Abstract
Neuroimmune dysfunction is a cardinal feature of neurodegenerative diseases. But how immune dysregulation in the brain and peripheral organs contribute to neurodegeneration remains unclear. Here, we discuss the recent advances highlighting neuroimmune dysfunction as a key disease-driving factor in frontotemporal dementia (FTD). We provide an overview of the clinical observations supporting a high prevalence of autoimmune diseases in FTD patients with mutations in GRN or C9orf72. We then focus on a myriad of evidence from human genetic studies, mouse models, in vitro assays, and multi-omics platform, which indicate that haploinsufficiency in GRN and C9orf72 promotes neuroimmune dysfunction and contributes to neurodegeneration and premature death. These compelling data provide key insights to disease mechanisms, biomarker discovery, and therapeutic interventions for FTD (120 words).
Introduction
Age is a common denominator for neurodegenerative diseases. One hallmark of aging is the development of persistent proinflammatory responses that contribute to systemic diseases, including atherosclerosis, metabolic syndrome, cancer, and fragility. In a similar vein, dysfunction in the brain’s innate immune system, characterized by the expansion of reactive microglia and astrocytes, has increasingly been recognized as a key factor that contributes to brain aging and neurodegeneration. Indeed, recent literature based on genome-wide association studies (GWAS), clinical studies, cell biology, animal models, and neuropathology indicate that perturbations in the expression of genes involved in the homeostasis of microglia and astrocytes may have critical role in the pathogenesis of late onset Alzheimer’s disease (AD) [1]. In addition to the dysfunction in the brain’s innate immune system in promoting neurodegeneration, there is compelling evidence that dysfunction in peripheral immunity can contribute to the pathophysiology of neurodegenerative diseases. This is supported by clinical and epidemiological studies that show elevated inflammatory markers increase the risk of developing AD [2].
This review focuses on the recent evidence supporting that dysfunction in the peripheral and brain’s immune systems also have essential roles in the pathophysiology of frontotemporal dementia (FTD), the second most common neurodegenerative disease that affects patients younger than 65-years old [3]. We begin with an overview of the seminal clinical observations and genetic studies that suggest a role for peripheral immune dysfunction in FTD patients. We then summarize data from animal models that provide critical insights into how loss-of-function in two most common FTD genes leads to age-dependent dysregulation in the peripheral and central immune functions. Finally, we discuss several critical next-steps and opportunities for future research that will translate this knowledge into therapeutic interventions.
Clinical features, genetics, and neuropathology of FTD
The manifestations of FTD are characterized by progressive deficits in behaviors, executive functions, or language [3]. The underlying neuropathological features of FTD, collectively known as frontotemporal lobar degeneration (FTLD), involve severe atrophy in the frontal and temporal lobes and the accumulation of microtubule-associated protein tau and two RNA binding proteins, TDP-43 and FUS [4]. Among these, FTLD-TDP is the most common type of FTLD, representing > 50% of all FTD cases. Based on the distribution and the morphology of the misfolded TDP-43 inclusions and dystrophic neurites in the frontal cortex, FTLD-TDP can be further classified into 5 different subtypes (Figure 1a) [5].
Figure 1. Neuropathology and autoimmune dysfunction in FTLD patients.
(a) Gross neuropathology of FTLD caused by GRN mutations (upper panel). Microscopic features of TDP-43 proteinopathy in FTLD that define type A, B and C. Although TDP-43 is located inside of nuclei in normal neurons, aggregates of TDP-43 are detected in the frontal cortex of FTLD-TDP patients. Based on the morphology of TDP-43 proteinopathy, FTLD-TDP can be classified into type A, which shows compact aggregation of TDP-43 in neuronal cytoplasm and TDP-43 thread-like inclusions in the neuropils or neurites in layers 2/3, type B, which shows diffuse cytoplasmic granular TDP-43 aggregates in layers 2–5, and type C, which contains long tortuous TDP-43-containing neurites and cytoplasmic TDP-43 aggregates in layer 2/3 in the frontal cortex (b) Case-control studies show a higher propensity for FTLD-TDP patients to have co-occurrence of systemic autoimmune diseases. Pie charts are modified based on the data previously published [12,13].
Two major genetic mutations for FTLD-TDP provide key insights to uncovering disease mechanisms. The first is dominant mutations in the Progranulin (GRN) gene on chromosome 17q21.31, which activates nonsense-mediated decay of GRN mRNA and leads to significant reductions in Progranulin (PGRN) protein levels [6,7]. Another involves hexanucleotide (GGGGCC)n repeat expansion located between exons 1a and 1b of the gene on chromosome 9 open reading frame 72 (C9orf72) [8,9]. Subsequent large-scale GWAS confirm the central roles of GRN and C9orf72 mutations in FTLD-TDP, each accounting for 13.9% and 25.5% of all cases [10]. Less frequent mutations in the gene encoding TANK-binding kinase 1 (TBK1) account for 1.5% of all FTLD-TDP patients. Other FTLD-TDP-associated mutations include TARDBP, DCNT1, VCP, ATXN2, UBQLN2, MATR3, HNRNPA1/B2, and OPTN. In addition, several genetic risk alleles for FTLD-TDP have been identified, including HLA-DQA2, DPP6, and UNC13A [10,11].
