Keywords: enteric, intestine, organoids, virology, virus
Abstract
Viruses are among the most prevalent enteric pathogens. Although virologists historically relied on cell lines and animal models, human intestinal organoids (HIOs) continue to grow in popularity. HIOs are nontransformed, stem cell-derived, ex vivo cell cultures that maintain the cell type diversity of the intestinal epithelium. They offer higher throughput than standard animal models while more accurately mimicking the native tissue of infection than transformed cell lines. Here, we review recent literature that highlights virological advances facilitated by HIOs. We discuss the variations and limitations of HIOs, how HIOs have allowed for the cultivation of previously uncultivatable viruses, and how they have offered insight into tropism, entry, replication kinetics, and host-pathogen interactions. In each case, we discuss exemplary viruses and archetypal studies. We discuss how the speed and flexibility of HIO-based studies contributed to our knowledge of SARS-CoV-2 and antiviral therapeutics. Finally, we discuss the current limitations of HIOs and future directions to overcome these.
INTRODUCTION
The human gastrointestinal epithelium is a remarkably dynamic, cellularly diverse, and histologically intricate system under tight regulation. It not only must absorb dietary nutrients to nourish every cell in the human body, but it must also form a protective barrier against the tremendous array of pathogens that make their way into the gut lumen. Viruses are among the most abundant and diverse pathogens and continue to drive the majority of gastrointestinal infections (1–3).
Historically, studies focused on the pathogenesis of enteric viruses relied on transformed cell lines that lack both the cellular diversity and coordinated function of the gastrointestinal epithelium (4). Although animal models provide a functional readout and a more holistic model of infection, they are costly, low throughput, and often not susceptible to, or exhibit different symptoms in response to, human enteric virus infection (5). Within the past decade, virologists have increasingly turned to human intestinal organoids (HIOs) as a tool to further elucidate viral-host interactions. HIOs are stem cell-derived ex vivo culture systems that are nontransformed, human origin, and recapitulate the cellular diversity of the native human intestinal epithelium (6, 7). They maintain a tight, polarized epithelial barrier and regulated secretory activity allowing for functional studies (8–10). As such, HIOs have become a valuable tool for virologists.
This review discusses recent advances in virology made possible by HIOs. We discuss how HIOs have allowed for the cultivation of previously uncultivatable viruses, facilitated studies of viral pathogenesis, and offered a fast and adaptable platform to study SARS-CoV-2. Finally, we discuss the current limitations of HIOs in virology and future directions to overcome these.
VARIATIONS AND TECHNIQUES USING HIOs
Although technological advances have allowed for the diversification of organoid models, the fundamental principle remains consistent. Broadly, organoids are derived from isolated stem cells. The term “enteroid” refers specifically to HIOs derived from multipotent stem cells isolated directly from the intestinal crypt. Although enteroids are still themselves HIOs, HIOs can also be derived from pluripotent or induced pluripotent stem cells (iPSCs; 11). HIOs derived from iPSCs maintain mesenchymal cells which are not found in enteroids (12).
It is possible to culture HIOs in a variety of platforms. Most frequently, HIOs are maintained as three-dimensional spheres in synthetic matrices (e.g., Matrigel) that support growth. Three-dimensional HIOs can be dissociated into single-cell suspensions and replated on collagen-coated culture dishes as monolayers. In addition, HIOs can be plated as monolayers on permeable membranes that separate the apical and basolateral compartments. Separation of the basolateral and apical media allows investigators to measure electrical potential across the monolayer, membrane permeability, migration, and the unique effects of compounds applied apically versus basolaterally.
Recent advances have also allowed for the enrichment of cell types that are typically rare in HIOs. Using a tetracycline-inducible promoter to overexpress neurogenin-3 allows for the enrichment of enteroendocrine cells, addition of retinoic acid and lymphotoxin promote M cell differentiation, and IL-22 increases the number of Paneth cells (13–15). Given the unique roles these cell types play in defense against pathogens, antiviral responses, and gut function, these techniques offer novel platforms to study cell type-specific contributions to pathogenesis.
HIOs FOR THE CULTIVATION OF VIRUSES
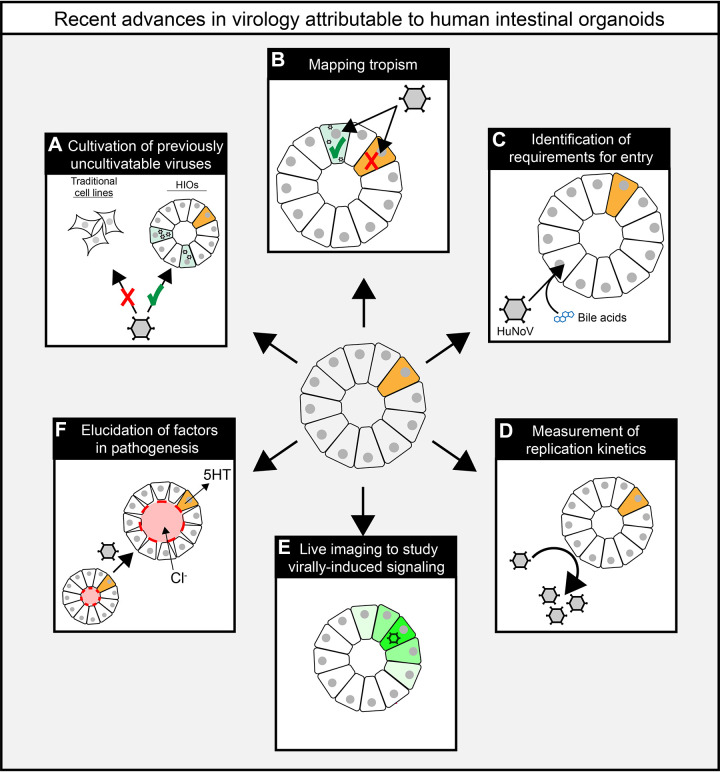
Arguably the most important advance HIOs have provided the field of virology is the ability to establish new infection models for previously uncultivatable viruses. The most notable example of this is human norovirus (HuNoV). HuNoV is a highly infectious pathogen that targets intestinal epithelial cells, causing acute gastroenteritis in immunocompetent patients and severe, chronic disease in the immunocompromised (16). Despite the significant global burden of HuNoV infection, researchers have made limited progress in the production of effective vaccines or antiviral therapies. This can be, in part, attributed to the lack of robust replication in conventional cell culture systems and animal models. Although cultured B cells were shown to support the replication of HuNoV, this system has proven difficult to consistently reproduce (17, 18). In 2016, the Estes group resolved nearly 50-years delay in establishing a robust human norovirus cultivation system by using HIOs, the first model to consistently enable replication of multiple HuNoV genotypes (Fig. 1A; 19). To date, GI.1, 11 genotypes of GII, and six variants of GII.4 HuNoV have shown productive replication in HIOs (20). Although multiple groups have attempted to use HIOs to cultivate other enteric caliciviruses, including human sapovirus and bovine norovirus, they have yet to establish optimal conditions (21).
