Abstract
Although real-time reverse transcriptase polymerase chain reaction (real-time RT-PCR) remains as a golden standard for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, it can not be easily expanded to large-scaled screening during outbreaks, and the positive results do not necessarily correlate with infectious status of the identified subjects. In this study, the performance of Vstrip® RV2 COVID-19 Antigen Rapid Test (RAT) and its correlation with virus infectivity was examined by virus culture using 163 sequential respiratory specimens collected from 26 SARS-CoV-2 infected patients. When the presence of cytopathic effects (CPE) in cell culture was used as a reference method for virus infectivity, the sensitivity, specificity and accuracy of Vstrip® RV2 COVID-19 Antigen Rapid Test was 96.43%, 89.63%, and 90.8%, respectively. The highest Ct value was 27.7 for RdRp gene and 25.79 for E gene within CPE-positive samples, and the highest Ct value was 31.9 for RdRp gene and 29.1 for E gene within RAT positive samples. When the Ct values of specimens were below 25, the CPE and RAT results had high degree of consistency. We concluded that the RAT could be a great alternative method for determining the infectious potential of individuals with high viral load.
Keywords: SARS-COV-2, Real-time RT-PCR, Rapid antigen test, Infectivity
1. Introduction
The Coronavirus disease 2019 (COVID-19) pandemic was caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [17]. It was mainly transmitted by droplets [8]. Common symptoms included headache, loss of smell and taste, nasal congestion and rhinorrhea, cough, muscle pain, sore throat, fever, diarrhea and difficulty breathing [2]. However, a non-neglectable portion of infected patients are asymptomatic, which would increase the transmission and spreading of SARS-CoV-2 [1]. To control the SARS-CoV-2 pandemic, quarantine is a passive and inevitable measure to block the connection of people but it exhausts a lot of resource. Because of that, a suitable diagnosis method for contagious COVID-19 patients is important to control the spread of SARS-CoV-2.
Real-time reverse transcriptase polymerase chain reaction (real-time RT-PCR) is considered as a golden standard for detection of SARS-CoV-2. However, the real-time RT-PCR requires not only professional operators, but also a qualified central diagnostic laboratory. Moreover, it takes relatively longer to have the diagnosis results with an averaged 4–6 h. In addition, when the specimen transportation time is considered, it might exceed 24 h [7]. Most important of all, because the real-time RT-PCR is too sensitive, studies have shown that positive PCR results may not represent the presence of culturable SARS-CoV-2 in the respiratory specimens [5,25]. Therefore, it is essential to find out a more practical and efficient way to confirm the infectivity of virus in COVID19 patient combined with highly sensitive PCR method to minimize the impact of quarantine.
Rapid antigen test (RAT) is an in-vitro diagnostic device (IVD) which is of low cost and easy to use by general population. Most of the RAT use immunochromatography to detect the interaction between antibody and antigen. For SARS-CoV-2, many RATs use SARS CoV-2 nucleocapsid protein as target antigen due to its abundance on virus particles and sequence conservation during virus evolution [19]. The relatively simple test procedure renders its quick adaption to identify infected individuals during COVID-19 pandemic, and further expand its application as a home-based self-test [15]. Vstrip® RV2 COVID-19 Antigen Rapid Test (Panion & BF Biotech Inc.) is a rapid qualitative detection test for detecting SARS-CoV-2 antigen in human nasopharyngeal specimens. In this study, the correlation of its performance with virus infectivity in the sequential nasopharyngeal specimens from SARS-CoV-2 confirmed cases was evaluated.
2. Materials and methods
2.1. Study design and participants
This prospective study was conducted at Taoyuan General Hospital, Ministry of Health and Welfare, Taoyuan, Taiwan between November and December of 2020. Eligible participants were confirmed with SARS-CoV-2 infection by realtime PCR and were ≥18 years of age. A standardized case record form was used to collect information on the patients’ demographics, comorbidity, treatment history, travel history, and laboratory data. The study was approved by the Research Ethics Committee or institutional review board at Taoyuan General Hospital (registration number TYGH109057). All patients gave written informed consent before enrollment to provide their samples and clinical and laboratory data for research.
A total of 26 patients were recruited for the study. Nasopharyngeal swabs were collected from each participant every day during their stay in the hospital until their real-time RT-PCR results became negative. For comparison, two nasopharyngeal swabs were collected from every participant at each time points. One was stored in universal transport medium (UTM) for real-time RT-PCR by Cobas Z480 and virus infection, the other was stored in lysis buffer and tested by Vstrip® RV2 COVID-19 Antigen Rapid Test (Panion & BF Biotech Inc.). A total of 326 nasopharyngeal swabs were collected for analysis in the study.
