Abstract
Background:
In December 2020, the COVID-19 disease was confirmed in 1,665,775 patients and caused 45,784 deaths in Spain. At that time, health decision support systems were identified as crucial against the pandemic.
Methods:
This study applies Deep Learning techniques for mortality prediction of COVID-19 patients. Two datasets with clinical information were used. They included 2,307 and 3,870 COVID-19 infected patients admitted to two Spanish hospitals. Firstly, we built a sequence of temporal events gathering all the clinical information for each patient, comparing different data representation methods. Next, we used the sequences to train a Recurrent Neural Network (RNN) model with an attention mechanism exploring interpretability. We conducted an extensive hyperparameter search and cross-validation. Finally, we ensembled the resulting RNNs to enhance sensitivity.
Results:
We assessed the performance of our models by averaging the performance across all the days in the sequences. Additionally, we evaluated day-by-day predictions starting from both the hospital admission day and the outcome day. We compared our models with two strong baselines, Support Vector Classifier and Random Forest, and in all cases our models were superior. Furthermore, we implemented an ensemble model that substantially increased the system’s sensitivity while producing more stable predictions.
Conclusions:
We have shown the feasibility of our approach to predicting the clinical outcome of patients. The result is an RNN-based model that can support decision-making in healthcare systems aiming at interpretability. The system is robust enough to deal with real-world data and can overcome the problems derived from the sparsity and heterogeneity of data.
Keywords: COVID-19, Mortality prediction, Time series, Recurrent Neural Network
1. Introduction
According to the daily report of the Coordination Center for Health Alerts and Emergencies [1], as of December 2, 2020, a total of 1,665,775 confirmed cases of COVID-19 and 45,784 deaths were reported in Spain. To effectively respond to a challenge of this magnitude and to optimize the hospitalization process for this emerging disease, decision support systems for clinical care and health services management are crucial. Artificial Intelligence and Deep Learning tools offer a range of possibilities to obtain predictive models trained with historical data that can predict future scenarios.
In this work, we apply Deep Learning techniques for predicting the clinical outcome of patients with COVID-19. The result is a model that predicts mortality risk to support decision-making. This study leverages two datasets with clinical information on 2,307 and 3,870 patients from HM Hospitales (HM) and the Hospital Universitario 12 de Octubre (H12O) in Spain.
We proposed a time series analysis system using an ensemble model based on Recurrent Neural Networks (RNN) suited to the situation hospitals faced during the pandemic. We developed a model for daily predictions in a hospital environment operating as an early warning system. The system predicts the daily mortality risk for each inpatient by considering all previously available data.
One relevant characteristic of our system is the effort devoted to the interpretability of the model, which is based on an attention mechanism. The attention mechanism combines the RNN’s output with respect to the previous ones to relevantly weight past information for daily prediction, aiming at providing explanatory capabilities in the temporal dimension. This collaborative work was the first approach for developing and training predictive models from real-world data in the Electronic Health Record of H12O. This project may thus have a remarkable impact, particularly on engineering and data science processes. The main contributions of this work are the following:
-
•
Exhaustive experimentation, exploration and evaluation of different data representation methods, culminating in a methodology for modelling, extracting and transforming EHR data into a transferable format that AI systems can consume.
-
•
Development of an RNN-based model that produces daily predictions, leveraging temporal and static patient data, endowed with an explicable attention mechanism to identify the relevant days for the final prediction.
-
•
Design of an ensemble system that encourages model diversity, that is, by combining RNN models with different predictions, that outperformed the best RNN model in performance and robustness.
-
•
Application of the focal loss to address class-imbalanced data by appropriately weighting examples based on the model’s confidence.
-
•
The results of the predictive algorithm have provided evidence of the value of real-world data beyond traditional uses. Consequently, they promoted a data culture at H12O by raising awareness among healthcare professionals, researchers and managers about the critical impact of recording high-quality data during routine healthcare activity. [2].
-
•
Finally, this work has been the basis for implementing an intelligent data platform for data collection and standardization, with the purpose of developing, validating and executing predictive algorithms.2
We believe that our system may lay the foundations for applicable systems that have a positive impact on the healthcare decision-making process.
2. Related works
Time series prediction represents a complex scenario in which predictive modelling via RNNs is well suited to capturing sequential information from temporal data. Specifically, RNNs learn long-term dependencies by incorporating a memory that preserves the sequence states over time. In the healthcare domain, RNNs have been applied to address Intensive Care Unit (ICU) mortality prediction based on Electronic Health Records (EHR), as in [3], [4], [5], [6].
[7] produced the first study using a Long Short Term Memory RNN [8] to classify diagnoses given multivariate time series. [7] produced the first study using a Long Short Term Memory RNN [8] to classify diagnoses given multivariate time series. In their experiment, the diagnostic labels had no timestamps and were used only as predictive labels. Similarly to our work, in the training phase, the authors used target replication (an output is generated at every sequenced step). However, at prediction time, the system considers only the output during the final step so that, although the system considers the whole series of events, it does not give a prediction until the end.
[9] developed Doctor AI, a predictive model that uses diagnosis, medication, or procedure codes as input for the RNN to predict the diagnosis and medication categories for a subsequent visit. In contrast to our work, they had enough data to use Skip-gram [10] to capture the latent representation of medical codes from the EHR.
[11], suggest a Reverse Time Attention Mechanism with an RNN to create an interpretable predictive model and to cope with long sequences of visits. They used a two-level neural attention model that detects influential past visits and relevant clinical variables within those visits. We also use the attention mechanism to focus on the time sequence trying to identify influential episodes, but we do not reverse sequences as, in our case, these are much shorter.