One intriguing clinical observation in FTLD-TDP patients is the co-occurrence of autoimmune diseases. Compared to AD patients and age-matched controls, FTLD-TDP cohorts, especially those with GRN and C9orf72 mutation carriers show an increased prevalence of non-thyroid autoimmune diseases, such as inflammatory arthritides, cutaneous disorders, and gastrointestinal conditions (Figure 1b) [12, 13]. Several lines of evidence support that dysfunction in the immunity in peripheral organs and central nervous system (CNS) may contribute to the pathogenesis of FTLD-TDP. First, GWAS studies identify polymorphisms in the 3’ untranslated region (UTR) of C9orf72 as one of the three risk alleles that influence whether patients with rheumatoid arthritis (RA) respond to anti-TNF treatment [14]. These results, reported years before C9orf72 is identified as a disease gene for familial FTD and ALS, suggest that the gene product of C9orf72 may modulate TNFα inflammatory pathway. Consistent with this idea, elevations in plasma TNFα were observed in FTD patients with GRN mutations [12]. Moreover, a recent study shows that patients with systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) have a high propensity to carry intermediate GGGGCC expansion repeats (9–30) in C9orf72 [15]. Several risk genes for FTLD-TDP identified via GWAS studies, including the HLA loci on chromosome 6, are well-known genes implicated in autoimmune disorders [11]. Finally, PET imaging using ligands for activated microglia and TDP-43 in FTD patients show positive correlation supporting the presence of microglial activation and TDP-43 accumulation [16,17].
Given the preponderance of new evidence supporting GRN and C9orf72 as the gatekeepers of the neuroimmune systems, we believe that a timely review of these studies will provide an integrative and deeper understanding of the physiological functions of these genes. We define “neuroimmune” broadly to avoid compartmentalizing the CNS from peripheral immunity and to highlight the intricate interactions between these systems and their impact on the entire organism.
Neuroimmune dysfunction in PGRN deficiency
Initially isolated as a secreted glycoprotein, PGRN has been shown to promote neuronal survival and neurite outgrowth similar to neurotrophic factors [18]. The mechanism by which PGRN exerts its neurotrophic function remains unclear. However, PGRN can bind to sortilin, which facilitates PGRN endocytosis and its subsequent degradation in the lysosomes [19](Figure 2a). It is conceivable that PGRN may bind to other membrane-bound receptor(s) to activate signaling pathways to support survival and neurite outgrowth. In addition to its role as a secreted protein, PGRN can regulate the trafficking of intracellular vesicles and organelle dynamics. These functions are sortilin-independent and involve PGRN binding with prosaposin (PSAP) and subsequent delivery of the PGRN-PSAP complex to lysosomes through mannose 6-phosphate receptor (M6PR) and low-density lipoprotein (LDL) receptor-related protein 1 (LRP1). Interestingly, GWAS shows that single nucleotide polymorphisms in the human PSAP gene affect the PGRN levels in the plasma presumably through the role PSAP plays in regulating dimerization and secretion of PGRN [20]. Once in the lysosomes, PGRN can regulate cathepsin D activity, maintain the acidification of lysosomes, and exocytosis of lysosomes (or exophagy) upon treatment with aggregated LDL [21-24]. Finally, loss of PGRN inhibits lysosome fusion and induces autophagosome accumulation in microglia and macrophages, whereas in cortical neurons PGRN appears to have modest effects in regulating autophagic flux [25].
Figure 2. Cell biology of Progranulin (PGRN) deficiency in microglia.
(a) Secretion of PGRN can be mediated via the secretory and exophagy pathways. Once secreted, PGRN is endocytosed by binding with sortilin and M6PR and eventually reaches the lysosomes where it interacts with PSAP and regulates lysosomal acidification and degradation of cargoes. PGRN can interact with CpG and TLR9 to promote TNFα expression in innate immune response. In addition, PGRN can use sortilin-independent mechanism to transport from Golgi to late endosomes where it regulates vesicle trafficking from late endosomes to early lysosomes. In PGRN deficient microglia, there are increases in lysosomal storage materials, complement C3 and its cleavage product C3b, TMEM106B, PSAP and lipids. The excessive production of complement proteins, cytokines, and lipids in Grn−/− microglia (b) Transcriptomic analyses in Grn−/− thalamus show age-dependent microglial activation, characterized by overproduction of proinflammatory cytokines and complement proteins, C1q and C3b, from 7 to 12 months old. During this period, Grn−/− microglia promote synaptic pruning and cause hyperexcitability in the thalamocortical circuit. From 12 to 24 months old, Grn−/− microglia exhibit ameboid morphology and continue to produce excessive C1q, C3b, bioreactive lipids, and other unknown factors to accelerate neuronal cell death by compromising nuclear pore integrity and TDP-43 proteinopathy.
Several lines of evidence show that loss-of-function in PGRN leads to dysfunction in innate immunity and host defense in various organs, including spleen, liver, and brain. In peripheral immunity, Grn−/− macrophages produce more proinflammatory cytokines which can be aggravated by LPS treatment. Interestingly, Grn−/− mice exhibit increased sensitivity to Listeria monocytogenes infection due to reduced clearance of pathogens by macrophages [25,26]. The mechanism for reduced clearance of pathogens by Grn−/− macrophages is likely due to the role of PGRN in controlling macrophage-associated inflammatory responses and defects in lysosome-mediated degradation pathway. For instance, PGRN binds to and enhances toll-like receptor 9 (TLR9) signaling upon the treatment of CpG oligonucleotides in macrophage to promote the TNFα secretion [27]. Furthermore, in collagen-induced inflammatory arthritis model, PGRN attenuates TNFα-induced MAPK and NF-κB signaling for anti-inflammatory effects by binding to tumor necrosis factor receptors (TNFRs) and antagonizing TNFα [28], though the interaction between PGRN and TNFR is disputed by other studies [29,30].
