Abstract
Background
Colorectal cancer (CRC) is the third most diagnosed cancer and the second most common cause of cancer deaths worldwide. CRC patients present with an increase in pathogens in their gut microbiota, such as polyketide synthase-positive bacteria (pks +) and enterotoxigenic Bacteroides fragilis (ETBF). The pks + Escherichia coli promotes carcinogenesis and facilitates CRC progression through the production of colibactin, a genotoxin that induces double-strand DNA breaks (DSBs). ETBF is a procarcinogenic bacterium producing the B. fragilis toxin (bft) that promotes colorectal carcinogenesis by modulating the mucosal immune response and inducing epithelial cell changes.
Methods
Fecal samples were collected from healthy controls (N = 62) and CRC patients (N = 94) from the province of Québec (Canada), and a bacterial DNA extraction was performed. Fecal DNA samples were then examined for the presence of the pks island gene and bft using conventional qualitative PCR.
Results
We found that a high proportion of healthy controls are colonized by pks + bacteria (42%) and that these levels were similar in CRC patients (46%). bft was detected in 21% of healthy controls and 32% of CRC patients, while double colonization by both pks + bacteria and ETBF occurred in 8% of the healthy controls and 13% of the CRC patients. Most importantly, we found that early-onset CRC (< 50 years) patients were significantly less colonized with pks + bacteria (20%) compared to late-onset CRC patients (52%).
Conclusions
Healthy controls had similar levels of pks + bacteria and ETBF colonization as CRC patients, and their elevated levels may place both groups at greater risk of developing CRC. Colonization with pks + bacteria was less prevalent in early-compared to late-onset CRC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13099-022-00523-y.
Keywords: Colibactin, Enterotoxigenic Bacteroides fragilis, Tumorigenesis, Colorectal cancer
Background
The composition and function of the gut microbiome have been shown to potentially influence the initiation and progression of colorectal cancer (CRC) [1]. Patients with CRC have an unbalanced gut microbiome, or dysbiosis, which is characterized by a decrease in beneficial bacteria and an increase in pathobionts, such as colibactin-producing Escherichia coli and enterotoxigenic Bacteroides fragilis (ETBF) [2].
While gut microbiota contains commensal E. coli strains, some strains may carry a pathogenic potential [3]. The pks genomic island contains the colibactin (clb) gene cluster, which encodes the genes required for colibactin synthesis [4]. Colibactin is a genotoxin that causes inter-strand cross-links (ICLs) [5] and double-strand DNA breaks (DSBs), cell cycle arrest, senescence, and chromosomal abnormalities in mammalian cells [6]. Murine models of pks + E. coli mono-colonization [7, 8] and colonization of adenomatous polyposis coli multiple intestinal neoplasia (ApcMin/+) mice with colibactin producing E. coli [9] revealed a causal link between the presence of colibactin and intestinal tumorigenicity. Other Enterobacteriaceae species, such as Klebsiella, inherited the pks island and some genes of the cluster from horizontal transfer and can also produce colibactin [10, 11]. Colonization with colibactin-producing bacteria in humans occurs mainly during early life [12], and the presence of the phylogroup of pks + E. coli is steadily increasing worldwide [13, 14].
Bacteroides strains such as EBTF have also been associated with CRC. ETBF which produces Bacteroides fragilis toxin (bft), has been shown to contribute to colon carcinogenesis [15] through induction of colonocyte proliferation [16], inhibition of apoptosis and promotion of proinflammatory signaling [17, 18]. Accordingly, ETBF colonization in a murine model of colitis-induced CRC increased the number of tumours [19], while in the ApcMin/+ CRC mouse model, it promoted the development of colon adenomas [20], further confirming its carcinogenic potential.
In this study we assessed the prevalence of pks + bacteria and ETBF in a cohort of 94 CRC patients and 62 healthy individuals from the province of Québec, Canada.
Methods
Patient recruitment and sample collection
Patients with CRC and healthy individuals were recruited at the Centre hospitalier de l’Université de Montréal (CHUM) (Additional file 1: Table S1). Individuals with inflammatory bowel disease (IBD), polyps or antibiotic treatment 6 months prior to sampling were excluded from the control group. Participants were requested to provide a fresh fecal sample collected at home following the International Human Microbiome Standards procedure [21]. Samples were collected in hermetic containers with an anaerobic sachet (BD BBL™ GasPak™ anaerobic indicator, BD, ON, Canada) and stored at −80 °C upon arrival at the laboratory within 24 h of sampling.
