Abstract
Microsporidia are unicellular eukaryotes occuring as obligate intracellular parasites which produce resistant spores. A unique motile process is represented by the sudden extrusion of the sporal polar tube for initiating entry of the parasite into a new host cell. The complete sequence of an acidic proline-rich polar tube protein (renamed PTP1) has been previously reported for Encephalitozoon cuniculi and E. hellem. Our immunological investigations provided evidence for an additional PTP in E. cuniculi, termed PTP2. The corresponding gene was sequenced and then expressed in Escherichia coli. As expected, mouse antibodies raised against the recombinant protein reacted specifically with the polar tube. The singlecopy ptp1 and ptp2 genes of E. cuniculi were tandemly arranged on chromosome VI. Polyadenylation of the mRNAs was demonstrated. Identification and sequencing of homologous genes in the two other human-infecting Encephalitozoon species (ptp2 in E. hellem and ptp1 and ptp2 in E. intestinalis) were facilitated by conserved gene clustering. PTP2 appears as a novel structural protein (30 kDa) with a basic lysine-rich core and an acidic tail. Unlike PTP1, this protein is devoid of large tandem repeats. The interspecies conservation of cysteine residues supports a major role of disulfide bridges in polar tube assembly. The two PTPs should serve as both molecular markers of spore differentiation and diagnostic tools.
Microsporidia (phylum Microspora Sprague, 1977) are small spore-forming unicellular eukaryotes with an obligate intracellular parasitic lifestyle. These parasites, characterized by 70S ribosomes and the absence of mitochondria, were thought to be very ancient (10). However, data accumulated from recent molecular phylogenies lend credit to a close relationship of these organisms with fungi (36). Several species are of medical and veterinary significance, infecting animals and humans (7). Three species from the Encephalitozoon genus (E. cuniculi, E. hellem, and E. intestinalis) are known to be involved in AIDS-associated pathologies (13). Disorders in immunocompetent individuals also have been reported. For example, E. intestinalis was found in travelers, not infected with human immunodeficiency virus, presenting with chronic diarrhea (33). Serological studies with blood donors and pregnant women revealed a prevalence of about 8% (37).
Microsporidia exhibit a remarkable invasion mechanism depending on the extrusion of a specific organelle called the polar tube, originally coiled within the spore. The polar tube discharges from the anterior pole of the spore like an everting glove finger (25) and then is used to transfer the sporoplasm inside a potential host cell. The whole process of in vitro spore germination is completed in less than 2 s (17). Very little information is available about the primary structure of polar tube proteins (PTPs) and the extent of interspecies sequence variability. Molecular characterization of the polar tube is therefore of importance for improving diagnostic and defining therapeutic strategies.
The polar tube resists dissociation in detergents, urea, and acids but dissociates in the presence of thiol-reducing agents, e.g. 2-mercaptoethanol or dithiothreitol (DTT) (19, 40). A Glugea americanus 43-kDa PTP, differentially solubilized with 2% DTT and purified by high-pressure liquid chromatography, was shown to contain a large amount of proline residues (19). Proline-rich PTPs were similarly isolated from Encephalitozoon species, with the apparent molecular sizes varying from 45 to 55 kDa (20). We previously described the first complete sequence of a proline-rich 55-kDa PTP in E. cuniculi (11). The predicted protein has 395 amino acids (aa), with a central region consisting of four 26-aa repeats, and shows no homology with known proteins. A similar ptp gene encoding a 453-aa protein in E. hellem has been also sequenced (21). The repeated region (six 20-aa repeats) is very divergent relative to that in E. cuniculi (22). Since the molecular sizes calculated from sequences (43 kDa in E. hellem and 37 kDa in E. cuniculi) are not consistent with those deduced from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) migrations (50 to 55 kDa), we propose the designation PTP1 for members of this new protein family.
We identified an E. cuniculi PTP (PTP2) assumed to be more conserved than PTP1 among microsporidian species, as judged by immunological cross-reactivity with a 34- to 35-kDa protein from a species of the genus Glugea (12). As reported in the present paper, the genes encoding PTP2 in the three human-infecting Encephalitozoon species were fully sequenced. To complete the comparison of the two different PTPs, the ptp1 gene was also cloned and then sequenced in E. intestinalis. The conservation of the PTP2 sequence within a microsporidian genus suggests that this protein plays a basic role in the construction of the polar tube and may be of interest for medical applications.
MATERIALS AND METHODS
Growth of parasites and isolation of DNA.
E. cuniculi, E. hellem, and E. intestinalis were grown in vitro in either Madin-Darby canine kidney (MDCK), human lung fibroblast (MRC-5), or rabbit kidney (RK13) cells as described elsewhere (2). Spores collected from supernatants were harvested (5,000 × g for 10 min), washed, purified as described previously (11), and stored in phosphate-buffered saline (PBS) at 4°C. Genomic DNA was released by boiling purified spores at 100°C for 10 min.
Antibody production.
Polyclonal antibodies (PAbs) and monoclonal antibodies (MAbs) to microsporidian proteins were described previously (11). BALB/c mice were immunized with the recombinant E. cuniculi (EcPTP2) expressed in E. coli. After expression, the recombinant protein was purified by chromatography on Ni-nitrilotriacetic acid (Ni-NTA) resin (Qiagen). Recombinant protein was then excised from Coomassie blue-stained gels and crushed in PBS with a Potter homogenizer. Mice were injected intraperitoneally with samples homogenized with Freund complete adjuvant, and identical injections were given on days 14 and 21 with Freund incomplete adjuvant. Sera were collected 1 week after the last injection and stored at −20°C.
SDS-PAGE and Western blotting analysis.
SDS-PAGE was performed using standard methods (24). Crude extracts from microsporidia or recombinant bacteria were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Western blot analysis was carried out using 10 to 12% polyacrylamide gels run under reducing conditions with 5 to 10% 2-mercaptoethanol in the loading samples. After electrophoresis, proteins were transferred to polyvinylidene difluoride (Immobilon-P; Polylabo). For detection, the membranes were incubated with MAbs or PAbs and then with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG), IgA, and IgM (Sigma), and bound antibodies were visualised using the ECL system (Amersham).
Indirect immunofluorescence (IFA).
Intracellular parasites grown for 24 to 48 h in MRC-5 cells on glass slides were fixed with 4% paraformaldehyde and 0.05% glutaraldehyde for 20 min at room temperature or with methanol for 10 min at −70°C. Cells were then permeabilized with 70% ethanol–0.5% Triton X-100 and blocked with 5% skim milk in PBS. Slides were incubated for 1 h with primary antibodies (PAbs or MAbs) diluted in PBS–0.1% Tween 20, washed, and incubated further for 1 h in fluorescein isothiocyanate-labeled goat anti-mouse IgG, IgA, and IgM (Sigma). Slides were mounted and preparations were examined with a BH2 Olympus epifluorescence microscope.