Pertinent to the role of PGRN deficiency in neurodegeneration, several groups report that Grn−/− mice show an age-dependent expansion of microglia and heightened microglial responses in toxin-induced injury [26,31]. The microgliosis phenotype in Grn−/− mice preferentially affects the thalamocortical circuit and cerebellar white matter, leading to obsessive-compulsive disorder (OCD)-like behaviors, poor motor coordination, and gait imbalance [32,33]. The expansion of Grn−/− microglia is accompanied by progressive morphological and transcriptomic changes, suggesting that Grn−/− microglia undergo phenotypic transition into a proinflammatory state. Consistent with these results, Grn−/− microglia produce abundant complements, C1q and C3b, to promote synaptic pruning in the ventral thalamus, which leads to an imbalance in the excitation-inhibition input to the thalamocortical circuit and OCD-like grooming behaviors in Grn−/− mice [33]. Interestingly, pre-symptomatic GRN mutation carriers also show hyperconnectivity in the thalamocortical network [34]. While these results suggest that the thalamus could be a highly evolutionarily conserved neural circuit selectively affected by PGRN deficiency, it is noteworthy that microglial activation phenotypes can also be detected in the striatum in Grn−/− mice, leading to increased TNFα production and activation of the NFκB signaling [32]. Moreover, deletion of C1q or Ikbkb genes mitigates these behavioral phenotypes in Grn−/− mice.
The robust microglial phenotypes in Grn−/− mice raise the intriguing hypothesis that persistent microglial activation may contribute to neurodegeneration and TDP-43 protein aggregation (TDP-43 proteinopathy), two key neuropathological features in FTLD-GRN patients. Indeed, single nucleus RNA-sequencing (snRNA-seq) using microdissected thalamus from an aging cohort of wild-type (Grn+/+) and Grn−/− mice shows that, among all the cell clusters in thalamus, microglia are the first to show transcriptomic changes as early as 7-months old, before the onset of behavioral phenotypes. As Grn−/− mice become older, Grn−/− microglia undergo further age-dependent transcriptomic changes indicating the transition of Grn−/− microglia from homeostatic state to disease state by downregulation of homeostasis-related genes and upregulation of genes related to lysosome functions, lipid metabolism, and trafficking [35,36]. Interestingly, snRNA-seq data show a selective loss of excitatory neurons in the thalamus of Grn−/− mice at 19-months old, which is preceded by the prominent accumulation of nuclear and cytoplasmic TDP-43 aggregation, nuclear pore defects, and neuronal cell death [35]. Together, these results support the hypothesis that microglial toxicity and neuronal vulnerability contribute to neuronal degeneration in PGRN deficiency (Figure 2b).
In support of the critical role of PGRN deficiency in microglial activation, large-scale GWAS studies identify multiple SNPs in the TMEM106B gene as potential modifiers for neurodegeneration in FTLD-GRN patients [37-39]. These TMEM106B variants appear to modify PGRN expression level in FTLD-GRN patients. From the cell biology perspective, TMEM106B has partial functional overlaps with PGRN in regulating lysosomal trafficking. Consistent with this idea, upregulation of TMEM106B mitigates the lysosomal defects caused by the reduction of PGRN level. To characterize the in vivo functions of TMEM106B and PGRN, several groups generate Grn−/−;Tmem106b−/− mice and show that the complete loss of TMEM106B and PGRN leads to more severe lysosomal abnormalities and neuroinflammation, characterized by early onset and drastic expansion of reactive microglia and astrocytes, and upregulation of many proinflammatory genes [40-42]. In addition, loss of TMEM106B and PGRN accelerates neuronal loss with prominent aggregation of ubiquitinated proteins and phospho-TDP-43 in the spinal cord leading to motor deficits [40,41]. Interestingly, partial loss of TMEM106B may provide some partial protection in lysosomal dysfunction caused by the loss of PGRN [43]. These findings provide critical insights into the dosage-dependent role of PGRN and TMEM106B in regulating lysosomal biogenesis and functions. They also underscore TMEM106B as a bona fide disease modifier for FTLD-GRN. Based on these studies, loss of TMEM106B appears to primarily affects the neuroinflammation. It remains unclear whether TMEM106B has any effect in the peripheral immunity.
Another prominent feature in the Grn−/− mouse brain and FTLD-GRN brain is the accumulation of lipofuscin, which consists of highly oxidized mixtures of proteins, lipids and carbohydrates [44-46]. Indeed, lipofuscin deposits in the retina have been proposed as an early manifestation of disease in GRN carriers [45]. Consistent with these results, lipidomic profiling using cortices in FTLD-GRN patients and aged Grn−/− mouse brain shows significant alteration of lipid composition, including glycolipids and phospholipids [47]. CRISPR-based genetic screening in BV2 cells identifies GRN as an important genetic modifier of lipid droplet accumulation that promotes pro-inflammatory states in microglia during brain aging [48]. Additional lipidomic profiling in aged Grn−/− mouse cortices shows lysosomal lipid dysregulation, including downregulation of bis-monoacylglycero-phosphate (BMP) and upregulation of glucosylsphingosine, which primarily affects Grn−/− microglia [49]. While these results strongly implicate PGRN as a key regulator of lipid metabolism, the exact mechanism remains unclear.