DNA extraction and polymerase chain reaction
Total DNA was extracted from human fecal samples with the PowerSoil® DNA Extraction Kit (Qiagen Inc., Toronto, ON, Canada) and polymerase chain reaction (PCR) was performed using PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) in the RG 3000A R PCR machine (Qiagen Inc.) using the following cycling conditions; 50 °C for 2 min, 95 °C for 2 min, 38 cycles of 15 s at 95 °C, followed by 1 min at 60 °C. Simultaneous amplification of colibactin A gene (clbA) and E. coli 16S rRNA were done with the following primers for clbA: Fw 5ʹ-CTCCACAGGAAGCTACTAAC-3ʹ, Rv 5ʹ-CGTGGTGATAAAGTTGGGAC-3ʹ [4] and Ecoli 16S: Fw 5ʹ-GTTAATTTTGCTCATTGA-3ʹ, Rv 5ʹ-ACCAGGGTATCTAATCCTGTT-3ʹ[22], with a 1:1:1:1 ratio. For the detection of ETBF, we performed a simultaneous PCR of the bft gene and B. fragilis 16S rRNA with the following primers for bft: Fw 5ʹ-GAACCTAAAACGGTATATGT-3’, Rv 5ʹ-GTTGTAGACATCCCACTGGC-3ʹ [8] and Bfr: Fw 5ʹ-CTGAACCAGCCAAGTAGCG-3ʹ, Rv 5ʹ-CCGCAAACTTTCACAACTGACTTA-3ʹ [23], with a 5:5:1:1 ratio. We used the E. coli NC101 strain (EcNC101 (a gift from Dr. Christian Jobin, Cancer Microbiota & Host Response, UF Health Cancer Center, University of Florida)) as a positive control for the presence of the pks island, and the ETBF strain (a gift from Dr. Cindy Sears, Johns Hopkins University School of Medicine [8]) as the positive control for the bft gene. The PCR products were then visualized on a 1.8% agarose gel containing Eco-stain plus (Bio Basic Inc., Markham, ON, Canada). The expected product sizes were: 330 bp for E. coli 16S rRNA; 300 bp for clbA; 230 bp for B. fragilis 16S rRNA; and 370 bp for bft.
Statistics
All data were analyzed using GraphPad Prism (Version 5.0, GraphPad Software, San Diego, CA, USA). χ2 tests were used to compare categorical variables, unless expected frequencies were < 5, in which case Fisher’s exact test was used. P < 0.05 were considered statistically significant.
Results and discussion
The presence of colibactin-producing bacteria in stool samples collected from participants (Additional file 1: Table S1) was detected by PCR using specific primers targeting the clbA gene encoded in the pks island, required for colibactin production [4]. In addition, as a positive control for the PCR reaction, primers universal for all strains of E. coli were used [22] (Fig. 1a). We found that 42% of healthy donors and 46% of CRC patients were colonized by a pks + bacteria (Fig. 2, Table 1). Interestingly, pks + bacteria were more prevalent in late-onset (40 out of 79; 52%) compared to early-onset CRC (3 out of 15; 20%; P < 0.05) (Fig. 2, Table 1). Overall, the levels of pks + bacteria colonization in our CRC patients were within the range previously reported in literature with 68% [8] and 66.7% [7] in two cohorts from the USA, 56.4% in Sweden [24], 43% in Japan [25], 23% in Iran [26], and 16.7% in Malaysia [27]. As for the healthy population, they approached levels reported in a Japanese cohort (46%) [25], whereas lower levels were found in other healthy cohorts: 22% [8] and 20.8% [7] in the USA; 18.5% in Sweden [24]; 7.1% in Iran [26] and 4.35% in Malaysia [27]. These disparities in prevalence around the world could be attributed to dietary differences. For example, the so-called Western diet [28] has been linked to a higher incidence of colorectal cancer containing pks + E. coli [29]. Our study indicates that colibactin-producing bacteria are less prevalent in early-onset compared to late-onset CRC, although this finding should be confirmed in larger cohorts. While this could indicate that colibactin-producing bacteria may not be involved in the etiology of early-onset CRC, we cannot rule out that pks + E. coli and other colibactin-producing bacteria may have been present during childhood and subsequently eliminated, with the effects of early mutagenic exposure manifesting later in life [30].
Fig. 1.
