Abstract
Plasmodium falciparum infections can be fatal, while P. vivax infections usually are not. A possible factor involved in the greater virulence of P. falciparum is that this parasite grows in red blood cells (RBCs) of all maturities whereas P. vivax is restricted to growth in reticulocytes, which represent only approximately 1% of total RBCs in the periphery. Two proteins, expressed at the apical end of the invasive merozoite stage from P. vivax, have been implicated in the targeting of reticulocytes for invasion by this parasite. A search of the P. falciparum genome databases has identified genes that are homologous to the P. vivax rbp-1 and -2 genes. Two of these genes are virtually identical over a large region of the 5′ end but are highly divergent at the 3′ end. They encode high-molecular-mass proteins of >300 kDa that are expressed in late schizonts and localized to the apical end of the merozoite. To test a potential role in merozoite invasion of RBCs, we analyzed the ability of these proteins to bind to mature RBCs and reticulocytes. No binding to mature RBCs or cell preparations enriched for reticulocytes was detected. We identified a parasite clone that lacks the gene for one of these proteins, showing that the gene is not required for normal in vitro growth. Antibodies to these proteins can inhibit merozoite invasion of RBCs.
A number of Plasmodium species cause malaria in humans. Plasmodium falciparum, the most virulent form that causes human malaria, invades both reticulocytes and mature red blood cells (RBCs), while P. vivax and P. ovale, which cause less severe disease, are both restricted to reticulocytes. Plasmodium species that infect rodents also show a preference for RBCs of different stages of development and maturity. For example, virulent strains of P. yoelii invade both mature and immature RBCs, while nonlethal strains show a preference for reticulocytes (9). Hence, members of the Plasmodium species can be divided into two groups: those that predominantly invade reticulocytes, and those which invade RBCs at all stages of maturity. The basis of this RBC specificity is presumably the presence of different ligands at the apical end of the invasive merozoite stage of the various species.
A 235-kDa rhoptry protein from P. yoelii has been suggested to be important in the ability of this parasite to invade mature RBCs (8). Passive transfer of monoclonal antibodies (MAbs) specific to this protein protect mice infected with the virulent YM strain, by restricting invasion of reticulocytes (4). In P. vivax, proteins termed P. vivax reticulocyte binding protein 1 (PvRBP-1) and PvRBP-2 have been shown to bind reticulocyte-enriched RBCs (5). PvRBP-1 and PvRBP-2 form a protein complex through noncovalent interactions and colocalize to the apical end of the merozoite. The PvRBP-1 and PvRBP-2 proteins have calculated molecular masses of 325 and 330 kDa, respectively, and share similar structures with a signal sequence at the N terminus and a putative transmembrane domain and cytoplasmic tail at the C terminus (6). Interestingly, the full sequence of one member of the Py235 family recently deposited in GenBank (accession no. U36927) encodes a protein with a predicted molecular mass of 325 kDa and the same structure as PvRBP-1 and -2 (7). PvRBP-2 and members of the Py235 family share a 500-amino-acid region which shows significant homology (9). The Py235 proteins are encoded by a multigene family of up to 50 members, with at least 11 distinct genes spread across different chromosomes of the P. yoelii genome (2). At least one member of this protein family has been shown to bind both mature and immature RBCs (11), a finding consistent with the fact that a Py235 MAb can restrict parasite invasion of reticulocytes (4).
More recently, it has been found that individual merozoites within a single developing schizont can have different Py235 genes transcribed in P. yoelii (14). It is not known if each Py235 protein has a distinct target cell specificity, but it is likely that the proteins are antigenically distinct. This would ensure that even with host anti-Py235 antibodies, some merozoites would be free to invade new RBCs at each cycle.
In this study, we describe two genes initially identified from the P. falciparum genome databases (P. falciparum sequencing group at the Sanger Centre [ftp://ftp.sanger.ac.uk/pub/ databases/P.falciparum_sequences], the Stanford DNA Sequencing and Technology Centre [http://sequence-www.stanford.edu/group/malaria], and The Institute for Genomic Research [ftp://ftp.tigr.org]) that are homologous to Pvrbp-2 and the Py235 family. We have analyzed the expression of these genes, and results of immunofluorescence assay (IFA) experiments are consistent with a subcellular localization at the apical end of the merozoite. By analogy with the role of the other members of this family, these proteins may be involved in the targeting of particular RBC subpopulations for invasion by P. falciparum merozoites.
MATERIALS AND METHODS
Parasites and nucleic acids.
Parasites were maintained (20) and synchronized by standard procedures. Genomic DNA (gDNA) was extracted from trophozoites as described elsewhere (21). Southern blotting was carried out using standard procedures. Poly(A)+ RNA was obtained from synchronized late-stage schizont cultures (Ambion Inc.) and then converted to cDNA using Superscript II (Gibco-BRL).
Antibodies.
Two Pfr2h fragments were amplified by PCR from 3D7 genomic DNA, subcloned into pGEX, and fusion protein affinity purified on glutathione-agarose. The fusion proteins were used to immunize both rabbits and mice. The primers used for production of the 2A9 antibody were 5′-GGATGGATCCGAATTACGTGAATTGTCTACGGC-3′ and 5′-TATTCTCGAGCATCTCTTCCATTTGAAATAATTTTTC-3′. The primers used for the 2A11 antibody were 5′-GAGGGATCCCTTAATATAAATAATATTATGAATGAAACG-3′ and 5′-TTGACTCGAGGTCATCTTTTTTTTCTTTAGATGTTATC-3′. Note that the latter primer pair could amplify only the Pfr2ha gene, but part of the product overlaps the Pfr2hb gene. Locations of the expressed fragments in relation to the complete PfR2H proteins are shown in Fig. 2C. Other antibodies used in the IFA experiments were a mouse MAb specific to P. falciparum apical membrane antigen 1 (PfAMA1), 2C5 (22), a mouse MAb specific to P. falciparum rhoptry-associated antigen 1 (PfRAP1), 7H8/50 (17), and a mouse anti-EBA175 (erythrocyte binding antigen 175) serum (16).
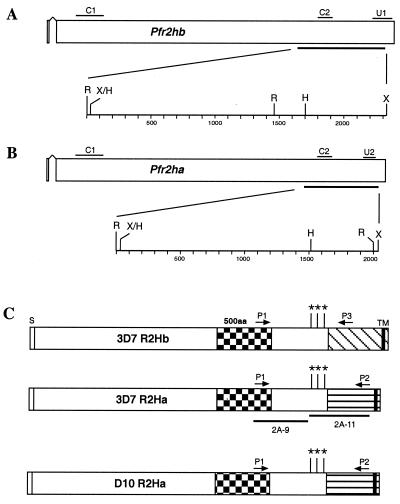
FIG. 2.