Neuroimmune dysfunction in C9orf72 deficiency
The identification of hexanucleotide (GGGGCC)n repeat expansion in C9orf72 gene represents a major milestone in FTD-amyotrophic lateral sclerosis (ALS) research [8,9]. Collectively, mutations in C9orf72 account for ~26% of FTLD-TDP cases [10] and a majority of familial ALS patients. At least three distinct mechanisms have been postulated to promote neurodegeneration caused by C9orf72 mutations, including the production of dipeptide repeat (DPR) proteins repeat-associated non-ATG (RAN) translation via sense and antisense (GGGGCC)n RNA transcripts [50,51], the presence of RNA foci in neuronal nuclei, and haploinsufficiency in C9orf72 protein function. An indepth discussion on the first two mechanisms for C9orf72 mutations is beyond the scope of this article. Readers interested in these topics will find many exciting discoveries regarding the role of C9orf72 mutations in nucleocytoplasmic transport [52-54], stress granule dynamics and heterochromatin organization [55,56], DNA damage repair [57], phase separation [58-60], mitochondrial functions [61], and in p53-mediated neuronal degeneration [62].
Aside from the gain-of-function properties associated with C9orf72 mutations, C9orf72 has critical roles in the autophagy-lysosome pathway and mitochondrial energy metabolism (Figure 3a). Related to the autophagy pathway, C9orf72 forms multiprotein complex with SMCR8 and WPR41, which acts as a GDP-GTP exchange factor, and control Rab8a and Rab39b GTPases activity for the recruitment them to organelle membrane to regulate the membrane trafficking. Besides, C9orf72 regulates autophagy initiation through controlling ULK1 expression and activity [63-66]. Moreover, it has been also reported that coactivator-associated arginine methyltransferase 1 (CARM1)-related autophagy-lysosome pathway is supported by C9orf72 protein, showing that C9orf72 deficiency leads to dysregulated autophagic digestion which induces endoplasmic reticulum-derived lipid droplets accumulation and free fatty acid secretion under nutrient stress [67]. In addition to its role in the initiation of autophagy, C9orf72 has been shown to regulate Rab5-mediated recycling of glutamate receptors. iPSC-derived motor neurons from patients with C9orf72 mutation show impaired lysosomal degradation of glutamate receptors, which render these neurons more vulnerable to excitotoxicity [68]. Finally, C9orf72 has a critical role in regulating oxidative phosphorylation and ATP production by the stabilizing translocase of inner mitochondrial membrane domain containing 1 (TIMMDC1)-associated mitochondrial complex I assembly [69].
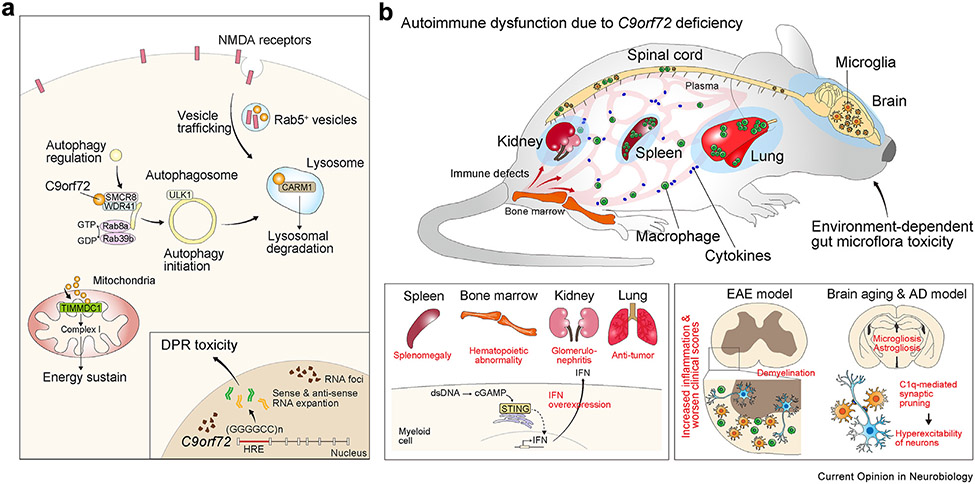
Figure 3. Cell biology of C9orf72 and its roles as a gatekeeper to peripheral immunity.
(a) Intracellular function of C9orf72 regulates the initiation of autophagy by forming protein complexes with SMCR8 and WDR41. In addition, C9orf72 regulates vesicle trafficking and autophagosome-lysosome fusion, and can interact with CARM1 to process lysosomal degradation. In addition, C9orf72 regulates energy homeostasis through mitochondrial complex I in oxidative phosphorylation. The (GGGGCC)n hexanucleotide repeat expansion (HRE) in C9orf72 causes accumulation of RNA foci and production of dipeptide repeat (DPR) that can have neurotoxic properties. (b) Loss-of-function in C9orf72 causes autoimmune dysfunction in spleen, kidney, and lung. Transcriptomic and functional analyses show that C9orf72 deficiency leads to the overexpression of IFN in myeloid cells and microglia through the STING-cGAS pathway, thereby disrupting immune homeostasis. In the central nervous system, C9orf72 mutation also induces immune defects showing microgliosis during aging and in disease models for experimental allergic encephalitis and Alzheimer disease (AD). Abbreviations: DPR, dipeptide repeat; EAE, experimental allergic encephalitis; HRE, hexanucleotide repeat expansion; IFN, interferon.