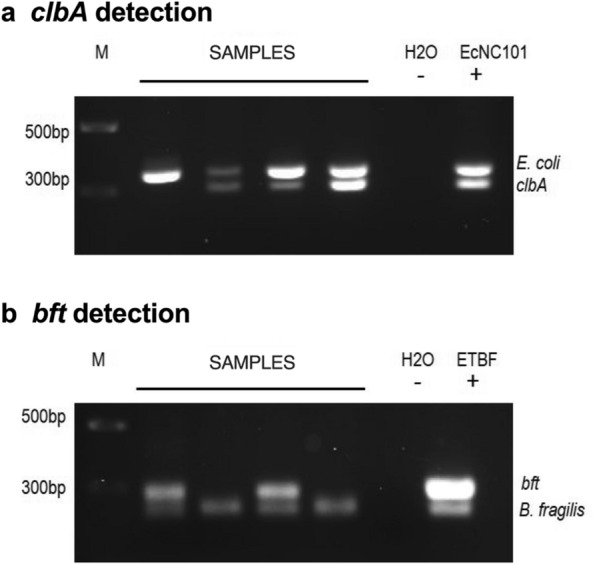
clbA and bft detection. a clbA and b bft presence in DNA extracted from fecal samples were detected using conventional qualitative PCR. M: marker; negative (−) control: water (H2O); positive control ( +): DNA extracted from pks + EcNC101 or ETBF bacteria
Fig. 2.
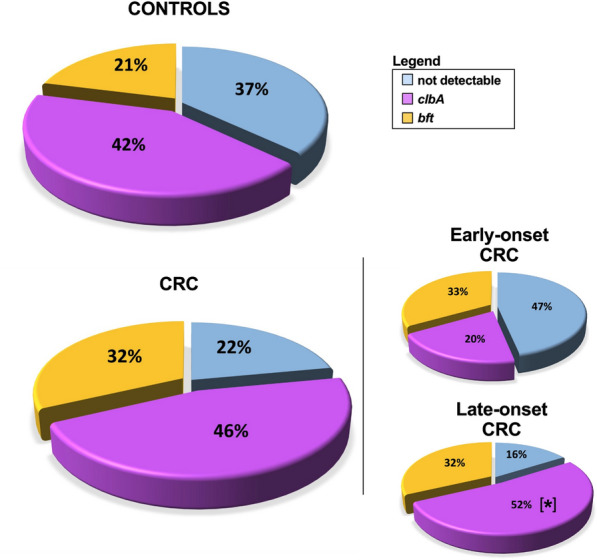
Prevalence of pks + and ETBF in the cohort. Controls – healthy participants; CRC – colorectal cancer patients. * P < 0.05, Fisher’s exact test
Table 1.
Prevalence of clbA and bft in controls and CRC patients
| Total (156) | clbA + | p-value | bft + | p-value | |
|---|---|---|---|---|---|
| Cohort (%) | |||||
| Controls | 62 (40) | 26 (42) | N.s | 13 (21) | N.s |
| CRC | 94 (60) | 43 (46) | 30 (32) | ||
| Controls | |||||
| Sex (%) | |||||
| Female | 33 (53) | 11 (33) | N.s | 6 (18) | N.s |
| Male | 29 (47) | 15 (52) | 7 (24) | ||
| CRC | |||||
| Onset (%) | |||||
| Early | 15 (16) | 3 (20) | 0.046 | 5 (33) | N.s |
| Late | 79 (84) | 40 (51) | 25 (32) | ||
| Sex (%) | |||||
| Female | 38 (40) | 14 (37) | N.s | 12 (32) | N.s |
| Male | 56 (60) | 29 (52) | 18 (32) | ||
| Location (%) | |||||
| Proximal | 26 (28) | 14 (54) | N.s | 7 (27) | N.s |
| Distal | 68 (72) | 29 (43) | 23 (34) | ||
| Stage (%) | |||||
| I | 13 (14) | 5 (38) | N.s | 1 (8) | N.s |
| II | 22 (23) | 13 (59) | 7 (32) | ||
| III | 48 (51) | 21 (44) | 16 (33) | ||
| IV | 10 (11) | 4 (40) | 4 (40) | ||
Other possible explanations for the increasing incidence rate of CRC in the younger population [31] could be related to early exposures to a deleterious lifestyle, environmental pollutants, a western diet [32], diets high in sugar [33], metabolic diseases during adolescence [34] or other components of the gut microbiota, such as the genera Fusobacterium and Flavonifractor [35].