Figure 1.
Graphic summary of virological insights from experiments using human intestinal organoids (HIOs). HIOs have allowed for the cultivation human norovirus (HuNoV; A) and determination of cell type-specific tropism for HuNoV, astrovirus, and rotavirus (RV), which infects preferentially infects differentiated enterocytes over undifferentiated stem cells (orange; B). HIOs also allowed investigators to determine that HuNoV requires both bile acids and ceramide for entry (C) and identify determinants of replication kinetics for enterovirus (D). Finally, genetically engineered HIOs allowed investigators to characterize a new role for paracrine signaling in rotavirus Ca2+ signaling (green) and pathogenesis (E) and understand contributors like serotonin (5HT) and chloride (Cl−) to luminal (red) fluid secretion and swelling (F).
Human astrovirus is another clinically relevant enteric pathogen and an important cause of acute gastroenteritis in children (22–24). Recent work identified divergent groups of astrovirus and documented their potential to cause extraintestinal infections (25–29). These studies underscore a critical need for a deeper understanding of astrovirus tropism and pathogenesis, but, similar to HuNoV, the lack of physiologically relevant cultivation system has hindered progress. In addition, although most investigators use the Caco-2 human colon adenocarcinoma cell line to propagate classical astrovirus serotypes, it does not support the replication of all nonclassical serotypes (30, 31). In 2019, Kolawole et al. (32) showed that HIOs could be used to replicate strains of human astrovirus from all three clades. This work has opened the door to study astroviruses more broadly and improve our understanding of pathogenesis.
THE BROAD CONTRIBUTIONS OF HIOs TO THE STUDY OF VIRAL PATHOGENESIS
Tropism
A key advantage of HIOs is that they recapitulate the diversity of epithelial cell types found in the gastrointestinal epithelial layer. In a proliferative state (i.e., undifferentiated), HIOs maintain a crypt-like profile of stem/progenitor and Paneth cells. Following differentiation, these cultures express mature cell types of the villus epithelium including enterocytes, goblet cells, enteroendocrine cells, and Tuft cells (7). This allows for a more precise determination of viral tropism (Fig. 1B). Both transcriptional studies and immunofluorescent microscopy (Fig. 2) have contributed to this (33–35). Using HIOs, investigators have determined that, in addition to infecting mature enterocytes, both rotavirus and norovirus are capable of infecting enteroendocrine cells (13, 34). The nonclassical human astrovirus, VA1, had previously undetermined tropism in the small intestine. Using HIOs, however, investigators were able to identify at least three distinct cell types that are infected with human astrovirus VA1: mature enterocytes, goblet cells, and intestinal progenitor cells (32). Importantly, these findings established VA1 as the first enteric virus capable of infecting progenitor cells and may provide the first hints of viral factors needed for broader tropism and extraintestinal spread. In a recent study, Triana et al. (36) used multiplexed in situ hybridization to show that astrovirus RNA colocalized with transcripts marking transiently amplifying cells and mature enterocytes, and to a lesser extent with those marking stem-like cells, goblet cells, and enteroendocrine cells.
Figure 2.
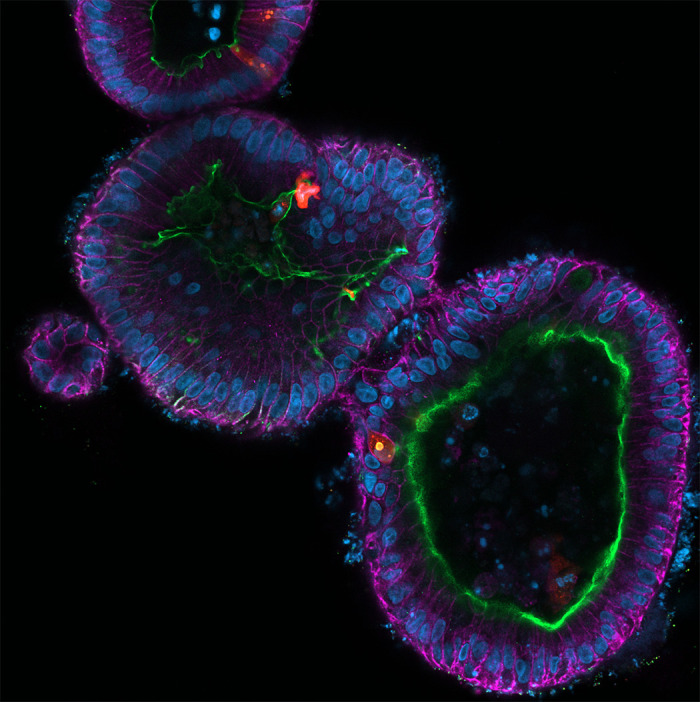
Fluorescent micrograph of multiple human intestinal organoids infected with rotavirus. The organoids were fixed and stained for E-cadherin (magenta), villin (green), nuclei (blue), and rotavirus (red). The image was acquired on a Zeiss LSM980 confocal microscope.
Beyond cellular tropism, Saxena et al. (33) also used HIOs to investigate host range restriction of different rotavirus strains, showing that HIOs are more susceptible to a human versus simian strain. In addition, using HIOs generated from the duodenum, jejunum, and ileum, they showed that HIOs generated from any segment are susceptible to rotavirus infection.
Entry
Before the establishment of a cell culture model for human norovirus, mechanistic studies relied upon replicon systems, which used transfection of self-replicating norovirus RNA (37). Although these early studies provided insight, they could not account for factors required for productive replication at all stages (binding, entry, genome replication, assembly, and release). Small animal models overcome some of these limitations, yet in these systems, it is difficult to manipulate and study host factors involved in susceptibility and permissibility using these models alone. HIOs, therefore, provided a new opportunity to robustly study host/pathogen interactions in the context of a native human norovirus infection. This breakthrough has led to the identification of components of the intestinal milieu, which are required for replication, and for the appreciation of strain-specific differences in these requirements. Ettayebi et al. (19) established that bile is required for the replication of GI.1, GII.3, and GII.17 strains of human noroviruses, and enhances the replication of GII.4 strains (Fig. 1C). Following this finding, Murakami et al. (38) went on to show that bile acids were the active component of bile that promoted GII.3 replication, and through specific interactions with the bile acid receptor, S1PR2, enhanced both entry and infection (38). These studies underlie the utility of the HIO model in understanding mechanisms of entry in physiologically relevant systems, critically broadening the potential for new antiviral therapeutic targets.
Replication Kinetics
Beyond tropism and entry, HIOs have offered valuable insight into the mechanisms underlying viral replication. This is exemplified by enteroviruses which, similar to HuNoV and astrovirus, lacked a broad physiologically relevant culture system before the advent of HIOs.