2.2. Real-time RT-PCR
All samples stored in universal transport medium (UTM) were extracted for viral RNA. Quantitative RT-PCR (qRT-PCR) was performed and analyzed using the Roche Cobas Z480 analyzer (Roche Molecular Systems, Rotkreuz, Switzerland) under the following conditions: 20 min at 50 °C and 20 s at 95 °C, followed by 45 cycles of 3 s at 95 °C and 30 s at 60 °C. Samples were considered SARS-CoV-2 positive if they tested positive for the E or RdRp genes [6].
2.3. Virus infection and isolation
The samples stored in UTM were propagated in Vero or VeroE6 cells in Dulbecco's modified Eagle medium (DMEM) supplemented with 2 μg/mL tosylsulfonyl phenylalantyl chloromethyl ketone (TPCK)‐trypsin (Sigma‐Aldrich). Culture supernatants were harvested when virus-induced cytopathic effects (CPE) were observed in more than 70% of cells. The full‐length genomic sequences of the derived clinical isolates from each patient were determined by Sanger sequencing and submitted, along with the patients’ travel history and basic information, to the GISAID database. The isolated virus strains were listed in Table 6 .
Table 6.
The lineage and Nucleocapsid (NP) mutations of isolated SARS-CoV-2 strains in the study.
| Accession ID | Lineage (Pango v.4.0.6 PANGO-v1.8) | Nucleocapsid (NP) mutations | |
|---|---|---|---|
| hCoV-19/Taiwan/NTU31/2020 | EPI_ISL_693,302 | B.1.36.16 | S194L |
| hCoV-19/Taiwan/NTU32/2020 | EPI_ISL_693,303 | B.1.320 | None |
| hCoV-19/Taiwan/NTU33/2020 | EPI_ISL_693,304 | B.1.1.222 | R203K, G204R |
| hCoV-19/Taiwan/NTU34/2020 | EPI_ISL_693,305 | B.1.1.315 (AD.2)a | R203K, G204R, D377Y, A398V |
| hCoV-19/Taiwan/NTU35/2020 | EPI_ISL_693,306 | B.1.232 | S194L |
| hCoV-19/Taiwan/NTU36/2020 | EPI_ISL_738,064 | B.1.470 | T205I |
| hCoV-19/Taiwan/NTU37/2020 | EPI_ISL_740,547 | B.1.459 | None |
| hCoV-19/Taiwan/NTU38/2020 | EPI_ISL_738,065 | B.1.1.29 (B.1.1.398)a | R203K, G204Q |
| hCoV-19/Taiwan/NTU39/2020 | EPI_ISL_872,591 | B.1.36 (B.1.456)a | S194L |
| hCoV-19/Taiwan/NTU40/2020 | EPI_ISL_872,592 | B.1.36 (B.1.456)a | S194L |
| hCoV-19/Taiwan/NTU41/2020 | EPI_ISL_872,593 | B.1.36 (B.1.456)a | S194L |
| hCoV-19/Taiwan/NTU42/2020 | EPI_ISL_872,594 | B.1.459 | A119S, T205I, D377Y |
| hCoV-19/Taiwan/NTU44/2020 | EPI_ISL_872,595 | B.1.1.263 | R203K, G204R |
| hCoV-19/Taiwan/NTU45/2020 | EPI_ISL_872,596 | B.1.2 | N67S, P199L |
| hCoV-19/Taiwan/NTU46/2020 | EPI_ISL_872,597 | B.1.160 | M234I, A376T |
| hCoV-19/Taiwan/NTU47/2020 | EPI_ISL_872,598 | B.1.459 | A119S, T205I, D377Y |
| hCoV-19/Taiwan/NTU48/2020 | EPI_ISL_872,599 | B.1.459 | A119S, T205I, D377Y |
The lineage was reclassified in 2022 (updated on June 15, 2022).
2.4. Rapid antigen test (RAT)
The nasopharyngeal swabs were placed in the lysis buffer and tested by Vstrip® RV2 COVID-19 Antigen Rapid Test (Panion & BF Biotech Inc.). This assay could detect the presence of SARS-CoV-2 nucleocapsid protein (N) by immunochromatography. After the test strip was placed in the lysis buffer for 10–15 min, the result could be interpreted. Samples were considered SARS-CoV-2 positive when both the test lines and control lines could be seen by the naked eye.
2.5. Statistical analysis
All subjects’ characteristics of demographic data (age, gender, and symptoms etc.) at study entry were listed for all subjects. Frequencies and percentages were reported for all categorical data.
The parameter including sensitivity, specificity, accuracy, positive percent agreement (PPA), negative percent agreement (NPA) and overall percent agreement (OPA) were analyzed according to the test results of CPE and RAT. Categorical variables were compared using chi-square test or Fisher's exact test. Continuous variables were compared using the Kruskal-Wallis one-way analysis of variance or Mann-Whitney U test. Unless otherwise specified, a two-tailed p value <0.05 was considered statistically significant. All statistical analysis programming was performed using SAS version 9.4 version.
