Abstract
Microfluidic diagnostic (μDX) technologies miniaturize sensors and actuators to the length-scales that are relevant to biology: the micrometer scale to interact with cells and the nanometer scale to interrogate biology’s molecular machinery. This miniaturization allows measurements of biomarkers of disease (cells, nanoscale vesicles, molecules) in clinical samples that are not detectable using conventional technologies. There has been steady progress in the field over the last three decades, and a recent burst of activity catalyzed by the COVID-19 pandemic. In this time, an impressive and ever-growing set of technologies have been successfully validated in their ability to measure biomarkers in clinical samples, such as blood and urine, with sensitivity and specificity not possible using conventional tests. Despite our field’s many accomplishments to date, very few of these technologies have been successfully commercialized and brought to clinical use where they can fulfill their promise to improve medical care. In this paper, we identify three major technological trends in our field that we believe will allow the next generation of μDx to have a major impact on the practice of medicine, and which present major opportunities for those entering the field from outside disciplines: 1. The combination of next generation, highly multiplexed μDx technologies with machine learning to allow complex patterns of multiple biomarkers to be decoded to inform clinical decision points, for which conventional biomarkers do not necessarily exist. 2. The use of micro/nano devices to overcome the limits of binding affinity in complex backgrounds in both the detection of sparse soluble proteins and nucleic acids in blood and rare circulating extracellular vesicles. 3. A suite of recent technologies that obviate the manual pre-processing and post-processing of samples before they are measured on a μDX chip. Additionally, we discuss economic and regulatory challenges that have stymied μDx translation to the clinic, and highlight strategies for successfully navigating this challenging space.
Graphical Abstract
Introduction
The history of microfluidic diagnostic (μDx) chips has striking parallels to the development of the complex microelectronic circuits that underpin the information age we live in today. The invention of the solid-state transistor was followed by decades of relentless innovation seeking to shrink the size of transistors and integrate billions of them into a single chip, resulting in versatile, inexpensive microprocessors and an exponential growth in computational power over time. In the 1990s, researchers embarked upon an analogous path to revolutionize the tools that we use to interact with biological systems and to diagnose and guide the treatment of disease. The pioneers of this field envisioned that sensors and actuators miniaturized to the relevant length scales of biology - the micrometer scale of cells, and the nanometer scale of biology’s molecular machinery - could offer dramatic advantages for sample manipulation and analysis compared to bulk techniques.1 Furthermore, just as electronic circuits could be densely integrated onto silicon chips, these miniaturized sensors and actuators could be interconnected using microscale fluidic channels to create complex “lab-on-a-chip” μDx systems.2 A particular goal of the μDx field was to make powerful medical diagnostics as ubiquitous as consumer electronics, fundamentally changing the way that we monitor and maintain our health.
In the 30 years since this field began, there has been a global effort to realize this vision, involving leaders in academia, industry, and national labs and leveraging advances in microfluidics, electronics, nanomaterials, photonics, and microelectromechanical systems (MEMS).3,4 From an engineering perspective, we are at an exciting stage marked by an acceleration in the capabilities of new μDx devices to detect, sort, and quantify rare and clinically useful biomarkers from complex samples.5-19 The progress has been all the more astonishing given the relative youth of the field, the immense challenges posed by the complexity of biological systems, and the diverse and multitudinous background of material present in patient samples, such as blood, from which biomarkers must be measured. In the last few years, μDx technology has begun to reach the level of maturity for commercialization, regulatory approval, and practical clinical use (Table 1). Amidst these encouraging signs, it must be noted that μDx technologies have often faced significant barriers to commercialization and, compared to the complete permeation of consumer electronics into everyday life, μDx devices have not yet transformed medicine in the way envisioned by the founders of the field.
Table. 1.
Selected products on the market or in late stage of development. This list is not intended to be comprehensive, but rather it demonstrates several key technologies that have successfully matured to the point of either being used, or close to being used, in the clinic. POC indicates whether the instrument can be used at the point of patient care.
| Product | Lead manufacturer | Technology | Disease | POC | Stage |
|---|---|---|---|---|---|
| Cellsearch | Menarini Silicon Biosystems | Immunostain | Cancer | No | Approved |
| Claros1 | Opko Diagnostics | Microfluidics | PSA | Yes | Approved |
| FilmArray | BioFire | PCR | Resp. infection | Yes | CLIA waived |
| GeneXpert | Cepheid | PCR | TB, others | No | FDA cleared for extragenital testing for chlamydia and gonorrhea. Authorized for COVID |
| GenieIII | Optigene | PCR | Yes | ||
| Prosigna | Nanostring | nCounter gene expression | Breast cancer | Yes | Approved |
| Simoa | Quanterix | Bead based digital imumunodetection | Many | No | FDA Breakthrough status for Alzheimer’s and authorization for COVID |
| Solana, Triage | Quidel | Infection, lab tests | Yes | Approved | |
| T2 Panels | T2Biosystems | Miniaturized T2MR | Fungal infection, sepsis, COVID | Approved | |
| xMAP | Luminex | Bead based immunodetection | Infection disease | Authorized for COVID |
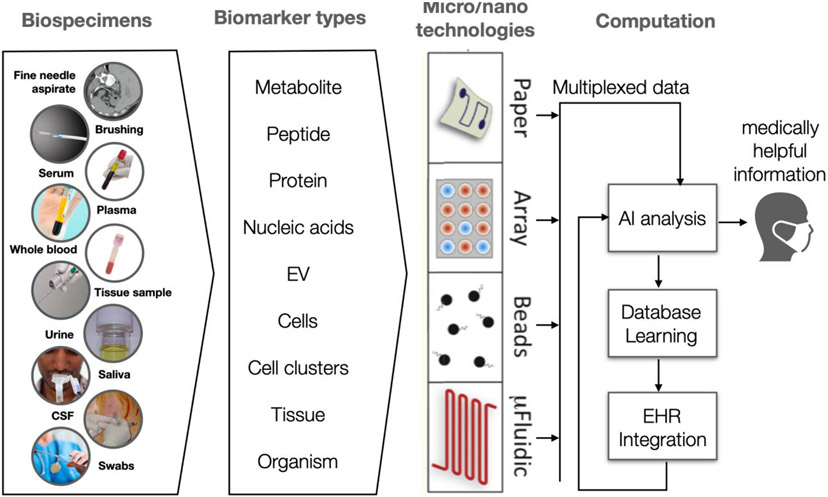
We believe that the recent advances in μDx technology represent a pivotal inflection point in its history, where μDx devices are no longer simply miniaturized versions of conventional instruments, but now have opened up new diagnostic possibilities by overcoming fundamental limitations in existing technologies.20 The latest generation of devices measure biomarkers and classify disease states that are not detectable using conventional technologies (e.g. sparse protein biomarkers for brain injury that are measured using digital ELISA21 or the early detection of pancreatic cancer using nanoscale vesicles isolated with a nanoscale device22). The emergence of these powerful technologies, along with an increased set of case studies of successful and unsuccessful attempts to commercialize μDx technology, offers hope that a μDx-driven revolution in medical practice may be imminent (Fig. 1).