HIOs have been established as a viable platform to study multiple species of enterovirus including coxsackievirus B, echovirus 11, enterovirus A71, and poliovirus type 3 (PV-3; 39, 40). Adapting this model, Drummond et al. (39) were able to characterize inflammatory responses in HIOs that more closely mimic those seen in vivo as compared with transformed cell lines. Given that inflammatory processes often modulate viral replication, this suggests that HIOs may offer advantages when studying replication kinetics. By assessing tissue culture infectious dose (TCID50) and viral transcript levels, Tsang et al. (40) leveraged HIOs to more thoroughly characterize the kinetics of enterovirus replication (Fig. 1D). Finally, they showed a high level of expression of the receptors for EV-A71, CVB2, and PV-3 in HIOs, but low expression of the receptor (ICAM-5) for a fourth enterovirus, EV-D68, which did not replicate in HIOs. This finding is consistent with and may explain the observation that EV-D68 primarily infects respiratory epithelium in humans (41).
HOST-VIRUS INTERACTIONS AND CELL SIGNALING
When aiming to study host-pathogen interactions, HIOs offer distinct advantages over both in vitro and in vivo model systems. Multiple groups have genetically engineered HIOs to express fluorescent biosensors (42), knockout or overexpress genes of interest (20, 42, 43), or favor differentiation of certain cell populations (13, 44). These manipulations have allowed for focused investigations of specific signaling pathways involved in enteric viral pathogenesis, detailed in this section. For a broader review of strategies to genetically engineer organoids, see the review by Menche et al. (45).
Calcium Signaling
Calcium (Ca2+) signaling is a crucial regulator of homeostasis within the intestinal epithelium (46–48). Multiple groups had shown that rotavirus exploits Ca2+ signaling for replication in immortalized cell lines, but the nature of these signals and their effect on the intestinal epithelium remained unclear (48–53). Our group generated HIOs expressing the genetically encoded cytosolic calcium indicator GCaMP6s to characterize novel signals induced upon rotavirus infection (Fig. 1E; 42). Not only did we find unique Ca2+ signatures in infected cells, but the HIO-GCaMP6s system allowed us to characterize intercellular Ca2+ waves that elevate Ca2+ signaling in surrounding, uninfected cells. This dysregulated Ca2+ signaling leads to increased fluid secretory activity and serotonin release from enterochromaffin cells, contributing to rotavirus pathogenesis. Not only did this identify a novel component of virus-induced signaling, but it also offered a potential new target for antidiarrheal therapeutics. Furthermore, although intercellular Ca2+ waves are rare in cell lines outside of the setting of infection, this work demonstrated that HIOs display a low level of Ca2+ waves at baseline. This suggests a broader regulatory role of intercellular Ca2+ waves in the intestinal epithelium, underscoring the importance of physiologically accurate models.
Innate Immunity and Interferon Signaling
The synthesis and secretion of interferons (IFNs) are key tenants of innate immunity and early antiviral responses (54, 55). Epithelial cells are an important component of IFN production and signaling and as such, must be appropriately modeled to understand the interplay between infection and host immune responses. Studies have demonstrated that baseline interferon production and the interferon response that is mounted to an infection can be altered in senescent or immortalized cells, complicating the interpretation of immune signaling pathways in primary and transformed cell lines (56–58). In addition, transfection of viral RNA in nonsusceptible or permissive cells often induces an immune response that differs from that of native virus infection. Interferon signaling is a key element of this response. A meta-analysis of RNA sequencing on organoids infected with astrovirus, norovirus, and rotavirus demonstrated that in all cases, interferons and interferon-stimulated genes make up an important part of the host response to infection (59). Type I and type III interferon predominate in the intestine, with type I associated with controlling systemic virus spread and type III associated with localized restriction of mucosal infection (60). Cell model-dependent discrepancies in the types of interferons stimulated by infection and the susceptibility of viruses to these factors have been demonstrated for two important enteric viruses, human norovirus, and human adenovirus (HAdV).
In HuNoV studies, transfection of immortalized cell lines with RNA from clinical HuNoV isolates (35) fails to stimulate interferon production (37, 61). This is at odds with findings from other human enteric viruses and murine norovirus, suggesting a potential limitation of the transfection-based model rather than a true absence of interferon signaling (62, 63). Studies using genetically modified HIOs overcame these limitations to elucidate the role of specific interferons in HuNoV infection. By performing RNA sequencing of HuNoV-infected HIOs, Hosmillo et al. (43) demonstrated that HuNoV indeed elicits expression of interferon-stimulated genes and downstream activation of JAK/STAT signaling. Pretreatment with either IFN-β1 or IFNγ1/2 reduced HuNoV replication in HIOs, supporting an inhibitory effect of IFNs. Likewise, HIOs engineered to constitutively express interferon inhibitors offered enhanced HuNoV replication, further supporting an antagonistic role of interferons in infection. Later studies using CRISPR-Cas9 mediated knockout of IFNα, IFNγ, MAVS, STAT1, and STAT2 in HIOs allowed Lin et al. (64) to characterize strain-specific effects of IFNs on HuNoV, which may underlie broader epidemiological differences.
Studies of human adenoviruses (HAdVs) also demonstrate the advantages of HIOs over standard cell lines with regard to innate immunity. In cancer cell lines, the interferon response fails to restrict wild-type HAdV replication (65, 66). This contrasts with in vivo data, which demonstrates enhanced adenovirus load in STAT2-knockout animals compared with wild-type and in vitro data in primary human cell lines, which shows significant inhibition of Ad5 replication with exogenous IFNα and IFNγ treatment (67, 68). To resolve these differences, Holly et al. (69) cultivated clinical HAdV isolates in human ileal enteroids and transformed A549 cells. These studies recapitulated important differences in adenovirus sensitivity to IFN treatment, demonstrating that IFNβ and IFNλ3 inhibit HAdV-5p replication in ileal organoids but do not affect HAdV-5 replication in A549 cells. These studies underlie the need to carefully consider how cell culture models may confound our understanding of host responses and virus replication.
In addition, studies of astrovirus infection in HIOs revealed lineage-specific differences in expression of interferon-stimulated genes. Shortly after infection, stem and goblet cells showed higher expression of CCL2, STAT1, IFNGR1, and IRF2 relative to enterocytes. Though expression of these markers was lower in uninfected HIOs, the cell-type specific differences in interferon-stimulated gene expression persisted, suggesting that they are intrinsic. Furthermore, the antiviral response was not limited to infected cells. Bystander cells also showed an upregulation of interferon expression, albeit to a lesser extent (36).