Peptide sequencing.
The 35-kDa protein isolated from two-dimensional gels was digested with 0.8 μg of endoprotease-LysC in 0.1 M Tris-HCl (pH 8.6)–0.5 M EDTA–0.03% SDS at 35°C for 18 h, and peptides were then separated using high-pressure liquid chromatography on DEAE-C18 columns with a gradient of acetonitrile–0.1% trifluoroacetic acid. One peptide of 15 aa (AVQGTDRCILAGIID) was sequenced using Edman degradation (Applied Biosystems 473A sequencer).
Cloning and sequencing of ptp genes.
The different PCR and single-specific primer PCR (SSP-PCR) amplification steps are described in Fig. 2. The primers used were A (5′-CAGGGIACIGAYMGITGYATHYTIGC-3′), B (5′-GTACTTGCGCTTGTTCACC-3′), C (5′-GAGGAGACAAGCTAATTGC-3′), D (5′-GACATACAGAAGACGGGG-3′), E (5′-CTTATCAGAGCAGATGTTC-3′), F (5′-CCATGCGAACCTAAGAAG-3′), G (5′-GGCTGAAGTCCATAGTCAAC-3′), H (5′-GAAGGAGATCAAGGAGAGCCC-3′), I (5′-ATGAAAGGTATTTCTAAG-3′), J (5′-GATTGTTTTTAGAGGGATCTG-3′), K (5′-CATTGTCATTGTCGACATCG-3′), L (5′-GGCGAGAAGTAACAACAT-3′), M (5′-GAGATTTCTAACGGCGAGG-3′), N (5′-ATRCAICKRTCIGTICCYTG-3′), and O (5′-GCAATGGTTCAAAGAGCC-3′). Amplified products were cloned into pGEM-T Easy Vector System I (Promega). Recombinant plasmids were sequenced using the ABI Prism Dye Terminator Cycle Sequencing kit according to the recommendations of the manufacturer (Perkin-Elmer). Thermocycling of the sequencing reactions and electrophoresis were carried out on a GeneAmp PCR system 2400 and a ABI Prism 377 sequencer (Perkin-Elmer), respectively. Gel readings were processed using the Staden package (35), and the resulting contigs were compared with databases using BLAST (1). Staden package and BLAST programs are available on the French molecular biology server Infobiogen.
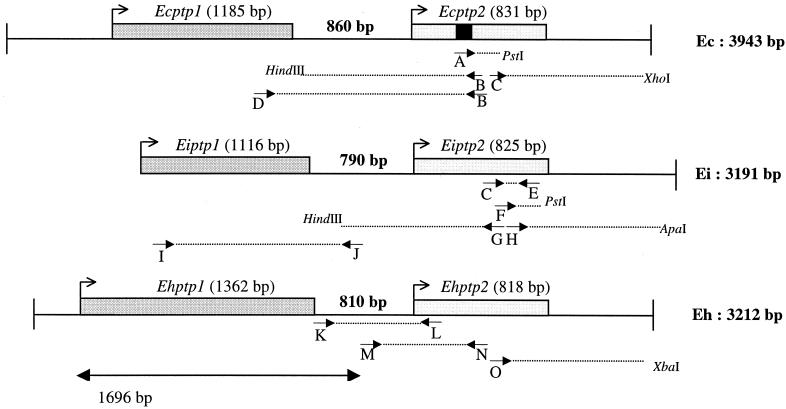
FIG. 2.
Schematic representation of ptp1 and ptp2 gene clusters with positions of PCR primers (A to O) in the three Encephalitozoon species, E. cuniculi (Ec), E. intestinalis (Ei), and E. hellem (Eh). The peptide obtained by microsequencing of EcPTP2 (AVQGTDRCILAGIID) is indicated by a black square. Sense primer D, designed 186 bp upstream from the stop codon of Ecptp1, combined with antisense primer B, designed 380 bp downstream from the initiation codon of Ecptp2, were used to amplify a 1.4-kbp DNA fragment. Two primers deduced from the Ecptp2 sequence (C and E, positions 419 to 699 in the Ecptp2 ORF) were used to amplify a 280-bp DNA fragment in E. intestinalis. The corresponding sequence shared about 90% identity with that of Ecptp2 but showed some differences that were useful to determine specific oligonucleotides. Downstream and upstream regions of the 280-bp known sequence were amplified with PstI and HindIII ligation products, respectively, using two specific primers (F and G) and the reverse vector primer. Thus, SSP-PCR experiments led to 120-bp (3′ with PstI) and 1,150-bp (5′ with HindIII) amplicons, respectively. The 3′ end of Eiptp2 was completed through another SSP-PCR step with primer H, resulting in a 570-bp amplification from the ApaI ligation product. The final sequence is 1,968 bp in length with a predicted 825-bp ORF coding for EiPTP2. The Eiptp1 gene was amplified using a combination of an antisense primer in the 5′ flanking region of Eiptp2 (J) and a sense primer (I) determined from the alignment of conserved regions encoding signal peptides of EcPTP1 and EhPTP1. For E. hellem, primer K, determined in the 3′ known flanking region of Ehptp1, was combined with the reverse primer L, determined by the alignment of the highly conserved N-terminal sequence encoding the PTP2 signal peptide in E. cuniculi and E. intestinalis. The whole coding sequence of Ehptp2 and its 3′ UTR were completed by SSP-PCR amplification with primer O. The 1,696-bp sequence is from reference 21.
For application of the SSP-PCR technique (34), digestion of genomic DNA (200 ng) for 4 h with 100 U of different restriction enzymes was followed by two phenol-chloroform-isoamyl alcohol extractions. A 1-μg sample of pBluescript-II SK(+) (Stratagene) vector was digested using the corresponding enzyme and dephosphorylated with 10 U of calf intestine alkaline phosphatase (Eurogentec). After two phenol-chloroform-isoamyl alcohol extractions, vector DNA was coprecipitated with the digested genomic DNA. Ligation was carried out overnight at 16°C in 12 μl of a mixture containing 6 U of T4 DNA ligase (Pharmacia) and 1 mM ATP in One Phor All buffer. PCR was carried out in 50 μl of reaction mixture containing 1 μl of diluted ligation mixture (1:10), 0.25 μM vector primer (universal or reverse), 0.25 μM specific primer, 20 μM each deoxynucleoside triphosphate in Taq DNA polymerase buffer (2.5 mM MgCl2), and 1 U of Taq DNA polymerase (Goldstar; Eurogentec).
