Abstract
Limited evidence exists on the link between inflammation and epigenetic aging. We aimed to (a) assess the cross-sectional and prospective associations of 22 inflammation-related plasma markers and a signature of inflammaging with epigenetic aging and (b) determine whether epigenetic aging and inflammaging are independently associated with mortality. Blood samples from 940 participants in the Melbourne Collaborative Cohort Study collected at baseline (1990–1994) and follow-up (2003–2007) were assayed for DNA methylation and 22 inflammation-related markers, including well-established markers (eg, interleukins and C-reactive protein) and metabolites of the tryptophan–kynurenine pathway. Four measures of epigenetic aging (PhenoAge, GrimAge, DunedinPoAm, and Zhang) and a signature of inflammaging were considered, adjusted for age, and transformed to Z scores. Associations were assessed using linear regression, and mortality hazard ratios (HR) and 95% confidence intervals (95% CI) were estimated using Cox regression. Cross-sectionally, most inflammation-related markers were associated with epigenetic aging measures, although with generally modest effect sizes (regression coefficients per SD ≤ 0.26) and explaining altogether between 1% and 11% of their variation. Prospectively, baseline inflammation-related markers were not, or only weakly, associated with epigenetic aging after 11 years of follow-up. Epigenetic aging and inflammaging were strongly and independently associated with mortality, for example, inflammaging: HR = 1.41, 95% CI = 1.27–1.56, p = 2 × 10−10, which was only slightly attenuated after adjustment for 4 epigenetic aging measures: HR = 1.35, 95% CI = 1.22–1.51, p = 7 × 10−9). Although cross-sectionally associated with epigenetic aging, inflammation-related markers accounted for a modest proportion of its variation. Inflammaging and epigenetic aging are essentially nonoverlapping markers of biological aging and may be used jointly to predict mortality.
Keywords: Biological aging, Epigenetic aging, Inflammation, Inflammaging, Kynurenines
The physiological processes of biological aging, the progressive decline in integrity of cells and organs with age, have been a major focus of gerontological research (1). Although substantial progress has been made in identifying and characterizing several of the phenotypic “hallmarks” of biological aging (2), such as inflammatory, transcriptomic, and epigenetic cellular changes, investigation of the interplay between them remains notably deficient (1). Recent candidate biological aging markers such as measures of epigenetic aging and of inflammation and “inflammaging” provide promising avenues for investigating these relationships (3).
Potentially the most promising of the candidate markers of biological aging are those based on DNA methylation (ie, epigenetic aging) (4–6). Epigenetic aging measures are developed by combining the DNA methylation values (variation in which is highly sensitive to age-related physiological changes) at cytosine-guanine-dinucleotides (CpGs) across the genome into a weighted average. These measures tend to be highly correlated with chronological age, and the phenomenon by which an individual’s epigenetic age differs from their chronological age is commonly referred to as epigenetic “age acceleration” (AgeAccel) in the epigenetic field. Positive AgeAccel has been found to be associated with increased risk of several chronic diseases and earlier mortality (4,7).
Another feature of biological aging is the increasingly inflammatory phenotype that tends to occur with age. This phenotype is often referred to as “inflammaging” and is recognized as an important risk factor for age-related morbidity and mortality (3). Most diseases of aging have an important inflammatory component (8). A related set of markers are those of the tryptophan–kynurenine (TK) pathway, the metabolic pathway that is the starting point for production of nicotinamide adenine dinucleotide (NAD+). The TK pathway is increasingly recognized as playing an important role in inflammation (9). Recently, we combined 10 age-associated inflammatory and TK pathway markers into a novel candidate marker of biological aging and a signature of inflammaging (10). This inflammaging signature was found to be strongly associated with mortality in the Melbourne Collaborative Cohort Study (MCCS) and the Hordaland Health Study (10).