Hormonal Signaling
Hormones produced in the gut are integral for the regulation of both the gut itself and many organ systems throughout the body (70–73). Although enteric pathogens perturb hormone signaling within the gut (74), these perturbations are unobservable in transformed cell lines that lack significant hormone production. HIOs offer a platform to study hormonal dysregulation, but the primary hormone-producing cell type, enteroendocrine cells, are typically quite rare in HIOs. This results in hormone levels that are difficult to detect through standard techniques. To overcome this, our group engineered a line of HIOs that expresses neurogenin-3 under a doxycycline-inducible promoter (13). Neurogenin-3 drives enteroendocrine cell differentiation, thus by adding doxycycline to induce neurogenin-3 overexpression, we can enrich for enteroendocrine cells. This system allowed us to detect the secretion of serotonin, monocyte chemoattractant protein-1, glucose-dependent insulinotropic peptide, peptide YY, and ghrelin upon rotavirus infection, suggesting it may be a useful model for hormone-related studies. Furthermore, this serves as a valuable proof-of-concept for those interested in studying other rare cell types in HIOs.
Functional Studies
HIOs grow as three-dimensional spheroids or as polarized monolayers, expanding their versatility for studying viral-host interactions. During viral infection, a three-dimensional HIO exhibits upregulated fluid secretion into its lumen, which increases the luminal volume (33). The change in the cross-sectional area of an infected HIO over time, measurable using standard bright-field microscopy, correlates with the degree of fluid secretion (33, 75). Given that fluid secretion is a critical driver of clinical disease, this technique, known as a swelling assay, provides means to assess factors that directly contribute to virally induced fluid loss. Using this technique, our group showed that paracrine purinergic signaling is an integral component of rotavirus-mediated fluid secretion (Fig. 1F; 42). Similarly, HIO monolayers can be plated on transwell filters, which offer independent access to both the apical and basolateral sides of a polarized monolayer without disruption of the monolayer itself (76, 77). These can be used to measure barrier integrity, solute transport, fluid secretion, and other aspects of epithelial physiology.
HIOs AS A PLATFORM FOR THE STUDY OF SARS-CoV-2
While maintaining the cellular diversity of the native intestinal epithelium, HIOs offer distinct speed and adaptability over traditional in vivo models. Establishing a novel genetic mouse model and using it to generate experimental data often takes two or more years (78). Genetic manipulation of HIOs presents challenges of its own, but an experienced user can establish a genetically engineered HIO line in as little as 2 mo (79). Furthermore, HIOs are of human origin. This is an important consideration given the host range restriction of most human viruses. SARS-CoV-2, for example, is unable to infect wild-type laboratory mice due to the incompatibility of the mouse angiotensin-converting enzyme (ACE)-2 receptor and the SARS-CoV-2 spike protein (80). As such, HIOs can expedite studies of viral pathogenesis while maintaining the benefits of nontransformed human tissue. Unsurprisingly, many groups capitalized on the speed and adaptability of HIOs to facilitate studies of SARS-CoV-2 (81–83).
Less than 2 mo after the World Health Organization declared COVID-19 a pandemic (84), three groups published work implementing HIOs to further our understanding of SARS-CoV-2 (81, 82, 85). Although clinical studies established that SARS-CoV-2 promoted gastrointestinal symptoms and was detected in the stool (86–88), studies in HIOs provided the first definitive evidence that SARS-CoV-2 could both infect and replicate in human intestinal epithelial cells (81). By using polarized HIO monolayers plated in transwells, Lamers et al. (81) also showed that SARS-CoV-2 both infected and exited enterocytes primarily at the apical membrane. Furthermore, they observed fusion of SARS-CoV-2-infected cells, suggesting that the virus is capable of inducing syncytia formation in enterocytes (82). Using HIOs from 13 different donors, Jang et al. (89) showed tremendous host-to-host variation in viral yield following SARS-CoV-2 infection. Viral titers in the culture media showed a 423-fold difference between the highest and lowest replicators. Interestingly, this variability correlated with the expression levels of the SARS-CoV-2 receptors ACE2 and TMPRSS2.
Beyond pathogenesis, HIOs were also rapidly implemented to study the activity of antiviral therapeutics against SARS-CoV-2 (90). Using HIOs in conjunction with lung organoids, Han et al. (90) showed that three different antiviral compounds inhibit replication of SARS-CoV-2 in organoids. In addition, they demonstrated that the mechanism of action of one of these compounds, Imatinib, is multifactorial, reducing receptor expression and altering expression of genes involved in lipid metabolism. In these studies, HIOs offered a physiologically relevant platform to rapidly investigate antiviral efficacy and mechanism of action at a time when speed and translatability were high priority.
CURRENT LIMITATIONS AND FUTURE DIRECTIONS OF HIOs IN VIROLOGY
Although HIOs offer many advantages, as with all model systems there are important limitations to consider. Perhaps the most salient limitation for virological studies is the lack of tissue type diversity. Although HIOs maintain the cell diversity of the intestinal epithelium, they lack the other cell types critical for gastrointestinal tract function, including immune cells, lymphatics, endothelial cells, neurons, smooth muscle, and fibroblasts, let alone integration with other tissues outside the gut. All of these tissue types play a role in host-pathogen interactions but are unobservable using organoids alone.
To overcome this, many have begun to develop systems in which HIOs are maintained in culture with other nonepithelial cell types (91). These coculture systems have allowed for broader investigation of the interactions between virally infected epithelial cells and other, nonepithelial cell types including monocytes, dendritic cells, lymphocytes, stromal cells, adipocytes, endothelial cells, and neurons. As coculture systems continue to improve, they will offer valuable platforms to study elements of pathogenesis that extend beyond the epithelium.
Furthermore, HIOs fail to maintain the intricate morphology of the intestinal epithelium. In vivo, the intestinal epithelium is neatly organized into villi and crypts with distinct cell types distributed predictably throughout (92). HIOs have crypt-like regions but generally lack villi and fail to show the same predictable distribution of cell types. Importantly, enteric viruses often show a pattern of infection that is spatially restricted along the villus-crypt axis. For example, rotavirus preferentially infects the fully differentiated enterocytes at the tips of villi (93). These morphology-dependent features of infection are difficult to assess in HIOs.
However, recent developments offer promising advancements. By using microfluidic chambers to mimic intestinal flow and peristalsis, or by inducing softening of the culture matrix in predefined patterns, investigators can push HIOs to develop the native villus-crypt architecture (94, 95). Groups have already utilized this technology for live imaging to assess epithelial responses (96). Adapting these systems for virological studies would allow for interrogation of the temporal and spatial dynamics of viral infections without the implicit difficulties of intravital imaging.
HIOs require specialized media that has yet to be universally standardized. Media supporting human intestinal organoids is typically enriched with Wnt, R-spondin, noggin, human epidermal growth factor, and serum. Although enriched media is commercially available, it is often more cost effective for investigators to create their own media using cell lines that have been engineered to express and secrete the aforementioned growth factors. Because of this, there is a high degree of variability between HIOs maintained in different laboratories (97). As HIOs increase in their popularity and accessibility, researchers must validate findings across intestinal segments, lines derived from distinct patients, and culture conditions.