EcPTP2 heterologous expression in E. coli.
The coding sequence of EcPTP2 was amplified by PCR to introduce a BamHI site at the start codon. The oligonucleotide 5′-GGATCCGCAGCACCTCTCCATG-3′ was combined with the antisense oligonucleotide primer 5′-CACTTGAAGATTCAATCC-3′ for PCR amplification. The PCR product was first cloned into pGEM-T Easy vector (Promega) and subcloned after BamHI-SacI digestion into the bacterial expression vector pQE-30 (pQE expression system from Qiagen). Expression of the recombinant protein was analyzed in E. coli strain M15 after induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
RNA extraction and RT-PCR.
Total RNA was extracted from E. cuniculi-infected MRC-5 cells. Cells were lysed in 5 ml of a lysis buffer containing 180 mM Tris, 90 mM LiCl, 4.5 mM EDTA, and 1% SDS (pH 8.2). After incubation at 55°C for 30 min in 1 volume of phenol-chloroform and three phenol-chloroform-isoamyl alcohol extractions, RNA was precipitated, resuspended in 200 μl of diethyl pyrocarbonate-distilled water, and stored at −80°C. DNA was eliminated by treatment with 10 U of DNase (Promega) in the presence of 40 U of RNAsine (Promega) for 1 h at 37°C. Reverse transcription (RT) was done using 5 μg of total RNA. After denaturation for 2 min at 65°C, RNA was incubated with 40 U of avian myeloblastosis virus reverse transcriptase (Promega) in 50 mM Tris (pH 8.3)–8 mM MgCl2–30 mM KCl–10 mM DTT–0.5 mM deoxynucleoside triphosphates–RNAsine–100 pmol of oligo(dT) (5′-GACTCACTATAGGGCATGCTTTTTTTTTTTTTTTTTT-3′). The reaction mix was incubated for 90 min at 42°C. PCR amplification of the cDNA 3′ end was done with a 1:20 dilution of the RT reaction mixture with a primer specific for either Ecptp1 (D, 5′-GACATACAGAAGACGGGG-3′) or Ecptp2 (C, 5′-GAGGAGACAAGCTAATTGC-3′) and the primer corresponding to the adapter sequence (5′-GACTCACTATAGGGCATGC-3′).
Sequence analysis.
Protein sequence alignments were done using the GeneStream alignment program, which is accessible via an electronic mail server (http://vega.igh.cnrs.fr/bin/nph-align_query.pl). The prediction of signal peptide cleavage sites was done using the algorithm of von Heijne (39) and the PSORT program (30).
Nucleotide sequence accession numbers.
The nucleotide sequences of EcPTP1, EcPTP2, EiPTP1, EiPTP2, and EhPTP2 have been deposited in GenBank under accession numbers AX007049 through AX007053, respectively (C. Vivarès, A. Danchin, and F. Delbac. July 1998. Patent WO0001724; FR no. 98/08692, 07.07.1998. Protéines de tube polaire de microsporidie, acides nucléiques codant pour ces protéines et leurs applications).
RESULTS
An E. cuniculi gene encodes a lysine-rich 30-kDa PTP (EcPTP2).
We previously showed that the MAb Ec102, directed against the polar tube of E. cuniculi and reacting with the proline-rich EcPTP1 (a protein with an apparent size of 55 kDa in SDS-PAGE), cross-reacted in Western blotting with two other protein bands of 35 and 28 kDa in size (12). In addition, a PAb (PAb anti-35) raised against an E. cuniculi 35-kDa protein band specifically labeled the polar tube mainly after extrusion.
As a prerequisite to the isolation of the gene encoding this putative 35-kDa PTP, two-dimensional electrophoresis and immunoblotting analysis (with MAb Ec102 and PAb anti-35) were performed. The reactive spot, with a basic pI close to 9, was used for an internal microsequencing after endolysine C digestion. One peptide (AVQGTDRCILAGIID) was chosen for designing oligonucleotide primers. In a first step of SSP-PCR using PstI-digested genomic DNA, with a degenerate primer A determined from the peptide sequence QGTDRCILA, a 150-bp DNA fragment was amplified. The DNA sequence was then extended in both directions using two specific primers (B and C). The new DNA fragments of about 900 and 800 bp were delimited by XhoI and HindIII restriction sites, respectively (see Fig. 2). The complete sequence (1,739 bp) included a 831-bp open reading frame (ORF) that encodes a 277-aa protein.
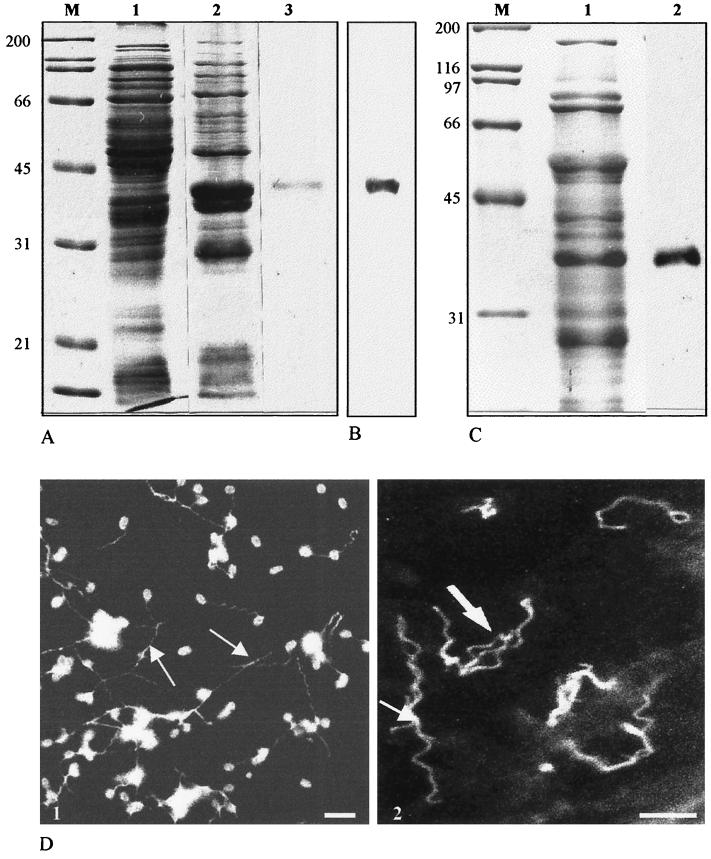
To assess polar tube localization, heterologous expression of EcPTP2 was done in Escherichia coli. First, the full-length ptp2 gene was PCR amplified and cloned into a bacterial expression plasmid (pQE30-ptp2) in frame with six histidine residues at the N terminus of the protein. After induction, the bacterial lysate was analyzed by SDS-PAGE. No expression was obtained when using the full ORF. Some toxicity of the protein may be assumed, because IPTG induction resulted in the inhibition of bacterial growth. In contrast, a high expression level was observed with a construction devoid of the first 60 nucleotides encoding a potential signal peptide. The recombinant PTP2, purified on an Ni-NTA column, migrated at 35 to 40 kDa, which is again beyond the predicted molecular mass (Fig. 1A). As expected, MAb Ec102 reacted in Western blotting with the recombinant proteins (Fig. 1B). Mouse PAbs raised against the recombinant protein recognized both a 35-kDa band in E. cuniculi protein extracts (Fig. 1C) and the polar tube in IFA (Fig. 1D), confirming the isolation of the expected ptp gene.