3. Results
3.1. Demographics of study participants
The demographic information of participated COVID-19 patients was shown in Table 1 . The median age of the participants was 32 years old. More than half of the participants were female (61.54%). By travel history, 61.53% of the participants had been to Asia, and among these people, the most had been to Indonesia. Cough (34.61%) was the most-reported symptom from participants, followed by sore throat (23.07%) and congestion or runny nose (23.07%). Besides, the proportion of asymptomatic patients (23.07%) was as same as that of sore throat, congestion or runny nose. The median time for recruitment since disease onset among the participants was 7 days, and median for duration of hospital stay was 37 days. Nevertheless, none of the participants stayed in an intensive care unit (ICU). During the hospitalization period, respiratory specimens were collected from each subject every day for real-time RT-PCR, virus infection and rapid antigen tests (RAT) until their real-time RT-PCR results became negative (Supplementary Figure 1).
Table 1.
Demographics of study subjects (N = 26).
| Male gender,%(n) | Male | 38.46 (10) |
|---|---|---|
| Age, years (Mean ± SD) | Mean | 34.54±10.8 |
| Travel history,%(n) | Asia | 61.53 (16) |
| Americas | 23.07 (6) | |
| Europe | 15.38 (4) | |
| Clinical symptoms,%(n) | No symptoms | 23.07% (6) |
| Shortness of breath or difficulty breathing | 0.00% (0) | |
| Cough | 34.61% (9) | |
| Sore throat | 23.07% (6) | |
| Congestion or runny nose | 23.07% (6) | |
| Headache | 3.84% (1) | |
| New loss of taste or smell | 19.23% (5) | |
| Fever or Chills | 19.23% (5) | |
| Nausea or vomiting | 0.00% (0) | |
| Diarrhea | 11.53% (3) | |
| Muscle or body aches | 15.38% (4) | |
| Fatigue | 0.00% (0) | |
| Recruitment since disease onset, Days (Mean ± SD) | Mean | 6.3 ± 5.33 |
| Duration of Hospital stay, Days (Mean ± SD) | Mean | 31.92±14.58 |
| ICU stay (%) | 0.00% (0) | |
*Abbreviation: SD, standard deviation; ICU, intensive care unit;.
3.2. The performance of rapid antigen test compared with real-time RT-PCR
The results of RAT and real-time RT-PCR were summarized in Table 2 . The results of real-time RT-PCR by RdRp gene was used as a reference method compared with RAT. The sensitivity, specificity, positive prediction rate, negative prediction rate and accuracy was 27.70% (41/148), 100% (15/15), 100% (41/41), 12.30% (15/122) and 34.36% (56/163), respectively (Table 3 ).
Table 2.
The clinical diagnostic performance of real-time RT-PCR and rapid antigen test (RAT).
| Real-time RT-PCR (RdRp gene) |
|||
|---|---|---|---|
| Positive | Negative | ||
| RAT | Positive | 41 | 0 |
| Negative | 107 | 15 | |
*The Performance of Vstrip® RV2 COVID-19 Antigen Rapid Test was determined using the real-time RT-PCR results of RdRp gene as a reference.
Table 3.
Comparison of the clinical diagnostic performance of real-time RT-PCR and rapid antigen test (RAT).
| Value(95%CI) | |
|---|---|
| Sensitivity (%) | 27.70 (20.67–35.65) |
| Specificity (%) | 100.00(78.20–100) |
| Positive Predictive Value (%) | 100 |
| Negative Predictive Value (%) | 12.3(11.26–13.41) |
| Accuracy (%) | 34.36(27.11–42.19) |
aThe performance of Vstrip® RV2 COVID-19 Antigen Rapid Test was determined using the real-time RT-PCR results of RdRp gene as a reference.
3.3. The correlation of rapid antigen test results with virus infectivity
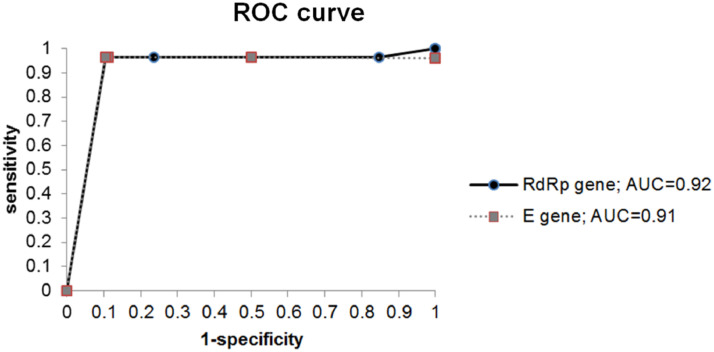
The correlation of rapid antigen test results with virus infectivity was shown in Table 4 . The presence of CPE was used as an indication of virus infectivity in the respiratory specimens. When CPE was used as a reference method, the sensitivity, specificity, positive prediction rate, negative prediction rate and accuracy of the RAT was 96.43% (27/28), 89.63% (121/135), 65.85% (27/41), 99.18% (121/122) and 90.8% (148/163), individually (Table 5 ). Besides, the area under the curve (AUC) values associated with RAT performance was 0.92 based on the real-time RT-PCR results of RdRp gene and 0.91 based on those of the E gene. (Fig. 1 )
Table 4.
