Abstract
Background
Myocardial infarction (MI) is characterized by the emergence of dead or dying cardiomyocytes and excessive immune cell infiltration after coronary vessel occlusion. However, the complex transcriptional profile, pathways, cellular interactome, and transcriptional regulators of immune subpopulations after MI remain elusive.
Methods and Results
Here, male C57BL/6 mice were subjected to MI surgery and monitored for 1 day and 7 days, or sham surgery for 7 days, then cardiac CD45‐positive immune cells were collected for single‐cell RNA sequencing to determine immune heterogeneity. A total of 30 135 CD45+ immune cells were partitioned into macrophages, monocytes, neutrophils, dendritic cells, and T or B cells for further analysis. We showed that macrophages enriched for Olr1 and differentially expressed Gpnmb represented 2 crucial ischemia‐associated macrophages with distinct proinflammatory and prophagocytic capabilities. In contrast to the proinflammatory subset of macrophages enriched for Olr1, Gpnmb‐positive macrophages exhibited higher phagocytosis and fatty acid oxidation preference, which could be abolished by etomoxir treatment. In addition to macrophages, MI triggered prompt recruitment of neutrophils into murine hearts, which constituted the sequential cell‐fate from naïve S100a4‐positive, to activated Sell‐high, to aging Icam1‐high neutrophils. In silico tools predicted that the excessively expanded neutrophils at 1 day were attributed to chemokine C‐C motif ligand/chemokine C‐X‐C motif ligand pathways, whereas CD80/inducible T‐cell costimulator (ICOS) signaling was responsible for the immunosuppressive response at day 7 after MI. Finally, the Fos/AP‐1 (activator protein 1) regulon was identified as the critical regulator of proinflammatory responses, which was significantly activated in patients with dilated cardiomyopathy and ischemic cardiomyopathy. We showed the enriched Fos/AP‐1 target gene loci in genome‐wide association study signals for coronary artery diseases and MI. Targeting Fos/AP‐1 with the selective inhibitor T5224 blunted leukocyte infiltration and alleviated cardiac dysfunction in the preclinical murine MI model.
Conclusions
Taken together, this single‐cell RNA sequencing data lay the groundwork for the understanding of immune cell heterogeneity and dynamics in murine ischemic hearts. Moreover, Fos/AP‐1 inhibition mitigates inflammatory responses and cardiac dysfunction, which might provide potential therapeutic benefits for heart failure intervention after MI.
Keywords: Fos/AP‐1, immune cell, macrophage, myocardial infarction, single‐cell RNA sequencing
Subject Categories: Myocardial Infarction, Remodeling, Inflammation
Nonstandard Abbreviations and Acronyms
- CCL
chemokine C‐C motif ligand
- CXCL
chemokine C‐X‐C motif ligand
- FAO
fatty acid oxidation
- PB
peripheral blood
- SASP
senescence‐associated secretory phenotype
Clinical Perspective.
What Is New?
Using single‐cell RNA sequencing of CD45+ leukocytes after myocardial infarction (MI), we demonstrated the dynamic influx of immune cells under ischemia and showed 2 ischemia‐associated macrophage subsets that were infiltrated at 1 and 7 days after myocardial infarctionwith discrete inflammatory effects and fuel preference.
The Fos/AP‐1 (activator protein 1) regulon was identified as the critical regulator of proinflammatory responses, which was significantly activated in patients with dilated cardiomyopathy and ischemic cardiomyopathy. Moreover, Fos/AP‐1 inhibition attenuated cardiac leukocyte infiltration and cardiac adverse remodeling.
What Are the Clinical Implications?
Immune cells coordinate cardiac healing processes after MI and play critical role in the pathology of MI; therefore, deciphering the diverse dynamics and functional patterns of specific immune clusters provides clues for precise targeting and yields therapeutic benefits in the treatment of MI.
Fos/AP‐1 acted as a key regulator of cardiac proinflammatory responses and inactivated Fos/AP‐1 signaling provide potential strategic insights for clinical MI intervention and heart failure prevention.
Myocardial infarction (MI), initiated by sudden and severe occlusion of coronary vessels, is the leading cause of mortality worldwide. 1 After ischemia, dynamic cell cascades ensue to limit tissue injury and promote healing. 2 Immune cells coordinate cardiomyocyte responses by scavenging dead materials 3 or non‐cardiomyocyte responses by activating fibroblasts and endothelial cells to promote scar formation and angiogenesis, 4 highlighting the critical role of immune cells in the pathology of MI.
Acute ischemic injury triggers the dynamic influx of innate immune cells, including mast cells, macrophages, neutrophils, and natural killer (NK) cells in the inflammatory phase (1–3 days post MI), and adaptive immune cells (T and B cells) in the reparative phase (3–7 days post MI). 5 However, rather than occurring as a homogenous population, these immune cells exhibit diverse ontogeny and pathological functions. Using genetic fate mapping, heart transplantation, and single‐cell RNA sequencing (scRNA‐seq), it is reported that CCR2− macrophages originate from the yolk sac, replenish via self‐proliferation, and take priority over antigen presentation and efferocytosis under ischemic conditions. In contrast, CCR2+ macrophages originate from circulating monocytes and are maintained via monocyte recruitment. 6 , 7 , 8 , 9 In line with macrophages, neutrophils are promptly recruited into the infarcted sites after MI and exhibit heterogeneous functional patterns. Horckmans et al revealed that neutrophils help modulate macrophage function toward a reparative phenotype. 10 The plasticity of neutrophils is evidenced by the prominent proinflammatory N1‐neutrophils 1 day post MI, along with increased anti‐inflammatory N2‐neutrophils 5 days post MI. 11 These findings suggest that immune cells exhibit diverse dynamics and functional patterns after MI, and discriminating specific immune clusters with marker genes can provide clues for precise targeting and yield therapeutic benefits in the treatment of MI.
Immune responses after ischemic injuries consist of early proinflammatory and late reparative phases, which are complex and highly coordinated processes. Consecutive proinflammatory responses can exacerbate ischemic injuries, leading to the progression of cardiac remodeling and heart failure. 12 In this regard, the underlying mechanisms of immune transition from proinflammatory to immunosuppressive responses after MI have become a topic of interest. It is reported that macrophages downregulate the expression of proinflammatory genes, such as interleukin‐6 (IL‐6), TNF (tumor necrosis factor), and MMP9 (matrix metalloproteinase 9), while upregulating TGF‐β (transforming growth factor‐beta) and VEGF (vascular endothelial growth factor) expression to foster angiogenesis and wound healing. 13 , 14 In addition, γδT cells may suppress the production and mobilization of neutrophils into the infarcted heart by secreting IL‐17A, which is responsible for the resolution of inflammation and improvement of cardiac remodeling. 15 , 16 Considering the prominent value of orchestrated inflammatory and immunosuppressive responses in cardiac remodeling and heart failure after MI, it is critical to understand the mechanisms of immune transition from activation to resolution. Using scRNA‐seq technology, researchers have identified novel immune cell clusters and regulatory networks among a series of cardiac diseases, including MI, 17 , 18 cardiac hypertrophy, 19 atherosclerosis, 20 and heart failure. 21 However, several issues remain unexplored: (1) Does ischemia induce specific immune dynamics and functional patterns during the early and late healing process? (2) Is any factor or cell‐to‐cell communication responsible for the initiation and suppression of inflammation responses? (3) Does any transcriptional regulon govern the proinflammatory responses and further cardiac dysfunction, and is any therapeutic benefit achieved by targeting these key regulons?
To address these questions, we performed scRNA‐seq analysis to explore the heterogeneities of CD45+ immune cells at 7 days post sham surgery and at 1 day and 7 days post MI. This analysis identified 2 ischemia‐associated macrophage subsets that were shared at 1 day and 7 days after MI with discrete inflammatory effects and fuel preference. Neutrophils emerged as an early responsive element of MI, displaying differential developmental states from mature to aging. We also confirmed that the chemokine C‐C motif ligand/chemokineC‐X‐C motif ligand (CCL/CXCL) pathways were responsible for the recruitment of myeloid cells 1 days after MI, whereas the CD80/inducible T‐cell costimulator (ICOS) interactome derived from dendritic cells (DCs) and Ly6c‐low monocytes contributed to the resolution of immune responses. Finally, we showed that Fos/AP‐1 (activator protein 1) acted as a key regulator of cardiac proinflammatory responses, inactivated Fos/AP‐1 signaling with its specific inhibitor, T5224, attenuated cardiac leukocyte recruitment, and mitigated cardiac dysfunction after MI, providing strategic insights for MI intervention and heart failure prevention.
METHODS
The data that support the findings of this study are available from the corresponding author upon reasonable request. All other supporting data are available in the article and supplemental material.
Availability of Data and Materials
All relevant data are available in the figures and supplemental material. Raw and analyzed RNA‐sequencing data generated during this study will be available in the Gene Expression Omnibus repository once this study is accepted.
Animal Studies
Eight‐week‐old male C57BL/6 mice used in this study were purchased from the Shanghai Laboratory Animal Center (Shanghai, China) and maintained under a 12:12 hour light–dark cycle. All animal experimental procedures were approved by the Animal Care Committee of Shanghai Jiao Tong University School of Medicine and were conducted according to the institutional guidelines. Further details can be found in Data S1.
Human Subjects
Peripheral blood (PB) used in this study was collected from controls and patients with acute coronary syndrome (ACS) before coronary angiography surgery. All enrolled patients gave written informed consent. This study was reviewed and approved by the Institutional Review Board of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (Ref. no: 2018‐183) and conformed to the ethical guidelines of the 1975 Declaration of Helsinki.
Detailed descriptions of experimental design and methods, including the MI model, single cell isolation and fluorescence‐activated cell sorting assay, RNA‐seq library preparation and scRNA‐seq, quality control of scRNA‐seq data, cell clustering and annotation, differentially expressed genes (DEGs) and pathway enrichment analyses, single‐cell regulatory network inference and clustering analysis, functional score calculation, cellular interactome analysis, flow cytometry assay, mouse bone marrow‐derived macrophage isolation and treatment, quantitative reverse transcription‐polymerase chain reaction, western blotting, fluorometric phagocytosis assay, and echocardiography are provided in Data S1.
Statistical Analysis
Data are presented as mean±SEM for preclinical data or mean ± SD for the clinical characteristics of enrolled patients and were statistically analyzed using SPSS software (version 23; SPSS Inc., Chicago, IL, USA) or Rstudio (version 1.4.1106) software. The Shapiro–Wilk test or the Kolmogorov–Smirnov test was conducted to assess the distribution of data normality. For data that passed the normality test, differences between 2 groups were compared using 2‐tailed unpaired Student's t‐tests, data with more than 2 groups were compared using 1‐way (1 variable) or 2‐way (more than 2 variables) ANOVA followed by least significant difference or Bonferroni's post hoc test. For data that did not meet the normality distribution, the Mann–Whitney test (2 groups) and the Kruskal‐Wallis test followed by Dunn post hoc test (more than 2 groups) were used. Ns referred to not significant. Statistical significance was considered at *P<0.05, **P<0.01, and ***P<0.001.