[12] use RNNs with a simple attention mechanism to interpret the results and to forecast health information of patients from the historical sequences of visits over time. They employ bidirectional RNNs to remember all the information about both past and future visits, and they introduce three attention mechanisms to measure the relationships between different visits for prediction. We also experiment with the attention mechanism, but we use unidirectional RNN as, in our setting, we want daily predictions, and thus we unroll the RNN.
[13] used an ensemble algorithm of LSTMs to deal with heterogeneous ICU data for mortality prediction. The eventual ensemble models make their predictions by merging the results of multiple parallel LSTM classifiers.
We also explore the use of ensemble methods to improve sensitivity. However, our approach is different as we did not use bootstrapping or random feature subspace on training data. Instead we designed a voting system and defined an algorithm that, starting with the best model in terms of cross-validated F1 score and sensitivity, heuristically chooses the most different models (i.e., the ones that make the most different predictions, to encourage model diversity in the ensemble).
More recently, a COVID-19 mortality risk study that used HM data, among others, was published by [14]. This is the first multi-centre COVID-19 mortality risk study that uses data from 3,062 patients across four different countries. They used the XGBoost algorithm to predict mortality using 22 features. Unlike us, the experiment does not consider the sequence of events but instead uses the data from the first event in its analysis.
Therefore, as a contribution to the aforementioned studies, our work constitutes one of the first effective predictive models of time series data with COVID-19 patient records. Unlike [14], [15], we apply full-time series inference and make daily predictions instead of using static data or collapsing the dynamic data into fixed-size vectors. To achieve this goal, we trained different RNN models and assembled them, prioritizing a given metric. Using this technique, we substantially increased the system’s sensitivity while producing more stable predictions.
Besides RNNs, other approaches for early diagnosis have also been developed, such as the recent work by [16], that proposed genetic programming as an evolutionary computation for the diagnosis of Alzheimer’s disease based on spontaneous speech data. On the data side, the authors in [17] employed GAN-based methods to handle the data imbalance through data augmentation. Furthermore, integrating available unstructured data from different sources, such as images and texts, represents another promising direction towards a better diagnosis that could be addressed with multimodal machine learning [18].
Nevertheless, in our work, we intentionally avoided sophisticated methods that rely on complex approaches which may complicate the deployment into hospital infrastructures and potentially reduce the explainability of the predictions.
3. Methods
3.1. Datasets
We used two different COVID datasets.3 One is the dataset made available by HM from April 25, 2020 in their “COVID Data Save Lives” initiative, which contains information on patients diagnosed with COVID until that date. The other one comes from H12O, and the cohort extends to October 14. In both cases, the data include two different types of variables: static variables that are constant throughout admissions, such as sex or age, and dynamic variables that are measured at different times during the hospital stay, such as medication, laboratory analysis and vital signs.
Although both datasets contain information about COVID patients, they have different characteristics which prevented us from joining the datasets into a single one. The disparity in coding (e.g., ICD-9 vs ICD-10) and the differences in the type of information contained made it difficult to aggregate data and would produce a smaller dataset due to the low overlap. Note, however, that having two different datasets allowed us to validate the robustness of the proposal. Further details about the datasets and data cleaning are given in the Additional Material.
3.2. Feature representation
Information in EHR needs to be transformed into appropriate data representations to be used by clinical machine learning. Learning informative representations is critical for performance improvement. Feature representation is data processing used to encode variables into numerical values, depending on the variable’s values. Our dataset includes:
-
•
Categorical variables: variables that have multiple values with no order, each one associated with a given case (e.g., medications).
-
•
Ordinal variables: variables taking discrete values with a given order. The discrete values are defined by comparing the original values of the variables with a given interval of reference (e.g., vital signs and some lab results).
-
•
Continuous variables: a sub-type of ordinal variables that take numerical values ordered along a continuous scale (e.g., temperature).
In general, categorical variables are one-hot encoded, where each value of the variable becomes a binary variable itself, indicating the absence or presence of the original variable’s value. Ordinal variables are encoded using binary values or 0, 0.5, 1 values depending on the reference interval or normalized in the [0, 1] interval. Finally, continuous variables are normalized in the [0, 1] interval.
For many variables, we found a considerable amount of missing values that can be classified into ‘zero values’ and ‘missing measurements’. Medications and diagnoses do typically have ‘zero values’ indicating that the patient was not prescribed a certain medication or was not diagnosed with a certain feature. On the other hand, laboratory results and vital signs do typically have ‘missing measurements’. Thus, a missing value in blood pressure does not mean that the patient has no blood pressure but only that it was not measured or recorded. To handle the missing values, we use the following criteria: (i) ‘zero values’ are assigned 0, whereas (ii) ‘missing measurements’ are replaced with their previous values for the same patient if they exist; otherwise, they are filled with the median of the original value over the whole sample.
For static variables, the gender variable was binarized and the age variable was normalized between [0, 1]. Diagnosis (HM and H12O) and procedures on admission (H12O) were included as one-hot-encoded values, taking the three first digits of the ICD-10 and ICD-9 for HM and H12O, respectively.
For dynamic variables, we applied similar methods for both datasets but with slight variations. For HM the information about ICU stay was used to create a binary variable indicating whether the patient is in the ICU or not each day of the hospital stay. In the case of H12O, information about ICU stay could not be extracted and was omitted. Diagnostics and procedure variables, encoded as ICD-10 (HM) or ICD-9 (H12O) codes, were one-hot-encoded, taking only the first three digits. For missing measurements, we experimented with two different ways of imputation: (i) we used the previous day values of the same patient, when available, and the median value of the dataset (excluding the test set) when not available and (ii) just used the median value for all cases. Additionally, we also used the extra binary missing value indicators and also generated extra datasets without this feature. For the laboratory determinations, we tried two methods: applying reference values and normalizing between [0, 1].