To quantify the association between epigenetic aging and inflammation/inflammaging, we considered 4 “second generation” measures of epigenetic aging: PhenoAge (11), GrimAge (12), Zhang (13), and DunedinPoAm (14) (note that we use the term “epigenetic aging” in the broad sense—inclusive of the Zhang measure that is not explicitly related to aging). PhenoAge is an epigenetic surrogate of a composite clinical measure of “phenotypic age,” which incorporates chronological age and nine physiological health markers (11). GrimAge is an epigenetic marker of lifespan formed as a weighted combination of chronological age, sex, and epigenetic signatures of plasma proteins and smoking pack years (12). The Zhang measure is an epigenetic score based on the methylation values of 10 CpGs whose methylation values strongly predicted mortality over 15 years of follow-up (13). Finally, DunedinPoAm (Dunedin pace of aging methylation) is an epigenetic signature that aims to measure the speed of biological aging, based on the longitudinal trajectory of several blood markers measured in 3 successive waves of the Dunedin Study (14).
Of previous studies investigating the association between epigenetic aging and inflammation, Levine et al. (11) found that positive PhenoAgeAccel was cross-sectionally associated with increased C-reactive protein (CRP) and increased transcription of inflammatory genes (such as tumor necrosis factor-mediated signaling pathways). Positive GrimAgeAccel has similarly been associated with increased CRP (12). Irvin et al. (15) found that “extrinsic AgeAccel,” related to the first-generation Hannum epigenetic age measure (16), was associated with interleukin-6 (IL-6), CRP, and tumor necrosis factor-alpha (TNF-α), albeit with modest effect sizes. Stevenson et al. (17), using longitudinal data from the Lothian Birth Cohort, found a positive cross-sectional association of extrinsic AgeAccel with CRP and IL-6, but little evidence of a prospective relationship.
Although preliminary evidence suggests some association between epigenetic aging and inflammation, most previous studies were limited to a few inflammatory markers (essentially CRP and IL-6). No study to date has assessed the association between TK pathway markers and epigenetic aging, nor has any study investigated the association between epigenetic aging and a recently developed signature of inflammaging (10). Addressing these gaps in the research may provide insight into the complexity of biological aging by identifying the degree to which markers of these distinct systems contain independent (or overlapping) information (3). This may have implications for the utility of these 2 sets of markers in predicting adverse health outcomes when used in combination. This study therefore had 2 aims: (a) to quantify the association of a panel of inflammatory and TK pathway markers, and a signature of inflammaging, with 4 measures of epigenetic aging and (b) to determine to what extent epigenetic aging and inflammaging are independently associated with all-cause mortality.
Method
Study Sample
The Melbourne Collaborative Cohort Study (MCCS) is a prospective cohort study that recruited 41 513 participants between 1990 and 1994 and was designed to investigate prospectively the role of diet and other lifestyle factors as risk factors for cancer and other chronic diseases (18). Participants were recruited via the electoral roll (registration to vote is obligatory in Australia) and via local media and community advertisements. The majority (99%) of MCCS participants were aged between 40 and 69 years and were free of cancer at baseline. Blood samples were taken at baseline and follow-up (wave 2), years 1990–1994 and 2003–2007, respectively, for 99% and 64% of participants, respectively.
DNA methylation was measured in Guthrie card samples taken at baseline and follow-up for 1 100 participants selected as controls in a series of nested case–control studies of DNA methylation and cancer risk within the MCCS, a subset of which (N = 976) had markers of inflammation and the kynurenine pathway also measured at baseline (1990–1994) and follow-up (2003–2007) (10,19). Control participants were matched to cancer cases based on sex, age at blood draw, and smoking status (the latter in the lung cancer study only) (20,21). A flowchart describing the sample selection from the MCCS cohort to the final study sample is shown in Supplementary Figure 1.
The study was approved by the Cancer Council Victoria’s Human Research Ethics Committee (IEC 9001), Melbourne, VIC, Australia, and all participants provided informed consent in accordance with the Declaration of Helsinki.
Demographic, Anthropometric, and Health Data
At baseline, participants completed detailed questionnaires on demographic variables, lifestyle and medical history. The socioeconomic index for areas (SEIFA), a postcode-based metric of socioeconomic status created by the Australian Bureau of Statistics, was used as a proxy for socioeconomic status (pseudocontinuous variable ranging from 1 to 10 according to deciles). Smoking was assessed as smoking status (current, former, never), and smoking pack years. Alcohol intake (grams/day) was estimated based on the frequency, alcohol content and volume consumed by type of drink over the past week (at baseline) or over the past 12 months (at follow-up) (22). Trained personnel additionally measured height (baseline only) and weight. Body mass index (BMI) in kg/m2 was calculated. Vital status is obtained through yearly record linkage to the Victorian Registry of Births, Deaths, and Marriages (via the Victorian Cancer Registry) and the Australian National Death Index (Australian Institute of Health and Welfare). Vital status information was considered to be virtually complete up to October 31, 2019.