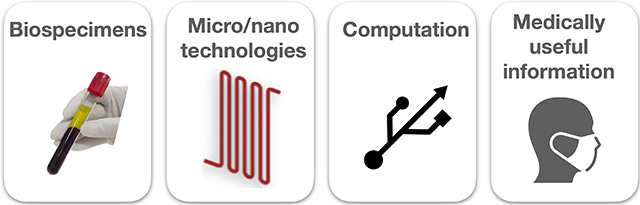
Figure 1.
A wide range of biological samples can be collected and each carry potentially valuable diagnostic information. A variety of μDx technologies can be leveraged for sensitive and specific measurements on these clinical samples to provide clinically actionable information. Multiplexed measurements made by μDX generate high dimension datasets that can be analyzed using artificial intelligence based analysis to identify patterns and extract medically useful information.
We organize this paper by trying to answer two overarching questions: 1. What barriers have kept μDx from significantly impacting the practice of medicine up to this point and 2. Which recent developments could overcome these barriers? First, we highlight recent technologies that have achieved commercial success. Subsequently, we identify three major research trends that will allow the next generation of μDx devices to have a major impact on the practice of medicine, and which present significant opportunities for those entering the fields from outside disciplines: 1. The combination of next generation, highly multiplexed μDx technologies with machine learning to allow complex patterns of multiple biomarkers to be decoded to inform a growing set of clinical decision points, for which conventional biomarkers do not necessarily exist. 2. The use of micro/nano devices to allow recent chips to overcome the limits of binding affinity in complex backgrounds in both the detection of sparse soluble proteins and nucleic acids in blood and rare circulating extracellular vesicles. 3. A suite of recent technologies that obviate the manual pre-processing and post-processing of samples before they are measured on a microfluidic chip, allowing these chips to operate directly from “sample-to-answer”. Finally, we highlight economic and regulatory challenges that have stymied μDx translation to the clinic, and highlight strategies for successfully navigating this challenging space.
The recent COVID-19 pandemic has brought particular attention to medical diagnostics, their enormous potential for improving the global responses to emerging infectious diseases, and the inadequacy that existing diagnostics had for meeting the moment. The discussion of COVID-19 diagnostics could be, and has been, a topic for an entire paper. As such, in this paper we focus our attention on the more general trends of microfluidic diagnostic chips and point the reader to several, excellent pieces that focus on COVID-19 diagnostics.23-25 This manuscript focuses on the challenges and opportunities of developing microfluidic chips for clinical applications. For a comprehensive review of state of the art in microfluidics technology, we refer the interested reader to other pieces that provide excellent summaries of the state of the field.26-31
New Technological Directions That Give μDx Significant Advantages Over Conventional Diagnostics
The combination of next generation, highly multiplexed μDx technologies with machine learning
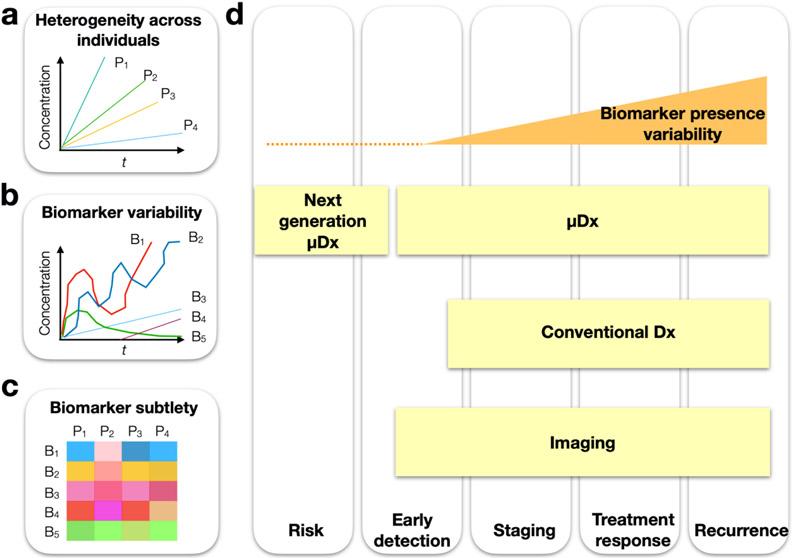
The fundamental challenges that current medical diagnostics have yet to overcome (Fig. 2) are i. the detection of subtle early disease signatures and ii. the identification of biomarkers that are insensitive to the inherent heterogeneity between individuals and their disease profile. These aspects have made it challenging to develop diagnostics with sufficient sensitivity and specificity to distinguish clinically useful states of disease (i.e does a particular patient have a version of a disease that would make them benefit most from taking drug A or drug B?). In particular, diagnostics that use measurements of a single biomarker to answer these questions face the following challenges32: i) a biomarker’s level at a single point in time may be a poor indicator of disease state, as levels can fluctuate in a patient over time, ii) the expression of a biomarker can vary between individuals, especially across diverse populations (e.g. age, gender), even if they have the same state of disease, iii) many diseases (e.g. cancers) can have heterogeneous phenotypes, which can make a single marker less reflective of the state of the disease, and iv) the limited sensitivity and specificity of current assays is not sufficient to measure some known biomarkers (e.g. EGFR over expression vs normal levels). Consequently, few individual biomarkers have had their performance successfully validated in large patient cohorts.
Figure 2.
Challenges of biological measurements can arise from biomarker heterogeneity and variability. a. An illustration of multiple individuals P1:5 with the same disease that have heterogeneous expression of a given individual biomarker. b. An illustration showing that within a given patient, individual biomarkers B1:5 can vary independently, with some having values that can fall below the noise floor. c. While individual biomarkers might show suboptimal correlation with a disease state, trends across multiple biomarkers may be more indicative and can be recognized using machine learning. d. We illustrate the stages at which next generation μDx and conventional diagnostics (Dx) and imaging have the potential to resolve disease states.
To overcome the limitations of individual markers, μDx technology development has focused on measuring panels of biomarkers to capture the state of the patient more comprehensively than would be possible with any single biomarker.33-35 Biomarker panel studies have generally shown better performance compared to single-marker measurements, since each marker is typically chosen to provide information that is as independent as possible from the others about the disease state, i.e. the selected biomarkers do not correlate strongly with one another, to avoid measuring redundant information. Additionally, multi-modal diagnostics have taken this approach a step further by combining multiple biomarkers from multiple technologies (e.g. by combining a blood based biomarker with imaging).36 In addition to improving diagnostics for disease states that already have biomarkers, biomarker panels can potentially create biomarkers for disease states for which it has not previously been possible to pair with relevant circulating biomarkers.32 In some cases, the relevant biomarkers may be expressed at levels too low to detect; in other cases there might not exist individual markers that specifically map to the relevant states of the diseases to suitably guide treatment. Despite these discoveries, validating these biomarker panels in large patient cohorts has been a challenge as this is often prohibitively expensive at early development stages37.