RESULTS
scRNA‐Seq Identified Immune Cells of Myocardial Infarction
To delineate the immune repertoire of MI in the early inflammatory and late reparative phases, we performed fluorescence‐activated cell sorting experiments to collect live cardiac CD45+ leukocytes at 7 days post sham or 1 day or 7 days post MI surgery (Figure S1A) and subjected them to scRNA‐seq using the 10× Genomics Chromium platform and reagents (Figure 1A). Doublets in each sample were detected and removed using the scrublet package (Figure S1B). After passing a quality‐control filter, the resulting cell library consisting of sham (n=9591), MI‐1D (n=10 943), and MI‐7D (n=9601) immune cells was merged for further analysis using the Seurat package (Figure S1C). Cell subpopulations were clustered according to their differentially expressed marker genes and annotated by matching their marker genes with the CellMarker website and previously published scRNA‐seq data sets. 19 , 20 Most immune cell types were identified, including macrophages, monocytes, neutrophils, NK cells, DCs, B cells, and T cells, with matched transcriptional profiles and putative biological roles (Figure 1B through 1E, Data S2, and Figure S1D). For example, macrophages with the highest expression of Mrc1, Arg1, Adgre1, Csf1r, Cx3cr1, and Ms4a7 (Figure 1D) were enriched in phagosome and lysosome pathways (Figure 1E). Neutrophils with high expression of S100a9, S100a8, Retnlg, and Mmp9 were enriched in TNF‐ and NF‐kappa B (nuclear factor‐kappa B) signaling pathways (Figure 1E). The T cells (highly expressed Cd3g, Cd3d, Cd28) exhibited effects of Th17, Th1, and Th2 cell differentiation and T‐cell receptor signaling pathways (Figure 1E). To exclude confounding cell clusters evoked by tissue dissociation or fluorescence‐activated cell sorting procedures as described previously, 22 we checked the expression of immediate early genes and did not observe significantly upregulated immediate early gene scores in identified cell clusters (Figure S1E). To gain further insight into the dynamics of immune subpopulations, we showed that the distribution of cell lineages was shared between sham and MI conditions while exhibiting differential cell proportion. Specifically, a remarkable infiltration of neutrophils was observed at 1 day after MI surgery, and macrophages expanded at 7 days post operation compared with the sham sample (Figure 1F), which was consistent with our previous reports using flow cytometry. 23 Taken together, these results revealed that murine hearts shared immune lineages under sham and ischemic conditions and that MI triggered dynamic immune flux in early inflammatory and late reparative phases.
Figure 1. scRNA‐seq identifying CD45+ immune cells under homeostatic and infarcted conditions.
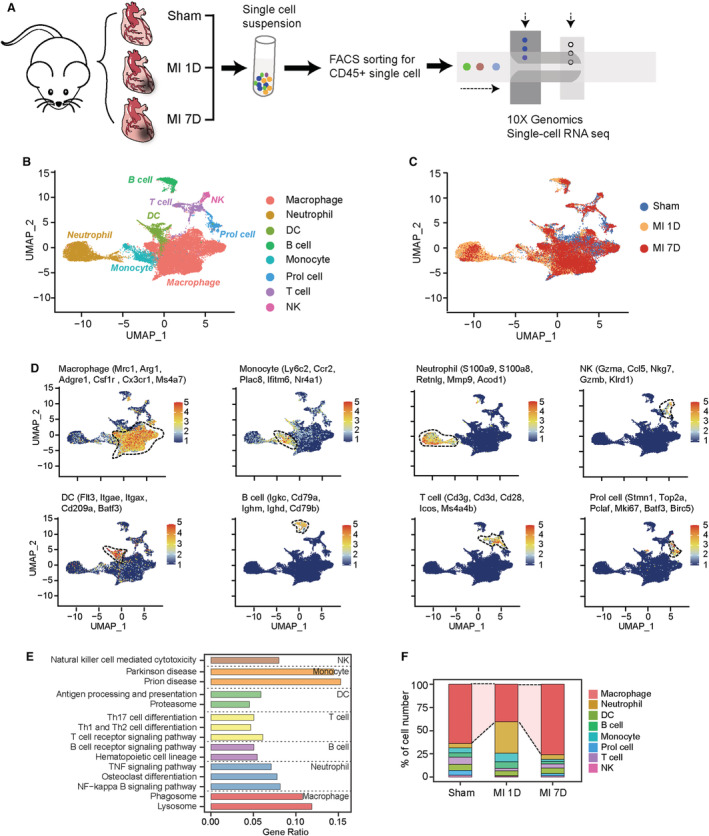
A, Schematic diagram of scRNA‐seq analysis to detect cardiac immune cells at 7 days post sham surgery, MI‐1D, and MI‐7D. B, UMAP plot showing included immune cells at 7 days post sham surgery, MI‐1D, MI‐7D. Cell lineages are denoted according to the expression of marker genes and colored accordingly. C, UMAP plot represented cells from sham, MI‐1D, and MI‐7D samples. D, Feature plots showing scaled expression of marker genes for macrophages, monocytes, neutrophils, NK, DCs, B and T cells, and proliferation‐like cells (Prol cell). E, Kyoto Encyclopedia of Genes and Genomes pathway analysis (KEGG) based on differentially expressed marker genes determined the biological function of different cell lineages. F, Proportion of immune cells at 7 days post sham surgery, MI‐1D, and MI‐7D. DC indicates dendritic cells; FACS, fluorescence‐activated cell sorting; MI, myocardial infarction; MI‐1D, 1 day after MI surgery; MI‐7D, 7 days after MI surgery; NF, nuclear factor; NK, natural killer; scRNA‐seq, single‐cell RNA sequencing; and UMAP, uniform manifold approximation and projection.
Two Ischemia‐Associated Macrophages Exhibited Differential Biological Functions and Energetic States
We further investigated the dynamics and functional discrepancy of monocytes and macrophages under ischemic conditions. Nine distinct monocyte/macrophage subclusters were identified in this scRNA‐seq data, including 3 tissue‐resident macrophages (MAC_TRs), 2 monocytes, and 4 ischemia‐associated macrophage subclusters with differentially expressed marker genes (Figure 2A and Figure S2A). Except for 3 MAC_TR subsets, the proportion of monocytes and ischemia‐associated macrophages was increased after MI surgery (Figure S2B). MAC_Olr1 (macrophages enriched for Olr1) were the largest subset at 1 day post MI (15% of all CD45+ immune cells), whereas the numbers of MAC_Gpnmb (differentially expressed Gpnmb) sharply increased at 7 days post MI (Figure 2B and Figure S2B). In line with their distinct dynamics, we observed consistently upregulated MAC_Olr1 expression at 1 day and MAC_Gpnmb level at 7 days post MI in scRNA‐seq and RNA‐seq data 24 (Figure 2C), suggesting their disparate functional patterns during MI progression.
Figure 2. Two ischemia‐associated macrophage subpopulations contributed to inflammation and phagocytosis.
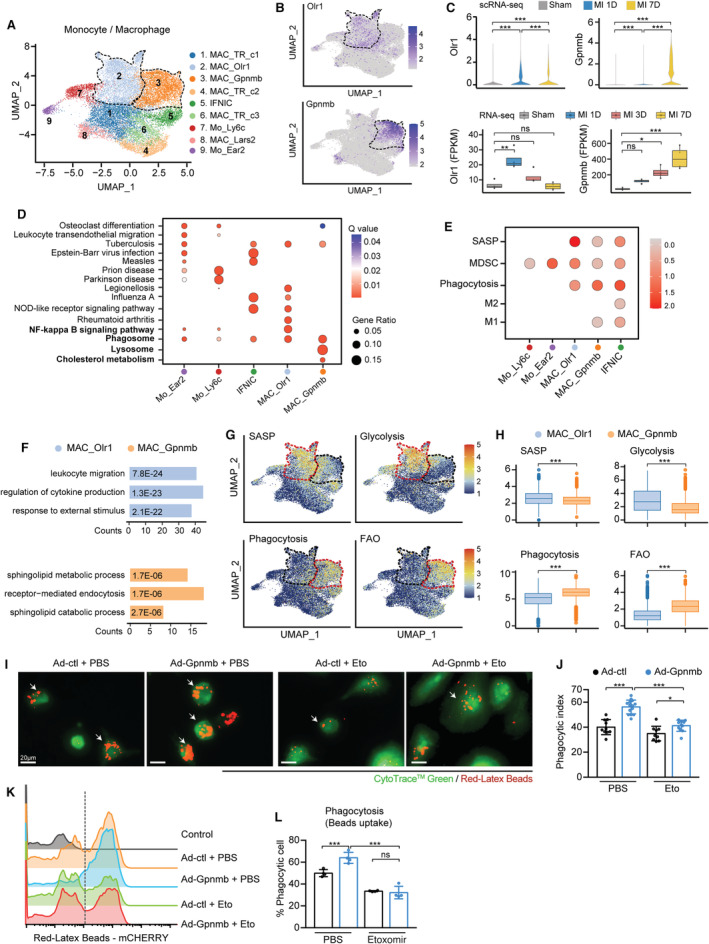
A, UMAP plot showing 9 monocyte/macrophage subpopulations at 7 days post sham surgery, MI‐1D, and MI‐7D. B, MAC_Olr1 and MAC_Gpnmb is distinguished by the differential expression of Olr1 and Gpnmb, as visualized by the feature plot. C, Top: Violin plots showing the scaled expression of macrophage Olr1 and Gpnmb in scRNA‐seq (n=6554, 5361, 7468 in sham, MI‐1D, and MI‐7D, respectively; Kruskal–Wallis followed by Dunn test). Bottom: Determining the transcriptional expression of macrophage Olr1 and Gpnmb in public RNA‐seq data sets after MI (n=4 for each group; 1‐way ANOVA followed by Bonferroni test). D, Gene ontology (GO) analysis with differentially expressed marker genes of monocytes (Mo_Ear2, Mo_Ly6c) and macrophages (IFNIC, MAC_Olr1, MAC_Gpnmb). E, Dot plot exhibiting scores of SASP, MDSC, phagocytosis, M2 and M1 macrophages in monocyte and macrophage subsets. F, GO pathway analysis of differentially upregulated genes in MAC_Olr1 and MAC_Gpnmb subsets. G, UMAP plots showing the scaled level of SASP, glycolysis, phagocytosis, and FAO in monocyte/macrophage subsets. H, Quantitative results of SASP, glycolysis, phagocytosis, and FAO in MAC_Olr1 and MAC_Gpnmb subclusters (n=5004, 3855 in MAC_Olr1 and MAC_Gpnmb; Mann–Whitney test). I, BMDMs with or without Gpnmb overexpression treated with etomoxir or PBS and subjected to fluorometric phagocytosis assay. Scale bar: 20 μm. J, Phagocytotic index referring to panel I, quantified by the ratio of macrophages with fluorometric red‐Latex beads among all CytoTrace Green‐positive cells (n=10, 14, 9, 11, respectively, 2‐way ANOVA followed by least significant difference test). K, Ability of macrophage phagocytosis, as determined by flow cytometry analysis. L, Quantification of panel K (n=4 for each group; 2‐way ANOVA followed by least significant differencetest). Ad indicates adenovirus; BMDM, bone marrow‐derived macrophage; FAO, fatty acid oxidation; IFNIC, macrophages with a type I interferon signature; MAC_Gpnmb, macrophage with differentially expressed Gpnmb; MAC_Olr1, macrophage enriched for Olr1; MDSC, myeloid‐derived suppressor cell; NF, nuclear factor; NOD, nucleotide‐binding and oligomerization domain; SASP, senescence‐associated secretory phenotype; scRNA‐seq, single‐cell RNA sequencing; and UMAP, uniform manifold approximation and projection.
To decipher the functional differences between MAC_Olr1 and MAC_Gpnmb, pathway enrichment analysis revealed that the MAC_Olr1 subcluster was associated with the NF‐kappaB signaling pathway; however, enrichment of lysosome and cholesterol metabolism pathway was found in the MAC_Gpnmb cluster (Figure 2D). Macrophages were previously classified into M1 or M2 groups; however, it has been reported that the heterogeneities of macrophages are too complex to fit in the simple M1/M2 model. 18 To further determine their functional states, we examined the gene signatures related to myeloid‐derived suppressor cells, 25 senescence‐associated secretory phenotype (SASP), 26 M2 and M1 macrophages, and phagocytosis in macrophage subsets. High SASP and myeloid‐derived suppressor cell scores were found in MAC_Olr1, whereas MAC_Gpnmb exhibited higher phagocytosis scores than the MAC_Olr1 clusters (Figure 2E). Compared with MAC_Gpnmb, MAC_Olr1 was associated with activated leukocyte migration and cytokine production driven by upregulated Cxcl2, Ccl9, Ccl24, Il1b, and Trem1 (Figure 2F and Figure S2C). In contrast, MAC_Gpnmb was enriched for lipid metabolism and endocytosis pathways, with increased levels of Gpnmb, Fabp5, and Trem2 (Figure 2F and Figure S2C). Previous publications have demonstrated that metabolic reprogramming contributed to macrophage plasticity and functional changes, highlighted the positive relationship between glycolysis and proinflammatory macrophages. 27 Indeed, we found MAC_Olr1 displayed higher SASP scores and glycolysis scores than MAC_Gpnmb clusters displayed (Figure 2G and 2H). By contrast, the phagocytosis and fatty acid oxidation (FAO) scores were consistently increased in MAC_Gpnmb clusters, indicating a positive link between phagocytosis and FAO (Figure 2G and 2H). To determine whether similar MAC_Olr1 and MAC_Gpnmb subsets, especially the functional patterns, occur in public scRNA‐seq data, we jointly analyzed the data sets of Farbehi et al 18 and King et al, 28 which included total non‐cardiomyocytes and CD45+ leukocytes after MI. Integrated analysis revealed that a distinct macrophage subset overlapped with the MAC_Gpnmb cluster identified in our scRNA‐seq data (Figure S3A). Notably, we also observed the uniform enrichment of phagocytosis and FAO signaling within Gpnmb‐positive macrophages in public scRNA‐seq data sets (Figure S3B).