FIG. 1.
(A) SDS-PAGE analysis of E. coli-expressed recombinant EcPTP2 (Coomassie blue staining). Lanes 1 and 2, total E. coli lysate before (lane 1) and after (lane 2) IPTG induction; lane 3, recombinant EcPTP2 after purification on an Ni-NTA column; lane M, molecular mass standards in kilodaltons. (B) Immunoblotting reactivity of MAb Ec102 with E. coli proteins 4 h after IPTG induction. The blot was probed with a 1:10,000 dilution of MAb Ec102 and developed using ECL (Amersham). (C) Lane 2, immunoblot of E. cuniculi whole-cell homogenates probed with anti-recombinant EcPTP2 antiserum. Lane 1, total E. cuniculi proteins stained with Coomassie blue. (D) Indirect immunofluorescence of E. cuniculi spores with extruded polar tubes (arrows). (1) Labeling with an antiserum against total proteins; (2) specific labeling of polar tubes with anti-recombinant PTP2. Bars, 5 μm.
The deduced amino acid sequence of EcPTP2 (see Fig. 4) represents a 277-residue polypeptide with a molecular mass of 30075 Da. The calculated pI (∼8.6) is in agreement with that deduced from two-dimensional polyacrylamide gels. Comparison with proteins from databases failed to reveal significant homology. The protein is mainly characterized by lysine (11.6%) and glutamate (9%) richness. Tryptophan is the only amino acid which is lacking. The proline (5.4%) and cysteine (2.9%) contents are below those of EcPTP1 (13.4 and 4.6%, respectively). The N-terminal part shows characteristics of a signal peptide (hydrophobic residues and an alpha-helix), with a putative cleavage site between residues 13 and 14. The central region contains a lysine-rich octapeptide motif (KPKKKKSK). In contrast, a C-terminal region of 27 residues, devoid of any basic residue, possesses 4 aspartate and 5 glutamate residues, thus forming an acidic tail (pI 3.3). One putative N-glycosylation site, NSTS (residues 134 to 137), and one RGD motif (residues 140 to 142), possibly involved in some protein-protein interactions, are present.
FIG. 4.
Alignment of the three complete PTP2 amino acid sequences. Amino acids are numbered on the right. Identical residues are indicated by asterisks. The common putative cleavage site for the signal peptide is shown by an arrow. The eight cysteine residues are shaded. The common putative N-glycosylation site and the RGD sequence are indicated in boldface. The lysine-rich central region and the acidic tail are underlined.
Flanking regions are AT rich, particularly a 100-bp sequence extending upstream from the ATG initiation codon (64% of AT nucleotides). A putative polyadenylation site (AATTAAA) is located 6 nucleotides downstream from the TAA stop codon. We also investigated the gene copy number and chromosomal location of Ecptp2 using Southern hybridization with a ptp2 probe to either E. cuniculi genomic DNA cut with different restriction enzymes or the molecular karyotype. Our data indicate that the Ecptp2 gene exists as a single copy per haploid genome and is located on the same chromosome as the Ecptp1 gene, i.e., chromosome VI (data not shown).
A ptp1-ptp2 gene cluster exists in the three Encephalitozoon species.
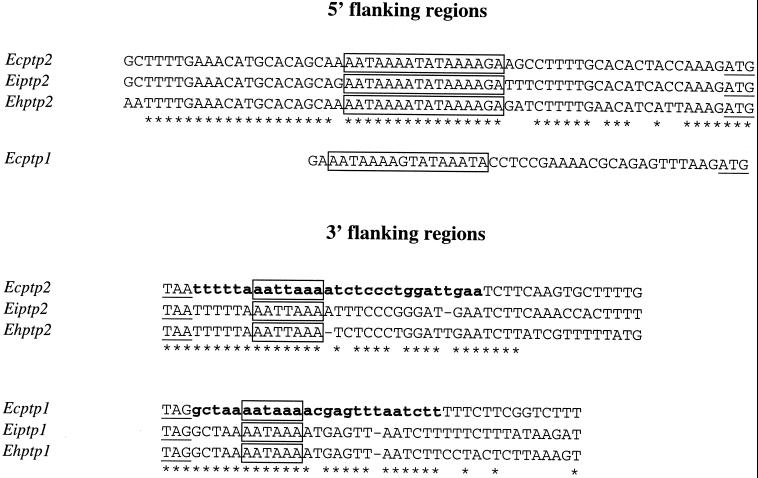
The chromosomal colocalization of the E. cuniculi ptp1 and ptp2 genes led us to test possible clustering of these genes, through PCR experiments with pairs of primers designed to correspond to the 5′ end of one gene and the 3′ end of the other gene. Sequencing of a 1.4-kbp amplified fragment showed that the Ecptp2 gene is located downstream of the Ecptp1 gene, with the respective ORFs being on the same DNA strand and separated by 860 nucleotides (Fig. 2). There is no sequence homology of this interval with known genes, while the 3′ flanking region of the ptp2 gene shared homology with an RNA-binding protein-encoding gene (on the complementary strand). RT of RNAs from E. cuniculi-infected MRC-5 cells was performed using a poly(T) oligonucleotide coupled with an adapter sequence in the 5′ region. Specific PCR amplification of 3′ regions of the cDNAs corresponding to Ecptp1 and Ecptp2 mRNAs was then done with primers D and C, respectively, and the primer corresponding to the adapter sequence. Sequencing of the corresponding PCR products (260 and 480 bp) provided evidence for short 3′ untranslated regions (UTRs) (25 nucleotides in Ecptp1 and 27 to 29 nucleotides in Ecptp2) with polyadenylation signals and poly(A) tails (Fig. 3). This confirms that the genomic sequence is intronless (at least in the 3′ end) and supports independent transcription of the two genes.
FIG. 3.