In μDx assay systems, microtechnology can be leveraged to expand the size of the biomarker panel at marginal cost.5,38 Further, the increased sensitivity of μDx decreases the volume of sample required per assay, allowing higher levels of multiplexing on a given volume of a patient sample18. Due to the potential for low-cost and minimally invasive measurements, μDx chips can also enable increased numbers of longitudinal measurements.39 Droplet microfluidics techniques, such as single cell sequencing and digital PCR, have found particular utility in multiplexing the profiling of nucleic acids often down to the single-cell level.40,41 Droplet-based multiplexing has been extended to include the profiling of proteins, as well as RNA, using DNA-barcoded antibodies.42 These assays have not been translated to clinical use yet, potentially due to cost and long run times. A number of these assays (10x Genomics, Bio-Rad ddPCR, Dolomite, Fluigent) have recently been commercialized and automated, paving the way for user-friendly instrumentation within clinical settings.
As more biomarkers are profiled and measured, analyzing and interpreting the increasingly multidimensional data to arrive at a clinical decision manually can be impractical. To this end, there has been recent work using machine learning algorithms to analyze panels of biomarkers measured by microscale chips, reducing the dimensionality of the raw data to an interpretable output that can be linked to a clinical decision (Fig. 1).7,43-45 Multiplexed biomarker panels typically generalize well outside of the discovery cohort since the variability that arises between patients and between centers can be taken into account by machine learning, resulting in more robust performance compared to single markers.46 These machine learning approaches may provide advantages in cases where disease information is contained within markers of different types, in dynamic changes of a biomarker over time, or where a single biomarker might not exist. To account for the limited number of patient samples available in early studies, work on transfer learning approaches have been explored in which panels of data measured in prior liquid biopsy data sets can be used to inform model and hyper-parameter selection and improve model fitting with limited available clinical samples.47 Machine learning as a tool for biomarker analysis is an ongoing area of research that requires those that are experts in μDx development, machine learning, biomarker discovery, and those that are on the front-lines interacting with patients to collaborate early in the development of these biomarker development strategies.
Using micro/nano devices to overcome the limits of binding affinity in complex backgrounds
Microfluidic-based diagnostics can overcome the fundamental challenge of detecting rare biological targets (e.g. cells, pathogens, vesicles, molecules) in the complex and vast background present in clinical samples such as blood. In this section we will highlight two general micro/nanoscale device approaches that have had particular success in tackling this problem: 1. Microdroplet-based digital assays that allow orders of magnitude improvements in limit of detection (LOD) compared to their conventional counterparts, 2. Micro/nano-devices that allow specific objects, such as cells or extracellular vesicles (EVs), to be isolated with high specificity, removing a large portion of background material for improved downstream measurements.
Converting conventional analog measurements of proteins and nucleic acids to digital assays
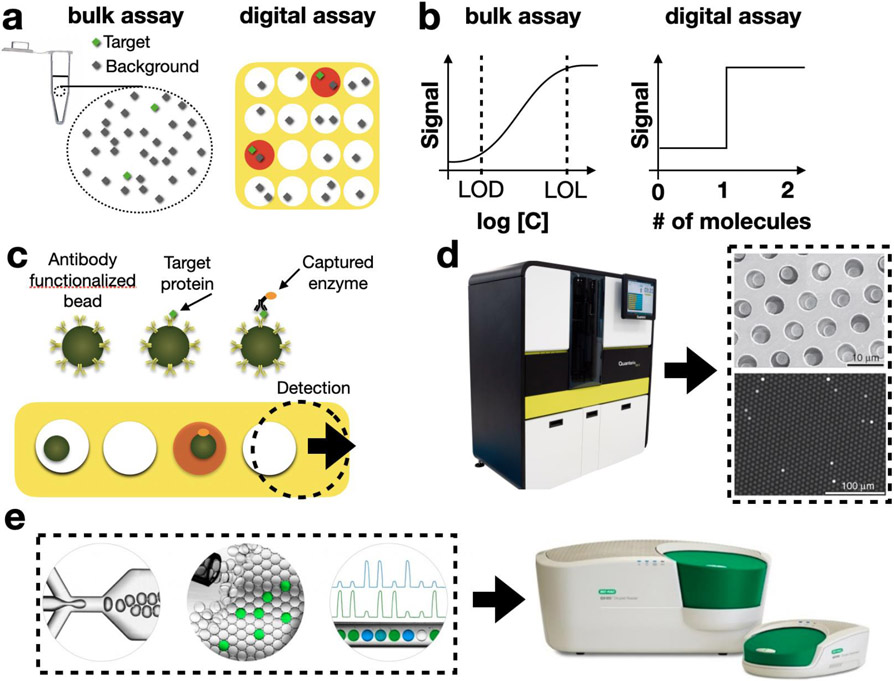
Digital assays push the sensitivity and specificity of analyte detection far beyond what is possible in bulk experiments. For example, digital ELISA (dELISA) has been able to achieve LODs on the order of 10−17 M in plasma, which contains a background of 10−3 M proteins,10,37,47 ~1,000x lower than possible using conventional ELISA. This performance is made possible by breaking the sample into many droplets or on-chip compartments, such that each individual container contains either one or zero copies of the target protein and orders of magnitude less background proteins than were present in the bulk sample.(Fig. 3a,b) Within these individual reactors, sandwich assays can be used to sequester an enzyme into compartments that contain a target protein. Digital assays have demonstrated utility as a platform for the ultrasensitive detection of proteins10,37,48 and nucleic acids49,50, as well as the analysis of single cells51,52 and single exosomes53-56. dELISA has been commercialized by Quanterix (Fig. 3c) and has found utility in several clinical applications, including traumatic brain injury and cancer diagnostics11,57,58. Digital PCR has also been commercialized (Fig. 3d)59 and achieved attogram per milliliter sensitivity and high levels of multiplexing for a broad range of targets.59 The improvement in LOD of digital assays over conventional assays allows measurement of ultrasparse clinical biomarkers below the LOD of conventional technology, enabling new potential biomarkers in applications such as traumatic brain injury, HIV, early cancer detection, and disease recurrence.11,57,60
Figure 3.