DNA Methylation Measurement and Calculation of Epigenetic Age
The methods relating to DNA extraction and bisulfite conversion, and processing of DNA methylation data have been described previously (23). In brief, for all samples included in this study, DNA was extracted from dried blood spots and the Illumina Infininum HumanMethylation450K Beadchip array, which targets 485 577 CpGs across the genome, was used to measure DNA methylation. Data normalization steps followed a well-established pipeline that has been described in detail previously (24). Data from a total of 946 participants were available after quality control of methylation data.
PhenoAge and GrimAge were calculated using Horvath’s online calculator at https://dnamage.genetics.ucla.edu/ (25). Zhang and DunedinPoAm were calculated using the weights provided in the respective original publications (13,14). Each epigenetic age variable was regressed on chronological age with the resulting residuals retained as AgeAccel estimates. As a sensitivity analysis, we considered that the relationship between epigenetic and chronological age could be nonlinear and fitted penalized cubic splines on each epigenetic age variable, showing associations were all virtually linear (Supplementary Figure 3).
Inflammation Marker Measurement
Plasma samples were collected at the same time-point as dried blood spots used for DNA methylation measurement and stored in liquid nitrogen at −180°C until transportation for laboratory analysis. Measurement of TK pathway markers was performed at Bevital laboratories in Bergen, Norway http://www.bevital.no/. TK pathway markers included tryptophan, kynurenine, 3-hydroxykynurenine, kynurenic acid, xanthurenic acid, anthranilic acid, 3-hydroxyanthranilic acid, picolinic acid, and quinolinic acid and were assayed using liquid chromatography–tandem mass spectrometry (LC–MC) (26). Neopterin was also assayed using LC–MC. The inflammatory markers CRP, serum amyloid A, calprotectin, and cystatin C were assayed using matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI–TOF MS), also at Bevital laboratories. The remaining inflammatory markers interleukin-6 (IL-6), IL8, IL-10, interferon-gamma (IFN-γ), and TNF-α were analyzed separately at the International Agency for Research on Cancer (IARC) laboratories in Lyon, France, using Meso Scale Discovery 6-Plex kits. Several derived markers related to tryptophan metabolism were calculated. These included the kynurenine to tryptophan ratio, a surrogate for the activity of the indoleamine 2,3-dioxygenase enzyme and reflective of immune activity (27); the ratio of pyridoxal 5′-phosphate to (pyridoxal + pyridoxic acid; PAr index), an index of vitamin B6 catabolism during inflammation (28); and the ratio of 3-hydroxykynurenine to xanthurenic acid (HK:XA) as a marker of vitamin B6 status (29).
The inflammaging signature was calculated as a linear combination of the log concentration of 10 inflammatory markers with weights obtained from the original publication (see Supplementary Table S1) (10). It was also regressed on age to obtain an age-adjusted measure (AAinflammaging).
Confounders
In regression models assessing the association between inflammatory/TK pathway markers and AgeAccel, we adjusted for confounder variables sex and country of birth (Australia/New Zealand, Southern Europe, Northern Europe). Previous investigations have found that sex strongly determines AgeAccel, particularly GrimAgeAccel (30). Country of birth was used as a proxy for genetic ancestry, which strongly determines patterns of DNA methylation. Note that age was not considered as a confounder since AgeAccel variables and AAinflammaging, are independent of chronological age. We additionally considered as potential confounders BMI, smoking (current, former, never), alcohol intake, and socioeconomic status (SEIFA decile score) as these factors may cause elevated inflammation and epigenetic aging (31–33). Other factors such as dietary habits and physical activity were not considered as previous studies indicate that they may have only minor influences on epigenetic aging (31).
