used Gene Ontology (GO) to characterize the biological functions of each of the clusters.
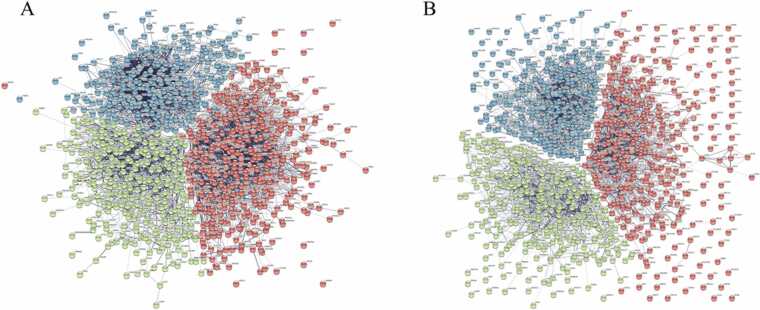
Fig. 4.
STRING k-means clustering of the top 1000 dysregulated genes in AD identified by the principal component analysis (PCA). (a) Frontal cortex principal component 1 (PC1) k-means clustering showed three distinct clusters of genes. Red n = 420, green n = 294, blue n = 286. (b) Cerebellum PC2 k-means clustering showed three distinct clusters of genes. Red n = 392, green n = 325, blue n = 283.
The first k-means cluster (red) for the frontal cortex was characterized by signaling processes (Supplementary Figure 1). Within this cluster, 15 central node genes were identified that play important roles in voltage-dependent calcium channels, guanine nucleotide-binding protein (G protein) formation and activity, AMPA receptor, cAMP-dependent protein kinase A (PKA), and the SNARE complex (Table 2; Supplementary Table 2). The second frontal cortical cluster (blue) was characterized by metabolic processes relating to macromolecules, proteins, and DNA (Supplementary Figure 2). Here, 26 genes were identified as being central nodes in the network with extensive roles in general cellular metabolic processes including DNA repair, transcriptional regulation, ubiquitin pathway regulation, and apoptosis (Table 2; Supplementary Table 2). The third k-means cluster identified in the frontal cortex (green) was related to mitochondrial processes. Within this cluster, there were two distinct sub-clusters identified based on biological function. The first was related to mitochondrial-specific energy, ATP, and oxidative phosphorylation and had 36 central nodes in the network (Supplementary Figure 3). These genes were all related to different mitochondrial subunits including: the F0-F1 ATP synthase (also known as mitochondrial complex V, ATP synthase), V-ATPase, mitochondrial complex I, and mitochondrial complex III (Table 2; Supplementary Table 2). The second sub-cluster within the green k-means cluster was defined by mitochondria-mediated cellular biosynthesis (Supplementary Figure 3) characterized by 10 central nodes where all genes were specific to the nuclear-encoded mitochondrial ribosomal 39 S or 28 S subunits, which play a central role in protein synthesis in mitochondria (Table 2; Supplementary Table 2).
Table 2.
Summary of the overall characterization of the k-means clusters identified in the frontal cortex principal component 1 (PC1).
| K-means Cluster | Functional Pathway Characterization | Number of Central Gene Nodes | Roles of Central Gene Nodes |
|---|---|---|---|
| 1 (red) | Signaling processes | 15 | Voltage-dependent calcium channels, G-protein formation and activity, AMPA receptor, cAMP-dependent protein kinase A, and SNARE complex |
| 2 (blue) | Metabolic processes (macromolecular, proteins, and DNA) | 26 | General cellular metabolic processes, DNA repair, transcriptional regulation, ubiquitin pathway regulation, and apoptosis |
| 3 A (green) | Mitochondria (energy, ATP, and oxidative phosphorylation) | 36 | ATP synthase, V-ATase, mitochondrial complex I, mitochondrial complex III |
| 3B (green) | Mitochondria (cellular biosynthesis) | 10 | Mitochondrial ribosomal 39 S and 28 S subunits, mitochondrial protein synthesis |
The characterization of the cerebellum k-means clusters revealed the same functional pathways to those in the frontal cortex (Table 3) albeit the specific genes and the number of central gene nodes were different. The metabolic processes pathway was characterized by 17 central gene nodes which plays a role in transcription and nucleic acid binding (Table 3; Supplementary Figure 4; Supplementary Table 3). The signaling processes pathway had 10 central gene nodes (Supplementary Figure 5) and there was some overlap here in terms of the roles of the genes with the frontal cortex signaling processes pathway including voltage-dependent calcium channels and SNARE complexes. Unique to the cerebellum signaling pathway, however, was the involvement of cell-cell junctions, actin filament and cytoskeleton, and vesicular transport (Table 3; Supplementary Table 3). Like the frontal cortex, the third (green) k-means cluster identified in the cerebellum was related to mitochondrial processes and was subdivided into two distinct sub-clusters: energy, ATP, and oxidative phosphorylation and cellular biosynthesis (Supplementary Figure 6). The energy, ATP, and oxidative phosphorylation sub-cluster was characterized by 18 central gene nodes involved in ATP synthase, V-ATPase, and mitochondrial complexes I and III (Table 3; Supplementary Table 3). The cellular biosynthesis sub-cluster included 11 central gene nodes that made up components of the mitochondrial ribosomal 39 S and 28 S subunits and were involved in mitochondrial ribosomal protein synthesis (Table 3; Supplementary Table 3).
Table 3.