Like Grn−/− mice, C9orf72−/− mice show shortened lifespan due in large part to autoimmune dysfunction (Figure 3b). In the cerebral cortex, C9orf72 is most abundantly detected in microglia, followed by neurons [70]. Consistent with these results, RNA-seq analyses in C9orf72−/− brain show transcriptomic features indicative of activation in the interferon signaling and lysosomal dysfunction [70]. In addition to the neuroinflammatory phenotypes, C9orf72−/− mice show prominent autoimmune dysfunction with marked lymphadenopathy, splenomegaly, and glomerulonephritis [70-72]. Interestingly, deleting C9orf72 in myeloid cells using the Cx3cr1-Cre drive is sufficient to recapitulate the autoimmune dysfunction in the C9orf72−/− mice. The peripheral immune phenotypes are worse in C9orf72−/− mice that also express the hexanucleotide repeat expansion (HRE) transgene [73]. Furthermore, loss of C9orf72 activates the STING pathway-mediated interferon in the myeloid cells and increases neuroinflammation in the experimental allergic encephalitis model and increases antitumor immunity [74]. In addition, during brain aging, loss of C9orf72 activates STING-mediated interferon pathways in microglia and promotes microglia-mediated synaptic pruning via C1q activation [75]. In the 5XFAD model, loss of C9orf72 increases lysosomal accumulation in microglia, reduces dendritic arborization and synaptic density in cortical neurons, and impairs motor function [75]. Together, these results support the critical role of C9orf72 in maintaining microglia homeostasis during brain aging and the innate response of microglia to amyloid deposits and amyloid-mediated toxicity. These results further underscore the similar roles of PGRN and C9orf72 in regulating microglial function during aging and in disease models.
Another critical manifestation of the systemic fatal autoimmune diseases in C9orf72−/− mice is the increased vulnerability to environment-associated gut bacteria microflora [76]. Specifically, increase in the abundance of bacterial species, such as Helicobacter spp and Tritrichomonas muris, is associated with severe immune defects and early lethality in C9orf72−/− mice. Conversely, therapeutic interventions, including antibiotic treatment and fecal transplantation, alter gut microflora abundance and drastically mitigate the production of proinflammatory cytokines, autoimmune dysfunction, and survival in C9orf72−/− mice [76]. Although it remains unclear how gut microflora aggravates the immune system in C9orf72−/− mice, there is evidence that changes in gut microflora can induce proliferation in the myeloid cells in the peripheral immune system and activate microgliosis in the spinal cord. Interestingly, the observation that gut microflora can impact the survival and disease progression in ALS are also detected in SOD1G93A mice. However, the specific types of microflora and how they impact survival of SOD1G93A mice are quite different from those in C9orf72−/− mice [77]. Unlike C9orf72−/− mice where gut microflora-induced inflammation appears to be a major pathogenic factor, the contribution of gut microflora to disease onset and progression in SOD1G93A mice is due in part to the metabolites made by the bacteria, including the level of nicotinamide. Together, these results provide intriguing and contrasting mechanisms by which gut microflora use diverse mechanisms to promote disease onset and progression in ALS.
Conclusion and outlook
The discovery of GRN and C9orf72 ushers in a new era of research that uncovers the disease mechanisms of FTD. It is now clear that PGRN and C9orf72 are not only required for maintaining the homeostasis of microglia during brain aging, but they can also regulate the intricate interaction between the innate and adaptive immunity in the peripheral organs. Collectively, these results provide new insights and bridge the gap between neuroinflammation and autoimmune dysfunction. There are, however, many unanswered questions as to how PGRN and C9orf72 regulate immune functions during the aging process. From cell biology perspectives, the current literature supports that loss of PGRN or C9orf72 leads to dysfunction in macrophages and other myeloid cells. Although it is well-established that PGRN regulates endolysosomal trafficking, it remains unclear how lysosomal defects caused by PGRN insufficiency activate microglia and/or other immune cell types, such as macrophages, lymphocytes, dendritic cells, or NK cells that ultimately contribute to neuroinflammation and autoimmune dysfunction. Interestingly, neuroinflammation similar to those in Grn−/− mouse brain has also been reported in several mouse models of lysosomal storage disease, including mucopolysaccharidosis (MPS), neuronal ceroid lipofuscinosis (NCL), and Niemann-Pick’s disease (NPC) [78-81]. Future studies will be required to elucidate the mechanism(s) of lysosomal dysfunction in immune activation. The higher prevalence of autoimmune disorders in FTLD-TDP patients further raises the intriguing question as to whether the manifestations of peripheral immune dysfunction and neuroinflammation can serve as effective biomarkers that predict, capture, and track the disease onset and progression in these patients. Finally, one major task for future studies is to determine how mutual interactions between the peripheral immune dysfunction and neuroinflammation might facilitate disease progression in FTLD-TDP patients. For FTLD caused by C9orf72 mutations, it will be important to investigate the potential contributions of RNA foci and DPR to neuroinflammation and disease pathogenesis.
Since haploinsufficiency of PGRN and C9orf72 is a key disease-driving factor in FTLD and ALS, restoring PGRN and C9orf72 to their physiological levels is a plausible therapeutic strategy. As proof-of-concept approach, adeno-associated virus (AAV)-mediated delivery of PGRN can mitigate neuroinflammation and behavioral phenotypes in Grn+/− and Grn−/− mice [82,83]. However, similar AAV-mediated approaches to restore PGRN expression trigger a robust proliferation in T-lymphocytes [84]. Another non-viral PGRN replacement approach is to combine PGRN with a modified Fc domain that binds human transferrin receptor, which increases the efficiency for PGRN to cross the blood brain barrier (BBB). Results from this study shows that this BBB-penetrant PGRN can ameliorates microgliosis in Grn−/− mice and mitigate lysosomal phenotypes, such as depletion of BMP lipid and restoration of lysosomal pH and integrity in Grn−/− microglia in vivo [49]. In addition, there are ongoing clinical trials to suppress the toxic gain-of-function effects of C9orf72 mutations with antisense oligonucleotides [85,86]. It is possible that combinatorial therapies that include C9orf72 replacement may have additional beneficial effects. Finally, given the implications of PGRN and C9orf72 in aging and AD [75,87-89], replacement therapy for PGRN and C9orf72 could have broader impacts in these conditions.