Considering the concomitant activation of phagocytosis and FAO in Gpnmb‐expressed macrophages, we investigated whether Gpnmb modulated phagocytotic capacity dependent on FAO activity. To address this hypothesis, we first examined the mRNA expression of phagocytosis‐related genes in bone marrow‐derived macrophages with or without the FAO inhibitor, etomoxir, and found markedly downregulated Marcks, Marco, and Vav1 expression after FAO inhibition (Figure S4A). The bone marrow‐derived macrophages were then transfected with Gpnmb adenovirus or the empty control (Figure S4B through S4D) and treated with etomoxir for 24 hours. Using fluorometric red latex beads, we showed that macrophages overexpressing Gpnmb were more efficient at phagocytosis than the control group; however, the inhibition of FAO abolished Gpnmb‐induced phagocytic potential (Figure 2I through 2L, Figure S4E). Taken together, these results suggest that 2 distinct macrophage subsets emerge after MI with distinctive functions and energetic states. The MAC_Olr1 cluster populated at 1 day post MI is characterized by upregulated SASP and glycolysis, whereas MAC_Gpnmb exhibits stronger phagocytosis and FAO preference.
Two Distinct Monocyte Subsets Exhibited Proangiogenesis or Immune Regulatory Capacities
We further determined the heterogeneity of 2 monocyte clusters, Mo_Ly6c and Mo_Ear2, with differentially expressed Ly6c2 and Ear2 (Figure S2A). Monocytes have been previously classified into classical proinflammatory (Ly6Chi monocytes) and nonclassical prohealing (Ly6Clo monocytes) groups. 29 Classical monocytes are recruited to the infarcted hearts 1 day post MI and play dominant roles in inflammation, tissue vascularization, and angiogenesis. 30 , 31 In contrast, Ly6Clo monocytes peak at 7 days after MI and play crucial roles in inflammatory resolution, angiogenesis, and adaptive immune response. 32 , 33 , 34 To clarify the effects of monocyte subsets, we performed DEG pathway enrichment analyses between Mo_Ly6c and Mo_Ear2 at 1 day and 7 days post‐MI, respectively. Compared with Mo_Ear2, Mo_Ly6c at 1 day post MI exhibited upregulated neutrophil migration and granulocyte chemotaxis genes, including Ly6c2, Ccl2, Ccr2, Lgals3, and Ccl9 (Figure S5A and S5B). Comparing the DEGs of 2 monocyte subsets at 7 days after MI, Mo_Ear2 exhibited distinctive functions in the immune effector process, whereas the upregulated genes in Mo_Ly6c mapped to VEGF production signaling (Figure S5C and S5D). These observations suggest that monocytes possess heterogeneous functions in the inflammatory and reparative phases. Mo_Ly6c was related to proinflammatory response and angiogenesis, and the Mo_Ear2 cluster likely participated in adaptive immune modulation after MI.
Identifying a Unique Folr2+ Tissue Resident Macrophage Cluster
Among 3 clusters of MAC_TRs, MAC_TR_c2 exclusively expressed Folr2, Timd4, and Lyve1 (Figure S2A; Figure 3B and 3C), similar to the previously described Timd4‐cluster. 8 Timd4+ macrophages were reported as the most conserved tissue resident macrophage subset among multiple organs in mice and humans, and were maintained solely by self‐proliferation without monocyte contribution. 35 Deciphering the functional diversity of this MAC_TR cluster with other macrophages would help elucidate its pathophysiological roles and optimize targeting strategies. Therefore, we assigned all macrophages as either Folr2+ and Folr2− macrophages (Figure 3A). Folr2− macrophages were defined by upregulated Ccr2 and Lgals3 expression and were positively related to proinflammatory NF‐kappa B signaling, apoptosis, and IL‐17 signaling pathways (Figure 3B through 3D). On the contrary, the hematopoietic cell lineage, lysosome, and endocytosis signaling was enriched in Folr2+ macrophages (Figure 3D). Furthermore, RNA velocity analysis was used to identify the developmental trajectory of macrophages and showed that monocytes contributed to ischemia‐associated macrophages; however, Folr2+ macrophages were self‐renewing with minimal monocyte input (Figure 3E), which was consistent with Dick et al's research. 8 To determine the transcriptional basis for its unique clonal expansion capacity, we interrogated their differences in proliferative gene expression and transcriptional regulons (TFs) activities. Multiple proliferation‐related genes and TFs controlled cell differentiation and proliferation (such as Jund, Tcf4, and Maf) were activated at Folr2+ macrophages rather than the Folr2− macrophages (Fosl1, Jdp2, Cebpb) (Figure 3F and 3G), suggesting distinct transcriptional patterns and TF networks endowed the self‐renewing property of Folr2+ MAC.
Figure 3. Isolation and validation of Folr2+ tissue‐resident macrophages.
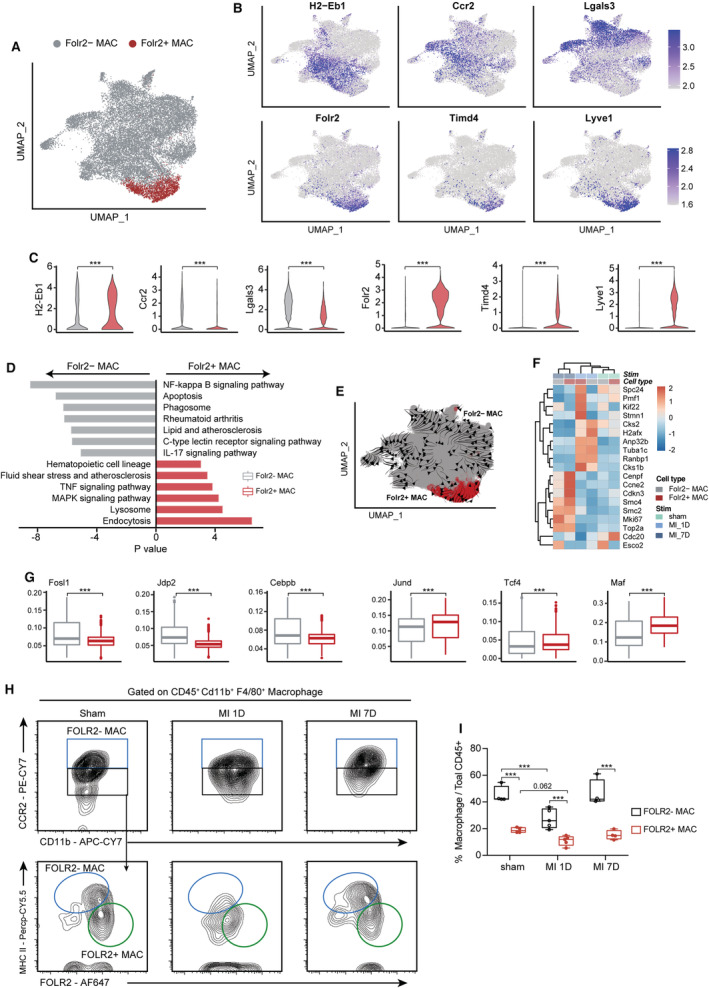
A, UMAP plot showing the reassigned Folr2+ and Folr2− MAC. B, Feature plot showing the differentially expressed marker genes in Folr2+ and Folr2− MAC. C, Quantification results of B (n=17 698, 1685 for Folr2− and Folr2+ MAC, respectively; Mann–Whitney test). D, KEGG analysis based on differentially expressed marker genes of Folr2+ and Folr2− MAC subsets. E, RNA velocity analysis derived from monocyte and macrophage subsets was projected into a UMAP‐based plot and visualized as streamlines. F, Heatmap showing the expression of proliferation‐related genes in Folr2+ and Folr2− MAC at 7 days post sham surgery, MI‐1D, and MI‐7D. G, The differential transcriptional regulons in Folr2+ and Folr2− MAC (n=17 698, 1685 for Folr2− and Folr2+ MAC, respectively; Mann–Whitney test). H, Flow cytometry analysis determined the Folr2+ and Folr2− MACs with CCR2, MHC II, and FOLR2 antibodies at 1 days and 7 days post MI, or 7 days post sham surgery. I, The frequency of Folr2+ and Folr2− MAC subsets under sham or ischemic conditions (n=4, 4, 5, 5, 4, 4, respectively; 2‐way ANOVA followed by Bonferroni test). KEGG indicates Kyoto Encyclopedia of Genes and Genomes pathway analysis; MAC, macrophage; MAPK, mitogen‑activated protein kinase; MHC II, major histocompatibility complex II; TNF, tumor necrosis factor; and UMAP, uniform manifold approximation and projection.
Based on the exclusive expression of Ccr2, Folr2, and MHC II (major histocompatibility complex II), we sought to identify the existence and dynamics of Folr2− macrophages (CD45+ CD11b+ F4/80+ FOLR2− MHC IIhi) and Folr2+ macrophages (CD45+ CD11b+ F4/80+ CCR2− FOLR2+ MHC IIint‐low) under homeostatic or ischemic conditions using flow cytometry (Figure S6). Using this gating strategy, we showed a clear Folr2+ macrophages subset at 7 days post sham surgery, the relative proportions of Folr2+ macrophages decreased at 1 day and restored at 7 days after MI (Figure 3H and 3I). Altogether, these findings suggest that hearts contain distinct Folr2+ tissue resident macrophages that exhibit differential biological functions, developmental trajectory, and TF activities. We also constructed a sorting strategy to isolate and identify this macrophage subset.
Neutrophils Exhibited Sequential Developmental State After MI
Three neutrophil clusters were identified in the scRNA‐seq data with differentially expressed marker genes and dynamic influx after MI: Neu_Sell with upregulated Sell, Retnlg, Cxcr2, and interferon‐stimulated genes (ISGs, including Ifit1, Ifit3, and Isg15); Neu_Icam1 with differentially expressed Icam1, Siglecf, Tnf, Il1b, and Gpr84; and Neu_S100a4, identified by S100a4, S100a10, and Gdf15 (Figure 4A and 4B; Figure S7A and S7B). Previous studies have identified various neutrophil subclusters in infection and MI. Xie et al performed scRNA‐seq analysis of mouse bone marrow, blood, and spleen neutrophils and described 3 circulating neutrophil clusters in the blood and spleen (G5a, G5b, and G5c). 36 By comparing our neutrophil subclusters with those of Xie et al, we found that Neu_Icam1 matched the G5c subcluster (highly expressed Il1b, Cxcr4), and Neu_Sell was related to G5b with upregulated ISGs. Vafadarnejad et al comprehensively analyzed cardiac LY6G positive neutrophils after MI and identified 6 neutrophil subpopulations (Neutro 1–6). 17 Neutro 1 highly expressed Tnf, Icam1, and Gpr84 and was similar to our Neu_Icam1 cluster; Neutro 4 (characterized by Retnlg and Sell expression) and Neutro 6 (high level of Gdf15) was related to Neu_Sell and Neu_S100a4 clusters in our data set. These studies confirmed the heterogeneity of neutrophil subclusters in circulating and hearts during infection or MI.