Summary of the overall characterization of the k-means clusters identified in the cerebellum across principal component (PC) 2.
| K-means Cluster | Functional Pathway Characterization | Number of Central Node Genes | Roles of Central Node Genes |
|---|---|---|---|
| 1 (green) | Metabolic processes (RNA and nucleic acid) | 17 | Transcription, nucleic acid binding (mRNA, RNA, DNA) |
| 2 (red) | Signaling processes | 10 | Voltage-dependent calcium channel, cell-cell junctions, actin filament, vesicular transport, actin cytoskeleton, and SNARE complex |
| 3 A (blue) | Mitochondria (energy, ATP, and oxidative phosphorylation | 18 | ATP synthase, V-ATPase, mitochondrial complex I, and outer mitochondrial membrane |
| 3B (blue) | Mitochondria (cellular biosynthesis) | 11 | Mitochondrial ribosomal 39 S and 28 S subunits, mitochondrial protein synthesis |
3.3. Supervised machine learning reveals the top signaling pathways for AD
The central gene nodes identified in the k-means clusters and sub-clusters within each brain structure were subsequently analyzed using supervised machine learning.
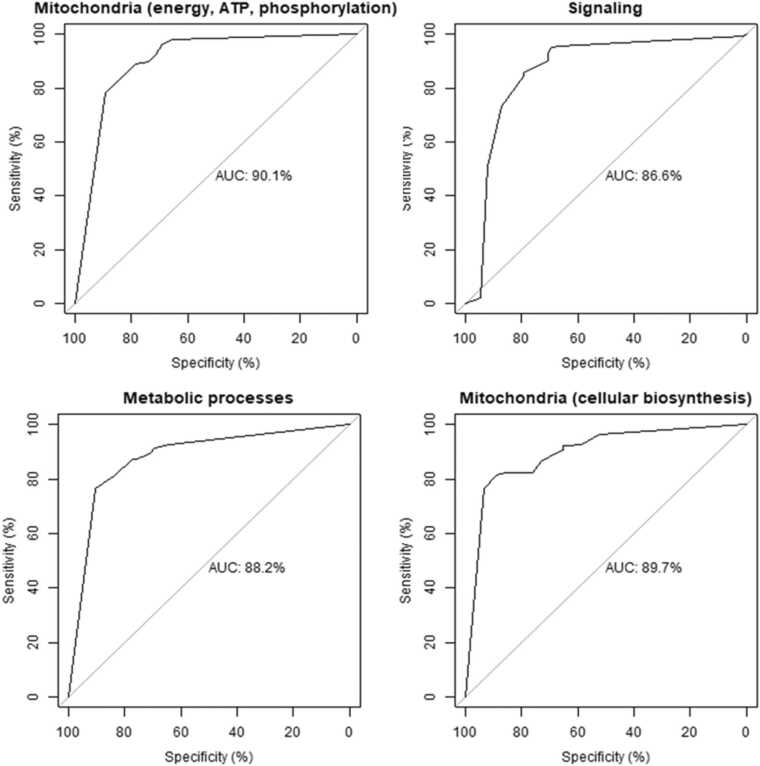
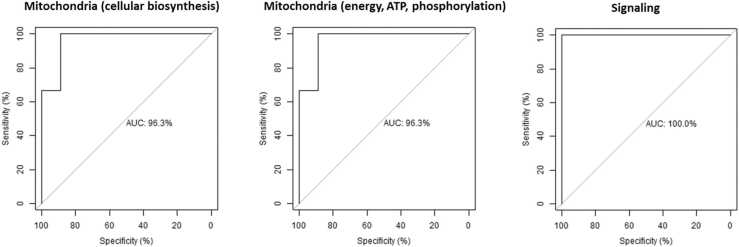
The top performing cluster for the frontal cortex was related to mitochondrial energy, ATP, and oxidative phosphorylation. This was followed very closely by the synaptic signaling, metabolic processes, and mitochondrial cellular biosynthesis clusters (Table 4, Fig. 5). While sensitivity was the same between both models, the mitochondrial energy, ATP, and oxidative phosphorylation cluster model had a slightly higher specificity and AUC, suggesting that this model was slightly better at identifying controls (Fig. 5).
Table 4.
CART performance metrics for the k-means clusters and sub-clusters in the AD frontal cortex. AUC: area under the curve.
| Cluster | Specificity | Sensitivity | Accuracy | AUC |
|---|---|---|---|---|
| Mitochondrial energy, ATP, and oxidative phosphorylation | 0.83 | 0.93 | 0.85 | 90.1 |
| Signaling | 0.83 | 0.92 | 0.86 | 86.6 |
| Metabolic processes | 0.82 | 0.91 | 0.84 | 88.2 |
| Mitochondrial cellular biosynthesis | 0.81 | 0.88 | 0.83 | 89.7 |
Fig. 5.
Receiver operating characteristic (ROC) curve of the CART models performance for each cluster in the frontal cortex. AUC: area under the curve.
The frontal cortex dataset was characterized by a large difference in ages between the AD and control donor samples. As listed in Table 1, the mean ages were 80 and 62, respectively, for AD and controls (Welch two sample t-test, t695 = 23.38, p = 2.2e−16). To determine if any of the models’ performance was due to this age difference, performed additional CARTs using the same central gene nodes and pathways on an age-normalized version of the frontal cortex dataset. Here, we removed donor samples aged below the 60th quantile (youngest) in the control group and above the 60th quantile (oldest) in the AD group. The final dataset had a total n of 396 (251 CE and 95 control) and mean ages of 74 and 73, respectively, for AD and control (Welch two sample t-test, t160.09 = 1.33, p = 0.186). Using the same procedure for CART, we found that all models performed similarly in the age-normalized dataset (Supplementary Figure 7).