Acknowledgements
This work has been supported by the postdoctoral fellowship from the Japanese Society for the Promotion of Science (K.H.), NIH grants Blueprint for Neuroscience Diversity Supplement R01 AA027074-04S1 (N.J.), F99 NS124183-01A1 (N.J.), K23 AG048291 (Z.A.M.), Blueprint for Neuroscience R01 AA027074-04 (E.J.H.), R01 AG057462-05 (E.J.H.), R01 AG068290-02 (E.J.H.), Department of Veterans Affairs Merit Award I01 BX001108-09, and the Bluefield Project to Cure FTD (E.J.H.).
Abbreviations
- FTD
Frontotemporal dementia
- FTLD
Frontotemporal lobar degeneration
- AD
Alzheimer’s disease
- GWAS
Genome-wide association studies
- TDP-43
TAR DNA binding protein 43
- FUS
Fused in sarcoma
- RA
Rheumatoid arthritis
- SLE
Systemic lupus erythematosus
- M6PR
Mannose 6-phosphate receptor
- LRP1
Low-density lipoprotein receptor-related protein 1
- TNF
Tumor necrosis factor
- TNFR
Tumor necrosis factor receptor
- SNP
Single nucleotide polymorphism
- NCL
Neuronal ceroid lipofuscinosis
- NPC
Niemann-Pick’s disease
- MPS
Mucopolysaccharidosis
- BBB
Blood-brain barrier
- ALS
Amyotrophic lateral sclerosis
- GRN
Progranulin(gene)
- PGRN
Progranulin (protein)
- PSAP
Prosaposin (protein)
- C9orf72
Chromosome 9 open reading frame 72
- OCD
Obsessive-compulsive disorder
Footnotes
Conflict of interest statement
None declared.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Karch CM, Cruchaga C, Goate AM: Alzheimer’s disease genetics: from the bench to the clinic. Neuron 2014, 83:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettcher BM, et al. : Peripheral and central immune system crosstalk in Alzheimer disease - a research prospectus. Nat Rev Neurol 2021, 17:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bang J, Spina S, Miller BL: Frontotemporal dementia. Lancet 2015, 386:1672–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann JW, Seeley WW, Huang EJ: RNA binding proteins and the pathogenesis of frontotemporal lobar degeneration. Annu Rev Pathol 2019, 14:469–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackenzie IR, et al. : A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol 2011, 122:111–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker M, et al. : Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006, 442:916–919 [DOI] [PubMed] [Google Scholar]
- 7.Cruts M, et al. : Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 2006, 442:920–924. [DOI] [PubMed] [Google Scholar]
- 8.DeJesus-Hernandez M, et al. : Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renton AE, et al. : A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011, 72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.**. Pottier C, et al. : Genome-wide analyses as part of the international FTLD-TDP whole-genome sequencing consortium reveals novel disease risk factors and increases support for immune dysfunction in FTLD. Acta Neuropathol 2019, 137:879–899. Large scale GWAS study showing GRN and C9orf72 mutations as the two most common mutations in FTLD-TDP patients. This study also reveals additional risk alleles for FTLD-TDP that could affect immune functions.
- 11.*. Broce I, et al. : Immune-related genetic enrichment in frontotemporal dementia: an analysis of genome-wide association studies. PLoS Med 2018, 15:e1002487. Large scale GWAS study that reveals several genetic risk alleles for FTD are implicated in immune functions.
- 12.*. Miller ZA, et al. : TDP-43 frontotemporal lobar degeneration and autoimmune disease. J Neurol Neurosurg Psychiatry 2013, 84:956–962. First study to show that semantic variant primary progressive aphasia and GRN mutation carriers have a high propensity to develop autoimmune diseases.
- 13.*. Miller ZA, et al. : Increased prevalence of autoimmune disease within C9 and FTD/MND cohorts: completing the picture. Neurol Neuroimmunol Neuroinflamm 2016, 3:e301. A follow-up study that further confirms the increased prevalence of autoimmune disorders in FTLD-TDP patients with C9orf72 mutations and FTD/ALS cases without known mutations.
