Abstract
We have successfully created induced pluripotent stem cells (iPSC) from patients carrying a heterozygous mutation in the gene encoding STING. The gain-of-function mutation leads to constitutive activation of STING which leads to the development of the disease STING-associated vasculopathy with onset in infancy (SAVI). The iPSC lines derived from the SAVI patitents are shown to be morphologically and phenotypically normal and have the potential to self renew and differentiate into the three germ layers. These iPSC provide a powerful tools to investigate the role of STING in the regulation of immune responses and vascular renegeration.
1. Resource Table
| Unique stem cell lines identifier | NIHTVBi024-A |
| NIHTVBi025-A | |
| NIHTVBi026-A | |
| NIHTVBi027-A | |
| Alternative names of stem cell lines | 182–5 (NIHTVBi024-A) |
| 189–2 (NIHTVBi025-A) | |
| 199–2 (NIHTVBi026-A) | |
| 1131 (NIHTVBi027-A) | |
| Institution | National Heart, Lung, and Blood Institute (NHLBI), |
| National Institutes of Health (NIH), Bethesda, | |
| Maryland, USA | |
| Contact information of distributor | Manfred Boehm; boehmm@nhlbi.nih.gov |
| Type of cell lines | iPSC |
| Origin | Human |
| Cell Source | Dermal fibroblasts and peripheral blood mononuclear cells |
| Clonality | Clonal cell lines |
| Method of reprogramming | Sendai-virus vectors containing the transcription factors Oct-4, Klf4, Sox2 and c-MYC |
| Multiline rationale | Lines derived from four individual |
| Gene modification | Yes |
| Type of modification | Hereditary |
| Associated disease | STING-associated vasculopathy with onset in infancy (SAVI) |
| Gene/locus | STING1 |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | July 1, 2016 |
| Cell line repository/bank | https://hpscreg.eu/cell-line/NIHTVBi024-A |
| https://hpscreg.eu/cell-line/NIHTVBi025-A | |
| https://hpscreg.eu/cell-line/NIHTVBi026-A | |
| https://hpscreg.eu/cell-line/NIHTVBi027-A | |
| Ethical approval | NCT02974595 |
1.1. Resource utility
Human induced pluripotent stem cells (hiPSC) carrying a heterozygous mutation in STING1 gene exhibit the potential of selfrenew and differentiating into variety of cell types including vascular cells and immune cells. The derivatives sustaining the mutation could be a powerful platform allowing for the investigation of the molecular mechanisms and identification of potential therapeutic targets in patients.
2. Resource details
STING-associated vasculopathy with onset in infancy (SAVI) is a rare monogenic autoinflammatory interferonopathy with a gain-of-function mutation in the stimulator of interferon response cGAMP interactor 1 (STING1) gene (Liu et al., 2014). This results in a constituently activated stimulator of interferon genes (STING) protein which is downstream to cyclic GMP-AMP synthase (cGAS), an innate cytosolic nucleic acid sensor (Liu et al., 2014, Zhang et al., 2022). The STING mutation is associated with high levels of type 1 interferon along with a spectrum of clinical features featuring fevers, a peripheral vasculopathy presenting as a violaceous scaling lesions that can progress to acral ulceration, necrosis, and autoamputation, and interstitial lung disease (including pulmonary fibrosis) with respiratory failure the most common cause of death (Liu et al., 2014). Patients can present as early as 6–8 weeks following birth (Liu et al, 2014). Specific treatments include the use of JAK-STAT 1/2 inhibitors which has shown to have partial benefits (Sanchez et al., 2018). While the cGAS-STING pathway has previously been explored (Zhang et al., 2022), it is unclear whether this exact pathway is the mechanistic driver of the changes seen in different cell types with constitutively activated STING.