To date, HIOs have facilitated many meaningful advancements in virology. As technology develops to support more versatile culture systems, HIOs will continue to play an integral role in studies ultimately aimed at lessening the burden of virological diseases worldwide.
GRANTS
This review was supported by the McNair Foundation MD/PhD Scholars Program (to J. T. Gebert); National Institute of Allergy and Infectious Diseases (NIAID), Division of Microbiology and Infectious Diseases Grant R01AI158683 (to J. M. Hyser); National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), Division of Diabetes, Endocrinology, and Metabolic Diseases (DEM) Grant R01DK115507 (to J. M. Hyser); and NIDDK Grants F31DK132942 (to F. Scribano), F32DK130288 (to K. A. Engevik), F30DK131828 (to J. T. Gebert), and NIAID Grant F31AI169983 (to J. L. Perry).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.T.G. prepared figures; J.T.G., F.S., K.A.E., and J.L.P. drafted manuscript; J.T.G., F.S., K.A.E., and J.M.H. edited and revised manuscript; J.T.G., F.S., K.A.E., J.L.P., and J.M.H. approved final version of manuscript.
REFERENCES
- 1. Troeger C, Blacker BF, Khalil IA, Rao PC, Cao S, Zimsen SR, Albertson SB, Stanaway JD, Deshpande A, Abebe Z, Alvis-Guzman N, Amare AT, Asgedom SW, Anteneh ZA, Antonio CAT, Aremu O, Asfaw ET, Atey TM, Atique S, Avokpaho EFGA, Awasthi A, Ayele HT, Barac A, Barreto ML, Bassat Q, Belay SA, Bensenor IM, Bhutta ZA, Bijani A, Bizuneh H. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 18: 1211–1228, 2018. doi: 10.1016/S1473-3099(18)30362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD Diarrhoeal Diseases Collaborators. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 17: 909–948, 2017. [Erratum in Lancet Infect Dis 17: 897, 2017]. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cohen AL, Platts-Mills JA, Nakamura T, Operario DJ, Antoni S, Mwenda JM, Weldegebriel G, Rey-Benito G, de Oliveira LH, Ortiz C, Daniels DS, Videbaek D, Singh S, Njambe E, Sharifuzzaman M, Grabovac V, Nyambat B, Logronio J, Armah G, Dennis FE, Seheri ML, Magagula N, Mphahlele J, Fumian TM, Maciel ITA, Gagliardi Leite JP, Esona MD, Bowen MD, Samoilovich E, Semeiko G. Aetiology and incidence of diarrhoea requiring hospitalisation in children under 5 years of age in 28 low-income and middle-income countries: findings from the Global Pediatric Diarrhea Surveillance network. BMJ Glob Health 7: e009548, 2022. doi: 10.1136/bmjgh-2022-009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dolskiy AA, Grishchenko I. V, Yudkin D. V. Cell cultures for virology: usability, advantages, and prospects. Int J Mol Sci 21: 7978, 2020. doi: 10.3390/ijms21217978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ayyavoo V. Modeling human viral diseases: trials and triumphs. Front Virol 1: 722297, 2021. doi: 10.3389/fviro.2021.722297. [DOI] [Google Scholar]
- 6. Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol 21: 571–584, 2020. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zachos NC, Kovbasnjuk O, Foulke-Abel J, In J, Blutt SE, de Jonge HR, Estes MK, Donowitz M. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J Biol Chem 291: 3759–3766, 2016. doi: 10.1074/jbc.R114.635995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265, 2009. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 9. Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141: 1762–1772, 2011. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 10. Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, Brandsma AM, de Jong NWM, Bijvelds MJC, Scholte BJ, Nieuwenhuis EES, van den Brink S, Clevers H, van der Ent CK, Middendorp S, Beekman JM. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med 19: 939–945, 2013. 2013 19:7 doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 11. Stelzner M, Helmrath M, Dunn JC, Henning SJ, Houchen CW, Kuo C, Lynch J, Li L, Magness ST, Martin MG, Wong MH, Yu J; NIH Intestinal Stem Cell Consortium. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol 302: G1359–G1363, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, Wells JM. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470: 105–109, 2011. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang-Graham AL, Danhof HA, Engevik MA, Tomaro-Duchesneau C, Karandikar UC, Estes MK, Versalovic J, Britton RA, Hyser JM. Human intestinal enteroids with inducible Neurogenin-3 expression as a novel model of gut hormone secretion. Cell Mol Gastroenterol Hepatol 8: 209–229, 2019. doi: 10.1016/j.jcmgh.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He G-W, Lin L, DeMartino J, Zheng X, Staliarova N, Dayton T, Begthel H, van de Wetering WJ, Bodewes E, van Zon J, Tans S, Lopez-Iglesias C, Peters PJ, Wu W, Kotlarz D, Klein C, Margaritis T, Holstege F, Clevers H. Optimized human intestinal organoid model reveals interleukin-22-dependency of paneth cell formation. Cell Stem Cell 29: 1333–1345.e6, 2022. [Erratum in Cell Stem Cell 29: 1718–1720, 2022]). doi: 10.1016/j.stem.2022.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding S, Song Y, Brulois KF, Pan J, Co JY, Ren L, Feng N, Yasukawa LL, Sánchez-Tacuba L, Wosen JE, Mellins ED, Monack DM, Amieva MR, Kuo CJ, Butcher EC, Greenberg HB. Retinoic acid and lymphotoxin signaling promote differentiation of human intestinal M cells. Gastroenterology 159: 214–226.e1, 2020. doi: 10.1053/j.gastro.2020.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Atmar RL, Ramani S, Estes MK. Human noroviruses: recent advances in a 50-year history. Curr Opin Infect Dis 31: 422–432, 2018. doi: 10.1097/QCO.0000000000000476. [DOI] [PubMed] [Google Scholar]
- 17. Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinjé J, Tibbetts SA, Wallet SM, Karst SM. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346: 755–759, 2014. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moore MD, Goulter RM, Jaykus LA. Human norovirus as a foodborne pathogen: challenges and developments. Annu Rev Food Sci Technol 6: 411–433, 2015. doi: 10.1146/annurev-food-022814-015643. [DOI] [PubMed] [Google Scholar]
- 19. Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. Replication of human noroviruses in stem cell-derived human enteroids. Science 353: 1387–1393, 2016. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ettayebi K, Tenge VR, Cortes-Penfield NW, Crawford SE, Neill FH, Zeng X-L, Yu X, Ayyar BV, Burrin D, Ramani S, Atmar RL, Estes MK. New insights and enhanced human norovirus cultivation in human intestinal enteroids. mSphere 6: e01136-20, 2021. doi: 10.1128/mSphere.01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oka T, Stoltzfus GT, Zhu C, Jung K, Wang Q, Saif LJ. Attempts to grow human noroviruses, a sapovirus, and a bovine norovirus in vitro. PLoS One 13: e0178157, 2018. doi: 10.1371/journal.pone.0178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herrmann JE, Taylor DN, Echeverri P, Blacklow NR. Astroviruses as a cause of gastroenteritis in children. N Engl J Med 324: 1757–1760, 1991. doi: 10.1056/NEJM199106203242501. [DOI] [PubMed] [Google Scholar]