We experimented and generated different dataset representations identified as follows:
-
•
base: when using the mean for missing measurement imputation.
-
•
imputation: when using the previous value of a feature for that patient (if available) and the mean of the sample (if not), for missing measurement imputation.
-
•
missing: a new binary feature is added to indicate if the original value is missing following [19].
-
•
reference: when using reference values in determination results.
Finally, for a given patient, we joined all static features together to form the final static feature vector . Similarly, for a given patient and a given day, we joined all the dynamic features together to create a sequence of dynamic feature vectors where are the hospital days. Note that, as mentioned above, while creating the dynamic feature vectors, a few extra features may be generated to identify ’missing measurements’.
3.3. Architecture
Motivated by the temporal nature of our problem and the effectiveness of the RNN models in the medical domain [20], [21], [22], we proposed an RNN-based model to monitor the mortality risk of the patient by producing a daily prediction during the patient’s hospital stay. For this purpose, we fed each daily record of temporal and static data day-by-day to output a prediction.
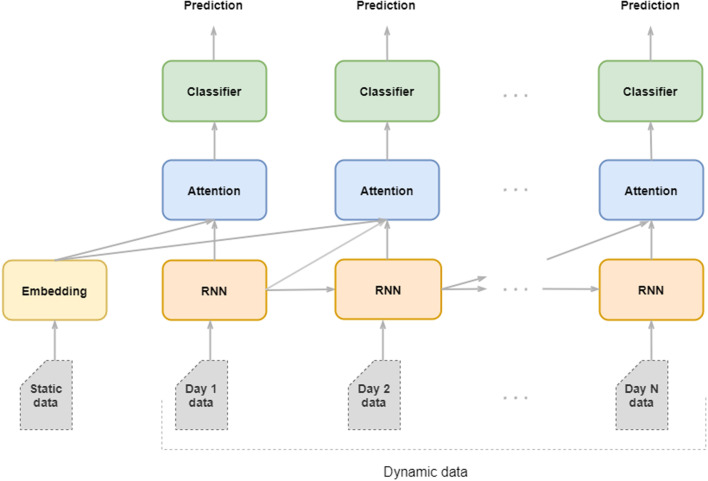
As Fig. 1 shows, we designed an Artificial Neural Network with four modules, namely, the embedding module, the recurrent module, the classifier module and, optionally, the attention modules.
Fig. 1.
Architecture of the RNN described in Section 3.3 showing how data are fed through the embedding, RNN, attention and classifier modules to produce daily predictions.
The embedding module feeds the static vector into a fully-connected layer to encode the high-dimensional sparse feature vector into a lower-dimensional dense vector.
The recurrent module accounts for the encoding and the memorization of the temporal information provided by the RNN’s hidden states. It consists of a unidirectional RNN, built with either an LSTM or a Gated Recurrent Unit (GRU) [23] that models the causal relationship between the clinical events occurring in the temporal order.4 On each day, the RNN cells processed the relevant information of the patient’s dynamic data and produced an output vector that captures the relevant clinical information to perform the daily prediction.
The attention module of the dynamic field finds the correlations of all previous RNN outputs and merges all the global relevant information of the sequence until a given day. First, the attention module creates a context vector as a linear combination of the RNN outputs using the dot-product of the attention scores as in [24]. Then, every day, the context vector and the current RNN output are concatenated and fed to a fully-connected layer to which it was applied a hyperbolic tangent function to get the final attention vector5 . The attention module on the static field finds correlations of unembedded static data with respect to the dynamic hidden layer. As we will discuss in the next sections, in our work, the attention module is the component responsible for the interpretability of the model.
Finally, the classifier module consists of two fully-connected layers followed by a sigmoid activation function to produce the binary mortality predictions. We placed the classifier module on top of the RNN cells and the static embeddings to predict the mortality risk each day.
3.4. Training and inference
Unlike standard training schemes for recurrent models, we designed an ad-hoc training scheme to produce daily predictions. Specifically, we did not feed the entire sequence of dynamic data into the RNN to output a single prediction. Instead, we fed each daily record of temporal and static data day-by-day and output a prediction for each day.
To mitigate the impact of class-imbalanced data on the learning process, we employed the e introduced in [25] besides the classical Vanilla Binary Cross-Entropy. Focal Loss was originally developed for computer vision as a strategy for countering class imbalance. Based on the assumption that the most frequent label will generate more confident predictions, Focal Loss reshapes the Vanilla Cross-Entropy through two parameters ( and ) such that well-classified samples have less weight than the incorrectly classified ones. During training, to 1 and used as a hyperparameter.
In addition, as detailed in the Additional Material, we experimented with other models’ hyperparameter configurations. All the layers are trained end-to-end with stochastic gradient descent.
At inference, the model is fed with the static vector and then, at each time step, we input the corresponding dynamic vector, and the model outputs the probability of patient mortality. To obtain the mortality predictions, the probability is discretized into a binary label using a threshold, as in logistic regression. We set the threshold to the default value of 0.5. Note that, after training, other values of the model’s threshold can be used, depending on how confident in the prediction we want to be. For the metrics used for evaluation, see Section 3.7.