The clinical diagnostic performance of virus isolation and rapid antigen test (RAT).
| Virus isolation | |||
|---|---|---|---|
| Positive | Negative | ||
| RAT | Positive | 27 | 14 |
| Negative | 1 | 121 | |
Table 5.
Comparison of the clinical diagnostic performance of virus isolation with rapid antigen test (RAT).
| Value(95%CI) | |
|---|---|
| Sensitivity (%) | 96.43(81.65–99.91) |
| Specificity (%) | 89.63(83.21 −94.21) |
| Positive Predictive Value (%) | 65.85(53.89–76.10) |
| Negative Predictive Value (%) | 99.18(94.63–99.88) |
| Accuracy (%) | 90.80(85.28–94.76) |
*The performance of Vstrip® RV2 COVID-19 Antigen Rapid Test was determined using the virus isolation results as a reference.
Fig. 1.
Receiver-operator curve (ROC) analyses for rapid antigen test (RAT) with real-time RT-PCR. The performance of RAT results was plotted with sensitivity on the y axis and 1-specificity on the x axis. The area under the curve (AUC) values were also indicated.
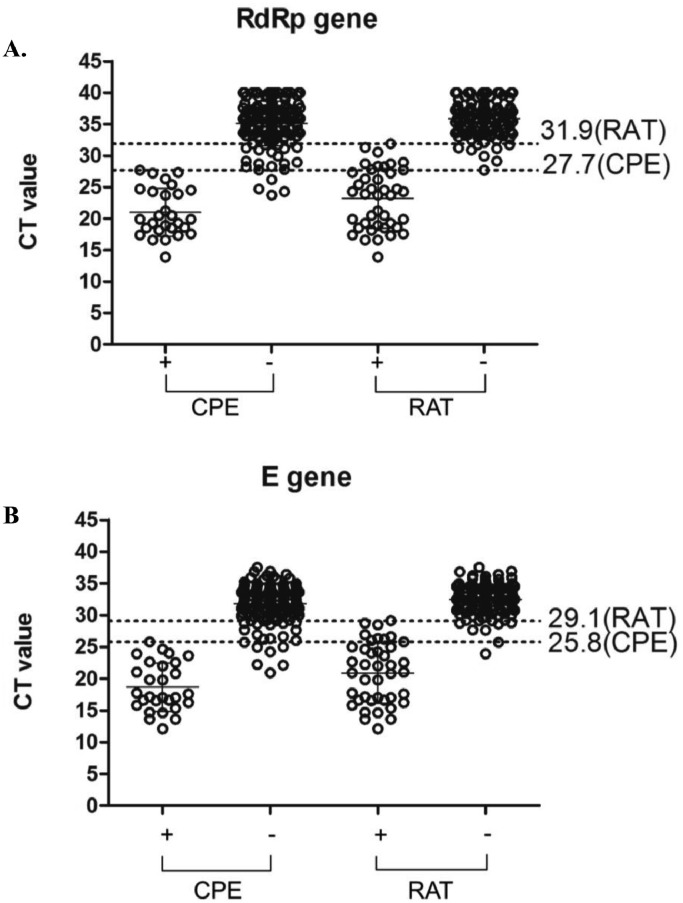
Further analysis and comparison of the correlation between the results of CPE, RAT and the Ct value of real-time RT-PCR was conducted (Fig. 2 ). The overall Ct values of the samples with positive RAT result was slightly higher than that with positive CPE results. Among the CPE-positive samples, the highest Ct value was 27.7 for RdRp gene and 25.8 for E gene, while for the RAT positive samples, the highest Ct value was 31.9 for RdRp gene and 29.1 for E gene. In general, the limit of detection for RAT is about 4 Ct values higher than that for CPE.
Fig. 2.