To detect the presence of ETBF among the cohort, PCR using specific primers targeting the bft gene [8] was performed. Additionally, as a positive control for the PCR reaction, primers universal for B. fragilis strains [23] were used (Fig. 1b). Bft was detected in 21% of healthy donors and 32% of CRC patients (Fig. 2, Table 1). Overall, the levels of ETBF colonization in our CRC patients were within the range previously reported from other cohorts with 6.1% in Japan [36], 31.6% [37] and 47% [38] in two Iranian cohorts, 38% in Turkey [16], 49.3% in New Zealand [39], and 60% in the USA [8]. Regarding healthy controls, prevalence in our cohort was higher than those reported in the Turkish cohort (12%) [16], and in two Iranian cohorts (3.8% and 8.3%) [37, 38], whereas higher levels were reported in a cohort from the USA (30%) [8].
Finally, double colonization with pks + bacteria and ETBF was detected in 8% of healthy individuals and 13% of CRC patients (Table 2). In a US cohort, higher levels of double colonization with pks + bacteria and ETBF were detected in the healthy population (22%), with even higher levels reported in patients with familial adenomatous polyposis (FAP) (52%) [8]. Of note, the presence of both pks + bacteria and ETBF may lead to higher colonic inflammation and tumorigenesis [8].
Table 2.
Prevalence of double colonization in controls and CRC patients
| Total (156) | clbA + /bft + | p-value | |
|---|---|---|---|
| Cohort (%) | |||
| Controls | 62 (40) | 5 (8) | N.s |
| CRC | 94 (60) | 12 (13) | |
| Controls | |||
| Sex (%) | |||
| Female | 33 (53) | 1 (3) | N.s |
| Male | 29 (47) | 4 (14) | |
| CRC | |||
| Onset (%) | |||
| Early | 15 (16) | 0 (0) | N.s |
| Late | 79 (84) | 12 (15) | |
| Sex (%) | |||
| Female | 38 (40) | 2 (5) | N.s |
| Male | 56 (60) | 10 (18) | |
| Location (%) | |||
| Proximal | 26 (28) | 3 (12) | N.s |
| Distal | 68 (72) | 9 (13) | |
| Stage (%) | |||
| I | 13 (14) | 0 (0) | N.s |
| II | 22 (23) | 6 (27) | |
| III | 48 (51) | 4 (8) | |
| IV | 10 (11) | 2 (20) | |
Conclusion
The prevalence of colibactin-producing bacteria and ETBF in CRC patients from our cohort was within the range reported in other studies. Nevertheless, we found that healthy controls had higher prevalence of pks + bacteria and ETBF when compared to most of the other cohorts. However, when comparing different reports, it should be taken into account that the type of tissue (mucosal vs. fecal samples) and measurement techniques (cultured vs. direct PCR) used to determine the prevalence of pro-carcinogenic bacteria may account for some of the variations between reported results. In any case, as these healthy individuals may be at a higher risk of developing CRC due to the potentially elevated levels of pks + bacteria and ETBF, it is critical to propose adapted dietary and medical interventions to regulate the abundance of these bacteria. A novel result of our study is the finding of a low prevalence of pks + bacteria in early-onset compared to late-onset CRC. Further studies are needed to understand the role of colibactin-bacteria in early-onset CRC.
Supplementary Information
Additional file 1: Table S1. Demographic and clinical characteristics of the cohort.
Acknowledgements
Banking was done in collaboration with the Réseau de Recherche sur le cancer (Fonds de la Recherche du Québec—Santé) that is affiliated with the Canadian Tumor Repository Network (CTRNet). We also thank Kishanda Vyboh for her help in editing the manuscript.
Abbreviations
- ApcMin/+
Adenomatous polyposis coli multiple intestinal neoplasia
- Bft
Bacteroides fragilis toxin
- Clb
Colibactin
- clbA
colibactin A gene
- CRC
Colorectal cancer
- DSBs
Double-strand DNA breaks
- ETBF
Enterotoxigenic Bacteroides fragilis
- FAP
Familial adenomatous polyposis
- IBD
Inflammatory bowel disease
- ICLs
Inter-strand cross links
- PCR
Polymerase chain reaction
- Pks
Polyketide synthase
Author contributions
The work reported in the paper was performed by the authors. MO and RH contributed to the investigation. MO, RH, TC, GF, AC, FD, RL, HS, FS, RW, RR, and EB contributed to the experiments. MO and MMS contributed to conceptualization, validation, formal analysis, data visualization and writing/reviewing the original draft. CSR and MMS contributed to the study's supervision, resources, and funding acquisition. All authors have read and approved the final manuscript.