3.5. Interpretability
The attention layer is the building block of interpretability in our model. The attention module of the dynamic field relates the RNN outputs to the RNN output history vectors to retrieve meaningful past dynamic information for daily prediction. The idea is to identify which days in the previous sequence are influencing most the prediction of the current day. The attention can be useful as a starting point to learn which clinical variables could have a critical role. See the animated figure6 to visualize how the attention scores dynamically change as the sequence evolves and focuses on the most relevant days for a given patient. See the Additional Material to learn about attention analysis.
3.6. Experimental framework
We conducted extensive experiments within an experimental setting that included: dataset splits, a feature selection mechanism, a proper model’s hyperparameter tuning and model selection, and two competitive baselines.
In particular, we would like to point out the role of model tuning as a necessary step considering our case. Since we designed our network from scratch and applied it to a dataset not yet studied, a suitable assessment of the model’s capability through a functional exploration of hyperparameters was required. Analogously, a thorough evaluation helped in testing the robustness of the model. See the Additional Material for detailed descriptions.
3.7. Metrics and evaluation
To evaluate the performance of the models and the consistency of the model’s predictions, we considered the following metrics:
-
•
Accuracy: measures the number of correct predictions over the total number of cases.
-
•
F1 score: harmonic mean of the model’s precision and recall. For this project, we use a macro F1 score, computing the metric for each label and then finding the unweighted mean. Macro F1 does not take label imbalance into account, and therefore the score for each class counts up to half of the total score.
-
•
Sensitivity: measures the capacity to correctly predict mortality, also known as recall.
-
•
Specificity: proportion of the capacity to correctly predict survivals.
-
•
Area Under the Curve (AUC): the probability of a random example with the true label of fatality receiving a higher score than a random example with the true label of survival.
When computing the global performances for each of the metrics above, we used the average metric computed every day in the patient’s sequence for all patients.
In addition, and more interestingly, we designed specific evaluation metrics to test the model better and reflect our intended use case by providing daily predictions. In this case, we take the prediction vectors already returned by the system and use them to calculate the daily performances as follows:
-
•
Daily performances from the admission computes the performance every day from the admission day.
-
•
Daily performances from the outcome computes the performance every day from the outcome day.
Note that, in daily performance calculations, the system ignores whether or not the patients are long-term patients and there is no leakage of other past or future events. We simply evaluate the predictions already performed by the system.
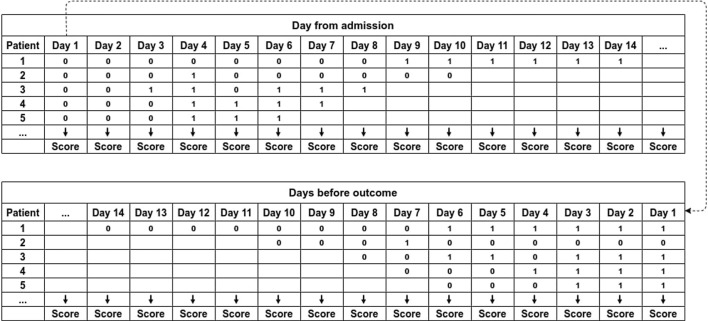
Fig. 2 shows an example of how day-by-day predictions are calculated. To compute the daily performances from admission, we use the predictions of each patient, starting from the left. To compute the daily performances from the outcome, first, we align all the predictions to the right, and then we use the predictions of each patient, starting from the right. In this case, Day 1 is the outcome day.
Fig. 2.
Example of system’s predictions from admission and from outcome.
4. Results and discussion
Table 1 shows the global evaluation results for HM on cross-validation, comparing the best SVC, RF, and the top RNN models (all of them selected as per results in the validation set).
Table 1.
HM, final evaluation results on cross-validations.
| Mod | Imp | Mis | Ref | FS | Met | Accuracy | AUC | Sensitivity | Specificity | F1 |
|---|---|---|---|---|---|---|---|---|---|---|
| RNN | ✔ | ✕ | ✕ | ✕ | F1 | 0.9072 ± 0.0034 | 0.9408 ± 0.0062 | 0.6775 ± 0.0228 | 0.9597 ± 0.0041 | 0.8374 ± 0.0073 |
| RF | ✕ | ✕ | ✕ | – | Acc | 0.9032 ± 0.0032 | 0.9361 ± 0.0009 | 0.6169 ± 0.0109 | 0.9687 ± 0.0018 | 0.8228 ± 0.0061 |
| SVC | ✔ | ✔ | ✕ | – | F1 | 0.8889 ± 0.0035 | 0.9458 ± 0.0035 | 0.7806 ± 0.0121 | 0.9137 ± 0.0028 | 0.8269 ± 0.0055 |
| RNN | Ensemble-F1 | Acc | 0.9054 ± 0.0045 | – | 0.7494 ± 0.0244 | 0.9411 ± 0.0106 | 0.8443 ± 0.0041 | |||
| RNN | Ensemble-SEN | Acc | 0.8896 ± 0.0054 | – | 0.8450 ± 0.0075 | 0.8998 ± 0.0073 | 0.8351 ± 0.0063 | |||
We ran one-tailed Student’s t-test and Wilcoxon signed-rank tests on sensitivity scores by folds. Student’s t-test shows that there is a significant statistical difference in the 5-fold cross-validation sensitivity scores for RNN (mean 0.8442, SD 0.0157) and RF (mean 0.6168, SD 0.0122), with . Also for the RNN with respect to the SVC (mean 0.7806, SD 0.0135) there is a significant difference, with . In the Wilcoxon signed-rank, the test results are positive and the same as the previous Wilcoxon test on accuracy, in both RNN over RF and RNN over SVC (with ).