- 23. Vu DL, Bosch A, Pintó RM, Guix S. Epidemiology of classic and novel human astrovirus: gastroenteritis and beyond. Viruses 9: 33, 2017. doi: 10.3390/v9020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeong HS, Jeong A, Cheon DS. Epidemiology of astrovirus infection in children. Korean J Pediatr 55: 77–82, 2012. doi: 10.3345/kjp.2012.55.3.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Finkbeiner SR, Kirkwood CD, Wang D. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol J 5: 117, 2008. doi: 10.1186/1743-422X-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Finkbeiner SR, Le BM, Holtz LR, Storch GA, Wang D. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg Infect Dis 15: 441–444, 2009. doi: 10.3201/eid1503.081213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 4: e1000011, 2008. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Finkbeiner SR, Li Y, Ruone S, Conrardy C, Gregoricus N, Toney D, Virgin HW, Anderson LJ, Vinjé J, Wang D, Tong S. Identification of a novel astrovirus (Astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol 83: 10836–10839, 2009. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, Shah D, Brooks T, Paine SML, Anderson G, Virasami A, Tong CYW, Clark DA, Plagnol V, Jacques TS, Qasim W, Hubank M, Breuer J. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis 60: 881–888, 2015. doi: 10.1093/cid/ciu940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vu D-L, Bosch A, Pintó RM, Ribes E, Guix S. Human astrovirus MLB replication in vitro: persistence in extraintestinal cell lines. J Virol 93: e00557-19, 2019. doi: 10.1128/JVI.00557-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Janowski AB, Bauer IK, Holtz LR, Wang D. Propagation of astrovirus VA1, a neurotropic human astrovirus, in cell culture. J Virol 91: e00740-17, 2017. doi: 10.1128/JVI.00740-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kolawole AO, Mirabelli C, Hill DR, Svoboda SA, Janowski AB, Passalacqua KD, Rodriguez BN, Dame MK, Freiden P, Berger RP, Vu D-L, Hosmillo M, O’Riordan MXD, Schultz-Cherry S, Guix S, Spence JR, Wang D, Wobus CE. Astrovirus replication in human intestinal enteroids reveals multi-cellular tropism and an intricate host innate immune landscape. PLoS Pathog 15: e1008057, 2019. doi: 10.1371/journal.ppat.1008057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saxena K, Blutt SE, Ettayebi K, Zeng X-L, Broughman JR, Crawford SE, Karandikar UC, Sastri NP, Conner ME, Opekun AR, Graham DY, Qureshi W, Sherman V, Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Donowitz M, Estes MK. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J Virol 90: 43–56, 2016. doi: 10.1128/JVI.01930-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Green KY, Kaufman SS, Nagata BM, Chaimongkol N, Kim DY, Levenson EA, Tin CM, Yardley AB, Johnson JA, Barletta ABF, Khan KM, Yazigi NA, Subramanian S, Moturi SR, Fishbein TM, Moore IN, Sosnovtsev SV. Human norovirus targets enteroendocrine epithelial cells in the small intestine. Nat Commun 11: 2759, 2020. doi: 10.1038/s41467-020-16491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bomidi C, Robertson M, Coarfa C, Estes MK, Blutt SE. Single-cell sequencing of rotavirus-infected intestinal epithelium reveals cell-type specific epithelial repair and tuft cell infection. Proc Natl Acad Sci USA 118: e2112814118, 2021. doi: 10.1073/pnas.2112814118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Triana S, Stanifer ML, Metz-Zumaran C, Shahraz M, Mukenhirn M, Kee C, Serger C, Koschny R, Ordoñez-Rueda D, Paulsen M, Benes V, Boulant S, Alexandrov T. Single-cell transcriptomics reveals immune response of intestinal cell types to viral infection. Mol Syst Biol 17: e9833, 2021. doi: 10.15252/msb.20209833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guix S, Asanaka M, Katayama K, Crawford SE, Neill FH, Atmar RL, Estes MK. Norwalk virus RNA is infectious in mammalian cells. J Virol 81: 12238–12248, 2007. doi: 10.1128/JVI.01489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murakami K, Tenge VR, Karandikar UC, Lin S-C, Ramani S, Ettayebi K, Crawford SE, Zeng X-L, Neill FH, Ayyar BV, Katayama K, Graham DY, Bieberich E, Atmar RL, Estes MK. Bile acids and ceramide overcome the entry restriction for GII.3 human norovirus replication in human intestinal enteroids. Proc Natl Acad Sci USA 117: 1700–1710, 2020. [Erratum in Proc Natl Acad Sci USA 117: 16084, 2020]. doi: 10.1073/pnas.1910138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Drummond CG, Bolock AM, Ma C, Luke CJ, Good M, Coyne CB. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc Natl Acad Sci USA 114: 1672–1677, 2017. doi: 10.1073/pnas.1617363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsang JO-L, Zhou J, Zhao X, Li C, Zou Z, Yin F, Yuan S, Yeung M-L, Chu H, Chan JF-W. Development of three-dimensional human intestinal organoids as a physiologically relevant model for characterizing the viral replication kinetics and antiviral susceptibility of enteroviruses. Biomedicines 9: 88–16, 2021. doi: 10.3390/biomedicines9010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiang Z, Wang J. Respiratory viral infections: enterovirus D68 and human respiratory infections. Semin Respir Crit Care Med 37: 578–585, 2016. doi: 10.1055/s-0036-1584795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chang-Graham AL, Perry JL, Engevik MA, Engevik KA, Scribano FJ, Gebert JT, Danhof HA, Nelson JC, Kellen JS, Strtak AC, Sastri NP, Estes MK, Britton RA, Versalovic J, Hyser JM. Rotavirus induces intercellular calcium waves through ADP signaling. Science (1979) 370: eabc3621, 2020. doi: 10.1126/science.abc3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hosmillo M, Chaudhry Y, Nayak K, Sorgeloos F, Koo B-K, Merenda A, Lillestol R, Drumright L, Zilbauer M, Goodfellow I. Norovirus replication in human intestinal epithelial cells is restricted by the interferon-induced JAK/STAT signaling pathway and RNA polymerase II-mediated transcriptional responses. mBio 11: e00215-20, 2020. doi: 10.1128/mBio.00215-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boonekamp KE, Dayton TL, Clevers H. Intestinal organoids as tools for enriching and studying specific and rare cell types: advances and future directions. J Mol Cell Biol 12: 562–568, 2020. doi: 10.1093/jmcb/mjaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Menche C, Farin HF. Strategies for genetic manipulation of adult stem cell-derived organoids. Exp Mol Med 53: 1483–1494, 2021. 2021 53:10 doi: 10.1038/s12276-021-00609-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Timar Peregrin AL. Effects of calcium channel blockade on intestinal fluid secretion: sites of action. Acta Physiol Scand 160: 379–386, 1997. [Erratum in Acta Physiol Scand 162: 523, 1998]. doi: 10.1046/j.1365-201X.1997.00173.x. [DOI] [PubMed] [Google Scholar]