Digital Assays. a. A schematic showing a bulk assay and a digital assay. In the digital assay every compartment has one or zero copies of the target molecule (green), and the signal to background (green / black) is greater in each individual droplet than it is in the bulk. b. on curves show the signal versus concentration C for a conventional bulk assay and for a digital assay. c. A schematic of digital ELISA, in which a sandwich assay is performed on antibody functionalized beads. Any bead that has captured a target protein, captures an enzyme that makes the droplet fluoresce. d. The SIMOA system (Quanterix) uses disposable cartridges, in which the dELISA assay is performed, and a benchtop instrument for dELISA readout. e. A commercial digital droplet PCR (ddPCR) system packages droplet generation, droplet processing, and fluorescence detection together within a benchtop instrument (BioRad).
One outstanding challenge in digital assays, which μDx are well poised to solve, is that the instrumentation to generate and process the many individual micrometer-scale reactors can be cumbersome. Current commercial systems are often of large size and cost >$100k. To address this challenge, several groups have developed miniaturized devices to make digital assays more accessible. In 2009, the Ismagilov group developed Slipchip, a pump-free microfluidic platform that minimizes the instrumentation for digital assays. Their startup company, Talis Biomedical, has commercialized this technology, incorporating on-chip operations such as sample preparation, addition of reagents, mixing, target amplification and the read-out of digital assays onto their low-cost platform. The Slipchip technology has been validated for applications in protein crystallization61, immunoassays,62 multiplexed nucleic acid amplification,13 and analysis of the gut microbiome.63
In addition to solving the challenge of generating and processing droplets, μDx chips are also being developed to read out the results of the millions of individual micrometer-scale reactors for digital assays. To this end, microfluidic-based techniques have been coupled successfully with miniaturized fluorescence detection. In one promising approach from the Tang Lab at Stanford,64 a hybrid CMOS/microfluidic chip was developed to detect droplets flowing in parallel channels for high throughput detection (254,000 droplets/sec). The Issadore lab recently reported rapid (1 million droplets/sec) droplet fluorescence measurement using a mobile phone-based imaging technique, which leveraged time-domain encoding.65 Subsequently, this technique was used to demonstrate multiplexed protein measurement from raw serum, with an LOD that matched that of a refrigerator-sized dELISA instrument commercialized by Quanterix.12 There are several other platforms that have been developed that use smaller numbers (<10,000) of nanoliter wells, compared to the femtoliter wells used in the ultrasensitive systems. These simpler systems have reduced sensitivity, dynamic range, and capability for multiplexing, but can be more readily formatted as portable devices.12,61,66
Improving sensitivity and specificity by first isolating specific subtypes of biomarkers
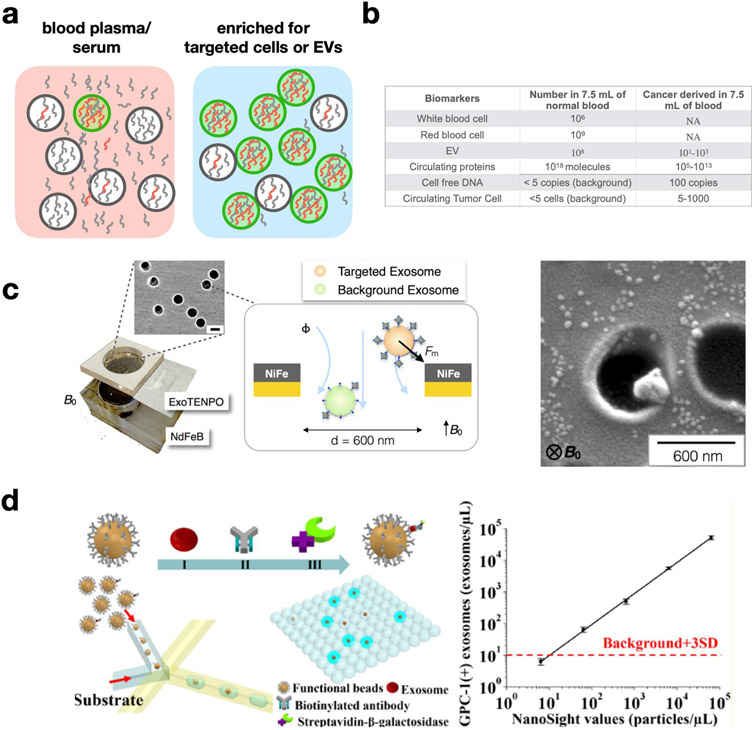
A particularly successful strategy to detect sparse biomarkers in complex samples is to pursue them within biological compartments, such as cells and EVs, where their concentration is effectively increased. (Fig. 4a,b) Cells and EVs found in blood and other fluid samples contain molecular cargo derived from their tissue of origin, due to their biogenesis. Quantifying the proteins and nucleic acids within these compartments gives insight into the molecular state of the tissue of origin without needing direct access to tissue samples. The surface marker profile of the circulating cells and EVs also reflects that of the tissue of origin, making it possible to label and isolate the compartments using affinity ligands. Moreover, cell and EV sub-types can be accurately distinguished from one another, either by quantifying the presence of a target surface molecule and comparing it to a threshold, or by using a combination of multiple surface markers.68,69
Figure 4.