Figure 4. Three neutrophil subsets with differential ontogeny from mature, activation, to aging.
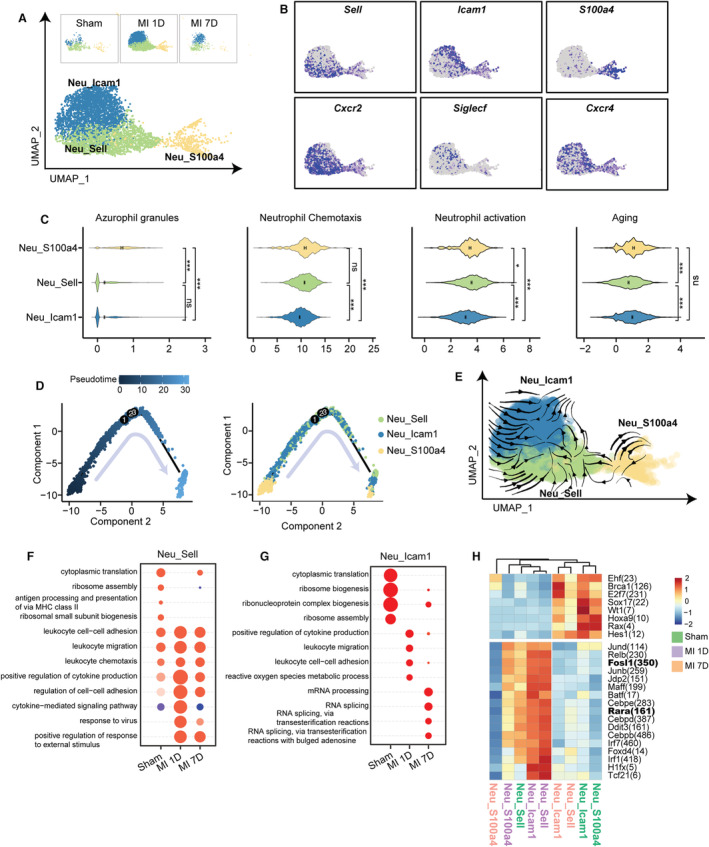
A, UMAP plot of 3 neutrophil subpopulations from sham, MI‐1D, and MI‐7D samples. B, Scaled expression of neutrophil marker genes. C, Scores of azurophil granules, neutrophil chemotaxis, activation, and aging in 3 neutrophil clusters (n=473, 1689, 2460 in Neu_S100a4, Neu_Sell, Neu_Icam1, respectively; 1‐way ANOVA followed by Bonferroni test). D, Analyzing developmental trajectories of 3 neutrophil clusters with monocle method, cells are reordered and colored by pseudotime or by cell type. E, RNA velocity analysis including all neutrophils were visualized as streamlines to determine potential differentiation directions. F, Selected top GO terms of Neu_Sell at 7 days post sham surgery, MI‐1D, and MI‐7D by comparing uniquely upregulated genes at 1 time point with those of the others. G, The enriched GO categories of Neu_Icam1 at sham, MI‐1D, and MI‐7D. H, The activities of transcriptional regulons among 3 neutrophil subsets at sham, MI‐1D, and MI‐7D. GO indicates gene ontology; MHC II, major histocompatibility complex II; and UMAP, uniform manifold approximation and projection.
The ontogeny of neutrophils involved granulopoiesis, maturation and tissue migration, and ubsequent aging and clearing processes. 37 In this regard, immature neutrophils were classified by the emergence of neutrophil granules (azurophil, specific, and gelatinase granules) and secretory vesicles, whereas mature neutrophils were characterized by elevated Cxcr2 expression, which contributed to their mobilization into blood and inflamed sites. After activation, upregulated Icam1 and Cxcr4 mediated homing or cleaning of aged neutrophils. 38 To discriminate maturation and developmental trajectory of 3 neutrophil subclusters, we first compared the transcriptional differences of granules, neutrophil activation, and aging genes, as previously described. 36 Neu_S100a4, with low expression of Cxcr2 and Cxcr4, had the highest expressed granule genes compared with the other neutrophil clusters (Figure 4C; Figure S7C, Data S3). In contrast, Neu_Sell had the highest neutrophil chemotaxis and activation score, and Neu_Icam1 represented the most aging neutrophil clusters, suggesting that the hearts consisted of 3 distinct neutrophil clusters with differential ontogenetic states (Figure 4C). To verify these results, we further examined cell fate by ordering the direction of cell lineages or by placing all neutrophil clusters along the continuous trajectory in pseudo‐time. Combining the results of monocle and RNA velocity analysis, we showed that neutrophil maturation started from the Neu_S100a4 clusters without obvious division, directed toward Neu_Sell, and ended at Neu_Icam1 (Figure 4D and 4E), along with upregulated chemokine (Ccl3, Ccl4, Cxcl2, and Cxcl10) and Mmp9 expression (Figure S7D and S7E).
We further investigated the transcriptional differences and regulons of neutrophils during MI. Compared with the sham group, all neutrophil clusters at 1 day exhibited activated immune pathways (such as leukocyte cell–cell adhesion, leukocyte migration, chemotaxis, and cytokine production), which were relatively resolved in MI‐7D (Figure 4F and 4G; Figure S8A and S8B). Notably, the activated immune responses in MI‐1D were associated with high Fosl1 and Rara TF activities (Figure 4H; Figure S8C). Taken together, these findings identified 3 neutrophil subsets with different ontogenetic states under homeostatic and ischemic conditions.
Verifying scRNA‐Seq Results in Murine Hearts and Human Peripheral Blood
The scRNA‐seq results suggested that ischemic heart consisted of 3 neutrophil subclusters; therefore, we validated the results of scRNA‐seq in murine hearts and PB using flow cytometry. The presence of Neu_Icam1 and Neu_Sell clusters was identified as CD45+ CD11b+ LY6G+ ICAM1+ or CD62L+ (Figure S9A and S9B). We found that Neu_Sell represented the largest neutrophil subcluster under homeostatic conditions (7 days post sham surgery), which was consistent with the scRNA‐seq results (Figure S7A; Figure 5A and 5B). MI led to striking infiltration of neutrophils, involving CD62L‐high neutrophils (10% versus 16%, sham versus MI‐1D, respectively) and ICAM1‐positive neutrophils (from 7.9% to 22.6% in sham and MI‐1D, respectively; Figure 5A and 5B). Furthermore, increased proportion of ICAM1‐high neutrophils after MI was also observed in murine PB (Figure 5C and 5D). To further determine the increased Icam1‐high neutrophils present in the PB of patients with ACS (basic clinical characteristics of patients were listed as Table S1), we identified different neutrophil subsets in the PB of controls and patients with ACS (Figure S9C) and revealed that the proportion of Neu_Icam1 was significantly higher in the PB of patients with ACS than in control patients (Figure 5E and 5F).
Figure 5. Validating the existence of discrete neutrophil subsets in murine hearts and human peripheral blood with flow cytometry.
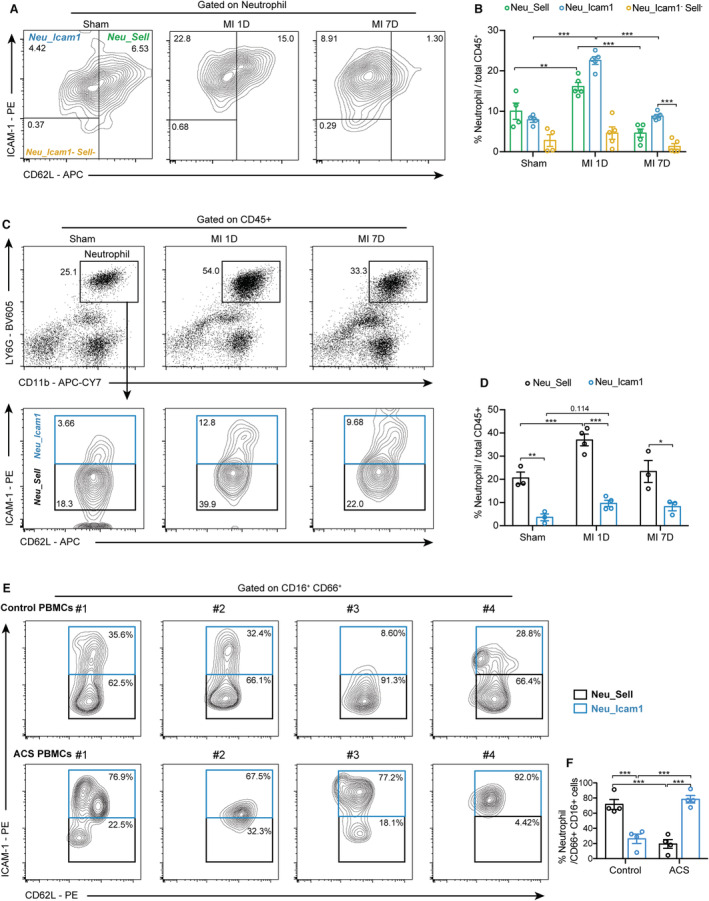
A, Wild‐type mice were subjected to MI surgery for 1 and 7 days, or sham surgery for 7 days. The neutrophil subsets were identified by the expression of ICAM1 and Sell/Cd62L using flow cytometry. B, Quantification of A (n=4, 4, 4, 5, 5, 5, 5, 5, 5, respectively; 2‐way ANOVA followed by Bonferroni test). C, Neutrophils of PB were collected from sham, MI‐1D, and MI‐7D mice, and the frequency of ICAM+ or CD62L+ neutrophils among CD45+ cells were calculated. D, Quantification results of panel C (n=3, 3, 4, 4, 3, 3, respectively; 2‐way ANOVA followed by Bonferroni test). E, PB neutrophils from patients of control subjects and ACS were collected for flow cytometry analysis. F, The proportions of Neu_Icam1 and Neu_Sell in patient PB (n=4 for each group, 2‐way ANOVA followed by least significant difference test). ACS indicates acute coronary syndrome; MI, myocardial infarction; PB, peripheral blood; and PBMC, peripheral blood mononuclear cell.
Heterogeneous DCs Were Associated With Activation and Resolution of Inflammation
DCs are a small subset of hematopoietic cells with crucial roles in antigen processing and initiation of adaptive immune responses. 39 Classical DCs in nonlymphoid tissues consisting of Cd103+ DCs excel in priming CD8+ T cells, whereas Cd11b+ DCs may participate in the activation of CD4+ T cells. In addition, plasmacytoid DCs display a superior ability to produce cytokines, such as interferon‐α for innate or adaptive immunity. 40 To decipher the heterogeneities of DCs in ischemic responses, we analyzed DCs in the scRNA‐seq data and identified 4 DC clusters (DC_Cd11b, DC_Cd209, DC_Cd103, and DC_Ccr7) with differentially expressed marker genes (Figure S10A and S10B). DC_Cd11b was the largest DC subset under sham conditions, whereas the proportion of DC_Cd209 was remarkably expanded in MI‐1D with highly expressed activation marker Cd69, and the frequency of DC_Ccr7 cells increased in MI‐7D compared with that in MI‐1D (Figure S10C and S10D). To understand the intrinsic heterogeneity of DC subpopulations, Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that the DC_Cd209 and DC_Ccr7 were prominently associated with IgA production and T‐cell differentiation compared with DC_Cd11b and DC_Cd103 (Figure S10E). Therefore, we focused on the functional differences of DC_Cd209 and DC_Ccr7 subsets. Compared with DC_Cd209, a series of immunomodulatory genes were increased in DC_Ccr7, including Cd274/PD‐L1, Icosl, Socs2, and Cd200 (Figure S10F), which were upregulated in MI‐7D compared with sham and MI‐1D (Figure S10G). In contrast to DC_Ccr7, DC_Cd209 differentially expressed interferon‐induced transmembrane proteins (IFITMs, Ifitm1, and Ifitm6) and cytotoxicity‐associated genes (Klrd1) (Figure S10F and S10G). This indicated the proinflammatory effect of DC_Cd209 and immunosuppressive role of DC_Ccr7 in ischemic responses. Furthermore, by analyzing the transcriptional regulons responsible for their distinctive immune phenotypes, we found that DC_Ccr7 had higher Foxp3 and Sox4 TF activities, whereas the activity of Fos and Fosb was markedly increased in DC_Cd209 subsets (Figure S10H). Altogether, these findings reveal that 2 distinctive DC subsets exhibit varied roles in activating or suppressing immune responses via Fos/Fosb or Foxp3/Sox4 TF activities.