Table 2 shows the global evaluation results for H12O on cross-validation, on cross-validation, comparing the best SVC, RF, and the top RNN models (all of them selected as per results in the validation set).
Table 2.
H12O, final evaluation results on cross-validations.
| Mod | Imp | Mis | Ref | FS | Met | Accuracy | AUC | Sensitivity | Specificity | F1 |
|---|---|---|---|---|---|---|---|---|---|---|
| RNN | ✔ | ✔ | ✕ | 150 | Acc | 0.9246 ± 0.0031 | 0.9466 ± 0.0025 | 0.6700 ± 0.0497 | 0.9630 ± 0.0101 | 0.8281 ± 0.0063 |
| RF | ✔ | ✔ | ✕ | – | Acc | 0.9341 ± 0.0010 | 0.9504 ± 0.0012 | 0.6255 ± 0.0102 | 0.9806 ± 0.0015 | 0.8380 ± 0.0030 |
| SVC | ✔ | ✔ | ✔ | – | Acc | 0.9203 ± 0.0005 | 0.9413 ± 0.0024 | 0.6397 ± 0.0087 | 0.9626 ± 0.0011 | 0.8161 ± 0.0021 |
| RNN | Ensemble-F1 | Acc | 0.9201 ± 0.0047 | – | 0.7434 ± 0.0294 | 0.9468 ± 0.0092 | 0.8315 ± 0.0052 | |||
| RNN | Ensemble-SEN | F1 | 0.9011 ± 0.0106 | – | 0.8082 ± 0.0327 | 0.9152 ± 0.0163 | 0.8119 ± 0.0115 | |||
We ran the same significance tests as in HM for H12O for sensitivity by folds. The one-tailed Student’s t-test shows that there is a significant statistical difference in the 5-fold cross-validation sensitivity scores for RNN (mean 0.8082, SD 0.0326) and RF (mean 0.6255, SD 0.0114), with and also for the RNN with respect to SVC (mean 0.6397, SD 0.0097) there is a significant difference, with . In the Wilcoxon signed-rank, the test results were positive in both RNN over RF and RNN over SVC (with ).
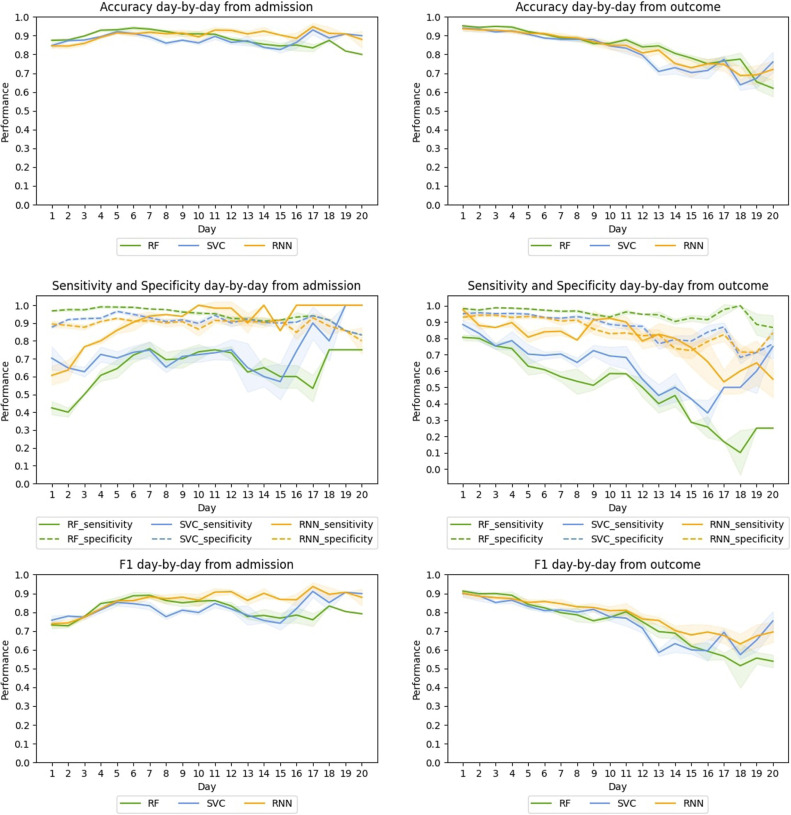
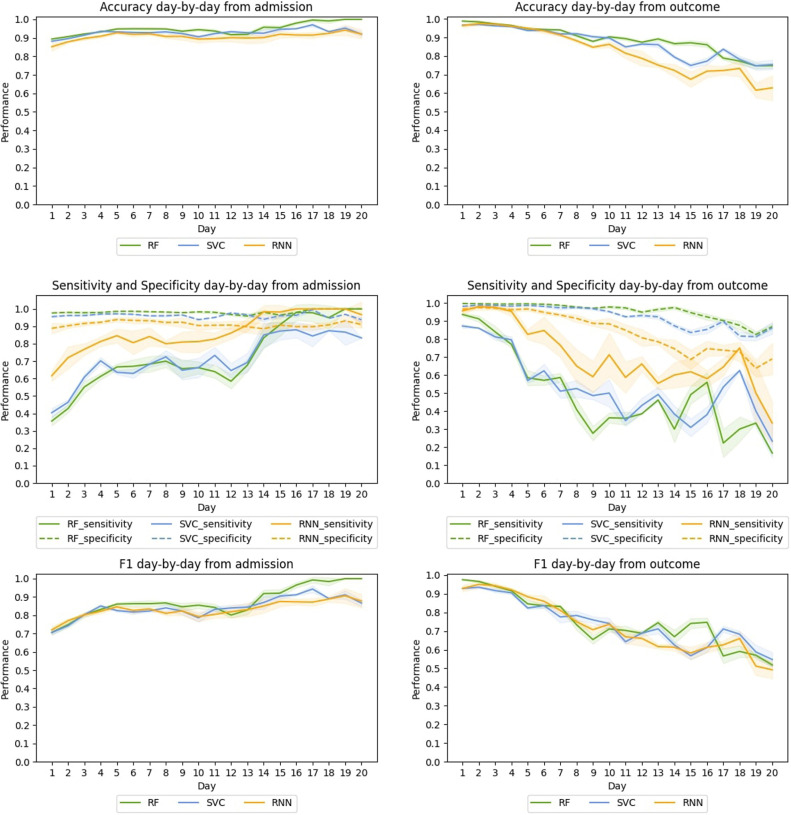
Complete results by folding in sensitivity in both datasets are provided in the Additional Material. There we also include the tables with the daily results for all the models. Here Fig. 3, Fig. 4 summarize the results.
