Abstract
Peninsular bighorn sheep (Ovis canadensis nelsoni) are found exclusively in Southern California and Baja Mexico. They are federally endangered due to multiple threats, including introduced infectious disease. From 1981 – 2017, we conducted surveillance for 16 pathogens and estimated population sizes, adult survival, and lamb survival. We used mixed effects regression models to assess disease patterns at the individual and population levels. Pathogen infection/exposure prevalence varied both spatially and temporally. Our findings indicate that the primary predictor of individual pathogen infection/exposure was the region in which an animal was captured, implying that transmission is driven by local ecological or behavioral factors. Higher Mycoplasma ovipneumoniae seropositivity was associated with lower lamb survival, consistent with lambs having high rates of pneumonia-associated mortality, which may be slowing population recovery. There was no association between M. ovipneumoniae and adult survival. Adult survival was positively associated with population size and parainfluenza-3 virus seroprevalence in the same year, and orf virus seroprevalence in the previous year. Peninsular bighorn sheep are recovering from small population sizes in a habitat of environmental extremes, compounded by infectious disease. Our research can help inform future pathogen surveillance and population monitoring for the long-term conservation of this population.
Keywords: endangered species, epidemic pneumonia, lamb recruitment, Mycoplasma ovipneumoniae, pathogen spillover, Peninsular Ranges, survival, wildlife-livestock interface
Introduction
Pneumonia epidemics are a source of mortality and decreased lamb survival in bighorn sheep (Ovis canadensis) throughout much of their range (Besser et al., 2013; DeForge et al., 1982; Nolen, 2010). Pathogens associated with severe pneumonia are introduced to bighorn sheep herds through contact with domestic sheep (Foreyt & Jessup, 1982) but can be maintained by carrier bighorn sheep for years without continued spillover from domestic animals (Raghavan et al., 2016), causing intermittent epidemics in lambs and suppressing recruitment (Cassirer et al., 2018). Bighorn sheep pneumonia is a disease complex involving co-infection with pathogens, environmental and immune factors, and host behavior (Besser et al., 2013; Wobeser, 2007). Recent research indicates that Mycoplasma ovipneumoniae infection can cause pneumonia by decreasing respiratory immune function and allowing colonization by other pathogens (Besser et al., 2012, 2014; Dassanayake et al., 2010). Numerous management tools including vaccination, population reduction, and supplemental feeding have failed to prevent or control pneumonia outbreaks in bighorn sheep (Cassirer et al., 2001, 2018; Ward et al., 1999) but recent efforts to test and remove chronic M. ovipneumoniae carriers demonstrate promising results, including improved lamb survival (Garwood et al., 2020).
Peninsular bighorn sheep (Ovis canadensis nelsoni) reside in the Peninsular Ranges of southern California and Baja Mexico, and are currently considered a genetically distinct metapopulation of desert bighorn sheep (Buchalski et al., 2016). Peninsular bighorn sheep were listed as federally endangered in 1998 due to a multitude of population threats, including habitat loss and fragmentation, infectious disease, predation, and drought (US Fish and Wildlife Service, 2000). The Peninsular metapopulation has been steadily increasing in size from ~300 at the time of listing to ~900 in 2016; however, infectious disease continues to threaten survival and recruitment (Colby & Botta, 2019).
Bighorn sheep behavior and spatial distribution plays a role in the transmission and maintenance of disease. The Peninsular bighorn sheep metapopulation consists of at least 19 herds that inhabit the desert slopes, alluvial fans, and washes of the Peninsular Ranges (Colby & Botta, 2019). While most individuals within each herd are philopatric, a subset of ewes and rams will disperse to neighboring herds on a seasonal basis (Bighorn Institute, 2018; Buchalski et al., 2015; Colby & Botta, 2019). The Peninsular mountains are divided into 9 “recovery regions” (hereafter, “regions”) defined for bighorn sheep population management (US Fish and Wildlife Service, 2000) (Fig. 1). Historically, these regions were thought to roughly correspond to different herds (Rubin et al., 1998) but some regions now contain multiple overlapping herds and inter-regional movements are regularly observed (Bighorn Institute, 2018; Colby & Botta, 2019).
Figure 1.
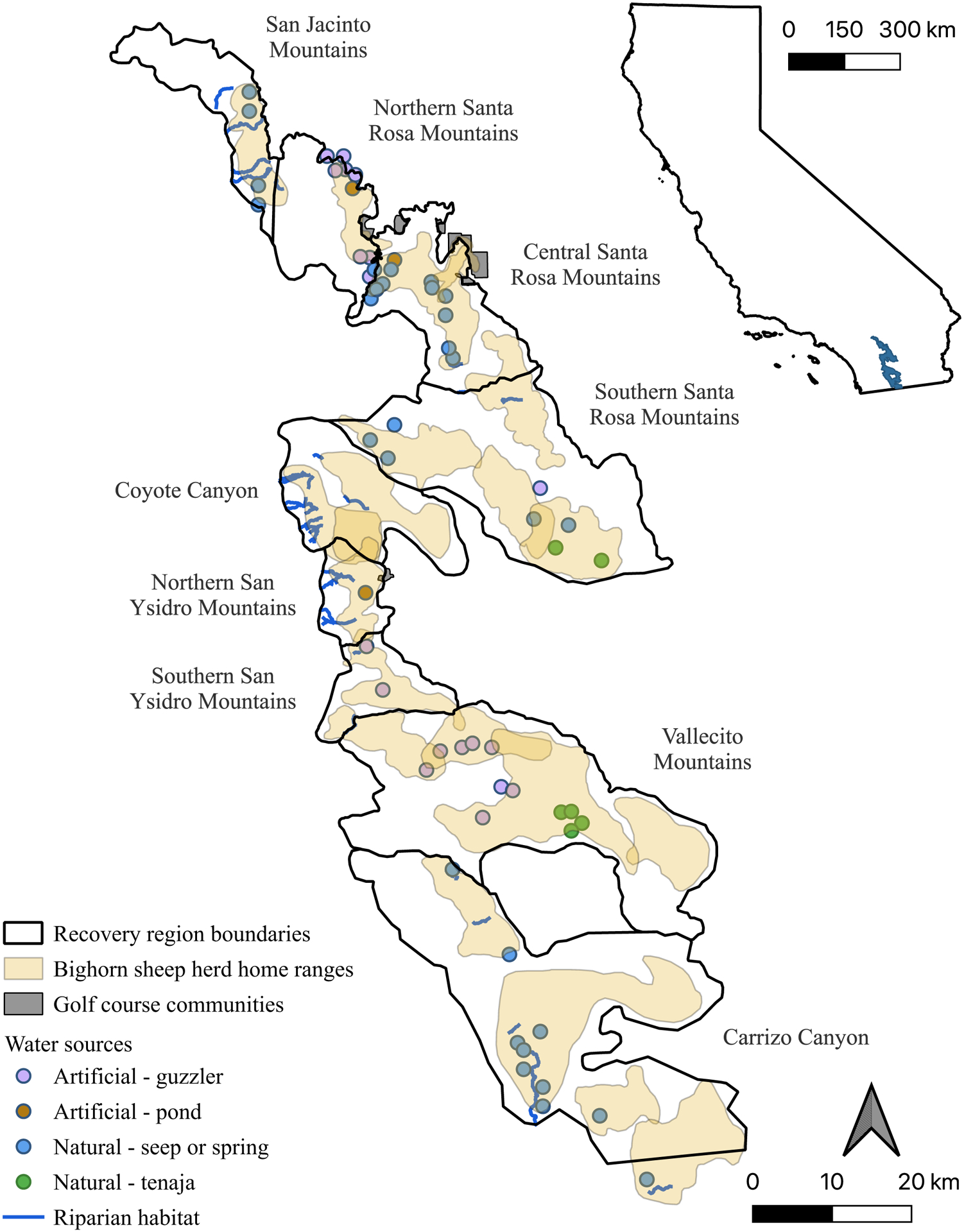
Map of the study area within the Peninsular Ranges of southern California, USA. Map depicts recovery region boundaries, bighorn sheep herd home ranges, golf course communities bordering or within bighorn sheep habitat, and primary water sources. Major riparian areas have perennial or intermittent creeks and relatively large amounts of vegetation, including canopy cover and a dense understory. These areas are also utilized by deer and sometimes mountain lions. Artificial ponds and guzzlers provide year-round water through municipal sources or by collecting rainwater then delivering them to a drinking area. Guzzlers tend to be elevated, while ponds are at ground level and therefore vulnerable to contamination by rain run-off and/or animal excrement. Natural seeps and springs are small point sources of water at ground level that contain variable quantities and quality of water throughout the year. Tenajas are small rock depressions that hold water at the bottom of drainages, tend to be poor water quality, and are not dependable during the summer months. Golf course communities shown are those that bighorn sheep utilize on a regular basis; they are in urban areas where human-wildlife conflict is likely, but also have highly nutritious forage and many dependable water sources such as ponds, creeks, canals, reservoirs, and swimming pools.
Bighorn sheep movements are driven by food and water availability, which are especially scarce in drought years. California has had chronically low rainfall for several decades, including a severe drought from 2012 – 2016 that significantly reduced the surface water available for wildlife in desert ecosystems where bighorn sheep are found (U. S. Geological Survey, 2017). Bighorn sheep congregate in high densities at natural and artificial water sources and urban areas where irrigation and landscaping provide resources (Bighorn Institute, 2018; Colby & Botta, 2019) (Fig. 1). This co-mingling of animals from different herds, age classes, and disease statuses increases the risk of pathogen transmission, and higher density herds are associated with an increased risk of respiratory disease outbreaks (Monello et al., 2001; Sells et al., 2015).
Peninsular bighorn sheep are recovering in a desert ecosystem that is evolving with climate change. The goal of our research is to identify key epidemiologic factors driving disease prevalence in bighorn sheep, with special attention paid to pathogens associated with epidemic pneumonia. We aim to: 1) Estimate pathogen prevalence by age, sex, and region; 2) Identify demographic and geographic predictors of pathogen infection/exposure in individual bighorn sheep; 3) Identify associations between pathogen infection/exposure prevalence and adult and lamb survival while controlling for important environmental variables associated with climate change; 4) Identify communities of pathogens that co-occur together within individual bighorn sheep and may impact fitness.