3D7 parasites have both Pfr2ha and Pfr2hb genes, while D10 has only Pfr2ha. (A) Schematic representation of the Pfr2hb gene in 3D7 parasites. The short signal peptide (exon 1) is followed by an intron and then exon 2. The C1 probe common to both Pfr2ha and Pfr2hb was amplified from D10 gDNA using primers 5′-ACAGGAAATATGTGAAAAACGG-3′ and 5′-TTATTATTATTAGTGTTTTTAC-3′. The C2 probe common to both Pfr2ha and Pfr2hb was amplified from 3D7 gDNA using primers 5′-CACCAAGATCCTTTATATCA-3′ and 5′-CTTAATATAAATAATATTATGAAT-3′. The U1 probe, unique to the Pfr2hb gene, was amplified from 3D7 gDNA using primers 5′-GAATTGATAGTACTGACCAACGT-3′ and 5′-CTTCATTTTCATCAAACACAATTTC-3′. A region of 2,312 bp bounded by RsaI (R) and XmnI (X) sites is shown expanded below the Pfr2hb gene. The HinfI (H) site is shown together with fragment sizes in base pairs. (B) Schematic representation of the Pfr2ha gene in D10 and 3D7 parasites. The C1 and C2 probes are as in panel A. The U2 probe was amplified from 3D7 gDNA using primers 5′-TAAACTAGAATCTGATATGGTGA-3′ and 5′-GTCATCTTTTTTTTCTTTAGATGT-3′. A region of 2,037 bp bounded by RsaI and XmnI sites is shown expanded below the Pfr2ha gene. (C) Schematic representation of the PfR2Ha and -Hb proteins in 3D7 and the PfR2Ha protein in D10 parasites. The three sequences are presumed to be nearly identical (see Results) but differ markedly from amino acid (aa) 2776 onward even though they are structurally similar, with a putative transmembrane domain (TM) and a short cytoplasmic tail at the C terminus. The unique regions are shown as diagonally hatched in R2Hb and horizontally hatched in R2Ha. The 500-amino-acid region showing some conservation in PvRBP-2 and Py235 as shown in Fig. 1 is indicated. The DNA corresponding to approximately 1,100 amino acids at the C terminus of the 3D7 r2ha and r2hb genes and approximately 800 amino acids at the C terminus of the D10 r2hb gene was sequenced. This encompassed the regions to which the 2A9 and 2A11 antibodies were made. For PCR amplification of the D10 and 3D7 r2ha genes, primers P1 (5′-AATTACGTGAATTGTCTACGGC-3′) and P2 (5′-GTCATCTTTTTTTTCTTTAGATGTTATC-3′) were used. For amplification of the 3D7 r2hb gene, primers P1 and P3 (5′-AAACAACATGATCATACGCATTG-3′) were used. The approximate locations of primers P1, P2, and P3 are shown. The PCR products were fully sequenced using internal primers. The amino acid differences within the regions of the three genes which are nearly identical are indicated by asterisks. The 5′-most change (amino acid 2546) is A (Ala) in 3D7 R2Ha but D (Asp) in the other proteins. The next change (position 2635) is E (Glu) in both R2Ha proteins but K (Lys) in 3D7 R2Hb. The 3′-most change (beginning at position 2719) is EEELRKK in 3D7 R2Ha but EALKKQ in the other proteins. The portions of PfR2Ha and -Hb used for production of rabbit antibodies 2A9 and 2A11 are also shown. The lengths of the signal sequence (S) and transmembrane domain (TM) are not shown to scale. The checkered shading represents the 500-amino-acid conserved region. The diagonal and horizontal shaded regions represent the unique regions of PfR2Hb and PfR2Ha, respectively.
Erythrocyte binding.
Metabolic labeling of parasites and erythrocyte binding assays were performed as described elsewhere (3). Synchronized parasite cultures at the trophozoite stage were enriched to greater than 80% parasitemia on Percoll gradients consisting of 80, 60, 50, and 40% steps. Purified trophozoites for production of unlabeled supernatant were put in a culture dish containing only one-fourth of the original volume of complete medium. For [35S]Met-labeled supernatant, purified trophozoites were washed in methionine- and cysteine-deficient medium and cultured in the same medium containing [35S]methionine-[35S]cysteine mixture (Trans35S; 200 μCi/ml; ICN Radiochemicals). Parasites were incubated for 16 h, and the supernatant was harvested by centrifugation at 12,000 × g for 20 min at 4°C. The supernatant was stored in aliquots at −70°C.
Erythrocyte binding assays were performed using 500 μl of unlabeled or radiolabeled culture supernatant which was mixed with 100 μl of packed RBCs for 30 min at 23°C. The cells were centrifuged at 12,000 × g for 30 s through 500 μl of silicone oil (Dow Corning 550). The supernatant depleted of RBC binding proteins was recovered for some experiments. Proteins bound to the RBCs were eluted by incubation with 20 μl of 1.5 M NaCl for 15 min at 23°C and then centrifugation at 12,000 × g for 30 s. An equal volume of 2× reducing sample buffer was added to the eluted proteins. Unlabeled salt-eluted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then processed as described below for Western blotting. Salt-eluted proteins labeled with 35S were immunoprecipitated as described below.
Reticulocyte enrichment.
Fresh human blood with a reticulocyte count of approximately 1% was used for production of reticulocyte-enriched or -depleted blood. Blood was initially passed over a Ficoll-Hypaque gradient to remove white blood cells. RBCs were then separated over Percoll gradients consisting of 60, 70, 80, and 90% steps. To determine the percentage of reticulocytes, samples were mixed with an equal volume of 0.5% brilliant cresyl blue for 15 min, smeared on a glass slide, and counted microscopically under 100× magnification.
Immunoprecipitation.
Immunoprecipitation of radiolabeled supernatants was performed as described previously (5), with minor modifications. Volumes of 100 μl were precleared with protein G-Sepharose beads for 30 min at 4°C; then 2 μl of mouse serum or 5 μl of affinity-purified rabbit serum was added for 1 h on ice. Immune complexes were precipitated with protein G-Sepharose beads (1 h at 4°C), and the beads were washed four times in TNET (0.5% Triton X-100, 150 mM NaCl, 10 mM EDTA, 50 mM Tris [pH 7.4]). Immunoprecipitates were analyzed by SDS-PAGE and fluorography.
