SUMMARY
Laboratory-generated hybrids between phage λ and related phages played a seminal role in establishment of the λ model system, which, in turn, served to develop many of the foundational concepts of molecular biology, including gene structure and control. Important λ hybrids with phages 21 and 434 were the earliest of such phages. To understand the biology of these hybrids in full detail, we determined the complete genome sequences of phages 21 and 434. Although both genomes are canonical members of the λ-like phage family, they both carry unsuspected bacterial virulence gene types not previously described in this group of phages. In addition, we determined the sequences of the hybrid phages λ imm21, λ imm434, and λ h434 imm21. These sequences show that the replacements of λ DNA by nonhomologous segments of 21 or 434 DNA occurred through homologous recombination in adjacent sequences that are nearly identical in the parental phages. These five genome sequences correct a number of errors in published sequence fragments of the 21 and 434 genomes, and they point out nine nucleotide differences from Sanger’s original λ sequence that are likely present in most extant λ strains in laboratory use today. We discuss the historical importance of these hybrid phages in the development of fundamental tenets of molecular biology and in some of the earliest gene cloning vectors. The 434 and 21 genomes reinforce the conclusion that the genomes of essentially all natural λ-like phages are mosaics of sequence modules from a pool of exchangeable segments.
KEYWORDS: bacteriophage, phage, hybrid phage, phage lambda, phage 21, phage 434, temperate phage, phage genetics, lysogeny
INTRODUCTION
The study of laboratory-generated hybrid phages, the viable products of genetic recombination between two distinct phages, contributed significantly to early genetic and molecular studies of the biology of temperate phages. These hybrid phages were critical players in the attainment of fundamental insights into the structure of genes and how their expression is controlled. The story starts with the isolation and choice of phage λ as a model system for genetic analysis. Phage λ was discovered by Esther Lederberg in 1950 when it was released from Escherichia coli K-12 (a 1922 clinical isolate from California) after UV irradiation (1). Various versions of this strain of E. coli K-12 and thus different strains of phage λ were in circulation for several years (2), until Dale Kaiser crossed two of the extant strains and isolated a derivative that had desirable properties, namely, the larger plaque size of the Pasadena (CalTech) strain and immunity features of the Paris (Institut Pasteur) smaller-plaque strain (3, 4). This recombinant, called λ PaPa for Paris and Pasadena, is the progenitor of nearly all currently extant laboratory strains and is often referred to as “wild-type” λ. The cause of the increased plaque size turned out to be a frameshift mutation in λ’s stf (side tail fiber) gene (4). The larger plaque-forming variant was better for genetic studies, because mutants with plaque morphology changes were easier to see and so comprised the first useful category of mutants for study. Mutants of λ PaPa were generated by UV mutagenesis that formed small (s), minute (mi), and three types of clear plaques (c, co1, and co2), and crosses between these isogenic mutants generated the first λ genetic map (3):
where the numbers indicate recombination frequency as a percentage of progeny viruses.
In the late 1950s, Jacob and Wollman (5) screened 500 natural isolates of Escherichia coli, contributed by the citizens of Paris, for spontaneously inducible prophages, i.e., strains whose culture supernatants contained plaque-forming phages. This screen yielded a collection of 54 temperate phages, both UV inducible and UV noninducible. The UV-inducible phages able to form plaques on E. coli K-12 included phages 21, 82, 424, and 434. Meanwhile, other E. coli phages, including phage ϕ80 (6), HK97 (7), HK022 (7), and Salmonella enterica phages P22 (8) and L (9) were isolated by others and found to have similar properties. These phages were important in the development of our current understanding of molecular genetics (10), and a number of them were used to make hybrid phages that were important in this process. Experimental “handles” were few at that time, so members of this group of phages were initially characterized for (i) the prophage’s ability to provide the host cell with immunity to infection by the same phage (“homoimmunity”) but not to other phages in the group (“heteroimmunity”), (ii) the ability to infect different bacterial strains (“host range”), and (iii) the prophage location on the bacterial chromosome. These characterizations were accomplished by using simple tests that showed that even by these relatively crude criteria, they were all different phages.
The immunity function of a prophage blocks the growth of a superinfecting homoimmune phage while having no effect on heteroimmune phages. Thus, the immunity type of a phage could be determined by simply asking if the phage of interest can form plaques on a panel of E. coli K-12 lysogens, each carrying one of the other phages as a prophage. It turned out that all of the above phages were heteroimmune to one another, except two homoimmune pairs, 21/P22 and λ/HK97, indicating a large diversity in the immunity function. A related important property was plaque morphology, namely, large or small, crisp or fuzzy, and clear or turbid. The latter was important in that wild-type temperate phages in this group produce turbid plaques, signifying successful lysogeny; the turbidity is due to homoimmune lysogens propagating within the plaque.
Phage host range was studied by asking if a phage of interest can form plaques on other host strains or bacterial species. Examination of the ability of the above-mentioned phages to grow on bacterial mutants that were resistant to other phages led to the identification of several different receptor molecules on the bacterial surface, thus revealing several host range classes. For example, the phage 21 and λ virions were found to possess the same host range, requiring the host LamB outer membrane protein (Omp) as the receptor, which turned out to be the maltodextrin porin of E. coli (11). In contrast, phage 434 and ϕ80 virions utilize other porins for their attachment, namely, the general porin OmpC (12) and the ferrichrome import porin FhuA (see reference 13 and references therein), respectively, while P22 requires the O-antigen surface polysaccharide (see reference 14 and references therein).
The third phage property examined at that time was the position of the prophage on the E. coli chromosome. In the 1950s, Jacob and Wollman were concurrently studying mating and recombination in E. coli. By using various lysogenic strains in interrupted mating experiments between Hfr male and F− E. coli female strains, they discovered that their prophages occupy specific and different positions on the E. coli chromosome. For example, prophages λ and 434 localized near the gal (galactose utilization) genes (5), whereas prophage 21 integrated at a different site, later shown to be inside the isocitrate dehydrogenase gene (icd) (15, 16).
These described properties of immunity, plaque morphology, host range, and prophage location were useful tools in the construction and characterization of hybrids and in very early genetic studies that included the study of hybrids between λ and 434 and between λ and 21. We recently reported the complete genome sequences of phages 21 and 434, as well as their hybrid phages λ imm434 (short for λ immunity 434, also referred to as λ i434), λ imm21, and λ h434 imm21 (h434 is 434 host range) (17). In the context of these sequences, we review the contributions of hybrid phages to the very early development of molecular genetics.
PHAGE 21 GENOME
Regions of Similarity with the Phage λ Genome
Because phage 21 has a long laboratory history, we sequenced complete wild-type genomes from two different sources (17). Both sequences were 42,931 bp long, and they were identical (GenBank accession number OL657228). About 45% (19,286 bp) of the phage 21 genome sequence had been previously reported in 11 partly overlapping GenBank entries (listed in Table S1A in the supplemental material); there are 71 mostly single-base-pair differences between our new sequence and these early sequences, a number of which affect reading frame integrity. Since our two phage 21 genome sequences are identical and because sequencing methods have become more accurate, we believe that the sequence analyzed here is accurate and corrects the previous errors. Seventy-three protein-encoding genes and two tRNA genes were annotated in the phage 21 genome, and a map of the genome is shown in Fig. S1.
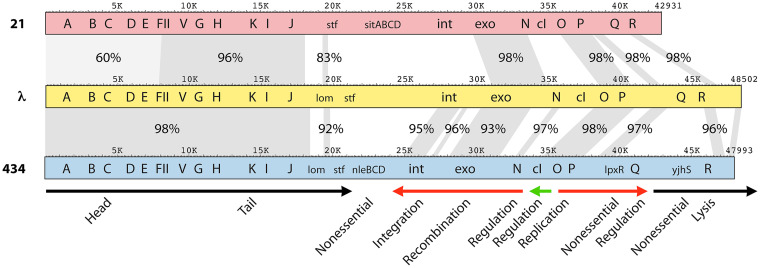
In 1971, the seminal publication by Simon et al. (18) used electron microscopic DNA heteroduplex mapping to show that the phage 21 chromosome has five regions that are sufficiently similar to phage λ DNA to enable the formation of heteroduplexes under their observation conditions. A comparison of the 21 and λ genome sequences confirms this highly mosaic relationship and shows five major regions of high similarity (>95% identity) (Fig. 1; Table S2A), which are in excellent agreement with the measured heteroduplex locations. Simon et al. (18) also noted that the region comprising bp 1 to 8000 gave a variable heteroduplex formation in 40% formamide, but the heteroduplex was never observed in 60% formamide, a stronger denaturing condition. This observation also agrees well with the fact that phage 21 and λ DNAs are about 60% identical across this region. The phage 21 genome sequence highlights the excellent accuracy of those early electron microscopic measurements, which, in that nondigital age, depended on projecting the electron micrograph onto a paper hung from a wall and using a manual planimeter to trace the length of thin (single-strand) and thick (duplex) DNA, normalized to standard DNAs included in the sample.
FIG 1.
Genome maps of phages 21 and 434. Maps of phages 21, λ, and 434 are shown with selected λ genes indicated; homologs in 21 and 434 are marked by the λ gene name. At the bottom, the major transcription units are indicated by horizontal arrows: red, early left and right operons; black, late operons; green, repressor transcript expressed in the lysogen. The universal order of gene functions in the λ-like phages is shown at the bottom. Gray areas between maps indicate regions of homology, with the percent identity values shown.
Virion Assembly Genes
Overall, the phage 21 virion assembly genes are homologous to and syntenic with those of λ but vary in their degree of nucleotide similarity to λ. The head genes (the leftmost ~8 kbp; see above) encode proteins that range downward from 65% identity to their cognate λ proteins, and at least some have partner binding specificities that are different from those of λ. A good example of the latter is that of terminase, the enzyme composed of small and large subunits TerS and TerL that is responsible for packaging phage DNA. The phage 21 terminase differs from λ terminase in its DNA target (cohesive end site or cos) specificity, TerS/TerL subunit interaction specificity, and TerL/portal protein binding specificity. The dissection of these properties using hybrid phages was important in gaining an understanding of the multiple interactions that occur during DNA packaging (reviewed in reference 19) (see Hybrid Phages and Protein Functional Domains DNA Packaging below). Complementation experiments between λ and 21 showed that the λ terminase proteins, gene product (gp)Nu1 and gpA, and procapsid proteins gpB, gpC, gpNu3, and gpE do not successfully replace their 21 homologs. On the other hand, gpW and gpFII (see following paragraph) do so weakly and strongly, respectively (20), and replacing the λ head-stabilizing decoration protein gpD with the 21 gpD homolog results in partial stabilization of the viral head shell (21).
The phage 21 tail proteins are very similar to those of λ, and their genes are largely included in a region of high similarity to λ, bp ~8000 to ~18150 (Fig. 1; Table S2A). For example, their major tail shaft subunits (λ gpV) and tape measure proteins (λ gpH) are 96% and 97% identical, respectively. The junction between moderate- and high-similarity regions transitions through a region of intermediate similarity between approximately bp 7900 and 8000. This transition is inside the FII gene, whose protein product determines the site on λ heads to which tails bind (22). The N-terminal 40 amino acids (aa) of gpFII are 52% identical to those of λ gpFII, while the C-terminal aa 76 to 117 are 81% identical. Such boundaries in phage virion assembly genes are almost certainly the result of past recombination between divergent parents that resulted in the currently observed mosaic genomes (23, 24). Such events can be intragenic, and when they are, they often occur between protein domain boundaries (25). Figure S2 shows the location of this transition zone in the λ gpFII protein structure. Juhala et al. (26) suggested that the C-terminal portion of λ-like phage HK022 gpFII, where there is an even more striking change in similarity to λ gpFII, binds tails and that the N-terminal portion binds heads (but see the report of Maxwell et al. [27]). Although separate peptide domain locations are ambiguous in this case, the N- and C-terminal parts of gpFII are physically separate in the folded protein, so these proteins accommodate exchanges of these two parts.
The right end of this high-similarity tail region includes the 5′-terminal two-thirds of the J gene. The 21 and λ J protein N-terminal 810 aa are 97% identical, while the remaining C-terminal regions (322 and 350 aa in λ and 21, respectively) are only 48% identical. The C-terminal 249-aa fragment of λ gpJ binds its LamB outer membrane protein receptor in vitro (28), and changes in aa 1040, 1077, or 1127 alter λ’s host range (29), so the C-terminal part of λ gpJ appears to determine the phage’s host range specificity by binding its bacterial surface receptor. Virions of phages 21 and λ both adsorb to LamB protein (5), so the substantial differences near their C termini suggest that the two proteins contact different parts of LamB or contact the same part in different ways.
To the right of the J gene, the λ-like phages often carry a side tail fiber stf gene, which is interrupted by a frameshift mutation in λ stocks typically in laboratory use today (4). The N-terminal 117 aa of the 21 Stf protein (encoded by its gene 22) are 86% identical to the parallel region of λ Stf, but the sequence of the remaining C-terminal portions of the two proteins are essentially unrelated. Since all known tail fibers bind to the virion through their N-terminal regions (30), this suggests that gene 22 protein is a side fiber on 21’s tail and that its dissimilar C-terminal domain may have different host secondary receptor-binding properties than the λ side fiber’s OmpC target (4).
Nonessential b Region
We define the “b region” of the λ-like genomes as the section between the virion assembly gene cluster and the integrase gene; this region is dispensable for λ lysogeny and lytic growth. It was originally named after “b” deletions that were missing parts of this region and hence produced functional virus particles with altered buoyant densities in CsCl density gradients (31). This region is exceptionally variable among the λ-like phages (our unpublished analysis). The three λ b region genes, ea47, ea31, and ea59, have not been studied in detail, but ea59 encodes an endonuclease (32, 33) and all three are apparently expressed early in infection from the early left promoter (34, 35). They do not appear to be expressed in a lysogen. Between its gene 23 (which encodes a tail fiber assembly chaperone) and its integrase gene, phage 21 carries 10 annotated genes that are unrelated to the b region genes of λ. Four of these, sitABCD (phage 21 genes 25 to 28), encode an apparently complete Fe/Mg ABC type transporter system. The sitABCD gene cluster was originally studied in Salmonella enterica serovar Typhimurium SL1344 (36), where it has been shown to be a Salmonella virulence factor (37). The phage 21 SitA protein, for example, is 75% identical to the Salmonella SL1344 protein, but the sitABCD genes do not appear to be in a prophage in this Salmonella strain. Of the six other genes in this phage 21 region, gene 30 may be a transposase pseudogene, and 24, 29, 31, 32, and 33 have no predicted function. Very similar 21-like “b region” genes are present at this location in λ-like prophages in a number of Enterobacteriaceae bacterial genomes. For example, in E. coli strain UT189, sitABCD and gene 29 homologs (locus tags UTI89_C1336 to UTI89_C1341) lie at the same location in a λ-like prophage.
Early Left Operon and Integration
There are 17 open reading frames, predicted genes 34 through 50, in the phage 21 early left operon, including homologs of λ genes int, xis, exo, bet, gam, kil, cIII, ea10, ral, and N. The 21 N (gene 50) product is 59% identical to that of λ and is known to have a different target specificity (38–41). As is typical of λ-like phages, this operon is highly mosaic in structure relative to other λ-like phages, and the remaining seven genes of unknown function all have closely related homologs in other λ-like phages. Phage 21 integration is known to be inside the icd gene of E. coli (15, 16).
Immunity Region
Previous studies have shown that the 21 prophage repressor (CI), encoded by its gene 51, has operator binding specificity that differs from that of λ repressor but is very similar (identity, 93%) to that of phage P22 repressor (42). The 21 repressor gene transcript appears to be monocistronic, whereas λ and a number of other λ-like phages (see phage 434 below) have a lysogenic conversion gene(s) immediately downstream of cI that is expressed from the prophage; for example, lysogenic conversion genes of λ are rexA, whose product binds CI repressor to assist the transition from lysogenic to lytic growth, and rexB, whose product modulates RexA function (43). The 21 Cro and CII regulatory proteins are 98% and 88% identical to their homoimmune P22 homologs, respectively, and presumably recognize the same or very similar target sequences in the two phages.