Methods
Pathogen infection and exposure prevalence
We sampled wild Peninsular bighorn sheep captured from 1981 – 2017. Animals were captured and sampled by the California Department of Fish and Wildlife (CDFW) or contractors following CDFW guidelines. Protocols were reviewed and approved by CDFW, or other land management agencies when appropriate, and objectives and goals have been directed by the Peninsular Bighorn Sheep Recovery Plan since 2000 (US Fish and Wildlife Service, 2000).
We tested blood samples (735 individuals; 844 sampling events) for infection or exposure to up to 15 pathogens, including: Anaplasma spp., bluetongue virus (BTV), bovine herpesvirus-1, bovine respiratory syncytial virus (BRSV), bovine viral diarrhea virus types 1 and 2, Brucella ovis, Chlamydia spp., epizootic hemorrhagic disease virus (EHDV), Leptospira spp., Mycoplasma ovipneumoniae, ovine progressive pneumonia virus, orf virus, parainfluenza-3 virus (PI-3), and Toxoplasma gondii (Table 1). Virus isolation was performed for BT and EHDV, but all other blood tests measured antibodies and more likely indicated previous exposure (Table 1). “Prevalence” hereafter refers to the proportion of positive tests, indicating infection or exposure depending on the test used.
Table 1.
Summary of pathogen infection/exposure prevalence in Peninsular bighorn sheep across the entire study period (1981 – 2017), stratified by sex, age, and recovery region (first capture event for each individual). Fractions represent the number of positive tests over the number of tests performed. Sample sizes within each stratification level may not sum to the same totals as in the “overall” column because age class and sex were not available for all samples. Bolded text indicates significant differences (p ≤ 0.05) among groups within a stratification level (i.e., females vs. males).
| Sex | Age | Recovery Region | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogen | Test type | Antibody (A) or pathogen (P) | Overall | Female | Male | Lamb/yearling | Adult | San Jacinto Mtns | Northern Santa Rosa Mtns | Central Santa Rosa Mtns | Southern Santa Rosa Mtns | Coyote Canyon | Northern San Ysidro Mtns | Southern San Ysidro Mtns | Vallecito Mtns | Carrizo Canyon |
| Anaplasma spp. | CA | A | 49.7% (158/318) | 48.1% (117/243) | 55.4% (41/74) | 34.6% (9/26) | 51.0% (148/290) | 42.9% (12/28) | 29.8% (14/47) | 48.3% (14/29) | 29.6% (8/27) | 57.1% (20/35) | 77.1% (27/35) | 65.9% (29/44) | 48.5% (16/33) | 45.0% (18/40) |
| Bovine herpesvirus-1 | SVN | A | 0.6% (3/537) | 0.2% (1/407) | 1.6% (2/129) | 0.0% (0/66) | 0.6% (3/469) | 0.0% (0/51) | 3.0% (3/100) | 0.0% (0/57) | 0.0% (0/32) | 0.0% (0/45) | 0.0% (0/51) | 0.0% (0/49) | 0.0% (0/45) | 0.0% (0/107) |
| Bovine respiratory syncytial virus | IFA | A | 39.3% (259/659) | 41.9% (222/530) | 28.9% (37/128) | 34.8% (23/66) | 39.9% (236/592) | 12.1% (8/66) | 48.7% (38/78) | 37.0% (30/81) | 72.9% (43/59) | 43.8% (21/48) | 28.1% (18/64) | 44.1% (26/59) | 40.0% (30/75) | 34.9% (45/129) |
| Bovine viral diarrhea virus type-1 | SVN | A | 0.7% (4/541) | 0.7% (3/411) | 0.8% (1/129) | 3.0% (2/67) | 0.4% (2/472) | 0.0% (0/51) | 1.0% (1/101) | 0.0% (0/57) | 0.0% (0/32) | 0.0% (0/45) | 0.0% (0/54) | 0.0% (0/49) | 0.0% (0/45) | 2.8% (3/107) |
| Bovine viral diarrhea virus type-2 | SVN | A | 0.0% (0/83) | 0.0% (0/65) | 0.0% (0/17) | 0.0% (0/7) | 0.0% (0/76) | 0.0% (0/7) | 0.0% (0/4) | NT | NT | 0.0% (0/7) | 0.0% (0/9) | 0.0% (0/12) | 0.0% (0/2) | 0.0% (0/42) |
| Brucella ovis | ELISA | A | 5.0% (23/459) | 5.9% (20/340) | 2.5% (3/118) | 3.4% (2/59) | 5.3% (21/399) | 0.0% (0/38) | 0.0% (0/87) | 0.0% (0/50) | 0.0% (0/26) | 5.3% (2/38) | 16.3% (7/43) | 10.9% (5/46) | 0.0% (0/38) | 9.7% (9/93) |
| Chlamydia spp. | CF | A | 42.8% (199/465) | 41.7% (145/348) | 45.7% (53/116) | 36.5% (19/52) | 43.4% (179/412) | 48.8% (20/41) | 17.0% (9/53) | 71.4% (40/56) | 12.9% (4/31) | 38.1% (16/42) | 30.8% (16/52) | 42.6% (20/47) | 42.2% (19/45) | 56.1% (55/98) |
| Leptospira spp. | MAT | A | 12.1% (38/313) | 12.7% (30/237) | 10.5% (8/76) | 8.1% (3/37) | 12.7% (35/275) | 3.2% (1/31) | 9.1% (6/66) | 5.4% (2/37) | 30.8% (8/26) | 16.0% (4/25) | 10.0% (3/30) | 33.3% (8/24) | 6.5% (2/31) | 9.3% (4/43) |
| Mycoplasma ovipneumoniae | PCR | P | 12.0% (38/316) | 11.9% (33/278) | 13.5% (5/37) | 14.3% (4/28) | 11.8% (34/288) | 13.3% (4/30) | 5.9% (2/34) | 3.1% (1/32) | 15.2% (5/33) | 7.7% (2/26) | 0.0% (0/30) | 20.0% (6/30) | 23.8% (10/42) | 13.6% (8/59) |
| cELISA | A | 60.2% (336/558) | 59.5% (267/449) | 63.0% (68/108) | 58.3% (28/48) | 60.3% (307/509) | 55.6% (30/54) | 71.2% (42/59) | 58.1% (36/62) | 55.8% (29/52) | 48.9% (22/45) | 56.7% (34/60) | 50.0% (25/50) | 71.0% (49/69) | 64.5% (69/107) | |
| Ovine progressive pneumonia virus | AGID | A | 0.0% (0/186) | 0.0% (0/138) | 0.0% (0/48) | 0.0% (0/14) | 0.0% (0/170) | 0.0% (0/26) | 0.0% (0/36) | 0.0% (0/9) | 0.0% (0/12) | 0.0% (0/13) | 0.0% (0/17) | 0.0% (0/21) | 0.0% (0/18) | 0.0% (0/34) |
| Orf virus | CF | A | 71.8% (319/444) | 70.4% (247/351) | 77.2% (71/92) | 57.1% (24/42) | 73.6% (295/401) | 70.3% (26/37) | 62.8% (49/78) | 50.0% (23/46) | 84.2% (32/38) | 69.2% (27/39) | 83.7% (36/43) | 82.2% (37/45) | 63.6% (28/44) | 82.4% (61/74) |
| Parainfluenza-3 virus | VI | P | 11.4% (4/35) | 13.0% (3/23) | 8.3% (1/12) | 23.1% (3/13) | 4.5% (1/22) | 0.0% (0/2) | 20.0% (4/20) | NT | NT | NT | NT | NT | NT | 0.0% (0/13) |
| HI | A | 21.2% (152/717) | 21.4% (123/576) | 20.7% (29/140) | 12.5% (10/80) | 22.2% (141/635) | 1.5% (1/66) | 32.5% (38/117) | 9.9% (8/81) | 30.5% (18/59) | 8.9% (5/56) | 12.3% (8/65) | 19.4% (12/62) | 35.4% (29/82) | 25.6% (33/129) | |
| Toxoplasma gondii | LA | A | 18.0% (16/89) | 9.1% (6/66) | 43.5% (10/23) | 14.3% (1/7) | 18.5% (15/81) | 20.0% (4/20) | 37.0% (10/27) | NT | NT | NT | 5.9% (1/17) | 7.7% (1/13) | NT | 0.0% (0/12) |
| Bluetongue virus | VI | P | 3.3% (3/91) | 4.5% (3/66) | 0.0% (0/25) | 11.1% (2/18) | 1.4% (1/72) | 0.0% (0/16) | 5.0% (2/40) | NT | 0.0% (0/4) | 0.0% (0/5) | NT | NT | 0.0% (0/5) | 4.8% (1/21) |
| cELISA | A | 10.4% (60/576) | 9.6% (45/467) | 13.9% (15/108) | 3.8% (2/52) | 11.1% (58/523) | 8.0% (4/50) | 5.2% (3/58) | 3.7% (3/81) | 8.5% (5/59) | 5.4% (3/56) | 22.9% (11/48) | 12.3% (7/57) | 18.3% (15/82) | 10.6% (9/85) | |
| Epizootic hemorrhagic disease virus | VI | P | 0.0% (0/22) | 0.0% (0/14) | 0.0% (0/8) | 0.0% (0/10) | 0.0% (0/12) | NT | 0.0% (0/18) | NT | NT | NT | NT | NT | NT | 0.0% (0/4) |
| SVN | A | 24.4% (10/41) | 20.0% (6/30) | 36.4% (4/11) | 17.6% (3/17) | 29.2% (7/24) | NT | 18.9% (7/37) | NT | NT | NT | NT | NT | NT | 75.0% (3/4) | |
| Orbivirus spp. | AGP/AGID | A | 21.6% (145/670) | 19.9% (107/538) | 29.0% (38/131) | 15.1% (11/73) | 22.5% (134/595) | 30.6% (19/62) | 31.9% (37/116) | 7.5% (4/53) | 12.1% (7/58) | 9.8% (5/51) | 21.5% (14/65) | 17.7% (11/62) | 20.3% (15/74) | 25.6% (33/129) |
| Mannheimia haemolytica betahemolytic | culture | P | 85.0% (119/140) | 86.5% (90/104) | 80.6% (29/36) | 100.0% (14/14) | 83.3% (105/126) | 70.0% (7/10) | 85.7% (18/21) | 87.0% (20/23) | 100.0% (12/12) | 90.0% (9/10) | 83.3% (15/18) | 82.4% (14/17) | 76.5% (13/17) | 91.7% (11/12) |
| Mannheimia haemolytica nonhemolytic | culture | P | 23.6% (33/140) | 24.0% (25/104) | 22.2% (8/36) | 21.4% (3/14) | 23.8% (30/126) | 0.0% (0/10) | 23.8% (5/21) | 34.8% (8/23) | 25.0% (3/12) | 10.0% (1/10) | 33.3% (6/18) | 17.6% (3/17) | 11.8% (2/17) | 41.7% (5/12) |
| Bibersteinia trehalosi betahemolytic | culture | P | 12.1% (17/140) | 15.4% (16/104) | 2.8% (1/36) | 7.1% (1/14) | 12.7% (16/126) | 0.0% (0/10) | 0.0% (0/21) | 0.0% (0/23) | 0.0% (0/12) | 30.0% (3/10) | 38.9% (7/18) | 0.0% (0/17) | 29.4% (5/17) | 16.7% (2/12) |
| Bibersteinia trehalosi nonhemolytic | culture | P | 77.9% (109/140) | 77.9% (81/104) | 77.8% (28/36) | 71.4% (10/14) | 78.6% (99/126) | 90.0% (9/10) | 90.5% (19/21) | 82.6% (19/23) | 83.3% (10/12) | 50.0% (5/10) | 61.1% (11/18) | 94.1% (16/17) | 64.7% (11/17) | 75.0% (9/12) |
AGP = agar gel precipitin, AGID = agar gel immunodiffusion, CA = card agglutination, CF = complement fixation, ELISA = enzyme-linked immunosorbent assay, cELISA = competitive ELISA, IFA = immunofluorescence assay, HI = hemagglutination inhibition, LA = latex agglutination, MAT = modified agglutination test, PCR = polymerase chain reaction, SVN = serum virus neutralization, VI = virus isolation, NT = not tested.