Western blotting.
Synchronized D10 parasites were sampled at 8-h intervals throughout the asexual life cycle. Parasite samples of each time point and from erythrocyte binding assays were analyzed by SDS-PAGE on 6% polyacrylamide gels and transferred to nitrocellulose as described elsewhere (21). The antibodies to P. falciparum hsp70 (Pfhsp70) have been previously described (1). Bound antibody was detected with horseradish peroxidase-coupled sheep anti-rabbit immunoglobulin G (IgG; Silenus Laboratories, Boronia, Victoria, Australia) and developed by the enhanced chemiluminescence method (Amersham International).
Immunofluorescence.
Synchronized late-stage D10 schizonts were smeared and air dried followed by fixation with 100% methanol for 2 min at −20°C. Smears were incubated with mouse antibodies to either PfAMA1, PfRAP1, or EBA175 and coincubated with the rabbit anti-PfR2Ha- and -Hb antibodies (2A9 and 2A11). Secondary antibodies were a mixture of rhodamine anti-mouse IgG (Chemicon), and fluorescein isothiocyanate (FITC) anti-rabbit IgG (Silenus Laboratories). Dual-color fluorescence images were captured using a digital camera (Zeiss, Jena, Germany).
Invasion inhibition assay.
Synchronized trophozoites were purified by passage over a Percoll gradient and counted in a hemocytometer; 8 × 105 schizonts were added to a final volume of 180 μl of medium with 4 × 107 uninfected human erythrocytes at 2% hematocrit. The anti-PfR2Ha- and -Hb antibodies (2A9 and 2A11) or normal rabbit serum (NRS) control were purified by affinity chromatography on a protein G column and then resuspended to a concentration of 5 mg of IgG per ml in phosphate-buffered saline. Antibodies were diluted in PBS to 2.5 and 1.25 mg/ml, and 20 μl was added per well. Experiments were done in duplicate in flat-bottom microtiter plates. Parasites were incubated for 24 h to allow for reinvasion. Smears of each well were taken, and parasitemia was calculated by counting ring-stage-infected erythrocytes. The slides were counted in a blind manner. Efficiency of invasion was calculated as percentage of the invasion of the NRS antibody control.
RESULTS
The P. falciparum genome encodes at least four genes related to the Pvrbp-2 and Py235 gene families.
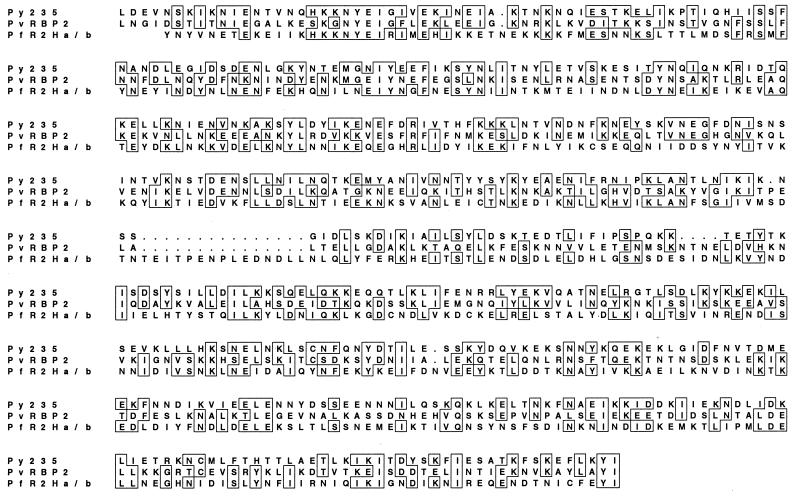
The PvRBP-2 and Py235 protein families have been demonstrated to bind reticulocytes or mature RBCs, and it was of interest to determine if P. falciparum expressed similar proteins. A 500-amino-acid region that showed homology between the Py235 and PvRBP-2 protein families (9) was used to search the P. falciparum (3D7 parasite) genome sequence databases. Five genes containing the homologous region were identified. The homologous regions from the proteins encoded by two of these genes (Pfr2ha and -hb) were aligned with the same region in PvRBP-2 and Py235 (Fig. 1). Although the overall homology is low, there are a number of short conserved blocks of amino acids. PfR2ha and -hb were predicted to encode proteins with molecular masses of 370 and 383 kDa, respectively, comparable in size to Py235 (325 kDa [accession no. U36927]) and PvRBP-2 (330 kDa). Both proteins have a putative signal sequence at the N terminus and a potential transmembrane domain followed by a short cytoplasmic tail at the C terminus (Fig. 2C), similar to the structures of Py235, PvRBP-1, and PvRBP-2 (7).
FIG. 1.
Comparison of the 500-amino-acid region conserved between PvRBP-2 and Py235 and its sequence in the P. falciparum homologues PfR2Ha and -Hb. Accession numbers for the sequence data shown: Py235, L27838; Pvrbp-2, Q00799; Pfr2ha/hb (AL049181, AF312916, and AF312917). Sequences were aligned using CLUSTAL V software. Boxes represent positions which have >50% identity; dots represent spaces inserted into the sequence to provide optimal homology.
Structurally, Pfr2ha and Pfr2hb consist of two exons. The first exon could encode the putative signal sequence, followed by the rest of the gene on the large second exon (Fig. 2A and B), a gene structure identical to that found for Py235 and Pvrbp-2 (6). Oligonucleotide primers designed to amplify a region of Pfr2ha and -hb across the putative introns confirmed expression of these genes in late schizonts of the 3D7 parasite line. Sequencing of these reverse transcription-PCR products also confirmed the intron-exon structure shown in Fig. 2 (data not shown). The chromosomal locations of Pfr2ha and -hb were determined using a P. falciparum linkage map (19; available at www.ncbi.nlm.nih.gov/Malaria/index.html). Pfr2ha and -hb are found in an internal location on the same contig on chromosome 13, in contrast to the subtelomeric location described for the Py235 genes in P. yoelii (12).
Identification of a P. falciparum parasite line that lacks the Pfr2hb gene.