Data Preparation
Six participants with missing values in more than 50% of the markers were excluded from the analysis. There were between 0% and 6% missing values for the inflammatory markers at either baseline or follow-up, which were commonly due to issues with biological samples (insufficient quantity or quality) or quantification issues in assay instruments and were assumed to be missing completely at random. Missing data for inflammation markers and a small number of confounder values were imputed using the random forest method missForest, which performs very well when the proportion of missing data is low (34).
Inflammatory marker variables occasionally had values below the limit of detection (<2% in all cases). These were replaced with the limit of detection divided by 2 for each marker. Subsequently, inflammatory markers were log2 transformed to obtain distributions closer to Gaussian, and winsorized at <−3 and >+3 SD from the mean to minimize the influence of potential outliers. AgeAccel variables and AAinflammaging, which were approximately normally distributed, were similarly winsorized at 3 SD from the mean. Prior to fitting regression models, inflammatory markers, AAinflammaging, and AgeAccel variables were standardized to Z scores to facilitate comparison of effect sizes.
Statistical Analysis
The associations between the inflammatory markers, AAinflammaging, and AgeAccel measures were assessed cross-sectionally (baseline and follow-up) and prospectively using multivariable linear regression, adjusted for sex and country of birth. The prospective models were additionally adjusted for baseline AgeAccel measures as appropriate. In a sensitivity analysis, we additionally adjusted for socioeconomic status (SEIFA decile score), BMI, daily alcohol intake, and smoking status. Regression assumptions of linearity and homoscedasticity were checked using component-residual and scale-location plots, respectively. Associations were approximately consistent with linearity and no marked heteroscedasticity was noted.
We additionally fitted multivariable models including all the inflammatory marker variables and AAinflammaging together as predictors in order to estimate the overall variance explained in AgeAccel. To reduce overfitting, we fitted these models in the Bayesian framework using the regularized “horseshoe” shrinkage prior (35,36). The horseshoe prior consists of a “global” shrinkage hyperparameter, which shrinks all coefficients toward zero, as well as a “local” shrinkage hyperparameter which allows for coefficients with strong signals to escape this shrinkage. Models were estimated with probabilistic programming language Stan via the brms interface (37). Four independent chains were run, each with 1 500 warm-up iterations and 2 500 saved iterations. According to a previous recommendation, priors on hyperparameters were set based on an a priori expectation that 1 in 5 predictors was important (36). Variance explained was estimated using leave-one-out adjusted Bayes R-squared (38).
Finally, we used Cox proportional hazards models to estimate hazard ratios (HR) and 95% confidence intervals (95% CI) for the association of follow-up AgeAccel measures and AAinflammaging with all-cause mortality. Time at risk was calculated from the date of follow-up visit to the date of death or end of follow-up (October 31, 2019). Three multivariable models were fitted, adjusting for sex and country of birth, and using age as the underlying time scale, including (a) each AgeAccel measure and AAinflammaging individually, (b) all AgeAccel measures in a multivariable model, and (c) all AgeAccel measures and AAinflammaging in a multivariable model. To evaluate the predictive performance of the models, concordance indices (c-index) were calculated (39,40). Model comparison was performed using the likelihood ratio test.
Statistical analyses were performed using R version 4.0.5. Figures were generated using the R package ggplot2.
Results
Sample Characteristics and Marker Correlations
The mean age of the 940 participants was 57.5 years (range: 40–70 years) at baseline and 68.9 years (range: 50–83 years) at follow-up. Most participants were male (69%) and 51% of the sample were never smokers, Table 1. Time between baseline and follow-up ranged from 9 to 14.5 years with a mean of 11.4 years (SD = 1.2 years). Concentrations of inflammation-related markers at baseline and follow-up are summarized in Supplementary Table S2.
Table 1.