Funding
This work was supported by grants from the Canadian Institutes of Health Research [CIHR, grant PJT-159775]. MO and TC are the recipients of the Canderel scholarship from the Institut du cancer de Montréal; RH received a scholarship from the Fonds de recherche du Québec-Santé [FRQ-S]/ Ministère de la Santé et des Services sociaux [MSSS; Resident Physician Health Research Career Training Program].
Availability of data and materials
The authors declare the data used to support the findings of this study are available from the corresponding author upon request.
Declarations
Ethical approval and consent to participate
The Research Ethics Board approved this study at the CHUM (Study numbers: 19.021, 21.153 and 21.368). Patients were asked to participate, and written consent was obtained from every patient who agreed to participate.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158(2):322–40. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2(2):123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 4.Homburg S, Oswald E, Hacker J, Dobrindt U. Expression analysis of the colibactin gene cluster coding for a novel polyketide in Escherichia coli. FEMS Microbiol Lett. 2007;275(2):255–62. doi: 10.1111/j.1574-6968.2007.00889.x. [DOI] [PubMed] [Google Scholar]
- 5.Xue M, Kim CS, Healy AR, Wernke KM, Wang Z, Frischling MC, et al. Structure elucidation of colibactin and its DNA cross-links. Science. 2019;365(6457):eaax2685. doi: 10.1126/science.aax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Secher T, Samba-Louaka A, Oswald E, Nougayrède J-P. Escherichia coli producing colibactin triggers premature and transmissible senescence in mammalian cells. PLOS ONE. 2013;8(10):e77157. doi: 10.1371/journal.pone.0077157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–3. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592–7. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopès A, Billard E, Casse AH, Villéger R, Veziant J, Roche G, et al. Colibactin-positive Escherichia coli induce a procarcinogenic immune environment leading to immunotherapy resistance in colorectal cancer. Int J Cancer. 2020;146(11):3147–3159. doi: 10.1002/ijc.32920. [DOI] [PubMed] [Google Scholar]
- 10.Putze J, Hennequin C, Nougayrede JP, Zhang W, Homburg S, Karch H, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun. 2009;77(11):4696–703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strakova N, Korena K, Karpiskova R. Klebsiella pneumoniae producing bacterial toxin colibactin as a risk of colorectal cancer development—a systematic review. Toxicon. 2021;197:126–35. doi: 10.1016/j.toxicon.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Tsunematsu Y, Hosomi K, Kunisawa J, Sato M, Shibuya N, Saito E, et al. Mother-to-infant transmission of the carcinogenic colibactin-producing bacteria. BMC Microbiol. 2021;21(1):235. doi: 10.1186/s12866-021-02292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8(3):207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 14.Stoppe NC, Silva JS, Carlos C, Sato MIZ, Saraiva AM, Ottoboni LMM, et al. Worldwide phylogenetic group patterns of Escherichia coli from commensal human and wastewater treatment plant isolates. Front Microbiol. 2017;8:2512. doi: 10.3389/fmicb.2017.02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22(2):349–69. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulger Toprak N, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12(8):782–6. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim JM, Lee JY, Kim Y-J. Inhibition of apoptosis in Bacteroides fragilis enterotoxin-stimulated intestinal epithelial cells through the induction of c-IAP-2. Eur J Immunol. 2008;38(8):2190–2199. doi: 10.1002/eji.200838191. [DOI] [PubMed] [Google Scholar]
- 18.Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124(2):392–400. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- 19.Hwang S, Lee CG, Jo M, Park CO, Gwon S-Y, Hwang S, et al. Enterotoxigenic Bacteroides fragilis infection exacerbates tumorigenesis in AOM/DSS mouse model. Int J Med Sci. 2020;17(2):145–52. doi: 10.7150/ijms.38371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–22. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.INRA. IHMS. 2015. http://www.human-microbiome.org/. Accessed 12 may 2021.
