Abstract
Single-guide RNAs can target exogenous CRISPR/Cas proteins to unique DNA locations, enabling genetic tools that are efficient, specific, and scalable. Here we show that short synthetic guide piRNAs (21 nucleotide sg-piRNAs) expressed from extra-chromosomal transgenes can, analogously, reprogram the endogenous piRNA pathway for gene-specific silencing in the hermaphrodite germline, sperm, and embryos of C. elegans. piRNA-mediated interference (“piRNAi”) is more efficient than RNAi, can be multiplexed, and auxin-mediated degradation of the piRNA-specific Argonaute PRG-1 allows conditional gene silencing. Silencing is target-specific and results in decreased mRNA levels, amplification of secondary small interfering RNAs, and repressive chromatin modifications. Short (300 bp) piRNAi transgenes amplified from arrayed oligo pools also induce silencing, potentially making piRNAi highly scalable. We show that piRNAi can induce transgenerational epigenetic silencing of two endogenous genes (him-5 and him-8). Silencing is inherited for four to six generations after target-specific sg-piRNAs are lost, whereas depleting PRG-1 leads to essentially permanent epigenetic silencing.
Introduction
CRISPR/Cas proteins have fundamentally changed our ability to manipulate genetic material in cells due to the precise targeting conferred by short spacer sequences (~20 nucleotides) within single guide RNAs (sgRNAs)1. The easy design and specificity of sgRNAs has enabled experiments ranging from single-gene genetic knock-outs to genome-wide genetic screens in mammalian cells by silencing genes with CRISPR interference (CRISPRi)2. In C. elegans, RNA interference (RNAi) is the primary method used for transient gene silencing of individual genes3 or genome-wide screens4, whereas CRISPRi is relatively inefficient5 and cosuppression6,7 is not widely used. RNAi is commonly induced by injecting animals with transcribed dsRNA3, by feeding bacteria expressing dsRNA8, or by expressing dsRNA from plasmids9. Despite its popularity, RNAi has several limitations: the more convenient and scalable feeding method is less efficient than dsRNA injections8, some genes and tissues (e.g., sperm) are resistant to silencing10, and multiplexed gene silencing is relatively inefficient11.
Can endogenous small RNA pathways be reprogrammed to silence genes, analogous to how sgRNAs target Cas9? In C. elegans, piRNAs constitute a large class of genes (>15,000) that result in expression of 21-nucleotide long RNAs in the female and male germline from short (~80 bp) independent transcriptional units12. C. elegans piRNAs act via the PRG-1 Argonaute and can silence transgenes via transcriptional and post-transcriptional mechanisms that result in semi-permanent inherited epigenetic silencing13–15. piRNAs bind to mRNAs with some target mismatch tolerance16–18 and it is becoming clear that piRNAs participate in regulating endogenous genes18, in addition to their canonical role in silencing transposons19. Exogenous expression of piRNAs from transgenes has been used to study piRNA biogenesis20,21 and silence gfp17 and endogenous piRNA loci have been modified by CRISPR/Cas9 to target transgenes17,18. These studies demonstrate the silencing potential of piRNAs but are of limited experimental use because endogenous genes were resistant to silencing17.
Here we describe an efficient method, piRNA-mediated interference (piRNAi), that allows specific and multiplexed silencing of endogenous genes in the hermaphrodite germline, sperm, and embryos of C. elegans. We use piRNAi to identify novel targets and conditions where epigenetic silencing is inherited transiently (4–5 generations) or essentially permanently (>15 generations).
Results
Synthetic guide piRNAs silence endogenous germline genes.
In order to be useful, a novel silencing method based on the piRNA pathway and expression of synthetic guide piRNAs (sg-piRNAs) would need to be efficient, relatively easy, specific, and preferably allow conditional silencing. As a first step, we wanted to determine whether sg-piRNAs could silence endogenous genes and, if so, how many sg-piRNAs were required. To express multiple sg-piRNAs from a transgene, we identified short genomic regions (1.5 kb) that contained at least six highly expressed piRNAs (referred to as “piRNA clusters”). As a first test for piRNA interference (piRNAi), we chose the gene him-5, which is resistant to RNAi-mediated silencing22 and has a loss-of-function phenotype that is easy to score (~35% males in population). We chose 16 piRNA loci that were amenable to gene synthesis for testing. Gene synthesis allowed fast and flexible transgene design as well as cloning-free transgenesis. For each piRNA transgene, six of the endogenous piRNAs in the locus were substituted with the same set of sg-piRNAs anti-sense to him-5 (Fig. 1a). We quantified piRNAi-mediated silencing by injecting the piRNA clusters and scoring transgenic animals with extra-chromosomal arrays for males in the population. For 14 of the 16 synthetic piRNA clusters, we observed a high male frequency, often comparable to the genetic mutant him-5(e1490) (Fig. 1b). In contrast, we observed no males in negative controls injected with an unmodified piRNA cluster nor from sg-piRNAs targeting the sense strand of him-5 or an unrelated transgene (gfp). We tested silencing of a second gene with the same phenotype, him-823, using one efficient piRNA cluster (clusterE) and observed a large increase in males in the population (Fig. 1c). For simplicity, we mainly used piRNA clusterE for all subsequent experiments. The strong silencing of him-5 and him-8 by sg-piRNAs was in sharp contrast to the limited efficiency of RNAi-induced silencing by feeding or injection (Fig. 1c and Supplementary Figure 1). Similarly, RNAi only partially reduced GFP expression in male sperm, whereas piRNAi resulted in complete silencing (Fig. 1d–e). Finally, we silenced a ubiquitously expressed gfp (Peft-3::gfp) by piRNAi and observed no fluorescence in the adult germline or developing embryos until the ~100-cell stage and no silencing in adult somatic tissues (Fig. 1f). We conclude that piRNAi efficiently silences genes in the hermaphrodite germline, male sperm, and developing embryos.
Figure 1 – Synthetic guide piRNAs silence endogenous genes.
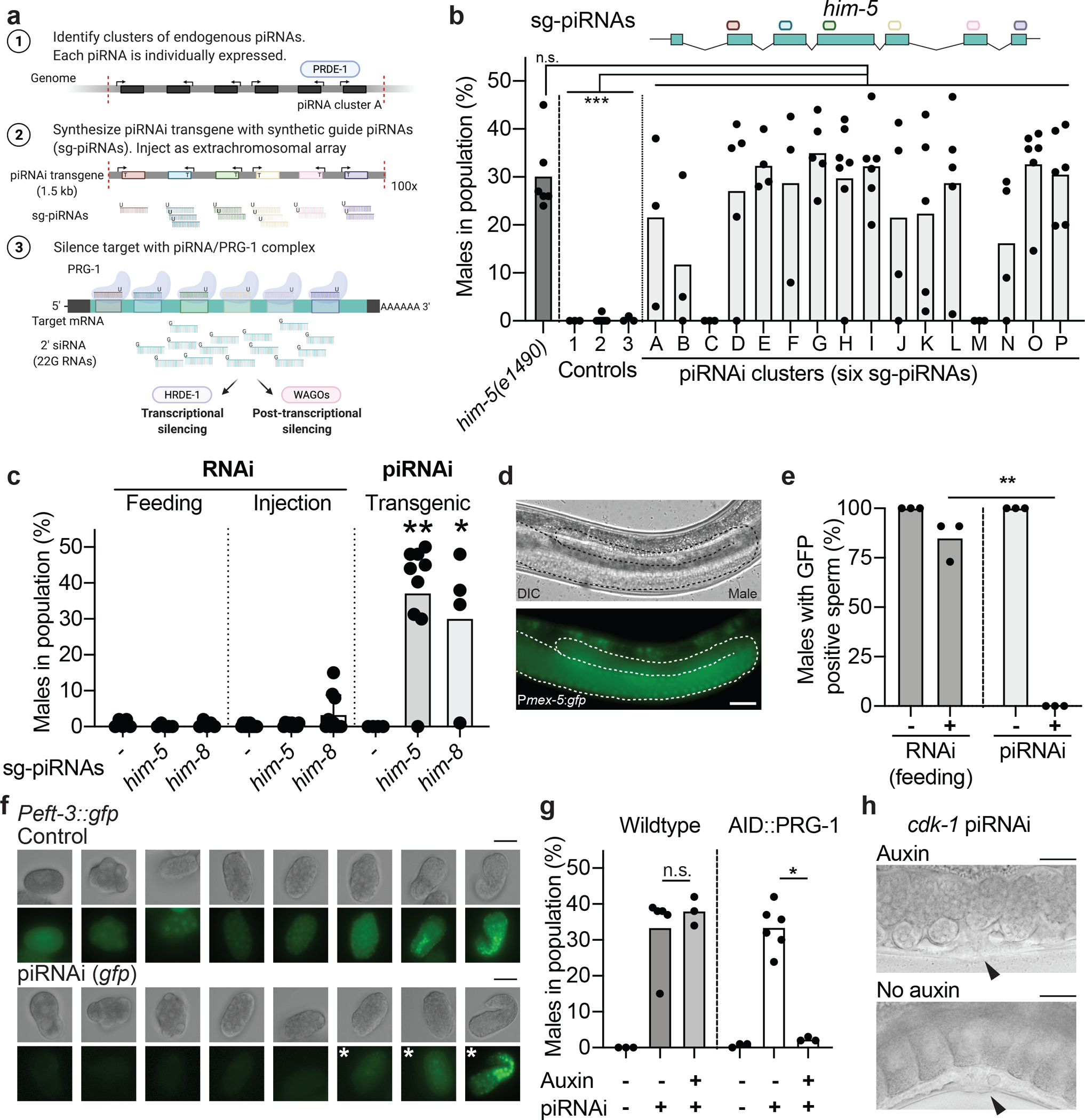
a. Schematic overview of piRNA-mediated RNA interference (piRNAi). Synthetic guide piRNAs (sg-piRNAs, 21 nt) are expressed from semi-stable extra-chromosomal arrays and silence target mRNAs via transcriptional and post-transcriptional mechanisms.
b. Silencing the endogenous him-5 gene with six sg-piRNAs expressed using 16 different recoded piRNA clusters (A to P) results in increased male frequency. The schematic above the him-5 locus indicates the location of antisense sg-piRNAs. Controls = him-5(e1490) mutant animals and negative controls: “1” = unmodified piRNA clusterA, “2” = him-5 sense sg-piRNAs, “3” = six sg-piRNAs targeting gfp. Kruskal-Wallis ANOVA P < 0.0001, Dunn’s multiple comparison. *** P = <0.001 and n.s. P > 0.999.
c. Gene silencing of him-5 or him-8 by RNAi by feeding (left), injection of in vitro transcribed dsRNA (middle), or piRNAi (right, clusterE) in Pmex-5::gfp transgenic animals. Controls (“−”) = empty RNAi vector (pL4440), dsRNA targeting gfp, and piRNA clusterE with randomized sg-piRNAs, respectively. Mann-Whitney test, comparison between RNAi and piRNAi condition. ** P = 0.0060 and * P = 0.0397.
d. Male sperm expressing GFP from a Pmex-5::gfp transgene. The male germline is outlined with a white dotted line. 40x magnification. Scale bar = 25 um.
e. Quantification of GFP expression in male sperm by visual inspection after silencing with RNAi (left) and piRNAi (right). Two-tailed t-test with Welch’s correction, ** P = 0.0049.
f. sg-piRNAs targeting a ubiquitous gfp (Peft-3::gfp) silences fluorescence in the germline (not shown) and in embryos until approximately the 100-cell stage (visible GFP fluorescence indicated by white asterisks). Representative images from more than ten embryos imaged. Scale bar = 25 um.
g. Conditional piRNAi using an auxin-inducible degron (AID) at the endogenous prg-1 locus26. In the presence of auxin (“+”), PRG-1 is degraded. “−” = un-injected strain. Mann-Whitney test, * P = 0.0238, n.s. = not significant.
h. Conditional piRNA-mediated silencing of cdk-1 in AID::PRG-1 animals result in single-cell embryonic arrest when transgenic animals are shifted to plates with no auxin. Arrowhead indicates the vulva. Representative images from more than ten adult animals. Scale bar = 25 um.
Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (b) n = 6 (him-5), n = 3 (“1”), n = 7 (“2”), n = 3 (“3”), n = 3 (A), n = 3 (B), n = 3 (C), n = 5 (D), n = 4 (E), n = 3 (F), n = 5 (G), n = 7 (H), n = 6 (I), n = 4 (J), n = 5 (K), n = 5 (L), n = 3 (M), n = 4 (N), n = 6 (O), n = 6 (P) biologically independent transgenic animals, (c) Feeding: n = 5 (all conditions) biologically independent animals; Injection: n = 10 (all conditions) biologically independent animals; piRNAi: n = 4 (“−”), n = 9 (him-5), n = 4 (him-8) biologically independent transgenic animals, (e) RNAi: n = 3 (both conditions) biologically independent animals, piRNAi: n = 3 (both conditions) biologically independent transgenic animals, (g) Wildtype: n = 3 (“−/−”), n = 5 (“−/+”), n = 3 (“+/+”), AID::PRG-1: n = 3 (“−/−”), n = 6 (“−/+”), n = 3 (“+/+”) biologically independent transgenic animals.
We tested and optimized piRNAi across a range of conditions. In summary, piRNAi was able to silence genes expressed exclusively in sperm (spe-12) and the hermaphrodite germline (mes-4, dcr-1, pie-1) (Extended Data Figure 1). Reproducible piRNAi required six sg-piRNAs when targeting him-5 or gfp and we observed very limited silencing when targeting non-coding regions (5’ UTR, 3’ UTR, or introns) (Extended Data Figure 2). Silencing was relatively insensitive to sg-piRNA sequences, did not require specific growth temperatures (15°C to 25°C), and silencing was stable or increased over 12 generations (Extended Data Figure 2). Furthermore, sg-piRNAs expressed from a C. elegans piRNA cluster could silence cbr-him-8 in C. briggsae (Extended Data Figure 2). piRNAi efficiency was poorly correlated to the expression level of the modified endogenous piRNAs (Supplementary Figure 2) and only linear piRNAi transgenes (synthetic genes or linearized plasmids) efficiently silenced him-5, likely due to their higher copy number in arrays (Extended Data Figure 3). These results demonstrate that piRNAi is robust and reproducible across a range of common experimental conditions.
Disruption of genes that act in the germline and early development frequently causes lethality or sterility4. To enable such experiments, a gene can be tagged with an auxin-inducible degron (AID) for conditional protein depletion by growing animals on auxin24. Although this approach is efficient, the AID-system requires tagging each target gene, preferably at the endogenous locus, which complicates the usage of auxin for genetic screens. We reasoned that the AID-system could be used as a proxy for conditional piRNAi-mediated gene silencing. piRNAs in C. elegans act via a single Argonaute (PRG-1) and prg-1 mutants are generally healthy, except for a progressive mortal phenotype that takes >15 generations to manifest25. Thus, in an AID-tagged prg-1 allele26, piRNAi-mediated silencing should only be active in the presence of auxin. Accordingly, when we targeted him-5 by piRNAi in the AID::PRG-1 strain, we observed only an insignificant increase in males when transgenic animals were grown on NGM plates with auxin (Fig. 1g). We tested the utility of conditional piRNAi by targeting two genes with well-characterized germline and embryonic phenotypes: cdk-1 (one-cell embryonic arrest) (Fig. 1h and Extended Data Figure 4) and pie-1 (sterility) (Extended Data Figure 1). We conclude that conditional piRNAi is efficient and can be used to silence genes with severe phenotypes.
Optimizing piRNA interference.
The ubiquitous usage of CRISPR/Cas has been accelerated by studies determining the mismatch tolerance of sgRNAs to minimize off-target effects, online tools to select optimal spacer sequences, and methods to generate large-scale sgRNA libraries by array-based oligo synthesis2. Here, we describe similar approaches to facilitate and expand the use of piRNAi.