Sample Characteristics of the Participants in the Melbourne Collaborative Cohort Study Included in the Analysis (n = 940)
| Characteristic | Baseline | Follow-up |
|---|---|---|
| Age, mean (SD) | 57.5 (7.9) | 68.9 (8.0) |
| Sex, n (%) | ||
| Male | 648 (68.9) | — |
| Female | 292 (31.1) | |
| BMI, mean (SD) | 26.7 (3.73) | 27.1 (4.15) |
| Country of birth, n (%) | ||
| Australia/New Zealand | 727 (77.3) | — |
| Northern Europe | 78 (8.30) | |
| Southern Europe | 135 (14.4) | |
| SEIFA decile, median (IQR) | 7 (3–9) | 6.8 (4–9) |
| Smoking, n (%) | ||
| Current | 96 (10.2) | 51 (5.4) |
| Former | 363 (38.6) | 409 (43.5) |
| Never | 481 (51.2) | 480 (51.0) |
| Alcohol intake (g/d), median (IQR) | 7.8 (0–21.7) | 10.2 (0–22.2) |
| Epigenetic age measures | ||
| DunedinAgeAccel, mean (SD) | 0 (0.08) | 0 (0.07) |
| GrimAgeAccel (y), mean (SD) | −0.02 (4.26) | −0.01 (4.08) |
| PhenoAgeAccel (y), mean (SD) | 0.01 (7.03) | −0.03 (6.78) |
| ZhangAgeAccel, mean (SD) | 0 (0.43) | 0 (0.41) |
Notes: BMI = body mass index; IQR = interquartile range; SEIFA = socioeconomic index for areas, a postcode-based metric of socioeconomic status.
The correlations between inflammatory markers, AAinflammaging, and AgeAccel variables at follow-up are shown in Supplementary Figure 2. Spearman’s correlation (ρ) between AgeAccel variables ranged from ρ = 0.26 (PhenoAgeAccel and DunedinAgeAccel) to ρ = 0.54 (ZhangAgeAccel and GrimAgeAccel). Scatter plots of the correlations of epigenetic aging measures and the inflammaging signature with chronological age at follow-up are shown in Supplementary Figure 3.
Cross-sectional Association Between Inflammation and Epigenetic Aging
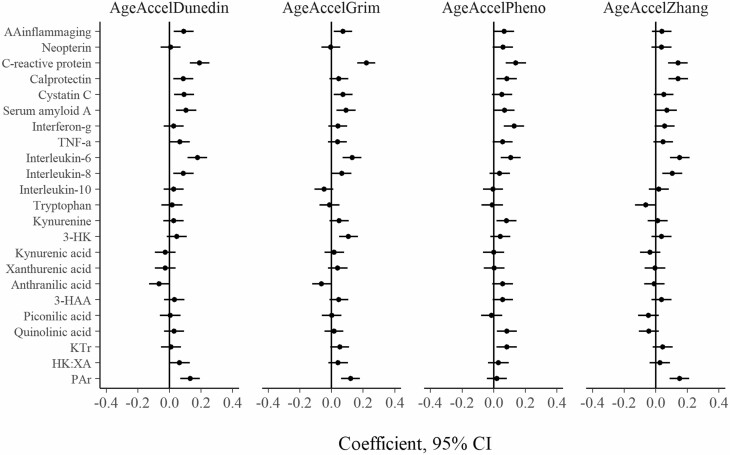
The cross-sectional associations between inflammation and AgeAccel variables at each of baseline and follow-up are shown in Figures 1 and 2, respectively. At baseline, the strongest associations were between CRP and GrimAgeAccel (coefficient per SD = 0.22; 95% CI: 0.16, 0.28), CRP and DunedinAgeAccel (0.19; 95% CI: 0.13, 0.25), and IL-6 and DunedinAgeAccel (0.18; 95% CI: 0.11, 0.24). The associations between AAinflammaging and the AgeAccel variables were relatively weaker, with coefficients per SD ranging from 0.04 (95% CI: −0.02, 0.10) for ZhangAgeAccel to 0.09 (95% CI: 0.03, 0.15) for DunedinAgeAccel.
Figure 1.
Cross-sectional association between inflammatory and kynurenine pathway markers and AgeAccel variables at baseline (N = 940). Each independent model is adjusted for sex and country of birth. Inflammatory markers are log-transformed. All variables are standardized to Z scores. 3-HK = 3-Hydroxykynurenine; 3-HAA = 3-Hydroxyanthranilic acid; KTR = kynurenine to tryptophan ratio; PAr index = ratio of pyridoxal 5′-phosphate to (pyridoxal + pyridoxic acid); HK:XA = ratio of 3-hydroxykynurenine to xanthurenic acid; AgeAccel = epigenetic age acceleration.