Early Right Operon
The 21 early right operon had been largely sequenced as multiple segments in previous studies (44–46), and our sequence corrects a number of apparent errors in those sequences. It is a typical λ-like right operon, where the two replication genes encode proteins that are 100% and 99% identical to the O and P proteins of λ, respectively, and the “nin region” between the P and Q genes contains seven predicted genes, all of which have closely related homologs in λ. In 2014, we examined the 81 λ-like phage genome sequences available at the time for putative λ Q-like proteins and found that they encoded five very different (in some cases apparently nonhomologous) late transcription antitermination proteins (47). The gene 63 phage 21 antitermination protein belongs to type 3, while that of λ is type 1 (they are only 13% identical in amino acid sequence). The 21 protein’s structure while bound to the qut site, the DNA site used for effecting antitermination, has been studied in detail (48).
Promoter-Proximal Late Operon and Lysis Genes
Just downstream from the transcription initiation site (bp 39541) (48) of the 21 late promoter, there are asparagine and threonine tRNA genes; tRNA genes in this location are not unusual among λ-like phages (see, for example, phages L [49] and Sf6 [50]). The roles of these tRNAs remain unclear, and it is not known if they have their own promoter or are cleaved from the late mRNA. The first protein-encoding genes of the late transcript in both λ and 21 are the lysis genes. Like λ’s S107-S105-R-Rz-Rz1 gene cluster, the 21 lysis genes 64 through 68 form a functional cassette, with genes encoding antiholin, holin, endolysin, i-spanin, and o-spanin, respectively. In λ, the S gene encodes two proteins from the same reading frame, but translation starts two codons apart, resulting in S107 and S105 proteins (51). S105 is the prototypic holin, which forms large micrometer-scale holes in the inner membrane (IM) that allow the release of the lambda endolysin R, a soluble transglycosylase, into the periplasm, where it attacks the cell wall peptidoglycan (PG) (52). In contrast, S107 acts as an inhibitor of S105 (53). At first glance, 21 lysis looks similar to that of λ, although the holins have very different sequences, in that genes 64 and 65 differ only in their start codons, with gp64 having an extra N-terminal Met1-Lys2-Ser3, which blocks the exit of the N-terminal transmembrane domain (TMD) from the bilayer, rather than blocking entry (54). Instead of forming micrometer-scale holes, gp65 forms small holes about 2 nm in diameter and has been dubbed “pinholin” (51, 55). The 21 gp66 PG hydrolytic enzyme is a “true” lysozyme (glycosyl hydrolase marked by an E-X8-D-X5-T catalytic triad), while the λ endolysin is a transglycosylase (56). In addition, gp66 has an N-terminal TMD that is metastable in the membrane (57). This SAR (signal anchor and release) endolysin is inactive as long as it remains inserted in the membrane, but when gp65 forms IM “pinholes” that collapse the membrane potential, it is released from the bilayer into the periplasmic space, where it becomes active and attacks the PG. After PG destruction, the final step in lysis is disruption of the outer membrane (OM) (58, 59). Here, λ and 21 are more alike mechanistically; the 21 gp67 IM protein i-spanin forms a complex with the gp68 OM-lipoprotein o-spanin that connects the two membranes (hence the term spanin). Once the PG is degraded, the spanin is thought to cause fusion of the IM and OM and, thus, removal of the final barrier for release of the progeny virions (60, 61). The λ o-spanin (Rz1) gene is embedded entirely within the i-spanin gene but in a different reading frame (59), while the 21 o-spanin gene begins within the coding region of 67 and extends beyond its 3′ end. Phage λ hybrids with the entire 21 lysis cassette were fundamental to the dissection of the 21 lysis pathway (51, 54).
Finally, to the right of the phage 21 lysis genes lies a bor-like gene (62), a tonB homolog, and three small genes, 70, 72, and 73, of unknown function. The tonB homolog (gene 71) encodes a 108-aa polypeptide with 33% identity to the C terminus of the 239-aa E. coli TonB protein (UniProt ID P02929) of E. coli K-12. The TonB protein is anchored in the inner membrane by its N terminus and has a periplasmic domain that interacts with a number of outer membrane siderophore transporters, including FhuA (UniProt ID P06971), supplying energy required for transporter function (63). As noted above in the Introduction, FhuA is a ferrichrome transporter that also serves as a receptor for a number of bacteriophages, including ϕ80, ES18, T1, and T5, and several colicins. Several TonB truncated fragments, including its C-terminal 117 aa, have been shown to block ferrichrome transport and ϕ80 infection (64). Although it is possible that the truncated TonB of phage 21 acts to block predation by some TonB-dependent phages, we found that a 21 prophage did not prevent efficient plaque formation by ϕ80 (M. Feiss, unpublished data). Perhaps high expression of the gene 71 protein during the lytic cycle acts to prevent superinfection by TonB-dependent phages.
PHAGE 434 GENOME
Regions of Similarity with the Phage λ Genome
Like phage 21, phage 434 has a very long laboratory history, so we sequenced two laboratory strains, the wild type and a clear plaque mutant, from different sources (17). There are seven previously reported 434 partially overlapping sequences present in the GenBank database that cover 8.5% (4,084 bp) of the genome (Table S1B). These sequences possess four differences from our sequence, and the 434 genome sequence reported here corrects these possible errors. Surprisingly, the two “phage 434” genome sequences have a major difference: the two phages, which we call 434 and 434B, have genomes that are 47,993 bp (GenBank accession no. OL657226) and 47,075 bp (GenBank accession no. OL657227) long, respectively. Analysis of these sequences allowed the deduction that the former is likely the original phage 434, as follows: Simon et al. (18) used electron microscopy to measure the 434 chromosome to be 100.1% the size of the λ chromosome, or about 48,550 bp. This value is significantly closer to the length of the 434 genome sequence (47,993 bp) than to the 434B sequence (47,075 bp). As with phage 21 above, a comparison of the 434 and λ sequences confirms their highly mosaic relationship and shows that the positions of the 11 patches of >89% sequence identity agree very well with heteroduplex locations observed by Simon et al. (18) (Fig. 1 and Table S2B). Analysis of their λ-434 heteroduplex results and our sequences shows that the major difference between 434 and 434B lies between the 9th and 10th homology sections (434’s bp 39715 to 46454), and the measured interval length of this section (6,700 bp) by Simon et al. is much closer to our observed 434 interval length (6,739 bp) than to the parallel 434B interval length (5,821 bp). We conclude that 434B underwent a genetic substitution in this region sometime during its laboratory passages over the past 60 years (details discussed In Phage 434B Clear Mutant Genemone below). Seventy-seven protein coding genes were annotated in the 434 genome, and a map of their locations is shown in Fig. S3.
Virion Assembly Genes
The 434 virion assembly genes are also syntenic to those of λ, and very high similarity extends from bp 389 to the left of the cos (near the right linearized virion chromosome end) across the cos site to bp 18450 within the J gene. For example, the large terminase subunit (homolog of λ gpA), major capsid protein (gpE), major tail protein (gpV), and the tape measure protein (gpH) are 99%, 100%, 96%, and 98% identical, respectively, to their λ counterparts. The N-terminal 973 aa of the 434 tail tip gene 21 protein are 98% identical to those of λ gpJ, but the high similarity ends abruptly at that point, the remaining C-terminal 165 aa being only 32.3% identical in the two phages. As discussed above, such a C-terminal difference agrees with the facts that this region controls λ host range and that 434 uses a different bacterial receptor (OmpC) from that of λ (12). Transcriptionally downstream and immediately to the right of this gene, phage 434’s gene 22 is homologous to λ lom (65), but the two Lom protein products are only 45% identical. The next downstream gene to the right, 23, encodes a putative side tail fiber protein whose first 117 aa are 93% and 92% identical to λ Stf and 21 Stf, respectively (see Phage 21 Genome, Virion Assembly Genes above); the remainder of these proteins have no recognizable similarity, which suggests that the 434 fiber may interact with a cell surface feature that is different from that of λ’s OmpC side fiber receptor or from the 21 Stf target. A curious feature of the C-terminal part of 434 Stf is a 111-codon stretch of GlyXY triplets, where X and Y are variable amino acids. Such extensive “collagen-like” triplet repeats have been noted before in trimeric phage tail fibers of distantly related phages (66), and a current BLASTp search reveals that this type of repeat is fairly common in side tail fibers of λ-like and other phages. We find such repeats of comparable length scattered among ~65 of the currently available 305 Enterobacterales-infecting phages (not including prophage sequences in bacterial genomes) that have λ-like gene organization. We also note that 434’s putative 437-aa long side tail fiber is considerably shorter than the 774-aa λ fiber (4). Phage 434 gene 24 encodes a tail fiber assembly chaperone (Tfa) protein.
Nonessential b Region
The 434 b region contains six annotated genes. Four of these, genes 25, 26, 27, and 29, are homologs of enterobacterial nleB, nleC, nleH, and nleD genes, respectively, and the short genes 28 and 30 in this region encode a transposase fragment and a protein of unknown function that is 100% identical to E. coli phage 933W gene 60 protein, respectively. The nle class of genes normally encodes bacterial virulence “effector” proteins (67), but here, nleB and nleH are apparently disabled by a nonsense codon and a truncation, respectively (neither of these mutations is a sequence error since they are both present in the 434 and 434B sequences). Where they have been studied, such effector proteins are injected into eukaryotic target cells by a bacterial type III protein secretion system. The NleB enzyme, a member of the NleB/SseK effector protein family, adds N-acetylglucosamine moieties to arginines of target proteins (68). One target protein that has been studied in detail is the Fas-associated death domain protein, and its modification may block apoptosis in the target cell (69). NleC and NleD are both metalloproteases; in homologous systems, NleC targets eukaryotic transcription factor NF-κB for degradation (70), and NleD inactivates the host mitogen-activated protein kinase signaling proteins JNK and p38 (71).
The Nle proteins are often encoded by genes present in other phages and prophages. For example, 434 NleB is 51% identical to λ-like contractile-tailed Salmonella phage ST64B gpSb26 protein (which has no internal stop codon) (68), and there are homologs of NleB, NleC, and NleD encoded in the possibly defective λ-like prophage CP-933K in E. coli EDL933 (GenBank accession no. NC_002655; locus tags Z0985, Z0986, and Z0990, respectively) (72).
Early Left Operon and Integration
Phage 434 integrates at the λ attB site (73), and its integrase gene is 99% identical to that of λ. However, the two sequences diverge greatly immediately to the left of the phage attP sites, so it is not clear that 434 regulates int gene expression with a “retroregulation” mechanism like that of λ (74, 75). The early left operon of 434 has 17 genes, 31 to 34 and 37 to 49, which include λ int, xis, exo, bet, gam, kil, cIII, ea10, and N homologs, all of which encode proteins that are >93% identical to their λ counterparts. The remaining early left operon genes are nearly all related to similarly located genes in other λ-like phages. In addition, there are two oppositely oriented genes, 35 and 36, of unknown function, embedded in the early left operon region.
Immunity Region
The operator binding specificity of 434 repressor (CI, encoded by gene 51) is known to be different from that of λ repressor (76), and early (genetically engineered) 434-λ hybrid repressor gene constructs were important in the determination of the part of DNA binding repressor proteins that is responsible for their target specificity (76, 77). The 434 repressor’s closest relatives in currently known phages are 99% identical and encoded by E. coli phages HK446 (GenBank accession no. JQ086372) and HK633 (accession no. JQ086377) and Shigella flexneri phages Sf6 (accession no. AF547987) and Sf101 (accession no. KJ832078), indicating that these phages almost certainly possess the same immunity as 434 (no other known phage repressor is closer than 60% identity to that of 434). Gene 50 of 434, which has previously been named hex (78, 79), is immediately left of 51, and its encoded protein is 99% identical to the gene 38 protein of the S. flexneri phage Sf6 (50). Hex protein has been reported to block CI repressor autocleavage (and thus prophage induction), which is orchestrated by the host RecA protein in response to SOS signals (78). The hex gene is likely cotranscribed with gene 51 from the 434 prophage and may function to lower the level of spontaneous induction of the 434 prophage and/or to require a stronger signal for efficient induction.
Early Right Operon
The early right operon of 434 contains 15 genes (52 to 64, 66, and 67), and its gene 52 encodes a Cro type repressor whose amino acid sequence is not easily recognizable as related to that of λ Cro, but the 434 gene 53, 54, and 55 proteins are 99%, 100%, and 97% identical to proteins CII, gpO, and gpP of λ, respectively. Since Cro binds the same operators as CI, it is not surprising that the putative 434 Cro protein is identical to those of the four phages mentioned above that are predicted to have 434 immunity. The 434 nin region between the λ P and Q homologs is very similar to that of λ, except that ninC and ninD are replaced by a putative adenine DNA methylase gene (gene 59) and there is no homolog of λ ninH. Unlike other known λ-like phages, this 434 region also contains a gene, 65, that is transcribed in the opposite direction (right to left on the standard phage map). It is a homolog of bacterial lpxR genes which encode a lipid A 3′-O-deacylase (80). For example, the Salmonella lpxR deacylase protein (gene STM1328 in strain LT2, apparently not in a prophage; GenBank accession no. AAL20253.1) is 71% identical to the 434 protein. LpxR action is thought to dampen the human immune response by lowering the recognition of lipid A by host TLR4-MD2-mediated immune surveillance (81) and is known to enhance the intracellular growth of Salmonella in macrophages (82), suggesting that the 434 lpxR is likely expressed in the lysogen. Although no lpxR genes are present in other currently known phages, very closely related homologs are present at this location in numerous λ-like prophages in sequenced bacterial genomes. For example, a lpxR gene (locus tag Z0955) lies in the same location in E. coli EDL933 prophage CP-933K (that also carries the nleBCD homologs; see Phage 434 Genome Nonessential b Region above). Although CP-933K carries bacterial virulence genes similar to those of 434, it has head genes that are very similar to those of Ctd-Iø (GenBank accession no. AB285204), an authentic λ-like phage with very different head genes (83). Finally, the 434 Q type late operon transcription antitermination protein, encoded by its gene 67, belongs to type 5 of Grose and Casjens’ classification (see Phage 21 Genome Early Right Operon above) (47), the group that also contains the phage 82 Q protein which was important in unraveling the mechanism of action of this type of transcription factor (84).
Promoter-Proximal Late Operon and Lysis Genes
The phage 82 Q gene and nearby late promoter were identified and sequenced by Goliger and Roberts (85, 86), and the high similarity of its sequence to the 434 sequence in this region (the Q proteins are 88% identical) indicates that the 434 late promoter should initiate mRNA synthesis at bp 42004. Phage 434 has no tRNA genes near the start of the late mRNA. Instead, between its late promoter and the lysis genes, its gene 71 is a homolog of the E. coli yjhS gene (homologs are also known as nanS). The E. coli phage 933W yjhS homolog encodes a 5-N-acetyl-9-O-acetyl O-acetylneuraminic acid monodeacetylase (87). This enzyme activity confers on the bacterial host the ability to use 5-N-acetyl-9-O-acetylneuraminic acid, a carbohydrate present in mammalian gut mucin, as a carbon source (88–90). The expression of 434 gene 71 has not been studied directly, although it seems likely that it is expressed from the prophage if it is to aid host metabolism. Although homologs of this gene often found at this location in other λ-like phage genomes are usually next to Shiga-like toxin genes, as in phage 933W (91), there are no such toxin genes in the 434 genome. The 434 YjhS-like protein is 95% identical to those encoded by E. coli phage YYZ-2008, which carries adjacent Shiga-like toxin genes (GenBank accession no. FJ184280), and phage JLK-2012 (accession no. JQ347801) (92), which does not. The YjhS type esterase encoded by phage 933W gene 42 is only 54% identical to the 434 protein, so it is possible that the two enzymes have different substrate specificities.
Transcriptionally downstream and to the right of gene 71, the 434 lysis gene cluster includes a pinholin gene whose product is 75% identical to the prototypical pinholin protein of phage 21 (see PHAGE 21 GENOME Promoter-Proximal Late Operon and Lysis Genes above), as well as SAR-endolysin and spanin genes, whose products are all ≥94% identical to those of 21. Finally, between the 434 lysis and phage head genes is a bor-like gene (see Phage 21 Genome Promoter-Proximal Late Operon and Lysis Genes above) whose encoded protein is 91% identical to that of λ (62).