Fig. 3.
HM, Average day-by-day test performance from the admission (left) and from the outcome (right) in the cross-validation for the best RF, SVC, and RNN-based models.
Fig. 4.
H12O, Average day-by-day test performance from the admission (left) and from the outcome (right) in the cross-validation for the best RF, SVC, and RNN-based models.
4.1. Global performance
For HM, the best ensemble RNN model is significantly better than the other models when considering sensitivity, reaching 0.84 in sensitivity with no penalty in F1 and a small impact on accuracy. These interesting results are achieved in all the folds, demonstrating their consistency and robustness (see sensitivity global results by folds in the Additional Material). All RNN models show slightly better scores in accuracy and F1 when compared to baselines, which is a positive result taking into account that F1 is especially useful when having an uneven class distribution. SVC results are surprisingly performant in sensitivity, clearly surpassing those of the RF, the non-ensemble RNN and the Ensemble-F1 models. All HM models (60) use attention and, as described in the attention peak analysis in the Additional Material, the results show that 56.59% of the patients had relevant peak days.
For H12O, the RNN ensemble model outperforms all models in sensitivity. In particular, the Ensemble-SEN model outperforms RF in 18 points and SVC in 16 points reaching 0.80 in sensitivity. Again, for sensitivity, the results are the same in all folds. Although RF is slightly better in accuracy and F1, the difference is minimal. As for attention, it is worth mentioning that 53 out of the top 60 RNN models use attention and the best non-ensemble RNN model had 66.36% patients with peak days.
4.2. Day-by-day performance
When evaluating day-by-day from the admission, we have longer sequences as we go on. Thus, at each new step, the model uses more information and gets closer to the outcome. As Fig. 3 shows, in HM, this has a small positive effect on RF and SVC: accuracy, specificity, and F1 curves are high and rather flat with no noticeable improvement. For sensitivity, the RF shows a moderate improvement until day 7, a slight fall from day 12 onward and a sudden peak on day 18. SVC shows a long and strong performance curve, starting at 0.70 with a sudden rise on day 17.
The RNN-model shows similar behaviour, with high, rather flat curves for accuracy, specificity and F1. However, the model performs better with respect to sensitivity, with a curve starting at 0.60, a peak of 1.00 on day 10, and then stabilizing between 0.85 and 1.00 until the last day.
When evaluating from the outcome, we have shorter sequences as we evaluate backwards. In this case, at each new step, the model has less information and is farther away from the outcome. Notice also that the farther we are from the outcome, the fewer examples we have. As shown in Table 1 from Additional Material, this reduces the number of patients labelled with death, which may cause fluctuations, especially in sensitivity.
For HM, all models show similar descending curves for accuracy, specificity and F1. For sensitivity, SVC has a sharper decrease. The RNN model shows a higher and flatter curve in sensitivity, starting at 0.97, ending at 0.55 and maintaining a performance of 0.79 or better until day 14. This shows that the system is efficient at making early predictions since it provides reliable mortality predictions up to 14 days before the outcome.
For H12O, the day-bay-day performance from the admission is similar to that of HM. Thus, in terms of sensitivity, the RNN shows a substantial improvement compared with the baselines. RNN mainly leads the sensitivity curve, and it is very performant on both F1 and accuracy, which confirms the enhancement.
When evaluating H12O from the outcome, RF and SVC show similar behaviour: the specificity curve stays high and flat, accuracy shows a moderate decrease, F1 suffers a sharper drop, and sensitivity plummets. Again RNN shows a higher curve and a better performance in sensitivity, maintaining a score of 0.64 or better until seven days before the outcome.
As in the case of HM, sensitivity curves from the outcome have greater fluctuations. As mentioned before, the gradual reduction in data and death labels (in absolute terms) as we move forward makes the sensitivity curves more unstable.
5. Conclusions
This work is a first approximation of an RNN-based model to predict the outcome of patients with COVID-19. The models were trained with two relatively small COVID-19 datasets and will be the basis of a future system in a larger setting. The models consume dynamic information to generate daily predictions. Unlike standard training schemes for recurrent models, we do not feed the entire sequence of dynamic data into the RNN to output a single prediction. Instead, we feed each daily record of temporal and static data day-by-day and output a prediction for each day.
We defined an ensemble method as a voting system developing an algorithm that, starting with the best model in terms of cross-validated accuracy or F1 score, heuristically chooses the most different models (i.e. the ones that make the most different predictions) to encourage model diversity in the ensemble but without penalizing the results. Results obtained with this ensemble method are not only superior, but they also reduce the standard deviation, producing less variation in all metrics among folds.