Correlation distribution between cytopathic effect (CPE) and rapid antigen test (RAT) results with SARS-CoV-2 real-time RT-PCR. Ct values of real-time RT-PCR targeting (A) RdRp and (B) E gene were shown on the y axis. Samples were drawn into a dot distribution graph based on CPE and RAT results. The four categories include positive CPE (n = 28), negative CPE (n = 135), positive RAT (n = 41) and negative RAT (n = 122). The dotted line represented the highest Ct value within the CPE and RAT positive samples.
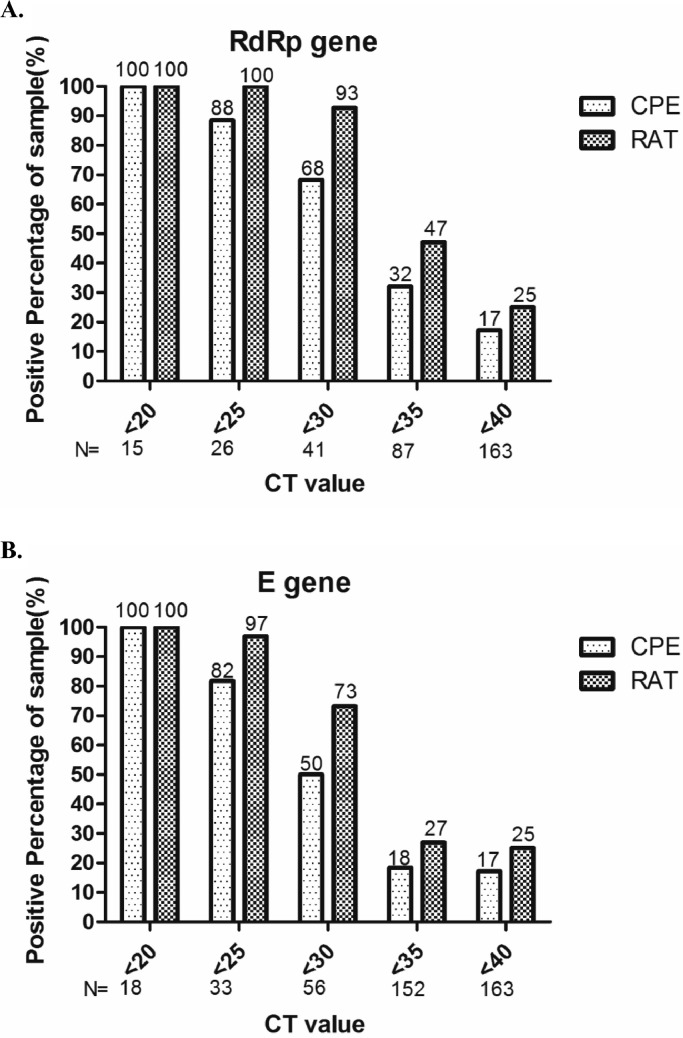
Next, we compared the correlation between the results of CPE and RAT with the amount of viral RNA. For samples with Ct values of RdRp and E genes lower than 20, both CPE and RAT were 100% positive (Fig. 3 ). The RAT remained 100% positivity when the Ct value was lower than 25 for RdRp gene. In general, when the Ct values of specimens were lower than 25, the CPE and RAT positive rate had high degree of consistency. When the Ct value of samples was lower than 30, the RAT still had over 70% positivity. However, when the Ct value was greater than 30, the positive rate of CPE and RAT dropped significantly, despite of higher positive rate of RAT in RdRp gene. In general, positive RAT results had great correlation with virus infectivity when the Ct value of specimen was lower than 25.
Fig. 3.
The relationship between the positive rate of cytopathic effect (CPE) and rapid antigen test (RAT) with real-time RT-PCR Ct values of real-time RT-PCR targeting (A) RdRp and (B) E gene were shown on the x axis. The positive rates of CPE and RAT were calculated according to the accumulative numbers categorized by Ct values.