In some years, nasal/pharyngeal swabs were also collected (316 individuals; 349 sampling events) and tested for combinations of M. ovipneumoniae via polymerase chain reaction (PCR), Pasteurellaceae spp. via culture, and PI-3 via virus isolation (VI) to detect active infection (or very recent exposure).
BTV and EHDV are both orbiviruses and cross-react on agar gel precipitin (AGP) and agar gel immunodiffusion (AGID), so we created an “Orbivirus spp.” group which included animals positive for BTV and/or EHDV via AGP/AGID. We classified animals as exposed to BTV if they tested positive on the more specific competitive enzyme-linked immunosorbent assay (cELISA).
We did not include Leptospira spp. serovars in analyses due to cross-reaction on the modified agglutination test, and an animal was considered positive if any serovar was detected at titers >1:100.
Age, sex, and region were recorded at the time of capture. Age was usually recorded categorically based on dentition and horn growth rings, with lambs and yearlings grouped together and older animals categorized as adults. The dataset was skewed towards adult females (80.3%, n = 590/735) since they were the target population for radio-collaring (Colby & Botta, 2019). Most individuals were only captured once (n = 641).
We summarized counts of animals that tested positive vs. negative for each pathogen, then stratified by age, sex, and region. We tested for differences among 2 groups using Fisher’s exact test and among ≥3 groups using one-way analysis of variance (significance at p ≤ 0.05). These calculations only included samples from first capture events to eliminate re-testing errors and biases due to persistent antibodies. We also calculated overall prevalence of each pathogen for each diagnostic test type, summarized for each recovery region and year, including all capture events. All statistics were performed in R version 4.0.4 (R Core Team, 2021).
Annual adult survival rates (Junet-1 – Mayt) for each region were previously calculated by CDFW and Bighorn Institute using Kaplan Meier estimates from radio-collared bighorn sheep, modified to allow for staggered entry (Bighorn Institute, 2018; Colby & Botta, 2019; Ostermann et al., 2001). Lamb survival is considered to be an excellent demographic predictor of health status in bighorn sheep populations (Cassirer et al., 2013). In the Peninsular Ranges, the majority of pneumonia-induced deaths in lambs occur between 8 and 10 weeks (Colby & Botta, 2013). Lamb survival for each region was evaluated based on the ratio of lambs to ewes (lamb:ewe) estimated from observations made during range-wide helicopter surveys, waterhole counts, or ground observations (Bighorn Institute, 2018; Colby & Botta, 2019). This was used as a proxy of lamb survival to ~3 – 9 months, depending on when surveys were performed. Pregnancy rates in the Peninsular Ranges are consistently high, with 94.3% of radio-collared ewes 2 – 19 years of age giving birth from 2005 – 2022, and twins are rare (CDFW, unpublished data). Therefore, lamb:ewe ratios are primarily a reflection of lamb survival rather than birth rates.
Population-level risk factors associated with adult and lamb survival
We created “population-level models” using Bayesian, multilevel, ordered beta regression to evaluate associations between annual adult survival or lamb:ewe ratios (outcomes), and pathogen prevalence, population size, and meteorologic covariates. We also evaluated bivariate relationships between model covariates, including year, as part of model building with univariable, ordered beta regression models. The unit of analysis was the year-region unit, and the random intercept was region. We selected weakly informative priors for intercept and beta parameters [Normal(0, 5)], and phi parameter [exp(0.1)] (Kubinec, 2020). Population size, meteorologic covariates, year, and lamb survival were min-max scaled as needed to match outcome variables, so values ranged from 0 – 1 but the relative differences between values were maintained. This was done for each variable by subtracting the minimum value from each x, then dividing by the range of the original variables.
We calculated pathogen prevalence for each year-region unit (i.e., “2010 – San Jacinto Mountains”) for which ≥5 samples were tested (all capture events included). Models included individual pathogens and combinations of respiratory pathogens (M. ovipneumoniae, BRSV, and PI-3) as covariates, and we tested prevalence lag times of −1-year to +1-years.
We interpolated missing annual population size estimates for each region by averaging values for yeart−1 and yeart+1, but only where estimates were missing for a single year.
Temperature and precipitation for each region were included to control for meteorologic factors influencing survival. We extracted rasters of daily meteorologic data (4×4 km resolution) from “gridMET” (Abatzoglou, 2013) using the “climateR” package, then cropped by the geographic extent of each recovery region and aggregated temporally as described below, resulting in a single summary value for each year-region unit.
We calculated temperature as the average daily maximum temperature (Celsius) from June – September for yeart, which have historically been the hottest months in the Peninsular Ranges (Rubin et al., 2000; Turner et al., 2004).
Precipitation in the Peninsular Ranges is bimodal, with the largest volume and most consistent rains occurring November – February, and more variable monsoons occurring July – September (Rubin et al., 2000). We calculated annual precipitation as the sum of daily precipitation (centimeters) from November of yeart−1 through October of yeart. We also aggregated precipitation annually for winter (Novembert−1 – Aprilt) and summer (Mayt – Octobert). Winter corresponded to the winter rains and bighorn sheep gestational and peak birthing period, while summer corresponded to summer monsoons, post-lambing, and the rut (Colby & Botta, 2017; Rubin, Boyce, Stermer, et al., 2002; Rubin et al., 2000).
Individual-level risk factors associated with pathogen exposure or infection
We created “individual-level models” using Bayesian, multilevel, logistic regression to evaluate risk factors for an animal being infected or exposed to a pathogen. Predictor variables included age class (lamb/yearling, adult), sex (female, male), and recovery region (categorical, n = 9). Reference groups were adults, males, and the San Jacinto Mountains. The unit of analysis was the individual animal, and the random intercept was animal ID (all capture events included). We selected weakly informative priors [Normal(0, 2.5)] for intercept and beta parameters to account for complete or quasi-separation of data (Ghosh et al., 2018).
All regression models were built in package “brms” (Bürkner, 2017, 2018). Models contained 4 chains with 10,000 iterations each. We calculated point estimates as the median value of the posterior and used the 95% highest density interval as the credible interval (CI). We only included models with ≥5 observations per covariate in results. We compared models with the same number of observations using leave-one-out cross-validation information criterion (LOO IC) in the “loo” package (Vehtari et al., 2020). Appendix S1 contains heatmaps illustrating model variables by year and region.
Pathogen co-occurrence network
We looked for “communities” of pathogens to which individual bighorn sheep were co-infected/co-exposed. Concurrent or subsequent infections can take a toll on host immune function and overall fitness, as has been demonstrated by the polymicrobial nature of pneumonia in bighorn sheep (Asghar et al., 2015; Besser et al., 2008; Jamieson et al., 2013). Clusters of pathogens that regularly co-occur together could be associated with clinical phenotypes and direct future disease surveillance.
Since the diagnostic tests used in this study can indicate infection or previous exposure, we use “co-occurrence” to mean that an animal was infected with a pair of pathogens during its life, but perhaps not concurrently. We generated a weighted, undirected network from the proportion of samples (from first capture events only) that were positive for 2 pathogens, given that they were tested for both pathogens. Density and betweenness centrality were used to describe how tightly pathogens clustered (package “sna”) (Butts, 2008). We calculated network modularity using the fast greedy modularity optimization algorithm (package “igraph”) (Clauset et al., 2004) and visualized the network in Gephi (Bastian et al., 2009).
Results
Pathogen infection and exposure prevalence
A total of 735 first-capture samples were collected from 1981 – 2017. Pathogen and antibody prevalence estimates are in Table 1, including stratifications by age, sex, and region. Not all diagnostic tests were performed in every year or region.
The pathogens with the highest seroprevalence were orf virus (71.8%), M. ovipneumoniae (cELISA; 60.2%), Anaplasma spp. (49.7%), Chlamydia spp. (42.8%), BRSV (39.3%), EHDV (serum virus neutralization [SVN]; 24.4%), Orbivirus spp. (21.6%), PI-3 (hemagglutination inhibition [HI]; 21.2%), and T. gondii (18.0%).