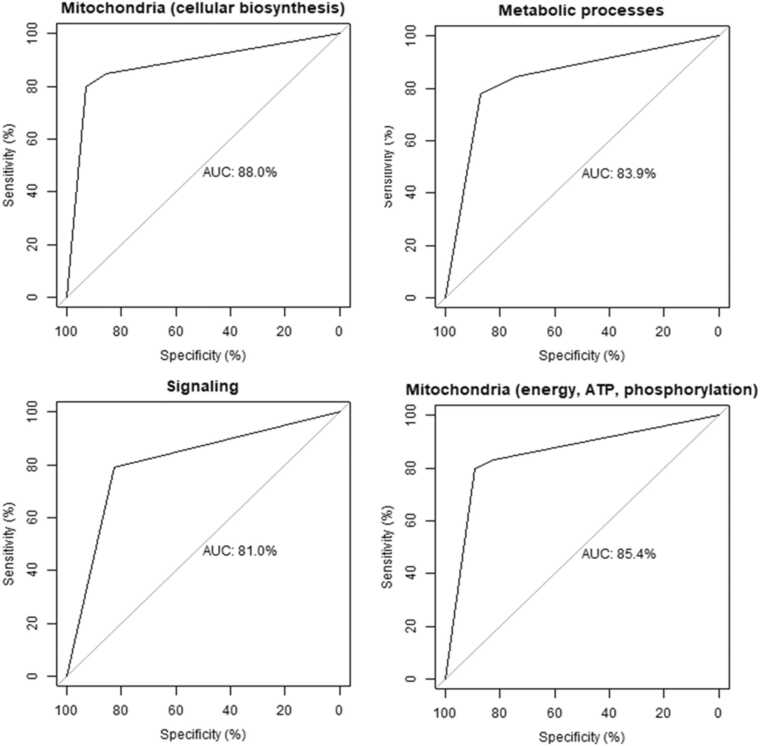
For the cerebellum, the top performing cluster was for mitochondrial cellular biosynthesis. This was closely followed by metabolic processes and signaling and finally by mitochondrial energy, ATP, and oxidative phosphorylation (Table 5, Fig. 6). Although the levels of sensitivity vary between pathways in each brain region, all four pathways were common to both brain regions. Unlike the frontal cortex, the low starting n for this dataset (total 230 donor samples) precluded a similar age-normalized analysis.
Table 5.
CART performance metrics for the k-means clusters and sub-clusters in the AD cerebellum.
| Cluster | Specificity | Sensitivity | Accuracy | AUC |
|---|---|---|---|---|
| Mitochondrial cellular biosynthesis | 0.80 | 0.85 | 0.76 | 88.0 |
| Metabolic processes | 0.76 | 0.89 | 0.82 | 83.9 |
| Signaling | 0.74 | 0.89 | 0.81 | 81.0 |
| Mitochondrial energy, ATP, and oxidative phosphorylation | 0.67 | 0.86 | 0.76 | 85.4 |
Fig. 6.
Receiver operating characteristic (ROC) curve of the CART models performance for each cluster in the cerebellum. AUC: area under the curve.
3.4. Analysis of the overlapping gene nodes between frontal cortex and cerebellum reveals the importance of ATP5L and ATP5H
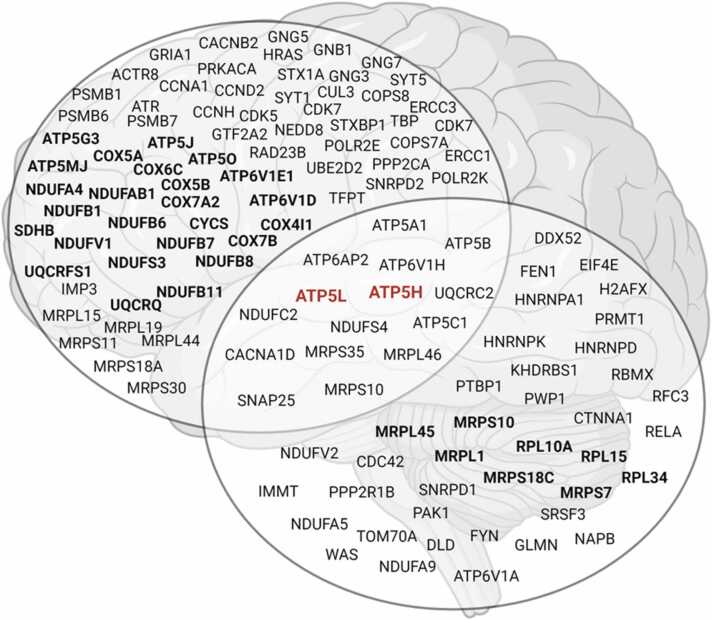
We then identified the common central node genes in the four overlapping pathways, (Table 6). Overlapping central gene nodes were disproportionately represented from the mitochondrial energy, ATP, and oxidative phosphorylation pathway, with 10 genes being dysregulated in AD across the frontal cortex and cerebellum. These genes largely comprised subunits of ATP synthase (n = 5), followed by the V-ATPase (n = 2), mitochondrial complex I (n = 2), and mitochondrial complex III (n = 1). Three central gene nodes from mitochondrial cellular biosynthesis pathway: one 39 S subunit ribosomal protein gene and two 28 S subunit ribosomal protein genes. Only two central gene nodes overlapped in the signaling pathway between the cerebellum and frontal cortex: a subunit for a voltage-dependent calcium channel and a SNARE complex member (Table 6).
Table 6.