μDx isolation of extracellular vesicles (EVs). a. A schematic showing how the signal-to-background of a molecular biomarker (red/grey) can be improved by first enriching for targeted vesicles (green) from the vast background of vesicles (grey) and molecules (grey) present in clinical samples. b. Order of magnitude numbers for the quantity of biomarker concentrations for the application of cancer diagnostics in 7.5 mL of blood. c. A schematic of the Track Etched Magnetic NanoPOre (TENPO) device, as well as an electron microscopy image of an immunomagnetically isolated vesicle.7 d. Droplet based technologies have recently emerged, which isolate single EVs into droplets where an enzymatic reaction, such as ELISA or PCR, can be carried out to amplify the signal from a single EV. These technologies offer the potential for single EVs to be rapidly counted and profiled.43
Micro- and nanofluidic devices, which match the size scale of the objects that they are processing, can be leveraged to isolate rare cells and EVs with much higher recovery rate and purity than is possible with conventional technologies such as centrifugation or flow cytometry.67 Inspired by the success of μDX devices for the analysis of circulating tumor cells (CTCs) in the diagnosis and monitoring of cancer, recent efforts have focused on developing analogous devices for EV capture and analysis.70 While EVs are much more abundant (in the range of 1010/mL) in blood and other patient-derived fluid samples than are CTCs (100-2/mL), EVs are much smaller (30 nm −1 μm, compared to 10 μm CTCs); furthermore, healthy patient samples contain large amounts of EVs from host cells, making the specific isolation of disease-associated EVs a major technological challenge for μDx systems.71 Nanoscale EV analysis devices based on scaling down CTC capture strategies have generally demonstrated high specificity but low sample throughput.6 Strategies such as chaotic mixing in microscale channels72 and parallelized nanopore-based immunomagnetic capture7 have been introduced recently to maintain throughput while increasing EV capture specificity and recovery rate. (Fig. 4c) Downstream PCR, RNA sequencing, Western blotting, and electrochemistry have been used to characterize the cargo of captured EVs6,9,73,74. Since these downstream techniques are susceptible to the background contributed by non-specific EVs, isolation of EV subtypes based on surface marker expression is a critical aspect of producing recognizable disease signatures using these technologies.8,73 One particularly compelling aspect of EVs is that they have been found to cross the blood-brain barrier, making μDx-based EV analysis a particularly compelling tool to diagnose brain-related diseases that do not typically have good blood-based cellular or molecular markers.75
Another rapidly growing area of interest for μDx is the analysis of EV surface markers and cargo at the single-vesicle level, in order to quantify heterogeneity within EV sub-populations and understand the effects of heterogeneity on disease state. Single-vesicle analysis based on traditional scanning-probe, fluorescence, or electron microscopy have a very limited practical throughput (104 particles) compared to the typical abundance of EVs.76-79 Very recently, a new generation of μDx technologies has emerged that uses droplet microfluidics to quantify and profile single EVs with high throughput.(Fig. 4d) In these assays, signals from scant quantities of protein on the surface of single EVs are amplified within each droplet using digital ELISA44, PCR80, or next-generation sequencing (NGS)81 into a measurable signal at high throughput. While these techniques are still in their infancy, further work on these devices could produce useful research and clinical tools to improve our understanding of EVs and their role in disease. Affinity-based EV isolation can be challenging because EVs have a 10,000x smaller surface area than cells and therefore much fewer surface proteins than cells, resulting in proportionally weaker trapping forces for methods such as immunomagnetic labeling. Moreover, next-generation μDX chips for EV isolation will not necessarily obviate established isolation techniques such as ultracentrifugation or column purification, which may continue to play a role in upstream sample pre-processing and benchmarking the performance of new assays. Furthermore, while many of the off-chip techniques used in the analysis of EV cargo have already been successfully integrated into clinical workflows, the hand-off between the on-chip isolation and the outside world is inherently slow and lossy. Further innovations in the on-chip analysis of EV cargo and their integration with microfluidic EV isolation will be a fruitful area for improvements in performance of multiplexed assays and analyses of small sample volumes.
Transitioning from a “Chip in a Lab” to a “Lab on a Chip”
One long-standing problem of micro- and nano-scale devices is that many of these chips cannot be directly used on clinical samples without access to laboratory equipment (e.g. centrifuges, microscopes, pipettes). Expert users are often required to pre-process samples, post-process the output of the chips for further analysis, or dynamically control the operation of the device based on feedback from the device (e.g. a microscope video feed of device operation). This challenge has been described as the “chip in a lab” problem because it often takes a laboratory worth of equipment to operate a “lab on a chip”. The inherent complexity of clinical samples and the sensitivity of devices with micro- or nano-scale features to failure modes such as clogging and bio-fouling have made this a formidable challenge to overcome. Another drawback brought by the “chip in a lab” problem lies in the increase in total assay times that come from having to transport the samples to specialized facilities where the chips can be operated and the samples can be pre- or post-processed.
One breakthrough that has been particularly successful in overcoming this challenge is parallelization, wherein chips incorporate many thousands of (or greater) replicate devices that operate in parallel to simultaneously increase throughput and device robustness. This strategy leverages a core benefit of μDx chips, which is that the marginal cost to add additional copies of a microfluidic device on a single chip is not significant, analogous to adding circuit elements to an integrated circuit in microelectronics. In a parallelized device, if the number of devices that fail is small compared to the total number then there is not a significant change in overall device performance, making the devices robust to clogging while keep its high throughput. This approach of parallelization has been successfully applied to flow cytometry,82 immunomagnetic sorting of cells83,84 and EVs,7 droplet generation,85,86 and on-chip digital assays12,64. In each of these applications, parallelization led to a >100x improvement in throughput compared to the operation on a single device chip.
Additionally, new device architectures and modalities that reduce the dependency of μDx devices on ancillary laboratory equipments is an active area of research. There have been several approaches to replace the use of centrifugation for sample preparation, such as using inertial flow to transfer cells and microparticles across laminar fluid streams87, magnetophoretic techniques to separate targeted cell populations from clinical samples88,89, acoustofluidics to separate targeted cells from clinical samples such as sputum90, and centrifugal microfluidics to separate pathogenic DNA91. Another major development that is helping untether microfluidic based diagnostics from laboratories and expert users are clever designs that make microfluidic devices less sensitive to flow rate, obviating the need for instrumentation such as high performance syringe pumps or microscopes. One example is the Millipede microfluidic droplet generator, which can produce droplets in a flow-rate insensitive manner.92 These droplet generators have enabled point-of-care (POC) devices that use droplet microfluidics.12 Additionally, others have used artificial intelligence to monitor and automatically control inherently unstable microfluidic processes.93,94
Alternatively, rather than pre-processing the sample to remove background, some groups have explored sensing schemes that are insensitive to background and can detect biological targets directly in unprocessed clinical samples. Magnetic nanoparticles (MNPs) can be designed to bind to specific biological entities of interest, including nucleic acids, proteins, viruses, bacteria, and cells. Targets that are magnetically labeled can be distinguished directly from the background material that confounds optical techniques, because of the intrinsically low magnetic susceptibility of biological material. This approach has been applied extensively for magnetic separation87 as well as magnetic sensing90 or a combination of both95. Magnetic sensing and sorting is particularly well suited to miniaturization and integration into monolithic chips because it obviates the need for bulky optical equipment.96-98 Moreover, the binding efficiency of MNP labeling is enhanced compared to that of individual recognition ligands, as MNPs provide multiple binding sites.95 In this area, T2 Diagnostics has commercialized NMR-based magnetic sensing and has developed an FDA-cleared rapid sepsis diagnostic being used in US and European hospitals. Additionally, Menarini Silicon Biosystems has commercialized the iChip, which removes white blood cells from suspension using magnetic sorting, negatively enriching for CTCs that can be analyzed off of the chip.15
Navigating The Economic and Regulatory Challenges of Bringing μDx to the Clinic
New technology must either be additive or disruptive to the conventional diagnostic market
The global diagnostic market is currently a 25 billion dollar industry and expected to grow to over 33 billion dollars by 2025.99 A large share of this market is in the US (~40%) and the expansion is driven by an aging population with high prevalence of chronic diseases (diabetes, heart and lung diseases, cancer). Much of current diagnostic testing is performed in central and established labs with high throughput capabilities, quality controls, and integration into electronic health care records and billing systems. One notable exception to this workflow is the POC diabetes market. The majority of existing centrally located technologies for clinical chemistry, immunoassays, histology/hematology, microbiology are well established and dominated by a few large enterprises servicing hospitals. We expect that these segments will likely not be replaced by μDx approaches in the near term. Rather, we expect that innovation and expansion in μDx technologies will first penetrate in novel application areas, which current technology does not address and where specific criteria, detailed below, are met. The largest market share in the US are currently the sale of instruments followed by reagents and then software. The services segment currently is the smallest but is expected to show rapid growth over the next few years, and is where μDx could play a significant role. It is worth noting that there already exist POC technologies used in the clinic based on non-microfluidic workflows. These tools automate the entire assay workflows to provide real-time clinical information. One example is a POC analysis system that performs a quantitative cardiac assays for a fast (< 15 minutes) multiplexed evaluation of patients presenting suspected of myocardial ischemia (Stratus® CS 200 Acute Care™ Analyzer, Siemens). To find clinical utility, new platforms have to offer either better performance for existing biomarkers, allow broader multiplexing, allow entirely new biomarkers to be measured, or be significantly less expensive.