- 14.Liu C, et al. : Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Mol Med 2008, 14:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredi M, et al. : C9orf72 intermediate alleles in patients with amyotrophic lateral sclerosis, systemic lupus erythematosus, and rheumatoid arthritis. Neuromolecular Med 2019, 21:150–159. [DOI] [PubMed] [Google Scholar]
- 16.Malpetti M, et al. : In vivo PET imaging of neuroinflammation in familial frontotemporal dementia. J Neurol Neurosurg Psychiatry 2021, 92:319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bevan-Jones WR, et al. : Neuroinflammation and protein aggregation co-localize across the frontotemporal dementia spectrum. Brain 2020, 143:1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Damme P, et al. : Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J Cell Biol 2008, 181:37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu F, et al. : Sortition-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 2010, 68:654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson AM, et al. : Prosaposin is a regulator of progranulin levels and oligomerization. Nat Commun 2016, 7:11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X, et al. : Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol 2015, 210:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valdez C, et al. : Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum Mol Genet 2017, 26:4861–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen AD, et al. : Progranulin in the hematopoietic compartment protects mice from atherosclerosis. Atherosclerosis 2018, 277:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka Y, et al. : Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum Mol Genet 2017, 26:969–988. [DOI] [PubMed] [Google Scholar]
- 25.Chang MC, et al. : Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J Exp Med 2017, 214:2611–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin F, et al. : Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J Exp Med 2010, 207:117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park B, et al. : Granulin is a soluble cofactor for toll-like receptor 9 signaling. Immunity 2011, 34:505–513. [DOI] [PubMed] [Google Scholar]
- 28.Tang W, et al. : The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science 2011, 332:478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, et al. : Progranulin does not bind tumor necrosis factor (TNF) receptors and is not a direct regulator of TNF-dependent signaling or bioactivity in immune or neuronal cells. J Neurosci 2013, 33:9202–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etemadi N, et al. : Progranulin does not inhibit TNF and lymphotoxin-alpha signalling through TNF receptor 1. Immunol Cell Biol 2013, 91:661–664. [DOI] [PubMed] [Google Scholar]
- 31.Martens LH, et al. : Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J Clin Invest 2012, 122:3955–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krabbe G, et al. : Microglial NFkappaB-TNFalpha hyper-activation induces obsessive-compulsive behavior in mouse models of progranulin-deficient frontotemporal dementia. Proc Natl Acad Sci U S A 2017, 114:5029–5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.**. Lui H, et al. : Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell 2016, 165:921–935. By combining transcriptomic, histopathology and electrophysiology in brain slice recording, this study shows Grn−/− mice develop age-dependent microgliosis that affects the sensory thalamus. In this brain region, Grn−/− microglia produce excessive complement protein C1q to promote synaptic pruning, which leads to hyperexcitability in the thalamocortical circuit.
- 34.*. Lee SE, et al. : Thalamo-cortical network hyperconnectivity in preclinical progranulin mutation carriers. Neuroimage Clin 2019, 22:101751. Using functional MRI, this study shows that pre-symptomatic GRN mutation carriers exhibit hyperconnectivity in the thalamocortical network.
- 35.**. Zhang J, et al. : Neurotoxic microglia promote TDP-43 proteinopathy in progranulin deficiency. Nature 2020, 588:459–465. The authors perform single-cell transcriptomics in microdissected thalamic from an aging cohort of Grn+/+ and Grn−/− mice to reveal how age-dependent microglial activation promotes neuronal cell death, nuclear pore defects, and TDP-43 proteinopathy in excitatory neurons.
- 36.Wu Y, et al. : Microglial lysosome dysfunction contributes to white matter pathology and TDP-43 proteinopathy in GRN-associated FTD. Cell Rep 2021, 36:109581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Deerlin VM, et al. : Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet 2010, 42:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finch N, et al. : TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology 2011, 76:467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pottier C, et al. : Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol 2018, 17:548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.*. Zhou X, et al. : Loss of Tmem106b exacerbates FTLD pathologies and causes motor deficits in progranulin-deficient mice. EMBO Rep 2020, 21:e50197. These three studies reveal that loss of TMEM106B exacerbates the neuropathological phenotypes in Grn−/− brain and spinal cord, leading to profound lysosomal dysfunction, TDP-43 proteinopathy, gliosis, and neuronal loss.
- 41.*. Feng T, et al. : Loss of TMEM106B and PGRN leads to severe lysosomal abnormalities and neurodegeneration in mice. EMBO Rep 2020, 21:e50219. These three studies reveal that loss of TMEM106B exacerbates the neuropathological phenotypes in Grn−/− brain and spinal cord, leading to profound lysosomal dysfunction, TDP-43 proteinopathy, gliosis, and neuronal loss.
- 42.*. Werner G, et al. : Loss of TMEM106B potentiates lysosomal and FTLD-like pathology in progranulin-deficient mice. EMBO Rep 2020, 21:e50241. These three studies reveal that loss of TMEM106B exacerbates the neuropathological phenotypes in Grn−/− brain and spinal cord, leading to profound lysosomal dysfunction, TDP-43 proteinopathy, gliosis, and neuronal loss.
- 43.Klein ZA, et al. : Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 2017, 95:281–296. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almeida MR, et al. : Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol Aging 2016, 41:200 e1–200 e5. [DOI] [PubMed] [Google Scholar]
- 45.Ward ME, et al. : Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci Transl Med 2017, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arrant AE, et al. : Reduction of microglial progranulin does not exacerbate pathology or behavioral deficits in neuronal progranulin-insufficient mice. Neurobiol Dis 2019, 124:152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evers BM, et al. : Lipidomic and transcriptomic basis of lysosomal dysfunction in progranulin deficiency. Cell Rep 2017, 20:2565–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.*. Marschallinger J, et al. : Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat Neurosci 2020, 23:194–208. The authors identify PGRN as a regulator of lipid metabolism in microglia and show that the microglia with prominent lipid accumulation indicate proinflammatory state in the aged brain.
- 49.**. Logan T, et al. : Rescue of a lysosomal storage disorder caused by Grn loss of function with a brain penetrant progranulin biologic. Cell 2021, 184:4651–4668 e25. This study shows that Progranulin replacement therapy can mitigate a number of phenotypes in Grn−/− microglia.