PHAGE 434B CLEAR MUTANT GENOME
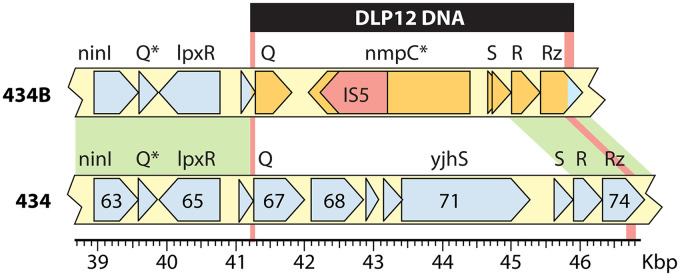
As mentioned above, the two “434” phage genomes that we sequenced are not identical. Figure 2 shows that they possess nonhomologous sequences in the 41- to 46-kbp (434 coordinates) region resulting in a nearly 1-kbp length difference. Examination of the 434B sequence shows that its substitution in this region is identical to a portion of the defective cryptic prophage DLP12 in the E. coli K-12 genome (93, 94) and that two homologous recombination events could have occurred between regions that are identical in 434 and DLP12 to create 434B (bp 41267 to 41274 at the left end of the substitution and bp 46775 to 46920 at the right end; 434 coordinates). The substitution replaces 5,499 bp of 434 DNA from gene 67 (homolog of λ Q) to a site within gene 74 (homolog of λ Rz) with 4,581 bp of DLP12 DNA. The DLP 12 outer membrane protein-encoding gene nmpC is inactivated by an IS5 insertion sequence that is also present in the 434B replacement (Fig. 2). The 434B late operon antitermination protein is a type 2 Q protein (47) that is essentially unrelated to that of 434; however, the 434B pinholin, endolysin, i-spanin, and o-spanin proteins are closely related to those of 434.
FIG 2.
Phage 434B genome structure. The region of difference between 434 and 434B is shown, with putative genes indicated as pointed boxes, where the points indicate the direction of transcription. Selected 434 gene names are indicated on the map, and λ gene names are shown above the maps. Asterisks mark nonfunctional genes. The solid horizontal black bar at the top indicates the extent of DLP12 DNA in 434B. Red vertical bars mark the regions of identity where homologous recombination occurred between 434 and DLP12 to create 434B (see the text). A kilobase pair scale is shown below the 434 map.
In addition, there are five single-base-pair differences in the homologous sequences shared between 434 and the 434B clear mutant that have accrued during their laboratory histories after separation of the two phages; the single-base-pair differences lie in genes homologous to λ C, I, J (two differences), and cI (listed in Table S3A). The cI difference is a transversion that changes 434 leucine codon 61 to an arginine codon in 434B. In the DNA binding N-terminal domain of 434, CI repressor leucine 61 lies at the end of α-helix 5 and contacts residues in the C terminus of α-helix 3, the helix that directly contacts the operator sequence. Replacement of this residue may impact the position of α-helix 3 or the overall folding of the domain (95). This is certainly the mutation that causes 434B to make clear plaques, since the other differences are all in genes that are not involved in lysogeny.
λ HYBRID PHAGES AND THEIR HISTORICAL IMPORTANCE
λ imm434, λ imm21, and λ h434 imm21
As stated above, phage λ was a central early player in the development of our current understanding of the molecular nature of genes, strategies by which gene expression is controlled, and other molecular processes such as recombination and DNA replication. Hybrids between λ and its close phage relatives were exceptionally powerful tools during those times when the availability of genetic markers and mutational types was extremely limited. For example, having hybrid phages with all λ DNA except for one or a few regulatory genes and their target sites from another phage meant that differences in behavior must be due to the differences between λ and the hybrid. In the first such hybrids studied, interest focused on understanding genes that control lysogeny, and early deductions included the ideas that controlling proteins could exert a negative effect, i.e., be repressors of gene function, and have different but critically important specificities in their interactions with nucleic acid sequences. In addition, since such hybrid phages carry markers as exchanged DNA regions rather than point mutations, mutational reversion could not easily confuse the experimental results. The clever use of such hybrids was in many ways crucial to the early development of λ into the informative and powerful genetic system it became in understanding the nature of the control of gene expression and protein-protein interactions.
The early electron microscopic examination of heteroduplex DNA molecules mapped the regions of high sequence similarity and sequence difference among the λ+ parent phage and its λ imm434 and λ imm21 derivatives and located the ends of their nonhomologous immunity segments (15, 86). These hybrid phage studies were among the very first to build physical maps of natural DNAs, where genes were placed in locations that reflect physical distances rather than locations determined by the frequency of recombination between alleles. This was a very important step forward in the attainment of a true understanding of genome structures.
In addition, these hybrid phages were central to the later development of the first DNA cloning vectors and so helped usher in modern genetic engineering technology. Utilization of the first λ cloning vector phages involved cutting the phage DNA with an appropriate restriction enzyme(s), ligating it with similarly cut foreign DNA, getting the phage genomes with DNA inserts into permissive cells where the phages could grow productively, and isolating the resulting phages whose genomes carry a foreign DNA insert. This strategy met with two major obstacles, both of which were overcome with the help of hybrid λ phages. First, the restriction enzymes utilized for cloning must cut only at a few, specific sequences that allow for insertion of foreign DNA without disrupting the viability of the phage genome, and second, the delivery of ligated DNA into cells by transfection was very inefficient. Since during those early days it was not possible to alter or move DNA sequences at will, Noreen Murray searched for and discovered λ hybrids that fortuitously had excluded specific restriction sites or, alternatively, possessed restriction sites that could be used for foreign gene insertion (96, 97). The second and more important problem was addressed by packaging the ligated DNA molecules into phage λ proheads in vitro. Kaiser and Masuda (98) developed the first such procedure by adding λ imm434 DNA to an extract of λ-infected cells, incubating the mixture to allow virion assembly, and subsequently plating the resulting mixture on an E. coli λ immλ lysogen. Only the exogenously added λ imm434 DNA could result in the formation of heteroimmune viruses capable of plaque formation. This system compensated for its relative inefficiency by possessing a powerful selection, namely, the in vitro-packaged λ imm434 virions could form a plaque on the λ immλ lysogen bacterial lawn. The amazing selectivity and specificity of in vitro packaging resulted in an improvement in recovery of phages that carry foreign DNA inserts to about 2 orders of magnitude better than that of transfection. Thus, the in vitro packaging of ligated phage vector/foreign DNA became the standard method of cloning foreign DNA into phage chromosomes at that time.
Because of their seminal roles in the origins of molecular biology, we determined the complete genome sequences of three important hybrids, λ imm434, λ imm21, and λ h434 imm21 (17), and discuss below their historical roles. The nonhomologous immunity “mosaic segments” or “modules” in λ imm434 and λ imm21 were already known to possess very different sequences from those of λ; however, as we discuss below, our sequences show that all four of the nonhomologous mosaic segment boundaries in these two hybrid phages were preexisting in the parental 434 and 21 genomes, and these hybrid phages were created by homologous recombination events on both sides of the immunity segments, occurring in adjacent, very similar sequences present in both parental phages. Thus, the mosaic segment boundaries—places where the 434 or 21 sequence diverges greatly from that of λ—are not the same as the sites where recombination happened to generate these hybrid phages; the former occurred in nature and were preexisting in the parental phages.
Construction of λ imm434, the First Laboratory-Created Hybrid between λ-Like Phages
Dale Kaiser, then the first postdoctoral fellow in Francois Jacob’s laboratory in the early 1950s, constructed the first simple genetic map of λ, using mutants that were altered for plaque size and morphology (see Introduction above) (3, 99). Wild-type λ phage produces turbid plaques on its E. coli host due to the growth of homoimmune lysogens in the plaque. Kaiser took advantage of this property by isolating clear plaque-forming mutants, defective in lysogen formation. He reasoned that these mutants would possess mutations in the systems that were necessary to form and/or maintain the stable prophage state. The clear mutations fell into three closely linked complementation groups, designated first as co2, c1, and co1 but later renamed genes cIII, cI, and cII, respectively (99). Subsequent studies by Kaiser showed that the cIII and cII mutant phages could form stable lysogens at a normal frequency, provided that the wild-type cII and cIII gene products (CII and CIII) were temporarily supplied in trans by a coinfecting heteroimmune cI+ phage. Hence, the cII and cIII gene products were shown to be needed only for the establishment of lysogeny, not for its maintenance.
In contrast, stable lysogens of cI mutants could not be isolated even in the presence of a c+ heteroimmune coinfecting phage, indicating that the λ cI product was required to maintain (and presumably establish) the lysogenic state. In addition, crosses between 434 and various λ mutants showed that most of the λ mutations available at the time could be easily crossed onto the 434 genome. For example, when λ mi, a minute plaque mutant, was crossed with 434 and the lysate was plated on a λ lysogen, recombinant phages with 434 immunity and the minute plaque phenotype were readily recovered, suggesting that 434 carried a mi+ allele analogous to that of λ. The sole exception was that mutations in the λ cI gene could not be crossed into 434 and vice versa. That is, the cross of 434 × λ cI− did not produce clear plaque recombinants able to form plaques on a λ lysogen, suggesting that cI mutations were in a λ genome segment that is an allelic alternative to 434’s immunity segment. In addition to demonstrating that the two immunities were allelic alternatives, this genetic analysis also suggested that the single gene, cI, was responsible for immunity. A final important conclusion was that the immunity segment likely occupied a relatively small part of the λ chromosome, since different cI clear mutations were closely linked to one another in comparison with the rest of the flanking markers on the λ genetic map.
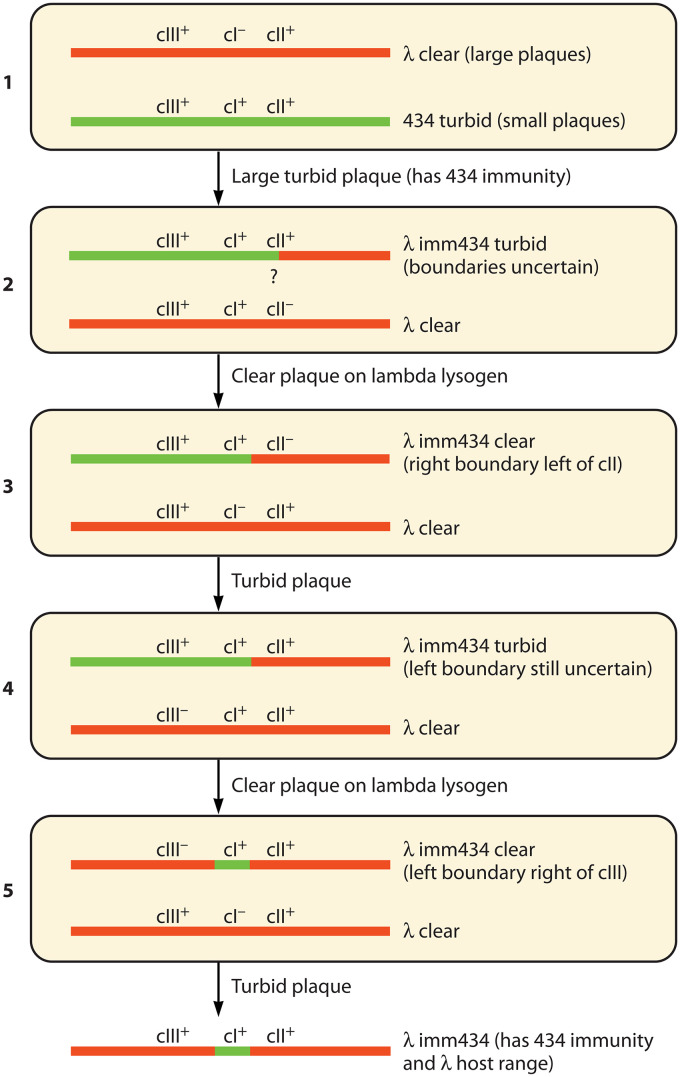
These clear plaque mutations were instrumental in the creation of a λ-434 hybrid phage that carries the immunity region from 434 but a bare minimum of other 434 DNA. To achieve this, Kaiser and Jacob (99) carried out a series of five sequential crosses in which λ-434 recombinants that had the immunity of 434 (designated here as imm434) and flanking λ DNA (Fig. 3) were selected. First, wild-type 434 (which naturally forms small turbid plaques) was crossed with a clear, large plaque-forming λ cI− mutant, and a rare recombinant that made large, turbid plaques was isolated. In this initial recombinant, the 434 DNA responsible for its small-plaque morphology was presumably replaced by λ DNA, while the λ cI− mutation was presumably replaced by 434 DNA, but the extent of the two different DNA exchanges were not known at that point. Four subsequent crosses were designed to modify the original hybrid so that DNA on both sides of the 434 immunity region was derived from λ (Fig. 3). To ensure that the right side of the immunity region was mostly λ DNA, the initial hybrid was crossed with λ cII− and a clear plaque recombinant with 434 immunity was isolated. Then, replacement of the λ cII− allele in this recombinant by wild-type λ cII+ DNA was accomplished with a backcross against λ cI−, and a turbid plaque-forming imm434 cII+ recombinant was selected. An analogous pair of genetic crosses on the left side of the immunity segment, using a λ cIII− clear mutant, completed the construction of λ imm434. The two crosses on each side of the immunity region made it highly likely that most, if not all, of the 434 DNA to the left and right of 434’s immunity segment was replaced with wild-type λ DNA sequences. Later, a series of crosses between λ and 21, analogous to those that generated λ imm434, produced the important hybrid phage 21hy1, which is now called λ imm21 (100), and subsequently, informative hybrids were also isolated between λ and ϕ80 (37, 38, 101–104), λ and P22 (42, 105, 106), λ and 82 (104), λ and HK022 (107, 108), λ and L (105), and P22 and L (79).
FIG 3.
Original λ imm434 construction. The five crosses used in the original construction of λ imm434 by Kaiser and Jacob (99) are shown in rounded boxes. λ DNA is shown as a red line, and 434 DNA is shown as a green line. A question mark indicates the initial boundary, whose location was uncertain. Selection used after each cross is shown between the boxes.
λ imm434 Genome Sequence
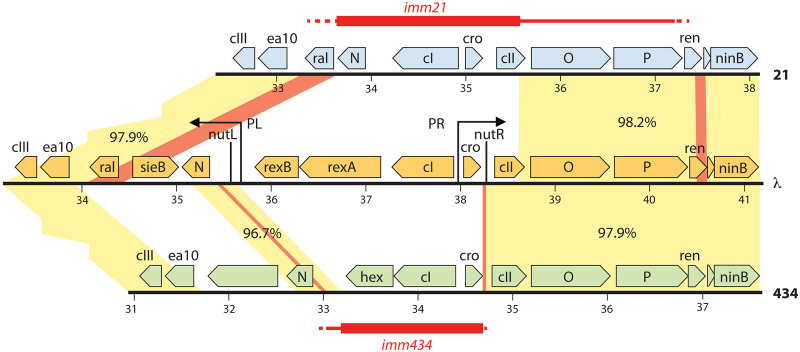
The λ imm434 genome sequence (GenBank accession no. OM418626) shows that 2,663 bp of λ DNA (bp 35584 to 38246) were replaced by 1,487 bp of nonhomologous DNA from 434 (bp 33198 to 34684). Since no other 434 DNA is present, Kaiser’s carefully done, multiple crosses (see previous section), designed to ensure that no extraneous 434 DNA persisted outside the immunity region in this hybrid, were successful. Pastrana (cited in reference 109) reported the location of the left end of the λ imm434 nonhomologous mosaic segment between the N gene and the early right promoter PL; our comparison of the 434 and λ imm434 genome sequences with that of λ agree in general but specify that it is 1 bp to the right of that location, between λ bp 35583 and 35584. Immediately to the left of the nonhomologous immunity region is a 459-bp section that is 96.7% identical in the two parental phages (λ bp 35125 to 35583; 434 bp 32739 to 33197) (Fig. 4). Comparisons of the 15 point differences in this region of the λ, 434, and λ imm434 genomes indicate that a homologous recombination event generated the right end of the 434 DNA in λ imm434, and it occurred in an 18-bp interval of identical sequence in λ and 434 (λ bp 35403 to 35420 and 434 bp 33017 to 33034) (diagrammed in Fig. S4 in the supplemental material). Similarly, a comparison of the λ and λ imm434 sequences confirms that the right end of the immunity imm434 nonhomology segment lies between λ bp 38246 and 38247 as previously reported (110, 111). This boundary lies between the 3′ end of the cro gene and the nutR site (see N Protein Function and Transcription Termination below), and immediately to its right lies a 2,838-bp section that is 97.9% identical in λ (bp 38247 to 41084) and 434 (bp 34685 to 37522), which includes genes cII-O-P-ren-ninA-ninB (Fig. 4). Comparison of the 61 point differences between λ and 434 in this region with the sequence of λ imm434 indicates that a homologous exchange occurred within the 15 leftmost base pairs of this region.