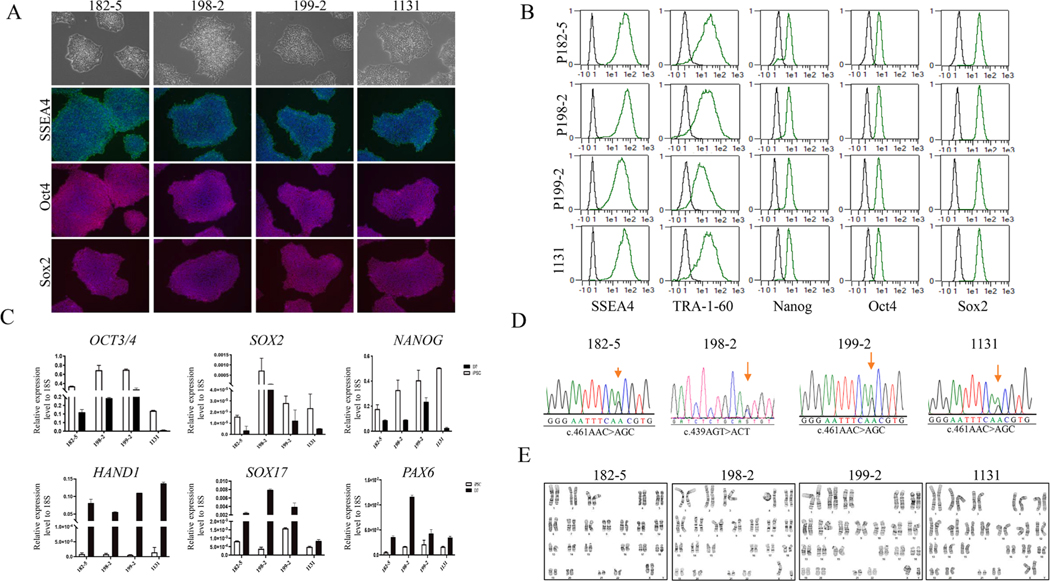
We initially identified four patients carrying the heterozygous mutation in the STING1 gene by Sanger sequencing after which they were enrolled into our NHLBI clinical protocols for further investigation. Information regarding clinical features of these patients were obtained using the standard clinical questionnaire (Table 1). Skin punch biopsy from all patients and peripheral blood sample from patient 199 were collected at the NIH Clinical Center. Using a Sendai-OKSM delivery system expressing four transcription factors (OCT4, SOX2, KLF4, and CMYC), iPSC lines were generated from skin fibroblasts (185–2, 198–2, and 1131) or peripheral blood mononuclear cells (PBMNC) (199–2) from the patients. Typical iPSC morphology was observed for all lines, and the generated iPSCs expressed the pluripotency markers of OCT4, NANOG, TRA-160, SSEA4, and SOX2 as shown by immunofluorescent staining (Fig. 1A), by FACS (Fig. 1B), and/or real-time (RT)-qPCR (Fig. 1C). The iPSC lines were free of Sendai virus as confirmed by RT-PCR at the 15th passage (Supplementary File1). Genotyping of the generated iPSC lines confirmed the heterozygous mutation in the STING1 gene was the same as their parental lines (Fig. 1D). To test the differentiation potential of the iPSC lines, we performed a monolayer differentiation assay to drive the cells towards the three germ layers in vitro. We showed that the expression of marker genes for the mesoderm (HAND1), endoderm (SOX17), and ectoderm (PAX6) germ layers by RT-qPCR confirmed that all four iPSC lines were able to differentiate into mesoderm, endoderm, and ectoderm cells (Fig. 1C). Short tandem repeat (STR) profiles indicated that the iPSC lines matched with its correspodning somatic cells completely in 15 amplified STR loci. All cultures were routinely tested for Mycoplasma contamination and were found to be Mycoplasma free (Supplementary File 2). All four iPSC lines demonstrated chromosomal stability and a normal karyotype with G-banding (Fig. 1E). Overall, the four patient derived iPSC lines demonstrated the pluripotent potential for self-renew and differentiation, suggesting the successful generation of iPSCs from the SAVI patients.
Table 1.
Summary of patients with a SAVI disease.
| iPSC line names | Abbreviation in figures | Gender | Age when cells harvested (years) | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
|
| ||||||
| NIHTVBi024-A | 182–5 | Female | 15 | White | STING1_mutation c.461AAC > AGC (p.N154S) | SAVI |
| NIHTVBi025-A | 198–2 | Male | 12 | Hispanic | STING1 mutation c.439AGT > ACT (p.V147L) | SAVI |
| NIHTVBi026-A | 199–2 | Male | 26 | White | STING1_mutation c.461AAC > AGC (p.N154S) | SAVI |
| NIHTVBi027-A | 1131 | Female | 10 | White | STING1_mutation c.461AAC > AGC (p.N154S) | SAVI |
Fig. 1.
Generation and characterization of human induced pluripotent stem cells (iPSCs) from four SAVI patients carried a heterozygous mutation in the STING gene.
3. Materials and methods
3.1. Subjects and derivation of fibroblasts and peripheral blood mononuclear cells (PBMNC)
The fibroblasts were derived from skin punch biopsy samples obtained from three of the SAVI patients with the heterozygous STING1 gene mutation and PBMNC was used for the remaining patient (199–2). Fibroblasts were propagated as previously described (Chen et al., 2019). PBMNC were obtained from the remaining patient which were isolated from whole blood using density gradient centrifugation via Ficoll-Paque according to the manufacturer’s direction. This study was approved by the NHLBI’s institutional review board, and samples were collected after obtaining informed written consent.
3.2. Generation and culture of human iPSC from fibroblasts or PBMNC
Fibroblasts or PBMNC from the SAVI patients were reprogrammed and expanded in a typical human embroynic stem cell/iPSC culture condition as previously described (Chen et al., 2019) and Supplementary File 3.