The pathway from bench to market for μDx
The development of new biomarkers, with or without μDx technology, is challenging. One reason is that large, expensive trials are required to assess a biomarker’s utility. Another reason is that the most biomarkers do not ultimately have the required sensitivity and specificity for clinical applications, and the performance tends to drop as the study is expanded to multiple study sites.100 Additionally, new biomarkers must gain regulatory approval. The United States FDA formally established a biomarker qualification program by publishing detailed guidance to facilitate industry-academic partnerships on biomarker development and regulatory approval.98 The qualification process for biomarkers is largely based on traditional biomarker research, wherein there is a direct relationship between a single biomarker concentration and clinical outcomes. Many microfluidic companies are measuring biomarkers already approved by the FDA to avoid the burden of validating the biomarker. For example, the Sangia, Claros1, and SIMOA platforms have been used to measure the already approved PSA biomarker for prostate cancer. For more effective diagnosis or treatment on life-threatening or irreversibly debilitating disease, FDA developed the Breakthrough Devices Program to accelerate the development, assessment, and review process. A recent breakthrough from this program is the first blood test to evaluate mild traumatic brain injury. The Banyan Brain Trauma Indicator was approved in fewer than 6 months. Biomarkers selected by statistical learning algorithms have also had some success in gaining regulatory approval, for example a 70 gene panel, processed using a machine learning algorithm for breast cancer recurrence prediction, was approved by the FDA in 2007.99 Recently, Quanterix’s SIMOA phospho-Tau 181 (pTau-181) blood test has been granted Breakthrough Device designation by the U.S. FDA as an aid in diagnostic evaluation of Alzheimer’s Disease (AD).
Catering directly to patients and interest groups
Hospitals and accredited laboratories currently capture the highest share of the US in vitro diagnostics market. Interestingly, other end-users (patients, small clinics, and office labs) currently make up at least 25% of the market. It is this segment of the market that is expected to grow and μDx will likely play a role in this development. Over the last years, the field of at-home testing has boomed and players in this space include EverlyWell, 23andMe, Color Genomics, Thorne, Habit, and Blueprint, although none of these represent accepted clinical diagnostic services. Despite the excitement about some of these companies and their ability to transform the healthcare market, there have been doubts about the marketed claims and the perceived gap between their hype and their technological capabilities (e.g. the demise of Theranos). A growth area for new biomarkers is in future therapeutics which will depend on identifying patients who are most likely to respond.101,103 It is increasingly evident that payers will require evidence that drugs are indeed working in a given patient. A slew of new protein biomarker panels, exosomes, multiplexed cellular fine needle aspirate (FNA) analysis, and genomic analyses will likely play a role in this space in addition to existing analyses.
New global markets
μDx systems are well suited to play an increasingly important role in new market segments, where use of CLIA lab-based diagnostics is not appropriate. Examples include markets in China or India with large population densities and lower number of centralized labs. There is also an unprecedented opportunity in bringing robust μDx devices to rural, resource-limited settings, where there are only rudimentary facilities with chronic shortages of equipment, supply chains, and adequate training. Integrated, automated μDx devices that are simple to use are particularly attractive for these resource limited settings.
The bench to market pathway for μDx is still complex, expensive but not well capitalized
Nearly two decades after introduction of microfluidics, commercialization remains plagued by problems. One issue is that μDx devices are not typically simple to design because of sensitivity/specificity requirements, the need for high durability of reagents and antibodies, demanding passivation and fouling design requirement, among others. Furthermore, once optimized, devices need to be tested in large-scale clinical trials, all of which are expensive. It is currently estimated that the average cost of development of a diagnostic ranges from $20-100M. Given the generally low reimbursement rates for diagnostics compared to therapeutics, the appetite to privately fund such developments has been muted. While exceptions exist, such as early investments in exosome technologies at nearly $400M, the venture capital (VC) portfolio willing to fund new μDx chips remains small. Very recently, following increasingly compelling data for the minimally-invasive detection of cancer, there have been reports of renewed intense interest in the VC community.104 Large diagnostic companies are also concerned about high development costs associated with miniaturization as it is often simpler and cheaper to add on to existing platforms. Conversely, academic laboratories well versed in technology development are often engineering focused and not expert in translation. Indeed only a few hubs with proven academic engineering/translational excellence exist. To address these issues, there are several promising pathways outlined below.
Establish interdisciplinary teams through federal funding
The development of microfluidic technology currently outstrips our ability to integrate multiple microfluidic components into a working system successfully. Interdisciplinary teams that partner technical experts with clinical, biological, and business strategic thinkers are essential to developing technological solutions that can make it to the clinic. Such teams can be challenging to form due to barriers between fields and misalignment of incentives in academia. Integration of a method into clinical workflows carries much fewer academic rewards than publishing original papers. Moreover, very few engineering teams have the expertise to develop a suitable clinical study to validate their technology or the knowledge to design their chips strategically to navigate a future clinical study successfully. There is a need to encourage genuinely interdisciplinary teams comprised of engineers, biologists, clinicians, and business leaders earlier in the development process and properly incentivize those teams. Close collaboration between engineers and biologists, in particular, can offer many advantages; namely, access to new tools to interrogate biological systems for the biologists and access to a deeper understanding of the biological systems that underly clinical applications for the engineers. There is an extraordinary opportunity for federal funding agencies to support such interdisciplinary teams for clinical translation.