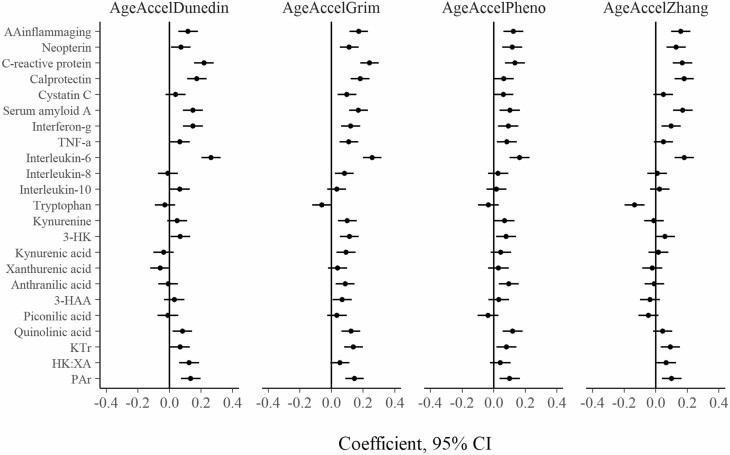
Figure 2.
Cross-sectional association between inflammatory and kynurenine pathway markers and AgeAccel variables at follow-up (N = 940). Each independent model is adjusted for sex and country of birth. Inflammatory markers are log-transformed. All variables are standardized to Z scores. 3-HK = 3-hydroxykynurenine; 3-HAA = 3-hydroxyanthranilic acid; KTR = kynurenine to tryptophan ratio; PAr index = ratio of pyridoxal 5′-phosphate to (pyridoxal + pyridoxic acid); HK:XA = ratio of 3-hydroxykynurenine to xanthurenic acid; AgeAccel = epigenetic age acceleration.
At follow-up, the strongest associations were between IL-6 and GrimAgeAccel (coefficient per SD = 0.26; 95% CI: 0.20, 0.32), IL-6 and DunedinAgeAccel (0.26; 95% CI: 0.20, 0.32), and CRP and GrimAgeAccel (0.24; 95% CI: 0.18, 0.30). Most coefficients were positive, except that of tryptophan with ZhangAgeAccel (−0.13; 95% CI: −0.20, −0.07). For AAinflammaging, the coefficients ranged from 0.12 (95% CI: 0.06, 0.18) for DunedinAgeAccel to 0.17 (95% CI: 0.11, 0.23) for GrimAgeAccel.
Notably, the regression coefficients for each inflammation marker were similar across AgeAccel variables. At follow-up, the correlations between the coefficients for each outcome variable ranged from ρ = 0.71 (PhenoAgeAccel and DunedinAgeAccel) to ρ = 0.90 (DunedinAgeAccel and ZhangAgeAccel; see Supplementary Figure 4). CRP and IL-6 tended to have the largest coefficients across all models (mean coefficient = .18 and .18, respectively), compared with the other inflammatory markers and AAinflammaging (|mean coefficient| < .12).
The estimated Bayes R-squared from the cross-sectional multivariable models, including all inflammation-related markers and AAinflammaging, are shown in Supplementary Figure 5. These cumulatively explained between 1.4% (PhenoAgeAccel) and 7.8% (GrimAgeAccel) of the variation in AgeAccel at baseline and between 2.1% (PhenoAgeAccel) and 11% (GrimAgeAccel) at follow-up.