Different T‐Cell and B‐Cell Subclusters Emerged After MI
A total of 1463 T/NK cells (highly expressing Cd3d, Cd4, Cd8, or Gzma) were further partitioned into 7 subpopulations based on uniquely expressed CD4 or CD8 genes (Figure S11A and S11B; Figure 6A). The genes related to NK cell‐mediated cytotoxicity, such as interferon gamma (Ifng), Gzma, and Klrb1c, were highly expressed in NK and γδT cells (Figure 6B; Figure S11C). CD4_C3_IL17A had the highest level of Th17 cytokine (Il17a) among all T‐cell clusters. Two naïve T‐cell subpopulations were identified with high Ccr7 and Lef1 expression (CD4_C1_CCR7 and CD8_C1_CCR7, Figure S11C). In addition, CD4_C2_CXCR3 and CD8_C2_Gzmk represented the effector T cells and were positively correlated with Th17, Th1, and Th2 cell differentiation, as well as PD‐L1 signaling pathways (Figure S11C and S11D). Consistent with their effector functions, the frequency of CD4_C2_CXCR3 and CD8_C2_Gzmk was expanded in MI‐7D (Figure 6C). To gain a deeper understanding of the transcriptional differences of T‐cell subclusters under ischemic conditions, we compared the DEGs and enriched pathways of CD4_C2_CXCR3 or CD8_C2_Gzmk between MI‐7D and sham groups. Compared with homeostasis, ischemia downregulated T‐cell mitochondrial oxidative phosphorylation levels, whereas T‐cell activation and Il‐2 production pathways were significantly activated at 7 days after MI (Figure 6D and 6E, and Figure S11E and S11F). Transcriptional regulon analysis revealed that ischemia induced CD4_C2_CXCR3 and CD8_C2_Gzmk activation were associated with decreased Jund, Junb, and Spib regulon activities, whereas increased Fos, Fosb, and Mef2c activities compared with the sham group (Figure 6F). Herein, we reported that the Cxcr3+ CD4 T cells and the Gzmk+ CD8 T cells represented the major adaptive immune cells with activated Fos and Fosb activities at 7 days post MI.
Figure 6. Single‐cell RNA sequencing identified the heterogeneity of cardiac T and B cells.
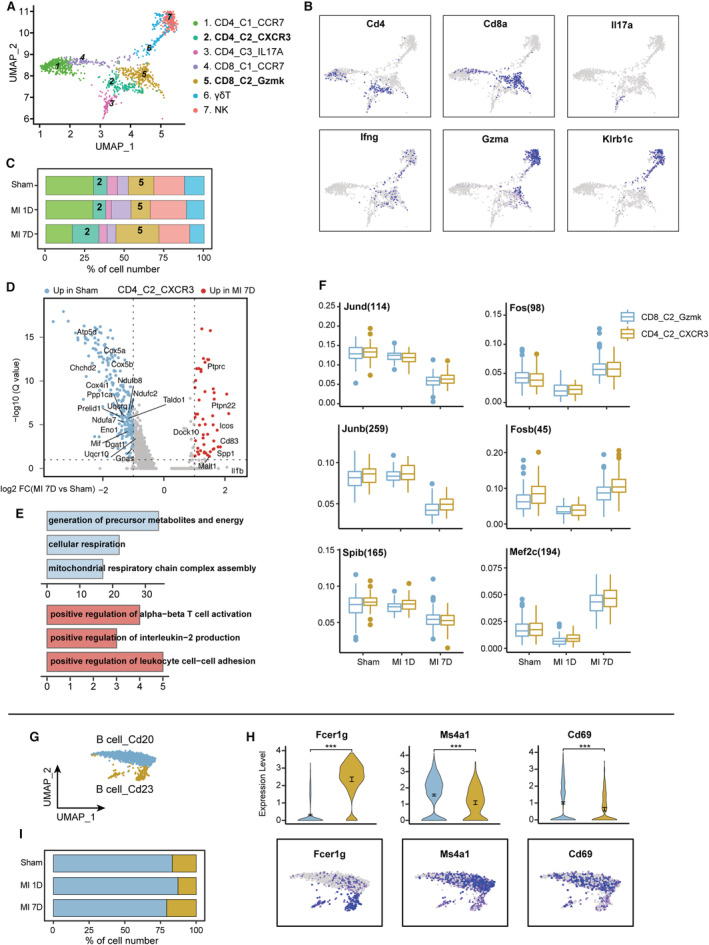
A, Cardiac T/natural killer (NK) cells were separated into 7 subsets and visualized as UMAP plots. Cell types are denoted as different colors. B, Feature plots exhibiting the scaled expression of T‐cell marker genes (Cd4, Cd8a), IL‐17 cytokines (Il17a), and cytotoxic genes (Ifng, Gzma, Klrb1c). C, Dynamic proportion of T/NK subpopulation at 7 days post sham surgery, MI‐1D, and MI‐7D in scRNA‐seq data. D, Volcano plot showing the transcriptional differences of CD4_C2_CXCR3 between MI‐7D and sham group. E, Top GO terms of CD4_C2_CXCR3 in sham (Blue) and MI‐7D (Red). F, Boxplots exhibiting the transcriptional regulons of CD8_C2_Gzmk and CD4_C2_CXCR3 in sham surgery, MI‐1D, and MI‐7D. G, Two B cell subsets were identified in scRNA‐seq data and projected into UMAP plot. H, Violin plots (top) and feature plots (bottom) showing the scaled expression of Fcer1g, Ms4a1, and B cell activation marker, Cd69 (n=167 and 1315 in B cell_Cd23, B cell_Cd20, respectively; Mann–Whitney test). I, Frequency of B‐cell subsets in sham surgery, MI‐1D, and MI‐7D. GO indicates gene ontology; IL‐17, interleukin‐17; scRNA‐seq, single‐cell RNA sequencing; and UMAP, uniform manifold approximation and projection.
A series of studies have described the functional diversity of B cells in immune homeostasis. In addition to their roles in antibody production, B cells can secrete cytokines for immune modulation or presentation to T cells for adaptive immune response. 41 Here, we identified 2 B‐cell clusters with differentially expressed Cd20 (Ms4a1) and Cd23 (Fcer1g) and denoted them B cell_Cd20 and B cell_Cd23 (Figure 6G and 6H). Cd20, a nonglycosylated protein, is closely related to B‐cell maturation, differentiation, and B‐cell receptor signaling pathways. Preclinical experiments have demonstrated that specific deletion of CD20+ B cells with anti‐CD20 antibody diminishes B‐cell mobilization and further limits infarct size and cardiac remodeling. 42 Moreover, a recent clinical study showed that deletion of B cells with a single infusion of rituximab in patients with ST‐segment–elevation MI decreases their circulating B‐cell count. 43 These studies confirmed the deleterious role of CD20+ B cells in cardiac remodeling. Consistent with the results of previous studies, we identified CD20+ B cells in our scRNA‐seq data, which exhibited slight expansion at 1 day (Figure 6I). Compared with B cell_Cd23, B cell_Cd20 highly expressed the B‐cell activation marker Cd69 (Figure 6H). Taken together, these observations suggest that CD4_C2_CXCR3, CD8_C2_Gzmk, and B cell_Cd20 cells represent activated adaptive immune subpopulations after MI.
Identifying Key Immune Interactome on Early Innate Immunity and Latter Adaptive Immunity Responses
Compared with homeostatic conditions, infarction leads to the extensive recruitment of innate or adaptive immune cells via redistributed (ligand–receptor) expression. Therefore, we investigated cell–cell communication under sham and ischemic conditions to understand the potential mechanisms of immune initiation and resolution in early inflammatory and latter reparative phases. First, we analyzed the interactions under homeostatic conditions (Figure S12A). Then, by calculating the number of ligand‐receptor pairs of all cell subclusters in sham, MI‐1D, and MI‐7D using the CellChat package, 44 we showed that the cell–cell communications were significantly enhanced after MI (Figure S12B). Specifically, CCL and CXCL chemokine signaling, which is critical for circulating myeloid and granulocyte recruitment, was remarkably enhanced in MI‐1D hearts compared with sham operated hearts. By investigating the differentially expressed ligand–receptor pairs between MI‐1D and MI‐7D, we found that regulatory (such as the CD80 and ICOS) and prohealing pathways (such as IL‐10 and insulin‐like growth factor signaling) were dramatically increased in MI‐7D (Figure S12C). These observations suggested that the CCL/CXCL pathways were responsible for the initiation of early proinflammatory responses, whereas the CD80/ICOS pathways were linked to immune resolution. Computational analysis identified neutrophils (Neu_Sell, Neu_Icam1, and Neu_S100a4) as the central communication hubs of inbound connections at 1 day (Figure S13A). Further analysis of the key interactomes of neutrophils showed that myeloid cell‐derived Cxcl1, Cxcl2, Cxcl3, Ccl6, Ccl7, and Ccl9, along with lymphocyte‐derived Ccl5, contributed to the early infiltration of neutrophils by interacting with their Cxcr2 and Ccr1 receptors (Figure S13B and S13C). By focusing on the differential interactomes between MI‐1D and MI‐7D, we revealed that DC_Ccr7 and CD4_C2_CXCR3 were the central outbound and inbound connections, respectively (Figure S13D). DC_Ccr7 and Mo_Ear2 monocytes mediated immune resolution by upregulating Cd80, Icosl, or Cd274 (PD‐L1) expression, which acted on CD4 or CD8 cells via Icos, Cd28, and Ctla4 receptors (Figure S13E and S13F), suggesting DC_Ccr7 and Mo_Ear2 played critical roles on late immune resolution after MI. Together, these findings suggested that the shift of proinflammatory to immunosuppressive responses was mediated by an orchestrated CCL/CXCL to CD80/ICOS pathways.
Inhibition of Fos/AP‐1 TF Activity Showed Potential Benefits for Cardiac Function Improvement After MI
Our scRNA‐seq data suggested that the abnormal activation of Fos/AP‐1 transcriptional activities was associated with proinflammatory responses. Specifically, the inflammatory Folr2−macrophage clusters displayed higher Fosl1/AP‐1 activities than the Folr2+ macrophage subset (Figure 3G). The extensive expansion of neutrophils in MI‐1D involved in cytokine production and leukocyte migration was positively related to higher AP‐1 transcriptional activities than those of the sham or MI‐7D groups (Figure 4H). Compared with the proresolutive DC_Ccr7 subset, the proinflammatory DC_Cd209 subclusters activated immune response accompanied by increased Fos/AP‐1 TF signaling (Figure S10H). Finally, we confirmed that 2 T‐cell subclusters were activated in MI‐7D with increased Fos and Fosb TF activities (Figure 6F).