- 50.Ash PE, et al. : Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 2013, 77:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mori K, et al. : The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 2013, 339:1335–1338. [DOI] [PubMed] [Google Scholar]
- 52.Freibaum BD, et al. : GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 2015, 525:129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K, et al. : The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015, 525:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jovicic A, et al. : Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 2015, 18:1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang YJ, et al. : Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat Med 2018, 24:1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang YJ, et al. : Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 2019, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez-Gonzalez R, et al. : Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron 2016, 92:383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boeynaems S, et al. : Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell 2017, 65:1044–1055 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boeynaems S, et al. : Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc Natl Acad Sci U S A 2019, 116:7889–7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen C, et al. : Phase separation and toxicity of C9orf72 poly(PR) depends on alternate distribution of arginine. J Cell Biol 2021, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi SY, et al. : C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nat Neurosci 2019, 22:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maor-Nof M, et al. : p53 is a central regulator driving neurodegeneration caused by C9orf72 poly(PR). Cell 2021, 184:689–708. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang M, et al. : A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv 2016, 2:e1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan PM, et al. : The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun 2016, 4:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sellier C, et al. : Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J 2016, 35:1276–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amick J, Roczniak-Ferguson A, Ferguson SM: C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell 2016, 27:3040–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, et al. : A C9orf72-CARM1 axis regulates lipid metabolism under glucose starvation-induced nutrient stress. Genes Dev 2018, 32:1380–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi Y, et al. : Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med 2018, 24:313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang T, et al. : C9orf72 regulates energy homeostasis by stabilizing mitochondrial complex I assembly. Cell Metab 2021, 33:531–546 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Rourke JG, et al. : C9orf72 is required for proper macrophage and microglial function in mice. Science 2016, 351:1324–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burberry A, et al. : Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med 2016, 8:347ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Atanasio A, et al. : C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep 2016, 6:23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.*. Zhu Q, et al. : Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat Neurosci 2020, 23:615–624. The authors indicate that aberrant DPR caused by C9orf72 mutation induces systemic autoimmune dysfunctions including splenomegaly and the reactivation of microglia and astrocytes.
- 74.**. McCauley ME, et al. : C9orf72 in myeloid cells suppresses STING-induced inflammation. Nature 2020, 585:96–101. Here, it is revealed that the loss of C9orf72 promotes STING-mediated inflammation in myeloid cells to promote neuroinflammation in the EAE model and anti-tumor properties mediated by the myeloid cells.
- 75.Lall D, et al. : C9orf72 deficiency promotes microglial-mediated synaptic loss in aging and amyloid accumulation. Neuron 2021, 109:2275–2291 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.**. Burberry A, et al. : C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature 2020, 582:89–94. The authors reveal that gut microflora propagates autoimmune disorders and shorten lifespan in C9orf72−/− mice by promoting myeloid cell infiltration and microglial reactivation in the nervous system.
- 77.Blacher E, et al. : Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 2019, 572:474–480. [DOI] [PubMed] [Google Scholar]
- 78.Partanen S, et al. : Synaptic changes in the thalamocortical system of cathepsin D-deficient mice: a model of human congenital neuronal ceroid-lipofuscinosis. J Neuropathol Exp Neurol 2008, 67:16–29. [DOI] [PubMed] [Google Scholar]
- 79.Colombo A, et al. : Loss of NPC1 enhances phagocytic uptake and impairs lipid trafficking in microglia. Nat Commun 2021, 12:1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.**. Chu TT, et al. : Tonic prime-boost of STING signalling mediates Niemann-Pick disease type C. Nature 2021, 596:570–575. This study uses a proximity labeling screen to show that Niemann-Pick type C1 (NPC1) is a cofactor that interacts with and recruits STING to lysosomes for degradation. Loss of NPC1 robustly activates STING pathway and promotes microglial activation.
- 81.Ohmi K, et al. : Activated microglia in cortex of mouse models of mucopolysaccharidoses I and IIIB. Proc Natl Acad Sci U S A 2003, 100:1902–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arrant AE, et al. : Restoring neuronal progranulin reverses deficits in a mouse model of frontotemporal dementia. Brain 2017, 140:1447–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arrant AE, et al. : Progranulin gene therapy improves lysosomal dysfunction and microglial pathology associated with frontotemporal dementia and neuronal ceroid lipofuscinosis. J Neurosci 2018, 38:2341–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amado DA, et al. : AAV-mediated progranulin delivery to a mouse model of progranulin deficiency causes T cell-mediated toxicity. Mol Ther 2019, 27:465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu Y, et al. : Variant-selective stereopure oligonucleotides protect against pathologies associated with C9orf72-repeat expansion in preclinical models. Nat Commun 2021, 12:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.**. Tran H, et al. : Suppression of mutant C9orf72 expression by a potent mixed backbone antisense oligonucleotide. Nat Med 2022, 28:117–124. (Tran H, Nature Med 2021) This is the first study to demonstrate the effect of antisense oligonucleotides (ASO) in suppressing DPR produced by C9orf72 mutations.
- 87.Minami SS, et al. : Progranulin protects against amyloid beta deposition and toxicity in Alzheimer’s disease mouse models. Nat Med 2014, 20:1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takahashi H, et al. : Opposing effects of progranulin deficiency on amyloid and tau pathologies via microglial TYROBP network. Acta Neuropathol 2017, 133:785–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Valenzano DR, et al. : The african turquoise killifish genome provides insights into evolution and genetic architecture of lifespan. Cell 2015, 163:1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]