Comparison of the nucleotide sequences of Pfr2ha and -hb suggested that they were identical over most of the sequences but diverged significantly at the 3′ end (Fig. 2C). To confirm that they represented two genes, we used PCR of genomic DNA from 3D7 with a 5′ primer (P1), contained within both genes, and two different 3′ primers specific to Pfr2ha and -hb (P2 and P3, respectively). These primers amplified DNA fragments of the expected lengths, and results of sequencing analysis were consistent with the presence of two genes that differed at the 3′ end (data not shown). To confirm these results, we hybridized a probe from the common region (C2 in Fig. 2A and B) to Southern hybridization filters containing gDNA from D10, 3D7, and HB3 digested with different restriction enzymes (Fig. 3A). Two fragments were detected in 3D7 and HB3 gDNA for three different restriction enzymes, which is consistent with the presence of two genes in these cloned lines.
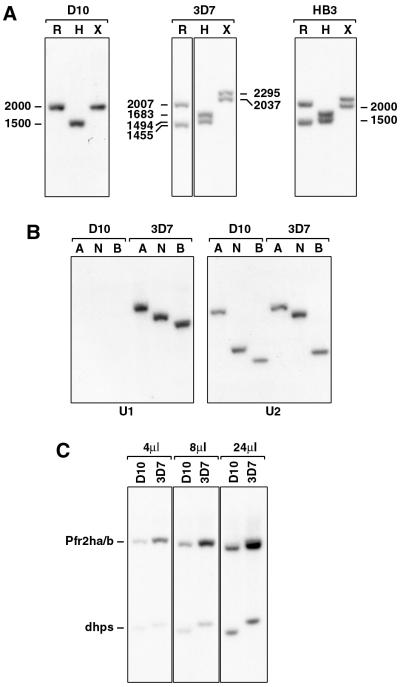
FIG. 3.
D10 parasites lack the Pfr2hb gene. (A) D10, 3D7, and HB3 gDNAs were digested with RsaI (R), HinfI (H), or XmnI (X), blotted to HybondN, and probed with the C2 probe. Sizes are in base pairs. (B) D10 and 3D7 gDNAs were digested with AccI (A), NsiI (N), or BstYI (B), blotted to HybondN, and probed with the Pfr2hb-specific probe U1 or the Pfr2ha-specific probe U2. (C) D10 and 3D7 gDNAs were double digested with NsiI/BglII, blotted to HybondN, and probed with a mixture of the single-copy gene dhps (21) and the C1 probe. Either 4, 8, or 24 μl of the digest was electrophoresed on the agarose gel. The dhps fragment was amplified from D10 gDNA using primers 5′-AAGATTAAATTTTCTTG-3′ and 5′-ATATAGAATTGTTACTTTTGTATA-3′. The copy number of the Pfr2ha and -hb genes in 3D7 relative to D10 was determined using a PhosphorImager (Molecular Dynamics).
Interestingly, hybridization of the C2 probe to D10 genomic DNA revealed only one hybridizing fragment with the different restriction enzymes, suggesting the presence of only one gene (Fig. 3A). This possibility was further analyzed by PCR using primers to the unique regions of Pfr2ha and -hb genes with genomic DNA from D10 and 14 other P. falciparum isolates. PCR products of the expected sizes were obtained for both the Pfr2ha and -hb genes for all parasite DNA samples tested except D10 (data not shown). D10 genomic DNA gave a PCR product for Pfr2ha but not for Pfr2hb. The absence of Pfr2hb in D10 was confirmed by Southern hybridization of genomic DNA from D10 and 3D7 probed with gene-specific probes U1 and U2 (Fig. 3B). The Pfr2hb-specific probe (U1) failed to hybridize to D10 but did hybridize to 3D7 genomic DNA, whereas the Pfr2ha-specific probe (U2) hybridized to both parasite DNAs. The presence of only one gene in D10 with the common 5′ end of Pfr2ha and -hb was also shown by quantitation of the gene copy number in D10 and 3D7 compared to the single-copy dhps (dihydropteroate synthase) gene (21). Restriction enzyme-digested genomic DNA from both D10 and 3D7 was probed with both the common region probe C1 and a dhps probe (Fig. 3C). Quantitation of the signal intensity of each hybridizing fragment using a PhosphorImager showed that 3D7 had two genes that shared the 5′ end whereas D10 had only a single copy. These results are consistent with the absence of Pfr2hb from the D10 genome.
Characterization of PfR2Ha and -Hb.
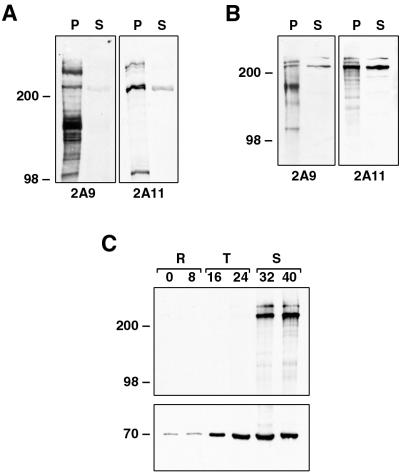
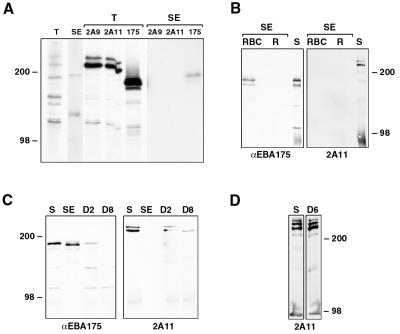
To characterize the proteins encoded by Pfr2ha and -hb, antibodies to two different regions of the 3D7 Pfr2ha gene were made. The 2A9 antibody was made to a region common to PfR2ha and -hb; the 2A11 antibody was to a region mostly within the unique 3′ end of Pfr2ha, although it did overlap into the common 5′ end (Fig. 2C). To confirm the specificities of both antibodies, total parasite proteins and culture supernatants from HB3 and D10 parasites were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with the two antisera. Both 2A9 and 2A11 antibodies detected three or more bands of the same size (>200 kDa) in the parasite pellet and the supernatant; however, the intensities of these bands varied with the different antibodies (Fig. 4A and B). In D10 parasites, which express only PfR2Ha, both the 2A9 and 2A11 antibodies react with essentially the same bands but with different intensities (Fig. 4B), which indicates that PfR2Ha is processed or degraded rapidly even in the pellet. Even in HB3 parasites, which express both PfR2Ha and -Hb, it is not possible to distinguish the two proteins since their sizes are 370 and 383 kDa, respectively. Smaller protein bands (<200 kDa) detected with both 2A9 and 2A11 antibodies may represent proteolytic degradation. The ability of 2A9 and 2A11 antibodies to recognize proteins of similar sizes confirms their reactivity with PfR2Ha and -Hb.