- 47. Peregrin AT, Ahlman H, Jodal M, Lundgren O. Involvement of serotonin and calcium channels in the intestinal fluid secretion evoked by bile salt and cholera toxin. Br J Pharmacol 127: 887–894, 1999. doi: 10.1038/sj.bjp.0702615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brunet JP, Cotte-Laffitte J, Linxe C, Quero AM, Géniteau-Legendre M, Servin A. Rotavirus infection induces an increase in intracellular calcium concentration in human intestinal epithelial cells: role in microvillar actin alteration. J Virol 74: 2323–2332, 2000. doi: 10.1128/jvi.74.5.2323-2332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hyser JM, Utama B, Crawford SE, Broughman JR, Estes MK. Activation of the endoplasmic reticulum calcium sensor STIM1 and store-operated calcium entry by rotavirus requires NSP4 viroporin activity. J Virol 87: 13579–13588, 2013. doi: 10.1128/JVI.02629-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tian P, Hu Y, Schilling WP, Lindsay DA, Eiden J, Estes MK. The nonstructural glycoprotein of rotavirus affects intracellular calcium levels. J Virol 68: 251–257, 1994. doi: 10.1128/jvi.68.1.251-257.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Michelangeli F, Ruiz MC, del Castillo JR, Ludert JE, Liprandi F. Effect of rotavirus infection on intracellular calcium homeostasis in cultured cells. Virology 181: 520–527, 1991. doi: 10.1016/0042-6822(91)90884-e. [DOI] [PubMed] [Google Scholar]
- 52. Pérez JF, Ruiz M-C, Chemello ME, Michelangeli F. Characterization of a membrane calcium pathway induced by rotavirus infection in cultured cells. J Virol 73: 2481–2490, 1999. doi: 10.1128/JVI.73.3.2481-2490.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dong Y, Zeng CQY, Ball JM, Estes MK, Morris AP. The rotavirus enterotoxin NSP4 mobilizes intracellular calcium in human intestinal cells by stimulating phospholipase C-mediated inositol 1,4,5-trisphosphate production. Proc Natl Acad Sci U S A 94: 3960–3965, 1997. doi: 10.1073/pnas.94.8.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee AJ, Ashkar AA. The dual nature of type I and type II interferons. Front Immunol 9: 2061, 2018. doi: 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol 15: 87–103, 2015. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hata N, Sato M, Takaoka A, Asagiri M, Tanaka N, Taniguchi T. Constitutive IFN-α/β signal for efficient IFN-α/β gene induction by virus. Biochem Biophys Res Commun 285: 518–525, 2001. doi: 10.1006/bbrc.2001.5159. [DOI] [PubMed] [Google Scholar]
- 57. Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, Sasaki S, Imai K, Shibue T, Honda K, Taniguchi T. Integration of interferon-α/β signalling to p53 responses in tumour suppression and antiviral defence. Nature 424: 516–523, 2003. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 58. Battcock SM, Collier TW, Zu D, Hirasawa K. Negative regulation of the alpha interferon-induced antiviral response by the Ras/Raf/MEK pathway. J Virol 80: 4422–4430, 2006. doi: 10.1128/JVI.80.9.4422-4430.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cieza RJ, Golob JL, Colacino JA, Wobus CE. Comparative analysis of public RNA-sequencing data from human intestinal enteroid (HIEs) infected with enteric RNA viruses identifies universal and virus-specific epithelial responses. Viruses 13: 1059, 2021. doi: 10.3390/v13061059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ingle H, Peterson ST, Baldridge MT. Distinct effects of Type I and III interferons on enteric viruses. Viruses 10: 46, 2018. doi: 10.3390/v10010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qu L, Murakami K, Broughman JR, Lay MK, Guix S, Tenge VR, Atmar RL, Estes MK. Replication of human norovirus RNA in mammalian cells reveals lack of interferon response. J Virol 90: 8906–8923, 2016. doi: 10.1128/JVI.01425-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. MDA-5 recognition of a murine norovirus. PLoS Pathog 4: e1000108, 2008. [Erratum in PLoS Pathog 4(10), 2008]. doi: 10.1371/annotation/3ce83911-9ccf-4452-a690-2816d0e94c10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thackray LB, Duan E, Lazear HM, Kambal A, Schreiber RD, Diamond MS, Virgin HW. Critical role for interferon regulatory factor 3 (IRF-3) and IRF-7 in type I interferon-mediated control of murine norovirus replication. J Virol 86: 13515–13523, 2012. doi: 10.1128/JVI.01824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lin S-C, Qu L, Ettayebi K, Crawford SE, Blutt SE, Robertson MJ, Zeng X-L, Tenge VR, Ayyar BV, Karandikar UC, Yu X, Coarfa C, Atmar RL, Ramani S, Estes MK. Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc Natl Acad Sci USA 117: 23782–23793, 2020. doi: 10.1073/pnas.2010834117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ullman AJ, Reich NC, Hearing P. Adenovirus E4 ORF3 protein inhibits the interferon-mediated antiviral response. J Virol 81: 4744–4752, 2007. doi: 10.1128/JVI.02385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anderson KP, Fennie EH. Adenovirus early region 1A modulation of interferon antiviral activity. J Virol 61: 787–795, 1987. doi: 10.1128/JVI.61.3.787-795.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zheng Y, Stamminger T, Hearing P. E2F/Rb family proteins mediate interferon induced repression of adenovirus immediate early transcription to promote persistent viral infection. PLoS Pathog 12: e1005415, 2016. doi: 10.1371/journal.ppat.1005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Toth K, Lee SR, Ying B, Spencer JF, Tollefson AE, Sagartz JE, Kong I-K, Wang Z, Wold WSM. STAT2 knockout syrian hamsters support enhanced replication and pathogenicity of human adenovirus, revealing an important role of type I interferon response in viral control. PLoS Pathog 11: e1005084, 2015. [Erratum in PLoS Pathog 12: e1005392, 2016]. doi: 10.1371/journal.ppat.1005084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Holly MK, Smith JG. Adenovirus infection of human enteroids reveals interferon sensitivity and preferential infection of goblet cells. J Virol 92: e00250-18, 2018. doi: 10.1128/JVI.00250-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Côté CD, Zadeh-Tahmasebi M, Rasmussen BA, Duca FA, Lam TKT. Hormonal signaling in the gut. J Biol Chem 289: 11642–11649, 2014. doi: 10.1074/jbc.O114.556068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Martin CR, Osadchiy V, Kalani A, Mayer EA. The brain–gut–microbiome axis. Cell Mol Gastroenterol Hepatol. 6: 133–148, 2018. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lu Y, Yuan X, Wang M, He Z, Li H, Wang J, Li Q. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol 15: 47, 2022. doi: 10.1186/s13045-022-01273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morais LH, Schreiber HL, Mazmanian SK. The gut microbiota–brain axis in behaviour and brain disorders. Nat Rev Microbiol 19: 241–255, 2021. doi: 10.1038/s41579-020-00460-0. [DOI] [PubMed] [Google Scholar]