The most common active infection was Pasteurellaceae spp., with all samples tested culturing at least one species. Mannheimia haemolytica betahemolytic and Bibersteinia trehalosi nonhemolytic were the most common Pasteurellaceae spp. (85.0% and 77.9%, respectively). Pasteurella multocida was not detected from first capture events, but was cultured in a single sample from a recaptured animal. Prevalence of active infection was much lower for M. ovipneumoniae (PCR; 12.0%) and PI-3 (VI; 11.4%). All other pathogens were relatively uncommon or absent (Table 1).
Leptospira was found in 12.1% of animals, across six serovars (although possible cross-reaction makes these diagnoses unreliable): Leptospira interrogans serovars bratislava (19.2%, n = 10/52), pomona (1.6%, n = 5/316), canicola (1.3 %, n = 4/316), and icterohaemorrhagiae (0.3%, n = 1/316); Leptospira kirschneri serovar grippotyphosa (5.7%, n = 18/316); Leptospira borgpetersenii serovar hardjo (1.6%, n = 5/310).
Females had higher rates of exposure to BRSV (p = 0.01), while males had higher exposure to T. gondii (p < 0.001) and Orbivirus spp. (p = 0.03). Lambs/yearlings tested positive for exposure to orf virus more often than adults (p = 0.03). There were differences among regions in the infection/exposure of Anaplasma spp. (p < 0.001), BRSV (p < 0.001), B. ovis (p < 0.001), Chlamydia spp. (p < 0.001), Leptospira spp. (p = 0.001), M. ovipneumoniae (PCR; p = 0.04), orf virus (p < 0.001), PI-3 (HI; p < 0.001), T. gondii (p = 0.02), BTV (cELISA; p = 0.01), EHDV (SVN; p = 0.01), Orbivirus spp. (p = 0.001), and B. trehalosi beta-hemolytic (p < 0.001).
All pathogens showed temporal changes in prevalence over the 36-year study period (Appendix S1). Some common pathogens (i.e., M. ovipneumoniae [cELISA] and Chlamydia spp.), were consistently present across the entire study period and all regions. BRSV, PI-3, and orf virus were more variable, sometimes ranging from 0% to 100% in the span of one year. B. ovis was only detected in the 1990s. M. ovipneumoniae (PCR) and BRSV showed increasing prevalence over time.
Population performance and meteorologic variables
Population size was positively associated (ß = 2.9, CI = 1.7 – 4.1) with adult survival (Appendix S2). There was no relationship between population size and lamb survival (ß = 0.5, CI = −0.4 – 1.4). Temperature and precipitation were not independently associated with adult or lamb survival (Appendix S2). Despite this lack of association, meteorologic covariates were included in population-level regression model building to test if they improved model fit because they have been previously established as important factors in bighorn sheep survival.
Population size (ß = 2.74, CI = 2.32 – 3.16) and temperature (ß = 0.52, CI = 0.35 – 0.69) were positively associated with year (Appendix S2). Annual (ß = −0.50, CI = −0.74 – −0.26), summer (ß = −0.67, CI = −0.98 – −0.37), and winter (ß = −0.50, CI = −0.76 – −0.24) precipitation were negatively associated with year (Appendix S2).
Pathogen prevalence and lamb survival
The following pathogens had enough datapoints to be included as regression model covariates to evaluate the impact of pathogens on adult and lamb survival: Anaplasma spp., BTV (cELISA), BRSV, Chlamydia spp., orf virus, Leptospira spp., M. ovipneumoniae (PCR, cELISA), Orbivirus spp., and PI-3 (HI).
M. ovipneumoniae exposure prevalence (cELISA) was negatively associated with current year lamb survival (no lag; ß = −1.29, CI = −2.48 – −0.07; Appendix S3). This relationship persisted with the addition of population size (ß = −1.56, CI = −2.76 – −0.39; Appendix S4), population size and temperature (ß = −1.45, CI = −2.66 – −0.29; Appendix S5), population size and annual precipitation (ß = −1.62, CI = -−2.84 – −0.38; Appendix S6), population size and summer precipitation (ß = −1.56, CI = −2.76 – −0.37; Appendix S7), and population size and winter precipitation (ß = −1.66, CI = −2.92 – −0.42; Appendix S8). In these models, population size and meteorologic covariates were not associated with lamb survival. M. ovipneumoniae exposure prevalence was also negatively associated with current year lamb survival (ß = −1.47, CI = −2.96 – −0.04) in the model including BRSV, although BRSV was not a significant predictor (Appendix S3).
Lamb survival was not associated with other pathogens, population size, temperature, or precipitation (Appendix S3–8). The inclusion of population size improved model fit in just 0.74% (n = 1/136) of models. LOO IC standard errors overlapped for all other models.
Pathogen prevalence and adult survival
Orf virus prevalence was positively associated with adult survival in the subsequent year (−1-year lag; ß = 1.6, CI = 0.4 – 2.9; Appendix S3). This relationship persisted with the addition of population size (ß = 1.2, CI = 0.0 – 2.5; Appendix S4), and population size with annual precipitation (ß = 1.2, CI = 0.0 – 2.4; Appendix S6).
PI-3 was associated with increases in current year survival once other covariates were accounted for: population size (ß = 1.8, CI = 0.2 – 4.1; Appendix S4), population size and temperature (ß = 1.8, CI = 0.3 – 3.9; Appendix S5), population size and annual precipitation (ß = 1.8, CI = 0.2 – 4.0; Appendix S6), population size and summer precipitation (ß = 1.5, CI = 0.1 – 3.7; Appendix S7), and population size and winter precipitation (ß = 1.8, CI = 1.2 – 4.1; Appendix S8). In these models, population size was positively associated with adult survival rates, but meteorologic covariates were not.
No other pathogens were predictors of adult survival, regardless of lag time or covariates (Appendix S3–8). Population size was associated with higher adult survival rates in 32.4% (n = 46/142) of models across all pathogens (Appendix S4–8). Higher summer temperatures were associated with lower adult survival rates in 7.1% (n = 2/28) of models (Appendix S5), and higher summer precipitation was associated with higher adult survival rates in 25.0% (n = 7/28) models (Appendix S7). Annual and winter precipitation were not significantly associated with adult survival in any models (Appendices S6, S8). Including population size and summer temperature improved model fit in 28.6% (n = 8/28) of models. LOO IC standard errors overlapped for all other models. The inconsistency in the significance of population size and meteorologic variables was likely because each model contained a different dataset; samples were not tested for every pathogen and adult survival rates were not available for every year-region unit.
Individual-level risk factors associated with pathogen exposure or infection
The following pathogens had enough datapoints to be included as regression model covariates to evaluate infection/exposure risk factors at the individual level: Anaplasma spp., BTV (cELISA), BRSV, B. ovis, Chlamydia spp., Leptospira spp., M. ovipneumoniae (PCR and cELISA), Orbivirus spp., orf virus, PI-3 (HI), and T. gondii (Fig. 2, Appendix S9).
Figure 2.
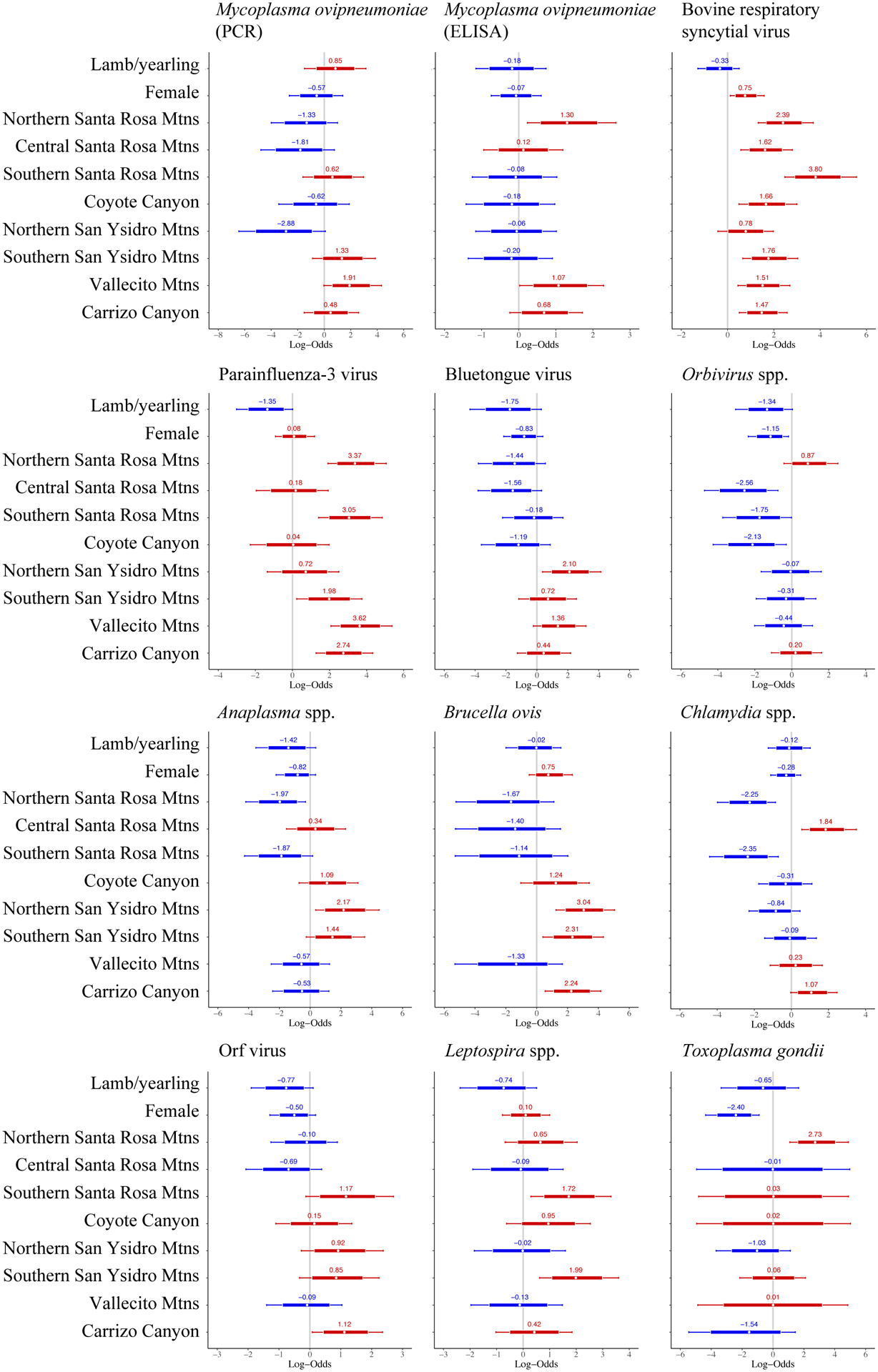
Forest plots demonstrating the relationship between pathogen status (positive, negative) and Peninsular bighorn sheep age class (lamb/yearling, adult), sex (female, male), and recovery region (categorical, n = 9). Reference categories were adults (for age), males (for sex), and the San Jacinto Mountains (for recovery region). Numbers and white circle represent the log odds of testing positive for a pathogen, relative to a reference category, using Bayesian, multilevel, logistic regression models. Log odds <0 (blue) indicate a lower risk of testing positive for a given pathogen, and log odds >0 (red) indicate a higher risk of testing positive for a given pathogen, relative to the reference category. Thick bars represent the 80% credible interval, and thin bars represented the 95% credible interval. A covariate was a significant predictor of pathogen status if the 95% credible interval did not cross 0.