To predict off-target effects of piRNAi, we tested sg-piRNA mismatch tolerance in the piRNA seed and non-seed regions17,18. him-5 silencing was relatively insensitive to one, two, and three sg-piRNA mismatches, whereas four and five mismatches abolished silencing (Fig. 2a), regardless of where mismatches were located (Extended Data Figure 5). sg-piRNAs with four mismatches to all other genes should, therefore, induce minimal off-target silencing. We developed an algorithm and online application (www.wormbuilder.org/piRNAi) to generate piRNAi transgenes that target genes with high specificity (Fig. 2b and Extended Data Figure 6). The app uses sg-piRNAs with the largest “edit distance” to any sequence in the genome (i.e., the Hamming distance) (Supplementary Information). The most stringent criteria (edit distance ≥ 4) allows targeting 91% of all genes, and less stringent criteria (edit distance ≥ 3) increase targeting to 96% (Supplementary Figure 3). Although this leaves some genes (e.g., histones, major sperm protein genes, and recent gene duplications, see Supplementary Table 1) refractory to direct silencing by piRNAi, we indirectly silenced one such gene with sg-piRNAs targeting gfp of an endogenously tagged allele (his-72::gfp) (Extended Data Figure 7).
Figure 2 – piRNAi tools for specific and scalable gene silencing.
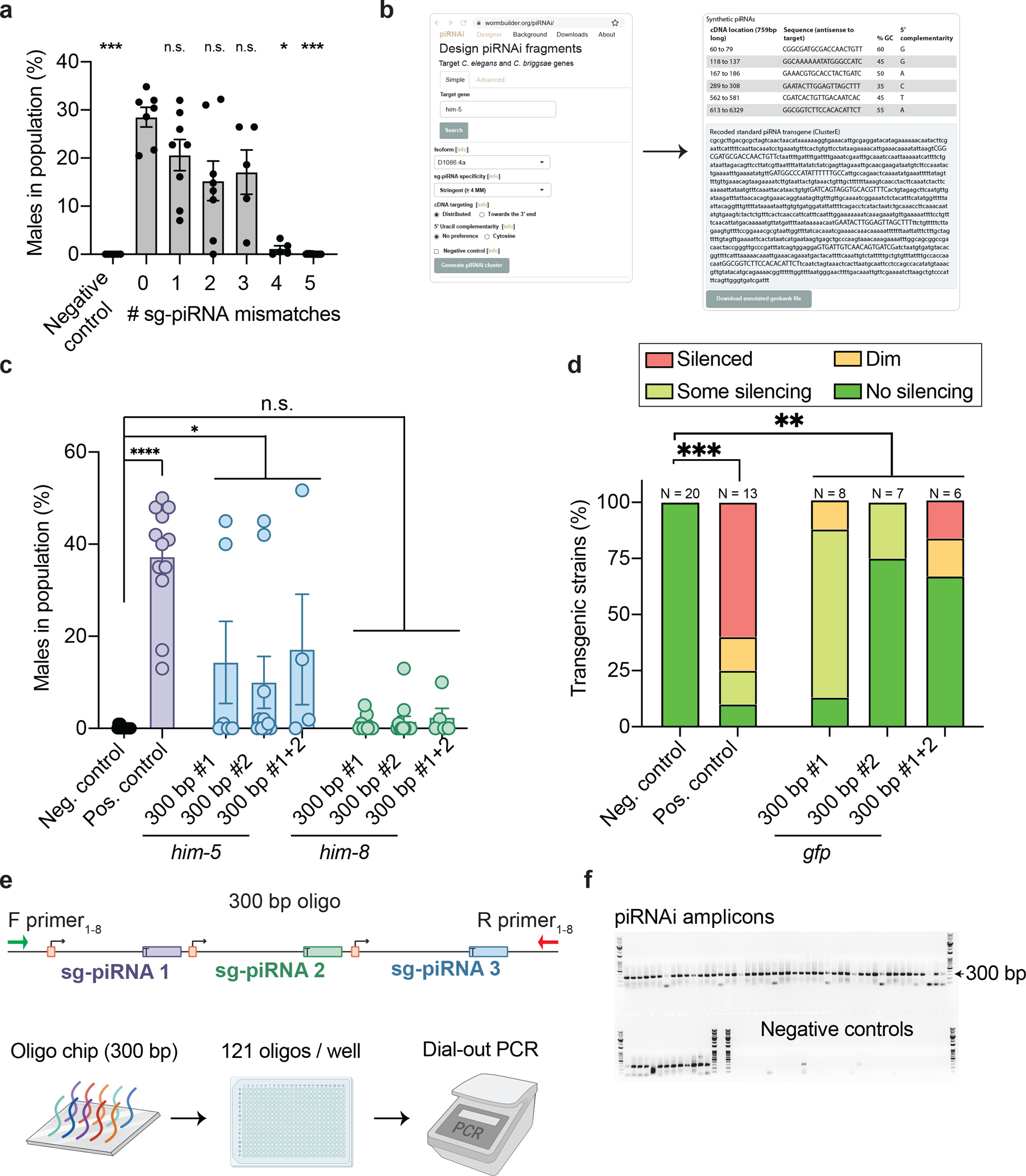
a. Mismatch tolerance of piRNAi transgenes targeting him-5 containing zero to five mismatches in each of the six sg-piRNAs (see Extended Data Figure 5). Negative control = inverted sg-piRNAs. Kruskal-Wallis ANOVA P < 0.0001, Dunn’s multiple comparisons (control columnn “0 mismatch”), *** P = <0.001, * P = <0.032, and n.s. P >0.999
b. Screenshots from interactive online application (wormbuilder.org/piRNAi) to generate synthetic piRNAi transgenes. The app generates sequences that target endogenous genes in C. elegans or C. briggsae with pre-computed 20-mers containing at least four mismatches to any off-target genomic sequence. The app also allows the generation of custom piRNAi transgenes targeting, for example, a transgene or specific gene isoforms (Extended Data Figure 6).
c. Two different short pairs of piRNAi transgenes (2× 300 bps) injected individually or together targeting him-5 or him-8. Kruskal-Wallis ANOVA, P < 0.0001, Dunn’s multiple comparison, *** P < 0.001, * P < 0.05, ns = not significant.
d. Two pairs of short (300 bp) piRNA transgenes targeting gfp for silencing in a strain with GFP expression in germline (Pmex-5::gfp). Data are normalized to the total number of independent biological experiments (indicated above bars). Chi-square test on “No silencing” vs some effect (“Dim”, “Some silencing”, or “Silenced”) and corrected for multiple comparisons. *** P < 0.001, ** P = 0.006.
e. Schematic of one piRNA transgene from a Matrix Oligo Pool (Twist Bioscience). 300 bp oligos synthesized at large scale on oligo chips are delivered as arrayed sub-pools with 121 unique oligos in each of 384 wells. The piRNAi transgenes include orthogonal primer sequences that allow selective amplification of any piRNAi transgene from the pool by “dial-out PCR”27.
f. PCR amplification of 2 × 60 piRNAi transgenes present in a single well (each 300 bp oligo encodes three sg-piRNAs). DNA ladder: 100–10,000 bp VersaLadder (GoldBio). All 12 individual piRNAi transgenes submitted for Sanger sequencing were correct (Extended Data Figure 8).
Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (a) n = 358 biologically independent transgenic strains; individual data points show average male frequency for all strains injected with a given transgene containing mismatches: n = 6 (negative control), n = 7 (0 mismatches), n = 8 (1 mismatch), n = 8 (2 mismatches), n = 5 (3 mismatches), n = 5 (4 mismatches), n = 12 (5 mismatches) biologically independent experiments, (c) n = 14 (Neg. control), n = 12 (Pos. control), him-5: n = 6 (300 bp #1), n = 10 (300 bp #2), n = 4 (300 bp #1+2), him-8: n = 9 (300 bp #1), n = 12 (300 bp #2), n = 5 (300 bp #1+2) biologically independent transgenic animals, (d) n = 13 (Neg. control), n = 20 (Pos. control), n = 8 (300 bp #1), n = 7 (300 bp #2), n = 6 (300 bp #1+2) biologically independent transgenic animals.
1.5 kb piRNAi transgenes targeting a few genes are relatively inexpensive (<$100/gene) and rapid (1 week) to generate by commercial gene synthesis yet this approach is not easily scaled to the genome-level. We, therefore, explored approaches to generate large-scale collections of piRNAi transgenes similar to how sgRNA pools are produced by oligonucleotide synthesis on gene-chips2. Large-scale oligo pools (>100K oligos) are useful for bulk cloning and cell transfection but are of limited use for animals that cannot be transfected (e.g., C. elegans). However, large-scale 300 bp oligo pools can be ordered as smaller, arrayed oligo pools (121 oligos per well). Individual piRNA loci are short (each ~ 85 bp long) and each oligo can, therefore, express three concatenated sg-piRNAs. We first generated such short piRNAi transgenes by standard gene synthesis and injected them pairwise (for a total of six sg-piRNAs) and observed significant silencing of him-5 and gfp but limited silencing of him-8 (Fig. 2c–d). We tested if the same 300 bp piRNAi transgenes could be recovered from oligo-pools. The oligo design included orthogonal primer sequences27 to allow selective PCR amplification (“dial-out-PCR”) of any piRNAi pair in the pool (Fig. 2e). With no optimization, we recovered 51 of 60 piRNAi transgene pairs in the pool and verified specific amplification by Sanger sequencing (12/12 correct) (Fig. 2f and Extended Data Figure 8). Further optimization is required (e.g., piRNA units, organization, or number of unique sg-piRNAs) but the results suggest that a single, easily distributed 384-well plate with 46,464 piRNAi transgenes could potentially target most C. elegans genes (>20,000).
In conclusion, we have identified rules, developed an application to silence individual genes, and have demonstrated that the scalability of oligo-pools, with some optimization, can be used for gene silencing in a multicellular organism.
piRNAi is specific and silencing can be multiplexed.
To quantify the effects of piRNAi at higher resolution than phenotypes and fluorophore silencing, we turned to high-throughput sequencing. First, we sequenced piRNAs and sg-piRNAs from transgenic piRNAi strains targeting the germline-expressed gene oma-128 or a mainly neuronal gene, unc-119. Endogenous piRNA expression was reproducible whereas sg-piRNAs expression was variable, as expected from variable transgene copy number in arrays (Fig. 3a, Supplementary Figure 4, and Supplementary Table 1). The sg-piRNAs constituted a relatively small fraction (from 0.02% to 0.47%) of all identified piRNAs and were expressed at two to ten-fold higher levels compared to the corresponding endogenous piRNAs (Fig. 3a and Supplementary Figure 5). Small RNA sequencing also identified a large increase in target-specific oma-1 secondary siRNAs (22G RNAs) compared to the negative control (unc-119) (Fig. 3b). At the mRNA level, oma-1 transcripts were depleted relative to the control locus (unc-119) with no apparent off-target silencing (Fig. 3d and Supplementary Figure 6) and piRNAi against him-5 and him-8 showed similar specific mRNAs depletion (Fig. 3e). The piRNAi constructs targeting oma-1, him-5, and him-8 were designed before developing the high-specificity algorithm and some sg-piRNAs, therefore, only had two or three mismatches to homologous mRNAs (Supplementary Information). However, even with these non-ideal piRNAi transgenes, we could not detect any obvious off-target increase in siRNAs (Supplementary Figure 7) or mRNA depletion (Supplementary Figure 8) near imperfectly matched sg-piRNAs.
Figure 3 – piRNAi is specific and can be multiplexed.
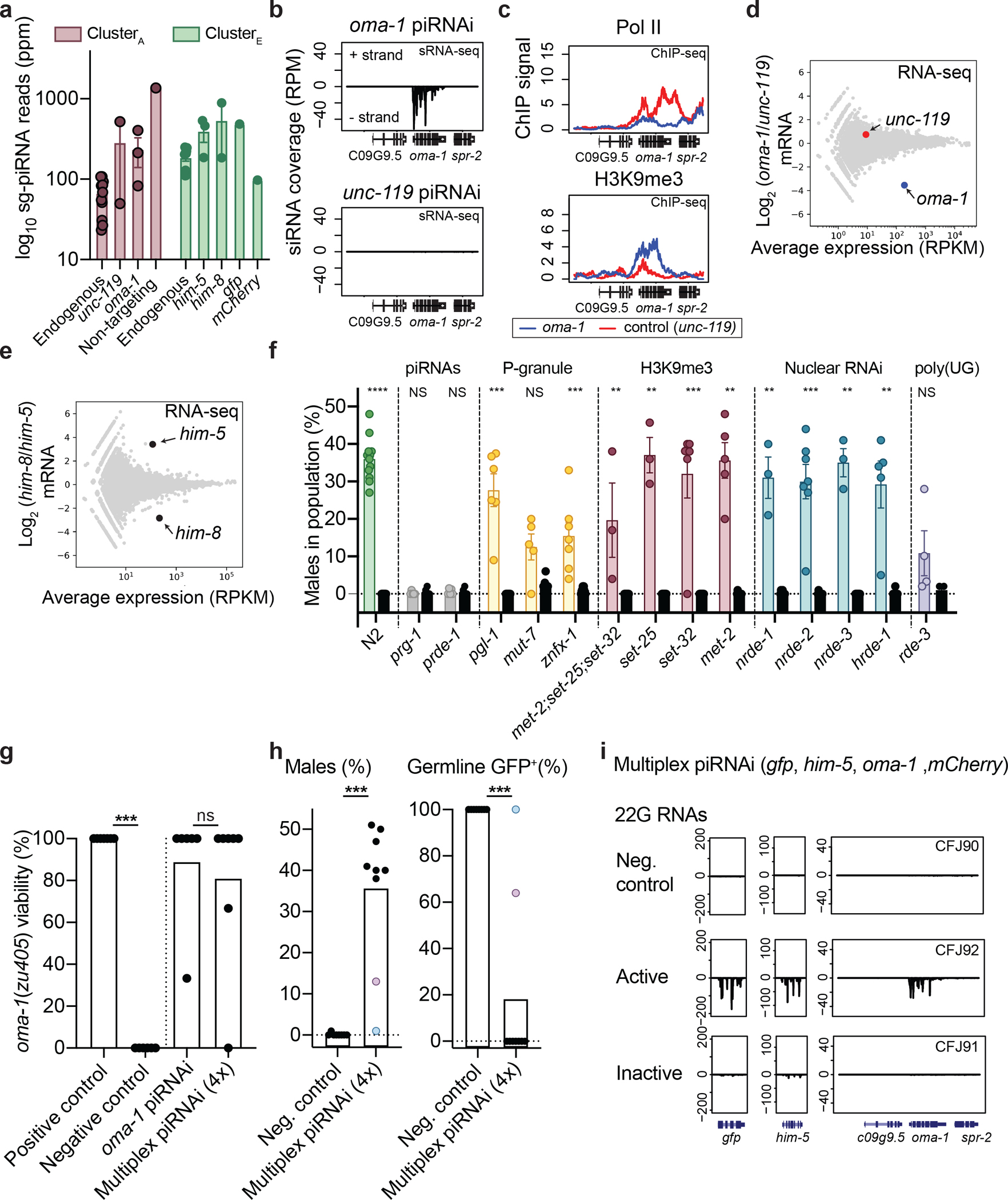
a. Total number of piRNA reads mapping to endogenous or sg-piRNAs from clusterA (centered on 21ur-199, purple) and the standard piRNA cluster (clusterE, centered on 21ur-1224, green). See also Supplementary Figures 4–5.
b. RNA-sequencing of secondary small RNAs (sRNA-seq) at the oma-1 locus in transgenic animals with piRNAi arrays targeting oma-1 or a control (unc-119).
c. Pol II (top) and H3K9me3 ChIP-seq (bottom) in strains with sg-piRNAs targeting oma-1 (blue) or unc-119 (red).
d. RNA sequencing (RNA-seq) of total RNA from young adult animals with piRNAi arrays targeting oma-1 (blue circle) or unc-119 (control, red circle).
e. Comparison of mRNA expression of a mixed population of young adult animals (hermaphrodites and males) with piRNAi against him-5 and him-8 (black circles).
f. Genetic requirements for silencing him-5 by piRNAi in N2 wildtype animals (green dots) or in mutant animals with primary effects on piRNAs (gray), P-granules (yellow), H3K9 methylation (red), nuclear RNAi (blue), and poly-UG tails (purple). Controls (black circles) are un-injected non-transgenic mutant strains. Mann-Whitney pairwise tests (injected vs un-injected). *** P < 0.0001, *** P < 0.001, ** P < 0.01, NS P > 0.05.
g. Multiplexed piRNAi-mediated silencing of a temperature-sensitive oma-1 allele using arrays with one (oma-1) or four (him-5, gfp, oma-1, and mCherry) piRNAi transgenes. Positive control = N2 injected with oma-1 piRNAi, negative control = oma-1(zu405ts). Mann-Whitney tests, P *** = 0.0006, ns = not significant (P = 0.853).
h. Multiplexed piRNAi against gfp and him-5 (in addition to oma-1 and mCherry) in transgenic Pmex-5::gfp animals. Neg. control = randomized sg-piRNAs. Purple and blue circles indicate matched data scored for males and GFP silencing in the same transgenic strain. Mann-Whitney test, P *** = 0.0001 (males) and *** = 0.0004 (GFP).
i. sRNA-seq from active (GFP negative and high male frequency) and inactive (GFP expression and no males) piRNAi arrays in Pmex-5::gfp transgenic animals (no mCherry transgene present in strain).
Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Negative controls = transgenic animals with randomized sg-piRNAs. Sense reads positive, antisense reads negative. Sample sizes (a) ClusterA: n = 11 (endogenous), n = 2 (unc-119), n = 3 (oma-1), n = 1 (random), ClusterA: n = 11 (endogenous), n = 3 (him-5), n = 2 (him-8), n = 1 (gfp), n = 1 (mCherry) small RNA libraries from biologically independent transgenic strains, (f) N2: n = 12 and n = 7, prg-1: n = 7 and n = 5, prde-1: n = 7 and n = 5, pgl-1: n = 6 and n = 7, mut-7: n = 5 and n = 7, znfx-1: n = 7 and n = 7, “met-2; set-25; set-32”: n = 5 and n = 7, set-25: n = 3 and n = 7, set-32: n = 3 and n = 7, met-2: n = 6 and n = 7, nrde-1: n = 3 and n = 7, nrde-2: n = 7 and n = 7, nrde-3: n = 3 and n = 7, hrde-1: n = 5 and n = 7, rde-3: n = 4 and n = 4 biologically independent transgenic strains (piRNAi and controls, respectively), (g) n = 7 (positive control), n = 6 (negative control), n = 6 (oma-1), n = 7 (multiplex) biologically independent transgenic strains, (h) n = 8 (negative control), n = 9 (multiplex) biologically independent transgenic strains.
RNAi depletes target mRNAs but also affects the corresponding genomic loci via chromatin modifications which may influence the expression of nearby genes29. To determine the effect of piRNAi on chromatin, and quantify possible off-target effects, we performed ChIP-seq against Pol II and the repressive chromatin mark H3K9me3 in strains with sg-piRNAs targeting oma-1, him-5, and him-8. We found that Pol II has a lower occupancy at target genes and increased H3K9me3 modifications, confirming that sg-piRNAs also act via transcriptional silencing (Fig. 3c, Supplementary Figure 6). him-8 is the last gene in an operon containing zim-1, zim-2, and zim-3; in this case, piRNAi against him-8 resulted in siRNAs (Supplementary Figure 9) and repressive chromatin modifications (Extended Data Figure 9) spreading into upstream regions. To determine if piRNAi spreading has functional effects, we silenced two single-copy germline-expressed fluorophores (mCherry and gfp) organized in an operon. Targeting either fluorophore by piRNAi led to complete or partial silencing of the adjacent fluorophore (Extended Data Figure 9). We conclude that piRNAi rarely induces direct off-target mRNA silencing but effects on nearby genes, particularly in operons, can be significant.
To define genetic pathways necessary for piRNAi, we silenced him-5 with piRNAi transgenes injected into mutants previously implicated in initiating or maintaining piRNA-mediated silencing15,16,30. We observed no silencing when injecting into mutants lacking the piRNA-specific Argonaute PRG-112 or a factor required for piRNA transcription (PRDE-1)31 (Fig. 3f). Most other genes were not strictly necessary to initiate silencing, even though silencing was not statistically significant for mutations affecting genes acting in phase-separated mutator foci (mut-7)32 or mRNA pUG-ylation (rde-3)33. In particular, mutations that abrogate H3K9 methylation (set-25, set-32, and met-2)34 or mutations in the nuclear (nrde-1, nrde-2, nrde-3) and heritable (hrde-1) RNAi pathway35 did not fully impair him-5 silencing (Fig. 3f).
To dissect genetic pathways and identify protein complexes, it is often useful to perturb multiple genes in parallel. The piRNA pathway is inherently highly multiplexed with more than 15,000 endogenous piRNAs12 and piRNAi might, therefore, allow multiplexed gene silencing. Transgenic strains with four piRNA transgenes targeting him-5, gfp, mCherry, and oma-1 induced silencing as efficiently as a single piRNA transgene, demonstrating that multiplexed silencing is possible (Fig. 3g–h and Supplementary Figure 10). In transgenic lines with incomplete gfp silencing, we also observed low frequencies of males (Fig. 3h and Extended Data Figure 3). This observation suggested an “all-or-none” effect which we verified by small RNA sequencing (Fig. 3i and Supplementary Figure 11). Thus, silencing a “neutral” gene by piRNAi (e.g., a germline-expressed gfp) can be used as a read-out for the activity of all sg-piRNAs during multiplexed silencing.
In conclusion, piRNAi is specific and can be multiplexed although effects on the genetic context of the targeted gene, e.g., genes in an operon, may perturb nearby gene expression.
Endogenous genes are transgenerationally silenced by piRNAi.
DNA excels at high-fidelity information transfer and gradual adaptive changes on long evolutionary time scales. However, some organisms also rely on, e.g., chromatin marks or small RNA populations to transfer short-term memories36. Transgenerational inheritance of individual genes in C. elegans is mainly studied by transient exposure to RNAi which results in permanent silencing of a germline-expressed gfp37 and 3–4 generations of epigenetic silencing of oma-128,35. Other endogenous targets with convenient phenotypes and clear inherited silencing have been difficult to identify38, although him-14 is a possible candidate based on co-suppression-induced inherited silencing7.
We reasoned that piRNAi might be a useful tool for identifying novel targets of inherited silencing and to determine what pathways influence the duration epigenetic silencing. First, we tested if piRNAi could induce inherited silencing using a canonical assay that relies on silencing a dominant, temperature-sensitive allele of oma-1(zu405ts), which lays dead eggs at higher temperatures (≥ 20°C)28. Loss-of-function oma-1 mutants have no obvious phenotype and inherited silencing is quantified by how many generations of viability are induced by transient RNAi exposure28. As expected, piRNAi against oma-1 suppressed oma-1(zu405ts) temperature-dependent embryonic lethality (Fig. 4a). We quantified epigenetic inheritance after losing the piRNAi array (identified by a fluorescent marker, Pmyo-2::mCherry) and, similar to RNAi28, observed three to four generations of inherited silencing using single or multiplexed sg-piRNAs (Fig. 4a). We proceeded to develop an assay to quantify him-5 and him-8 inherited silencing by counting males after losing the piRNAi transgene. him-5 silencing was reproducibly inherited for three to four generations whereas him-8 silencing was inherited for five to six generations (Fig. 4b, Extended Data Figure 10, and Supplementary Figure 13). The nuclear Argonaute hrde-1 is required for inherited silencing35. him-5 and him-8 piRNAi clusters injected into hrde-1 mutants showed moderately reduced initial silencing but no transgenerational inheritance, demonstrating that inheritance acts via the nuclear RNAi pathway (Fig. 4c and Extended Data Figure 10). We note that for some silenced genes, spe-8 and spe-12, we did not observe inherited silencing phenotypes (Extended Data Figure 10).
Figure 4 – piRNAi induces transgenerational silencing of endogenous genes.
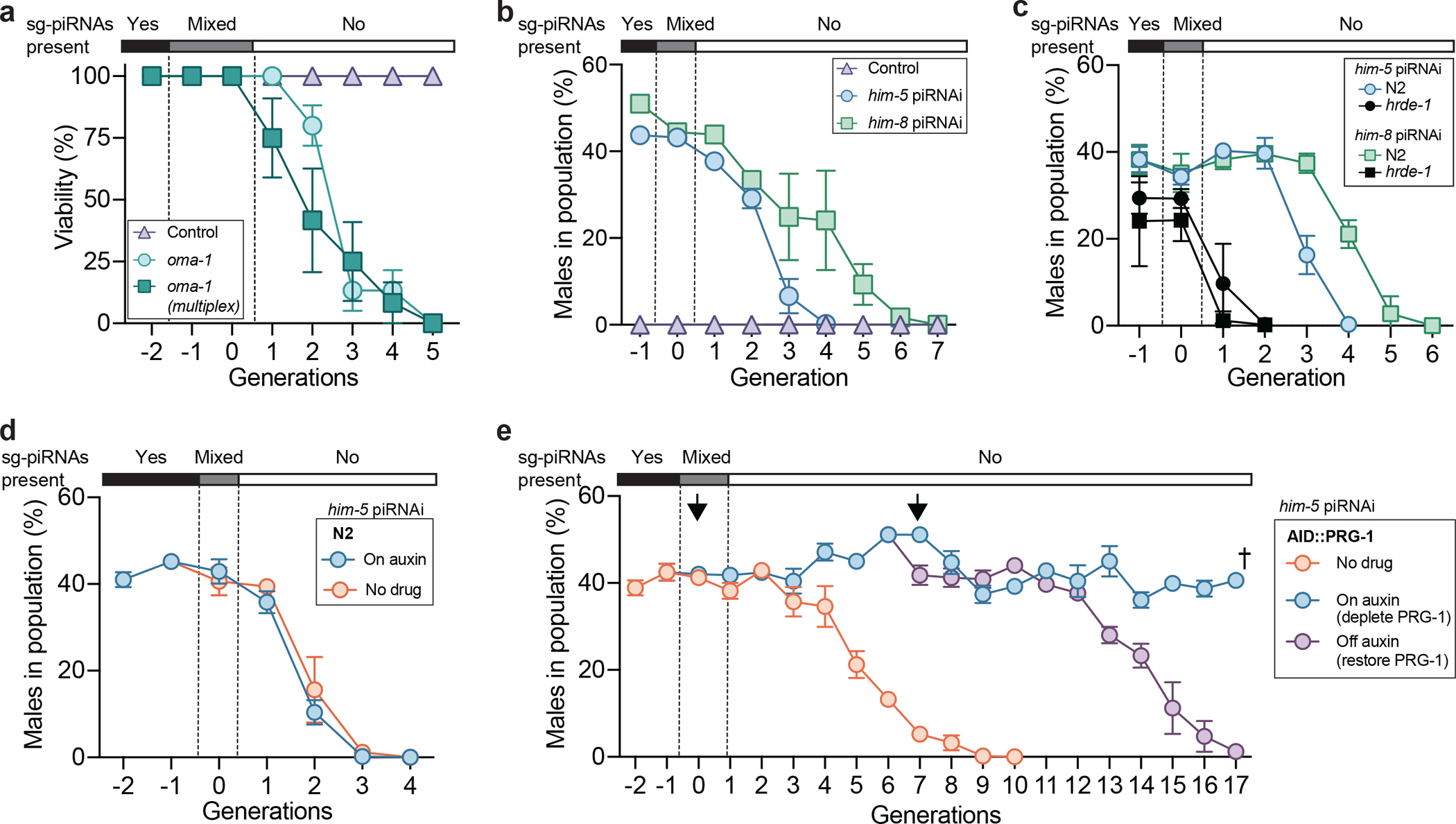
a. Transgenerational inheritance of oma-1 silencing. oma-1(zu405ts) animals have a gain-of-function temperature-sensitive mutation inducing embryonic lethality at temperatures above 15°C (here, 20°C). Animals with oma-1 loss-of-function alleles are overtly normal, and oma-1 silencing results in viable animals at the non-permissive temperature. Transgenic strains with sg-piRNAs targeting oma-1 were established (G−2), allowed to lose the array (G−1 to G0), and monitored for viable progeny in the absence of sg-piRNAs targeting oma-1 using an assay modified from Alcazar et al. (2008). Control = N2 with oma-1 sg-piRNAs.
b. Inheritance of him-5 and him-8 silencing. Frequency of males in strains exposed to sg-piRNAs targeting him-5 or him-8 before and after the loss of the extra-chromosomal piRNAi-array (see also Extended Data Figure 10). Control = N2 animals with a piRNA clusterA encoding randomized sg-piRNAs.
c. Assays for inherited him-5 and him-8 silencing in a nuclear heritable RNAi mutant (hrde-1). Control = N2 animals with piRNAi-arrays initially targeting him-5 or him-8.
d. Inherited silencing of him-5 piRNAi is unaffected by auxin in wildtype (N2) animals.
e. Inherited silencing of him-5 piRNAi becomes essentially permanent (RNA epigenetic) when PRG-1 is depleted by auxin. Arrows indicate transitions where animals are transferred between auxin and non-auxin plates leading to depletion and restoration of PRG-1, respectively. Dagger symbol indicates when AID::PRG-1 strains became sterile from the progressive mortal germline phenotype that is characteristic of prg-1 mutants. AID::PRG-1 animals have reduced levels of PRG-1 (approx. 20% of normal) in the absence of auxin26.
Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (a) n = 5 (control), n = 5 (oma-1), n = 4 (oma-1 multiplex), (b) n = 3 (control), n = 4 (him-5), n = 4 (him-8), (c) him-5: n = 3 (N2), n = 3 (hrde-1), him-8: n = 4 (N2), n = 4 (hrde-1), (d) n = 3 (no drug), n = 3 (on auxin), (e) n = 3 (all conditions) biologically independent transgenic strains. See Supplementary Figure 13 for biological replicates.
piRNA-mediated silencing has been shown to induce near-permanent inherited silencing of fluorescent transgenes (termed RNA epigenetic, RNAe)13,15,30, in contrast to the transient silencing we observed with oma-1, him-5, and him-8. RNAe was induced by homology to one or two piRNAs in the 3’ UTR and maintenance of inherited silencing was assayed by crossing silenced alleles to prg-1 mutants, which lack all endogenous piRNAs13,15,30. To understand these seemingly contradictory observations, we tested whether loss of individual sg-piRNAs was equivalent to impairing the entire piRNA pathway by auxin-mediated degradation of PRG-1. In wildtype animals, auxin had no effect on piRNAi-induced silencing or inherited him-5 silencing (Fig. 4d). In AID::PRG-1 animals, however, him-5 inherited silencing was prolonged (~ eight generations) even in the absence of auxin exposure (Fig. 4e); this extended silencing was likely caused by a reduction in the baseline level of PRG-1 (~20% of wildtype) in the absence of auxin in the AID::PRG-1 strain26. Even more striking, when silenced animals were transferred to auxin plates (and simultaneously lost the piRNAi array), inherited him-5 silencing became essentially permanent (Fig. 4e). We observed 17 generations of inherited silencing until PRG-1-depleted animals became progressively sick and sterile, the expected prg-1 null phenotype25,26. Indefinite silencing was not caused by changes to the him-5 locus because removing auxin (i.e., restoring PRG-1) resulted in gradual de-silencing (Fig. 4e). Furthermore, we could not detect any genomic changes at him-5 or elsewhere by whole-genome sequencing. These results show that PRG-1 plays dual roles in gene silencing: PRG-1 is required to initiate piRNA-mediated silencing but also prevents indefinite silencing. A similar role for PRG-1 in preventing permanent silencing after RNAi-mediated silencing of gfp and oma-1 was recently described39.
In conclusion, piRNAi can induce both transient and semi-permanent inherited silencing and we have identified two genes, him-5 and him-8, that are convenient novel targets for studying epigenetic inheritance.
Discussion
We have demonstrated that exogenous expression of synthetic guide piRNAs can reproducibly silence transgenes and endogenous genes in the hermaphrodite germline, male sperm, and in embryos of C. elegans. Although we cannot exclude subtle effects, we have not observed any apparent phenotypes (e.g., progressive sterility, small brood sizes, or morphological phenotypes) to suggest that piRNAi perturbs the endogenous piRNA pathway. Silencing a specific gene is intuitively simple and analogous to targeting genes using sgRNAs and CRISPR/Cas: a set of simple design rules and a web application can generate piRNA transgenes that stably silence genes with high efficiency and specificity. piRNAi is, to our knowledge, the only C. elegans method that allows efficient targeting in sperm, multiplexed gene silencing, and conditional silencing.