Outsource production and clinical trials
Given the high manufacturing and clinical trial costs in the US, international partnerships for developing μDx manufacturing and creating clinical trials are becoming increasingly attractive. Interestingly, the FDA has recognized that clinical research is becoming increasingly global and that sponsors may conduct multinational clinical studies, including domestic or foreign sites, but which can still require US patients. Sponsors may then decide to use the data that is obtained from non-IND (Investigational New Drug application) foreign sites to support marketing approval in the United States. The FDA has recently revised its guidelines to clarify acceptable documentation for the performance of such studies. Non-US markets, especially those in Asia, may be particularly attractive for certain emerging new technology platforms. Similarly, Asian VC communities have been much more open to fund new technologies that could find applications in their markets.
Outlook
We have articulated our view of the state of μDx technology and its enormous potential, as well as the formidable challenges, to leverage its unique capabilities to improve human health. We recommend that above all else that the technology development community continue its transition towards being ‘customer obsessed’ and strive to develop technology that can offer information that patients, doctors, pharmaceutical companies, and payers actually want and that can be leveraged to concretely improve healthcare. μDx technologies have enormous potential to measure biomarkers not measurable using conventional technologies. However, it remains non-trivial to develop this technology to a level of maturity where it can significantly improve patient care, be commercialized, and win regulatory approval successfully. Continued major investments are required to drive collaborative approaches that bring together engineers, physician-scientists, physicians, and business development experts for the technology to reach its full potential to revolutionize how diseases are treated and health is maintained.
Acknowledgments
We are grateful to Heiner Dreismann, former CEO and President of Roche Molecular Systems, and Josh Buser, Vice President of Technology Development at Chip Diagnostics, for reading this paper and providing their expert feedback. Funding was provided by the New Jersey Commission on Brain Injury Research (CSCR14IRG005), Allen Foundation, Health Research Formula Fund 4100077073 from Commonwealth of Pennsylvania, the Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program, the National Institute of Health 1R21CA182336-01A1, American Cancer Society-CEOs Against Cancer-CA Division Research Scholar Grant, (RSG-15-227-01-CSM), the National Science Foundation's CAREER Award (#1554200), The Hartwell Individual Research Award, and The Congressionally Directed Medical Research Program award (W81XWH-19-2-0002).
Works Cited
- 1.Tay FEH Microfluidics and BioMEMS applications (Springer, 2002). [Google Scholar]
- 2.Harrison DJ et al. Science, 1993, 261, 895–897. [DOI] [PubMed] [Google Scholar]
- 3.Lee H, Ham D & Westervelt RM CMOS biotechnology (Springer, 2007). [Google Scholar]
- 4.Issadore D & Westervelt RM Point-of-care Diagnostics on a Chip (Springer Science \& Business Media, 2013). [Google Scholar]
- 5.Im H et al. Nature biotechnology, 2014, 32, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanwar SS, Dunlay CJ, Simeone DM & Nagrath S. Lab on a Chip, 2014, 14, 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko J et al. ACS nano, 2017, 11, 11182–11193. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y et al. ACS applied materials & interfaces, 2018, 10, 43375–43386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang P et al. Nature biomedical engineering, 2019, 3, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rissin DM & Walt DR. Nano letters, 2006, 6, 520–523. [DOI] [PubMed] [Google Scholar]
- 11.Schubert SM et al. Scientific reports, 2015, 5, 11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yelleswarapu V et al. Proceedings of the National Academy of Sciences, 2019, 116, 4489–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen F et al. Analytical chemistry, 2010, 82, 4606–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giedt RJ et al. Nature communications, 2018, 9, 4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozkumur E et al. Science translational medicine, 2013, 5, 179ra47–179ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalinich M et al. Proceedings of the National Academy of Sciences, 2017, 114, 1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Dougan TJ, and Walt DR, ACS Nano, 2022, 16, 1, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, et al. Nature Biomedical Engineering, 2022, 6,108–117. [DOI] [PubMed] [Google Scholar]
- 19.Ko J, et al. , ACS Nano, 2021, 15.3, 5631–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin CD, Linder V & Sia SK. Lab on a Chip, 2012, 12, 2118–2134. [DOI] [PubMed] [Google Scholar]
- 21.Korley FK et al. Journal of neurotrauma, 2018, 36, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z et al. Clinical Cancer Research, 2020, [Google Scholar]
- 23.Weissleder R, Lee H, Ko J & Pittet MJ. Sci Transl Med, 2020, 12, [DOI] [PubMed] [Google Scholar]
- 24.Allam M et al. Diagnostics (Basel), 2020, 10, [Google Scholar]
- 25.Vandenberg O, Martiny D, Rochas O, van Belkum A & Kozlakidis Z. Nat Rev Microbiol, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sackmann EK, Fulton AL, & Beebe DJ. Nature, 2014, 507.7491, 181–189. [DOI] [PubMed] [Google Scholar]
- 27.Berlanda SF, Breitfeld M, Dietsche CL, & Dittrich PS, Analytical chemistry, 2020, 93, 311–331. [DOI] [PubMed] [Google Scholar]
- 28.Battat S, Weitz DA, and Whitesides GM. Lab on a Chip, 2022, 22, 530–536. [DOI] [PubMed] [Google Scholar]
- 29.Chin CD, Linder V, & Sia SK. Lab on a Chip, 2012, 12.12, 2118–2134. [DOI] [PubMed] [Google Scholar]
- 30.Sachdeva S, Davis RW, & Saha AK. Frontiers in Bioengineering and Biotechnology, 2021, 1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee WG, et al. Advanced drug delivery reviews, 2010, 62.4-5, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selleck MJ, Senthil M & Wall NR. Biomarker insights, 2017, 12, 1177271917715236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yurkovetsky ZR, Linkov FY, E Malehorn D & Lokshin AE. Future Medicine,2,6, 2006. [DOI] [PubMed] [Google Scholar]
- 34.Scher HI et al. Journal of clinical oncology, 33, 1348, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko J et al. Lab on a Chip, 18, 3617–3630, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shende P, Augustine S, Prabhakar B & Gaud RS. Expert review of molecular diagnostics, 2019, 19, 409–417. [DOI] [PubMed] [Google Scholar]
- 37.Catenacci DVT. Molecular oncology, 2015, 9, 967–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rissin DM et al. Nature biotechnology, 2010, 28, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazzeri M, Guazzoni G & Montorsi F 83–90 (Elsevier, 2016). [Google Scholar]