Prospective Association Between Baseline Inflammation and Epigenetic Aging
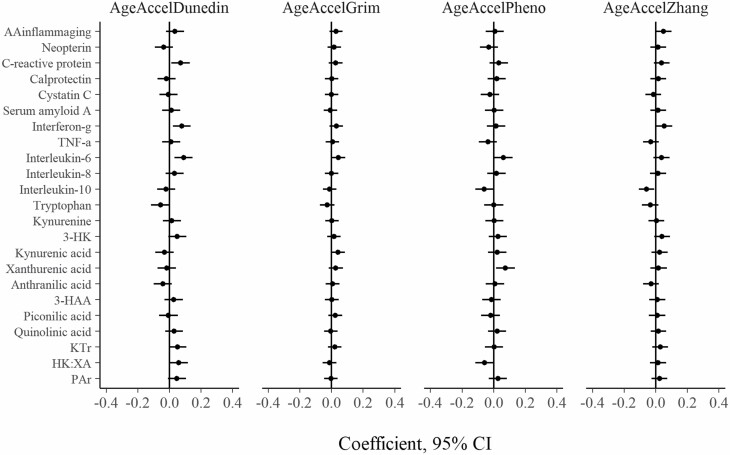
The prospective associations of the inflammatory markers and AAinflammaging at baseline with AgeAccel at follow-up are shown in Figure 3. The associations were considerably weaker than those observed in the cross-sectional analyses, with the majority of them compatible with no association. The strongest associations were for IL-6 and DunedinAgeAccel (coefficient per SD = 0.09, 95% CI: 0.03, 0.15), IFN-γ and DunedinAgeAccel (0.08, 95% CI: 0.02, 0.14), and xanthurenic acid and PhenoAgeAccel (0.08, 95% CI: 0.02, 0.13). For AAinflammaging, the coefficients ranged from 0.01 (95% CI: −0.05, 0.07) for PhenoAgeAccel to 0.05 (95% CI: −0.00, 0.10) for ZhangAgeAccel.
Figure 3.
Prospective association between baseline inflammatory and kynurenine pathway markers and follow-up AgeAccel variables (N = 940). Each independent model is adjusted for baseline AgeAccel, sex, and country of birth. Inflammatory markers are log-transformed. All variables are standardized to Z scores. 3-HK = 3-hydroxykynurenine; 3-HAA = 3-hydroxyanthranilic acid; KTR = kynurenine to tryptophan ratio; PAr index = ratio of pyridoxal 5′-phosphate to (pyridoxal + pyridoxic acid); HK:XA = ratio of 3-hydroxykynurenine to xanthurenic acid; AgeAccel = epigenetic age acceleration.
Sensitivity Analysis
The results of sensitivity analyses using models additionally adjusted for BMI, alcohol intake, smoking, and socioeconomic status are summarized in Supplementary Figures 6–8. Averaged across all 4 outcomes, the mean coefficient was modestly attenuated by 4% in the baseline cross-sectional analysis and 9% at follow-up, respectively. The largest absolute coefficient change was noted for IL-6 and CRP. Averaged across all 4 outcomes, the coefficients for IL-6 and CRP were 0.03 per SD smaller (23% reduction) and 0.03 per SD smaller (16% reduction), respectively, for the baseline cross-sectional model, 0.05 per SD smaller (24% reduction) and 0.04 per SD smaller (22% reduction), respectively, for the follow-up cross-sectional model.
Association of Inflammation, Inflammaging, and AgeAccel With All-Cause Mortality
Three models investigating the association between AAinflammaging, AgeAccel, and all-cause mortality are given in Table 2. A total of 299 deaths were observed over a median follow-up time of 14.4 years (IQR: 12.7–15.3 years). In Model 1, each individual AgeAccel measure individually was associated with greater mortality, with HRs per SD ranging from 1.21 (95% CI: 1.08–1.36, p = .001; PhenoAgeAccel) to 1.44 (95% CI: 1.27–1.63, p = 5 × 10−9; GrimAgeAccel). AAinflammaging was strongly associated with mortality (HR = 1.41, 95% CI: 1.27–1.56, p = 2 × 10−10). In Model 2, all of the AgeAccel variables were included together in the same model (AAinflammaging excluded), adjusted for sex and country of birth. The HRs for each AgeAccel variable were attenuated, with only GrimAgeAccel remaining clearly associated with mortality (HR = 1.31, 95% CI: 1.12–1.53; Table 2). The c-index for Model 2 was 0.61. In Model 3, adding AAinflammaging to Model 2, the HRs for the AgeAccel variables were largely unchanged, and AAinflammaging remained strongly associated with mortality (HR = 1.35, 95% CI: 1.22–1.51, p = 7 × 10−9; Table 2). The c-index for Model 3 was 0.64, and there was strong evidence that the addition of AAinflammaging improved model fit (likelihood ratio test; p = 5 × 10–8).