FIG 4.
Structures of the λ imm434 and λ imm21 genomes. Maps of the immunity regions of λ, 434, and 21 are shown, with genes indicated by pointed boxes that denote the direction of transcription. Light yellow-shaded areas with percent identity indications represent regions of similarity. Red connecting trapezoids between genomes mark the identical regions within these regions of similarity, where the homologous recombination events that created λ imm434 and λ imm21 occurred. Above and below the maps, thick red horizontal bars denote the locations of the nonhomologous 434 and 21 immunity mosaic sections or modules, thin horizontal red lines mark the further extent that 434 or 21 DNA replaced λ DNA in λ imm434 and λ imm21, and dashed red lines mark the regions in which the homologous recombination events occurred.
λ imm21 Genome Sequence
The λ imm21 genome sequence (GenBank accession no. OM418625) shows that 4,209 bp of λ DNA (bp 34382 to 38590) were replaced by 1,863 bp of nonhomologous DNA from phage 21 (bp 33658 to 35520). We note that the previously unanalyzed immunity region sequence of the λ cloning vector λ DE3, which carries imm21 (112), is identical to that in our λ imm21 genome. The precise location of the imm21 left mosaic section boundary has not been previously reported. Immediately to the left of the nonhomologous region is a block of 3,980 bp (λ bp 30402 to 34381; phage 21 bp 29678 to 33657) that is 97.6% identical between the two phages (Fig. 4). Analysis of the pattern of the 96 point differences between λ and 21 in this region shows that a left-side homologous recombination event occurred within a 341-bp section of identical sequence largely within the ral gene (λ bp 34026 to 34366; phage 21 bp 33301 to 33641). The right imm21 nonhomology boundary was previously reported to be between λ bp 38617 and bp 38618 (in codon 89 of the 97 codons present in the cII gene) (113, 114). Our λ imm21 genome sequence agrees with that sequence, but we note that although there is a 2-bp difference between λ and λ imm21 at bp 38617, the actual lack of 21 homology here ends at λ bp 38590. Again, comparison of point differences between the λ and 21 sequences shows that the actual location of the right-side homologous recombination event lies in a homologous sequence some distance to the right of this point within the adjacent block of λ 2,494 bp that is 98.0% identical in the two phages (λ bp 38591 to 41084; phage 21 bp 35521 to 38017). The pattern of the 50 single-base-pair differences between λ and 21 and this region of the λ imm21 sequence clearly shows that the actual crossover occurred within a region of 78 identical base pairs (λ bp 40503 to 40580; phage 21 bp 37436 to 37513) that lies within the ren gene. Thus, in addition to the 21 immunity region, λ imm21 actually carries the phage 21 cII, O, P genes and part of the ren gene (Fig. 4). We believe that this should not have affected interpretation of past experiments, since these phage 21 genes are all extremely similar to their λ homologs.
λ h434 imm21 Genome Sequence
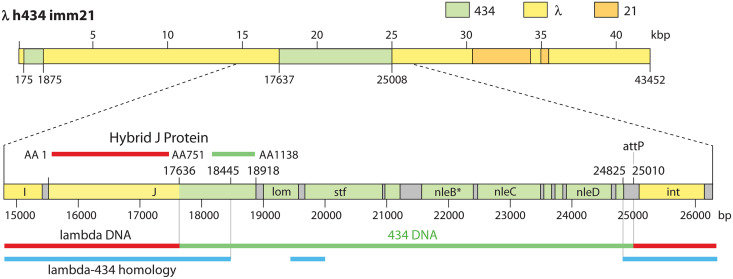
The sequence of the λ h434 imm21 genome (GenBank accession no. OM418627) shows that in addition to the imm21 region (identical to that in λ imm21), it contains 434 DNA that includes the 3′ portion of gene J, the stf gene, and the b region (bp 17637 through 25010) (Fig. 5). The left recombination event between λ and 434 was within the J gene and occurred within the bp 17620 to 17626 (λ coordinates) region of λ-434 identity. The resulting hybrid gpJ protein’s aa 1 to 751 are identical to those of λ, while its C-terminal 387 aa (aa 752 to 1138) are from 434 (there are two point differences at aa 1050 and aa 1114 from our 434 genome’s predicted gpJ protein) (details are given in Table S3D in the supplemental material). This result again supports the idea that 434 receptor specificity lies in the C-terminal portion of the hybrid protein (see Phage 21 Genome, Virion Assembly Genes above), but we note that this hybrid phage’s 434 gene side tail fiber gene (23) could also contribute to host recognition. The right-side recombination event that generated the h434 substitution appears to have been catalyzed by integrase, as it occurred within the bp 25011 to 25032 region of λ-434 identity (λ h434 imm21 coordinates), which is the core of the attachment (attP) site.
FIG 5.
Structure of the λ h434 imm21 genome. The λ h434 imm21 genome is shown at the top, with DNAs from different parents indicated by different colors. See the text for discussion of the following unexpected sequences: 434 DNA in the left end terS gene region and a patch of λ DNA in the imm21 region. The exchange that generated the h434 allele is shown at the bottom, with the genes in the region indicated.
The λ h434 imm21 genome shows two additional differences from λ (GenBank accession no. J02459) and λ imm21 which should not have had any major consequences in previous uses of this hybrid. First, 21 single-nucleotide differences from λ, λ imm21, and λ imm434 in the bp 75 to 1874 region (3′-end part of terS and 5′-end part of terL) show clearly that this region is in fact derived from 434. It is unlikely that these differences (minor in terms of terminase amino acid sequence) significantly affect terminase function, and they are not expected to affect experimental conclusions about the h434 region. Second, the phage 21 portion of λ h434 imm21 is identical to that of λ imm21, except for bp 34205 through 34451 (λ h434 imm21 coordinates). This section includes the 3′-end part of gene O and the 5′-end part of P and is within the λ-homologous 21 DNA immediately to the right of the immunity nonhomology section and again should not affect conclusions regarding host range (Fig. 4). In this λ h434 imm21 region, there are 22 differences from λ imm21, and 20 of these are the cognate λ base pairs. The source of this apparently λ DNA is not known, but it could have been introduced from λ h434 during the construction of the h434 imm21 double mutant from the single-mutant parents.
Hybrid λ Phage Summary and Relationship to the Originally Published λ Genome Sequence
The λ imm434, λ imm21, and λ h434 imm21 genome sequences show that each has the expected combination of mosaic DNA sections and pinpoint the actual recombination events that created them as having occurred in adjacent regions of DNA that have nearly identical sequences in the parental phages. Importantly, these hybrid genome sequences also show that these genomes do not contain other sections of 434 or 21 DNA sequences that might have affected the results of experiments that used these hybrid phages or conclusions derived from them.
We also note that the λ DNA portions of these hybrid phages have a few small differences from the originally reported λ genome sequence. The genome sequences of λ imm434, λ imm21, and λ h434 imm21 carry 9, 12, and 5 differences, respectively, from the published λ genome sequence (listed in Tables S3B, C, and E). Interestingly, these comprise a total of 13 differences, 9 of which are universally present in the λ DNA portions of these three hybrids. In addition, 6 of these 9 differences are present in the λ portions of the λ DE3 cloning vector (GenBank accession no. EU078592) (112), and two are present in λ O276, an HK022 hybrid whose genome has been sequenced (107, 108) (GenBank accession no. MH547045) (diagrammed in Fig. S5). These differences from the original λ sequence of Sanger et al. (115) are likely to be common if not universally present in laboratory strains of λ (indeed, we have noticed that the bp 138 short deletion is present in our own strains [M. Feiss, unpublished data]).
λ HYBRID PHAGES AND THE DAWN OF MOLECULAR BIOLOGY
λ imm434 and λ imm21 and the Concepts of Repression and Activation of Transcription Initiation
The λ imm434 and λ imm21 nonhomologous segments were found early on to contain alternative genes that confer the different immunity characteristics of the two viruses (see Introduction above). It was correctly hypothesized that these immunity genes encode prophage repressors with different target specificities. We now know that they bind to the left and right operators (OL and OR), thereby repressing the lytic promoters (PL and PR) within the immunity mosaic sections, which also contain the cro gene. Cro protein is the virus-specific lytic repressor that also acts at the OL and OR operators to block transcription of the cI gene from PRM on the one hand and to moderate expression of the PL and PR early promoters during lytic infection on the other hand. Detailed reviews of the lytic versus lysogenic decision have been published (116–118).
As mentioned above, the phage λ CII protein plays a central role in the lysis/lysogeny decision. Early genetic experiments and later biochemical analysis showed that CII is a transcriptional activator. One might thus expect that CII function would be tightly regulated in interesting ways, and one would not be disappointed. We now know that CII activates RNA polymerase to initiate mRNA synthesis more frequently at targeted promoters as follows: CII monomers assemble into a tetrameric transcription activator that acts at three promoters on the λ chromosome, i.e., PRE, PI, and PaQ, all of which promote lysogeny. First, the transcript from the CII-activated PI promoter encodes the prophage integrase (119). A second transcript, from the immunity establishment promoter PRE, produces high CI repressor levels during the establishment phase of lysogeny (120). Third, a short antisense transcript from the PaQ promoter delays production of the Q protein, the late gene activator, thus delaying the production of virion structural and host cell lysis proteins and thus favoring establishment of repression and integration of the circular phage DNA into the bacterial chromosome (121). The three CII-activated promoters are intrinsically weak, and CII activates these promoters by binding to TTGC repeats that flank their 6-bp-long “minus 35” segments, enhancing the affinity of RNA polymerase and subsequently promoting open complex formation (121–124). Interestingly, λ and P22 CIIs appear to recognize the same DNA sequence but will not cross-activate the other’s PRE promoter, so questions remain about the details of the mechanism of CII-mediated activation (summarized by Mondal et al. [125]). Early experiments by Liedke-Kulke and Kaiser (100) indicated that phage λ and 21 CII proteins are different, and Fien et al.’s (126) demonstration that they have different target site specificity used λ imm21 hybrid phages. We note that the DNA binding portion of the cII gene and PRE are within the imm21 nonhomology DNA segment (123, 124, 127), but PI and PaQ are not, and hence these λ promoters may not be properly activated in the λ imm21 hybrid phage. The 434 and λ CIIs are essentially identical and thus have the same target specificity, so even though the λ imm434 nonhomology section does not include cII, all the CII-dependent promoters should be activated normally in the hybrid.
CII activity in λ is also regulated at several different posttranscriptional levels. Host factors impact the stability of both the cII transcript and the CII protein itself. A short regulatory antisense RNA, the OOP RNA, accelerates cII mRNA decay by annealing with the cII mRNA originating at PR, forming a double-stranded RNA (dsRNA) that is subject to digestion by the host’s RNase III enzyme (128). In addition, the CII protein itself is metabolically unstable: CII’s C-terminal domain possesses a proteolysis signal recognized by the host FtsH protease (129–131). In turn, the CIII protein is an inhibitor of FtsH (129, 130). Finally, IHF, an E. coli DNA bending protein that also participates in the site-specific recombination process of prophage integration during lysogeny establishment, is required for CII expression by an unknown mechanism (129). Interactions of CII and CIII with host factors are not known to be phage specific, so these regulations likely function normally in the hybrids discussed here.
N Protein Function and Transcription Termination
Mutations in the λ N gene were found among the amber mutants in Allan Campbell’s set of λ essential gene nonsense mutants (132), and these were found to be defective for expression of the lytic genes (e.g., endolysin, since it was among the few gene functions that could be measured at that time) (133, 134). The simplest explanation of this observation was that N protein somehow acts as a transcription factor that activates expression of λ’s lytic genes. Early complementation and physiological studies showed that the gene N proteins of λ and 21, while carrying out analogous functions, are not functionally interchangeable; the λ, 21, ϕ80, and P22 N proteins were all shown to have species-specific targets of action (40, 135, 136), while λ and 434 N’s have the same target specificity (137). Thus, the facts that λ imm21 region contains the regulatory N gene but λ imm434 does not (Fig. 4) supported the idea that the critical targets of phage 21 N action must also be in the imm21 region.
Later, but still early in the study of λ, mRNA measurements showed that transcription initiated at PL or PR terminates shortly thereafter unless N protein is present (116, 138–140). Specifically, the defective N mutant phages transcribe only the genes cro from PR and N itself from PL but not distal genes (141, 142). Thus, a model was put forward in which, unlike activators that stimulate transcription initiation by RNA polymerase at a promoter (like CII), N acts by inhibiting downstream transcription termination signals—a novel and exciting concept at the time of its formulation in the late 1960s—thereby favoring expression of downstream genes. Strong transcription termination sites were discovered by the isolation and characterization of λ mutations, including deletions (byp, nin, etc.) that allowed lytic growth in the absence of N protein. These mutations turned out to inactivate or remove the downstream terminators in the early right operon (143–145).
Although PL and PR lie within the region of nonhomology between λ and λ imm434, the two phages have the same N protein specificity. On the one hand, this was one of the first indications that the site of N action might not be at the promoters (137, 146). On the other hand, experiments with the hybrid phage λ imm21 showed that the downstream terminators (in λ DNA) could not be the species-specific target of N action, since although it expresses only 21 N, transcription proceeds through the λ terminators to express the late genes (141). To solve this puzzle, it was proposed that the sites of N action, the downstream terminators, are separate and distinct from N’s recognition targets called nut (N utilization) sites (141, 142, 147). The nut sites were found to be in nonhomologous portions of the λ and 21 chromosomes (141, 142), and Salstrom et al. (148) used a triple λ-21-434 hybrid phage to show that the early left operon N specificity target site, nutL, is outside the promoter-operator region. Since then, the two nut sites in each phage, nutL and nutR, have been found to be located between the PL and PR promoters, respectively, and their downstream terminators (Fig. 4). In addition, studies of naturally isolated gal and trp operon fusions (phage-bacterium hybrids) driven by PL and PR that contain the promoter-proximal portions of the early right or left mRNAs showed that even foreign bacterial terminators were suppressed by λ N when a properly positioned λ nut site was present upstream of them (136, 142, 149).
We note that λ-like phage HK022 is unusual in that its N gene is replaced by the nun gene, whose protein product is expressed from the prophage and serves to block λ growth by inhibiting its N protein function (150). HK022’s early mRNA antitermination function is supplied by the special properties of parts of the early left and early right mRNAs called put RNAs (151). λ-HK022 hybrids were critical in gaining an understanding of the N-independent transcription antitermination that controls phage HK022 delayed early mRNA synthesis (107, 152).
Hybrid Phages and the Campbell Model for Prophage Insertion
Interrupted mating experiments with Hfr × F− E. coli strains initially showed that prophages, including λ, 434, and 21, were located at specific sites on the bacterial chromosome, but the basis for the positioning was unclear. Two basic ideas were entertained at that time to explain how the prophage was associated with the circular bacterial chromosome. The prophage chromosome was somehow anchored so that the prophage resides at its chromosomal site while remaining a separate genetic element (5), or alternatively, as proposed by Allan Campbell, a site-specific reciprocal recombination event could be responsible for integrating a circularized phage DNA into the bacterial DNA, thus allowing the prophage to be replicated passively as part of the bacterial chromosome (153). Early support for the integration model included bacterial mating experiments between lysogens with genetic markers both in bacterial DNA and within the prophage itself (154). Such matings indicated that the gene order in the prophage was a circular permutation of that obtained from lytic phage crosses (155), which is a central prediction of the Campbell model, since the integration (attP) and cos sites lie at significantly different places in the phage genome. Additional evidence came from analysis of the genetic content of defective galactose-transducing phages, called λdg’s, which are hybrids between λ and the E. coli galactose utilization operon. Rationalizing the gene content of the λdg’s which carry the bacterial gal genes also required that attP on the phage DNA be located near the center of the linear lytic genetic map while cleavage at cos created the two ends of the lytic map (156, 157).