- 22.Gao W, Zhang W, Meldrum DR. RT-qPCR based quantitative analysis of gene expression in single bacterial cells. J Microbiol Methods. 2011;85(3):221–7. doi: 10.1016/j.mimet.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Song Y, McTeague M, Vu AW, Wexler H, Finegold SM. Rapid identification of the species of the Bacteroides fragilis group by multiplex PCR assays using group- and species-specific primers. FEMS Microbiol Lett. 2003;222(1):9–16. doi: 10.1016/S0378-1097(03)00296-9. [DOI] [PubMed] [Google Scholar]
- 24.Eklof V, Lofgren-Burstrom A, Zingmark C, Edin S, Larsson P, Karling P, et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141(12):2528–36. doi: 10.1002/ijc.31011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimpoh T, Hirata Y, Ihara S, Suzuki N, Kinoshita H, Hayakawa Y, et al. Prevalence of pks-positive Escherichia coli in Japanese patients with or without colorectal cancer. Gut Pathog. 2017;9:35. doi: 10.1186/s13099-017-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nouri R, Hasani A, Masnadi Shirazi K, Alivand MR, Sepehri B, Sotoudeh S, et al. Mucosa-associated <i>Escherichia coli</i> in colorectal cancer patients and control subjects: variations in the prevalence and attributing features. Can J Infect Dis Med Microbiol. 2021;2021:2131787. doi: 10.1155/2021/2131787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyadorai T, Mariappan V, Vellasamy KM, Wanyiri JW, Roslani AC, Lee GK, et al. Prevalence and association of pks+ Escherichia coli with colorectal cancer in patients at the University Malaya Medical Centre, Malaysia. PLoS ONE. 2020;15(1):e0228217. doi: 10.1371/journal.pone.0228217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vernia F, Longo S, Stefanelli G, Viscido A, Latella G. Dietary factors modulating colorectal carcinogenesis. Nutrients. 2021;13(1):143. doi: 10.3390/nu13010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arima K, Zhong R, Ugai T, Zhao M, Haruki K, Akimoto N, et al. Western-style diet, pks Island-carrying Escherichia coli, and colorectal cancer: analyses from two large prospective cohort studies. Gastroenterology. 2022;163(4):862–74. doi: 10.1053/j.gastro.2022.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee-Six H, Olafsson S, Ellis P, Osborne RJ, Sanders MA, Moore L, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574(7779):532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 31.O’Sullivan DE, Sutherland RL, Town S, Chow K, Fan J, Forbes N, et al. Risk Factors for Early-Onset Colorectal Cancer: A Systematic Review and Meta-analysis. Clinical Gastroenterology and Hepatology. 2022;20(6):1229-40.e5. https://www.sciencedirect.com/science/article/pii/S1542356521000872 [DOI] [PubMed]
- 32.Puzzono M, Mannucci A, Grannò S, Zuppardo RA, Galli A, Danese S, et al. The role of diet and lifestyle in early-onset colorectal cancer: a systematic review. Cancers (Basel) 2021;13(23):5933. doi: 10.3390/cancers13235933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hur J, Otegbeye E, Joh H-K, Nimptsch K, Ng K, Ogino S, et al. Sugar-sweetened beverage intake in adulthood and adolescence and risk of early-onset colorectal cancer among women. Gut. 2021;70(12):2330. doi: 10.1136/gutjnl-2020-323450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinicrope FA. Increasing incidence of early-onset colorectal cancer. N Engl J Med. 2022;386(16):1547–1558. doi: 10.1056/NEJMra2200869. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Du L, Shi D, Kong C, Liu J, Liu G, et al. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat Commun. 2021;12(1):6757. doi: 10.1038/s41467-021-27112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumiya Y, Suenaga M, Ishikawa T, Hanaoka M, Iwata N, Masuda T, et al. Clinical significance of Bacteroides fragilis as potential prognostic factor in colorectal cancer patients. J Clin Oncol. 2022;40:137. doi: 10.1200/JCO.2022.40.4_suppl.137. [DOI] [Google Scholar]
- 37.Haghi F, Goli E, Mirzaei B, Zeighami H. The association between fecal enterotoxigenic B. fragilis with colorectal cancer. BMC cancer. 2019;19(1):879. doi: 10.1186/s12885-019-6115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamani S, Taslimi R, Sarabi A, Jasemi S, Sechi LA, Feizabadi MM. Enterotoxigenic Bacteroides fragilis: a possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front Cell Infect Microbiol. 2020;9:449. doi: 10.3389/fcimb.2019.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell RV, Pearson J, Aitchison A, Dixon L, Frizelle FA, Keenan JI. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017;12(2):e0171602. doi: 10.1371/journal.pone.0171602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Demographic and clinical characteristics of the cohort.
Data Availability Statement
The authors declare the data used to support the findings of this study are available from the corresponding author upon request.