To further determine the roles of Fos/AP‐1 signaling in human MI and heart failure, we curated a list of Fos/AP‐1 target genes, which were differentially expressed in this scRNA‐seq or reported on the cistrome website (http://cistrome.org/). We interrogated the activity of Fos/AP‐1 by checking the single nucleotide polymorphisms in CARDIOGRAMPLUSC4D genome‐wide association meta‐analysis study, which identified risk loci for coronary atherosclerotic diseases and MI. 45 The genes and gene‐set based analysis revealed that Fos/AP‐1 signaling target genes were significantly associated with the incidence of coronary atherosclerotic diseases or MI (Figure 7A, Multi‐marker Analysis of GenoMic Annotation competitive P=3.6×10−32). Multiple Fos/AP‐1 target genes contained MI‐associated risk loci, such as FAM117B and BBOX1 (Figure 7B, C, and Data S4). Furthermore, we analyzed the RNA‐seq data of patients with dilated cardiomyopathy (n=50) and normal patients (n=51, Figure S14), 46 and observed the Fos/AP‐1 target genes were extensively dysregulated in dilated cardiomyopathy hearts (Figure 7D). Computational analysis indicated that, compared with normal patients, the activity of Fos/AP‐1 signaling was markedly increased in patients with dilated cardiomyopathy and ischemic cardiomyopathy 46 , 47 (Figure 7E and 7F). Additionally, we observed that Fos/AP‐1 signaling was slightly increased in the hearts of patients with ischemic cardiomyopathy, meanwhile subjected with left ventricular assist devices partially repressed the activity of Fos/AP‐1 signaling 48 (Figure 7G). These RNA‐seq data highlighted the positive relationship between Fos/AP‐1 signaling with the progression of heart failure. Together, these data indicated that Fos/AP‐1 acted as a critical factor of MI and heart failure, and we hypothesized that selective Fos/AP‐1 inhibition may confer potential benefits for MI treatment.
Figure 7. Fos/AP‐1 signaling activation was associated with clinical CAD/MI risk and heart failure.
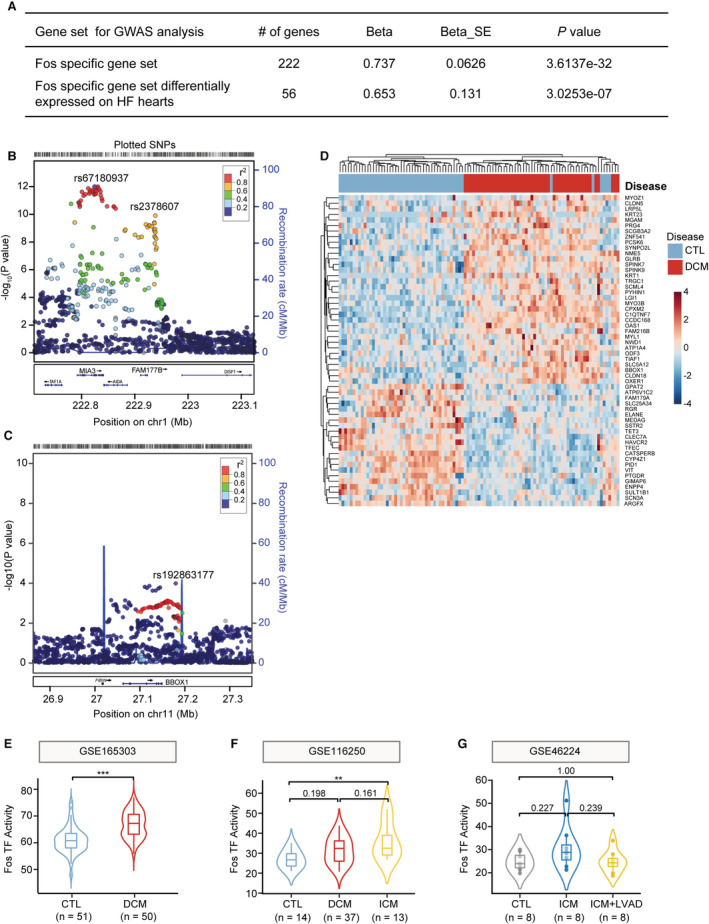
A, Gene‐set based analysis of CAD/MI risk signaling in CARDIOGRAMPLUSC4D genome‐wide association meta‐analysis study (GWAS). Fos/AP‐1 target genes (222 genes), and Fos/AP‐1 target genes that were differentially expressed in patients with heart failure in RNA‐seq data set (56 genes/222 genes, GSE165303) were used as queried gene lists for magma analysis. B and C, Regional association plots showing CAD/MI risk‐loci. Fos/AP‐1 target genes, FAM177B (B) and BBOX1 (C) were identified as potential risk loci. Linkage disequilibrium (r 2) was calculated based on the combined 1000 genomes. D, Reanalyzing RNA‐seq data (GSE165303) of dilated cardiomyopathy (DCM) and control patients (CTL) to determine the dysregulated expression of Fos/AP‐1 signaling in heart failure. E through G, Using 222 Fos/AP‐1 target genes, the Fos/AP‐1 activity was determined in RNA‐seq data sets of control and DCM patients (E, GSE165303, n=51, 50 in CTL and DCM, respectively; Student's t tests), or control, DCM, and ischemic cardiomyopathy (ICM) patients (F, GSE116250, n=14, 37, 13, respectively; 1‐way ANOVA followed by Bonferroni test), or the normal, patients with ICM before and after left ventricular assist device (LVAD) treatment (G, GSE46224, n=8 for each group; 1‐way ANOVA followed by Bonferroni test). AP‐1 indicates activator protein 1; CAD, coronary atherosclerosis diseases; MI, myocardial infarction; SNP, single nucleotide polymorphism; and TF, transcriptional regulon.
To address this hypothesis, mice after MI surgeries were randomly treated with the selective Fos/AP‐1 inhibitor T5224, or its vehicle, orally for 7 consecutive days (Figure S15A). Echocardiographic analysis revealed that, compared with the vehicle group, T5224 treatment improved cardiac function at 1 and 4 weeks post MI, with increased left ventricular ejection fraction and left ventricular fraction shortening, as well as rescued left ventricular end‐diastolic volume and left ventricular end‐systolic volume (Figure 8A and 8B). Consistent with the echocardiographic results, scar size was significantly decreased in T5224‐treated mice 4 weeks post MI compared with that of the vehicle group (Figure 8C and 8D, and Figure S15B). Intriguingly, T5224 supplementation attenuated cardiac leukocyte infiltration (CD45+ cells) compared with that of vehicle treated mice using flow cytometry analysis. Moreover, the abundance of neutrophils (CD45+ Cd11b+ LY6G+) and macrophages (CD45+ Cd11b+ LY6G− F4/80+) was reduced in T5224‐treated hearts, in contrast to that seen in the hearts of vehicle mice (Figure 8E and 8F). Altogether, these observations revealed that the specific inhibition of Fos/AP‐1 TF activities with T5224 effectively diminished ischemia‐induced immune responses and yielded therapeutic benefits by mitigating adverse cardiac remodeling and heart failure.
Figure 8. The selective Fos/AP‐1 inhibition mitigated cardiac leukocyte infiltration and cardiac dysfunction.
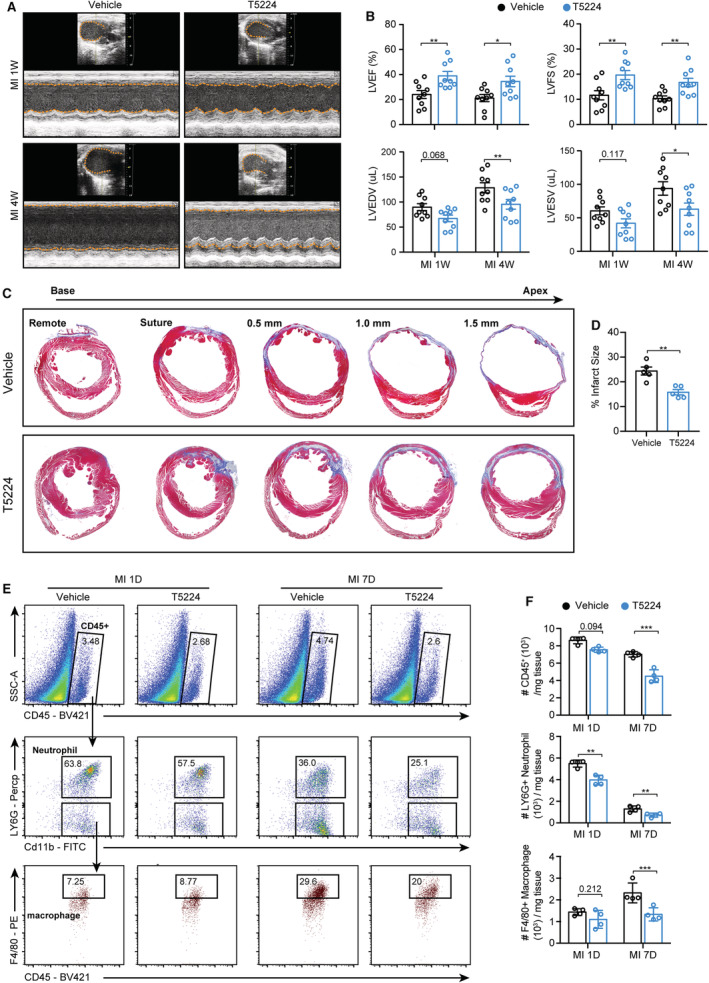
A, Wild‐type mice were subjected to MI surgery and randomly treated with Fos/AP‐1 inhibitor (T5224) or its vehicle for 7 consecutive days, then the echocardiography was used to compare the cardiac function at 1 and 4 weeks post MI. B, Quantification of A (n=9 for each group; 2‐way ANOVA followed by least significant difference test). C, Masson trichrome staining of vehicle‐ or T5224‐treated mice to determine the infarct size at 4 weeks post MI. D, Quantification results of C (n=5 for each group; Student's t‐tests). E, Number of leukocytes, neutrophils, and macrophages at 1 and 7 days after MI in vehicle‐ or T5224‐treated mice, as measured by flow cytometry analysis. F, Quantification results of panel E (n=4 for each group; 2‐way ANOVA followed by least significant difference test). AP‐1 indicates activator protein 1; LVEDV, left ventricular end diastolic volume; LVEF, left ventricular ejection fraction; LVESV, left ventricular end systolic volume; LVFS, left ventricular fraction shortening; and MI, myocardial infarction.
DISCUSSION
In this study, we used scRNA‐seq technology to investigate cardiac CD45+ immune cell subsets under the early inflammatory and late reparative phases to understand the underlying mechanisms of immune initiation and resolution after MI. We showed that 2 ischemia‐associated macrophage groups (MAC_Olr1 and MAC_Gpnmb) were extensively expanded after MI, with distinct immune phenotypes and energy preferences. Proinflammatory MAC_Olr1 clusters with high myeloid‐derived suppressor cell and SASP scores were related to high cellular glycolysis activity. On the other hand, MAC_Gpnmb clusters exhibited increased phagocytic capacity and FAO properties. We further revealed the requirement of FAO for Gpnmb‐mediated phagocytosis. Additionally, 3 neutrophil subsets were identified in the scRNA‐seq data with differential ontogenetic states, which were validated in murine hearts and in the PB of patients with ACS.
We also comprehensively analyzed the transcriptional profiles and regulons of DC, T/NK cells, and B cells in the scRNA‐seq data to elucidate their specific cellular responses after MI. Cardiac healing after MI is an orchestrated process that requires timely leukocyte recruitment and immunosuppressive responses for immune resolution. By focusing on cellular crosstalk, we revealed that CCL/CXCL pathways were critical for myeloid cell infiltration, while DC_Ccr7 and Mo_Ear2 subgroups were responsible for the resolution of cardiac immunity via CD80/ICOS signaling. Finally, we revealed that Fos/AP‐1 governed the cardiac inflammatory responses and was responsible for the progression of cardiac dysfunction after MI. Targeting Fos/AP‐1 with its specific inhibitor, T5224, produced therapeutic benefits for MI treatment.