More detailed support for the Campbell model came from studies that utilized a ϕ80-λ hybrid prophage, as follows. In 1963, Matsushiro isolated the E. coli λ-like phage ϕ80, whose prophage is located near the tryptophan operon (6). In addition to having a different prophage location site from λ, ϕ80 differed from λ with respect to immunity and host range (6). The E. coli tonB gene (see Phage 21 Genome Promoter-Proximal Late Operon and Lysis Genes above), which activates the FhuA bacterial cell receptor for phages T1 and ϕ80, was also found to be near the ϕ80 attachment site and the trp operon. Naomi Franklin noted that some T1-resistant tonB– deletions extended into the trp genes and that hybrid λ-ϕ80 phages could be readily obtained (38, 158, 159). Putting this information together, Franklin et al. (38) isolated tonB deletions of a lysogen of a λ attP80 imm80 hybrid, and each deletion mutant could be rationalized as removing a single contiguous block of DNA provided that (i) the hybrid phage DNA was inserted into the bacterial chromosome and (ii) the crossover site for the insertional recombination event was located on the λ DNA molecule between genes J and N. These results with hybrid phages provided very strong credence for the idea that the genetic map of the lytically grown hybrid virion chromosome is a circular permutation of the prophage genome. The permuted map of the wild-type λ prophage was confirmed by characterizing deletions that removed parts of a λ prophage located at attB, its natural location between the gal and bio operons of E. coli. The chlD and chlA genes also flank attB—the gene order is gal-chlD-attB-bio-chlA. The chlD and chlA genes, since renamed modC and moaA, are involved in molybdate transport and molybdopterin biosynthesis. Enzymes dependent on the molybdopterin cofactor reduce nitrate and chlorate. Chlorate reduction generates a lethally toxic derivative (160). Chlorate-resistant mutants with deletions extending into the prophage confirmed the prophage’s circular permutation relative to the virion chromosome and defined the integration sites in the phage and host chromosomes (161).
HYBRID PHAGES AND PROTEIN FUNCTIONAL DOMAINS
Other hybrid phages selected or constructed in various laboratories allowed the discovery and subsequent analyses of specificity domains in phage proteins. Here we present several examples.
DNA Replication
Working out the basics of λ DNA replication paralleled studies with E. coli as well as phages T7 and T4. Infections of E. coli replication mutants under nonpermissive conditions showed that λ replication was dependent on the host’s DNA synthesis elongation functions but that the initiation proteins DnaA and DnaC were not required (162). In addition, a survey of Campbell’s (132) nonsense mutants turned up two essential genes, O and P, whose products were absolutely essential for λ DNA replication (163). Both λ and E. coli initiate bidirectional DNA replication at a unique chromosomal site, oriλ and oriC, respectively (164, 165). E. coli’s replication starts when oriC is bound by DnaA initiator protein. Following this, DnaC escorts the replicative helicase, DnaB, to the oriC-DnaA complex, thus allowing the DnaG primase, bound to DnaB, to synthesize RNA primers that are extended by the PolIII polymerase throughout the DNA elongation stage (166).
λ's replication strategy involves hijacking E. coli’s replication elongation machinery, DnaB, DnaG, and PolIII as well as other proteins (166). Clever genetic selection for prophages with replication defects generated prophage mutants defective in oriλ, and early sequencing showed that oriλ resides in the 5′ portion of the O gene (reviewed in reference 167). Hybrid phages were critical for understanding how λ commandeers DnaB and the rest of E. coli’s replication machinery, and a landmark example is the analysis of the replication initiation O protein of λ (104). Hybrids of λ with either ϕ80 or phage 82 enabled the teasing out of species-specific domains in the O protein, as follows. Although neither ϕ80 nor 82 O protein can complement a λ O mutant, viable hybrids formed by a crossover within O could be selected. For example, λ imm80 hy42 contains ϕ80 DNA from the immunity region to a point within the O gene that includes oriϕ80 (168). λ imm80 hy42 does have a functional O gene, because growth of λ imm80 hy42 O–amber29 requires a host with an amber suppressor for plaque formation, and complementation experiments with ϕ80, λ, and their hybrids showed that λ imm80 hy42 O–amber29 could only be helped to replicate under nonsuppressing conditions if the helper phage carried the N-terminal half of ϕ80’s O gene. Since the N-terminal half of the O gene contains the ori site in both phages, these results indicated a phage species-specific interaction between oriϕ80 and the N-terminal half of the ϕ80 O-like protein.
What about the C-terminal portion of O protein? Some background observations follow. (i) Tomizawa showed that a mutation causing the O protein to be thermolabile could be suppressed by allele-specific missense mutations affecting the P protein, suggesting that O and P proteins physically interact (169). (ii) λ P protein recruits the host DnaB to the replication complex (170). Finally, (iii) λ’s O protein can function with the P protein of ϕ80, but λ’s P protein cannot function with ϕ80’s O protein (168). An informative hybrid phage, λ immλ repλ:80, was the reciprocal of λ imm80 hy42 in that it produced a chimeric O protein whose N-terminal half, including ori, was from λ and the C-terminal half was from ϕ80. The ϕ80 segment in the hybrid also contained ϕ80’s P gene. Given that λ's O protein cannot function with ϕ80’s P protein, the results indicated the presence of a phage-specific P-interacting domain near the O protein’s C terminus (168). These hybrid phage studies demonstrated two separable specificity domains in O protein, an N-terminal ori-interacting domain and a C-terminal domain for binding P protein (Fig. 6A).
FIG 6.
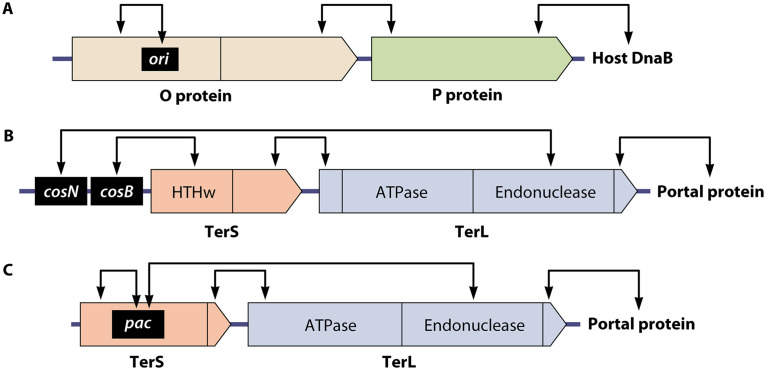
Phage-specific protein domain interactions deduced through use of hybrid phages. Pointed boxes represent the genes involved and their encoded proteins with their N termini on the left (not drawn to scale). The points on the boxes indicate the direction of transcription. Vertical bars within the boxes denote the boundaries between protein functional domains, and arrows above each map indicate species-specific protein-protein or protein-DNA interactions. (A) Specificity domains in λ O protein. The black rectangle indicates the location of the replication origin (ori). The N-terminal portion of λ O protein specifically binds to the phage replication origin, ori, and the C-terminal part interacts with λ P protein, which in turn interacts with the host’s replication machinery (adapted from Furth et al. [104]). (B) Specificity domains in λ terminase. The TerS N-terminal portion contains a winged helix-turn-helix DNA binding domain (HTHw) that interacts with the DNA packaging recognition site cosB. The TerS C-terminal domain interacts in a species-specific manner with the N-terminal region of TerL, and the C terminus of TerL, in turn, is a specificity domain for interaction with the procapsid’s portal protein (190–192). (C) Specificity domains of the P22 terminase. The black rectangle indicates the packaging recognition site pac (~20 bp, codons 90 to 96 of the 162-codon gene). The interactions parallel those of λ, except that the pac site is inside the terS gene (174, 187; E. Gilcrease and S. Casjens, unpublished results).
DNA Packaging
Although phages λ and 21 share 52% and 64% sequence identity in TerS and TerL, respectively, neither virus packages the other’s DNA, and the TerS-TerL as well as TerL-procapsid interactions are both phage specific (20). Study of λ-21 hybrids with chimeric TerS or TerL subunits identified four phage-specific protein domains. The λ TerS binds to an approximately 15-bp-long DNA recognition sequence, which is repeated three times in cosB, which is adjacent to cosN, the site where cohesive ends are generated. A λ-21 hybrid phage was isolated with a chimeric TerS subunit, and it showed that cosB specificity resides in TerS’s N-terminal half (171) (Fig. 6B). TerS’s N-terminal domain was later found to contain a winged helix-turn-helix DNA binding motif (HTHw) (172). The λ and 21 cosB sequences and TerS HTHw motifs contain nucleotide sequence differences that account for their phage-specific DNA recognition (173). The C-terminal half of TerS likewise has species-specific interactions with TerL, and other hybrid phages showed that a chimeric TerS with its N terminus from 21 and its C terminus from λ formed a functional terminase complex with λ TerL but not 21 TerL (171). Still other hybrid phages showed that the TerL’s N-terminal 48 aa contain a specificity domain for interacting with TerS (174), and TerL’s C-terminal 32 aa make a phage-specific interaction with the portal protein of the procapsid (175–177). These λ hybrid phages were selected by taking advantage of natural homologous recombination processes between similar but nonidentical sequences in the two parents. Similar functional hybrids between phages P22 and Sf6, which have much more divergent TerS and TerL proteins (only 11.7% and 13.4% identity between the two phages, respectively), have been engineered in the laboratory and have given very similar conclusions (see Serendipity or “Evolutionary Rationale”? below).
OTHER USES OF HYBRID PHAGES
Phage λ hybrids had many important purely practical uses in addition to their roles in the studies described above, and one example follows. Like any other phage, λ depends heavily upon host functions for carrying out the intracellular steps of its life cycle. Understanding such host functions was critical in deducing the mechanism by which N causes transcription antitermination. The host factors required for N protein function were initially discovered by identifying and characterizing mutations that affect host functions needed for λ lytic growth (reviewed by Friedman et al. [146] and Georgopoulos [178]). Such mutations revealed that host proteins NusA, NusB, NusC, NusE, and NusG are all required for N protein to act on RNA polymerase. In hunts for host mutants that block λ intracellular development, more common genetic changes that block phage adsorption must be avoided, and in the first successful strategies, these were avoided by screening for bacteria that disallow infection by both λ and a λ hybrid that utilizes a different receptor (152, 179). Phage λ adsorbs to host LamB protein, while λ hybrids h434 and hHK022 adsorb to the host cell surface OmpC and FhuA protein, respectively. The latter two phages were essentially identical to λ except for their host adsorption properties. Thus, any host mutation that affected intracellular lytic growth would affect both phages, but it would require a very rare double mutant to inactivate the receptors of both phages. This is but one example of the many ways in which λ hybrid phages were used as practical tools to attain the ends desired by experimenters.
WILD HYBRID PHAGES IN NATURE
Formation and Exchange of Mosaic Sections
New mosaic boundaries (novel sequence joints between mosaic sections or modules) in phage genomes are thought to arise by rare random nonhomologous recombination between divergent but related phage genomes (23, 24). If the parental phages have syntenic genomes and if such a recombination occurs “in register” on the genetic map, the hybrid phage resulting from a single crossover could be a fully functional virus with all the necessary genes. The number of phages on Earth is large enough that the loss of all “out-of-register” (incomplete or too large a genome to be packaged) recombinants, and perhaps the loss of most of the “in-phase” recombinants by negative selection, still leaves functional hybrids that can survive in the environment. One might imagine that such functional hybrids can also be generated by imperfect, “nearly in-register” recombination events that result in functional hybrid genomes with relic duplicated genes or gene fragments at the newly formed mosaic boundary. Juhala et al. (26) pointed out apparently defective, truncated N gene and head-tail joining gene relics in the genomes of HK97 and HK022, respectively, and we note here that phage 434 has two such entities, nun and Q gene fragments, annotated as genes 48 and 64, respectively. It cannot be absolutely proven that these gene fragments arose by out-of-register nonhomologous recombination rather than by other types of rearrangement. However, the fact that each of the above gene fragments is adjacent to a (divergent) cognate whole gene (note that N and nun are allelic alternatives) suggests that the former is likely to be the case. Thus, new mosaic boundaries form faster than ameliorative processes remove the excess nonfunctional DNA associated with imprecise module boundary formation events. Once formed, mosaic sections can be rapidly shuffled within a group of related phages such as the λ-like phages by homologous recombination between patches of similar sequence. The hybrid phages discussed above, λ imm21 and λ imm434, were clearly formed by events of the latter, homologous type.
Mosaic Section/Module Diversity
The chromosomes of λ-like phages isolated from the environment are mosaics of functional modules assembled from a large and extremely varied global pool of modules (180–183). For example, comparison of the 15 contiguous virion assembly genes of 700 P22-like phages found that they contain 27 naturally occurring mosaic sections that correspond to whole genes, gene fragments, and clusters of genes (25). Most of the 27 segments have multiple, very different “allele” types. For example, in the Salmonella P22-like phages, the most diverse segment, the receptor binding domain of the tail spike, has 18 extremely different sequence types that correspond to different host surface polysaccharide receptors (184), and there are over 80 (!) essentially unrelated sequence types if P22-like phages that infect other Enterobacterales host species are considered (S. Casjens, unpublished data). Finally, there are 47 different combinations of mosaic segment allele types in this gene cluster among 57 P22 genomes that were examined in detail (25). Although they have not been examined in such detail, the λ-like phage genomes also have highly mosaic relationships in their early regions (26, 47, 181).
There are currently over 300 λ-like phage genomes in the public database, and a majority of them are mosaically related to each other and have unique mosaic section allele combinations (S. Casjens, unpublished data). These alleles appear to move among the λ-like phages rather freely and are projected to create a huge number of potential allele combinations. This population structure and these dynamics make it seem likely that exchange of mosaic sections is beneficial to these phages; however, it is not known whether a newly formed particular mosaic segment allele combination confers a functional advantage in one of many specific (and perhaps changing?) environmental niches or whether diversity per se is valuable, but we note that the latter could help avoid CRISPR attack. We also note that if this population’s long-term survival depends on mosaic sectional shuffling, it would be advantageous if all participating members had the same gene order, since in that case a single recombination event can create a new functional hybrid, so module exchange could be one of the pressures to retain synteny. In such an evolutionary landscape, the λ-like phages are continuously exchanging modules through recombination with divergent relatives, and they can be viewed as a complex population of natural hybrid phages.
SERENDIPITY OR “EVOLUTIONARY RATIONALE”?
Do new mosaic boundary generation and shuffling events depend on luck to create hybrids with functional module combinations, or have modules evolved to be exchangeable? We approach this question by way of an example, the terminase genes of the P22-like phages. P22 and its relatives are headful DNA packaging phages (185, 186). Their packaging recognition (pac) site is recognized by the TerS N-terminal domain, and the pac site is embedded within the terS N-terminal domain coding sequence (Fig. 6C) (187). This arrangement, like the placement of oriλ within the O gene’s N-terminal half (see above), is parsimonious since the DNA binding domain of the protein and its target site cannot be separated by a recombination event to produce a TerS-pac specificity mismatch. Among 152 P22-like phage genomes examined, Leavitt et al. (187) found that the TerS pac site selecting domains of the P22-like phages comprise six very different sequence types. The smaller C-terminal regions of these TerS’s are also diverse, with four sequence types. No other mosaic segment boundaries were found within terS, suggesting that there are only two exchangeable parts in TerS. Structural analyses of TerS proteins from two of the groups, those typified by phages P22 and Sf6, confirmed that mosaic boundaries correspond to protein structural domain boundaries (188, 189). P22 and Sf6 TerS and TerL proteins are very different in the two phages, have different DNA packaging specificities, and cannot substitute for one another. Experiments with P22-Sf6 hybrid phages (constructed by DNA engineering) showed that the C-terminal 26 aa of P22 TerS are sufficient to allow the phage Sf6 TerS N-terminal pac site-selecting domain to function with P22 TerL (187), and a reciprocal construct showed that the C-terminal portion of Sf6 TerS allows the N-terminal domain of P22 TerS to function with Sf6 TerL (E. Gilcrease and S. Casjens, unpublished data). Similarly, a natural mosaic boundary was identified within and near the C terminus of the terL gene of the P22-like phages, and experimental tests similar to those described above used engineered hybrid phages to show that the C-terminal 24 aa of P22 TerL are sufficient to allow Sf6 TerL to function with P22 portal protein in P22 procapsids (E. Gilcrease and S. Casjens, unpublished data). The apparently random shuffling of the different types of TerS and TerL domains in nature (187) and the properties of the above-mentioned reciprocal functional TerS constructs (combinations not yet found in nature) suggest that these mosaic sections may have been evolutionarily “designed” to be able to function autonomously and thus increase the probability that they will be able to function properly with very different partners. This notion agrees with the above-mentioned postulate that it is evolutionarily useful for these phages to be able to exchange modules.