Alignments of ptp1 and ptp2 gene 5′ and 3′ flanking regions showing conserved signals. Start and stop codons are underlined. Identical nucleotides are indicated by asterisks. The AT-rich consensus sequence in the 5′ region and potential site of polyadenylation are boxed. Partial E. cuniculi cDNAs with short 3′ UTRs (less than 30 nucleotides) are in boldface lowercase letters.
Several anti-polar tube MAbs, including Ec102, reacted with some proteins of E. intestinalis and E. hellem and specifically decorated their respective polar tubes in immunofluorescence assays and electron microscope immunocytochemistry (reference 27 and unpublished data). In addition, anti-E. cuniculi recombinant PTP2 antibodies cross-reacted by immunoblotting with a 30-to 35-kDa band and the polar tubes from the two above-mentioned Encephalitozoon species, suggesting the presence of PTP2 homologues. Initially, based on the use of primers designed to correspond to E. cuniculi PTP sequences, a combination of PCR and SSP-PCR experiments with E. intestinalis genomic DNA provided a contig of 3,191 bp with tandemly arranged ptp1 and ptp2 ORFs as in E. cuniculi, with the noncoding interval being reduced to 790 bp (Fig. 2). The two predicted ORFs encoding EiPTP2 and EiPTP1 are 825 and 1,116 bp in length, respectively.
Assuming a similar gene organization in E. hellem, we designed primers in the 3′ flanking region of Ehptp1 and succeeded in amplifying both the intergenic and Ehptp2 coding regions. The sequencing of a 668-bp amplified product showed that the Ehptp2 ORF (818 bp) was located 810 bp downstream of the Ehptp1 ORF (1,362 bp). The whole coding sequence of Ehptp2 and the 3′ flanking region were completed by SSP-PCR amplifications, as shown in Fig. 2. Southern blotting indicated that this conserved ptp1-ptp2 gene pair (contig 3,212 bp) can also be assigned to a single chromosome (data not shown).
Sequence comparison of the 5′ and 3′ flanking parts of the ptp genes reveals some highly conserved AT-rich motifs (Fig. 3). For the ptp2 genes, an AT-rich region (67%) extends over 60 nucleotides upstream of the translation start codon. As the transcription initiation site has not been determined, this may include 5′ leader and promoter elements. There is more than 80% identity in this region shared by the three ptp2 genes. Ecptp1 exhibits an AT-rich 5′ region with a motif 23 nucleotides upstream of the start codon (boxed in Fig. 3) similar to that of ptp2 genes. However, this motif is absent in the sequence reported for Ehptp1 (21). The typical polyadenylation signal, usually seen in higher eukaryotes (AATAAA), is found 5 nucleotides downstream of the TAG stop codon of each ptp1 gene in the three Encephalitozoon species. The TAA stop codon, conserved for the three ptp2 genes, is followed by a similar motif, AATTAAA (Fig. 3). In E. intestinalis, as in E. cuniculi, an additional ORF having significant homologies with a gene encoding a RNA-binding protein is associated with the complementary DNA strand of the 3′ flanking region of ptp2.
Comparison of PTP2-coding regions.
PTP2s are basic proteins of similar size (close to 30 kDa), with the maximal difference between EcPTP2 and EhPTP2 being only five residues (Table 1). The degree of conservation at the amino acid level is higher than for PTP1 and extends throughout the entire coding region (Fig. 4). There is more than 80% identity between EcPTP2 and EiPTP2, 58% identity between EcPTP2 and EhPTP2, and 60% identity between EiPTP2 and EhPTP2. Thus, the PTP2 of E. intestinalis is more closely related to that of E. cuniculi than to that of E. hellem.
TABLE 1.
Major characteristics of PTP1 and PTP2 from E. cuniculi, E. intestinalis, and E. hellema
| Protein | Total aa | pI | %
|
C-terminal residue | Presence of tandem repeats (20–28 aa) | No. of N-glycosylation sites | ||
|---|---|---|---|---|---|---|---|---|
| Pro | Lys | Cys | ||||||
| EcPTP1 | 395 | 4.9 | 13.4 | 1.6 | 4.6 | C | + | 3 |
| EiPTP1 | 371 | 4.9 | 12.9 | 2.7 | 4.6 | C | + | 0 |
| EhPTP1 | 453 | 4.9 | 14.6 | 1.4 | 4.9 | C | + | 9 |
| EcPTP2 | 277 | 8.6 | 5.4 | 11.6 | 2.9 | E | − | 1 |
| EiPTP2 | 275 | 8.6 | 5.5 | 11.3 | 2.9 | E | − | 2 |
| EhPTP2 | 272 | 8.6 | 5.5 | 10.3 | 2.9 | E | − | 1 |
Three different regions can be distinguished: an N-terminal part of ∼50 noncharged residues, an internal basic region including a central lysine-rich hexapeptide (consensus K/VKKKS/TK), and an acidic C-terminal part (24 to 27 aa). An N-terminal signal peptide forming an α-helix is predicted, but cleavage at position 13 remains to be demonstrated. The major residue is lysine (Table 1), and one glutamate residue is observed at the C terminus of each PTP2. The other predominant amino acids are glutamate (5.9 to 9.0%), glycine (7.6 to 9.2%), serine (4.8 to 6.9%), and glutamine (5.8 to 7.3%). The eight cysteine residues are similarly located in the three PTP2 sequences (Fig. 4). Putative N-glycosylation sites were also deduced from the sequences: two in E. intestinalis (positions 132 to 135 and 248 to 251) and one each in E. cuniculi (134 to 137) and E. hellem (134 to 137); just downstream of this site, an RGD motif (for cell attachment?) is found in E. cuniculi and E. intestinalis but is replaced by RGN in E. hellem.
A shorter PTP1 with degenerate repeats in E. intestinalis.