Overlapping central gene nodes between the frontal cortex and cerebellum.
| Gene | Protein Name | Functional Pathway | Role / Function |
|---|---|---|---|
| ATP5A1 | ATP synthase F1 subunit α | Mitochondrial energy, ATP, and oxidative phosphorylation | F1 catalytic core of mitochondrial ATP synthase |
| ATP5B | ATP synthase F1 subunit β | F1 catalytic core of mitochondrial ATP synthase | |
| ATP5C1 | ATP synthase F1 subunit γ | F1 catalytic core of mitochondrial ATP synthase | |
| ATP5H | ATP synthase peripheral stalk subunit d | F0 complex of mitochondrial ATP synthase | |
| ATP5L | ATP synthase membrane subunit g | F0 complex of mitochondrial ATP synthase | |
| ATP6AP2 | ATPase H+ transporting accessory protein 2 | Transmembrane sector of mitochondrial V-ATPase | |
| ATP6V1H | ATPase H+ transporting V1 subunit H | Component of V-ATPase | |
| NDUFC2 | NADH:ubiquinone oxidoreductase subunit C11 | Mitochondrial complex I | |
| NDUFS4 | NADH:ubiquinone oxidoreductase subunit S4 | Mitochondrial complex I | |
| UQCRC2 | Ubiquinol-cytochrome c reductase core protein 2 | Mitochondrial complex III | |
| MRPL46 | Mitochondrial ribosomal protein L46 | Mitochondrial cellular biosynthesis | 39 S subunit mitochondrial ribosomal protein |
| MRPS10 | Mitochondrial ribosomal protein S10 | 28 S subunit mitochondrial ribosomal protein | |
| MRPS35 | Mitochondrial ribosomal protein S35 | 28 S subunit mitochondrial ribosomal protein | |
| CACNA1D | Calcium voltage-gated channel subunit α1D | Signaling | Voltage-dependent calcium channel |
| SNAP25 | Synaptosome associated protein 25 | SNARE complex |
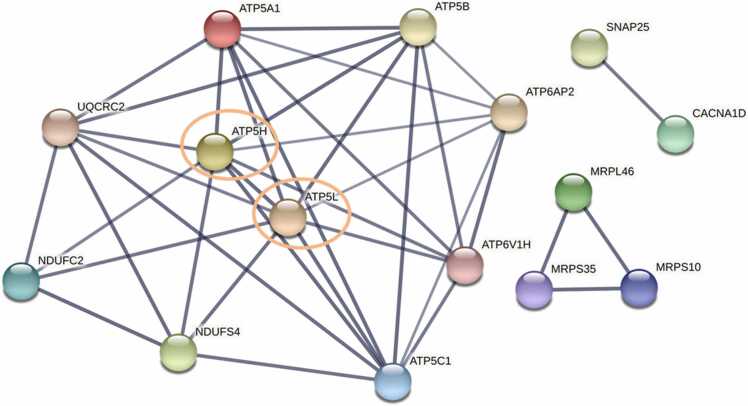
Given these central gene nodes overlapped in both the cerebellum and frontal cortex, we then sought to identify if any of these genes were themselves central nodes (i.e. had the most connections) within a network using STRING. We found that two of these genes had the most connections with the others and therefore were likely central nodes within this sub-group. They were ATP5L and ATP5H, each with nine connections across the other mitochondrial energy, ATP, and oxidative phosphorylation genes (Fig. 7). The biological processes that were enriched in this network included oxidative phosphorylation (FDR = 1.87e-10), mitochondrial ATP synthesis coupled proton transport (FDR = 3.50e-8), cristae formation (FDR = 9.98e-8), and mitochondrial transport (FDR = 6.76e-6).
Fig. 7.
STRING functional network analysis of the overlapping central gene nodes between the frontal cortex and cerebellum of AD patients. The STRING network analysis includes the following interaction sources: experiments, databases, co-expression, neighborhood, and gene fusion. The minimum interaction score was set to 0.7 (high confidence). Thickness of the line indicates the confidence in the interaction.
3.5. Importance of overlapping functional pathways and gene nodes generalizes to the hippocampus
Following the identification of overlapping gene nodes and pathways between the frontal cortex and cerebellum (Table 6), we sought to confirm if these findings were generalizable to another brain region implicated in AD, the hippocampus. To do this, we tested the performance of three shrinkage discriminant analysis (SDA) models each consisting of the pathways and respective genes identified in Table 6. The models were first trained on the large frontal cortex dataset prior to being tested on a novel hippocampal dataset (n = 15 total samples). All models performed similarly well on the novel hippocampal dataset (Table 7, Fig. 8). While all models correctly predicted all AD patients (sensitivity), there was some difference in their ability to predict controls (specificity). It is important to note, however, that sample size was very small (n = 10 controls) therefore the difference between the models was the correct prediction of only one healthy control.
Table 7.
SDA performance metrics for the overlapping gene nodes and pathways between frontal cortex and cerebellum on a hippocampal testing dataset.
| Cluster | Specificity | Sensitivity | Accuracy | AUC |
|---|---|---|---|---|
| Mitochondrial energy, ATP, and oxidative phosphorylation | 0.6 | 1 | 0.73 | 0.96 |
| Mitochondrial cellular biosynthesis | 0.75 | 1 | 0.87 | 0.96 |
| Signaling | 0.67 | 1 | 0.80 | 1 |
Fig. 8.
Receiver operating characteristic (ROC) curve of the SDA models performance for the overlapping central gene nodes and pathways on a novel hippocampal testing dataset. AUC: area under the curve.
4. Discussion
Identifying novel gene candidates in AD is challenging. We approached this challenge by performing a meta-analysis of microarray data from the frontal cortex and cerebellum of patients with AD. We then used an artificial intelligence-driven method, specifically a combination of unsupervised and supervised machine learning, to identify novel gene candidates for future study.