- 74. Zhou A. Crosstalk between the gut microbiota and epithelial cells under physiological and infectious conditions. Front Cell Infect Microbiol 12: 832672, 2022. doi: 10.3389/fcimb.2022.832672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Boj SF, Vonk AM, Statia M, Su J, Vries RR, Beekman JM, Clevers H. Forskolin-induced swelling in intestinal organoids: an in vitro assay for assessing drug response in cystic fibrosis patients. J Vis Exp 55159, 2017. doi: 10.3791/55159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Roodsant T, Navis M, Aknouch I, Renes IB, van Elburg RM, Pajkrt D, Wolthers KC, Schultsz C, van der Ark KCH, Sridhar A, Muncan V. A human 2D primary organoid-derived epithelial monolayer model to study host-pathogen interaction in the small intestine. Front Cell Infect Microbiol 10: 272, 2020. doi: 10.3389/fcimb.2020.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Altay G, Larrañaga E, Tosi S, Barriga FM, Batlle E, Fernández-Majada V, Martínez E. Self-organized intestinal epithelial monolayers in crypt and villus-like domains show effective barrier function. Sci Rep 9: 10140, 2019. [Erratum in Sci Rep 9: 18822, 2019]. doi: 10.1038/s41598-019-46497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gurumurthy CB, Saunders TL, Ohtsuka M. Designing and generating a mouse model: frequently asked questions. J Biomed Res 35: 76, 2021. doi: 10.7555/JBR.35.20200197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lin SC, Haga K, Zeng XL, Estes MK. Generation of CRISPR–Cas9-mediated genetic knockout human intestinal tissue–derived enteroid lines by lentivirus transduction and single-cell cloning. Nat Protoc 17: 1004–1027, 2022. doi: 10.1038/s41596-021-00669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dinnon KH, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586: 560–566, 2020. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science 369: 50–54, 2020. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zang R. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 5: eabc3582, 2020. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhao X, Li C, Liu X, Chiu MC, Wang D, Wei Y, Chu H, Cai J-P, Hau-Yee Chan I, Kak-Yuen Wong K, Fuk-Woo Chan J, Kai-Wang To K, Yuen KY, Zhou J. Human intestinal organoids recapitulate enteric infections of enterovirus and coronavirus. Stem Cell Reports 16: 493–504, 2021. doi: 10.1016/j.stemcr.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.World Health Organization. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19 – 11 March 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-11-march-2020. [last accessed 13 Dec 2022].
- 85. Zhou J, Li C, Liu X, Chiu MC, Zhao X, Wang D, Wei Y, Lee A, Zhang AJ, Chu H, Cai J-P, Yip CC-Y, Chan IH-Y, Wong KK-Y, Tsang OT-Y, Chan K-H, Chan JF-W, To KK-W, Chen H, Yuen KY. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med 26: 1077–1083, 2020. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 86. Cholankeril G, Podboy A, Aivaliotis VI, Tarlow B, Pham EA, Spencer SP, Kim D, Hsing A, Ahmed A. High prevalence of concurrent gastrointestinal manifestations in patients with severe acute respiratory syndrome coronavirus 2: early experience from California. Gastroenterology 159: 775–777, 2020. doi: 10.1053/j.gastro.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 158: 1831–1833.e3, 2020. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323: 1843–1844, 2020. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jang KK, Kaczmarek ME, Dallari S, Chen Y-H, Tada T, Axelrad J, Landau NR, Stapleford KA, Cadwell K. Variable susceptibility of intestinal organoid–derived monolayers to SARS-CoV-2 infection. PLoS Biol 20: e3001592, 2022. doi: 10.1371/journal.pbio.3001592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, Tang X, Yaron TM, Zhang T, Uhl S, Bram Y, Richardson C, Zhu J, Zhao Z, Redmond D, Houghton S, Nguyen D-HT, Xu D, Wang X, Jessurun J, Borczuk A, Huang Y, Johnson JL, Liu Y, Xiang J, Wang H, Cantley LC, tenOever BR, Ho DD, Pan FC, Evans T, Chen HJ, Schwartz RE, Chen S. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589: 270–275, 2021. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hentschel V, Seufferlein T, Armacki M. Intestinal organoids in coculture: redefining the boundaries of gut mucosa ex vivo modeling. Am J Physiol Gastrointest Liver Physiol 321: G693–G704, 2021. doi: 10.1152/ajpgi.00043.2021. [DOI] [PubMed] [Google Scholar]
- 92. Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14: 141–153, 2014. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 93. Crawford SE, Ramani S, Tate JE, Parashar UD, Svensson L, Hagbom M, Franco MA, Greenberg HB, O'Ryan M, Kang G, Desselberger U, Estes MK. Rotavirus infection. Nat Rev Dis Primers, 3: 17083, 2017. doi: 10.1038/nrdp.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gjorevski N, Nikolaev M, Brown TE, Mitrofanova O, Brandenberg N, DelRio FW, Yavitt FM, Liberali P, Anseth KS, Lutolf MP. Tissue geometry drives deterministic organoid patterning. Science 375: eaaw9021, 2022. doi: 10.1126/science.aaw9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kim HJ, Ingber DE. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb) 5: 1130–1140, 2013. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 96. Khan I, Prabhakar A, Delepine C, Tsang H, Pham V, Sur M. A low-cost 3D printed microfluidic bioreactor and imaging chamber for live-organoid imaging. Biomicrofluidics 15: 024105, 2021. doi: 10.1063/5.0041027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Criss ZK, Bhasin N, Di Rienzi SC, Rajan A, Deans-Fielder K, Swaminathan G, Kamyabi N, Zeng X-L, Doddapaneni H, Menon VK, Chakravarti D, Estrella C, Yu X, Patil K, Petrosino JF, Fleet JC, Verzi MP, Christakos S, Helmrath MA, Arimura S, DePinho RA, Britton RA, Maresso AW, Grande-Allen KJ, Blutt SE, Crawford SE, Estes MK, Ramani S, Shroyer NF. Drivers of transcriptional variance in human intestinal epithelial organoids. Physiol Genomics 53: 486–508, 2021. doi: 10.1152/physiolgenomics.00061.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]