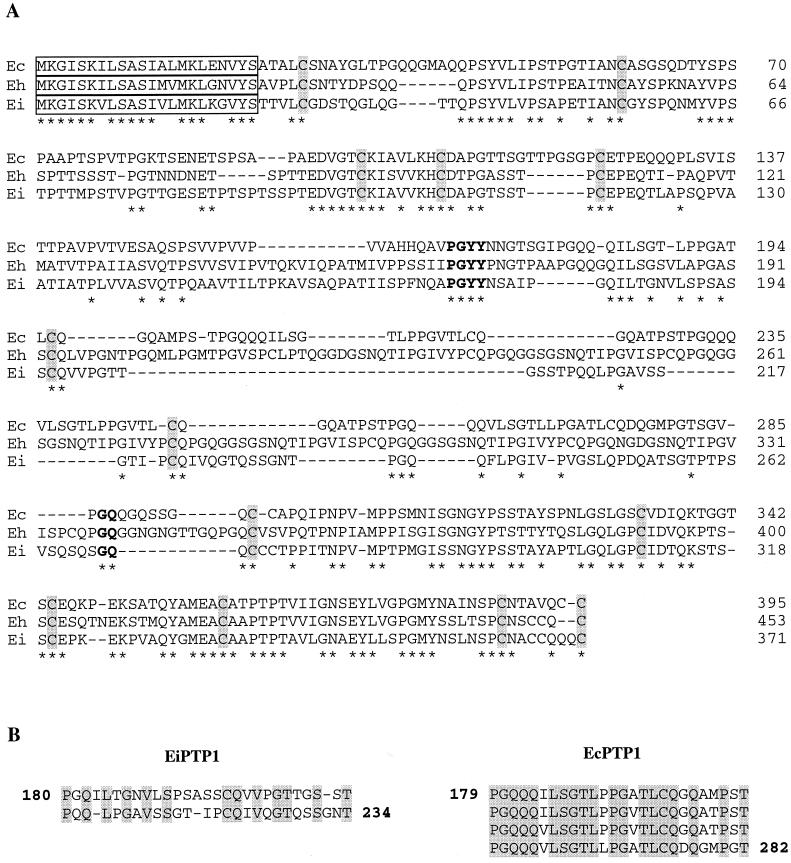
The PTP1-coding region in E. intestinalis, representing a 371-residue polypeptide (35 kDa), is shorter than those in other species (43 kDa in E. hellem and 37 kDa in E. cuniculi). Sequence alignment confirmed the highest homology in the N- and C-terminal domains (Fig. 5A). The N-terminal signal peptide is remarkably conserved (17 identical residues over 22). The whole sequence of EiPTP1 showed only 48 to 49% identity with those of EhPTP1 and EcPTP1, mainly because of the divergent central domain (delimited by common boundaries PGYY/GQ in Fig. 5A) The repetitive character of this core is less evident in E. intestinalis. Only two major, highly degenerated repeats of 27 or 28 aa were indeed distinguishable (Fig. 5B), contrasting with the nearly perfect repeats seen in EhPTP1 (six 20-aa repeats) and EcPTP1 (four 26-aa repeats) (11, 21). This suggests a rather minor role of such repeats in PTP1 organization and function.
FIG. 5.
(A) Alignment of the PTP1 amino acid sequences from E. cuniculi (Ec), E. hellem (Eh), and E. intestinalis (Ei). Amino acids are numbered on the right. Identical amino acids are indicated by asterisks. The highly conserved 22-aa signal peptide is boxed. The three PTP1s share common N- and C-terminal parts but diverge in their tandemly repeated sequences, forming a central core delimited by common boundaries (in boldface). Conserved cysteine residues are shaded. (B) Alignment of the tandemly repeated sequences of EiPTP1 (two repeats) and EcPTP1 (four repeats). Identical amino acids are shaded.
As expected, proline is the most predominant residue (Table 1). The glycine content is high (11.8%), and the two hydroxylated amino acids represent 21.7%. Tryptophan and arginine residues are lacking. As shown in Fig. 5A, at least 13 cysteine residues are at conserved positions, including the one at the extreme C terminus. Unlike in EcPTP1 and EhPTP1, no N-glycosylation site was found in EiPTP1.
DISCUSSION
The polar tube is a typical microsporidian spore structure whose extrusion is absolutely required for the invasion of a host cell. Its protein heterogeneity has been supported by biochemical and immunological data (3, 12, 20). However, only one PTP (here referred to as PTP1) was defined at the primary structure level, after isolation of the corresponding single-copy gene in two species of the family Encephalitozoonidae, E. cuniculi (11) and E. hellem (21). We describe here a gene encoding another antigenic protein (PTP2) located in the polar tubes of three Encephalitozoon species. For a better comparison, we have also cloned and sequenced the ptp1 gene of E. intestinalis, a major human-infecting microsporidian mainly responsible for intestinal disorders. Alignment of the deduced amino acid sequences confirmed that the three species exhibit two different PTPs reflecting two novel structural protein families. While PTP1 is a proline-rich acidic protein with a highly variable repeat-containing core, PTP2 is a more conserved lysine-rich basic protein with an acidic tail. The conserved charged residues in PTP sequences can be thought to be involved in some protein-protein ionic interactions required for the assembly of the polar tube. The role of disulfide bonds in protein-protein interactions has been postulated through in vitro polymerization of a purified PTP (23 kDa) in Ameson michaelis (40) and dissociation of the polar tube in the presence of thiol-reducing agents (19, 40). It seems very likely, therefore, that conserved cysteine residues in PTP2 as well as PTP1 can form intermolecular disulfide bridges of primary importance for the high tensile strength and functioning of the polar tube. This somewhat resembles the case of proline-rich minicollagens found in extrusive organelles (nematocysts) involved in prey capture and locomotion of cnidarians (23). However, it should be stressed that no evident homology exists between these unusual collagens and known PTPs, despite the fact that PTP1 is rich in glycine and proline residues.
The two PTPs are characterized by hydrophobic leader sequences suggestive of secreted proteins. The N terminus of PTP1 consists of a signal sequence of 22 aa cleaved during maturation, probably for targeting to the endoplasmic reticulum, in E. cuniculi and E. hellem (11, 21). This peptide also exists in E. intestinalis PTP1, and the high sequence conservation strongly argues for a similar processing. The signal peptide for PTP2 is predicted to be cleaved between the 13th and 14th aa. The reduced length relative to that in PTP1 cannot be due to a misplaced start codon. No in-frame ATG is found in the upstream region, and an A occupies the −3 position as observed in 5′ UTR of protozoa (41). Interestingly, the start codon is preceded by a trinucleotide, AAG, which is common to both ptp genes (Fig. 3) and the recently characterized 5′ UTR of an E. cuniculi gene encoding the spore wall protein SWP1 (6).
Several potential glycosylation sites can be deduced from PTP1 and PTP2 sequence analysis, suggesting their glycoprotein nature, which is in agreement with cytochemical data indicating the presence of glycoconjugates in the polar tube (38). Recent electron microscope studies indicate that the microsporidian spore can attach to the host cell membrane prior to the invasion and that entry of sporoplasm occurs by phagocytosis like for other intracellular parasites (8, 28). In addition, the basolateral domain of enterocytes would be the preferential site of penetration for E. intestinalis, as suggested by confocal microscopy observations of the colocalization of the parasite and F-actin at the periphery of host cells (16). Some N-linked oligosaccharides of PTPs might be essential for early interactions between the polar tube and the host cell surface.