Across both the frontal cortex and cerebellum there were four functional pathways found to be dysfunctional in AD. These included 1) signaling, 2) metabolic processes, 3) mitochondrial energy, ATP, and oxidative phosphorylation, and 4) mitochondrial cellular biosynthesis. Using an age-normalized frontal cortex dataset, we also demonstrated that the importance of these pathways was not dependent on age. Despite the overlapping functional pathways between the two regions, there were pronounced differences in the relative importance of these pathways and the central gene nodes identified within each (Fig. 6). First, the supervised machine learning analysis using CART identified that in the frontal cortex the mitochondrial energy, ATP, and oxidative phosphorylation pathway was the best predictor of AD. In the cerebellum, however, the mitochondrial cellular biosynthesis pathway was the best predictor of AD. There was little overlap (15/143) in the central gene nodes between the frontal cortex and cerebellum where fifteen central gene nodes were identical and most (10/15) were members of the mitochondrial energy, ATP, and oxidative phosphorylation functional pathway (Fig. 9). A STRING functional network analysis of these overlapping genes found they were enriched in mitochondrial-related biological processes, including oxidative phosphorylation, mitochondrial ATP synthesis coupled proton transport, cristae formation, and mitochondrial transport. Further, two genes in this network were the most highly connected: ATP5L and ATP5H and therefore represented central gene nodes within this overlapping sub-network. We also demonstrated that the overlapping gene nodes from the mitochondrial energy, ATP, and oxidative phosphorylation and mitochondrial cellular biosynthesis pathways, respectively, both predicted AD using hippocampal gene expression data. This further highlights the generalizability of our models and the relative importance of these genes in AD.
Fig. 9.
Summary of the central gene nodes identified in the frontal cortex and cerebellum, including the overlap in central gene nodes between the two regions. Black bolded genes represent those identified by the classification and regression trees (CART) as being significant predictors of AD. Red bolded genes represent the two central gene nodes of the overlapping genes between the frontal cortex and cerebellum.
Mitochondria are increasingly being shown to contribute to the development and progression of AD, with evidence for both primary and secondary dysfunctional mitochondrial cascades (for reviews see [42], [43]). Specifically, mitochondrial dysfunction not only affects AD pathology, including APP activity and β amyloid (Aβ) accumulation, but AD pathology also leads to further mitochondrial dysfunction [43]. Our findings are consistent with these studies showing that oxidative phosphorylation and mitochondrial ribosomal subunit mRNA are decreased in the bloods of patients with MCI who were at risk of developing AD as well as the AD patients themselves [44], [45]. This highlights an early pattern of systemic mitochondrial dysfunction that both precedes AD pathology and persists across the disease trajectory. These findings are further confirmed by a recent proteome analysis of the AD brain tissue showing that dysregulated mitochondrial complexes, including ATP synthase, are potential drivers for AD pathology [46]. The results of the present study also suggest that the role of the mitochondria in AD may be more nuanced. Specifically, our supervised machine learning analysis found that dysregulated oxidative phosphorylation and ATP synthesis genes are altered in the frontal cortex whereas dysregulated mitochondrial ribosomal genes were altered in the cerebellum. This suggests that although the mitochondria, broadly, are important, there may be key differences in specific mitochondrial mechanisms underlying pathology across brain regions. The reason underlying these differences, however, is unclear. It is also important to consider that this finding may be due to mitochondrial dysfunction resulting from varying levels of disease load across brain regions. For example, amyloid plaque deposition spreads to the cerebellum only in the end stages of AD [47] and perhaps amyloid β leads to mitochondrial ribosomal dysfunction. As discussed above, however, there is growing evidence that mitochondrial dysfunction precedes AD pathology. Future research would strongly benefit from examining these possibilities further.
Despite the finding of different mitochondrial mechanisms in the frontal cortex and cerebellum, we did find that two ATP synthase genes (ATP5L and ATP5H) represented a potential common mechanism underlying AD pathology in both regions. ATP5L encodes the g subunit and ATP5H encodes subunit d of the F0 membrane-spanning component of ATP synthase. ATP synthase, also known as mitochondrial complex V, is the final step in the oxidative phosphorylation pathway and the site of adenosine diphosphate (ADP) to adenosine triphosphate (ATP) conversion. It also plays a significant role in the formation of the mitochondrial inner membrane cristae [48]. There has been some research into the role of ATP synthase in AD. Dysfunctional ATP synthase has been shown to sensitize mitochondrial permeability transition pore formation and lead to AD pathology, including amyloid β [49]. AD pathology has also been found to lead to further decreases in ATP synthase activity via modifications to the α subunit of the F1 catalytic core [48], [50], [51]. There is also some evidence that patients with AD have serum anti-ATP synthase β subunit autoantibodies, suggesting that mitochondrial dysfunction may be driven by autoimmunity [52]. Despite the growing evidence that ATP synthase plays a role in AD, there have been no studies that specifically examine a role for the g (ATP5L) and d (ATP5H) subunits. Interestingly, however, two genome-wide association study (GWAS) meta-analysis of approximately 25,000 and 50,000 people, respectively, identified a shared ATP5H/KCTD2 locus for AD risk [53], [54]. In line with this finding, a recent analysis of polygenic risk scores for patients with AD found that oxidative phosphorylation genes, including ATP synthase, were strongly associated with risk of AD [55]. Given these genetic findings and our own that ATP5L and ATP5H dysregulation represents a possible brain-wide mechanism in AD pathology, future research would benefit from studying these ATP synthase subunits further. Although these two GWAS meta-analyses have identified this risk locus, it is worth noting that other AD GWAS have identified myriad risk loci, with little consensus between studies [56]. We would like to stress therefore, that both our results and the importance of the ATP5H/KCTD2 risk locus in AD have limited interpretation without confirmatory experimental evidence.