The successes of laboratory uses of the hybrid phages discussed above may seem somewhat serendipitous, since, for example, if the repressors’ operators, the cro gene or the N gene, had been located elsewhere, either the immunity sections of such hybrids would have to have been much larger and thus been less informative or the hybrids would not have been able to establish and maintain lysogeny and so would not have been isolated as useful temperate hybrids. But, if individual mosaic sections or modules such as the immunity segments evolved to be functionally autonomous, the exact placement of the phage’s regulatory components may not be due to simple luck but at least in part to evolutionary forces that ensure that mosaically related, naturally formed hybrids are often fully functional. And if this is true, it is no surprise that, for example, the original λ imm21 and λ imm434 recombinants are fully functional phages.
ACKNOWLEDGMENTS
We dedicate this publication to our late friend and colleague Roger Hendrix and to the late A. Dale Kaiser, Noreen E. Murray, Naomi Franklin, and Allan M. Campbell, without whose pioneering insights and studies the use of phage λ as an early model for molecular biological processes may never have happened.
We thank all of our colleagues for decades of productive and enjoyable scientific interactions in the phage research field and, in particular, Alan Davidson for providing unpublished information on the start codon of phage λ gene K. We regret that due to space limitations, valuable contributions have been left undiscussed.
This work was supported by grants NIH R01 GM114817 to S.R.C., NIH R35 GM136396 to R.Y., and NIH G35 GM131729 to G.F.H. M.F. was supported by the Temperate Phage Fund of the University of Iowa Foundation, and S.A. was partially supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute, and the Center for Cancer Research.
We dedicate this publication to our late friend and colleague Roger Hendrix and to the late A. Dale Kaiser, Noreen E. Murray, Naomi Franklin, and Allan M. Campbell, without whose pioneering insights and studies the use of phage λ as an early model for molecular biological processes may never have happened.
Footnotes
Supplemental material is available online only.
Contributor Information
Michael Feiss, Email: michael-feiss@uiowa.edu.
Sherwood R. Casjens, Email: sherwood.casjens@path.utah.edu.
REFERENCES
- 1.Lederberg EM. 1950. Lysogenicity in Escherichia coli strain K-12. Microb Genet Bull 1:5–9. [Google Scholar]
- 2.Dove WF. 1969. Strains of phage lambda in current use. Virology 38:349–351. doi: 10.1016/0042-6822(69)90378-x. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser AD. 1955. A genetic study of the temperate coliphage. Virology 1:424–443. doi: 10.1016/0042-6822(55)90036-2. [DOI] [PubMed] [Google Scholar]
- 4.Hendrix RW, Duda RL. 1992. Bacteriophage lambda PaPa: not the mother of all lambda phages. Science 258:1145–1148. doi: 10.1126/science.1439823. [DOI] [PubMed] [Google Scholar]
- 5.Jacob F, Wollman EL. 1961. Sexuality and the genetics of bacteria. Academic Press, New York, NY. [Google Scholar]
- 6.Matsushiro A. 1963. Specialized transduction of tryptophan markers in Escherichia coli K12 by bacteriophage ϕ80. Virology 19:475–482. doi: 10.1016/0042-6822(63)90041-2. [DOI] [PubMed] [Google Scholar]
- 7.Dhillon TS, Dhillon EK. 1972. Studies on bacteriophage distribution. II. Isolation and host range based classification of phages active on three species of Enterobacteriaceae. Jpn J Microbiol 16:297–306. doi: 10.1111/j.1348-0421.1972.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 8.Zinder ND, Lederberg J. 1952. Genetic exchange in Salmonella. J Bacteriol 64:679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bezdeh M, Amati P. 1967. Properties of P22 and a related Salmonella typhimurium phage. I. General features and host specificity. Virology 31:272–278. doi: 10.1016/0042-6822(67)90171-7. [DOI] [PubMed] [Google Scholar]
- 10.Casjens SR, Hendrix RW. 2015. Bacteriophage lambda: early pioneer and still relevant. Virology 479–480:310–330. doi: 10.1016/j.virol.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randall-Hazelbauer L, Schwartz M. 1973. Isolation of the bacteriophage lambda receptor from Escherichia coli. J Bacteriol 116:1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hantke K. 1978. Major outer membrane proteins of E. coli K12 serve as receptors for the phages T2 (protein Ia) and 434 (protein Ib). Mol Gen Genet 164:131–135. doi: 10.1007/BF00267377. [DOI] [PubMed] [Google Scholar]
- 13.Destoumieux-Garzón D, Duquesne S, Peduzzi J, Goulard C, Desmadril M, Letellier L, Rebuffat S, Boulanger P. 2005. The iron-siderophore transporter FhuA is the receptor for the antimicrobial peptide microcin J25: role of the microcin Val11-Pro16 beta-hairpin region in the recognition mechanism. Biochem J 389:869–876. doi: 10.1042/BJ20042107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwashita S, Kanegasaki S. 1976. Enzymic and molecular properties of base-plate parts of bacteriophage P22. Eur J Biochem 65:87–94. doi: 10.1111/j.1432-1033.1976.tb10392.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Yang CH, Lee G, Chang F, Wilson H, del Campillo-Campbell A, Campbell A. 1997. Integration specificities of two lambdoid phages (21 and e14) that insert at the same attB site. J Bacteriol 179:5705–5711. doi: 10.1128/jb.179.18.5705-5711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider SJ. 1992. Site-specific recombination of lambdoid phage 21 into the icd gene of Escherichia coli. Ph.D. thesis. Stanford University, Stanford, CA. [Google Scholar]
- 17.Feiss M, Young R, Ramsey J, Adhya S, Georgopoulos C, Hendrix R, Hatfull G, Gilcrease E, Casjens S. 2022. The complete genome sequences of lambdoid phages 21, 434, 434B, and several lambda hybrids. Microbiol Resour Announc 11:e00120-22. doi: 10.1128/mra.00120-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon M, Davis R, Davidson N. 1971. Heteroduplexes of DNA molecules of lambdoid phages: physical mapping of their base sequence relationships by electron microscopy, p 313–328. In Hershey AD (ed), The bacteriophage lambda. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 19.Feiss M. 1986. Terminase and the recognition, cutting and packaging of λ chromosomes. Trends Genet 2:100–104. doi: 10.1016/0168-9525(86)90193-9. [DOI] [Google Scholar]
- 20.Siegele DA, Frackman S, Sippy J, Momany T, Howard TM, Tilly K, Georgopoulos C, Feiss M. 1983. The head genes of bacteriophage 21. Virology 129:484–489. doi: 10.1016/0042-6822(83)90187-3. [DOI] [PubMed] [Google Scholar]
- 21.Wendt JL, Feiss M. 2004. A fragile lattice: replacing bacteriophage lambda's head stability gene D with the shp gene of phage 21 generates the Mg2+-dependent virus, lambda shp. Virology 326:41–46. doi: 10.1016/j.virol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 22.Casjens S. 1974. Bacteriophage lambda FII gene protein: role in head assembly. J Mol Biol 90:1–20. doi: 10.1016/0022-2836(74)90252-6. [DOI] [PubMed] [Google Scholar]
- 23.Casjens SR. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol 8:451–458. doi: 10.1016/j.mib.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Hendrix RW. 2002. Bacteriophages: evolution of the majority. Theor Popul Biol 61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- 25.Casjens SR, Thuman-Commike PA. 2011. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- 26.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol 299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell KL, Yee AA, Arrowsmith CH, Gold M, Davidson AR. 2002. The solution structure of the bacteriophage lambda head-tail joining protein, gpFII. J Mol Biol 318:1395–1404. doi: 10.1016/s0022-2836(02)00276-0. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Hofnung M, Charbit A. 2000. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J Bacteriol 182:508–512. doi: 10.1128/JB.182.2.508-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Werts C, Michel V, Hofnung M, Charbit A. 1994. Adsorption of bacteriophage lambda on the LamB protein of Escherichia coli K-12: point mutations in gene J of lambda responsible for extended host range. J Bacteriol 176:941–947. doi: 10.1128/jb.176.4.941-947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casjens S, Hendrix R. 1988. Control mechanisms in dsDNA bacteriophage assembly, p 15–91. In Calendar R (ed), The bacteriophages. Plenum Press, New York, NY. [Google Scholar]
- 31.Kellenberger G, Zichichi M, Weigle J. 1960. Mutations affecting the density of bacteriophage λ. Nature 187:161–162. doi: 10.1038/187161a0. [DOI] [Google Scholar]
- 32.Benchimol S, Lucko H, Becker A. 1982. A novel endonuclease specified by bacteriophage lambda. Purification and properties of the enzyme. J Biol Chem 257:5211–5219. doi: 10.1016/S0021-9258(18)34657-X. [DOI] [PubMed] [Google Scholar]
- 33.Benchimol S, Becker A, Murialdo H, Gold M. 1978. Role of the bacteriophage-lambda FI gene product during phage head assembly in vitro. Virology 91:205–221. doi: 10.1016/0042-6822(78)90370-7. [DOI] [PubMed] [Google Scholar]
- 34.Hendrix R. 1970. Proteins of bacteriophage lambda. Ph.D. thesis. Harvard University, Cambridge MA. [Google Scholar]
- 35.Oppenheim A, Oppenheim AB. 1978. Regulation of the int gene of bacteriophage lambda: activation by the cII and cIII gene products and the role of the PI and Pl promoters. Mol Gen Genet 165:39–46. doi: 10.1007/BF00270374. [DOI] [PubMed] [Google Scholar]
- 36.Zhou D, Hardt W-D, Galán JE. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect Immun 67:1974–1981. doi: 10.1128/IAI.67.4.1974-1981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaharik ML, Cullen VL, Fung AM, Libby SJ, Kujat Choy SL, Coburn B, Kehres DG, Maguire ME, Fang FC, Finlay BB. 2004. The Salmonella enterica serovar Typhimurium divalent cation transport systems MntH and SitABCD are essential for virulence in an Nramp1 G169 murine typhoid model. Infect Immun 72:5522–5525. doi: 10.1128/IAI.72.9.5522-5525.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franklin NC, Dove WF, Yanofsky C. 1965. The linear insertion of a prophage into the chromosome of E. coli shown by deletion mapping. Biochem Biophys Res Commun 18:910–923. doi: 10.1016/0006-291X(65)90868-5. [DOI] [Google Scholar]
- 39.Gottesman M, Weisberg RA. 1971. Prophage insertion and excision. p 113–138. In Hershey AD (ed), The bacteriophage lambda. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40.Hilliker S, Botstein D. 1976. Specificity of genetic elements controlling regulation of early functions in temperate bacteriophages. J Mol Biol 106:537–566. doi: 10.1016/0022-2836(76)90251-5. [DOI] [PubMed] [Google Scholar]
- 41.Couturier M, Dambly C, Thomas R. 1973. Control of development in temperate bacteriophages. Mol Gen Genet 120:231–252. doi: 10.1007/BF00267155. [DOI] [PubMed] [Google Scholar]
- 42.Botstein D, Herskowitz I. 1974. Properties of hybrids between Salmonella phage P22 and coliphage λ. Nature 251:584–589. doi: 10.1038/251584a0. [DOI] [PubMed] [Google Scholar]
- 43.Thomason LC, Schiltz CJ, Court C, Hosford CJ, Adams MC, Chappie JS, Court DL. 2021. Bacteriophage λ RexA and RexB functions assist the transition from lysogeny to lytic growth. Mol Microbiol 116:1044–1063. doi: 10.1111/mmi.14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karch H, Schmidt H, Janetzki-Mittmann C, Scheef J, Kröger M. 1999. Shiga toxins even when different are encoded at identical positions in the genomes of related temperate bacteriophages. Mol Gen Genet 262:600–607. doi: 10.1007/s004380051122. [DOI] [PubMed] [Google Scholar]
- 45.Guo H-C, Kainz M, Roberts J. 1991. Characterization of the late-gene regulatory region of phage 21. J Bacteriol 173:1554–1560. doi: 10.1128/jb.173.4.1554-1560.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonovich MT, Young R. 1991. Dual start motif in two lambdoid S genes unrelated to lambda S. J Bacteriol 173:2897–2905. doi: 10.1128/jb.173.9.2897-2905.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grose JH, Casjens SR. 2014. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology 468–470:421–443. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin Z, Kaelber JT, Ebright RH. 2019. Structural basis of Q-dependent antitermination. Proc Natl Acad Sci USA 116:18384–18390. doi: 10.1073/pnas.1909801116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaworski J, McClung C, Ruse C, Weigele PR, Hendrix RW, Ko C-C, Edgar R, Hatfull GF, Casjens SR, Raleigh EA. 2021. Genome analysis of Salmonella enterica serovar Typhimurium bacteriophage L, indicator for StySA (StyLT2III) restriction-modification system action. G3 (Bethesda) 11:jkaa037. doi: 10.1093/g3journal/jkaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casjens S, Winn-Stapley DA, Gilcrease EB, Morona R, Kühlewein C, Chua JE, Manning PA, Inwood W, Clark AJ. 2004. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J Mol Biol 339:379–394. doi: 10.1016/j.jmb.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 51.Park T, Struck DK, Dankenbring CA, Young R. 2007. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J Bacteriol 189:9135–9139. doi: 10.1128/JB.00847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dewey JS, Savva CG, White RL, Vitha S, Holzenburg A, Young R. 2010. Micron-scale holes terminate the phage infection cycle. Proc Natl Acad Sci USA 107:2219–2223. doi: 10.1073/pnas.0914030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White R, Tran TA, Dankenbring CA, Deaton J, Young R. 2010. The N-terminal transmembrane domain of lambda S is required for holin but not antiholin function. J Bacteriol 192:725–733. doi: 10.1128/JB.01263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park T, Struck DK, Deaton JF, Young R. 2006. Topological dynamics of holins in programmed bacterial lysis. Proc Natl Acad Sci USA 103:19713–19718. doi: 10.1073/pnas.0600943103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pang T, Savva CG, Fleming KG, Struck DK, Young R. 2009. Structure of the lethal phage pinhole. Proc Natl Acad Sci USA 106:18966–18971. doi: 10.1073/pnas.0907941106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evrard C, Fastrez J, Declercq JP. 1998. Crystal structure of the lysozyme from bacteriophage lambda and its relationship with V and C-type lysozymes. J Mol Biol 276:151–164. doi: 10.1006/jmbi.1997.1499. [DOI] [PubMed] [Google Scholar]
- 57.Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R. 2005. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 307:113–7117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- 58.Berry J, Rajaure M, Pang T, Young R. 2012. The spanin complex is essential for lambda lysis. J Bacteriol 194:5667–5674. doi: 10.1128/JB.01245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Summer EJ, Berry J, Tran TA, Niu L, Struck DK, Young R. 2007. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J Mol Biol 373:1098–1112. doi: 10.1016/j.jmb.2007.08.045. [DOI] [PubMed] [Google Scholar]