The system is robust enough to deal with real-world data, and it can overcome the problems derived from the sparsity and heterogeneity of the data. In addition, the approach was validated using two datasets showing substantial differences. Although both datasets contain information from COVID patients, they have different characteristics, which made it impossible to join the datasets into a single one. The disparity in coding conventions and the differences in the type of information contained precluded aggregating data, as this would produce a reduced dataset due to the low overlap. Instead, we generated a model for each data set, following the same method and using the same architecture. This not only validates the robustness of the proposal but also meets the requirements of a real scenario where interoperability between hospitals’ datasets is challenging to achieve.
This ended up in exhaustive experimentation, maximizing the potential of real-world data. We explored and evaluated different data representation methods, and this information is reported in detail in the article and in the Additional Material.
Regarding interpretability, when using the attention mechanism on the dynamic data vector, we show that there are relevant days that the model puts the focus on.
For future work, we are collaborating with two other reference Spanish hospitals (Hospital Clinic de Barcelona and Hospital Virgen del Rocio) to include them in a national initiative following the methodology described in this paper. We have already agreed on a common data model, and we will continue validating and improving predictive algorithms with these additional data. We also plan to include historical clinical data in the predictive model and to continue monitoring patients at discharge to predict the risk of hospital readmission.
Code availability
The code is available7 under MIT licence.
CRediT authorship contribution statement
Marta Villegas: Study design, Organized the authors, Synthesized the writing, Led the abstract, introduction, results, discussion sections. Aitor Gonzalez-Agirre: Study design, Data analysis and preparation, Manuscript preparation, Led the experimental framework and evaluation. Asier Gutiérrez-Fandiño: Study design, Data analysis and preparation, Manuscript preparation, Led the experimental framework and evaluation. Jordi Armengol-Estapé: Study design, Manuscript preparation, Led the experimental framework and evaluation. Casimiro Pio Carrino: Study design, Data analysis and preparation, Experimental framework, Models evaluation and manuscript preparation. David Pérez-Fernández: Study design, Data analysis and preparation. Felipe Soares: Experimental framework and evaluation. Pablo Serrano: Oversaw and advised the work. Miguel Pedrera: Data extraction, Data analysis and preparation. Noelia García: Data extraction, Data analysis and preparation. Alfonso Valencia: Oversaw and advised the work.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank José Antonio López-Martin from Hospital Universitario 12 de Octubre; Xavier Pastor and M. Jesús Betran from Hospital Clínic de Barcelona; and Carlos Luis Parra and Sara González from Hospital Virgen del Rocio for their interest in and support for the project; HM Hospitales for sharing the data with the scientific community; and finally, our colleague Eulàlia Farré i Maduell, a medical doctor and researcher at the Text Mining Unit at BSC, for her support for medical issues.
This work was funded by the Spanish State Secretariat for Digitalization and Artificial Intelligence to carry out support activities in supercomputing within the framework of the Plan- TL8 signed on 14 December 2018. All authors contributed significant amendments to the final manuscript.
Hospital 12 de Octubre is supported by PI18/00981 “Arquitectura normalizada de datos clínicos para la generación de infobancos su uso secundario en investigación: caso de uso cáncer de mama, cérvix útero, evaluación” funded by the Carlos III Health Institute from the Spanish National plan for Scientific and Technical Research and Innovation 2017–2020 and the European Regional Development Funds (FEDER).
Involved hospitals obtained informed consent from all participants.
For this reason we discarded bidirectional RNNs architectures.
The implementation is available in PyTorch: https://pytorchnlp.readthedocs.io/en/latest/_modules/torchnlp/nn/attention.html.
Attention mechanism animation: https://github.com/PlanTL-SANIDAD/covid-predictive-model/blob/main/documentation/images/patient_dynamic_attention.gif.
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.cmpbup.2022.100089.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
.
Data availability
HM raw data can be accessed by submitting an application1 for access. The data sharing agreement with H12O does not permit us to make the raw data available to third parties.
References
- 1.Spanish Health Ministry . 2020. COVID-19 update no. 263. https://www.mscbs.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/Actualizacion_263_COVID-19.pdf. [Online; Accessed 15 January 2020] [Google Scholar]
- 2.Pedrera-Jiménez M., García-Barrio N., Cruz-Rojo J., Terriza-Torres A.I., López-Jiménez E.A., Calvo-Boyero F., Jiménez-Cerezo M.J., Blanco-Martínez A.J., Roig-Domínguez G., Cruz-Bermúdez J.L., Bernal-Sobrino J.L., Serrano-Balazote P., Muñoz-Carrero A. Obtaining EHR-derived datasets for COVID-19 research within a short time: a flexible methodology based on Detailed Clinical Models. J. Biomed. Inform. 2021;115 doi: 10.1016/J.JBI.2021.103697. URL: https://pubmed.ncbi.nlm.nih.gov/33548541/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge W., Huh J.-W., Park Y.R., Lee J.-H., Kim Y.-H., Turchin A. AMIA Annual Symposium Proceedings. Vol. 2018. American Medical Informatics Association; 2018. An interpretable ICU mortality prediction model based on logistic regression and recurrent neural networks with LSTM units; pp. 460–469. URL: https://pubmed.ncbi.nlm.nih.gov/30815086. [PMC free article] [PubMed] [Google Scholar]
- 4.Teoh D. Towards stroke prediction using electronic health records. BMC Med. Inf. Decis. Mak. 2018;18 doi: 10.1186/s12911-018-0702-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D. Chang, D. Chang, M. Pourhomayoun, Risk Prediction of Critical Vital Signs for ICU Patients Using Recurrent Neural Network, in: 2019 International Conference on Computational Science and Computational Intelligence, CSCI, 2019, pp. 1003–1006.