Odds of exposure to M. ovipneumoniae (cELISA) were higher in the northern Santa Rosa Mountains and Vallecito Mountains (OR = 3.7 and 2.9, respectively), compared to the San Jacinto Mountains. Age, sex, and region were not significant predictors of active infection with M. ovipneumoniae (PCR).
BRSV and PI-3 had similar distributions, with most regions having higher odds of exposure compared to the San Jacinto Mountains (OR = 4.4 – 44.7; Fig. 2). Females were more likely to be exposed to BRSV than males (OR = 2.1).
Orbivirus spp. had lower odds of exposure in females (OR = 0.3) and in the northern half of the range (Fig. 2). The northern San Ysidro Mountains, in the middle of the range, had higher odds BTV exposure (OR = 8.2). The discrepancies in risk between BTV and Orbivirus spp. are likely due to Orbivirus spp. models including animals exposed to EHDV and/or BTV, and differences in the spatial/temporal testing for each pathogen (Table 1, Appendix S1).
B. ovis exposure was greater in the southern half of the range, including the northern (OR = 21.0) and southern San Ysidro Mountains (OR = 10.1), and Carrizo Canyon (OR = 9.4). Positive samples were limited to 1990 – 1997, with 17 of 24 positive results occurring in 1992 in the northern San Ysidro Mountains and Carrizo Canyon. Carrizo Canyon also had higher odds of exposure to orf virus (OR = 3.1).
Anaplasma spp. and Chlamydia spp. had patchy geographic distributions. The risk of exposure to Anaplasma spp. was lower in the northern Santa Rosa Mountains (OR = 0.1) and higher in the northern San Ysidro Mountains (OR 8.7). Odds of exposure to Chlamydia spp. were higher in the central Santa Rosa Mountains (OR = 6.3) but lower in the bordering northern and southern Santa Rosa Mountains (OR = 0.1 in both).
Exposure to Leptospira spp. was also scattered, with higher exposure odds in the southern Santa Rosa Mountains (OR = 5.6) and southern San Ysidro Mountains (OR = 7.3).
Odds of exposure to T. gondii was lower for females (OR = 0.1) and higher for animals in the northern Santa Rosa Mountains (OR = 15.3).
Pathogen co-occurrence network
All pathogens were connected to several other pathogens in the network (mean = 13, median = 14, range = 7 – 15 out of 15 possible connections) and there was minimal clustering, as evidenced by low modularity (0.07), low betweenness centrality (0.01), and high density (0.86) of the network (Fig. 3, Appendix S10).
Figure 3.
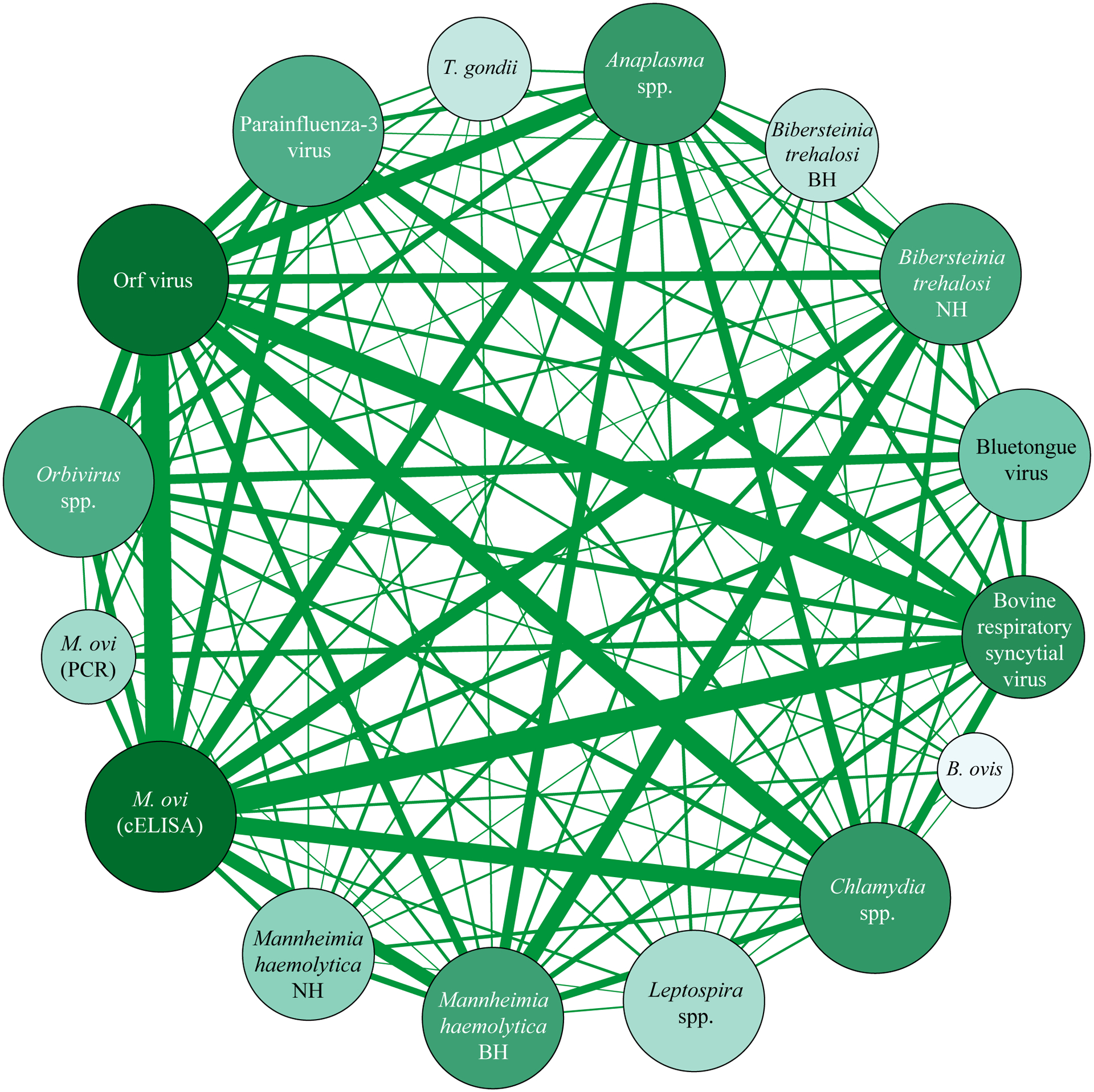
A weighted, undirected network of pathogens that co-occurred together within an individual bighorn sheep. Size of nodes (circles) is relative to the number of other pathogens that node is connected too (larger nodes are linked to more pathogens; range 7 – 15). Width of edges (lines) between nodes is relative to the proportion of bighorn sheep samples which were positive for a pair of pathogens, given that both pathogens were tested for. The color shade of the nodes corresponds to the weighted degree, calculated as the sum of the edges leading into a node, so that darker nodes are linked to a larger number of other nodes and also tested positive in a larger proportion of samples. B. ovis = Brucella ovis, M. ovi = Mycoplasma ovipneumoniae, T. gondii = Toxoplasma gondii, cELISA = competitive enzyme-linked immunosorbent assay, PCR = polymerase chain reaction, BH = beta-hemolytic, NH = non-hemolytic.
Discussion
Pathogen prevalence and bighorn sheep survival
Bighorn sheep infected with M. ovipneumoniae can die, clear the infection, or become carriers that persistently or intermittently shed bacteria (Cassirer et al., 2013). After the initial epidemic, adult M. ovipneumoniae PCR prevalence tends to be low because most animals stop shedding after <1 year (Plowright et al., 2017). Conversely, M. ovipneumoniae seropositivity (cELISA) is a measure of past disease exposure and could be indicative of the amount of transmission occurring during the spring lambing season in that year. Lamb survival in this study was lower in years with higher M. ovipneumoniae exposure prevalence (cELISA). The highest prevalence and odds of M. ovipneumoniae exposure was in the northern Santa Rosa Mountains, which also consistently had the lowest lamb survival across the study period (Table 1, Appendix S1).
Lambs in the Peninsular Ranges have been observed with clinical signs consistent with pneumonia in all regions, and almost all of the uncollared bighorn mortalities attributed to disease from 2002 – 2019 have been the result of bacterial pneumonia in lambs (Colby & Botta, 2019). Most of these mortalities have been detected in the central Santa Rosa Mountains and northern San Ysidro Mountains, where proximity to urban spaces and areas of heavy recreational use by humans results in decreased predation and increased detection of sick lambs, even though these regions do not consistently have the highest prevalence of respiratory pathogens (Table 1; Appendix S1).
Increased lamb mortality is associated with the presence of even a few ewes shedding M. ovipneumoniae and epidemics of pneumonia affect lamb survival and recruitment to a greater degree than adult survival (Cassirer et al., 2013; Manlove et al., 2014; Monello et al., 2001; Plowright et al., 2013). The extinction risk of Peninsular bighorn sheep is inversely related to adult female survival (Rubin, Boyce, & Caswell-Chen, 2002), which was not found to be associated with M. ovipneumoniae infection/exposure in this study. However, by reducing lamb recruitment, M. ovipneumoniae could both slow population recovery and potentially increase extinction risk by leading to a reduction in adult survival in an aging population.