How does piRNAi compare to other gene-silencing methods mediated by transgenes? One method induces RNAi by expressing sense/antisense RNA using two opposed promoters flanking coding sequences9. Plasmid-based RNAi is mainly used for tissue-specific silencing in the soma, particularly in neurons which are refractory to RNAi by feeding4. The method is, however, not used in the germline and early development because most transgenes are silenced in the germline40. Germline transgene-silencing can instead be used experimentally to silence homologous genes, a process called co-suppression6,7. Functionally, co-suppression is similar to piRNAi and has been used to induce inherited silencing7. Co-suppression is less amenable to “programmable silencing” than piRNAi because the genetic pathways mediating co-suppression are not well-established and larger gene fragments are required for silencing. Furthermore, co-suppression also partly acts in somatic cells with variable loss-of-function phenotypes observed in muscle41. The most analogous method to piRNAi is CRISPR interference (CRISPRi), where genes are silenced by expression of multiple target-specific sgRNAs2. In principle, CRISPRi is preferable to piRNAi because a fully exogenous Cas machinery would enable experiments unrelated to gene silencing (e.g., gene activation and RNA/DNA labeling) but CRISPRi is inefficient in C. elegans5. Future experiment may enable the use of catalytically inactive Cas proteins in C. elegans, which could perhaps use low-complexity oligo-pools to encode sgRNAs.
What are the main limitations of piRNAi? In wild-type animals, piRNAs are only expressed in the germline and early embryonic development12. Consistent with this tissue-specific role for piRNAs, we observed no somatic phenotypes, e.g., no Unc animals from sg-piRNAs targeting unc-119 or gfp silencing past the 100-cell embryo. Although this is clearly a limitation, this tissue and stage-specificity may be useful for separating a gene functions in soma and germline or early development. A second limitation is the requirement for transgene injections. For systems biology, RNAi by feeding is the most feasible way to conduct genome-wide screens in C. elegans4. Injections are challenging, although possible42, to scale beyond tens or hundreds of genes but stable array lines require minimal effort to propagate once generated. We propose that further optimization of chip-based oligo libraries will enable genetic screens at medium to high-throughput scales.
We expect that piRNAi will be particularly useful for studying transgenerational inheritance. As a proof-of-principle, we identified two endogenous genes, him-5 and him-8, that inherit silencing for four and six generations, respectively. We used the AID-system to reconcile this transient inherited silencing with prior descriptions of permanent piRNA-induced silencing13–15: impairing the piRNA pathway by degrading PRG-1 once silencing is established leads to permanent silencing, at least for him-5. These results are in line with recent results describing perpetual silencing after exposure to RNAi in prg-1 mutants, a process that is mediated by unrestricted amplification of secondary siRNAs in the poly(UG) pathway33,39. We propose that piRNAi will be powerful tool for studying biological processes in the germline and early embryo and will enable novel insights into piRNAi biology and epigenetic silencing.
Methods
Strains
All experiments were performed in accordance with KAUST Institutional Biosafety and BioEthics Committee (IBEC), approval #17IBEC34. All strains were grown according to standard protocols43 and are listed in Supplementary Table 1.
Transgenesis
Transgenic animals with piRNAi transgenes were generated according to standard protocols44 by DNA injection and extra-chromosomal array formation. The injection mix consisted of 15–20 ng/ul of synthetic non-clonal dsDNA piRNAi transgenes with adapters that were not further purified (Twist Bioscience), 2 ng/ul co-injection marker plasmid pCFJ90 (Pmyo2::mCherry::unc-54 3’UTR), 12.5 ng/ul hygromycin resistance plasmid pCFJ782 (Prps-0::hygroR::rps-27 3’UTR), and 1kb DNA ladder (1 Kb Plus DNA Ladder, Cat. no. #10787-018, Life Technologies) to a final total DNA concentration of 100 ng/ul. 36–48 hours post-injection, 500ul of a stock solution with 4 mg/ml hygromycin (Gold Biotechnology, Cat. no. H-270-1) was added to plates with injected animals to select for transgenic (F1) progeny. A maxium of one single transgenic clonal lines was established from each injected animal by picking a single F2 adult based on co-injection marker fluorescence and hygromycin resistance.
Molecular biology
All plasmids were generated by standard methods or by gene synthesis (Twist Bioscience). Plasmids were verified by restriction digests and Sanger sequencing. Transgenes for Mos1 mediated single-copy insertion (mosSCI) were inserted into pCFJ15045. Plasmid for RNAi contained 1kb to 1.9kb genomic regions of the respective endogenous target genes (unc-54, unc-22, him-5, him-8) cloned into pL44408 and T777T46 using Gibson assembly47. All transgenes and oligos are listed in Supplementary Table 1. Full DNA sequences of all plasmids are included as annotated GenBank files in Supplementary Data 1.
piRNAi transgenes
We identified relatively short (~1.5 kb) genomic locations with at least six highly expressed piRNAs. For fifteen such genomic loci, we modified the sequence in silico to express sg-piRNAs targeting him-5 in place of endogenous piRNAs and ordered the piRNA clusters by commercial gene synthesis (non-clonal genes from Twist Bioscience). We injected the dsDNA gene fragments as supplied by the vendor with no further purification, or, by PCR amplification when the initial supply had been exhausted. For silencing additional genes, we used the cluster centered on 21ur-1224 (ClusterE). See Supplementary Information for a detailed description piRNA transgene design.
piRNAi Matrix Oligo pools
We generated short synthetic 300 bp piRNAi scaffolds by concatenating three highly expressed endogenous piRNA loci (head to tail). Genes were targeted for silencing by recoding a pair of synthetic piRNAi scaffolds (for a total of six sg-piRNAs). Functional testing was done by gene synthesis of individual fragments (Twist Bioscience) and transgene injection, similar to experiments with full-lenght piRNA transgenes. The amplification of identical DNA fragments was performed on Matrix Oligo Pools (Twist Bioscience), which contain 121 different 300 bp oligos supplied in 384 well plates. Individual pairs of piRNAi oligos were amplified using a KAPA HiFi polymerase (Roche, cat. no. KK2502) with 16 orthogonal primers optimized for amplification of individual members of large-scale oligo pools27. The amplified DNA sequences were verified by Sanger sequencing. See Supplementary Table 1 for 300 bp oligo pool sequences and amplification primers. See Supplementary Information for a detailed amplification protocol.
Methods for high-throughput sequencing experiments (Gu lab)
Worm culture
The worms were cultured at 20°C on 10 cm plates with hygromycin (0.2 mg/ml)-containing NGM media and E. coli (OP50 strain) as a food source. Developmental synchronization was achieved by treating mix-staged animals with hypochlorite-NaOH solution, hatching embryos in M9 buffer without food, and releasing the starved L1 animals onto plates with OP50. Young adult animals were collected 68–70 hours after the L1 release, passed through M9 with 10% sucrose cushion to remove bacteria, and pulverized by grinding with a mortar and pestle in liquid nitrogen. The samples were stored at −80°C.
Library construction (mRNA-seq)
Total RNA was extracted from approximately 5000 animals using Trizol reagent (Life Technologies). Ribosomal RNA was removed by using an RNaseH-based approach as described previously34. mRNA-seq libraries were constructed using a linker ligation-based approach as described previously48 for the oma-1 and unc-119 piRNAi samples or the SMARTer stranded RNA-seq kit (Takara Bio USA) for the multiplexed piRNAi experiment.
Library construction (sRNA-seq)
Small RNA extraction was performed using the mirVana microRNA isolation kit (Thermo Fisher Scientific). The small RNA-libraries for secondary siRNA analysis were constructed using a 5’-monophosphate-independent method49. Small RNA libraries for piRNA analysis were constructed using a 5’-monophosphate-dependent method, which is identical to the 5’-monophosphate-independent method except that the steps of phosphatase and T4 polynucleotide kinase treatment were skipped to enrich for piRNAs.
Library construction (ChIP-seq)
Antibodies used for ChIP-seq: Anti-Histone H3 (tri methyl K9) - ChIP Grade, Abcam, catalog# 8898, lot# GR285794-2, Anti-RNA Pol II (S2), Abcam, catalog #: ab5095, lot#: GR231750-1, and Anti-H3K23me3, Active Motif, xatalog # 61500, lot #31913001. 1:200 dilution was used in ChIP-seq for all antibodies. Chromatin immunoprecipitation (ChIP) and libraries were constructed as previously described50. DNA libraries were sequenced on an Illumina HiSeq-X instrument using the 1×50 nucleotide (oma-1 and unc-119 piRNAi) or 1×150 nucleotide (multiplexed piRNAi) mode. No peaks were called. High-throughput sequencing data generated for this study have been deposited in NCBI Gene Expression Omnibus (accession number GSE165210).
Bioinformatics
Sequencing reads were aligned to the C. elegans genome or cDNA database (WS190) using Bowtie-1.2.351. Only perfect alignments were used for analysis. DEseq2 was used to calculate the fold change and adjusted p-values when comparing two different RNA-seq profiles for all protein-coding genes52. For the coverage plot analysis at individual loci, siRNA or ChIP coverage levels were calculated by summing the number of siRNAs at any given base pair position and normalized by the total number (in millions) of sequencing reads aligned to the whole genome. Plots were made by custom Python or R scripts.
Methods for high-throughput sequencing experiments (Froekjaer-Jensen lab)
Custom scripts are available at https://doi.org/10.5281/zenodo.5599659 and custom software apps at https://github.com/AmhedVargas/piRNAi_reprogramming.
Genomic DNA isolation
Total genomic DNA was isolated from six NGM plates containing mixed stage worms grown at 20°C and freeze-cracked by storing at −80°C. Total genomic DNA was isolated using DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s protocol. The abundance of DNA sequences in extrachromosomal arrays was estimated by dividing their mean read depth over the average read depth seen in C. elegans chromosomes.
Library construction (sRNA-seq)
Total RNA was isolated using Tri-reagent (Sigma) from a mixed-stage population of transgenic animals with piRNAi arrays grown on hygromycin. Small RNA libraries were generated and sequenced to a depth of 20 million reads for each strain by a commercial provider (Novogene) using a NEB Next® Multiplex Small RNA Library Prep kit.
Identification of piRNA expression levels in 5’-monophosphate-dependent cloning DNA libraries
Trimmed sequencing reads were aligned to an indexed reference fasta file, containing the C. elegans WS235/ce11 genome sequence and synthetic piRNAi transgene sequences, via bowtie v 1.2.251 with the following parameters: “-S -5 4 -v 0”. Subsequently, the resultant alignment was processed using GNU awk in order to keep only 21 bp long reads that mapped and whose sequence started with thymine. Coverage and plots were made by custom Python or R scripts available at https://doi.org/10.5281/zenodo.5599659.
Abundance analysis of piRNAi transgenes in C. elegans strains
Paired end sequencing reads were mapped to an indexed reference fasta file containing the C. elegans WS235/ce11 genome sequence, synthetic piRNAi transgene sequences, protein tags, and plasmid DNA sequences (bacterial backbone removed) via bwa (v.7.17-r1188) mem algorithm with default parameters. Duplicated reads were filtered out of the alignments using picard tools v2.23.6 and coverage analysis were performed using samtools (v.1.9) coverage function.
Examination of C. elegans and C. briggsae transcriptome for piRNAi targets
Genomes and annotations files for C. elegans and C. briggsae were downloaded from WormBase version WS270 and WS190 (ftp.wormbase.org). For each species, CDS sequences were extracted from each respective genome using bedtools v2.27.1, reverse-complemented, and converted in 20bp-long subsequences. Unique subsequences, i.e., DNA sequences that appear only once, were identified by custom Perl scripts and filtered to remove sequences containing thymine within their first 3bp as a measure to limit erroneous transcriptional starts. Two methods were used to identify the non-perfect complementary binding of these subsequences (off-targets), namely a heuristic approach based on BLAST, and an exact algorithm based on the hamming distance between each pair mRNA fragment and unique CDS subsequences. For a detailed discussion, please see Supplementary Information.
Blast approach
Each CDS subsequence was mapped back to their reference genome using BWA v.7.17-r1188 and Samtools v.1.9 as a proxy to retrieve their genomic coordinates. This set of unique sequences (0 mismatches to the genome) was then used iteratively to find targeting sequences with base pairs mispaired, i.e., targeting sequences that allow n mismatches, using the blast-short tool of the blastn-suite with a cut-off e-value of 1. Targeting sequences that had only a single alignment of size 20 - n were considered as unique for n mismatches and their genomic coordinates were extracted as previously described. The code used for this approach can be found at https://github.com/AmhedVargas/piRNAi-DB.
Hamming distance algorithm
In order to calculate the hamming distance, i.e. the number of different base pairs in a sequence alignment, between any mRNA and each unique CDS subsequence, a word dictionary composed of all the possible 20 bp long DNA sequences in mRNA transcripts was generated using custom awk scripts per species. Furthermore, an implementation to calculate hamming distances was developed in c++ to allow parallelization via the OpenMP library. The full implementation can be found at: https://github.com/AmhedVargas/CelegansHammingAlignments
Online app development
Development of a web-app to produce piRNAi constructs
We developed a web app to generate custom piRNAi transgenes that can be accessed via this link: www.wormbuilder.org/piRNAi. The app helps in the construction of synthetic piRNAi transgenes by manual or automatic selection of the best targeting sites per gene isoform. This app is produced via the “shiny” R package version 1.4.0.210 using R v3.6.3, which in turns runs through an Amazon Web Services (AWS) Elastic Computing Cloud (EC2) instance that uses Ubuntu 18.04.3 LTS as the operating system. The code and documentation of this app can be found at https://github.com/AmhedVargas/piRNAiBuilder.
RNAi experiments
RNAi by bacterial feeding
RNAi was performed by bacterial feeding according to standard protocols4 using HT115 E. coli bacteria expressing dsRNA from the RNAi vector pL44408 or an optimized RNAi vector T777T46. Negative controls were performed with empty vectors, and positive controls targeted unc-54, unc-22, and gfp.
RNAi by dsRNA injection
dsRNAs were amplified from RNAi plasmids pL4440 (gfp, him-5 and him-8) with the MEGAShortscript T7 transcription kit (ThermoFisher, cat. no. AM1354), column purified with the RNeasy kit (Qiagen, cat. no. 74004), and injected at a concentration of 500ng/ul into the gonads of the adult hermaphrodite animals. The adult F1 progeny from injected animals were scored for GFP silencing or male frequency, respectively.
Quantifying phenotypes and transgenerational assays
Please see Supplementary Information for detailed assays.
Male frequency (him-5 and him-8)
The male frequency was scored visually on a dissection microscope in a semi-synchronous population derived from four to six virgen L4 animals maintained on hygromycin. Transgenerational assays are shown schematically in Extended Data Figure 10. Briefly, transgenerational epigenetic inheritance was assayed by losing the piRNAi transgene that triggers silencing (extra-chromosomal arrays are unstably transmitted and are, therefore, easily lost in every generation as determined by the lack of fluorescent co-injection markers). We quantified the number of males in the population after losing the array until no males were observed as a proxy for him-5 or him-8 silencing.
Transgenerational inheritance after depleting AID::PRG-1
We silenced him-5 using piRNAi transgenes injected into a strain with auxin-inducible PRG-1 degradation (PD3523: prg-1(cc3523[AID-Flag-HA::prg-1]) I; unc-119(ed3) III; ieSi38[Psun-1::TIR1::mRuby::sun-1 3’utr, cbr-unc-119(+)] IV)26. The effect of the piRNA pathway on transgenerational epigenetic inheritance was assayed by depleting PRG-1 using auxin according to standard protocols24 and quantifying the male frequency in the population after loss of the trigger piRNAi transgene. Note that this strain expresses only 20% of wildtype levels of PRG-1 in the absence of auxin and long-term propagation on auxin results in the mortal germline phenotype common to prg-1 mutant animals26.
Embryonic lethality (oma-1)
oma-1(zu405) mutants produce 50% viable progeny at 15°C and 100% dead embryos at temperatures 20°C and above28 which can be used to test transgenerational epigenetic inheritance28. We tested piRNAi-mediated silencing by injecting piRNAi transgenes targeting oma-1 into zu405 mutant animals reared at the permissive temperature and propagating transgenic animals at the non-permissive temperature. We tested transgenerational epigenetic inheritance after loss of the piRNAi array based on loss of co-injection marker fluorescence.