Compared with the MAC_Olr1 subcluster, MAC_Gpnmb represented the restorative subset with higher phagocytic capacity and FAO. Macrophages have been described as key factors limiting cardiac inflammation via their phagocytic capacity. After engulfing apoptotic cardiomyocytes or debris, macrophages show a proresolutive phenotype by upregulating IL‐10 and TGFβ expression, while downregulating IL‐1β and TNF expression. 14 In addition, a series of publications have linked Gpnmb with macrophage phagocytosis, indicating that Gpnmb is a biomarker of lysosome and phagocytosis dysfunction. 49 Li et al reported that Gpnmb expression was markedly upregulated in macrophages during kidney ischemia and was critical for phagosome formation and LC3 recruitment. 50 By examining macrophage heterogeneity in a CCL4‐induced hepatic fibrosis model, researchers have pointed out that abundant proreparative macrophages emerged during immune resolutive phases with upregulated Gpnmb, Igf1, and phagocytosis‐related genes, 51 which confirmed the linkage of Gpnmb with phagocytosis and the restorative macrophage phenotype. Beyond being an intrinsic trigger of phagocytosis, Gpnmb is associated with lipid metabolism and metabolic disorders. In a preclinical high‐cholesterol diet model 52 and human obesity diseases, 53 Gpnmb expression was significantly elevated. Importantly, Gpnmb contributes to lipogenesis of white adipose tissue, suggesting a relationship between Gpnmb and lipid metabolism. 54 Here, we observed that MAC_Gpnmb was expanded in MI‐7D and was positively correlated with phagocytosis and cellular FAO levels. Overexpressing Gpnmb using adenovirus transfection increased macrophage phagocytic capacities, while inhibiting FAO with etomoxir impaired Gpnmb‐induced phagocytosis. Considering the intriguing effects of phagocytosis and FAO on the switching of macrophage to the proreparative phenotype, targeting Gpnmb might be a novel therapeutic approach for the treatment of MI.
Furthermore, through single‐cell regulatory network inference and clustering regulon analysis, we identified the Fos/AP‐1 TF as the key regulator of proinflammatory responses. Increased Fos/AP‐1 activities were observed in Folr2− negative inflammatory macrophage, neutrophil, DC_Cd209, and T‐cell subclusters (CD4_C2_CXCR3 and CD8_C2_Gzmk) under ischemic conditions. The AP‐1 transcriptional regulon comprises the JUN (c‐Jun, JunB, JunD), FOS (c‐Fos, Fosb, Fosl1), ATF (activating transcription factor, ATF2, ATF3), and MAF (musculoaponeurotic fibrosarcoma, c‐Maf, MafB, MafA) families, and each member can form homodimers or heterodimers with other members to modulate the transcriptional events of different genes. The AP‐1 complex controls a broad range of cellular processes, including cell proliferation, inflammation, differentiation, and tumorigenesis. 55 , 56 A series of published works have reported that Fos/AP‐1 contributes to the activation and maintaining of inflammatory responses in DCs 57 and macrophages. 58 Consistently, our studies reported that the specific transactivation of c‐Fos, Fosl1, and Fosb resulted in a broad‐spectrum upregulation of proinflammatory gene expression and the increase in local inflammatory macrophage recruitment after MI. 59 Palomer et al showed that Fos protein deficit attenuated cardiac inflammatory responses by downregulating IL‐6 and MCP1 (monocyte chemoattractant protein‐1) expression. 60 These studies suggest inhibiting Fos/AP‐1 activity as a potential anti‐inflammatory therapeutic treatment for attenuating cardiac remodeling and dysfunction after MI. To further determine the effects of Fos/AP‐1 signaling in MI, we used the Fos/AP‐1 inhibitor T5224 in this study. T5224 was first designed for arthritis treatment by Aikawa et al, who described that T5224 administration significantly suppressed the levels of inflammatory cytokines (IL‐1b, IL‐6, and COMP) and MMP3 in vivo and in vitro, indicating its intriguing role in immune supression. 61 Similarly, Fos/AP‐1 inhibition with T5224 played anti‐inflammatory roles in endotoxin‐induced acute kidney injury 62 and in lipopolysaccharide‐induced liver injury models. 63 For the first time, our study explored the effects and underlying mechanisms of Fos/AP‐1 inhibition during MI and demonstrated that T5224 administration effectively suppressed cardiac inflammatory responses, reduced infarct size, and mitigated cardiac remodeling and heart failure.
Although our study comprehensively explored the transcriptional profile, functional changes, and regulons of cardiac immune cells under early inflammatory and late reparative phases, there are some limitations that should be acknowledged. First, this study was designed to explore the heterogeneity of immune cells after MI; therefore, our scRNA‐seq data only included CD45+ immune cells. Nonimmune cells, including fibroblasts, endothelial cells, and cardiomyocytes, were not detected in this data set. Therefore, we could not compare the DEGs of nonimmune cells under homeostatic or ischemic conditions, or determine the cellular crosstalk between nonimmune and immune cells. Second, our scRNA‐seq data detected only a minor proportion of adaptive immune cells under sham or ischemic conditions, which impeded the efficiency to dissect and explore the heterogeneity of T/B cells and their T‐cell receptor or B‐cell receptor changes. Comprehensive scRNA‐seq studies with enriched T/B cells are needed in the future. Finally, in this study, we used male mice for all preclinical experiments, which are widely used as an ideal research tool for cardiac diseases. However, potential sexual dimorphism can exist in specific cell lineages or novel cell subclusters, and further scRNA‐seq studies that include female mice should be conducted to survey any sexual dimorphism of immune cells under homeostatic or ischemic conditions.
CONCLUSIONS
In conclusion, this study provides a potential resource for the understanding of cardiac immune responses in early inflammatory and late reparative phases of MI. We also identified Fos/AP‐1 as the central regulator of cardiac immune responses. Insights involving their regulatory effects and functional results after inhibitor administration provide preclinical data for MI treatment.
Sources of Funding
This study was supported by the National Natural Science Foundation of China (82070510 to R.Z.; 81922007, 91939103, and 82120108003 to X.Y.), Shanghai Municipal Education Commission‐Gaofeng Clinical Medicine Grant (20191803 to X.Y.), the “Shuguang Program”, supported by the Shanghai Education Development Foundation and Shanghai Municipal Education Commission (20SG15 to X.Y.), the Shanghai Science and Technology Supporting Project (19411963400 to X.Y.), and the Program of Shanghai Academic/Technology Research Leader (21XD1422500).
Disclosures
None.
Supporting information
Data S1–S4
Table S1
Figures S1–S15
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.122.027228
For Sources of Funding and Disclosures, see page 19.
Contributor Information
Jun Liu, Email: liujun52@126.com.
Xiaoxiang Yan, Email: cardexyanxx@hotmail.com.
References
- 1. Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA, White HD, Executive group on behalf of the joint European Society of Cardiology/American College of Cardiology/American Heart Association/World Heart Federation Task Force for the Universal Definition of Myocardial I . Fourth Universal definition of myocardial infarction (2018). J Am Coll Cardiol. 2018;72:2231–2264. doi: 10.1016/j.jacc.2018.08.1038 [DOI] [PubMed] [Google Scholar]
- 2. Swirski FK, Nahrendorf M. Cardioimmunology: the immune system in cardiac homeostasis and disease. Nat Rev Immunol. 2018;18:733–744. doi: 10.1038/s41577-018-0065-8 [DOI] [PubMed] [Google Scholar]
- 3. Gerlach BD, Ampomah PB, Yurdagul A Jr, Liu C, Lauring MC, Wang X, Kasikara C, Kong N, Shi J, Tao W, et al. Efferocytosis induces macrophage proliferation to help resolve tissue injury. Cell Metab. 2021;33:2445–2463. doi: 10.1016/j.cmet.2021.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yan X, Zhang H, Fan Q, Hu J, Tao R, Chen Q, Iwakura Y, Shen W, Lu L, Zhang Q, et al. Dectin‐2 deficiency modulates Th1 differentiation and improves wound healing after myocardial infarction. Circ Res. 2017;120:1116–1129. doi: 10.1161/CIRCRESAHA.116.310260 [DOI] [PubMed] [Google Scholar]
- 5. Epelman S, Liu PP, Mann DL. Role of innate and adaptive immune mechanisms in cardiac injury and repair. Nat Rev Immunol. 2015;15:117–129. doi: 10.1038/nri3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bajpai G, Schneider C, Wong N, Bredemeyer A, Hulsmans M, Nahrendorf M, Epelman S, Kreisel D, Liu Y, Itoh A, et al. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat Med. 2018;24:1234–1245. doi: 10.1038/s41591-018-0059-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bajpai G, Bredemeyer A, Li W, Zaitsev K, Koenig AL, Lokshina I, Mohan J, Ivey B, Hsiao HM, Weinheimer C, et al. Tissue resident CCR2− and CCR2+ cardiac macrophages differentially orchestrate monocyte recruitment and fate specification following myocardial injury. Circ Res. 2019;124:263–278. doi: 10.1161/CIRCRESAHA.118.314028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dick SA, Macklin JA, Nejat S, Momen A, Clemente‐Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh S, Aronoff L, et al. Self‐renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol. 2019;20:29–39. doi: 10.1038/s41590-018-0272-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, et al. Embryonic and adult‐derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40:91–104. doi: 10.1016/j.immuni.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horckmans M, Ring L, Duchene J, Santovito D, Schloss MJ, Drechsler M, Weber C, Soehnlein O, Steffens S. Neutrophils orchestrate post‐myocardial infarction healing by polarizing macrophages towards a reparative phenotype. Eur Heart J. 2017;38:187–197. doi: 10.1093/eurheartj/ehw002 [DOI] [PubMed] [Google Scholar]
- 11. Ma Y, Yabluchanskiy A, Iyer RP, Cannon PL, Flynn ER, Jung M, Henry J, Cates CA, Deleon‐Pennell KY, Lindsey ML. Temporal neutrophil polarization following myocardial infarction. Cardiovasc Res. 2016;110:51–61. doi: 10.1093/cvr/cvw024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ong SB, Hernandez‐Resendiz S, Crespo‐Avilan GE, Mukhametshina RT, Kwek XY, Cabrera‐Fuentes HA, Hausenloy DJ. Inflammation following acute myocardial infarction: multiple players, dynamic roles, and novel therapeutic opportunities. Pharmacol Ther. 2018;186:73–87. doi: 10.1016/j.pharmthera.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hilgendorf I, Gerhardt LM, Tan TC, Winter C, Holderried TA, Chousterman BG, Iwamoto Y, Liao R, Zirlik A, Scherer‐Crosbie M, et al. Ly‐6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ Res. 2014;114:1611–1622. doi: 10.1161/CIRCRESAHA.114.303204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF‐beta, PGE2, and PAF. J Clin Invest. 1998;101:890–898. doi: 10.1172/JCI1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liao YH, Xia N, Zhou SF, Tang TT, Yan XX, Lv BJ, Nie SF, Wang J, Iwakura Y, Xiao H, et al. Interleukin‐17A contributes to myocardial ischemia/reperfusion injury by regulating cardiomyocyte apoptosis and neutrophil infiltration. J Am Coll Cardiol. 2012;59:420–429. doi: 10.1016/j.jacc.2011.10.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yan X, Shichita T, Katsumata Y, Matsuhashi T, Ito H, Ito K, Anzai A, Endo J, Tamura Y, Kimura K, et al. Deleterious effect of the IL‐23/IL‐17A axis and gammadeltaT cells on left ventricular remodeling after myocardial infarction. J Am Heart Assoc. 2012;1:e004408. doi: 10.1161/JAHA.112.004408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vafadarnejad E, Rizzo G, Krampert L, Arampatzi P, Arias‐Loza AP, Nazzal Y, Rizakou A, Knochenhauer T, Bandi SR, Nugroho VA, et al. Dynamics of cardiac neutrophil diversity in murine myocardial infarction. Circ Res. 2020;127:e232–e249. doi: 10.1161/CIRCRESAHA.120.317200 [DOI] [PubMed] [Google Scholar]