- 40.Kang HM et al. Nature biotechnology, 2018, 36, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macosko EZ et al. Cell, 2015, 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gierahn TM et al. Nature methods, 2017, 14, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grissa D et al. Frontiers in molecular biosciences, 2016, 3, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo X et al. Nature Electronics, 2021, 4, 615–624. [Google Scholar]
- 45.Kuypers S et al. Small, 2021, 17, 2006786. [DOI] [PubMed] [Google Scholar]
- 46.Shadfan BH et al. Cancer Prevention Research, 2015, 8, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen H, et al. Lab on a Chip, 2020, 20.12, 2166–2174. [DOI] [PubMed] [Google Scholar]
- 48.Leirs K et al. Analytical chemistry, 2016, 88, 8450–8458. [DOI] [PubMed] [Google Scholar]
- 49.Pekin D et al. Lab on a chip, 2011, 11, 2156–2166. [DOI] [PubMed] [Google Scholar]
- 50.Hindson CM et al. Nature methods, 2013, 10, 1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shembekar N, Hu H, Eustace D & Merten CA. Cell reports, 2018, 22, 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eastburn DJ, Sciambi A & Abate AR. Analytical chemistry, 2013, 85, 8016–8021. [DOI] [PubMed] [Google Scholar]
- 53.Liu C et al. Nano letters, 2018, 18, 4226–4232. [DOI] [PubMed] [Google Scholar]
- 54.Yang KS et al. Gastroenterology, 2021, 160, 1345–1358. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoh KE et al. Journal of Immunological Methods, 2021, 490, 112936. [DOI] [PubMed] [Google Scholar]
- 56.Wu F et al. Translational Lung Cancer Research, 2021, 10, 2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shahim P et al. JAMA neurology, 2014, 71, 684–692. [DOI] [PubMed] [Google Scholar]
- 58.Beard K et al. Brain communications, 2021, 3, fcab151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baker M, Nature Methods, 2012, 9.6, 541–544. [DOI] [PubMed] [Google Scholar]
- 60.Passaes CPB et al. Journal of virology, 2017, 91, e02296–16.28077644 [Google Scholar]
- 61.Li L, Du W & Ismagilov RF. Journal of the American Chemical Society, 2009, 132, 112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W, Chen D, Du W, Nichols KP & Ismagilov RF. Analytical chemistry, 2010, 82, 3276–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma L et al. Proceedings of the National Academy of Sciences, 2014, 111, 9768–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim M et al. Lab on a Chip, 2015, 15, 1417–1423. [DOI] [PubMed] [Google Scholar]
- 65.Yelleswarapu VR, Jeong H-H, Yadavali S & Issadore D. Lab on a Chip, 2017, 17, 1083–1094. [DOI] [PubMed] [Google Scholar]
- 66.Hatch AC et al. Lab on a chip, 2011, 11, 3838–3845. [DOI] [PubMed] [Google Scholar]
- 67.Shields IV, Wyatt C, Reyes CD & L{\’o}pez GP. Lab on a Chip, 2015, 15, 1230–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lei C et al. Biomedical optics express, 2016, 7, 2703–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou M, Weber SR, Zhao Y, Chen H & Sundstrom JM 23–38 (Elsevier, 2020). [Google Scholar]
- 70.Shao H et al. Chemical reviews, 2018, 118, 1917–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnsen KB, Gudbergsson JM, Andresen TL & Simonsen JB. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 2018, [DOI] [PubMed] [Google Scholar]
- 72.Reategui E et al. Nature communications, 2018, 9, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park J et al. Nature Biomedical Engineering, 2021, 5, 678–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ko J et al. Cancer research, 2018, 78, 3688–3697. [DOI] [PubMed] [Google Scholar]
- 75.Ko J, Carpenter E & Issadore D. Analyst, 2016, 141, 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beekman P, et al. Lab Chip, 2019,19, doi: 10.1039/c9lc00081j. [DOI] [PubMed] [Google Scholar]
- 77.Lee K, et al. ACS Nano, 2018, 12, doi: 10.1021/acsnano.7b07060. [DOI] [PubMed] [Google Scholar]
- 78.Carney RP, et al. Anal. Chem, 2017, 89, doi: 10.1021/acs.analchem.7b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith ZJ, et al. , J. Extracell. Vesicles, 2015, 4, doi: 10.3402/jev.v4.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ko J et al. Advanced Biosystems, 2020, 1900307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ko J, Wang Y, Sheng K, Weitz DA & Weissleder R. ACS nano, 2021, 15, 5631–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McKenna BK, Evans JG, Cheung MC & Ehrlich DJ. Nature methods, 2011, 8, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yung CW, Fiering J, Mueller AJ & Ingber DE. Lab on a Chip, 2009, 9, 1171–1177. [DOI] [PubMed] [Google Scholar]
- 84.Ko J et al. Lab on a chip, 2017, 17, 3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nisisako T & Torii T. Lab on a Chip, 2008, 8, 287–293. [DOI] [PubMed] [Google Scholar]
- 86.Jeong H-H, Yelleswarapu VR, Yadavali S, Issadore D & Lee D. Lab on a Chip, 2015, 15, 4387–4392. [DOI] [PubMed] [Google Scholar]
- 87.Gossett DR et al. Small, 2012, 8, 2757–2764. [DOI] [PubMed] [Google Scholar]
- 88.Adams JD, Kim U & Soh HT. Proceedings of the National Academy of Sciences, 2008, 105, 18165–18170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia N et al. Biomedical microdevices, 2006, 8, 299. [DOI] [PubMed] [Google Scholar]
- 90.Li S et al. Analytical chemistry, 2016, 88, 5655–5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cho Y-K et al. Lab on a Chip, 2007, 7, 565–573. [DOI] [PubMed] [Google Scholar]
- 92.Stolovicki E, Ziblat R & Weitz DA. Lab on a Chip, 2018, 18, 132–138. [DOI] [PubMed] [Google Scholar]
- 93.Dressler OJ, Howes PD, Choo J & deMello AJ. ACS Omega, 2018, 3, 10084–10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perez JM, Josephson L, O’Loughlin T, H{\”o}gemann D & Weissleder R. Nature biotechnology, 2002, 20, 816. [DOI] [PubMed] [Google Scholar]
- 95.Haun JB et al. Science translational medicine, 2011, 3, 71ra16–71ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin H-Y et al. ACS nano, 2017, 11, 10062–10069. [DOI] [PubMed] [Google Scholar]
- 97.Issadore D et al. Lab on a Chip, 2011, 11, 2282–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gaster RS, Hall DA & Wang SX. Lab on a Chip, 2011, 11, 950–956. [DOI] [PubMed] [Google Scholar]
- 99.prnewswire. [Google Scholar]
- 100.Mandrekar SJ & Sargent DJ. Journal of Clinical Oncology, 2009, 27, 4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Amur S, LaVange L, Zineh I, Buckman-Garner S & Woodcock J. Clinical Pharmacology & Therapeutics, 2015, 98, 34–46. [DOI] [PubMed] [Google Scholar]
- 102.Van’t Veer LJ et al. nature, 2002, 415, 530. [DOI] [PubMed] [Google Scholar]
- 103.Emens LA, Butterfield LH, Hodi FS, Marincola FM & Kaufman HL. Journal for immunotherapy of cancer, 2016, 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sheridan C. Nature biotechnology, 2019, 37, 972. [DOI] [PubMed] [Google Scholar]