FIG. 4.
(A and B) Specificities of anti-PfRH antibodies. Two different anti-PfRH antibodies show similar specificities on HB3 (A) and D10 (B) parasites. For production of parasite pellets (P), cultures were sorbitol synchronized, cultured until the late-schizont stage, and saponin lysed, and then the pellet was resuspended in reducing sample buffer. Production of the EBA175 supernatant (S) is as described in Materials and Methods. Protein samples were separated by SDS-PAGE, transferred to nitrocellulose, and probed with both the 2A9 and 2A11 antibodies. (C) Protein samples from synchronized D10 parasites taken at 8-h intervals were separated by SDS-PAGE, transferred to nitrocellulose, and probed with both the 2A11 (top) and anti-Pfhsp70 (bottom) antibodies. During development, parasites transform from rings (R), through trophozoites (T), and to schizonts (S). Sizes are in kilodaltons.
Both Py235 and PvRBP-2 are expressed at the late schizont stage in developing merozoites (5, 9); therefore, it would be expected that P. falciparum homologues of these proteins would also be expressed late in schizogony. To determine the stage specificity of PfR2Ha/Hb expression, parasites were synchronized at the ring stage and protein samples were taken at 8-h intervals throughout the asexual life cycle. Equal numbers of parasites were separated by SDS-PAGE, and an immunoblot of the transferred proteins was probed with the 2A11 antibody (Fig. 4C, top). Two high-molecular-weight protein bands in the region of 250 to 300 kDa were detected with this antibody exclusively in late-schizont stages; no expression was seen in rings or trophozoites. An identical immunoblot probed with antibodies to Pfhsp70 showed, as expected, a band of 70 kDa throughout all life cycle stages. The pattern of expression of PfR2Ha and -Hb is consistent with a function in the invasion of merozoites into RBCs.
PfR2Ha and Hb are expressed at the apical end of the merozoite.
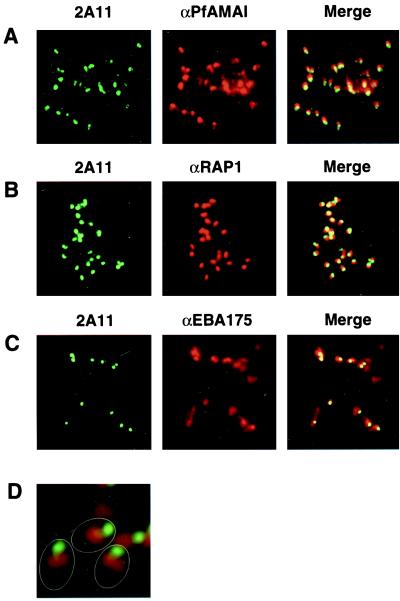
To determine the subcellular localization of PfR2Ha and -Hb, IFA experiments were performed with schizonts of D10 parasites smeared on glass slides (Fig. 5). The 2A11 antibody gave a punctate pattern in schizonts typical of the apical end of the merozoite (data not shown). This was confirmed in IFA analysis of free merozoites with the 2A11 antibody, where clear labeling of the apical end was observed (Fig. 5A). The same pattern of apical merozoite labeling was observed with the 2A11 and 2A9 antibodies for the 3D7 cloned line (data not shown). To further characterize the subcellular localization of PfR2Ha, we used IFA to colocalize this protein with respect to the merozoite proteins PfAMA1 (Fig. 5A), PfRAP1 (Fig. 5B), and EBA175 (Fig. 5C), which are believed to play a role in RBC invasion. PfAMA1 has previously been localized to the neck of the rhoptries, although there are some indications that it may be a micronemal protein. PfR2Ha is localized more apically within the merozoite compared to PfAMA1. EBA175 has been definitively localized to the micronemes, and IFA experiments with antibodies to PfR2Ha and EBA175 show that the two proteins do not exactly colocalize, suggesting that PfR2Ha is not located in the micronemes. PfRAP1 has been localized to the body of the rhoptries by immunoelectron microscopy. Localization of PfR2Ha and PfRAP1 shows that they overlap to some extent, although there is significant labeling of PfR2Ha that is more apical than PfRAP1. This is especially evident in Fig. 5D, which shows that PfR2Ha is located at the apical end of the merozoite and separate from PfRAP1, which is present within the body of the rhoptries.
FIG. 5.
PfR2Ha and -Hb do not colocalize with either PfAMA1, EBA175, or PfRAP1 by IFA analysis. Smears of free merozoites following schizont rupture of the D10 parasite are shown. Parasites were reacted with mouse antibodies to PfAMA1 (A), PfRAP1 (B), EBA175 (C), or PfRAP1 together with a rabbit antibody to PfR2Ha/Hb (2A11) (D). For images in the first column, the second antibody used was a sheep FITC-labeled anti-rabbit antibody; for images in the second column, the second antibody used was a goat rhodamine-labeled anti-mouse antibody; the Merge column shows the red and green images overlaid. Magnifications: A to C, ×1,000; D, ×3,000. The outline of three merozoites is shown in panel D.
Do PfR2Ha and Hb bind human RBCs?
The protein encoded by PvRBP-2 has been demonstrated to bind reticulocytes (5), and a member of the Py235 protein family can bind to mature RBCs (11). Therefore, we wished to determine if PfR2Ha and b could bind mature RBCs and/or reticulocytes. Purified trophozoites were grown to schizonts either in the presence of [35S]methionine or with no radiolabel; after schizont rupture, the supernatant was collected and used for erythrocyte binding assays. The [35S]Methionine-labeled total supernatant contained a large number of labeled proteins that included PfR2Ha and -Hb, as demonstrated by immunoprecipitation using both 2A9 and 2A11 antibodies (Fig. 6A). Similarly, anti-EBA175 antibodies precipitated a 175-kDa protein corresponding to EBA175 from the radiolabeled supernatant as has previously been shown. When the total supernatant was used in an erythrocyte binding assay, we detected a subset of proteins including EBA175, as shown by immunoprecipitation. Neither the 2A9 nor 2A11 antibody detected any PfR2Ha or -Hb in the proteins eluted from RBCs in binding assays, suggesting that the proteins released into the supernatant do not bind RBCs.
FIG. 6.