Hermaphrodite sterility (spe-8, spe-12)
Mutations in spe-8 or spe-12 cause sterility due to sperm defects in hermaphrodites; however, male sperm is functional. To propagate strong loss-of-function spe-8 and spe-12 phenotypes caused by piRNAi we, therefore, injected piRNA transgenes into a him-5(e1450) V mutant strain and maintained transgenic strains by male fertilization of hermaphrodites. We scored sterilty by counting the number of progeny laid by five virgin hermaphrodites picked at the L4 stage in 12 hours. Transgenerational assays were performed using transgenic animals with high penetrance of spe-8 or spe-12 silencing (i.e., low brood size). Inheritance was scored by quantifying the brood size before and after losing the extrachromosomal piRNAi transgene array based on co-injection marker fluorescence.
Single-cell embryonic arrest (cdk-1)
We injected piRNAi transgenes targeting cdk-1 into the AID::prg-1 strain maintained on auxin (1 mM) to prevent selection against active piRNAi arrays. cdk-1 arrested embryos were determined by quantifying the number of surviving progeny (viable progeny compared to number of eggs laid in a 12-hour window) from six transgenic L4 animals grown on non-auxin plates from early larval stages. The time-course experiments to determine the action of auxin was done by transferring larval stage animals (L1, L2, L3, and L4) or young adult animals to non-auxin plates and scoring internal embryo development at high magnification (40x oil immersion objective).
Maternal effect sterility (mes-4)
We injected piRNAi transgenes targeting mes-4 and him-5 into wildtype animals (him-5 was included to identify active arrays) and scored the frequency of sterile F2 animals from a semi-synchronous population derived from five transgenic adults. We note that only partially penetrant piRNAi arrays would be possible to maintain in this way and there would be strong selection against full mes-4 silencing.
Lethality (pie-1)
We injected piRNAi transgenes targeting pie-1 into the AID::prg-1 strain maintained on auxin (1 mM) to prevent selection against active piRNAi arrays. We scored lethality from a semi-synchronous population derived from five L4-stage animals transferred to non-auxin plates.
Temperature-dependent sterility (dcr-1)
We injected piRNAi transgenes targeting dcr-1 and him-5 (included to identify active arrays) into wild-type animals grown at 20°C. Transgenic animals were tested for temperature-sensitive sterility by determining brood sizes at 20°C and 25°C derived from five L4-stage transgenic animals.
Imaging and fluorescence quantification
All quantitative imaging was performed using 40x magnification (oil immersion objective) on a fluorescence microscope (Leica DM2500) with an LED-based fluorescence light-source (Leica SFL4000).
Quantification of GFP silencing in male sperm
We imaged transgenic animals with a single-copy transgene (Pmex-5::gfp) at ttTi5605 by mosSCI53. The gfp transgene was optimized for C. elegans expression54 and was expressed exclusively in the female and male germline under the mex-5 promoter. Transgenic animals were either subjected to RNAi-mediated silencing by feeding or to piRNAi-mediated silencing by injecting piRNA transgenes targeting gfp. We visually scored at least 11 animals for GFP expression in male sperm.
Perdurance of piRNAi silencing in embryos
We imaged animals with a single-copy ubiquitous transgene (Peft-3::gfp) inserted at ttTi5605 by mosSCI53. Transgenic animals were exposed to piRNAi by injecting recoded piRNA clusters targeting gfp or sequences not present in the genome (randomized sg-piRNAs). We bleached gravid adults to release embryos and imaged various stages.
Quantifying endogenous his-72::gfp silencing
We quantified GFP intensity in unfertilized oocytes of an animal with the native his-72 locus tagged with gfp by CRISPR55. For each biological replicate, 15–20 young adult animals were imaged and the fluorescence intensity in the last three oocytes nuclei before the spermatheca was quantified using FIJI software (NIH). The genotype of all animals was blinded to the experimenter.
Commercial software and statistical analysis
Statistical analysis was performed using GraphPad Prism for MacOS v9.0.0. “A plasmid Editor” (ApE) v3.0.6 was used for molecular biology design. The manuscript was written using Microsoft Word v16.54, Source Data tables and Supplementary Table 1 were generated using Microsoft Excel v16.54, and figures were generated with Adobe Illustrator v26.0. Some figure panels were generated with Biorender.com.
Extended Data
Extended Data Fig. 1 |. piRNAi can silence a variety of germline-expressed genes.
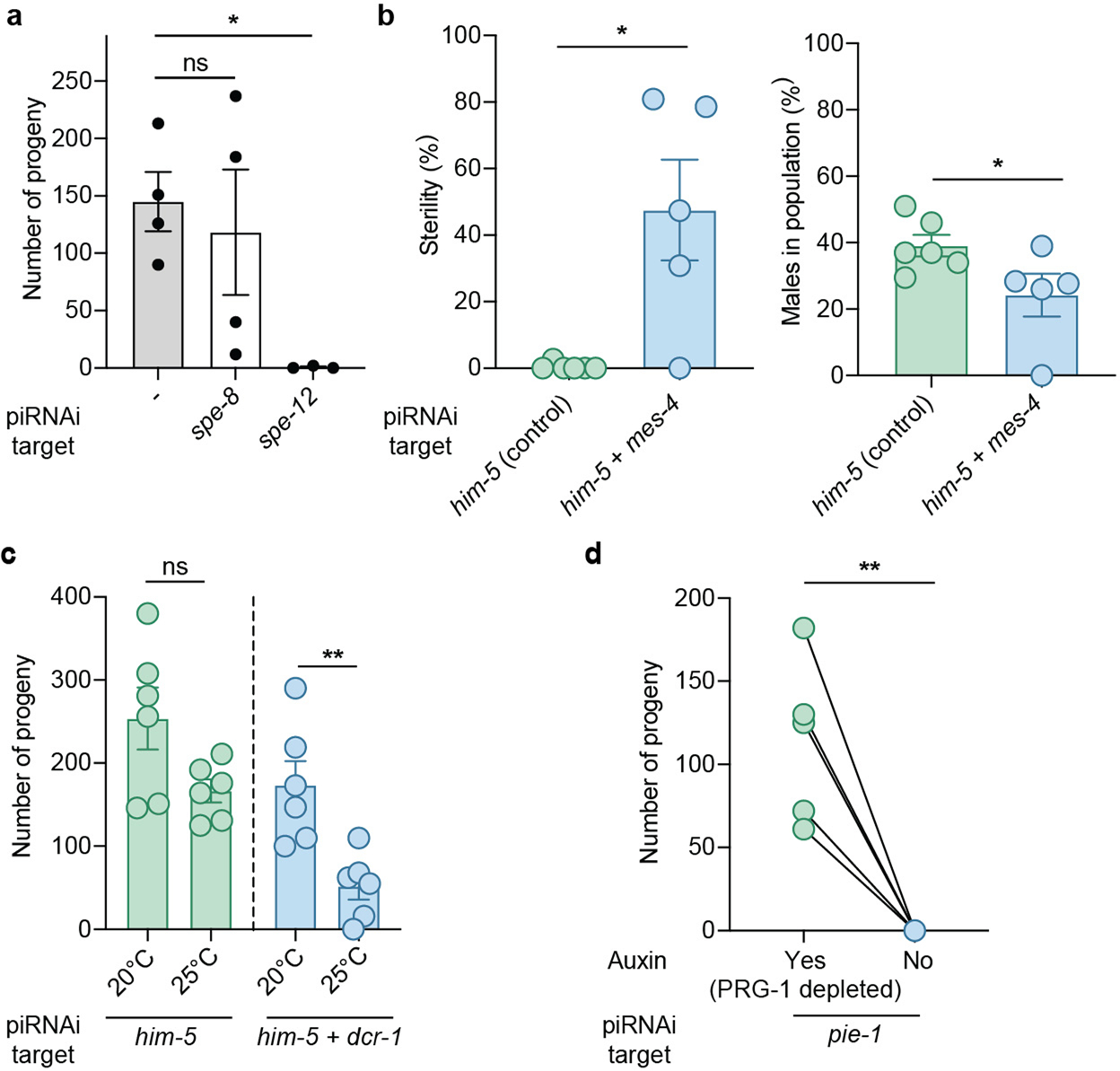
a. Quantification of the number of self-progeny in C. elegans strains with sg-piRNAs targeting the sperm specific genes spe-8 and spe-12. Fertility was assayed from unmated L4 hermaphrodites in a him-5(e1490) mutant background. Control (‘−’) = randomized sg-piRNAs in clusterA. Kruskal–Wallis ANOVA P = 0.0376, Dunn’s multiple comparison, * P = 0.0458, ns = P > 0.99. b. piRNAi against mes-4 and him-5 in wildtype (N2) animals. Transgenic animals were scored for sterile animals and for males (to identify ‘active arrays’). Two-tailed Mann-Whitney, * P = 0.0281 (sterility) and * P = 0.0455 (male frequency). c. piRNAi against dcr-1 and him-5 in wildtype animals (N2). Transgenic L4 stage animals were incubated at 20 °C and 25 °C and their progeny scored for lethality and males. dcr-1 genetic mutants are temperature-sensitive sterile at high temperatures. Two-tailed Mann-Whitney, ns P = 0.1320 and ** P = 0.0065. d. piRNAi against pie-1 and him-5 in the AID::PRG-1 strain maintained on 1 mM auxin (PRG-1 depleted). Larval stage animals from stable, independent transgenic lines were transferred to plates with or without auxin and scored for the number of progeny at 25 °C. Two-tailed Mann-Whitney, ** P = 0.0079. Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (a) n = 4 (‘−’), n = 4 (spe-8), n = 3 (spe-12), (b) n = 6 (him-5), n = 5 (him-5 + mes-4), (c) n = 6 (all conditions), (d) n = 5 biologically independent transgenic strains.
Extended Data Fig. 2 |. Rules for efficient piRNA silencing.
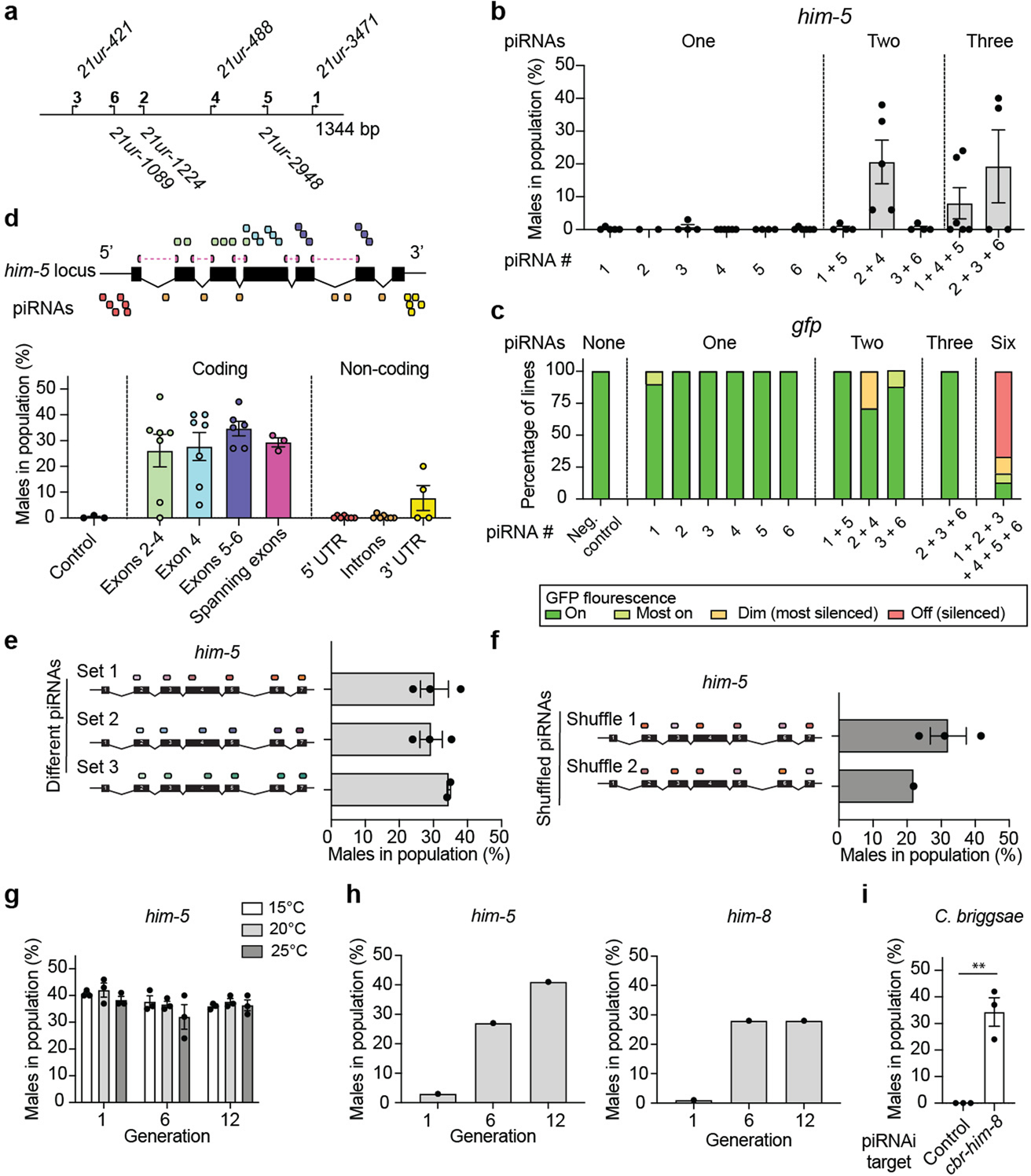
a. Schematic of sg-piRNA clusterE. b. Graph showing the effect of silencing him-5 with one, two, or three sg-piRNAs recoded in clusterE. c. Bar graph showing the effect of silencing a codon-optimized gfp expressed in the germline (Pmex-5::gfp) with zero, one, two, three, or six sg-piRNAs from clusterE. Independent biological strains (at least 11 animals per strain) were scored qualitatively on a dissection microscope blinded to genotype. d. Top. Schematic of the him-5 (D1086.4a.1) gene structure and the location of sg-piRNAs. Bottom. piRNAi using clusterE targeting him-5 exons or 5’ and 3’ untranslated regions (UTRs) or introns (‘non-coding’). Control = randomized sg-piRNAs. e. Three versions of piRNA clusterE using different sg-piRNAs were tested for him-5 silencing. Set 1 corresponds to Fig. 1b and the piRNAi transgenes in set 2 and set 3 target the same exons as set 1 but use different sg-piRNAs. f. The six sg-piRNAs in ‘set 1’ were shuffled (‘shuffle 1’ and ‘shuffle 2’), so each sg-piRNA was expressed by a different promoter in the piRNA cluster. The guide piRNA target locations in the him-5 transcript are shown as colored ovals. The strains were cultured at 25 °C. g. Transgenic animals were tested for him-5 silencing by piRNAi (piRNA clusterE) over 12 generations at various temperatures (15 °C, 20 °C, 25 °C). h. Propagation of two transgenic animals with sg-piRNAs targeting him-5 (left) and him-8 (right) that initially had a low frequency of males in the population. The two strains were propagated for 12 generations, and the male frequency was quantified every six generations. i. C. elegans piRNA clusterE targeting the gene cbr-him-8 in C. briggsae (AF16). Control (‘−’) = un-injected C. briggsae. Two-tailed t-test with Welch’s correction. ** P = 0.0236. Data are presented as mean values+/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (b) n = 5 (‘1’), n = 2 (‘2’), n = 4 (‘3’), n = 6 (‘4’), n = 4 (‘5’), n = 6 (‘6’), n = 4 (‘1 + 5’), n = 5 (‘2 + 4’), n = 4 (‘3 + 6’), n = 6 (‘1 + 4 + 5’), n = 4 (‘2 + 3 + 6’), (c) n = 4 (Neg. control), n = 6 (‘1’), n = 8 (‘2’), n = 10 (‘3’), n = 9 (‘4’), n = 7 (‘5’), n = 10 (‘6’), n = 8 (‘1 + 5’), n = 7 (‘3 + 4’), n = 5 (‘5 + 6’), n = 2 (‘2 + 3 + 6’), (d) n = 3 (control), n = 6 (Exons 2–4), n = 6 (exon 4), n = 5 (exons 5–6), n = 3 (spanning exons), n = 6 (5’ UTR), n = 6 (introns), n = 4 (3’ UTR), (e) n = 3 (set 1), n = 3 (set 2), n = 2 (set 3), (f) n = 3 (shuffle 1), n = 1 (shuffle 2), (g) n = 3 (all conditions), (h) n = 1 (all conditions), n = 3 (control), n = 3 (cbr-him-8) biologically independent transgenic strains.