Using a novel testing dataset, we were able to show that the overlapping gene nodes and pathways between the frontal cortex and cerebellum were generalizable to another brain region: the hippocampus. Both the mitochondrial energy, ATP, and oxidative phosphorylation and the mitochondrial cellular biosynthesis pathways were able to predict the presence of AD. This finding has several important implications. First, it suggests that mitochondrial dysfunction is likely brain-wide and is not dependent on the relative pathology present. Although all brain regions are involved in AD, the cerebellum typically has less pathology than the hippocampus and frontal cortex [57], [58]. It may be the case, therefore, that mitochondrial dysfunction occurs early in the neuropathological sequelae of AD. Indeed, there is a growing body of experimental evidence in both mice and humans that this is the case [42], [43], [44], [45], [59], [60]. We accept that this approach, however, could be biased to detecting more germline altered pathways that may result in AD. Second, the frontal cortex, cerebellum, and hippocampus are all made up of different cell populations (e.g. Purkinje cells are predominately in the cerebellum). Given that our findings of mitochondrial dysregulation are generalizable to all three brain regions, it suggests that our findings are not cell type dependent. Further research using single cell RNA-sequencing and spatial transcriptomics are required to confirm this. Third, while the frontal cortex and cerebellum datasets were made up of Caucasian donors the hippocampal dataset, on the other hand, consisted of Japanese donors. Given that both mitochondrial pathway models had a strong predictive performance, it suggests that mitochondrial dysfunction is unlikely to be dependent on ethnicity. Finally, the hippocampal dataset was also made up of donors who were substantially older than those of the frontal cortex and cerebellum datasets. In fact, both the AD and healthy control donors were 10–15 years older in the hippocampal dataset. This age difference further highlights that our findings are unlikely to be dependent on age and a simple artefact from age-related changes in mitochondrial function and health [61].
Although our work presents a strong case for a key role of novel genes in AD, there are some limitations that should be taken into consideration. First, based on our inclusion and exclusion criteria, we only identified three datasets that were appropriate for inclusion in the current study: two from the frontal cortex and one from the cerebellum. Although we did seek to include data from other brain regions, including the hippocampus and entorhinal cortex, the microarray datasets for these did not have the sample size to support our machine learning-based analyses. As such, we are unable to include a comparison of the genes and pathways that are involved in these regions to those discussed here. Future research would benefit from focusing on expanding gene expression data for these regions to enable large-scale analyses and the application of artificial intelligence methods. Our current findings, however, present a strong case for key overlapping genes across the frontal cortex and cerebellum despite differences in AD pathophysiology [57], which highlight the possibility of common underlying mechanisms. A second consideration is that our study has used established databases (e.g. STRING [34], Gene Ontology [35]) to identify functional networks and pathways in AD. An inherent limitation of these databases is that they rely on existing interactions between proteins and networks that have been previously identified in experimental literature. As such, the use of these databases precludes the possibility of other central gene node(s) that do not yet have a wealth of experimental data. Although there is no way to overcome this limitation, we strongly suggest that future experimental work confirm the findings presented here. A final consideration is that the two original datasets for the frontal cortex (GSE44770 and GSE33000) both used donor samples sourced from the Harvard Brain Tissue Resource Centre (HBTRC). Although we checked for duplicate samples when combining the datasets using z-scores, without the original HBTRC donor identification numbers we cannot rule out the possibility that some donor samples may be represented twice. This highlights the importance of performing high throughput analyses on donor samples sourced from different tissue banks.
In summary, our study reports an artificial intelligence-driven identification of novel gene candidates in the frontal cortex and cerebellum of patients with AD using a relatively unbiased methodology. Our findings highlight the importance of nuclear-encoded mitochondrial genes involved in oxidative phosphorylation in the frontal cortex and mitochondrial ribosomal protein synthesis in the cerebellum. Further, we found ATP synthase function as a possible common mechanism underlying AD pathology across brain regions. Together, these findings highlight the possibility that mitochondrial dysfunction and pathophysiological mechanisms in AD may be brain region specific and that ATP synthase subunit dysregulation is a common mechanism. These candidates should be investigated further as they may have significant implications for understanding the etiology of AD and future therapeutic strategies.
CRediT authorship contribution statement
Caitlin A. Finney: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing. Fabien Delerue: Methodology, Writing – original draft, Writing – review & editing. Wendy A. Gold: Methodology, Writing – original draft, Writing – review & editing. David A. Brown: Methodology, Writing – original draft, Writing – review & editing. Artur Shvetcov: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgements
This work was supported by a Macquarie University Research Acceleration Scheme grant to C.A.F. and A.S. (# 5224420).
Contributions
C.A.F. and A.S. conceived the study, acquired funding, performed the machine learning and STRING network analyses, and made the figures. C.A.F. curated the data for analysis. C.A.F., F.D., W.A.G., D.A.B., and A.S. contributed to the methodology and wrote, read, and approved the final manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2022.12.018.
Contributor Information
Caitlin A. Finney, Email: caitlin.finney@wimr.org.au.
Artur Shvetcov, Email: a.shvetcov@blackdog.org.au.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Data Availability
This meta-analysis did not generate any new data and used four publicly accessible datasets from the Gene Expression Omnibus database (GSE44770, GSE33000, GSE44768, and GSE36980). The top 1000 genes identified by the principal component analyses are available in Supplementary Table 1.
References
- 1.World Health Organization. Global action plan on the public health response to dementia 2017–2025. (ed Organization WH) (2017).