Do PfR2Ha and -Hb bind human RBCs? (A) 35S-labeled supernatant from D10 parasites was either immunoprecipitated directly with the anti-PfRh antibodies 2A9 and 2A11 or anti-EBA175 antibody or immunoprecipitated with the same antibodies after proteins were bound to RBCs and eluted with 300 mM NaCl. Total labeled proteins (T) or proteins eluted from RBCs (SE [salt eluted]) together with immunoprecipitated proteins were separated by SDS-PAGE and then detected by standard fluorographic methods. (B) Unlabeled supernatant from 3D7 parasites (200 μl) was bound to equal numbers of Percoll-purified reticulocytes enriched to 10% (R) and RBCs depleted of reticulocytes (RBC) by passage over Percoll. Bound proteins were eluted with 300 mM NaCl, separated by SDS-PAGE, transferred to nitrocellulose, and then probed with antibodies to EBA175 and PfRh (2A11). Two microliters of the supernatant (S) was also run as a positive control. SE, salt eluted. (C) Unlabeled supernatant from 3D7 parasites (100 μl) was bound to 20 μl of RBC and then eluted with 300 mM NaCl (SE). The supernatant was further depleted either two times (D2) or eight times (D8) by repeated addition of 20 μl of RBC for 30 min. Either 2 μl of the original supernatant (S) or between 2.5 and 3.2 μl (to allow for volume increases during repeated RBC additions) of the depleted supernatants, together with the salt-eluted proteins, was separated by SDS-PAGE, transferred to nitrocellulose, and then probed with antibodies to EBA175 and PfRh (2A11). (D) Unlabeled supernatant from 3D7 parasites (100 μl) which had been ultracentrifuged at 100,000 for 30 min was bound to 20 μl of RBC. The supernatant was depleted six times (D6) by repeated addition of 20 μl of RBC for 30 min. Either 2 μl of the original supernatant (S) or 2.6 μl of the depleted supernatant was separated by SDS-PAGE, transferred to nitrocellulose, and probed with the 2A11 antibody.
P. falciparum can preferentially invade younger RBCs and reticulocytes (10, 13), and it was possible that PfR2Ha and -Hb bind reticulocytes. To test this, we used unlabeled supernatants containing PfR2Ha/Hb and EBA175 to test binding to reticulocytes that had been enriched to 10% compared to RBCs depleted of reticulocytes. Bound proteins were salt eluted, separated by SDS-PAGE, immunoblotted, and probed with the EBA175 or 2A11 antibody (Fig. 6B). No PfR2Ha or -Hb could be detected in the fraction eluted from reticulocytes or RBCs; EBA175 could bind to RBCs, but we detected no binding to the fraction that was able to bind the cells enriched for reticulocytes. The explanation for this may be that the Percoll purification resulted in a mixture of reticulocytes and young RBC, neither of which could bind EBA175. Nevertheless, these results suggest that PfR2Ha and -Hb cannot bind to reticulocytes, although it was possible that the proteins could not be eluted by the conditions used or alternatively binds with very low affinity.
To assess the possibility that PfR2Ha and -Hb were able to bind to RBCs but could not be eluted under the conditions used, we increased the NaCl concentration stepwise up to 1.5 M for elution. However, we could not detect any PfR2Ha/Hb binding to RBCs at any NaCl concentration (data not shown). To confirm that PfR2Ha and -Hb could not bind RBCs, we did sequential depletion experiments where the same supernatant was incubated with fresh RBCs up to eight times to deplete any binding proteins (Fig. 6C). As expected, EBA175 was depleted from the supernatant after two cycles, and no protein was detected after eight rounds of incubation with RBCs; in contrast, PfR2Ha/Hb was still present after eight rounds of depletion on RBCs, although the amount, especially of the higher-molecular-weight protein, appeared to decrease (Fig. 6C).
This finding suggested that either very little binding occurred with each addition of fresh RBCs or each centrifugation may have depleted the PfR2Ha/Hb content. To test this, we used ultracentrifugation to remove PfR2Ha/Hb complexes and membrane-associated forms. About 10% of the PfR2Ha/Hb protein was lost from the supernatant (data not shown). When this supernatant, which now contained truly soluble PfR2Ha/Hb, was used in depletion experiments with six rounds of RBC incubation, no decrease in the presence of PfR2Ha/Hb was detected (Fig. 6D). These results suggest that the soluble PfR2Ha and -Hb do not bind RBCs under the conditions tested.
Antibodies to PfR2Ha and -Hb inhibit merozoite invasion.
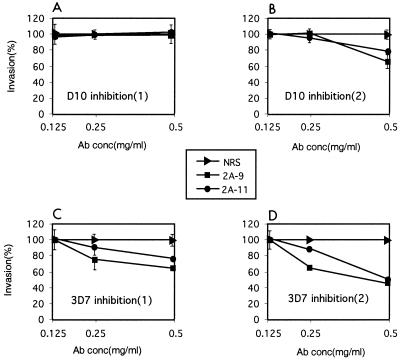
We tested the ability of anti-PfR2Ha/Hb rabbit antibodies to inhibit merozoite invasion of RBCs. Protein G-purified 2A9 and 2A11 antibodies and NRS were incubated in the presence of invading merozoites, and the ability to invade RBCs was assessed microscopically following Giemsa staining of smears. In two identical experiments, there was little inhibition of D10 merozites with either the 2A9 or 2A11 antibody, although in the second experiment there was a small level of inhibition (Fig. 7A and B). However, more significant inhibition of merozoite invasion was observed with the 3D7 parasite in two independent experiments (Fig. 7C and D). There was a marked dose response for 3D7 as the concentrations of both 2A9 and 2A11 antibodies were increased to 0.5 mg/ml compared to the same antibody concentration for NRS (Fig. 7). Both antisera were able to inhibit merozoite invasion in 3D7 to levels between 40 and 55% of control values with NRS. This suggests that PfR2Ha and -Hb are important for the invasion of merozites into RBCs.
FIG. 7.
Antibodies to PfR2Ha and -Hb result in some invasion inhibition. Purified schizonts from D10 and 3D7 parasites were plated in human erythrocytes to determine the ability of released merozoites to invade in the presence or absence of protein G-purified 2A9 and 2A11 antibodies (0.125, 0.25, and 0.50 mg/ml) for each parasite, results of two independent experiments done in duplicate are shown. Invasion in the presence of 0.125, 0.25 and 0.50 mg of protein G-purified normal NRS per ml was adjusted to 100%. Percent invasion was determined microscopically by counting at least 1,000 RBCs. The parasitemias at the end of the experiment were between 9 and 12% for the control wells containing NRS. Standard errors are shown except those less than 4% (e.g., panel D); the standard errors ranged from 0 to 14%.