Extended Data Fig. 3 |. piRNAi depends on plasmid structure and copy number.
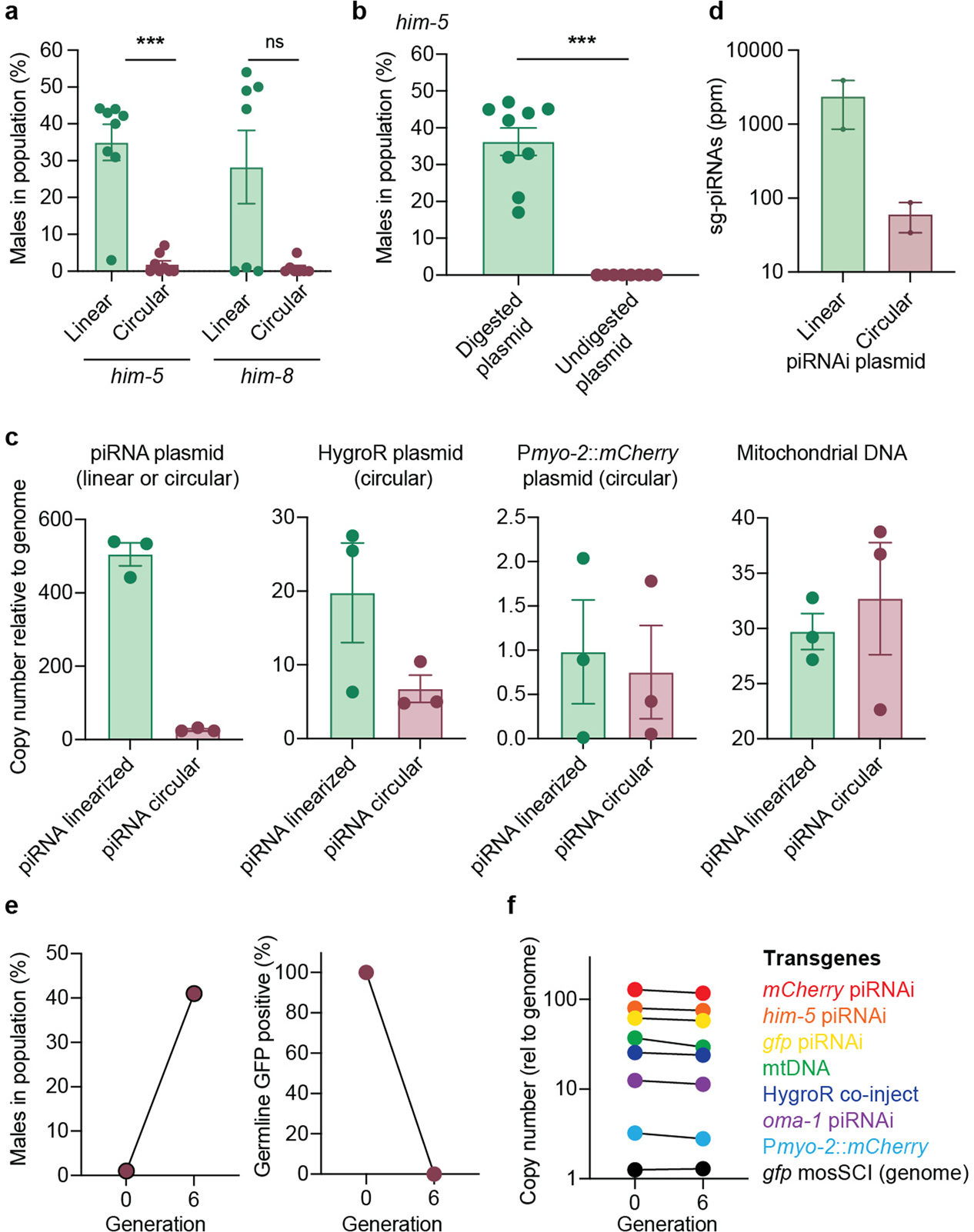
a. Transgenic lines generated by injecting a piRNA cluster either as linear dsDNA (1.5 kb) or the same DNA cloned into a plasmid backbone (high copy ampicillin pTwist vector). The DNA transgenes were generated from clusterE targeting him-5 and him-8 with six sg-piRNAs. Two-tailed Mann-Whitney tests. *** P = 0.0006, ns = not significant (0.0659). b. him-5 piRNAi plasmid (from panel a) digested with restriction enzymes that cuts in the bacterial vector backbone (left) or undigested (right) was used to induce silencing. The plasmid samples were treated identically (incubated in restriction enzyme buffer with or without restriction enzymes and purified over spin column) and injected at the same concentration. Two-tailed Mann-Whitney test. *** P = < 0.0001. c. Comparison of transgene copy number by whole genome sequencing of transgenic lines with extrachromosomal arrays formed from linear (green) or circular (purple) plasmids. d. Comparison of sg-piRNA expression targeting him-5 from transgenic strains carrying linearized and circular piRNAi transgenes. e. One ‘inactive’ multiplexed piRNAi strain (blue circle in Fig. 3h) was propagated for six generations and scored for males in the population and silencing of germline GFP fluorescence. ‘Generation 0’ corresponds to 2–3 generations after the initial injection. f. Whole-genome sequencing on the strain shown in panel e to determine the copy number of all plasmids in the extra-chromosomal array at the early (generation 1) and late (generation 6) timepoints. Mitochondrial DNA (mtDNA) copy number was used as a control. The copy number was calculated relative to the average sequencing coverage across the entire C. elegans genome. Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (a) him-5: n = 8 (linear), n = 8 (circular), him-8: n = 7 (linear), n = 7 (circular), (b) n = 6 (digested), n = 6 (undigested), (c) n = 3 (all conditions), (d) n = 2 (linear), n = 2 (circular), (e) n = 1, (f) n = 1 biologically independent transgenic strains.
Extended Data Fig. 4 |. time-course of conditional cdk-1 silencing using auxin.
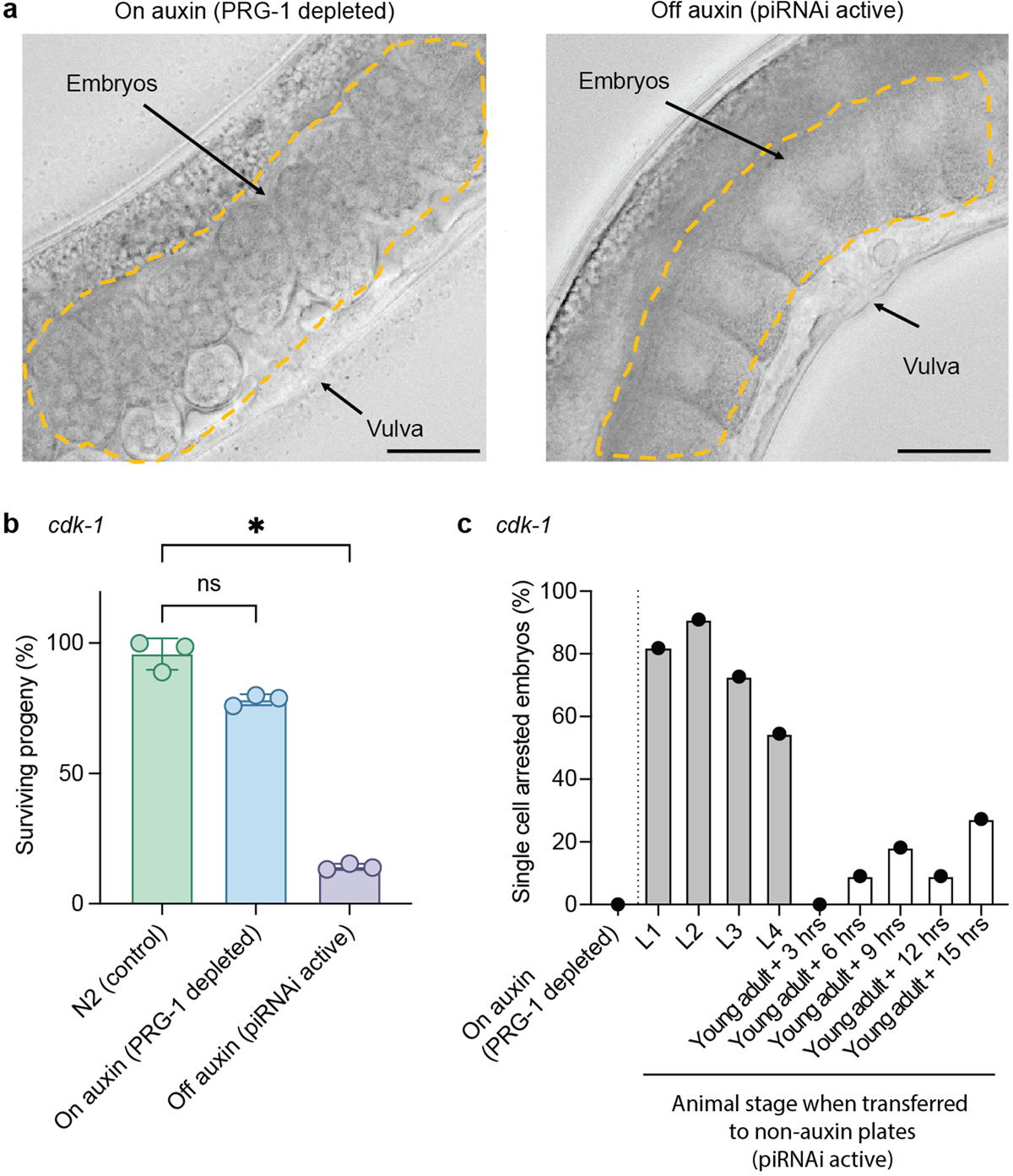
a. Brightfield image of animals with piRNAi targeting cdk-1 in the AID::PRG-1 strain on auxin (left panel) or off auxin (right panel). The dotted yellow line outlines fertilized embryos in the hermaphrodite uterus. Arrows indicate the vulva and embryos. cdk-1 encodes a cyclin-dependent kinase that is required for cell division and embryos arrest at the single-cell stage in cdk-1 mutants. Representative images from more than ten embryos imaged. Scale bar = 25 μm. b. piRNAi against cdk-1 in the AID:PRG-1 strain. Injected animals were maintained on 1 mM auxin plates (to deplete PRG-1). L4 stage animals were transferred to plates with or without auxin and surviving progeny was scored by first counting eggs and three days later counting the total number of adult progeny. Negative control = wildtype animals (N2). Kruskal-Wallis ANOVA P = 0.0036, Dunn’s multiple comparison * P = 0.0146, ns P = 0.3594. c. piRNAi activation after removal from auxin. To determine how long it takes to ‘turn on’ piRNA-mediated silencing after auxin removal, animals were transferred to non-auxin plates at each larval stage and at 3-hour intervals as young adults. The uteruses of adult animals (at least 11 animals) were scored for the presence of single-cell arrested embryos. Data are presented as mean values+/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (b) n = 3 (N2), n = 3 (auxin, both conditions), (c) n = 1 (all conditions) biologically independent transgenic strains.
Extended Data Fig. 5 |. piRNAi silencing tolerates up to three mismatches.
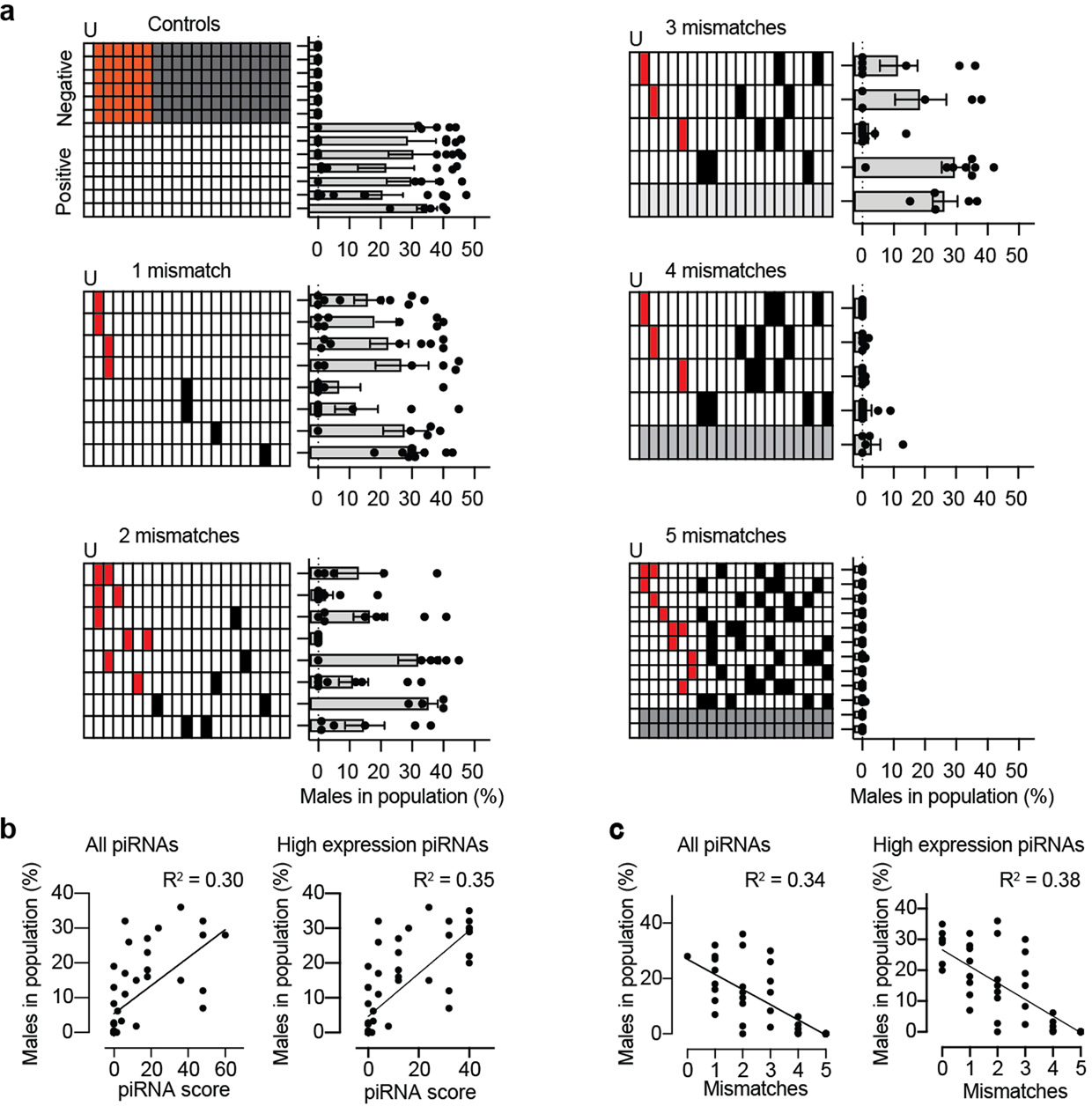
a. sg-piRNA mismatch tolerance using piRNA clusterE. The six sg-piRNAs targeting him-5 each contained from zero to five mismatches. The schematics show the location of mismatches in the piRNA seed sequence (red boxes, nucleotides in positions 2 to 7) or in the remainder of the piRNA (white boxes, positions 8 to 21). For all sg-piRNAs (including controls), the leading ‘U’ was not modified as this base is required for piRNA transcription. In some transgenes the overall number of mismatches was constant but the location in each sg-piRNA was randomized (indicated by gray shading). The negative control contains inverted him-5 sg-piRNAs. b. Relationship between male frequency and the aggregate piRNA score calculated based on Wu et al. (2018), which takes the location of mismatch and wobble base pairing into account. Silencing data from panel a. Left: all piRNAs. Right: four highest expressed guide piRNA in clusterE (the two remaining sg-piRNAs were rarely detectable by small RNA sequencing). Simple linear regression. R2 = goodness of fit. c. Relationship between male frequency and the number of mismatches in each guide piRNA. Silencing data from panel a. Left: all piRNAs. Right: four highest expressed guide piRNA in clusterE. Simple linear regression. R2 = goodness of fit. Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (a) Controls: n = 6, n = 6, n = 7, n = 8, n = 8, n = 5 (negative), n = 5, n = 9, n = 7, n = 7, n = 6, n = 6, n = 9 (positive), 1 mismatch: n = 9, n = 7, n = 8, n = 7, n = 6, n = 10 (technical and biological replicate), n = 7, n = 8, 2 mismatches: n = 5, n = 10, n = 8, n = 7, n = 6, n = 8, n = 6, n = 6, 3 mismatches: n = 7, n = 5, n = 8, n = 9, n = 5, 4 mismatches: n = 9, n = 6, n = 8, n = 8, n = 5, 5 mismatches: n = 7, n = 6, n = 7, n = 5, n = 7, n = 9, n = 8, n = 6, n = 9, n = 7, n = 5, n = 5, all values top to bottom, (b) n = 382, (c) n = 343 biologically independent transgenic strains.