3.4. The lineage of viruses isolated from the clinical samples
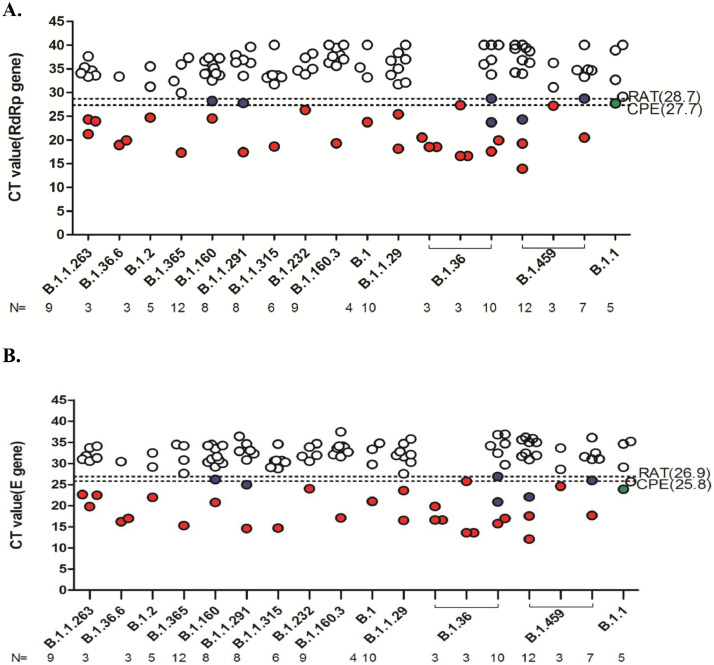
For those CPE positive specimens, the full-length virus genome sequence PCR-amplified from the culture supernatants and the virus lineage was determined. All viruses isolated in the study had D614G mutation on the S protein, which was the predominant around the world when the clinical specimens were collected. The correlation between results from virus isolation, RAT, and real time RT-PCR across the SARS-CoV-2 lineages was shown in Fig. 4 . Within all RAT positive samples, B.1.36 was the most predominant, and it had the highest Ct value within all RAT positive samples. Besides B.1.36, B.1.160, B.1.1.291 and B.1.459 had samples with negative CPE and positive RAT results. B.1.1 was the only sample with positive CPE, yet negative RAT results.
Fig. 4.
Correlation distribution between virus isolation, rapid antigen test (RAT), and real-time RT-PCR across the SARS-CoV-2 lineages from the study samples. The samples of participants who had evidence of virus infectivity (n = 115) were drawn into a dot distribution g raph based on the virus lineage and Ct value. The SARS-CoV-2 lineage for those successfully isolated from the respiratory specimens of recruited participants was shown on the x axis. The Ct values of real-time RT-PCR using (A) RdRP and (B) E gene as targets was shown on the y axis. The white and red dots represented negative and positive results for both cytopathic effect (CPE) and RAT respectively. The green dots represented positive for CPE only, and the blue dots represented positive for RAT only. The two dotted lines represented the highest Ct values for CPE and RAT positive samples, respectively.
4. Discussion
In this study, we evaluated the performance of Vstrip® RV2 COVID-19 Antigen Rapid Test (Panion & BF Biotech Inc.) in correlation with virus infectivity using sequential respiratory specimens collected from SARS-CoV-2 infected individuals and compared the results with those from real-time RT-PCR. The assay showed great performance on confirming virus infectivity by high sensitivity, specificity and accuracy, and the AUC was also greater than 0.9. The study results indicated that Vstrip® RV2 COVID-19 Antigen Rapid Test can serve as a convenient tool to identify the infectivity status of the COVID-19 suspected subjects or confirmed patients and can help to optimize the constrained resource during the pandemic.
In our study, the sensitivity of RAT is lower than the gold-standard method, real-time RT-PCR, with only 41 of 163 real-time RT-PCR positive samples being positive by the RAT. Our results were similar to many previous studies, which had shown that the RAT had a relatively poor sensitivity than that of real-time RT-PCR [10,14,20]. However, a very high consistence was observed between virus isolation and RAT when the Ct values of specimens were lower than 25. Also, the sensitivity of RAT versus virus isolation results was increased to 96.43%. Besides assay sensitivity, the possibility of lower sensitivity in RAT and virus isolation versus real-time RT-PCR might be due to viral RNA shedding during SARS-CoV-2 infection. In the clinical course of disease progression, increase of viral RNA in the respiratory specimens could be detected before the symptom onset, and could persist for many weeks even after disappearance of symptoms [11,16,25,27]. It has been reported that viral RNA could be detected in specimens via real-time RT-PCR with high Ct value but failed to isolate SARS-CoV-2 in cell culture [4,5,9,23]. Our study also confirmed that SARS-CoV-2 viruses could not be successfully isolated from the specimens with Ct value above 27.7 of RdRp gene and 25.8 of E gene (Fig. 2). Besides, the collection time point and disease severity are also related to the results of RAT, real-time PCR and virus isolation [3,12,13,24]. It has been reported that the successful rate for virus isolation from specimens collected at later time points after symptom onset would decrease even though the Ct was low [21,23,27]. Similar results were also observed in our study. Among the four specimens with low Ct values and no development of CPE in virus isolation, two were collected greater than 8 days after symptom onset. Nevertheless, our results demonstrated that RAT has a good sensitivity versus virus isolation in asymptomatic patients. Although only 6 asymptomatic patients were enrolled in our study, the viruses could be detected via RAT in 5 patients’ specimens, and their viruses could also be successfully isolated. Therefore, our study indicates that RAT could be used to distinguish contagious people from non-contagious population.