It was interesting that the 2A9 and 2A11 antibodies were able to inhibit invasion to some extent in 3D7 but showed little effect for D10. To determine if there were polymorphisms that may reduce the ability of the antisera raised to 3D7 PfR2Ha to inhibit the D10 parasite line, we sequenced the region of the Pfr2ha and -hb genes in D10 and 3D7 used to raise the 2A9 and 2A11 antibodies. There were six nucleotide changes resulting in amino acid differences between 3D7 and D10 Pfr2ha genes, and these occurred in the region used to raise the 2A11 antibodies (Fig. 2C). There were no polymorphisms in the region used to raise the 2A9 antibody between the 3D7 Pfr2ha and -hb genes and the D10 Pfr2ha gene (Fig. 2C).
DISCUSSION
Erythrocyte invasion by the merozoite form of P. falciparum is an ordered process requiring sequential steps that involve specific adhesive interactions of parasite ligands with receptors on the host cell. In P. vivax, PvRBP-1 and PvRBP-2 form a complex that binds to reticulocytes, and it has been hypothesized that this confers specificity for this cell type (6). Members of the Py235 protein family in P. yoelii are homologous to PvRBP-1 and -2, and one of these proteins has been shown to bind to erythrocytes (11). The identification of related protein families in P. vivax and P. yoelii involved in adhesion to reticulocytes and erythrocytes suggested that P. falciparum may express homologous proteins with similar functions. A search of the P. falciparum genome databases has identified two genes encoding proteins, which we have called PfR2Ha and PfR2Hb, that are related to the PvRBP and Py235 families.
Comparisons of gene structure, timing of expression, and subcellular localization suggest that PfR2Ha and -Hb are functional homologues of the PvRBP and Py235 protein families. Pfr2ha and -hb, Pvrbp-1 and -2, and Py235 genes have the same exon/intron structure (6, 7). The short intron at the 5′ end encodes a putative signal sequence region, and the second large exon contains a putative transmembrane domain near the C terminus of each protein with a very short cytoplasmic tail. This structure is found in all three gene families of the different Plasmodium species. Additionally, subcellular localization of PfR2Ha/Hb suggests that it is located at the apical end of the merozoite, similar to that found for the homologous proteins in P. vivax and P. yoelii.
Because of the similarity of PfR2Ha/Hb with PvRBP-1/2 and Py235, it was expected that the P. falciparum proteins would be able to bind to erythrocytes or reticulocytes. Experiments with both mature erythrocytes and enriched reticulocytes, using PfR2Ha/Hb protein released into the supernatant, failed to detect binding, suggesting that these proteins are unable to bind directly to either erythrocytes or reticulocytes. This observation was further confirmed by incubating an ultracentrifuged extract multiple times with erythrocytes, in which case no depletion of these proteins was detected. It is possible that the proteins found in the supernatant were processed and the RBC binding domains had been removed. This would be consistent with the multiple forms of PfR2Ha and -Hb seen in Western blots. This inference suggested that the full-length protein at the apical end of the merozoite could bind to RBCs but subsequent cleavage would remove this binding domain. Interestingly, we have identified in the P. falciparum genome databases a further three homologues of the Pfr2ha and -hb genes described here, some of which may be able to interact with erythrocytes (unpublished results). Second, it is possible that the proteins found in the supernatant were not processed but that the PfR2Ha/Hb complex had been disrupted such that it could no longer bind RBCs. Third, it is possible that both proteolytic processing and disruption of a binding complex occur rapidly in P. falciparum merozoites in the absence of fresh RBCs for invasion. It will be important to test the ability of the other proteins encoded by genes homologous to PfR2ha and -hb for binding to RBCs and reticulocytes.
The ability of anti-PfR2Ha/Hb antibodies to inhibit merozoite invasion in 3D7 suggests that these proteins play some role in this process. MAbs to Py235 are able to specifically inhibit invasion of reticulocytes in P. yoelii (4). It was surprising that the antibodies to PfR2Ha were not able to inhibit merozoite invasion in the D10 cloned line, and sequencing of the equivalent gene showed some polymorphism that may reduce the ability of antibodies raised to the 3D7 protein to bind and inhibit. However, there were no polymorphisms in the 2A9 portion of the protein, and it is possible that specific PfRH proteins function in different P. falciparum parasite lines. The demonstration in P. yoelii that different Py235 genes are expressed in individual merozoites within a schizont provides some support for this possibility (14).
It is clear that the D10 parasite genome does not contain the Pfr2hb gene, showing that this gene is not essential for parasite invasion and growth. However, this parasite does express the Pfr2ha gene, which is nearly identical to Pfr2hb throughout most of its sequence. Pfr2ha and -hb appear to lie next to each other on chromosome 13 (data not shown), suggesting they have arisen by a recent gene duplication. It is known for the P. yoelii parasite that individual merozoites express a different Py235 gene (14). From analysis of the two Py235 genes for which sequence is available (e3 and e8 types), it appears that there are extensive differences at the protein level with approximately 25% sequence variation (18). In P. falciparum, the much more closely related proteins PfR2Ha and -Hb may be expressed differentially in individual merozoites. The explanation for the absence of the Pfr2hb gene in D10 is unknown. The Pfr2ha/hb locus is in an internal location on the chromosome, and it is unlikely that loss of the gene has occurred by removal of the subtelomeric region of the chromosome that is common in P. falciparum. It is apparent that the other three Pfrh homologues in the D10 parasite are sufficient to compensate for the lack of the PfR2Hb protein.
Here we have described in detail two high-molecular-weight proteins (PfR2Ha and -Hb) which are expressed at the apical end of the P. falciparum merozoite. These proteins belong to a family of proteins found in other plasmodia which are involved in targeting of RBC populations prior to invasion by the merozoite. Following review of this report, work from another group describing analysis of this gene family was published (15). The conclusions drawn by these authors are in broad agreement to the work presented here. Importantly, they also have not been able to demonstrate interaction of PfR2Ha and -Hb with the RBC surface. A P. falciparum database search has identified a further three genes potentially belonging to this family (Pfrh3, Pfr1h, and Pfrh4). Pfrh3 appears to be a pseudogene (H. M. Taylor, T. Triglia, J. Thompson, M. Sajid, R. Fowler, M. E. Wickham, A. F. Cowman, and A. A. Holder, unpublished data). Pfr1h appears to be more closely related to Pvrbp-1 and may be the P. falciparum homologue. Pfrh4 has some features of other Py235/Pvrbp-2/Pfrh family members but appears to be more distantly related. The generation of specific antibodies to individual PfRH proteins should enable dissection of the roles of different members in RBC targeting by P. falciparum.