- 2.Cummings J. Lessons learned from Alzheimer's disease: Clinical trials with negative outcomes. Clin Trans Sci. 2018;11:147–152. doi: 10.1111/cts.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elder G.A., Gama Sosa M.A., De Gasperi R. Transgenic mouse models of Alzheimer's disease. Mount Sinai J Med. 2010;77:69–81. doi: 10.1002/msj.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jankowsky J.L., Zheng H. Practical considerations for choosing a mouse model of Alzheimer's disease. Mol Neurodegener. 2017;12 doi: 10.1186/s13024-017-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeTure M.A., Dickson D.W. The neuropathological diagnosis of Alzheimer's disease. Mol Neurodegener. 2019;14 doi: 10.1186/s13024-019-0333-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bali J., Gheinani A.H., Zurbriggen S., Rajendran L. Role of genes linked to sporadic Alzheimer's disease risk in the production of β-amyloid peptides. PNAS. 2012;109:15307–15311. doi: 10.1073/pnas.1201632109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stears R.L., Martinsky T., Schena M. Trends in microarray analysis. Nat Med. 2003;9:140–145. doi: 10.1038/nm0103-140. [DOI] [PubMed] [Google Scholar]
- 8.Freyhult E., Landfors M., Onskog J., Hvidsten T.R., Ryden P. Challenges in microarray class discovery: a comprehensive examination of normalization, gene selection and clustering. BMC Bioinform. 2010;11 doi: 10.1186/1471-2105-11-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers L.R.K. de los Campos G, Mias GI. Micorarray gene expression dataset re-analysis reveals variability in influenza infection and vaccination. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.02616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J., Yang Y. Potential genes and pathways along with immune cells infiltration in the progression of atherosclerosis identified via microarray gene expression dataset re-analysis. Vascular. 2020;28:643–654. doi: 10.1177/1708538120922700. [DOI] [PubMed] [Google Scholar]
- 11.La-Croix-Fralish M.L., Austin S.-L., Zheng F.Y., Levitin D.J., Mogil J.S. Patterns of pain: Meta-analysis of microarray studies of pain. Pain. 2011;152:1888–1898. doi: 10.1016/j.pain.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Grutzmann R., et al. Meta-anlaysis of microarray data on pancreatic cancer defines a set of commonly dysregulated genes. Oncogene. 2005;24:5079–5088. doi: 10.1038/sj.onc.1208696. [DOI] [PubMed] [Google Scholar]
- 13.Rhodes D.R., Barrette T.R., Rubin M.A., Ghosh D., Chinnaiyan A.M. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 14.Kelly J., Moyeed R., Carroll C., Albani D., Li X. Gene expression meta-analysis of Parksinson's disease and its relationship with Alzheimer's disease. Molecul Brain. 2019;12 doi: 10.1186/s13041-019-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noori A., Mezlini A.M., Hyman B.T., Serrano-Pozo A., Das S. Systematic review and meta-analysis of human transcriptomics reveals neuroinflammation, deficient energy metabolism, and proteostasis failure across neurodegeneration. Neurobiol Dis. 2021;149 doi: 10.1016/j.nbd.2020.105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan Y.-W., et al. Meta-analysis of the Alzheimer's disease human brain transcriptome and functional dissection in mouse models. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Signaevsky M., et al. Artificial intelligence in neuropathology: deep learning-based assessment of tauopathy. Lab Investig. 2019;99:1019–1029. doi: 10.1038/s41374-019-0202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su C., Tong J., Wang F. Mining genetic and transcriptomic data using machine learning approaches in Parksinson's disease. npj Parkinson's Dis. 2020;6 doi: 10.1038/s41531-020-00127-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahoo D., et al. Artificial intelligence guided discovery of a barrier-protective therapy in inflammatory bowel disease. Nat Commun. 2021;12 doi: 10.1038/s41467-021-24470-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma A., Dey P. A machine learning approach to unmask novel gene signatures and prediction of Alzheimer's disease with different brain regions. Genomics. 2021;113:1778–1789. doi: 10.1016/j.ygeno.2021.04.028. [DOI] [PubMed] [Google Scholar]
- 21.Luo J., et al. A comparison of batch effect removal methods for enhancement of prediction performance using MAQC-II microarray gene expression data. Pharmacogenet J. 2010:278–291. doi: 10.1038/tpj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claverie J.M. Computational methods for the identification of differential and coordinated gene expression. Human Mol Genet. 1999;8:1821–1832. doi: 10.1093/hmg/8.10.1821. [DOI] [PubMed] [Google Scholar]
- 23.Mutch D.M., Berger A., Mansourian R., Rytz A., Roberts M.-A. The limit fold change model: a practical approach for selecting differentially expressed genes from microarray data. BMC Bioinform. 2002;3 doi: 10.1186/1471-2105-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halsey L.G. The reign of the p-value is over: what alternative analyses could we employ to fill the power vacuum? Biol Lett. 2019;15 doi: 10.1098/rsbl.2019.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altman N., Krzywinski M. P values and the search for significance. Nat Methods. 2016;14 [Google Scholar]
- 26.Nuzzo R. Scientific method: statistical errors. Nature. 2014;506:150–152. doi: 10.1038/506150a. [DOI] [PubMed] [Google Scholar]
- 27.Verleysen M., Francois D. The curse of dimensionality in data mining and time series prediction. Springer (2005).
- 28.Tarca A.L., Carey V.J., Chen X.-W., Romero R., Draghici S. Machine learning and its application to biology. PLOS Comput Bio. 2007;3 doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jolliffe I.T., Cadima J. Principal component analysis: a review and recent developments. Philos Trans R Soc A. 2016;374 doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ringer M. What is principal component analysis? Nat Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 31.Yeung K.Y., Ruzzo W.L. Principal component analysis for clustering gene expression data. Bioinformatics. 2001;17:763–774. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- 32.Reich D., Price A.L., Patterson N. Principal component analysis of genetic data. Nat Genet. 2008;40:491–492. doi: 10.1038/ng0508-491. [DOI] [PubMed] [Google Scholar]
- 33.Szklarczyk D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Gene Ontology Consortium The gene ontology resource: 20 years and still going strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan J. Classification and regression tree analysis. Boston University (2014).