3.3. Immunofluorescent staining and flow cytometry analysis (FACS)
iPSC colonies (passage 12) were fixed with 4% paraformaldehyde and stained for NANOG, SOX2, SSEA4, and TRA-1–60 (Table 3) following the previous protocol (Chen et al., 2019).
Table 3.
Reagents Antibodies used for immunocytochemistry and FACS.
| Antibody | Dilution | Company | Cat# | RRID | |
|---|---|---|---|---|---|
|
| |||||
| Primary antibodies | Mouse anti-SSEA4 | 1:100 | MilliporeSigma | MAB4304 | AB_177629 |
| Mouse anti-TRA-1-60 | 1:150 | MilliporeSigma | MAB4360 | AB_2119183 | |
| Alexa Fluor 488 anti-SSEA4 Antibody | 1:10 | BioLegend | 330412 | AB_1089198 | |
| PE anti-TRA-1-60 Antibody | 1:10 | BioLegend | 330610 | AB_2119065 | |
| Alexa Fluor 488 anti-SOX2 Antibody | 1:10 | BioLegend | 656110 | AB_2563957 | |
| Alexa Fluor 488 anti-OCT4 Antibody | 1:10 | BioLegend | 653,708 | AB_2563184 | |
| Alexa Fluor 647 anti-NANOG Antibody | 1:10 | BioLegend | 674210 | AB_2650619 | |
| Secondary antibodies | Alexa Fluor 594 Donkey anti-rabbit | 1:300 | Life Technologies | A21207 | AB_141637 |
| Alexa Fluor 594 Donkey anti-mouse | 1:300 | Life Technologies | A21203 | AB_141633 | |
| Alexa Fluor 488 Donkey anti-mouse | 1:300 | Life Technologies | A21202 | AB_141607 | |
|
| |||||
| Primers used for RT-qPCR and PCR Target | Forward/reverse primer (5′–3′) | ||||
|
| |||||
| NANOG | AGG GAA ACA ACC CAC TTC T/CCT TCT GCG TCA CAC CAT T | ||||
| OCT3/4 | AGC GAA CCA GTA TCG AGA AC/TTA CAG AAC CAC ACT CGG AC | ||||
| SOX2 | CCC AGC AGA CTT CAC ATG T/CCT CCC ATT TCC CTC GTT TT | ||||
| HAND1 | CCA AGG ATG CAC AGT CTG G/AGG AGG AAA ACC TTC GTG CTG | ||||
| SOX17 | GTG GAC CGC ACG GAA TTT G/GGA GAT TCA CAC CGG AGT CA | ||||
| PAX6 | TGG GCA GGT ATT ACG AGA CTG/ACT CCC GCT TAT ACT GGG CTA | ||||
| STING | CAT GCC TTG GGA TTA AAG GA/CTG GGA GTG GGA CTG GTT TA | ||||
FACS data acquisition and analysis was performed on the iPSCs (passage 12) for the designed antibodies (Table 3) as previously described (Chen et al., 2019).
3.4. Monolayer differentiation assay
The in vitro assessment of iPSC differentiation ability at the 12th passage was performed as per the previous protocol with harvesting of cells for analysis at day 7 (Chen et al., 2019).
3.5. Gene expression analysis by RT-PCR
Total RNA isolation, cDNA synthesis, and PCR using the primers indicated in Table 2 was performed as previously described (Chen et al., 2019) with the iPSC obtained at passage 5 serving as the positive control (Pos) (Supplementary File 1).
Table 2.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
|
| |||
| Morphology | Phase-contrast microscope | Normal | Fig. 1A |
| Phenotype | Qualitative analysis (immunofluorescence staining) | Expression of pluripotency markers: OCT4, NANOG, SSEA4 and TRA-1–60 | Fig. 1A |
| Quantitative analysis (RT-qPCR) Qualitative analysis (FACS) | Expression of pluripotency markers: SOX2 and NANOG Expression of pluripotency markers: NANOG, SOX2, SSEA4, TRA-1–60, OCT4 |
Fig. 1C
Fig. 1B |
|
| Genotype | Karyotype (G-banding) and resolution | 46, XY or 46, XX; resolution 450–500 bands | Fig. 1E |
| Identity | Microsatellite PCR OR STR analysis | Not performed 15 sites tested, 100% match | N/A |
| Mutation analysis (IF APPLICABLE) | DNA sequencing | Heterozygous, STING gene point mutation | Fig. 1D |
| Southern blot OR WGS | Not performed | N/A | |
| Microbiology and virology | Mycoplasma testing by luminescence | Negative | Supplementary Table 2 |
| Differentiation potential | Monolayer differentiation assay | Differentiating cells are expression of HAND1, SOX17, and PAX6; iPSC were able to differentiate into three germ layers | Fig. 1C |
| Donor screening (OPTIONAL) | HIV1 + HIV2, hepatitis B virus, hepatitis C virus | Not performed | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping HLA tissue typing | Not performed Not performed |
N/A N/A |
After 15 passages, iPSCs were tested for Sendai virus (SeV) residues as outlined before (Alonso-Barroso et al., 2017).