Extended Data Fig. 6 |. Web application allows easy piRNAi transgene design.
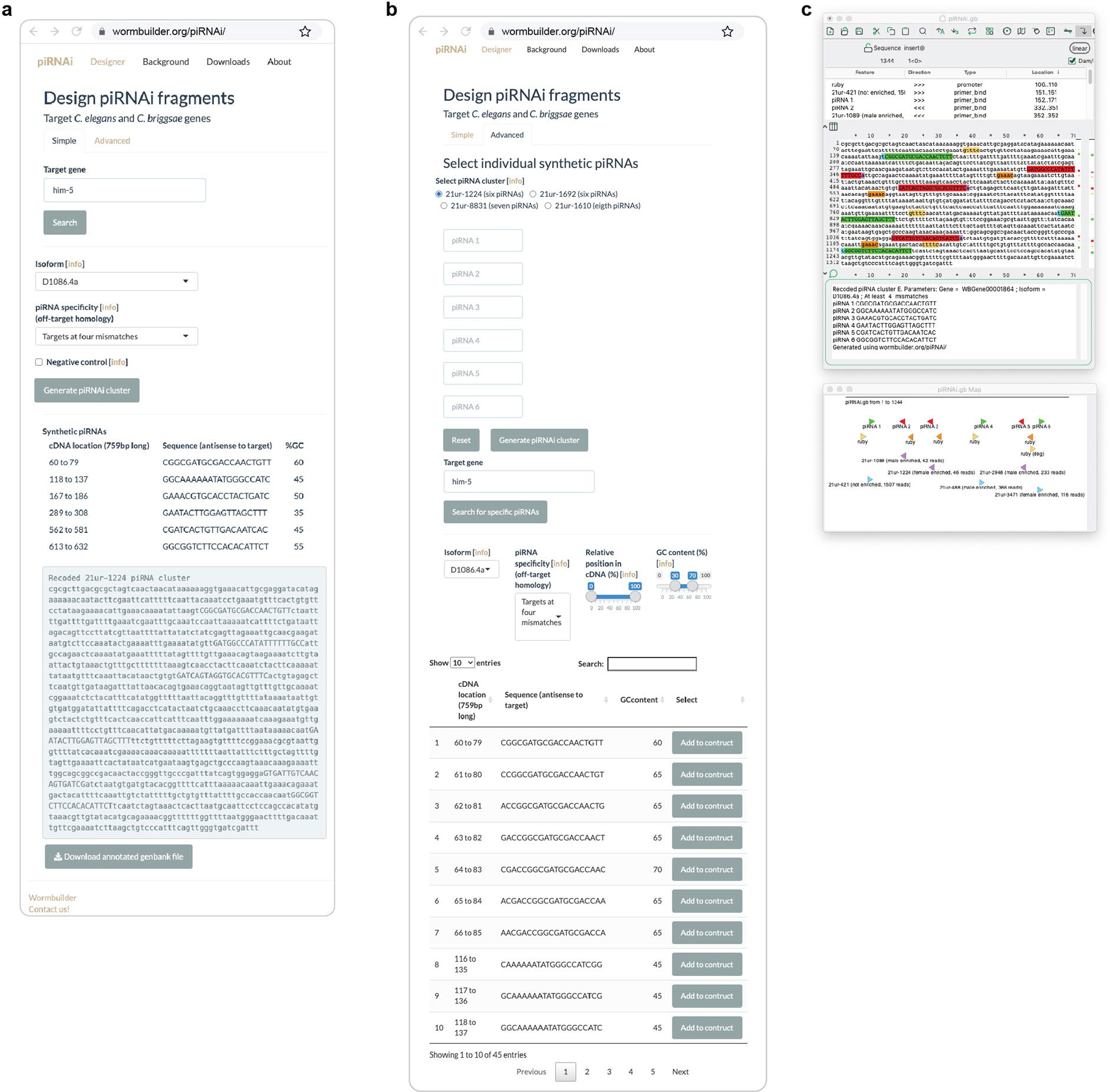
a. Screenshot from www.wormbuilder.org/piRNAi. Simple mode to target a single gene with pre-defined criteria. b. Advanced mode. In the advanced mode, several different piRNA clusters can be re-coded with either custom 20-mers (for example, targeting gfp) or by selecting all 20-mers at a given edit distance mapping to a selected gene isoform. c. Output file with the piRNAi transgene annotated in GenBank format. The sequence is displayed in ‘A plasmid Editor’ (ApE).
Extended Data Fig. 7 |. Silencing endogenously gfp-tagged his-72 with piRNAi.
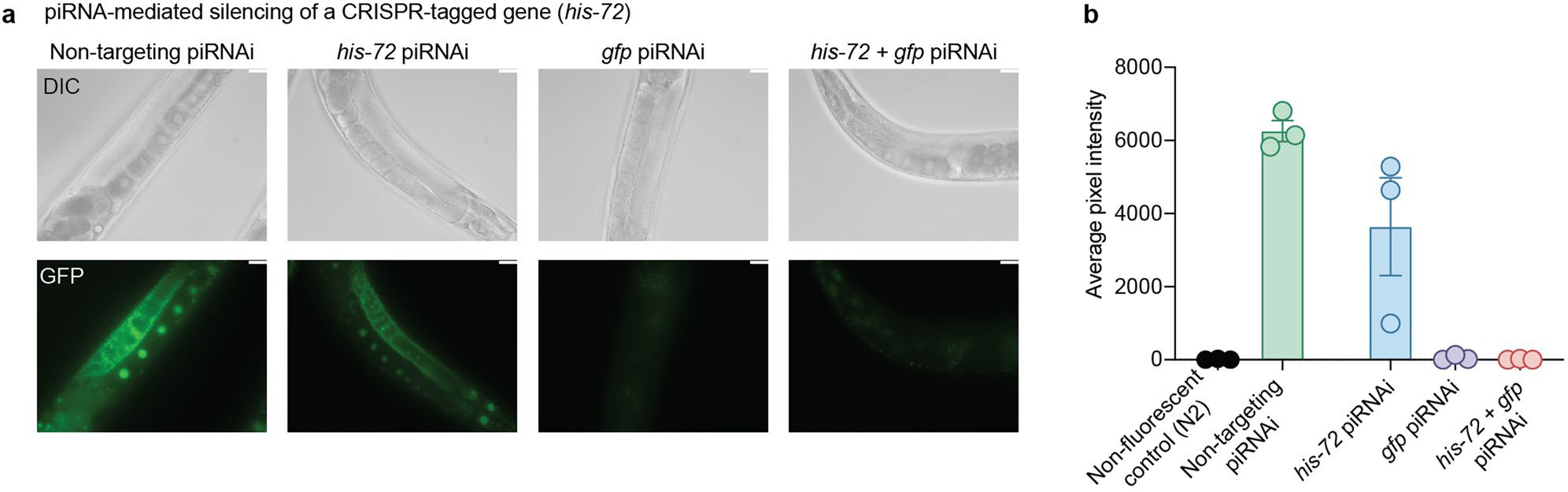
a. Brightfield and fluorescence images of a strain with an endogenously gfp-tagged his-72 locus. Images show a non-targeting control (mCherry), or sg-piRNAs targeting his-72, gfp, or his-72 + gfp. Images were acquired at 40x magnification using an oil immersion objective. Representative image from more than 50 adult animals imaged. White scale bar = 25 μm. b. We quantified GFP fluorescence in the nucleus of the ‘last’ three oocytes before the spermatheca using ImageJ (NIH). Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (b) n = 62 (N2), n = 59 (non-targeting), n = 64 (his-72), n = 62 (gfp), n = 62 (his-72 + gfp) fluorescent images from biologically identical transgenic strains across three technical replicates for each condition.
Extended Data Fig. 8 |. Amplification of short piRNAi transgenes from Matrix oligo Pools.
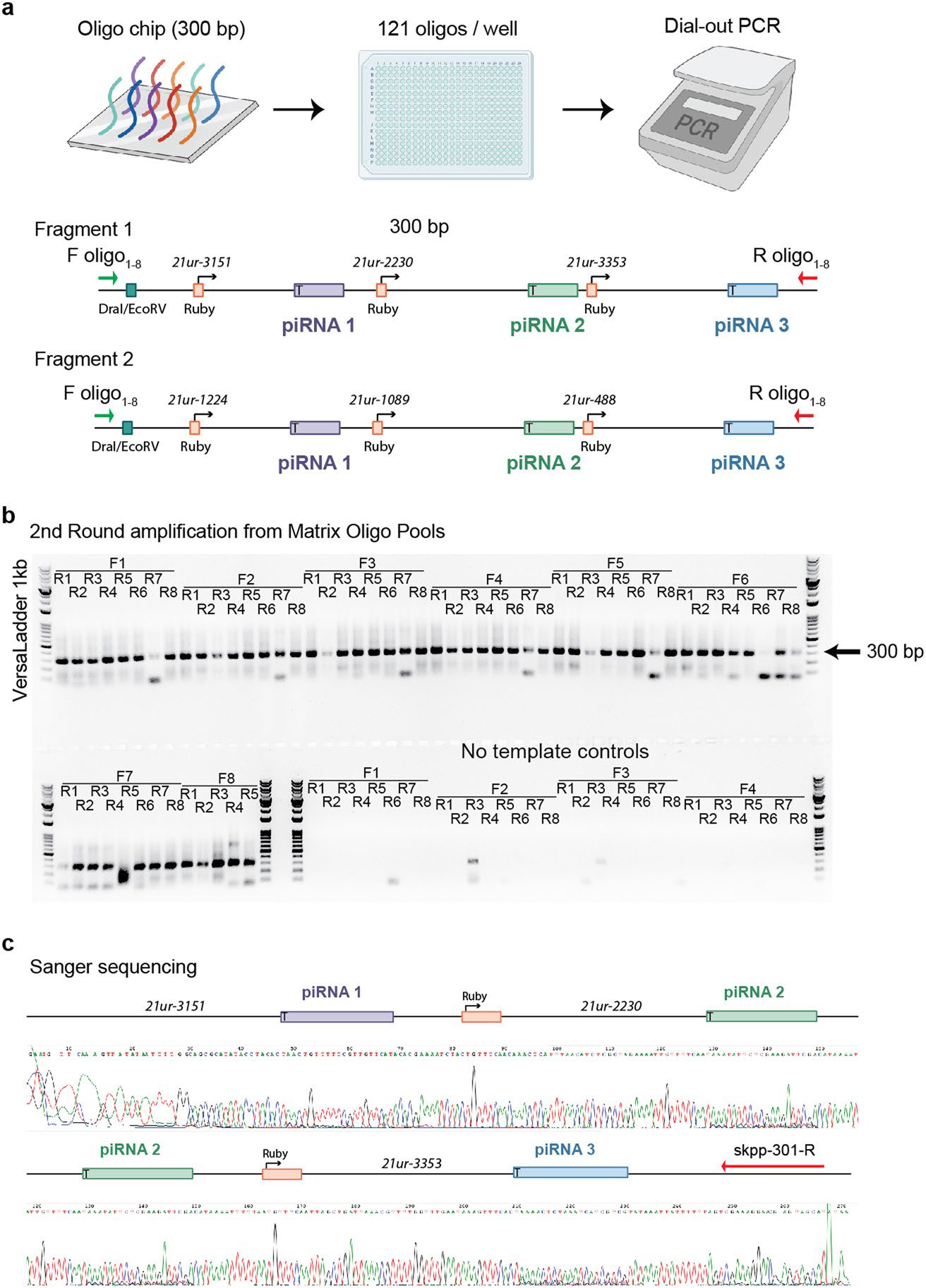
a. Schematic of two different 300 bp piRNAi transgenes, each encoding three sg-piRNAs. 300 bp oligos synthesized at large scale on oligo chips are delivered as arrayed sub-pools with 121 unique oligos in each of 384 wells. The piRNA transgenes are flanked by orthogonal forward and reverse primer sites that allow ‘dial-out’ PCR of transgenes from complex oligo pools (Kosuri et al., 2010). Each oligo pool contains 121 different oligos with a maximum length of 300 base pairs (Matrix Oligo Pools, Twist Bioscience). Two piRNAi transgenes (encoding six sg-piRNAs) are required for silencing. Each pool can, therefore, target 60 genes and individual piRNAi transgenes can be ‘dialed out’ using 16 orthogonal primers (8 forward * 8 reverse = 64 unique combinations). Restriction sites (DraI or EcoRV) allow Sanger sequencing of pair-wise amplified piRNA transgenes. b. 96-well amplification of 60 different piRNA transgene pairs targeting him-5 using two rounds of PCR. The first PCR was a bulk amplification of all oligos in the pool using all 16 amplification primers concurrently. 60 specific piRNA transgenes were amplified in a second round of PCR performed with pair-wise orthogonal primers listed above wells (expected size = 300 bp, indicated by arrow). Control reactions contained no template to assess background amplification of contaminants. With no optimization, we were able to amplify 51 of 60 piRNA transgenes (nine wells with weak or no band at 300 bp). Ladder = 100–10,000 bp VersaLadder (GoldBio). For a large-scale library (for example, a whole-genome library) a subcloning step after the first bulk amplification and transformation of the amplified oligo pool into bacteria would maintain long-term integrity and facilitate distribution of the oligo-pool as lyophilized plasmid pools. c. Representative example of Sanger sequencing of a PCR-amplified piRNA transgene from the oligo pool. 12 of 12 sequenced PCR products contained the expected three unique sg-piRNAs. From the sequencing trace, a low level of cross-talk is visible (minor peaks below the three sg-piRNAs), which can likely be minimized by reducing the number of PCR cycles and using a sub-cloned library as a PCR template.
Extended Data Fig. 9 |. Repressive chromatin modifications spreads in response to piRNAi.