Encephalitozoon species possess the smallest nuclear genomes so far identified: 2.9, 2.6, and 2.3 Mb in E. cuniculi, E. hellem, and E. intestinalis, respectively (4, 5). A report on a 4.3-kb chromosomal region of E. cuniculi has revealed that some intergenic regions can be less than 50 bp (15). Our data provide additional information about gene organization and transcription of microsporidian genes. All Encephalitozoon ptp1 and ptp2 genes are present as a single copy with a common clustering on one chromosome, with conserved orientation and spacing. This is the first example of conservation of synteny between microsporidian genomes. The products of the two contiguous genes are involved in the construction of the same cellular structure, the polar tube. This supports the conception that a strong selection pressure maintains the evolutionary conservation of the order of genes whose products are physically associated, as frequently invoked for prokaryotic gene clusters. Conserved synteny has been also reported among the chromosomes of all four species of human malaria parasites (9). We demonstrated that E. cuniculi ptp1 and ptp2 mRNAs are polyadenylated and display reduced 3′ UTRs. Short 3′ UTRs have been described for Giardia, an amitochondriate parasitic flagellate with a small genome (26, 29). It seems likely that the 5′ UTRs of ptp mRNAs are very short, as is the case for the SWP1 gene (6). In Trypanosoma, genes are transcribed polycistronically, with the pre-mRNA being processed by 3′ -end formation and trans splicing to create conventional eukaryotic monocistronic mRNAs (18). Caenorhabditis elegans also contains numerous polycistronic clusters with intergenic regions less than 200 bp in length (31). Considering the rather large interval between the ptp1 and ptp2 ORFs, short untranslated regions, and mRNA polyadenylation, we conclude that there are two separate transcription units. Studying the differential expression of PTP1 and PTP2 during development of the parasite within parasitophorous vacuoles is a future challenge for a better understanding of sporogenesis events.
Microsporidia are ubiquitous parasites, suggested to be waterborne pathogens (14). There is need for new detection techniques. Serological diagnosis using recombinant PTPs as antigens, in Western blotting or enzyme-linked immunosorbent assay, might be a potential tool to evaluate the prevalence of microsporidia. Through PCR amplifications of the PTP1 repeated regions in different E. cuniculi isolates, we provided the first data about the variability in both the sequence and repeat number of this protein (32). Further search for PTP1 and PTP2 homologues in species from other microsporidian genera should be undertaken to identify the most characteristic signatures. Immunological cross-reactions with proteins of the fish microsporidian species Glugea atherinae have been observed (12). This species, characterized by a larger genome (19.5 Mb), could be used to determine whether the synteny for the two ptp genes is maintained. Cloning and molecular characterization of new ptp genes, in conjunction with development of techniques for genetically manipulating microsporidia, will be required for elucidation of the structure of the microsporidian polar tube and its extrusion mechanism. Are PTPs truly unique to the phylum Microspora? The identification of sequences having homologies with ptp genes in lower metazoa, especially the myxozoa, which exhibit polar tube-like extrusomes, might help us to understand the evolutionary history of PTPs.
ACKNOWLEDGMENTS
We thank J. D'Alayer for peptide sequencing (Laboratoire de Microséquençage des Protéines, Institut Pasteur, Paris, France), P. Peyret for helpful discussions, and B. Chebance for technical assistance.
F. D. and I. P. were supported by a grant from the Ministére de l'Education Nationale de la Recherche et de la Technologie.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beauvais B, Sarfati C, Challier S, Derouin F. In vitro model to assess effect of antimicrobial agents on Encephalitozoon cuniculi. Antimicrob Agents Chemother. 1994;38:2440–2448. doi: 10.1128/aac.38.10.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckers P J A, Derks G J M M, Van Gool T, Rietveld F J R, Sauerwein R W. Encephalitozoon intestinalis-specific monoclonal antibodies for laboratory diagnosis of microsporidiosis. J Clin Microbiol. 1996;34:282–285. doi: 10.1128/jcm.34.2.282-285.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biderre C, Pagès M, Méténier G, Canning E U, Vivarès C P. Evidence for the smallest nuclear genome (2, 9 Mb) in the microsporidium Encephalitozoon cuniculi. Mol Biochem Parasitol. 1995;74:229–231. doi: 10.1016/0166-6851(95)02495-6. [DOI] [PubMed] [Google Scholar]
- 5.Biderre C, Canning E U, Méténier P G, Vivarès C P. Comparison of two isolates of Encephalitozoon hellem and E. intestinalis (Microspora) by pulsed field gel electrophoresis. Eur J Protistol. 1999;35:194–196. [Google Scholar]
- 6.Bohne W, Ferguson D J, Kohler K, Gross U. Developmental expression of a tandemly repeated, glycine- and serine-rich spore wall protein in the microsporidian pathogen Encephalitozoon cuniculi. Infect Immun. 2000;68:2268–2275. doi: 10.1128/iai.68.4.2268-2275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canning E U, Hollister W S. Microsporidia of mammals: widespread pathogens or opportunistic curiosities? Parasitol Today. 1987;3:267–272. doi: 10.1016/0169-4758(87)90103-7. [DOI] [PubMed] [Google Scholar]
- 8.Canning E U, Curry A, Lacey C J N, Fenwick J D. Ultrastructure of Encephalitozoon sp. infecting the conjunctival, corneal and nasal epithelia of a patient with AIDS. Eur J Protistol. 1992;28:226–237. doi: 10.1016/S0932-4739(11)80052-0. [DOI] [PubMed] [Google Scholar]
- 9.Carlton J M, Galinski M R, Barnwell J W, Dame J B. Karyotype and synteny among the chromosomes of all four species of human malaria parasite. Mol Biochem Parasitol. 1999;101:23–32. doi: 10.1016/s0166-6851(99)00045-6. [DOI] [PubMed] [Google Scholar]
- 10.Cavalier-Smith T. Eukaryotes with no mitochondria. Nature. 1987;326:332–333. doi: 10.1038/326332a0. [DOI] [PubMed] [Google Scholar]
- 11.Delbac F, Peyret P, Méténier G, David D, Danchin A, Vivarès C P. On proteins of the microsporidian invasive apparatus: complete sequence of a polar tube protein of Encephalitozoon cuniculi. Mol Microbiol. 1998;29:825–834. doi: 10.1046/j.1365-2958.1998.00975.x. [DOI] [PubMed] [Google Scholar]