- 18. Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub‐Lis K, Ho JW, Nordon RE, Harvey RP. Single‐cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife. 2019;8:e43882. doi: 10.7554/eLife.43882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martini E, Kunderfranco P, Peano C, Carullo P, Cremonesi M, Schorn T, Carriero R, Termanini A, Colombo FS, Jachetti E, et al. Single‐cell sequencing of mouse heart immune infiltrate in pressure overload‐driven heart failure reveals extent of immune activation. Circulation. 2019;140:2089–2107. doi: 10.1161/CIRCULATIONAHA.119.041694 [DOI] [PubMed] [Google Scholar]
- 20. Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A. Single‐cell RNA‐Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res. 2018;122:1661–1674. doi: 10.1161/CIRCRESAHA.117.312509 [DOI] [PubMed] [Google Scholar]
- 21. Wang L, Yu P, Zhou B, Song J, Li Z, Zhang M, Guo G, Wang Y, Chen X, Han L, et al. Single‐cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat Cell Biol. 2020;22:108–119. doi: 10.1038/s41556-019-0446-7 [DOI] [PubMed] [Google Scholar]
- 22. van den Brink SC, Sage F, Vertesy A, Spanjaard B, Peterson‐Maduro J, Baron CS, Robin C, van Oudenaarden A. Single‐cell sequencing reveals dissociation‐induced gene expression in tissue subpopulations. Nat Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437 [DOI] [PubMed] [Google Scholar]
- 23. Yan X, Anzai A, Katsumata Y, Matsuhashi T, Ito K, Endo J, Yamamoto T, Takeshima A, Shinmura K, Shen W, et al. Temporal dynamics of cardiac immune cell accumulation following acute myocardial infarction. J Mol Cell Cardiol. 2013;62:24–35. doi: 10.1016/j.yjmcc.2013.04.023 [DOI] [PubMed] [Google Scholar]
- 24. Mouton AJ, DeLeon‐Pennell KY, Rivera Gonzalez OJ, Flynn ER, Freeman TC, Saucerman JJ, Garrett MR, Ma Y, Harmancey R, Lindsey ML. Mapping macrophage polarization over the myocardial infarction time continuum. Basic Res Cardiol. 2018;113:26. doi: 10.1007/s00395-018-0686-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, Modak M, Carotta S, Haslinger C, Kind D, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845. doi: 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 26. Yousefzadeh MJ, Flores RR, Zhu Y, Schmiechen ZC, Brooks RW, Trussoni CE, Cui Y, Angelini L, Lee KA, McGowan SJ, et al. An aged immune system drives senescence and ageing of solid organs. Nature. 2021;594:100–105. doi: 10.1038/s41586-021-03547-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soto‐Heredero G, de Las G, Heras MM, Gabande‐Rodriguez E, Oller J, Mittelbrunn M. Glycolysis ‐ a key player in the inflammatory response. FEBS J. 2020;287:3350–3369. doi: 10.1111/febs.15327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. King KR, Aguirre AD, Ye YX, Sun Y, Roh JD, Ng RP Jr, Kohler RH, Arlauckas SP, Iwamoto Y, Savol A, et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat Med. 2017;23:1481–1487. doi: 10.1038/nm.4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, Pittet MJ. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Willenborg S, Lucas T, van Loo G, Knipper JA, Krieg T, Haase I, Brachvogel B, Hammerschmidt M, Nagy A, Ferrara N, et al. CCR2 recruits an inflammatory macrophage subpopulation critical for angiogenesis in tissue repair. Blood. 2012;120:613–625. doi: 10.1182/blood-2012-01-403386 [DOI] [PubMed] [Google Scholar]
- 31. Pedragosa J, Miro‐Mur F, Otxoa‐de‐Amezaga A, Justicia C, Ruiz‐Jaen F, Ponsaerts P, Pasparakis M, Planas AM. CCR2 deficiency in monocytes impairs angiogenesis and functional recovery after ischemic stroke in mice. J Cereb Blood Flow Metab. 2020;40:S98–S116. doi: 10.1177/0271678X20909055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu W, Ma G, Sheng Z, Wang Q, Chen L, Qi J, Shi R, Ji J, Ji Z, Dai Q. MSCs contribute to the conversion of Ly6C(high) monocytes into Ly6C(low) subsets under AMI. Stem Cells Int. 2020;2020:2460158–2460110. doi: 10.1155/2020/2460158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung K, Heishi T, Incio J, Huang Y, Beech EY, Pinter M, Ho WW, Kawaguchi K, Rahbari NN, Chung E, et al. Targeting CXCR4‐dependent immunosuppressive Ly6C(low) monocytes improves antiangiogenic therapy in colorectal cancer. Proc Natl Acad Sci USA. 2017;114:10455–10460. doi: 10.1073/pnas.1710754114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Narasimhan PB, Marcovecchio P, Hamers AAJ, Hedrick CC. Nonclassical monocytes in health and disease. Annu Rev Immunol. 2019;37:439–456. doi: 10.1146/annurev-immunol-042617-053119 [DOI] [PubMed] [Google Scholar]
- 35. Dick SA, Wong A, Hamidzada H, Nejat S, Nechanitzky R, Vohra S, Mueller B, Zaman R, Kantores C, Aronoff L, et al. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci Immunol. 2022;7:eabf7777. doi: 10.1126/sciimmunol.abf7777 [DOI] [PubMed] [Google Scholar]
- 36. Xie X, Shi Q, Wu P, Zhang X, Kambara H, Su J, Yu H, Park SY, Guo R, Ren Q, et al. Single‐cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat Immunol. 2020;21:1119–1133. doi: 10.1038/s41590-020-0736-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lawrence SM, Corriden R, Nizet V. The ontogeny of a neutrophil: mechanisms of granulopoiesis and homeostasis. Microbiol Mol Biol Rev. 2018;82: e00057‐17. doi: 10.1128/MMBR.00057-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grieshaber‐Bouyer R, Nigrovic PA. Neutrophil heterogeneity as therapeutic opportunity in immune‐mediated disease. Front Immunol. 2019;10:346. doi: 10.3389/fimmu.2019.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heath WR, Carbone FR. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat Immunol. 2009;10:1237–1244. doi: 10.1038/ni.1822 [DOI] [PubMed] [Google Scholar]
- 41. LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112:1570–1580. doi: 10.1182/blood-2008-02-078071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zouggari Y, Ait‐Oufella H, Bonnin P, Simon T, Sage AP, Guerin C, Vilar J, Caligiuri G, Tsiantoulas D, Laurans L, et al. B lymphocytes trigger monocyte mobilization and impair heart function after acute myocardial infarction. Nat Med. 2013;19:1273–1280. doi: 10.1038/nm.3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao TX, Ur‐Rahman MA, Sage AP, Victor S, Kurian R, Fielding S, Ait‐Oufella H, Chiu YD, Binder CJ, McKie M, et al. Rituximab in patients with acute ST‐elevation myocardial infarction (RITA‐MI): an experimental medicine safety study. Cardiovasc Res. 2021;118:872–882. doi: 10.1093/cvr/cvab113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jin S, Guerrero‐Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q. Inference and analysis of cell‐cell communication using CellChat. Nat Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1000 genomes‐based genome‐wide association meta‐analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feng Y, Cai L, Hong W, Zhang C, Tan N, Wang M, Wang C, Liu F, Wang X, Ma J, et al. Rewiring of 3D chromatin topology orchestrates transcriptional reprogramming and the development of human dilated cardiomyopathy. Circulation. 2022;145:1663–1683. doi: 10.1161/CIRCULATIONAHA.121.055781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sweet ME, Cocciolo A, Slavov D, Jones KL, Sweet JR, Graw SL, Reece TB, Ambardekar AV, Bristow MR, Mestroni L, et al. Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genomics. 2018;19:812. doi: 10.1186/s12864-018-5213-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, Nerbonne JM. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129:1009–1021. doi: 10.1161/CIRCULATIONAHA.113.003863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van der Lienden MJC, Gaspar P, Boot R, Aerts J, van Eijk M. Glycoprotein non‐metastatic protein B: an emerging biomarker for lysosomal dysfunction in macrophages. Int J Mol Sci. 2018;20:66. doi: 10.3390/ijms20010066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li B, Castano AP, Hudson TE, Nowlin BT, Lin SL, Bonventre JV, Swanson KD, Duffield JS. The melanoma‐associated transmembrane glycoprotein Gpnmb controls trafficking of cellular debris for degradation and is essential for tissue repair. FASEB J. 2010;24:4767–4781. doi: 10.1096/fj.10-154757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ramachandran P, Pellicoro A, Vernon MA, Boulter L, Aucott RL, Ali A, Hartland SN, Snowdon VK, Cappon A, Gordon‐Walker TT, et al. Differential Ly‐6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci USA. 2012;109:E3186–E3195. doi: 10.1073/pnas.1119964109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu J, Jullig M, Middleditch MJ, Cooper GJ. Modelling atherosclerosis by proteomics: molecular changes in the ascending aortas of cholesterol‐fed rabbits. Atherosclerosis. 2015;242:268–276. doi: 10.1016/j.atherosclerosis.2015.07.001 [DOI] [PubMed] [Google Scholar]
- 53. Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW Jr. Obesity activates a program of lysosomal‐dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gong XM, Li YF, Luo J, Wang JQ, Wei J, Wang JQ, Xiao T, Xie C, Hong J, Ning G, et al. GPNMB secreted from liver promotes lipogenesis in white adipose tissue and aggravates obesity and insulin resistance. Nat Metab. 2019;1:570–583. doi: 10.1038/s42255-019-0065-4 [DOI] [PubMed] [Google Scholar]
- 55. Shaulian E, Karin M. AP‐1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131 [DOI] [PubMed] [Google Scholar]
- 56. Eferl R, Wagner EF. AP‐1: a double‐edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- 57. Zhu J, Yao K, Guo J, Shi H, Ma L, Wang Q, Liu H, Gao W, Sun A, Zou Y, et al. miR‐181a and miR‐150 regulate dendritic cell immune inflammatory responses and cardiomyocyte apoptosis via targeting JAK1‐STAT1/c‐Fos pathway. J Cell Mol Med. 2017;21:2884–2895. doi: 10.1111/jcmm.13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hannemann N, Cao S, Eriksson D, Schnelzer A, Jordan J, Eberhardt M, Schleicher U, Rech J, Ramming A, Uebe S, et al. Transcription factor Fra‐1 targets arginase‐1 to enhance macrophage‐mediated inflammation in arthritis. J Clin Invest. 2019;129:2669–2684. doi: 10.1172/JCI96832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang CK, Dai D, Xie H, Zhu Z, Hu J, Su M, Liu M, Lu L, Shen W, Ning G, et al. Lgr4 governs a pro‐inflammatory program in macrophages to antagonize post‐infarction cardiac repair. Circ Res. 2020;127:953–973. doi: 10.1161/CIRCRESAHA.119.315807 [DOI] [PubMed] [Google Scholar]
- 60. Palomer X, Roman‐Azcona MS, Pizarro‐Delgado J, Planavila A, Villarroya F, Valenzuela‐Alcaraz B, Crispi F, Sepulveda‐Martinez A, Miguel‐Escalada I, Ferrer J, et al. SIRT3‐mediated inhibition of FOS through histone H3 deacetylation prevents cardiac fibrosis and inflammation. Signal Transduct Target Ther. 2020;5:14. doi: 10.1038/s41392-020-0114-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Aikawa Y, Morimoto K, Yamamoto T, Chaki H, Hashiramoto A, Narita H, Hirono S, Shiozawa S. Treatment of arthritis with a selective inhibitor of c‐Fos/activator protein‐1. Nat Biotechnol. 2008;26:817–823. doi: 10.1038/nbt1412 [DOI] [PubMed] [Google Scholar]
- 62. Miyazaki H, Morishita J, Ueki M, Nishina K, Shiozawa S, Maekawa N. The effects of a selective inhibitor of c‐Fos/activator protein‐1 on endotoxin‐induced acute kidney injury in mice. BMC Nephrol. 2012;13:153. doi: 10.1186/1471-2369-13-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Izuta S, Ueki M, Ueno M, Nishina K, Shiozawa S, Maekawa N. T‐5224, a selective inhibitor of c‐Fos/activator protein‐1, attenuates lipopolysaccharide‐induced liver injury in mice. Biotechnol Lett. 2012;34:2175–2182. doi: 10.1007/s10529-012-1022-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1–S4
Table S1
Figures S1–S15
Data Availability Statement
All relevant data are available in the figures and supplemental material. Raw and analyzed RNA‐sequencing data generated during this study will be available in the Gene Expression Omnibus repository once this study is accepted.