Comparing to real time RT-PCR and virus isolation, RAT had many advantages, including turn-around time, price, and easy-to-operate. It is also easier to be applied to large-scaled screening. However, availability of treatment regimen, capacity of medical support, effectiveness of quarantine strategy and duration of isolation will be affected by the false positive and negative prediction rates of detection method [22]. When wide-spread screening is required, negative predictive value is more important than positive predictive value in the aspect to set up a threshold to rule out infection [26]. Our results demonstrated that RAT had a relatively good negative predictive value (99.18%, Table 5) as compared to virus isolation, with only one sample having unexpected results. That sample, with a Ct value of 23.9 for E gene, was positive by virus isolation but negative by RAT. As expected, the negative predictive value of RAT declined to 12.3% when comparing with real time RT-PCR. The infection status, especially the early infection (incubation period) stage, might result in positive real-time RT-PCR but negative RAT. The possibility that the mutations on the epitope of Nucleocapsid (NP) protein would contribute to the false negative results of RAT could not be totally excluded. NP protein is the most abundant protein in coronavirus [19]. Previous studies showed that most of the dominant B cell epitopes are located between amino acid residues 76–82 or 176–206 [18]. This region also harbored the highest prevalence of NP mutations across lineages (Supplementary Figure 2). Two mutations, R203K and G204R, had approximate 100% prevalence in Alpha and Omicron variants. In our study, the sensitivity of different variants of concern (VOC) could not be determined owing to the participates were enrolled in April of 2021, before outbreaks of Alpha and Omicron variants in Taiwan. However, among the 5 specimens used in this study with R203K and G204R mutations on N gene, they could be recognized via RAT, expect lineage B.1.1 (Fig. 4). The results implicated that the recognition of VOC by RAT is similar to the wild type virus. Noteworthy, there are other non-tested high prevalence mutations existing in the VOC (Supplementary Figure 2), and their influences on the sensitivity, positive predictive value and negative predictive value of RAT to VOC need to be further evaluated in the future. The positive predictive value of RAT versus virus isolation was 65.85%. Fourteen specimens were positive by both RAT and real-time PCR, but negative by virus isolation. This phenomenon was likely due to the collection of specimens at later time points or inappropriate storage of specimens.
Overall, Vstrip® RV2 COVID-19 Antigen Rapid Test had good performance to confirm the contagious patients than real-time RT-PCR. The convenient and easy-to-use features of RAT make it the most suitable screening tool to restrict the spreading of asymptomatic infections during the outbreak.
5. Author contribution
TLC, WHL, YCC, MP, CYC, and SYC prepared the manuscript; HCH, YCL, CPC, SHC, and CYC helped recruitment of study subjects; WHL, SYH, HHC, YMT, RDF, and SML conducted experiments.
Declaration of Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This study and rapid antigen tests were supported by Panion & BF Biotech Inc. We also acknowledge the technical assistance and all of the health care workers and the patients for consenting to participate from Taoyuan General Hospital, Ministry of Health and Welfare and National Taiwan University Hospital. We would like to acknowledge the services provided by the Biosafety Level-3 Laboratory of the First Core Laboratory and the Transgenic Mouse Core Facility from National Taiwan University College of Medicine; the Biosafety Level-3 Laboratory from National Taiwan University Hospital.
Funding
This work was supported by Oneness Biotech Company and grants from the Ministry of Science and Technology, Taiwan (MOST-111–2740-B-002–006, MOST111–2321-B-002–017)
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcvp.2022.100133.
Appendix. Supplementary materials
References
- 1.Rothe C., et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abduljalil J.M., Abduljalil B.M. Epidemiology, genome, and clinical features of the pandemic SARS-CoV-2: a recent view. New Microbes New Infect. 2020;35 doi: 10.1016/j.nmni.2020.100672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abusrewil Z., Alhudiri I.M., Kaal H.H., El Meshri S.E., Ebrahim F.O., Dalyoum T., Efrefer A.A., Ibrahim K., Elfghi M.B., Abusrewil S., Elzagheid A. Time scale performance of rapid antigen testing for SARS-CoV-2: evaluation of 10 rapid antigen assays. J. Med. Virol. 2021;93:6512–6518. doi: 10.1002/jmv.27186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., Harney J., Chisty Z., Bell J.M., Methner M., Paul P., Carlson C.M., McLaughlin H.P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L.C., Kay M., Lewis J., Montgomery P., Stone N.D., Clark T.A., Honein M.A., Duchin J.S., Jernigan J.A., Public H.-.S., King C., Team C.C.-I. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., Doan K., Bastien N., Li Y., Van Caeseele P.G., Poliquin G. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin. Infect. Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green K, G.S., Turner P., Fanshawe T., Allen J. Oxford and Team, C.-E.S., Oxford COVID-19 Evidence Service: molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. Oxford COVID-19 Evid Serv Team, 2020.