Endogenous mRNA expression levels of OCT4, NANOG, SOX2, SOX17, PAX6, and HAND1 were determined in iPSCs and in differentiating cells at day 7 (D7) (Fig. 1C) via RT-qPCR as previously described (Chen et al., 2019) using the primers catalogued in Table 3.
3.6. Karyotyping assay
The iPSC karyotypes were evaluated (182–5 at passage 15, 198–2 at passage 34, 199–2 at passage 6, and 1131 at passage 30) by the WiCell Research Institute using G-banding metaphase karyotype analysis.
3.7. DNA sequencing and STR
DNA sequencing and STR analysis was derived from previous methods (Chen et al., 2019). PCR was performed using specific primers listed in Table 3 to amplify the corresponding deletion positions in the STING1 gene.
3.8. Mycoplasma detection
The supernatant from the iPSCs underwent mycoplasma testing as per the protocol detailed in Chen et al., 2019 using the MycoAlert™ Mycoplasma Detection Kit (Lonza, LT27–224) (Supplementary File 2).
Supplementary Material
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2022.102974.
References
- Alonso-Barroso E, Brasil S, Briso-Montiano Á, Navarrete R, Pérez-Cerdá C, Ugarte M, Pérez B, Desviat LR, Richard E, 2017. Generation and characterization of a human iPSC line from a patient with propionic acidemia due to defects in the PCCA gene. Stem Cell Res. 23, 173–177. 10.1016/j.scr.2017.07.021. [DOI] [PubMed] [Google Scholar]
- Chen G, Jin H, Yu Z, Liu Y, Li Z, Navarengom K, Schwartzbeck R, Dmitrieva N, Cudrici C, Ferrante EA, Biesecker LG, Yang D, Boehm M, 2019. Generation of human induced pluripotent stem cells from individuals with a homozygous CCR5Δ32 mutation. Stem Cell Res. 38, 101481 10.1016/j.scr.2019.101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, Lee C-C, DiMattia MA, Cowen EW, Gonzalez B, Palmer I, DiGiovanna JJ, Biancotto A, Kim H, Tsai WL, Trier AM, Huang Y, Stone DL, Hill S, Kim HJ, St. Hilaire C, Gurprasad S, Plass N, Chapelle D, Horkayne-Szakaly I, Foell D, Barysenka A, Candotti F, Holland SM, Hughes JD, Mehmet H, Issekutz AC, Raffeld M, McElwee J, Fontana JR, Minniti CP, Moir S, Kastner DL, Gadina M, Steven AC, Wingfield PT, Brooks SR, Rosenzweig SD, Fleisher TA, Deng Z, Boehm M, Paller AS, Goldbach-Mansky R, 2014. Activated STING in a vascular and pulmonary syndrome. New Engl. J. Med 371 (6), 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez GAM, Reinhardt A, Ramsey S, Wittkowski H, Hashkes PJ, Berkun Y, Schalm S, Murias S, Dare JA, Brown D, Stone DL, Gao L, Klausmeier T, Foell D, de Jesus AA, Chapelle DC, Kim H, Dill S, Colbert RA, Failla L, Kost B, O’Brien M, Reynolds JC, Folio LR, Calvo KR, Paul SM, Weir N, Brofferio A, Soldatos A, Biancotto A, Cowen EW, Digiovanna JJ, Gadina M, Lipton AJ, Hadigan C, Holland SM, Fontana J, Alawad AS, Brown RJ, Rother KI, Heller T, Brooks KM, Kumar P, Brooks SR, Waldman M, Singh HK, Nickeleit V, Silk M, Prakash A, Janes JM, Ozen S, Wakim PG, Brogan PA, Macias WL, Goldbach-Mansky R, 2018. JAK1/2 inhibition with baricitinib in the treatment of autoinflammatory interferonopathies. J. Clin. Investig 128 (7), 3041–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liu Y, Zhu Y, Zhang Q, Guan H, Liu S, Chen S, Mei C, Chen C, Liao Z, Xi Y, Ouyang S, Feng XH, Liang T, Shen L, Xu P, 2022. A non-canonical cGAS-STING-PERK pathway facilitates the translational program critical for senescence and organ fibrosis. Nat. Cell Biol 24 (5), 766–782. 10.1038/s41556-022-00894-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.