- 12.Delbac F, Duffieux F, David D, Méténier G, Vivarès C P. Immunocytochemical identification of spore proteins in two microsporidia, with emphasis on extrusion apparatus. J Eukaryot Microbiol. 1998;45:224–231. doi: 10.1111/j.1550-7408.1998.tb04529.x. [DOI] [PubMed] [Google Scholar]
- 13.Didier E S. Microsporidiosis. Clin Infect Dis. 1998;27:1–8. doi: 10.1086/514607. [DOI] [PubMed] [Google Scholar]
- 14.Dowd S E, Gerba C P, Pepper I L. Confirmation of the human-pathogenic microsporidia Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Vittaforma corneae in water. Appl Environ Microbiol. 1998;64:3332–3335. doi: 10.1128/aem.64.9.3332-3335.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffieux F, Peyret P, Roe B A, Vivarès C P. First report on the systematic sequencing of the small genome of Encephalitozoon cuniculi (Protozoa, Microspora): gene organization of a 4.3 kbp region on chromosome I. Microb Comp Genomics. 1998;3:1–11. doi: 10.1089/omi.1.1998.3.1. [DOI] [PubMed] [Google Scholar]
- 16.Foucault C, Drancourt M. Actin mediates Encephalitozoon intestinalis entry into the human enterocyte-like cell line, Caco-2. Microb Pathog. 2000;28:51–58. doi: 10.1006/mpat.1999.0329. [DOI] [PubMed] [Google Scholar]
- 17.Frixione E, Ruiz L, Santillan M, de Vargas L V, Tejero J M, Undeen A H. Dynamics of polar filament discharge and sporoplasm expulsion by microsporidian spores. Cell Motil Cytoskel. 1992;22:38–50. [Google Scholar]
- 18.Johnson P J, Kooter J M, Borst P. Inactivation of transcription by UV irradiation of T. brucei provide a multicistronic transcription unit including a VSG gene. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- 19.Keohane E M, Orr G A, Takvorian P M, Cali A, Tanowitz H B, Wittner M, Weiss L M. Purification and characterization of a microsporidian polar tube protein. Mol Biochem Parasitol. 1996;79:255–259. doi: 10.1016/0166-6851(96)02666-7. [DOI] [PubMed] [Google Scholar]
- 20.Keohane E M, Orr G A, Takvorian P M, Cali A, Tanowitz H B, Wittner M, Weiss L M. Purification and characterization of human microsporidian polar tube proteins. J Eukaryot Microbiol. 1996;43:100S. doi: 10.1111/j.1550-7408.1996.tb05023.x. [DOI] [PubMed] [Google Scholar]
- 21.Keohane E M, Orr G A, Zhang H S, Takvorian P M, Cali A, Tanowitz H B, Wittner M, Weiss L M. The molecular characterization of the major polar tube protein gene from Encephalitozoon hellem, a microsporidian parasite of humans. Mol Biochem Parasitol. 1998;94:227–236. doi: 10.1016/s0166-6851(98)00071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keohane E M, Orr G A, Takvorian P M, Cali A, Tanowitz H B, Wittner M, Weiss L M. Analysis of the major microsporidian polar tube proteins. J Eukaryot Microbiol. 1999;46:29S–30S. [PubMed] [Google Scholar]
- 23.Kurz E M, Holstein T W, Petri B M, Engel J, David C N. Mini-collagens in hydra nematocytes. J Cell Biol. 1991;115:1159–1169. doi: 10.1083/jcb.115.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lom J, Vavra J. The mode of sporoplasm extrusion in microsporidian spores. Acta Protozool. 1963;1:81–92. [Google Scholar]
- 26.Lujan H D, Mowatt M R, Conrad J T, Bowers B, Nash T E. Identification of a novel Giardia lamblia cyst wall protein with leucine-rich repeats. J Biol Chem. 1995;270:29307–29313. doi: 10.1074/jbc.270.49.29307. [DOI] [PubMed] [Google Scholar]
- 27.Lujan H D, Conrad J T, Clark C G, Touz M C, Delbac F, Vivarès C P, Nash T E. Detection of microsporidia spore-specific antigens by monoclonal antibodies. Hybridoma. 1998;17:237–243. doi: 10.1089/hyb.1998.17.237. [DOI] [PubMed] [Google Scholar]
- 28.Magaud A, Achbarou A, Desportes-Livage I. Cell invasion by the microsporidium Encephalitozoon intestinalis. J Eukaryot Microbiol. 1997;44:81S. doi: 10.1111/j.1550-7408.1997.tb05795.x. [DOI] [PubMed] [Google Scholar]
- 29.Mowatt M R, Lujan H D, Cotten D B, Bowers B, Yee J, Nash T E, Stibbs H H. Developmentally regulated expression of a Giardia lamblia cyst wall protein gene. Mol Microbiol. 1995;15:955–963. doi: 10.1111/j.1365-2958.1995.tb02364.x. [DOI] [PubMed] [Google Scholar]
- 30.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 31.Page A P. A highly conserved nematode protein folding operon in Caenorhabditis elegans and Caenorhabditis briggsae. Gene. 1999;230:267–275. doi: 10.1016/s0378-1119(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 32.Peuvel, I., F. Delbac, G. Méténier, P. Peyret and C. P. Vivarès. Polymorphism of the gene encoding a major polar tube protein PTP1 in two microsporidia of the genus Encephalitozoon. Parasitology, in press. [DOI] [PubMed]
- 33.Raynaud L, Delbac F, Broussolle V, Rabodonirina M, Girault V, Wallon M, Cozon G, Vivarès C P, Peyron F. Identification of Encephalitozoon intestinalis in travelers with chronic diarrhea by specific PCR amplification. J Clin Microbiol. 1998;36:37–40. doi: 10.1128/jcm.36.1.37-40.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shyamala V, Ames G F-L. Genome walking by single specific primer-polymerase chain reaction (SSP-PCR) Methods Enzymol. 1993;217:436–446. doi: 10.1016/0076-6879(93)17082-g. [DOI] [PubMed] [Google Scholar]
- 35.Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 36.Van de Peer Y, Ben Ali A, Meyer A. Microsporidia: accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi. Gene. 2000;246:1–8. doi: 10.1016/s0378-1119(00)00063-9. [DOI] [PubMed] [Google Scholar]
- 37.Van Gool T, Vetter J C M, Weinmayr B, Van Dam A, Derouin F, Dankert J. High seroprevalence of Encephalitozoon species in immunocompetent subjects. J Infect Dis. 1997;175:1020–1024. doi: 10.1086/513963. [DOI] [PubMed] [Google Scholar]
- 38.Vavra J. Detection of polysaccharides in microsporidian spores by means of the periodic acid-thio-semicarbazide-silver proteinate test. J Microsc. 1972;14:357–360. [Google Scholar]
- 39.Von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weidner E. The microsporidian spore invasion tube; the ultrastructure, isolation, and characterization of the protein comprising the tube. J Cell Biol. 1976;71:23–34. doi: 10.1083/jcb.71.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamauchi K. The sequence flanking translational initiation site in protozoa. Nucleic Acids Res. 1991;19:2715–2720. doi: 10.1093/nar/19.10.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]