- 6.Yu K., Zhang M., Cui T., Hauskrecht M. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. Vol. 25. 2020. Monitoring ICU mortality risk with A long short-term memory recurrent neural network; pp. 103–114. URL: https://pubmed.ncbi.nlm.nih.gov/31797590. [PMC free article] [PubMed] [Google Scholar]
- 7.Lipton Z.C., Kale D.C., Elkan C., Wetzel R. 2015. Learning to diagnose with LSTM recurrent neural networks. arXiv:1511.03677. [Google Scholar]
- 8.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 9.Choi E., Bahadori M.T., Sun J. 2015. Doctor AI: predicting clinical events via recurrent neural networks. CoRR, arXiv:1511.05942. [PMC free article] [PubMed] [Google Scholar]
- 10.Mikolov T., Sutskever I., Chen K., Corrado G., Dean J. 2013. Distributed representations of words and phrases and their compositionality. arXiv:1310.4546. [Google Scholar]
- 11.Choi E., Bahadori M.T., Sun J., Kulas J.A., Schuetz A., Stewart W.F. NIPS. 2016. RETAIN: An interpretable predictive model for healthcare using reverse time attention mechanism; pp. 3512–3520. [Google Scholar]
- 12.F. Ma, R. Chitta, J. Zhou, Q. You, T. Sun, J. Gao, Dipole: Diagnosis Prediction in Healthcare via Attention-based Bidirectional Recurrent Neural Networks, in: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2017.
- 13.Xia J., Pan S., Zhu M., Cai G., Yan M., Su Q., Yan J., Ning G. A long short-term memory ensemble approach for improving the outcome prediction in intensive care unit. Comput. Math. Methods Med. 2019;2019 doi: 10.1155/2019/8152713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertsimas D., Lukin G., Mingardi L., Nohadani O., Orfanoudaki A., Stellato B., Wiberg H., Gonzalez-Garcia S., Parra-Calderon C.L., Robinson K., Schneider M., Stein B., Estirado A., a Beccara L., Canino R., Dal Bello M., Pezzetti F., Pan A. Cold Spring Harbor Laboratory Press; 2020. COVID-19 Mortality Risk Assessment: An International Multi-Center Study. MedRxiv. URL: https://www.medrxiv.org/content/early/2020/07/09/2020.07.07.20148304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., Sun C., Tang X., Jing L., Zhang M., Huang X., Xiao Y., Cao H., Chen Y., Ren T., Wang F., Xiao Y., Huang S., Tan X., Huang N., Jiao B., Cheng C., Zhang Y., Luo A., Mombaerts L., Jin J., Cao Z., Li S., Xu H., Yuan Y. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020;2(5):283–288. doi: 10.1038/s42256-020-0180-7. [DOI] [Google Scholar]
- 16.Nasrolahzadeh M., Rahnamayan S., Haddadnia J. Alzheimer’s disease diagnosis using genetic programming based on higher order spectra features. Mach. Learn. Appl. 2022;7 doi: 10.1016/J.MLWA.2021.100225. [DOI] [Google Scholar]
- 17.Chaudhari P., Agrawal H., Kotecha K. Data augmentation using MG-GAN for improved cancer classification on gene expression data. Soft Comput. 2019 24:15. 2019;24:11381–11391. doi: 10.1007/S00500-019-04602-2. URL: https://link.springer.com/article/10.1007/s00500-019-04602-2. [DOI] [Google Scholar]
- 18.Baltrušaitis T., Ahuja C., Morency L.-P. Multimodal machine learning: A survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2019;41(2):423–443. doi: 10.1109/TPAMI.2018.2798607. [DOI] [PubMed] [Google Scholar]
- 19.Che Z., Purushotham S., Cho K., Sontag D., Liu Y. Recurrent neural networks for multivariate time series with missing values. Sci. Rep. 2018;8(1):6085. doi: 10.1038/s41598-018-24271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi E., Schuetz A., Stewart W.F., Sun J. Using recurrent neural network models for early detection of heart failure onset. J. Am. Med. Inf. Assoc. JAMIA. 2017;24(2):361–370. doi: 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T., Qiu R.G., Yu M. Predictive modeling of the progression of Alzheimer’s disease with recurrent neural networks. Sci. Rep. 2018;8(1):9161. doi: 10.1038/s41598-018-27337-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beeksma M., Verberne S., van den Bosch A., Das E., Hendrickx I., Groenewoud S. Predicting life expectancy with a long short-term memory recurrent neural network using electronic medical records. BMC Med. Inf. Decis. Mak. 2019;19(1):36. doi: 10.1186/s12911-019-0775-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho K., van Merrienboer B., Gülçehre Ç., Bougares F., Schwenk H., Bengio Y. 2014. Learning phrase representations using RNN encoder-decoder for statistical machine translation. CoRR, arXiv:1406.1078. [Google Scholar]
- 24.Bahdanau D., Cho K., Bengio Y. 2014. Neural machine translation by jointly learning to align and translate. arXiv:1409.0473. [Google Scholar]
- 25.Lin T., Goyal P., Girshick R.B., He K., Dollár P. 2017. Focal loss for dense object detection. CoRR, arXiv:1708.02002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.
Data Availability Statement
HM raw data can be accessed by submitting an application1 for access. The data sharing agreement with H12O does not permit us to make the raw data available to third parties.