Poor lamb survival in some regions may be improved by conducting “test and removal” of M. ovipneumoniae carrier females, as has been evidenced through other recent studies (Garwood et al., 2020). The relatively linear north-south orientation of these populations may assist in the implementation of this method. However, “test and removal” efforts are costly, requiring the capture and testing of nearly all adult females within a herd, and herds where chronic shedders have been removed are still susceptible to reintroduction of M. ovipneumoniae, which could result in epidemics and all age die-offs.
We did not find evidence that orf virus was associated with decreased survival, but it is extremely common and could play a role in individual fitness. Contagious ecthyma (the disease caused by orf virus) is generally self-limiting and resolves within a few months, but can lead to secondary infections and mortality in young animals or those with co-morbidities (Colby & Botta, 2018; Jones et al., 2018; Michelsen & Smith, 2009). Infectious carrier states and reinfections are observed in domestic sheep (Lewis, 1996; Nandi et al., 2011), suggesting the virus may not fadeout from herd immunity.
Antibody prevalence of PI-3 was associated with higher adult survival, potentially indicating that age may be associated with cumulative exposure risk. PI-3 generally causes subclinical to mild respiratory signs as a sole agent in domestic sheep, but can predispose animals to fatal secondary bacterial pneumonia, especially from Pasteurellaceae spp. (Woolums et al., 2009).
We found a positive relationship between population size and adult survival but could not establish directionality. Larger population sizes may be the result of improving survival rates as the population recovers or there may be a survival benefit to larger groups, such as vigilance against predators.
Environmental impacts on survival
We found trends towards higher adult survival rates with lower summer temperatures and higher summer rainfall, once pathogen prevalence and population size were accounted for. Unfortunately, work evaluating a subset of the Peninsular Mountains (overlapping with regions 3 – 9) over the same time period as this study (1984 – 2017) determined that increases in summer temperatures and decreases in precipitation (Octobert−1 to Septembert) were associated with widespread declines in perennial vegetation cover, with a stronger magnitude of effect at the lower elevations (<500 m) preferred by bighorn sheep (Hantson et al., 2021). The increasing temperatures and decreasing precipitation we observed over the course of this study are expected to worsen in the southwestern desert regions as climate change continues (Hess et al., 2008), potentially affecting bighorn sheep through resource limitation and subsequent behavioral adaptations that may alter disease transmission.
While drought and increasing temperatures may drive sheep to aggregated at limited water sources, it may also lead to lower densities and contact rates if sheep are driven to disperse in search of increasingly sparse vegetation (Epps et al., 2004). This has been anecdotally observed recently, with declines in the density of spring vegetation and smaller bighorn sheep nursery groups occurring in the same years as fewer observations of lambs with clinical respiratory disease and lower M. ovipneumoniae PCR prevalence (CDFW, unpublished data).
Geography is a greater risk factor for pathogen infection/exposure than demographics
The primary risk factor for individual bighorn sheep pathogen infection/exposure was region, which is likely a proxy for local ecological and behavioral factors. The type and number of water sources in a region could influence rates of direct contact between sheep, and contamination of food and/or water with feces and urine could spread pathogens such as T. gondii and Leptospira spp. (Adler & de la Peña Moctezuma, 2010; Dubey, 2009). Behavioral observations and genetic data has demonstrated a strong matrilineal structure between bighorn sheep herds (Boyce et al., 1999), which could lead to greater disease transmission within herds compared to between herds.
Age was not a significant predictor of an animal’s pathogen status, perhaps because lambs and yearlings were categorized together and sampling generally happened in the fall, after the critical window when most pneumonia-associated lamb mortalities occur (Cassirer et al., 2018). Similarly, the relatively low numbers of both lambs/yearlings (11.0%, n = 81/735) and males (19.6%, n = 144/735) in the dataset may have decreased our power to detect differences among groups.
Potential impacts of co-infections and multiple pathogen strains
Bighorn sheep epidemic pneumonia is a complex disease process involving multiple pathogens and non-infectious stressors (Besser et al., 2013). We found that respiratory pathogens were relatively common in Peninsular bighorn sheep across their range, especially M. ovipneumoniae and BRSV. Although we found limited evidence for negative population-level effects of pathogens other than M. ovipneumoniae, the long-term circulation of multiple pathogens may have a subclinical effect or exacerbate concurrent, non-disease stressors. The co-occurrence network showed that all pathogens co-occurred with numerous other pathogens, and there were no communities that clustered together which could inform future targeted surveillance. This network may have had biases towards detecting highly prevalent pathogens with higher survival rates, long-lasting antibodies, and consistent testing. However, the potential synergism of co-infections has implications for individual fitness and long-term population resiliency. For example, lambs that are not feeding well due to painful orf sores around their mouth may be more likely to succumb to pneumonia.
Bighorn sheep do not appear to gain protective cross-immunity against different strains of M. ovipneumoniae after infection (Cassirer et al., 2017), and different M. ovipneumoniae strains have been associated with varying levels of morbidity/mortality (Besser et al., 2017). To date, 23 M. ovipneumoniae samples from Peninsular bighorn sheep have been genotyped using multi-locus sequence typing, and 2 distinct ovine strains have been identified, possibly representing distinct spillover events. One strain, most closely related to bighorn sheep samples from the nearby Orocopia Mountains, is found throughout all recovery regions (Cassirer et al., 2018). A second strain, most similar to samples from Joshua Tree National Park, was identified in 2020 from sheep in 2 northern regions (San Jacinto Mountains, central Santa Rosa Mountains; CDFW, unpublished data). Continued monitoring and strain typing will be important to detect the introduction and spread of novel strains which could lead to new outbreaks of pneumonia and all age class mortality.
Limitations and opportunities for future surveillance
The lack of associations in this study between most pathogens and bighorn sheep survival may be due to data limitations resulting from the shifting priorities and capabilities of this multi-decade recovery project. Most diagnostic tests measured previous exposure and we do not know the duration of seropositivity for many of these diseases. Our results might differ if we measured clinical disease, active infections, or directly observed lamb survival within the first few months of life. Not measuring lamb survival to a consistent age across years and regions may have masked age-related differences in survival. Using recovery region as the unit of analysis may have masked herd-level differences. More importantly, disease-induced mortality is a multifactorial process that includes variables not included in our models, such as immune function, pathogen virulence, and dynamic behaviors such as contact rates among hosts.
As with many wildlife studies, Peninsular bighorn sheep monitoring suffers from limited funding, staff, and access to rugged and remote habitats. While regular, frequent, and comprehensive molecular surveillance combined with observations of clinical disease is ideal for understanding detailed disease dynamics, it is not always attainable or sustainable. Long-term projects such as this one often need to spread out resources, electing for a lower intensity monitoring plan that can be sustained over many years to detect population-level changes. So, the question becomes, what is the optimal monitoring strategy to detect disease and assess overall population health in a low-density, highly mobile species occurring in remote and difficult to access habitats?
Pairing diagnostic tests that distinguish active from previous infection provides the most information on which diseases could be causing current mortalities vs. explain historical population performance. Diseases prioritized for testing should be those that have the greatest potential to negatively impact survival and recruitment, such as the pneumonia-associated pathogens: M. ovipneumoniae, BRSV, and PI-3 (WAFWA Wildlife Health Committee, 2016). Although Pasteurellaceae spp. have long been implicated as causal organisms in pneumonia, the strength of association is weak and they are common commensal organisms in healthy sheep, so the utility of regular testing is limited (Besser et al., 2013). Highly prevalent pathogens may also be of interest, even if there is no current evidence that they pose a threat to the population. Orf virus is not currently associated with reductions in survival and skin lesions can be detected visually, but it is very common in Peninsular bighorn sheep and active disease may be missed due to the short duration of clinical signs. Continuing to monitor seroprevalence will help us understand how many animals are suffering morbidity or mortality as the population grows.
Throughout most of the Peninsular Ranges, it is not possible to directly observe bighorn sheep on a regular basis due to remote and rugged terrain combined with extreme weather conditions. Continuing to collar a subset of animals for survival monitoring and quick carcass recovery should be a mainstay of bighorn sheep monitoring, especially in areas where directly observing animals is difficult. Collaring young lambs would provide better estimates of lamb survival, an important metric of population health that varies considerably among regions and years, and improve our ability to determine causes of death among juveniles. The large area and hot, dry weather of the Peninsular Ranges make it difficult to retrieve carcasses quickly enough to reliably determine the cause of death, even when animals are radio-collared, limiting our ability to detect disease outbreaks or other population threats. Future monitoring may focus on alternative methods, although they carry detection biases compared to radio-collaring.
As an alternative, remote cameras placed in areas where animals reliably congregate could be used to detect visible signs of morbidity, such as nasal/ocular discharge, postural changes, muscle wasting, lameness, and skin lesions (Brewster et al., 2017; Brown & Elmer, 2019; Carricondo-Sanchez et al., 2017; Muneza et al., 2019). Advancements in machine learning could aid in image processing and automated detection of species, posture, and potentially even disease lesions (Tuia et al., 2022), “Physiologgers,” implantable transmitters that record physiologic variables such as heart rate, body temperature, and respiratory rhythms, are a rapidly developing technology that could one day be utilized to monitor bighorn sheep for signs of stress and sickness that may not be detected by visual observations alone (Hawkes et al., 2021). If these methods detected signs of morbidity or a decrease in apparent bighorn abundance, a more detailed investigation could be triggered. Combining these tools with survival and movement data from radio-collared animals would provide more comprehensive insights into emerging health threats, bighorn sheep responses to environmental changes, and the effectiveness of management efforts.
Conclusions
Peninsular bighorn sheep are recovering from critically small population sizes in an ecosystem which includes natural and urban habitats at environmental extremes. This study demonstrates that M. ovipneumoniae is associated with lower lamb survival and identified regions with elevated risk of pathogen infection/exposure to guide future surveillance. Changes in bighorn sheep behavior and distribution in response to climate change and anthropogenic development may play a role in the maintenance or amplification of disease, especially where bighorn sheep congregate, such as around limited water sources and on lambing grounds. Long-term, consistent, range-wide pathogen testing and population surveys will be critical to advance our understanding of pathogen transmission and the role of disease in Peninsular bighorn sheep population recovery. Consideration of environmental factors and incorporation of novel technologies will also be important to adjust management strategies in the face of climate change and dynamic disease risks.