ACKNOWLEDGMENTS
This research was supported by the Australian National Health and Medical Research Council, the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR), and the National Institutes of Health. Sequence data for P. falciparum chromosome 12 were obtained as part of the Malaria Genome Project with support from the Burroughs Wellcome Fund.
We thank the Red Cross Blood Service (Melbourne, Victoria, Australia) for supplies of RBCs and serum.
REFERENCES
- 1.Bianco A E, Favaloro J M, Burkot T R, Culvenor J G, Crewther P E, Brown G V, Anders R F, Coppel R L, Kemp D J. A repetitive antigen of Plasmodium falciparum that is homologous to heat shock protein 70 of Drosophila melanogaster. Proc Natl Acad Sci USA. 1986;83:8713–8717. doi: 10.1073/pnas.83.22.8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borre M B, Owen C A, Keen J K, Sinha K A, Holder A A. Multiple genes code for high-molecular-mass rhoptry proteins of Plasmodium yoelii. Mol Biochem Parasitol. 1995;70:149–155. doi: 10.1016/0166-6851(95)00025-v. [DOI] [PubMed] [Google Scholar]
- 3.Dolan S A, Proctor J L, Alling D W, Okubo Y, Wellems T E, Miller L H. Glycophorin B as an EBA-175 independent Plasmodium falciparum receptor of human erythrocytes. Mol Biochem Parasitol. 1994;64:55–63. doi: 10.1016/0166-6851(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 4.Freeman R R, Trejdosiewicz A J, Cross G A. Protective monoclonal antibodies recognising stage-specific merozoite antigens of a rodent malaria parasite. Nature. 1980;284:366–368. doi: 10.1038/284366a0. [DOI] [PubMed] [Google Scholar]
- 5.Galinski M R, Medina C C, Ingravallo P, Barnwell J W. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell. 1992;69:1213–1226. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- 6.Galinski M R, Mengyao X, Barnwell J W. Plasmodium vivax reticulocyte binding protein-2 (PvRBP-2) shares structural features with PvRBP-1 and the Plasmodium yoelii 235kDa rhoptry protein family. Mol Biochem Parasitol. 2000;108:257–262. doi: 10.1016/s0166-6851(00)00219-x. [DOI] [PubMed] [Google Scholar]
- 7.Green J L, Holder A A. Structure of the E8 gene encoding a high molecular mass rhoptry protein of Plasmodium yoelii. Mol Biochem Parasitol. 2000;110:167–169. doi: 10.1016/s0166-6851(00)00251-6. [DOI] [PubMed] [Google Scholar]
- 8.Holder A A, Freeman R R, Uni S, Aikawa M. Isolation of a Plasmodium falciparum rhoptry protein. Mol Biochem Parasitol. 1985;14:293–303. doi: 10.1016/0166-6851(85)90057-x. [DOI] [PubMed] [Google Scholar]
- 9.Keen J K, Sinha K A, Brown K N, Holder A A. A gene coding for a high molecular mass rhoptry protein of Plasmodium yoelii. Mol Biochem Parasitol. 1994;65:171–177. doi: 10.1016/0166-6851(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 10.Mons B. Preferential invasion of malarial merozoites into young red blood cells. Blood Cells. 1990;16:299–312. [PubMed] [Google Scholar]
- 11.Ogun S A, Holder A A. A high molecular mass Plasmodium yoelii rhoptry protein binds to erythrocytes. Mol Biochem Parasitol. 1996;76:321–324. doi: 10.1016/0166-6851(95)02540-5. [DOI] [PubMed] [Google Scholar]
- 12.Owen C A, Sinha K A, Keen J K, Ogun S A, Holder A A. Chromosomal organisation of a gene family encoding rhoptry proteins in Plasmodium yoelii. Mol Biochem Parasitol. 1999;99:183–192. doi: 10.1016/s0166-6851(99)00015-8. [DOI] [PubMed] [Google Scholar]
- 13.Pasvol G, Weatherall D J, Wilson R J. The increased susceptibility of young red cells to invasion by the malarial parasite Plasmodium falciparum. Br J Haematol. 1980;45:285–295. doi: 10.1111/j.1365-2141.1980.tb07148.x. [DOI] [PubMed] [Google Scholar]
- 14.Preiser P R, Jarra W, Capiod T, Snounou G. A rhoptry-protein-associated mechanism of clonal phenotypic variation in rodent malaria. Nature. 1999;398:618–622. doi: 10.1038/19309. [DOI] [PubMed] [Google Scholar]
- 15.Rayner J C, Galinski M R, Ingravallo P, Barnwell J W. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc Natl Acad Sci USA. 2000;97:9648–9653. doi: 10.1073/pnas.160469097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed M B, Caruana S R, Batchelor A H, Thompson J K, Crabb B S, Cowman A F. Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid independent pathway of invasion. Proc Natl Acad Sci USA. 2000;97:7509–7514. doi: 10.1073/pnas.97.13.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schofield L, Bushell G R, Cooper J A, Saul A J, Upcroft J A, Kidson C. A rhoptry antigen of Plasmodium falciparum contains conserved and variable epitopes recognized by inhibitory monoclonal antibodies. Mol Biochem Parasitol. 1986;18:183–195. doi: 10.1016/0166-6851(86)90037-x. [DOI] [PubMed] [Google Scholar]
- 18.Sinha K A, Keen J K, Solabomi A, Ogun A O, Holder A A. Comparison of two members of a multigene family coding for high-molecular mass rhoptry proteins of Plasmodium yoelii. Mol Biochem Parasitol. 1996;76:329–332. doi: 10.1016/0166-6851(95)02546-4. [DOI] [PubMed] [Google Scholar]
- 19.Su X, Ferdig M T, Huang Y, Huynh C Q, Liu A, You J, Wootton J C, Wellems T E. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 20.Trager W, Jensen J B. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 21.Triglia T, Cowman A F. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1994;91:7149–7153. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Triglia, T., J. Healer, S. R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol., in press. [DOI] [PubMed]