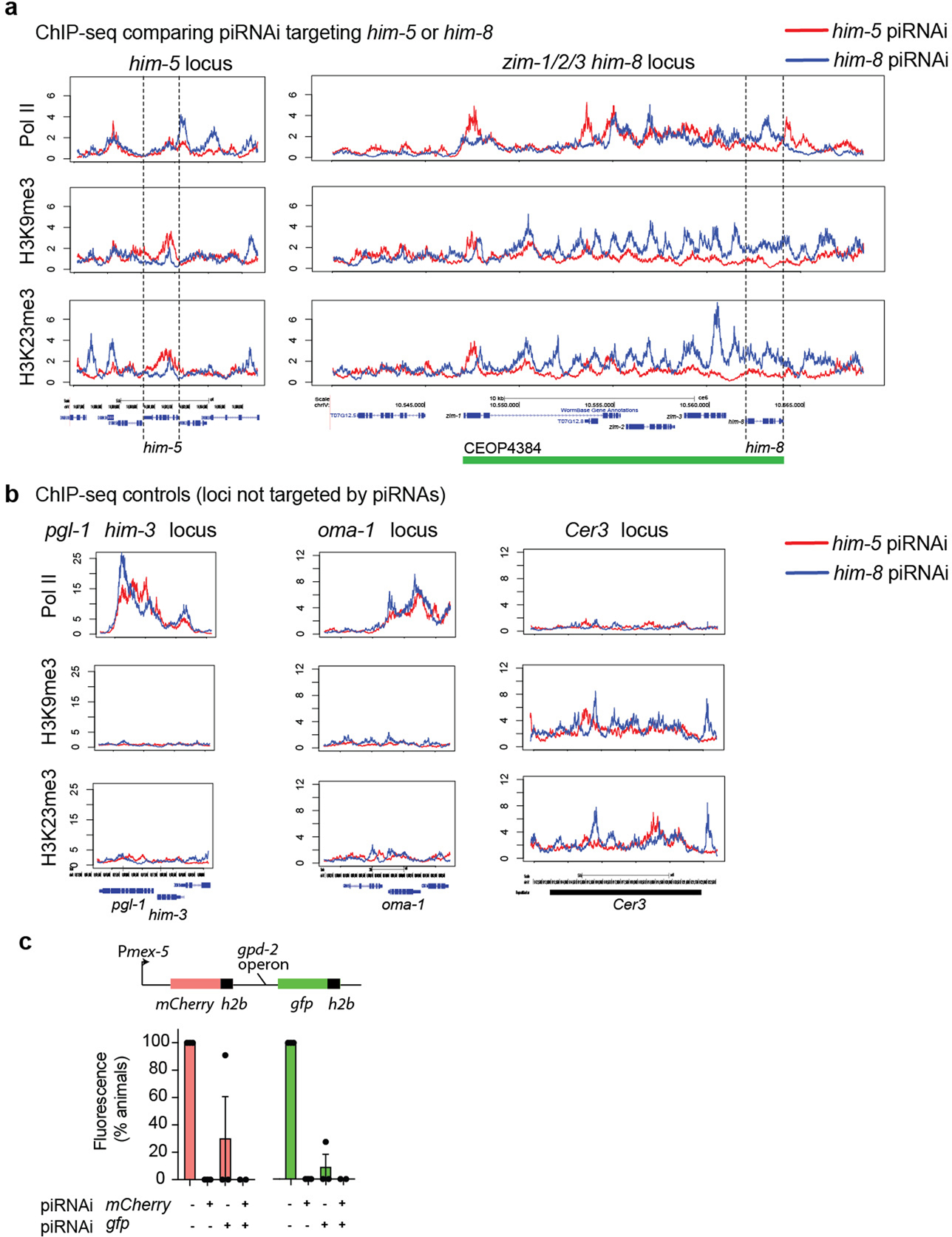
a. ChIP-seq with antibodies against Pol II, H3K9me3, and H3K23me3 in two strains carrying piRNAi transgenes targeting him-5 (red trace) and him-8 (blue trace). The piRNAi target genes are indicated by dotted lines. zim-1, zim-2, zim-3, and him-8 are part of a single operon (CEOP4384, green bar). b. Control loci not targeted by piRNAi. c. mCherry and gfp were co-expressed in an operon under the mex-5 promoter (Pmex-5::mCherry::H2B - gpd-2 operon – gfp::h2b::cye-1 3’UTR) as a single copy insertion at ttTi5605 (Frøkjær-Jensen et al., 2008). The fluorophores were targeted individually or together (control) for silencing with piRNA transgenes using piRNA clusterE. Data are presented as mean values+/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (c) mCherry: n = 3 (−/−), n = 5 (+/−), n = 7 (−/−), n = 5 (+/+), gfp: n = 3 (−/−), n = 5 (+/−), n = 7 (−/+), n = 5 (+/+) biologically independent transgenic strains.
Extended Data Fig. 10 |. piRNAi-induced transgenerational silencing.
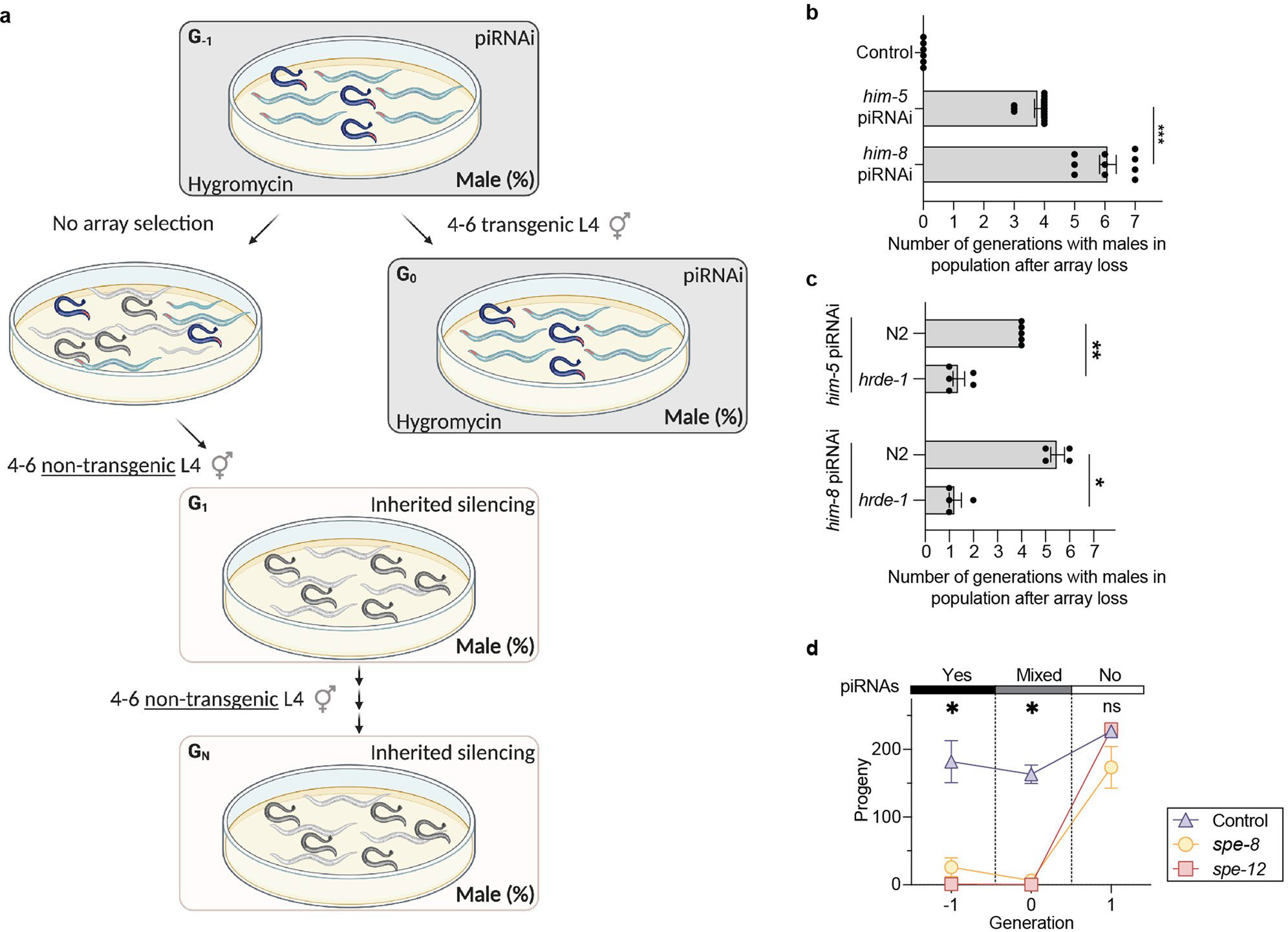
a. Schematic showing assay to determine him-5 and him-8 inherited silencing. To quantify the proportion of males generated from self-fertilization (as opposed to male cross progeny generated by mating), we picked four to six unmated L4 animals in every generation to new plates and scored their progeny for males (out of 100 animals). The assay has a ‘transition generation’ (G0) where a subset of animals are allowed to lose the extra-chromosomal piRNAi-array. Due to the shared germline cytoplasm, non-transgenic animals in the following G1 generation were exposed to sg-piRNAs in early embryonic development. The strains were not exposed to de novo sg-piRNAs generated during the L4 molt when sperm are made and female oocyte production is initiated. b. Duration of him-5 and him-8 transgenerational inheritance. We quantified transgenerational inheritance by counting the number of generations after losing the piRNA array before the fraction of males in the population was <= 1%. Data aggregated from panels Fig. 4bc and Supplementary Fig. 13. Kruskal-Wallis ANOVA, P=<0.0001, Dunn’s post hoc test *** P = 0.0007. c. Duration of transgenerational silencing in hrde-1 mutants. Two-tailed Mann-Whitney test, * P = 0.0286, ** P = 0.0079. d. Inherited silencing of piRNAi targeting spe-8 and spe-12 in a him-5(e1490) mutant background. Unmated L4 hermaphrodites were scored for viable progeny in the presence (solid black bar) or absence (solid white bar) of sg-piRNAs targeting spe-8 and spe-12, respectively. Control = him-5(e1490) animals with a randomized piRNAi cluster. Hygromycin was used to select transgenic animals, including controls, causing the increase in brood size of controls after the animals were grown on non-selective plates. Statistics: One-way ANOVA at each generation. * P = 0.0057 (generation −1), * P = 0.0127 (generation 0), ns P = 0.8312 (generation +1). Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (b) n = 6 (control), n = 14 (him-5), n = 8 (him-8), (c) him-5: n = 5 (N2), n = 5 (hrde-1), him-8: n = 4 (N2), n = 4 (hrde-1), (d) n = 2 (control), n = 2 (spe-8), n = 2 (spe-12) biologically independent transgenic strains.
Supplementary Material
Acknowledgments
We thank L. Wahba and A. Fire (Stanford University) for sharing reagents prior to publication. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). The research was funded by KAUST Office of Sponsored Research OSR-CRG2019-4016 (C.F.-J.), a Rutgers University Busch Biomedical Grant (S.G.G), and the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM111752 (S.G.G). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing Interests
The authors declare no competing interests.
Code availability
The authors’ custom scripts are accessible at https://doi.org/10.5281/zenodo.5599659. Custom software apps are described at https://github.com/AmhedVargas/piRNAi_reprogramming. All code is available with no restrictions.
Data availability
The sequencing data that support the findings of this study are available in NCBI Gene Expression Omnibus (GSE165210). Figures 1, 2, 3, 4 and Extended Data Figures 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 and Supplementary Figures 1, 2, 3, 4, 5, 7, 8, 9, 10, 11, 13 have associated raw data, available in Source Data files. The authors declare that all data supporting the findings of this study are available within the paper (and its supplementary information files). Genomes and annotations files for C. elegans and C. briggsae were downloaded from WormBase version WS270 and WS190 (ftp.wormbase.org). All data is available with no restrictions.
References
- 1.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shalem O, Sanjana NE & Zhang F High-throughput functional genomics using CRISPR–Cas9. Nature Reviews Genetics 16, 299–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fire A et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Kamath RS et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Long L et al. Regulation of transcriptionally active genes via the catalytically inactive Cas9 in C. elegans and D. rerio. Cell Research 25, 638–641 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ketting RF & Plasterk RH A genetic link between co-suppression and RNA interference in C. elegans. Nature 404, 296–298 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Dernburg AF, Zalevsky J, Colaiácovo MP & Villeneuve AM Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev. 14, 1578–1583 (2000). [PMC free article] [PubMed] [Google Scholar]
- 8.Timmons L & Fire A Specific interference by ingested dsRNA. Nature 395, 854–854 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A & Driscoll M Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nature Genetics 24, 180–183 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Bezler A et al. Tissue- and sex-specific small RNAomes reveal sex differences in response to the environment. PLoS Genet. 15, e1007905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG & Ahringer J Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biology 2, research0002.1 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batista PJ et al. PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Molecular Cell 31, 67–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashe A et al. piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell 150, 88–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H-C et al. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luteijn MJ et al. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31, 3422–3430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagijn MP et al. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D et al. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359, 587–592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen E-Z et al. Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 172, 937–951.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozata DM, Gainetdinov I, Zoch A, O’Carroll D & Zamore PD PIWI-interacting RNAs: small RNAs with big functions. Nature Reviews Genetics 1 (2018) doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- 20.Cecere G, Zheng GXY, Mansisidor AR, Klymko KE & Grishok A Promoters recognized by forkhead proteins exist for individual 21U-RNAs. Mol. Cell 47, 734–745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Billi AC et al. A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of Caenorhabditis elegans piRNAs. PLOS Genetics 9, e1003392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meneely PM, McGovern OL, Heinis FI & Yanowitz JL Crossover Distribution and Frequency Are Regulated by him-5 in Caenorhabditis elegans. Genetics 190, 1251–1266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips CM et al. HIM-8 Binds to the X Chromosome Pairing Center and Mediates Chromosome-Specific Meiotic Synapsis. Cell 123, 1051–1063 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Ward JD, Cheng Z & Dernburg AF The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon M et al. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep 7, 762–773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wahba L, Hansen L & Fire AZ An essential role for the piRNA pathway in regulating the ribosomal RNA pool in C. elegans. Developmental Cell 56, 2295–2312.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosuri S et al. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat Biotechnol 28, 1295–1299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alcazar RM, Lin R & Fire AZ Transmission Dynamics of Heritable Silencing Induced by Double-Stranded RNA in Caenorhabditis elegans. Genetics 180, 1275–1288 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu SG et al. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet 44, 157–164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirayama M et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weick E-M et al. PRDE-1 is a nuclear factor essential for the biogenesis of Ruby motif-dependent piRNAs in C. elegans. Genes Dev. 28, 783–796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips CM, Montgomery TA, Breen PC & Ruvkun G MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433–1444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shukla A et al. poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature 1–6 (2020) doi: 10.1038/s41586-020-2323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalinava N, Ni JZ, Peterman K, Chen E & Gu SG Decoupling the downstream effects of germline nuclear RNAi reveals that H3K9me3 is dispensable for heritable RNAi and the maintenance of endogenous siRNA-mediated transcriptional silencing in Caenorhabditis elegans. Epigenetics & Chromatin 10, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buckley BA et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez MF & Lehner B Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 21, 143–151 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Vastenhouw NL et al. Long-term gene silencing by RNAi. Nature 442, 882 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Lev I, Gingold H & Rechavi O H3K9me3 is required for inheritance of small RNAs that target a unique subset of newly evolved genes. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla A, Perales R & Kennedy S piRNAs coordinate poly(UG) tailing to prevent aberrant and perpetual gene silencing. Curr Biol 31, 4473–4485.e3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly WG, Xu S, Montgomery MK & Fire A Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227–238 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fire A, Albertson D, Harrison SW & Moerman DG Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle. Development 113, 503–514 (1991). [DOI] [PubMed] [Google Scholar]
- 42.Gönczy P et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408, 331–336 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Brenner S The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mello CC, Kramer JM, Stinchcomb D & Ambros V Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10, 3959–3970 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frøkjær-Jensen C et al. An Abundant Class of Non-coding DNA Can Prevent Stochastic Gene Silencing in the C. elegans Germline. Cell 166, 343–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sturm Á, Saskői É, Tibor K, Weinhardt N & Vellai T Highly efficient RNAi and Cas9-based auto-cloning systems for C. elegans research. Nucleic Acids Res 46, e105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Ni JZ, Chen E & Gu SG Complex coding of endogenous siRNA, transcriptional silencing and H3K9 methylation on native targets of germline nuclear RNAi in C. elegans. BMC Genomics 15, 1157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gent JI et al. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics 183, 1297–1314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz-Orbach L et al. Caenorhabditis elegans nuclear RNAi factor SET-32 deposits the transgenerational histone modification, H3K23me3. eLife 9, e54309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frøkjær-Jensen C et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40, 1375–1383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aljohani MD, El Mouridi S, Priyadarshini M, Vargas-Velazquez AM & Frøkjær-Jensen C Engineering rules that minimize germline silencing of transgenes in simple extrachromosomal arrays in C. elegans. Nat Commun 11, 6300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dickinson DJ, Ward JD, Reiner DJ & Goldstein B Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data that support the findings of this study are available in NCBI Gene Expression Omnibus (GSE165210). Figures 1, 2, 3, 4 and Extended Data Figures 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 and Supplementary Figures 1, 2, 3, 4, 5, 7, 8, 9, 10, 11, 13 have associated raw data, available in Source Data files. The authors declare that all data supporting the findings of this study are available within the paper (and its supplementary information files). Genomes and annotations files for C. elegans and C. briggsae were downloaded from WormBase version WS270 and WS190 (ftp.wormbase.org). All data is available with no restrictions.