- 37.Lemon S.C., Roy J., Clark M.A., Friedman P.D., Rakowski W. Classification and regression tree analysis in public health: methodological review and comparison with logistic regression. Ann Behav Med. 2003;26:172–181. doi: 10.1207/S15324796ABM2603_02. [DOI] [PubMed] [Google Scholar]
- 38.Fushiki T. Estimation of prediction error by using K-fold cross-validation. Statist Comput. 2011;21:137–146. [Google Scholar]
- 39.Zhang B., et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;25:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayanan M., et al. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol Syst Biol. 2014;10 doi: 10.15252/msb.20145304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hokama M., et al. Altered expression of diabetes-related genes in Alzheimer's disease brains: the Hisayama study. Cereb Cortex. 2014;24:2476–2488. doi: 10.1093/cercor/bht101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swerdlow R.H. Mitochondria and mitochondrial cascades in Alzheimer's disease. J Alzheimer's Dis. 2018;62:1403–1416. doi: 10.3233/JAD-170585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swerdlow R.H., Burns JM S.M.K. The Alzheimer's disease mitochondrial cascade hypothesis: Progress and perspectives. Biochimica et Biophysica Acta. 2014;1842:1219–1231. doi: 10.1016/j.bbadis.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lunnon K., et al. Mitochondrial dysfunction and immune activation are detectable in early Alzheimer's disease blood. J Alzheimer's Dis. 2012;30:685–710. doi: 10.3233/JAD-2012-111592. [DOI] [PubMed] [Google Scholar]
- 45.Lunnon K., et al. Mitochondrial genes are altered in blood early in Alzheimer's disease. Neurobiol Aging. 2017;53:36–47. doi: 10.1016/j.neurobiolaging.2016.12.029. [DOI] [PubMed] [Google Scholar]
- 46.Adav S., Park J.E., Sze S.K. Quantitative profiling brain proteomes revealed mitochondrial dysfunction in Alzheimer's disease. Molecul Brain. 2019;12 doi: 10.1186/s13041-019-0430-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thal D.R., Rub U., Orantes M., Braak H. Phases of Aβ-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58 doi: 10.1212/wnl.58.12.1791. [DOI] [PubMed] [Google Scholar]
- 48.Garone C., Pietra A., Nesci S. From the structural and (dys)function of ATP synthase to deficiency in age-related diseases. Life. 2022;12 doi: 10.3390/life12030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beck S.J., et al. Dysregulation of mitochondrial F1F0-ATP synthase via OSCP in Alzheimer's disease. Nat Commun. 2016;7 doi: 10.1038/ncomms11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terni B., Boada J., Portero-Otin M., Pamplona R., Ferrer I. Mitochondrial ATP-synthase in the entorhinal cortex is a target of oxidative stress at stages I/II of Alzheimer's disease pathology. Brain Pathol. 2010;20:222–233. doi: 10.1111/j.1750-3639.2009.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt C., et al. Amyloid precursor protein and amyloid β-peptide bind to ATP synthase and regulate its activity at the surface of neural cells. Mol Psychiatry. 2008;13:953–969. doi: 10.1038/sj.mp.4002077. [DOI] [PubMed] [Google Scholar]
- 52.Vacirca D., et al. Autoantibodies to the adenosine triphosphate synthase play a pathogenic role in Alzheimer's disease. Neurobiol Aging. 2012;33:753–766. doi: 10.1016/j.neurobiolaging.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 53.Boada M., et al. ATP5H/KCTD2 locus is associated with Alzheimer's disease risk. Mol Psychiatry. 2013;19:682–687. doi: 10.1038/mp.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traylor M., et al. Shared genetic contribution to ischaemic stroke and Alzheimer's disease. Ann Neurol. 2016;79:739–747. doi: 10.1002/ana.24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paliwal D., et al. Mitochondrial pathway polygenic risk scores are associated with Alzheimer's disease. Neurobiol Aging. 2021;108:213–222. doi: 10.1016/j.neurobiolaging.2021.08.005. [DOI] [PubMed] [Google Scholar]
- 56.Andrews S.J., Fulton-Howard B., Goate A. Interpretation of risk loci from genome-wide association studies of Alzheimer's disease. Lancet Neurol. 2020;19:326–335. doi: 10.1016/S1474-4422(19)30435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacobs H.I.L., et al. The cerebellum in Alzheimer's disease: evaluating its role in cognitive decline. Brain. 2018;141:37–47. doi: 10.1093/brain/awx194. [DOI] [PubMed] [Google Scholar]
- 58.Trejo-Lopez J.A., Yachnis A.T., Prokop S. Neuropathology of Alzheimer's disease. Neurotherapeutics. 2021;19:173–185. doi: 10.1007/s13311-021-01146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao J., Irwin R.W., Zhao L., Nilsen J., Hamilton R.T., Brinton R.D. Mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease. PNAS. 2009;106:14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez Ortiz J.M., Swerdlow R.H. Mitochondrial dysfunction in Alzheimer's disease: role in pathogenesis and novel therapeutic opportunities. Brit J Pharmacol. 2019;176:3489–3507. doi: 10.1111/bph.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jang J.Y., Blum A., Liu J., Finkel T. The role of mitochondria in aging. J Clin Investig. 2018;128:3662–3670. doi: 10.1172/JCI120842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Data Availability Statement
This meta-analysis did not generate any new data and used four publicly accessible datasets from the Gene Expression Omnibus database (GSE44770, GSE33000, GSE44768, and GSE36980). The top 1000 genes identified by the principal component analyses are available in Supplementary Table 1.