- 8.Harapan H., N.I., Yufika A., Winardi W., Keam S., Te H., Megawati D., Hayati Z., L A., Wagner M.M. Coronavirus disease 2019 (COVID-19): a literature review. J. Infect. Public Health. 2020;13:667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C.G., Lee K.M., Hsiao M.J., Yang S.L., Huang P.N., Gong Y.N., Hsieh T.H., Huang P.W., Lin Y.J., Liu Y.C., Tsao K.C., Shih S.R. Culture-based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID-19. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01068-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohmer N., Toptan T., Pallas C., Karaca O., Pfeiffer A., Westhaus S., Widera M., Berger A., Hoehl S., Kammel M., Ciesek S., Rabenau H.F. The comparative clinical performance of four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. J. Clin. Med. 2021;10 doi: 10.3390/jcm10020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W.D., Chang S.Y., Wang J.T., Tsai M.J., Hung C.C., Hsu C.L., Chang S.C. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J. Infect. 2020;81:318–356. doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Liao W., Wan L., Xiang T., Zhang W. Correlation between relative nasopharyngeal virus RNA Load and lymphocyte count disease severity in patients with COVID-19. Viral Immunol. 2020 doi: 10.1089/vim.2020.0062. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mak G.C., Lau S.S., Wong K.K., Chow N.L., Lau C.S., Lam E.T., Chan R.C., Tsang D.N. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;133 doi: 10.1016/j.jcv.2020.104684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manabe Y.C., Sharfstein J.S., Armstrong K. The need for more and better testing for COVID-19. JAMA. 2020;324:2153–2154. doi: 10.1001/jama.2020.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer T.R., Salit M. Testing at scale during the COVID-19 pandemic. Nat. Rev. Genet. 2021;22:415–426. doi: 10.1038/s41576-021-00360-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oberfeld B., Achanta A., Carpenter K., Chen P., Gilette N.M., Langat P., Said J.T., Schiff A.E., Zhou A.S., Barczak A.K., Pillai S. SnapShot: COVID-19. CellCell. 2020;181:e1. doi: 10.1016/j.cell.2020.04.013. 954-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira S.C., de Magalhaes M.T.Q., Homan E.J. Immunoinformatic analysis of SARS-CoV-2 nucleocapsid protein and identification of COVID-19 vaccine targets. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.587615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020;51:482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scohy A., Anantharajah A., Bodeus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., Ladhani S., Zambon M., Gopal R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Syal K. Guidelines on newly identified limitations of diagnostic tools for COVID-19 and consequences. J. Med. Virol. 2021;93:1837–1842. doi: 10.1002/jmv.26673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Kampen J.J.A., van de Vijver D., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N., van den Akker J.P.C., Endeman H., Gommers D., Cornelissen J.J., Hoek R.A.S., van der Eerden M.M., Hesselink D.A., Metselaar H.J., Verbon A., de Steenwinkel J.E.M., Aron G.I., van Gorp E.C.M., van Boheemen S., Voermans J.C., Boucher C.A.B., Molenkamp R., Koopmans M.P.G., Geurtsvankessel C., van der Eijk A.A. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat. Commun. 2021;12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Zhang L., Sang L., Ye F., Ruan S., Zhong B., Song T., Alshukairi A.N., Chen R., Zhang Z., Gan M., Zhu A., Huang Y., Luo L., Mok C.K.P., Al Gethamy M.M., Tan H., Li Z., Huang X., Li F., Sun J., Zhang Y., Wen L., Li Y., Chen Z., Zhuang Z., Zhuo J., Chen C., Kuang L., Wang J., Lv H., Jiang Y., Li M., Lin Y., Deng Y., Tang L., Liang J., Huang J., Perlman S., Zhong N., Zhao J., Malik Peiris J.S., Li Y., Zhao J. Kinetics of viral load and antibody response in relation to COVID-19 severity. J. Clin. Invest. 2020;130:5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 26.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection - challenges and implications. N. Engl. J. Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 27.Young B.E., Ong S.W.X., Ng L.F.P., Anderson D.E., Chia W.N., Chia P.Y., Ang L.W., Mak T.M., Kalimuddin S., Chai L.Y.A., Pada S., Tan S.Y., Sun L., Parthasarathy P., Fong S.W., Chan Y.H., Tan C.W., Lee B., Rotzschke O., Ding Y., Tambyah P., Low J.G.H., Cui L., Barkham T., Lin R.T.P., Leo Y.S., Renia L., Wang L.F., Lye D.C., Singapore Novel Coronavirus Outbreak Research, T Viral dynamics and immune correlates of coronavirus disease 2019 (COVID-19) Severity. Clin. Infect. Dis. 2021;73:e2932–e2942. doi: 10.1093/cid/ciaa1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.