- 60.Rajaure M, Berry J, Kongari R, Cahill J, Young R. 2015. Membrane fusion during phage lysis. Proc Natl Acad Sci USA 112:5497–5502. doi: 10.1073/pnas.1420588112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kongari R, Rajaure M, Cahill J, Rasche E, Mijalis E, Berry J, Young R. 2018. Phage spanins: diversity, topological dynamics and gene convergence. BMC Bioinformatics 19:326. doi: 10.1186/s12859-018-2342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barondess JJ, Beckwith J. 1995. bor gene of phage lambda, involved in serum resistance, encodes a widely conserved outer membrane lipoprotein. J Bacteriol 177:1247–1253. doi: 10.1128/jb.177.5.1247-1253.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarver JL, Zhang M, Liu L, Nyenhuis D, Cafiso DS. 2018. A dynamic protein-protein coupling between the TonB-dependent transporter FhuA and TonB. Biochemistry 57:1045–1053. doi: 10.1021/acs.biochem.7b01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howard SP, Herrmann C, Stratilo CW, Braun V. 2001. In vivo synthesis of the periplasmic domain of TonB inhibits transport through the FecA and FhuA iron siderophore transporters of Escherichia coli. J Bacteriol 183:5885–5895. doi: 10.1128/JB.183.20.5885-5895.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vica Pacheco S, García González O, Paniagua Contreras GL. 1997. The lom gene of bacteriophage λ is involved in Escherichia coli K12 adhesion to human buccal epithelial cells. FEMS Microbiol Lett 156:129–132. doi: 10.1016/S0378-1097(97)00415-1. [DOI] [PubMed] [Google Scholar]
- 66.Smith MC, Burns N, Sayers JR, Sorrell JA, Casjens SR, Hendrix RW, Engel J. 1998. Bacteriophage collagen. Science 279:1831–1831. doi: 10.1126/science.279.5358.1831g. [DOI] [PubMed] [Google Scholar]
- 67.Wickham ME, Lupp C, Mascarenhas M, Vazquez A, Coombes BK, Brown NF, Coburn BA, Deng W, Puente JL, Karmali MA, Finlay BB. 2006. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J Infect Dis 194:819–827. doi: 10.1086/506620. [DOI] [PubMed] [Google Scholar]
- 68.Brown NF, Coombes BK, Bishop JL, Wickham ME, Lowden MJ, Gal-Mor O, Goode DL, Boyle EC, Sanderson KL, Finlay BB. 2011. Salmonella phage ST64B encodes a member of the SseK/NleB effector family. PLoS One 6:e17824. doi: 10.1371/journal.pone.0017824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scott NE, Giogha C, Pollock GL, Kennedy CL, Webb AI, Williamson NA, Pearson JS, Hartland EL. 2017. The bacterial arginine glycosyltransferase effector NleB preferentially modifies Fas-associated death domain protein (FADD). J Biol Chem 292:17337–17350. doi: 10.1074/jbc.M117.805036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pallett MA, Berger CN, Pearson JS, Hartland EL, Frankel G. 2014. The type III secretion effector NleF of enteropathogenic Escherichia coli activates NF-κB early during infection. Infect Immun 82:4878–4888. doi: 10.1128/IAI.02131-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Creuzburg K, Giogha C, Wong Fok Lung T, Scott NE, Mühlen S, Hartland EL, Pearson JS. 2017. The type III effector NleD from enteropathogenic Escherichia coli differentiates between host substrates p38 and JNK. Infect Immun 85:e00620-16. doi: 10.1128/IAI.00620-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marchés O, Wiles S, Dziva F, La Ragione RM, Schüller S, Best A, Phillips AD, Hartland EL, Woodward MJ, Stevens MP, Frankel G. 2005. Characterization of two non-locus of enterocyte effacement-encoded type III-translocated effectors, NleC and NleD, in attaching and effacing pathogens. Infect Immun 73:8411–8417. doi: 10.1128/IAI.73.12.8411-8417.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mascarenhas D, Kelley R, Campbell A. 1981. DNA sequence of the att region of coliphage 434. Gene 15:151–156. doi: 10.1016/0378-1119(81)90124-4. [DOI] [PubMed] [Google Scholar]
- 74.Schindler D, Echols H. 1981. Retroregulation of the int gene of bacteriophage lambda: control of translation completion. Proc Natl Acad Sci USA 78:4475–4479. doi: 10.1073/pnas.78.7.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guarneros G, Montañez C, Hernandez T, Court D. 1982. Posttranscriptional control of bacteriophage lambda gene expression from a site distal to the gene. Proc Natl Acad Sci USA 79:238–242. doi: 10.1073/pnas.79.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anderson JE, Ptashne M, Harrison SC. 1985. A phage repressor-operator complex at 7 Å resolution. Nature 316:596–601. doi: 10.1038/316596a0. [DOI] [PubMed] [Google Scholar]
- 77.Wharton RP, Brown EL, Ptashne M. 1984. Substituting an alpha-helix switches the sequence-specific DNA interactions of a repressor. Cell 38:361–369. doi: 10.1016/0092-8674(84)90491-4. [DOI] [PubMed] [Google Scholar]
- 78.Shkilnyj P, Colon MP, Koudelka GB. 2013. Bacteriophage 434 Hex protein prevents recA-mediated repressor autocleavage. Viruses 5:111–126. doi: 10.3390/v5010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Susskind MM, Botstein D. 1980. Superinfection exclusion by lambda prophage in lysogens of Salmonella typhimurium. Virology 100:212–216. doi: 10.1016/0042-6822(80)90571-1. [DOI] [PubMed] [Google Scholar]
- 80.Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CRH, Trent MS. 2006. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3'-acyloxyacyl moiety of lipid A. J Biol Chem 281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawasaki K, Ernst RK, Miller SI. 2004. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J Biol Chem 279:20044–20048. doi: 10.1074/jbc.M401275200. [DOI] [PubMed] [Google Scholar]
- 82.Kawano M, Manabe T, Kawasaki K. 2010. Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation enhances its intracellular growth within macrophages. FEBS Lett 584:207–212. doi: 10.1016/j.febslet.2009.11.062. [DOI] [PubMed] [Google Scholar]
- 83.Asakura M, Hinenoya A, Alam MS, Shima K, Zahid SH, Shi L, Sugimoto N, Ghosh AN, Ramamurthy T, Faruque SM, Nair GB, Yamasaki S. 2007. An inducible lambdoid prophage encoding cytolethal distending toxin (Cdt-I) and a type III effector protein in enteropathogenic Escherichia coli. Proc Natl Acad Sci USA 104:14483–14488. doi: 10.1073/pnas.0706695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang XJ, Goliger JA, Roberts JW. 1989. Specificity and mechanism of antitermination by Q proteins of bacteriophages lambda and 82. J Mol Biol 210:453–460. doi: 10.1016/0022-2836(89)90122-8. [DOI] [PubMed] [Google Scholar]
- 85.Goliger JA, Roberts JW. 1989. Sequences required for antitermination by phage 82 Q protein. J Mol Biol 210:461–471. doi: 10.1016/0022-2836(89)90123-X. [DOI] [PubMed] [Google Scholar]
- 86.Goliger JA, Roberts JW. 1987. Bacteriophage 82 gene Q and Q protein. Sequence, overproduction, and activity as a transcription antiterminator in vitro. J Biol Chem 262:11721–11725. doi: 10.1016/S0021-9258(18)60870-1. [DOI] [PubMed] [Google Scholar]
- 87.Nübling S, Eisele T, Stöber H, Funk J, Polzin S, Fischer L, Schmidt H. 2014. Bacteriophage 933W encodes a functional esterase downstream of the Shiga toxin 2a operon. Int J Med Microbiol 304:269–274. doi: 10.1016/j.ijmm.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 88.Saile N, Schwarz L, Eißenberger K, Klumpp J, Fricke FW, Schmidt H. 2018. Growth advantage of Escherichia coli O104:H4 strains on 5-N-acetyl-9-O-acetyl neuraminic acid as a carbon source is dependent on heterogeneous phage-borne nanS-p esterases. Int J Med Microbiol 308:459–468. doi: 10.1016/j.ijmm.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 89.Feuerbaum S, Saile N, Pohlentz G, Müthing J, Schmidt H. 2018. De-O-acetylation of mucin-derived sialic acids by recombinant NanS-p esterases of Escherichia coli O157:H7 strain EDL933. Int J Med Microbiol 308:1113–1120. doi: 10.1016/j.ijmm.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saile N, Voigt A, Kessler S, Stressler T, Klumpp J, Fischer L, Schmidt H. 2016. Escherichia coli O157:H7 strain EDL933 harbors multiple functional prophage-associated genes necessary for the utilization of 5-N-acetyl-9-O-acetyl neuraminic acid as a growth substrate. Appl Environ Microbiol 82:5940–5950. doi: 10.1128/AEM.01671-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Plunkett G, III, Rose DJ, Durfee TJ, Blattner FR. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J Bacteriol 181:1767–1778. doi: 10.1128/JB.181.6.1767-1778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kyle JL, Cummings CA, Parker CT, Quiñones B, Vatta P, Newton E, Huynh S, Swimley M, Degoricija L, Barker M, Fontanoz S, Nguyen K, Patel R, Fang R, Tebbs R, Petrauskene O, Furtado M, Mandrell RE. 2012. Escherichia coli serotype O55:H7 diversity supports parallel acquisition of bacteriophage at Shiga toxin phage insertion sites during evolution of the O157:H7 lineage. J Bacteriol 194:1885–1896. doi: 10.1128/JB.00120-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lindsey DF, Mullin DA, Walker JR. 1989. Characterization of the cryptic lambdoid prophage DLP12 of Escherichia coli and overlap of the DLP12 integrase gene with the tRNA gene argU. J Bacteriol 171:6197–6205. doi: 10.1128/jb.171.11.6197-6205.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Highton PJ, Chang Y, Marcotte WR, Jr, Schnaitman CA. 1985. Evidence that the outer membrane protein gene nmpC of Escherichia coli K-12 lies within the defective qsr' prophage. J Bacteriol 162:256–262. doi: 10.1128/jb.162.1.256-262.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mondragón A, Subbiah S, Almo SC, Drottar M, Harrison SC. 1989. Structure of the amino-terminal domain of phage 434 repressor at 2.0 Å resolution. J Mol Biol 205:189–200. doi: 10.1016/0022-2836(89)90375-6. [DOI] [PubMed] [Google Scholar]
- 96.Hohn B, Murray K. 1977. Packaging recombinant DNA molecules into bacteriophage particles in vitro. Proc Natl Acad Sci USA 74:3259–3263. doi: 10.1073/pnas.74.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Murray NE, Brammar WJ, Murray K. 1977. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet 150:53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- 98.Kaiser D, Masuda T. 1973. In vitro assembly of bacteriophage Lambda heads. Proc Natl Acad Sci USA 70:260–264. doi: 10.1073/pnas.70.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaiser AD, Jacob F. 1957. Recombination between related temperate bacteriophages and the genetic control of immunity and prophage localization. Virology 4:509–521. doi: 10.1016/0042-6822(57)90083-1. [DOI] [PubMed] [Google Scholar]
- 100.Liedke-Kulke M, Kaiser AD. 1967. The c-region of coliphage 21. Virology 32:475–481. doi: 10.1016/0042-6822(67)90299-1. [DOI] [PubMed] [Google Scholar]
- 101.Signer ER. 1964. Recombination between coliphages Lambda and ϕ80. Virology 22:650–651. doi: 10.1016/0042-6822(64)90090-X. [DOI] [PubMed] [Google Scholar]
- 102.Szpirer J, Thomas R, Radding CM. 1969. Hybrids of bacteriophages lambda and ϕ80: a study of nonvegetative functions. Virology 37:585–596. doi: 10.1016/0042-6822(69)90276-1. [DOI] [PubMed] [Google Scholar]
- 103.Tomita T. 1969. On the transcription of the ϕ80 phage genome. Mol Gen Genet 103:313–325. doi: 10.1007/BF00383481. [DOI] [PubMed] [Google Scholar]
- 104.Furth ME, McLeester C, Dove WF. 1978. Specificity determinants for bacteriophage lambda DNA replication. I. A chain of interactions that controls the initiation of replication. J Mol Biol 126:195–225. doi: 10.1016/0022-2836(78)90359-5. [DOI] [PubMed] [Google Scholar]
- 105.Hilliker S. 1981. Characterization of the Salmonella phage L early genes using λimmL hybrid phages. Virology 114:161–174. doi: 10.1016/0042-6822(81)90262-2. [DOI] [PubMed] [Google Scholar]
- 106.Gemski P, Jr, Baron LS, Yamamoto N. 1972. Formation of hybrids between coliphage lambda and Salmonella phage P22 with a Salmonella typhimurium hybrid sensitive to these phages. Proc Natl Acad Sci USA 69:3110–3114. doi: 10.1073/pnas.69.11.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oberto J, Clerget M, Ditto M, Cam K, Weisberg RA. 1993. Antitermination of early transcription in phage HK022. Absence of a phage-encoded antitermination factor. J Mol Biol 229:368–381. doi: 10.1006/jmbi.1993.1040. [DOI] [PubMed] [Google Scholar]
- 108.King RA, Perez MT. 2018. Sequence analysis of a hybrid HK022/λ bacteriophage and the precise identification of the λ b515 and λ b519 deletion endpoints. Microbiol Res Announc 7:e01118-18. doi: 10.1128/MRA.01118-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Landsmann J, Kroger M, Hobom G. 1982. The rex region of bacteriophage lambda: two genes under three-way control. Gene 20:11–24. doi: 10.1016/0378-1119(82)90083-x. [DOI] [PubMed] [Google Scholar]
- 110.Grosschedl R, Schwarz E. 1979. Nucleotide sequence of the cro-cII-oop region of bacteriophage 434 DNA. Nucleic Acids Res 6:867–881. doi: 10.1093/nar/6.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ovchinnikov YA, Guryev S, Krayev A, Monastyrskaya G, Skryabin K, Sverdlov E, Zakharyev V, Bayev A. 1979. Primary structure of an EcoRI fragment of λimm434 DNA containing regions cI-cro of phage 434 and cII-O of phage lambda. Gene 6:235–249. doi: 10.1016/0378-1119(79)90060-X. [DOI] [PubMed] [Google Scholar]
- 112.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 113.Rosenberg M, Court D, Shimatake H, Brady C, Wulff DL. 1978. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature 272:414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- 114.Schwarz E, Scherer G, Hobom G, Kössel H. 1978. Nucleotide sequence of cro, cII and part of the O gene in phage λ DNA. Nature 272:410–414. doi: 10.1038/272410a0. [DOI] [PubMed] [Google Scholar]
- 115.Sanger F, Coulson AR, Hong G, Hill D, Petersen G. 1982. Nucleotide sequence of bacteriophage λ DNA. J Mol Biol 162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 116.Herskowitz I, Hagen D. 1980. The lysis-lysogeny decision of phage λ: explicit programming and responsiveness. Annu Rev Genet 14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- 117.Little JW, Shepley DP, Wert DW. 1999. Robustness of a gene regulatory circuit. EMBO J 18:4299–4307. doi: 10.1093/emboj/18.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Court DL, Oppenheim AB, Adhya SL. 2007. A new look at bacteriophage λ genetic networks. J Bacteriol 189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ho Y-S, Wulff DL, Rosenberg M. 1983. Bacteriophage λ protein CII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature 304:703–708. doi: 10.1038/304703a0. [DOI] [PubMed] [Google Scholar]
- 120.Schmeissner U, Court D, Shimatake H, Rosenberg M. 1980. Promoter for the establishment of repressor synthesis in bacteriophage lambda. Proc Natl Acad Sci USA 77:3191–3195. doi: 10.1073/pnas.77.6.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hoopes BC, McClure WR. 1985. A CII-dependent promoter is located within the Q gene of bacteriophage lambda. Proc Natl Acad Sci USA 82:3134–3138. doi: 10.1073/pnas.82.10.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shih M-C, Gussin GN. 1984. Role of CII protein in stimulating transcription initiation at the λ PRE promoter: enhanced formation and stabilization of open complexes. J Mol Biol 172:489–506. doi: 10.1016/s0022-2836(84)80019-4. [DOI] [PubMed] [Google Scholar]