Supplementary Material
Acknowledgements
Numerous colleagues provided advice, expertise, and support throughout this study: Walter M. Boyce, David A. Jessup, Danielle J. Harvey, Daniel J. Tancredi, Gabriele Maier, E. Robert Atwill, Woutrina A. Smith, Kevin Keel, Beatriz Martínez López, Peregrine L. Wolff, Clinton W. Epps, T. Winston Vickers, Ashley E. Hill, Gabriel A. Reyes, Sarah T. Abusaa, Laura H. Backus, Peter J. Sebastian, Christine T. Chang, Joe Smith, and Shane M. Sanchez. This work was supported by the United States National Institutes of Health (grant number T32 OD 011147).
Footnotes
Conflict of Interest Statement
The authors declare they have no conflicts of interest.
Data Accessibility Statement
All data used in this manuscript are presented in Appendices S11–13.
Literature Cited
- Abatzoglou JT (2013). Development of gridded surface meteorological data for ecological applications and modelling. International Journal of Climatology, 33(1), 121–131. 10.1002/joc.3413 [DOI] [Google Scholar]
- Adler B, & de la Peña Moctezuma A (2010). Leptospira and leptospirosis. Veterinary Microbiology, 140(3), 287–296. 10.1016/j.vetmic.2009.03.012 [DOI] [PubMed] [Google Scholar]
- Asghar M, Hasselquist D, Hansson B, Zehtindjiev P, Westerdahl H, & Bensch S (2015). Hidden costs of infection: Chronic malaria accelerates telomere degradation and senescence in wild birds. Science, 347(6220), 436–438. 10.1126/science.1261121 [DOI] [PubMed] [Google Scholar]
- Bastian M, Heymann S, & Jacomy M (2009). Gephi: An open source software for exploring and manipulating networks. 3(1). [Google Scholar]
- Besser TE, Cassirer EF, Highland MA, Wolff P, Justice-Allen A, Mansfield K, Davis MA, & Foreyt W (2013). Bighorn sheep pneumonia: Sorting out the cause of a polymicrobial disease. Preventive Veterinary Medicine, 109(3–4), 185–185. 10.1016/j.prevetmed.2013.02.020 [DOI] [PubMed] [Google Scholar]
- Besser TE, Cassirer EF, Potter KA, & Foreyt WJ (2017). Exposure of bighorn sheep to domestic goats colonized with Mycoplasma ovipneumoniae induces sub-lethal pneumonia. PLoS ONE, 12(6), e0178707. 10.1371/journal.pone.0178707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser TE, Cassirer EF, Potter KA, Lahmers K, Oaks JL, Shanthalingam S, Srikumaran S, & Foreyt WJ (2014). Epizootic pneumonia of bighorn sheep following experimental exposure to Mycoplasma ovipneumoniae. PLoS ONE, 9(10). 10.1371/journal.pone.0110039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser TE, Cassirer EF, Potter KA, VanderSchalie J, Fischer A, Knowles DP, Herndon DR, Rurangirwa FR, Weiser GC, & Srikumaran S (2008). Association of Mycoplasma ovipneumoniae infection with population-limiting respiratory disease in free-ranging Rocky Mountain bighorn sheep (Ovis canadensis canadensis). Journal of Clinical Microbiology, 46(2), 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser TE, Highland MA, Baker K, Cassirer EF, Anderson NJ, Ramsey JM, Mansfield K, Bruning DL, Wolff P, Smith JB, & Jenks JA (2012). Causes of pneumonia epizootics among bighorn sheep, western United States, 2008–2010. Emerging Infectious Diseases, 18(3), 406–414. 10.3201/eid1803.111554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bighorn Institute. (2018). Bighorn Institute 2018 year-end report: Investigations of Peninsular bighorn sheep in the Santa Rosa Mountains and San Jacito Mountains of California. (p. 15). Bighorn Institute. [Google Scholar]
- Boyce WM, Ramey R, Rodwell T, Rubin E, & Singer R (1999). Population subdivision among desert bighorn sheep (Ovis canadensis) ewes revealed by mitochondrial DNA analysis. Molecular Ecology, 8(1), 99–106. 10.1046/j.1365-294X.1999.00536.x [DOI] [PubMed] [Google Scholar]
- Brewster K, Henke SE, Hilton C, & Ortega-S A Jr (2017). Use of remote cameras to monitor the potential prevalence of sarcoptic mange in southern Texas, USA. Journal of Wildlife Diseases, 53(2), 377–381. 10.7589/2016-08-180 [DOI] [PubMed] [Google Scholar]
- Brown WE, & Elmer J (2019). Remote detection and monitoring methods for Tasmanian Devils. In Hogg CJ, Fox S, Pemberton D, & Belov K (Eds.), Saving the Tasmanian devil: Recovery through science-based management (pp. 139–155). CSIRO Publishing. 10.1071/9781486307197 [DOI] [Google Scholar]
- Buchalski MR, Navarro AY, Boyce WM, Winston Vickers T, Tobler MW, Nordstrom LA, García JA, Gille DA, Penedo MCT, Ryder OA, & Ernest HB (2015). Genetic population structure of Peninsular bighorn sheep (Ovis canadensis nelsoni) indicates substantial gene flow across US–Mexico border. Biological Conservation, 184, 218–228. 10.1016/j.biocon.2015.01.006 [DOI] [Google Scholar]
- Buchalski MR, Sacks BN, Gille DA, Penedo MCT, Ernest HB, Morrison SA, & Boyce WM (2016). Phylogeographic and population genetic structure of bighorn sheep (Ovis canadensis) in North American deserts. Journal of Mammalogy, 97(3), 823–838. 10.1093/jmammal/gyw011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkner P-C (2017). brms: An R package for Bayesian multilevel models using Stan. Journal of Statistical Software, 80(1), 1–28. 10.18637/jss.v080.i01 [DOI] [Google Scholar]
- Bürkner P-C (2018). Advanced Bayesian multilevel modeling with the R package brms. The R Journal, 10(1), 395–411. [Google Scholar]
- Butts CT (2008). Social network analysis with sna. Journal of Statistical Software, 24(6), 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carricondo-Sanchez D, Odden M, Linnell JDC, & Odden J (2017). The range of the mange: Spatiotemporal patterns of sarcoptic mange in red foxes (Vulpes vulpes) as revealed by camera trapping. PLOS ONE, 12(4), e0176200. 10.1371/journal.pone.0176200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassirer EF, Manlove KR, Almberg ES, Kamath PL, Cox M, Wolff P, Roug A, Shannon J, Robinson R, Harris RB, Gonzales BJ, Plowright RK, Hudson PJ, Cross PC, Dobson A, & Besser TE (2018). Pneumonia in bighorn sheep: Risk and resilience. Journal of Wildlife Management, 82(1), 32–45. 10.1002/jwmg.21309 [DOI] [Google Scholar]
- Cassirer EF, Manlove KR, Plowright RK, & Besser TE (2017). Evidence for strain-specific immunity to pneumonia in bighorn sheep. The Journal of Wildlife Management, 81(1), 133–143. 10.1002/jwmg.21172 [DOI] [Google Scholar]
- Cassirer EF, Plowright RK, Manlove KR, Cross PC, Dobson AP, Potter KA, & Hudson PJ (2013). Spatio-temporal dynamics of pneumonia in bighorn sheep. Journal of Animal Ecology, 82(3), 518–528. 10.1111/1365-2656.12031 [DOI] [PubMed] [Google Scholar]
- Cassirer EF, Rudolph KM, Fowler P, Coggins VL, Hunter DL, & Miller MW (2001). Evaluation of ewe vaccination as a tool for increasing bighorn lamb survival following pasteurellosis epizootics. Journal of Wildlife Diseases, 37(1), 49–57. 10.7589/0090-3558-37.1.49 [DOI] [PubMed] [Google Scholar]
- Clauset A, Newman MEJ, & Moore C (2004). Finding community structure in very large networks. Physical Review E, 70(6), 066111. 10.1103/PhysRevE.70.066111 [DOI] [PubMed] [Google Scholar]
- Colby J, & Botta R (2013). California Department of Fish and Wildlife, Peninsular Bighorn Sheep 2013 Annual Report (p. 16). California Department of Fish and Wildlife. [Google Scholar]
- Colby J, & Botta R (2017). California Department of Fish and Wildlife, Peninsular Bighorn Sheep 2016–2017 Annual Report (p. 29). California Department of Fish and Wildlife. [Google Scholar]
- Colby J, & Botta R (2018). California Department of Fish and Wildlife Peninsular Bighorn Sheep 2017–18 Annual Report (p. 25). California Department of Fish and Wildlife. [Google Scholar]
- Colby J, & Botta R (2019). California Department of Fish and Wildlife Peninsular Bighorn Sheep 2018–19 Annual Report and Recovery Program Review 1992—2019 (p. 24). California Department of Fish and Wildlife. [Google Scholar]
- Dassanayake RP, Shanthalingam S, Herndon CN, Subramaniam R, Lawrence PK, Bavananthasivam J, Cassirer EF, Haldorson GJ, Foreyt WJ, Rurangirwa FR, Knowles DP, Besser TE, & Srikumaran S (2010). Mycoplasma ovipneumoniae can predispose bighorn sheep to fatal Mannheimia haemolytica pneumonia. Veterinary Microbiology, 145(3–4), 354–359. 10.1016/j.vetmic.2010.04.011 [DOI] [PubMed] [Google Scholar]
- DeForge JR, Jessup DA, Jenner CW, & Scott JE (1982). Disease investigations into high lamb mortality of desert bighorn in the Santa Rosa Mountains, California. Desert Bighorn Council Transactions, 26, 76–81. [Google Scholar]
- Di Luzio M, Johnson GL, Daly C, Eischeid JK, & Arnold JG (2008). Constructing retrospective gridded daily precipitation and temperature datasets for the conterminous United States. Journal of Applied Meteorology and Climatology, 47(2), 475–497. 10.1175/2007JAMC1356.1 [DOI] [Google Scholar]
- Dubey JP (2009). Toxoplasmosis in sheep—The last 20 years. Veterinary Parasitology, 163(1), 1–14. 10.1016/j.vetpar.2009.02.026 [DOI] [PubMed] [Google Scholar]
- Epps CW, McCullough DR, Wehausen JD, Bleich VC, & RECHEL L, J. (2004). Effects of climate change on population persistence of desert-dwelling mountain sheep in California. Conservation Biology, 18(1), 102–113. 10.1111/j.1523-1739.2004.00023.x [DOI] [Google Scholar]
- Foreyt WJ, & Jessup DA (1982). Fatal pneumonia of bighorn sheep following association with domestic sheep. Journal of Wildlife Diseases, 18(2), 163–168. 10.7589/0090-3558-18.2.163 [DOI] [PubMed] [Google Scholar]
- Garwood TJ, Lehman CP, Walsh DP, Cassirer EF, Besser TE, & Jenks JA (2020). Removal of chronic Mycoplasma ovipneumoniae carrier ewes eliminates pneumonia in a bighorn sheep population. Ecology and Evolution, 10(7), 3491–3502. 10.1002/ece3.6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J, Li Y, & Mitra R (2018). On the use of Cauchy prior distributions for Bayesian logistic regression. Bayesian Analysis, 13(2), 359–383. 10.1214/17-BA1051 [DOI] [Google Scholar]
- Hantson S, Huxman TE, Kimball S, Randerson JT, & Goulden ML (2021). Warming as a Driver of Vegetation Loss in the Sonoran Desert of California. Journal of Geophysical Research: Biogeosciences, 126(6), e2020JG005942. 10.1029/2020JG005942 [DOI] [Google Scholar]
- Hawkes LA, Fahlman A, & Sato K (2021). Introduction to the theme issue: Measuring physiology in free-living animals. Philosophical Transactions of the Royal Society B: Biological Sciences, 376(1830), 20200210. 10.1098/rstb.2020.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JJ, Malilay JN, & Parkinson AJ (2008). Climate change: The importance of place. American Journal of Preventive Medicine, 35(5), 468–478. 10.1016/j.amepre.2008.08.024 [DOI] [PubMed] [Google Scholar]
- Jamieson AM, Pasman L, Yu S, Gamradt P, Homer RJ, Decker T, & Medzhitov R (2013). Role of Tissue Protection in Lethal Respiratory Viral-Bacterial Coinfection. Science, 340(6137), 1230–1234. 10.1126/science.1233632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Gasper DJ, & Mitchell E (2018). Bovidae, Antilocapridae, Giraffidae, Tragulidae, Hippopotamidae. In Terio K, McAloose D, & St. Leger J (Eds.), Pathology of Wildlife and Zoo Animals (First, pp. 117–147). Elsevier. [Google Scholar]
- Kubinec R (2020). Ordered beta regression: A parsimonious, well-fitting model for survey sliders and visual analog scales. SocArXiv. 10.31235/osf.io/2sx6y [DOI] [Google Scholar]
- Lewis C (1996). Update on orf. In Practice, 18(8), 376–381. 10.1136/inpract.18.8.376 [DOI] [Google Scholar]
- Manlove KR, Cassirer EF, Cross PC, Plowright RK, & Hudson PJ (2014). Costs and benefits of group living with disease: A case study of pneumonia in bighorn lambs (Ovis canadensis). Proceedings of the Royal Society B: Biological Sciences, 281(1797), 20142331. 10.1098/rspb.2014.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manlove KR, Cassirer EF, Plowright RK, Cross PC, & Hudson PJ (2017). Contact and contagion: Probability of transmission given contact varies with demographic state in bighorn sheep. Journal of Animal Ecology, 86(4), 908–920. 10.1111/1365-2656.12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen PGP, & Smith BP (2009). Contagious ecthyma (sore mouth, orf, contagious pustular dermatitis, scabby mouth). In Smith BP (Ed.), Large Animal Internal Medicine (Fourth, pp. 780–790). Elsevier Health Sciences. [Google Scholar]
- Monello RJ, Murray DL, & Cassirer EF (2001). Ecological correlates of pneumonia epizootics in bighorn sheep herds. Canadian Journal of Zoology, 79(8), 1423–1432. 10.1139/z01-103 [DOI] [Google Scholar]
- Muneza AB, Ortiz-Calo W, Packer C, Cusack JJ, Jones T, Palmer MS, Swanson A, Kosmala M, Dickman AJ, & Macdonald DW (2019). Quantifying the severity of giraffe skin disease via photogrammetry analysis of camera trap data. Journal of Wildlife Diseases, 55(4), 770–781. 10.7589/2018-06-149 [DOI] [PubMed] [Google Scholar]
- Nandi S, De UK, & Chowdhury S (2011). Current status of contagious ecthyma or orf disease in goat and sheep—A global perspective. Small Ruminant Research, 96(2), 73–82. 10.1016/j.smallrumres.2010.11.018 [DOI] [Google Scholar]
- Nolen RS (2010). Severe pneumonia outbreak kills bighorn sheep. Javma-Journal of the American Veterinary Medical Association, 236(9), 936–936. [Google Scholar]
- Ostermann SD, Deforge JR, & Edge WD (2001). Captive breeding and reintroduction evaluation criteria: A case study of peninsular bighorn sheep. Conservation Biology, 15(3), 749–760. 10.1046/j.1523-1739.2001.015003749.x [DOI] [Google Scholar]
- Plowright RK, Manlove K, Cassirer EF, Cross PC, Besser TE, & Hudson PJ (2013). Use of exposure history to identify patterns of immunity to pneumonia in bighorn sheep (Ovis canadensis). PLoS ONE, 8(4), e61919. 10.1371/journal.pone.0061919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Manlove KR, Besser TE, Páez DJ, Andrews KR, Matthews PE, Waits LP, Hudson PJ, & Cassirer EF (2017). Age-specific infectious period shapes dynamics of pneumonia in bighorn sheep. Ecology Letters, 20(10), 1325–1336. 10.1111/ele.12829 [DOI] [PubMed] [Google Scholar]
- PRISM Climate Group, Oregon State University. (2004). http://prism.oregonstate.edu
- R Core Team. (2021). R: a language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/ [Google Scholar]
- Raghavan B, Erickson K, Kugadas A, Batra SA, Call DR, Davis MA, Foreyt WJ, & Srikumaran S (2016). Role of carriers in the transmission of pneumonia in bighorn sheep (Ovis canadensis). Biology Open, 5(6), 745–755. 10.1242/bio.018234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin ES, Boyce WM, & Bleich VC (2000). Reproductive strategies of desert bighorn sheep. Journal of Mammalogy, 81(3), 769–786. [DOI] [Google Scholar]
- Rubin ES, Boyce WM, & Caswell-Chen EP (2002). Modeling demographic processes in an endangered population of bighorn sheep. The Journal of Wildlife Management, 66(3), 796–810. 10.2307/3803144 [DOI] [Google Scholar]
- Rubin ES, Boyce WM, Jorgensen MC, Torres SG, Hayes CL, O’Brien CS, & Jessup DA (1998). Distribution and abundance of bighorn sheep in the Peninsular Ranges, California. Wildlife Society Bulletin, 26(3), 539–551. [Google Scholar]
- Rubin ES, Boyce WM, Stermer CJ, & Torres SG (2002). Bighorn sheep habitat use and selection near an urban environment. Biological Conservation, 104(2), 251–263. 10.1016/S0006-3207(01)00171-9 [DOI] [Google Scholar]
- Sells SN, Mitchell MS, Nowak JJ, Lukacs PM, Anderson NJ, Ramsey JM, Gude JA, & Krausman PR (2015). Modeling risk of pneumonia epizootics in bighorn sheep. The Journal of Wildlife Management, 79(2), 195–210. 10.1002/jwmg.824 [DOI] [Google Scholar]
- Tuia D, Kellenberger B, Beery S, Costelloe BR, Zuffi S, Risse B, Mathis A, Mathis MW, van Langevelde F, Burghardt T, Kays R, Klinck H, Wikelski M, Couzin ID, van Horn G, Crofoot MC, Stewart CV, & Berger-Wolf T (2022). Perspectives in machine learning for wildlife conservation. Nature Communications, 13(1), 792. 10.1038/s41467-022-27980-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JC, Douglas CL, Hallum CR, Krausman PR, & Ramey RR (2004). Determination of critical habitat for the endangered Nelson’s bighorn sheep in southern California. Wildlife Society Bulletin, 32(2), 427–448. 10.2193/0091-7648(2004)32[427:DOCHFT]2.0.CO;2 [DOI] [Google Scholar]
- U. S. Geological Survey. (2017, November 29). 2012–2016 California drought: Historical perspective.
- US Fish and Wildlife Service. (2000). Recovery plan for bighorn sheep in the Peninsular Ranges, California (Portland, OR, p. 251). [Google Scholar]
- Vehtari A, Gabry J, Magnusson M, Yao Y, Bürkner P, Paananen T, & Gelman A (2020). loo: Efficient leave-one-out cross-validation and WAIC for Bayesian models (2.3.1) [R package].
- WAFWA Wildlife Health Committee. (2016). Bighorn sheep herd health monitoring recommendations.
- Ward ACS, Hunter DL, Rudolph KM, DeLong WJ, Bulgin JM, Cowan LM, McNeil HJ, & Miller MW (1999). Immunologic responses of domestic and bighorn sheep to a multivalent Pasteurella haemolytica vaccine. Journal of Wildlife Diseases, 35(2), 285–296. 10.7589/0090-3558-35.2.285 [DOI] [PubMed] [Google Scholar]
- Wobeser GA (2007). Disease and epizootiology – basic principles. Disease in Wild Animals: Investigation and Management, 3–16. 10.1007/978-1-4757-5609-8_1 [DOI] [Google Scholar]
- Woolums AR, Ames TR, & Baker JC (2009). The bronchopneumonias (respiratory disease complex of cattle, sheep, and goats). In Smith BP (Ed.), Large Animal Internal Medicine (Fourth, pp. 602–643). Elsevier Health Sciences. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this manuscript are presented in Appendices S11–13.