- 123.Datta AB, Panjikar S, Weiss MS, Chakrabarti P, Parrack P. 2005. Structure of lambda CII: implications for recognition of direct-repeat DNA by an unusual tetrameric organization. Proc Natl Acad Sci USA 102:11242–11247. doi: 10.1073/pnas.0504535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jain D, Kim Y, Maxwell KL, Beasley S, Zhang R, Gussin GN, Edwards AM, Darst SA. 2005. Crystal structure of bacteriophage lambda CII and its DNA complex. Mol Cell 19:259–269. doi: 10.1016/j.molcel.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 125.Mondal A, Chattopadhyaya R, Datta AB, Parrack P. 2015. Crystallization and X-ray analysis of the transcription-activator protein C1 of bacteriophage P22 in complex with the PRE promoter element. Acta Crystallogr F Struct Biol Commun 71:1286–1291. doi: 10.1107/S2053230X15015708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fien K, Turck A, Kang I, Kielty S, Wulff DL, McKenney K, Rosenberg M. 1984. CII-dependent activation of the pRE promoter of coliphage lambda fused to the Escherichia coli galK gene. Gene 32:141–150. doi: 10.1016/0378-1119(84)90042-8. [DOI] [PubMed] [Google Scholar]
- 127.Kobiler O, Koby S, Teff D, Court D, Oppenheim AB. 2002. The phage lambda CII transcriptional activator carries a C-terminal domain signaling for rapid proteolysis. Proc Natl Acad Sci USA 99:14964–14969. doi: 10.1073/pnas.222172499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Krinke L, Wulff DL. 1990. RNase III-dependent hydrolysis of lambda cII-O gene mRNA mediated by lambda OOP antisense RNA. Genes Dev 4:2223–2233. doi: 10.1101/gad.4.12a.2223. [DOI] [PubMed] [Google Scholar]
- 129.Hoyt MA, Knight DM, Das A, Miller HI, Echols H. 1982. Control of phage lambda development by stability and synthesis of CII protein: role of the viral cIII and host hflA, himA and himD genes. Cell 31:565–573. doi: 10.1016/0092-8674(82)90312-9. [DOI] [PubMed] [Google Scholar]
- 130.Kihara A, Akiyama Y, Ito K. 1997. Host regulation of lysogenic decision in bacteriophage lambda: transmembrane modulation of FtsH (HflB), the cII degrading protease, by HflKC (HflA). Proc Natl Acad Sci USA 94:5544–5549. doi: 10.1073/pnas.94.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Banuett F, Hoyt MA, McFarlane L, Echols H, Herskowitz I. 1986. hflB, a new Escherichia coli locus regulating lysogeny and the level of bacteriophage lambda cII protein. J Mol Biol 187:213–224. doi: 10.1016/0022-2836(86)90229-9. [DOI] [PubMed] [Google Scholar]
- 132.Campbell A. 1961. Sensitive mutants of bacteriophage lambda. Virology 14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- 133.Kourilsky P, Bourguignon M-F, Bouquet M, Gros F. 1971. Early transcription controls after induction of prophage λ, p 305–314. In Hershey AD (ed), The bacteriophage lambda. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. doi: 10.1101/SQB.1970.035.01.040. [DOI] [Google Scholar]
- 134.Szybalski W, Bøvre K, Fiandt M, Hayes S, Hradecna Z, Kumar S, Lozeron H, Nijkamp H, Stevens W. 1971. Transcriptional units and their controls in Escherichia coli phage λ: operons and scriptons, p 341–353. In Hershey AD (ed), The bacteriophage lambda. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. doi: 10.1101/SQB.1970.035.01.045. [DOI] [Google Scholar]
- 135.Friedman DI, Jolly CT, Mural RJ. 1973. Interference with the expression of the N gene function of phage lambda in a mutant of Escherichia coli. Virology 51:216–226. doi: 10.1016/0042-6822(73)90381-4. [DOI] [PubMed] [Google Scholar]
- 136.Franklin NC. 1974. Altered reading of genetic signals fused to the N operon of bacteriophage lambda: genetic evidence for modification of polymerase by the protein product of the N gene. J Mol Biol 89:33–48. doi: 10.1016/0022-2836(74)90161-2. [DOI] [PubMed] [Google Scholar]
- 137.Friedman DI. 1983. Lytic mode of lambda development, p 21–51. In Hendrix R, Roberts J, Stahl F, Weisberg R (ed), Lambda II. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- 138.Thomas R. 1966. Control of development in temperate bacteriophages. I. Induction of prophage genes following hetero-immune super-infection. J Mol Biol 22:79–95. doi: 10.1016/0022-2836(66)90181-1. [DOI] [Google Scholar]
- 139.Dambly C, Couturier M, Thomas R. 1968. Control of development in temperate bacteriophages, II. Control of lysozyme synthesis. J Mol Biol 32:67–81. doi: 10.1016/0022-2836(68)90146-0. [DOI] [PubMed] [Google Scholar]
- 140.Luzzati D. 1970. Regulation of lambda exonuclease synthesis: role of the N gene product and lambda repressor. J Mol Biol 49:515–519. doi: 10.1016/0022-2836(70)90261-5. [DOI] [PubMed] [Google Scholar]
- 141.Friedman DI, Wilgus GS, Mural RJ. 1973. Gene N regulator function of phage lambda imm21: evidence that a site of N action differs from a site of N recognition. J Mol Biol 81:505–516. doi: 10.1016/0022-2836(73)90519-6. [DOI] [PubMed] [Google Scholar]
- 142.Adhya S, Gottesman M, De Crombrugghe B. 1974. Release of polarity in Escherichia coli by gene N of phage lambda: termination and antitermination of transcription. Proc Natl Acad Sci USA 71:2534–2538. doi: 10.1073/pnas.71.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Court D, Sato K. 1969. Studies of novel transducing variants of lambda: dispensability of genes N and Q. Virology 39:348–352. doi: 10.1016/0042-6822(69)90060-9. [DOI] [PubMed] [Google Scholar]
- 144.Hopkins N. 1970. Bypassing a positive regulator: isolation of a lambda mutant that does not require N product to grow. Virology 40:223–229. doi: 10.1016/0042-6822(70)90397-1. [DOI] [PubMed] [Google Scholar]
- 145.Butler B, Echols H. 1970. Regulation of bacteriophage lambda development by gene N: properties of a mutation that bypasses N control of late protein synthesis. Virology 40:212–222. doi: 10.1016/0042-6822(70)90396-x. [DOI] [PubMed] [Google Scholar]
- 146.Friedman DI, Olson ER, Georgopoulos C, Tilly K, Herskowitz I, Banuett F. 1984. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol Rev 48:299–325. doi: 10.1128/mr.48.4.299-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Salstrom JS, Szybalski W. 1978. Coliphage lambda nutL−: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol 124:195–221. doi: 10.1016/0022-2836(78)90156-0. [DOI] [PubMed] [Google Scholar]
- 148.Salstrom JS, Fiandt M, Szybalski W. 1979. The site controlling the specificity of N action is outside the promoter-operator region: a triple hybrid phage lambda N21 imm434 nin5. Gene 5:305–327. doi: 10.1016/0378-1119(79)90105-7. [DOI] [PubMed] [Google Scholar]
- 149.Segawa T, Imamoto F. 1974. Diversity of regulation of genetic transcription. II. Specific relaxation of polarity in read-through transcription of the translocated trp operon in bacteriophage lambda trp. J Mol Biol 87:741–754. doi: 10.1016/0022-2836(74)90082-5. [DOI] [PubMed] [Google Scholar]
- 150.King RA, Madsen PL, Weisberg RA. 2000. Constitutive expression of a transcription termination factor by a repressed prophage: promoters for transcribing the phage HK022 nun gene. J Bacteriol 182:456–462. doi: 10.1128/JB.182.2.456-462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sloan S, Rutkai E, King RA, Velikodvorskaya T, Weisberg RA. 2007. Protection of antiterminator RNA by the transcript elongation complex. Mol Microbiol 63:1197–1208. doi: 10.1111/j.1365-2958.2006.05579.x. [DOI] [PubMed] [Google Scholar]
- 152.Clerget M, Jin DJ, Weisberg RA. 1995. A zinc-binding region in the beta′ subunit of RNA polymerase is involved in antitermination of early transcription of phage HK022. J Mol Biol 248:768–780. doi: 10.1006/jmbi.1995.0259. [DOI] [PubMed] [Google Scholar]
- 153.Campbell AM. 1963. Episomes, p 101–145. In Caspari EW, Thoday JM (ed), Advances in genetics, vol 11. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 154.Calef E, Licciardello G. 1960. Recombination experiments on prophage host relationships. Virology 12:81–103. doi: 10.1016/0042-6822(60)90151-3. [DOI] [Google Scholar]
- 155.Kaiser AD. 1957. Mutations in a temperate bacteriophage affecting its ability to lysogenize Escherichia coli. Virology 3:42–61. doi: 10.1016/0042-6822(57)90022-3. [DOI] [PubMed] [Google Scholar]
- 156.Arber W. 1958. Transduction des caractères Gal par le bactériophage lambda. Arch Sci (Geneva) 11:259–337. [Google Scholar]
- 157.Adler J, Templeton B. 1963. The amount of galactose genetic material in λdg bacteriophage with different densities. J Mol Biol 7:710–720. doi: 10.1016/S0022-2836(63)80118-7. [DOI] [PubMed] [Google Scholar]
- 158.Signer E, Echols H, Weil J, Radding C, Shulman M, Moore L, Manly K. 1971. The general recombination system of bacteriophage λ. p 711–714. In Hershey AD (ed), The bacteriophage lambda. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. doi: 10.1101/SQB.1968.033.01.080. [DOI] [PubMed] [Google Scholar]
- 159.Gratia JP. 1966. Studies on defective lysogeny due to chromosomal deletion in Escherichia coli. I. Single lysogens. Biken J 9:77–87. [PubMed] [Google Scholar]
- 160.Leimkuhler S. 2020. The biosynthesis of the molybdenum cofactor in Escherichia coli. Environ Microbiol 22:2007–2026. doi: 10.1111/1462-2920.15003. [DOI] [PubMed] [Google Scholar]
- 161.Adhya S, Cleary P, Campbell A. 1968. A deletion analysis of prophage lambda and adjacent genetic regions. Proc Natl Acad Sci USA 61:956–962. doi: 10.1073/pnas.61.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Skalka AM. 1977. DNA replication—bacteriophage lambda. Curr Top Microbiol Immunol 78:201–237. [PubMed] [Google Scholar]
- 163.Brooks K. 1965. Studies in the physiological genetics of some suppressor-sensitive mutants of bacteriophage λ. Virology 26:489–499. doi: 10.1016/0042-6822(65)90011-5. [DOI] [PubMed] [Google Scholar]
- 164.Bird RE, Louarn J, Martuscelli J, Caro L. 1972. Origin and sequence of chromosome replication in Escherichia coli. J Mol Biol 70:549–566. doi: 10.1016/0022-2836(72)90559-1. [DOI] [PubMed] [Google Scholar]
- 165.Schnös M, Inman RB. 1970. Position of branch points in replicating λ DNA. J Mol Biol 51:61–73. doi: 10.1016/0022-2836(70)90270-6. [DOI] [PubMed] [Google Scholar]
- 166.Kornberg A, Baker T. 1992. DNA replication, 2nd ed W.H. Freeman, New York, NY. [Google Scholar]
- 167.Furth M, Wickner S. 1983. Lambda DNA replication, p 145–173. In Hendrix R, Roberts J, Stahl F, Weisberg R (ed), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 168.Furth M, Yates J. 1978. Specificity determinants for bacteriophage lambda DNA replication. II. Structure of O proteins of l-f80 and l-82 hybrid phages and of a l mutant defective in the origin of replication. J Mol Biol 126:227–240. doi: 10.1016/0022-2836(78)90360-1. [DOI] [PubMed] [Google Scholar]
- 169.Tomizawa J. 1971. Functional cooperation of genes O and P, p 549–552. In Hershey AD (ed), The bacteriophage lambda. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 170.Wickner SH. 1979. DNA replication proteins of Escherichia coli and phage lambda. Cold Spring Harbor Symp Quant Biol 43:303–310. doi: 10.1101/sqb.1979.043.01.037. [DOI] [PubMed] [Google Scholar]
- 171.Frackman S, Siegele DA, Feiss M. 1985. The terminase of bacteriophage lambda. Functional domains for cosB binding and multimer assembly. J Mol Biol 183:225–238. doi: 10.1016/0022-2836(85)90215-3. [DOI] [PubMed] [Google Scholar]
- 172.de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. 2002. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol Cell 9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 173.Sippy J, Patel P, Vahanian N, Sippy R, Feiss M. 2015. Genetics of critical contacts and clashes in the DNA packaging specificities of bacteriophages lambda and 21. Virology 476:115–123. doi: 10.1016/j.virol.2014.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wu WF, Christiansen S, Feiss M. 1988. Domains for protein-protein interactions at the N and C termini of the large subunit of bacteriophage lambda terminase. Genetics 119:477–484. doi: 10.1093/genetics/119.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Frackman S, Siegele DA, Feiss M. 1984. A functional domain of bacteriophage lambda terminase for prohead binding. J Mol Biol 180:283–300. doi: 10.1016/S0022-2836(84)80005-4. [DOI] [PubMed] [Google Scholar]
- 176.Yeo A, Feiss M. 1995. Mutational analysis of the prohead binding domain of the large subunit of terminase, the bacteriophage lambda DNA packaging enzyme. J Mol Biol 245:126–140. [PubMed] [Google Scholar]
- 177.Yeo A, Feiss M. 1995. Specific interaction of terminase, the DNA packaging enzyme of bacteriophage lambda, with the portal protein of the prohead. J Mol Biol 245:141–150. doi: 10.1006/jmbi.1994.0013. [DOI] [PubMed] [Google Scholar]
- 178.Georgopoulos C. 2006. Toothpicks, serendipity and the emergence of the Escherichia coli DnaK (Hsp70) and GroEL (Hsp60) chaperone machines. Genetics 174:1699–1707. doi: 10.1534/genetics.104.68262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Georgopoulos C. 1971. A bacterial mutation affecting N function, p 341–353. In Hershey AD (ed), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 180.Botstein D. 1980. A theory of modular evolution for bacteriophages. Ann N Y Acad Sci 354:484–490. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- 181.Casjens S, Hatfull G, Hendrix R. 1992. Evolution of dsDNA tailed-bacteriophage genomes. Semin Virol 3:383–397. [Google Scholar]
- 182.Campbell A, Botstein D. 1983. Evolution of the lambdoid phages, p 365–380. In Hendrix R, Roberts J, Stahl F, Weisberg R (ed), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 183.Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. 2000. The origins and ongoing evolution of viruses. Trends Microbiol 8:504–508. doi: 10.1016/s0966-842x(00)01863-1. [DOI] [PubMed] [Google Scholar]
- 184.Casjens SR, Grose JH. 2016. Contributions of P2-and P22-like prophages to understanding the enormous diversity and abundance of tailed bacteriophages. Virology 496:255–276. doi: 10.1016/j.virol.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jackson EN, Jackson DA, Deans RJ. 1978. EcoRI analysis of bacteriophage P22 DNA packaging. J Mol Biol 118:365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- 186.Tye B-K, Huberman JA, Botstein D. 1974. Non-random circular permutation of phage P22 DNA. J Mol Biol 85:501–28528. doi: 10.1016/0022-2836(74)90312-X. [DOI] [PubMed] [Google Scholar]
- 187.Leavitt JC, Gilcrease EB, Wilson K, Casjens SR. 2013. Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor. Virology 440:117–133. doi: 10.1016/j.virol.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, Tang L. 2010. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc Natl Acad Sci USA 107:1971–1976. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Roy A, Bhardwaj A, Cingolani G. 2011. Crystallization of the nonameric small terminase subunit of bacteriophage P22. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:104–110. doi: 10.1107/S174430911004697X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang JC, Kaiser A. 1973. Evidence that the cohesive ends of mature λ DNA are generated by the gene A product. Nat New Biol 241:16–17. doi: 10.1038/newbio241016a0. [DOI] [PubMed] [Google Scholar]
- 191.Davidson AR, Gold M. 1992. Mutations abolishing the endonuclease activity of bacteriophage lambda terminase lie in two distinct regions of the A gene, one of which may encode a “leucine zipper” DNA-binding domain. Virology 189:21–30. doi: 10.1016/0042-6822(92)90677-h. [DOI] [PubMed] [Google Scholar]
- 192.Arens JS, Duffy C, Feiss M. 2019. Acidic residues and a predicted, highly conserved α-helix are critical for the endonuclease/strand separation functions of bacteriophage λ's TerL. Mol Microbiol 112:1483–1498. doi: 10.1111/mmi.14373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download mmbr.00124-21-s0001.pdf, PDF file, 1.0 MB (1.1MB, pdf)