Table 2.
Epigenetic Age Acceleration, Age-Adjusted Inflammaging Signature, and All-Cause Mortality (N = 940 Melbourne Collaborative Cohort Study Participants)
| Model 1: Separate Models | Model 2: All AgeAccel Measures | Model 3: All AgeAccel Measures and AAinflammaging | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
N = 940 Deaths = 299 |
HR | 95% CI | p | HR | 95% CI | p | HR | 95% CI | p |
| PhenoAgeAccel (per SD) | 1.21 | 1.08–1.36 | .001 | 1.06 | 0.93–1.20 | .38 | 1.04 | 0.91–1.18 | .58 |
| GrimAgeAccel (per SD) | 1.44 | 1.27–1.63 | 5 × 10−9 | 1.31 | 1.12–1.53 | 9 × 10−4 | 1.27 | 1.09–1.49 | .003 |
| DunedinAgeAccel (per SD) | 1.30 | 1.16–1.47 | 7 × 10−6 | 1.12 | 0.98–1.28 | .10 | 1.12 | 0.98–1.28 | .10 |
| ZhangAgeAccel (per SD) | 1.26 | 1.12–1.42 | 1 × 10−4 | 1.04 | 0.90–1.20 | .60 | 1.02 | 0.88–1.18 | .77 |
| AAinflammaging (per SD) | 1.41 | 1.27–1.56 | 2 × 10−10 | — | 1.35 | 1.22–1.51 | 7 × 10−9 | ||
Notes: AgeAccel = epigenetic age acceleration; AAinflammaging = age-adjusted inflammaging signature; CI = confidence interval; HR = hazard ratio. Model 2 concordance index = 0.61; Model 3 concordance index = 0.64.
Similar analyses replacing the inflammaging signature with CRP and IL-6, respectively, produced the same conclusions, that is, a modest attenuation of the HR after adjustment for 4 measures of AgeAccel (Supplementary Tables S3 and S4).
Discussion
We have evaluated the associations between the circulating concentrations of 22 plasma markers of inflammation and the tryptophan/kynurenine pathway, a signature of inflammaging, and 4 epigenetic predictors of aging or mortality. Cross-sectionally, many of these markers were associated with AgeAccel, particularly IL-6 and CRP, and these associations were only modestly attenuated after accounting for BMI, alcohol intake, smoking, and socioeconomic status. Cross-sectional associations were similar at baseline (mean age: 58 years) and follow-up (mean age 69 years). Combined, the inflammatory markers and age-adjusted inflammaging signature explained a relatively small proportion of the variance in AgeAccel (<12% in all cases), with generally similar associations across the 4 AgeAccel variables. The interdependence of inflammation and epigenetic aging appeared slightly greater at older ages, which might reflect a putative copotentiation of these 2 phenotypes in the biological aging process (3), but the differences in variance explained between baseline and follow-up were generally quite small (<5%). In the prospective analysis, we found that baseline inflammation markers and inflammaging contained little predictive information about epigenetic aging after an average of 11 years of follow-up. Finally, the epigenetic aging variables and the inflammaging signature were independently associated with mortality, with improved mortality prediction when the measures were used conjointly.
The finding that markers of inflammation are associated with AgeAccel cross-sectionally is consistent with previous research. Using data from the Genetics Of Lipid Lowering Drugs and Diet Network study, Irvin et al. (15) found strong evidence that extrinsic AgeAccel was cross-sectionally associated with IL-6, CRP, and TNF-α, although with small effect sizes. Similar findings were reported for extrinsic AgeAccel in a sample of adolescents, in this case with CRP, IFNγ-inducible protein of 10 kDa, and soluble TNF receptor 2 (41). Of the second-generation epigenetic age measures, PhenoAgeAccel has been cross-sectionally associated with increased CRP and increased transcription of genes within a number of inflammation-related pathways (such as response to lipopolysaccharide) (11). GrimAgeAccel has also been associated with increased concentrations of CRP cross-sectionally (12).
To the best of our knowledge, only the study by Stevenson et al. (17), using data from the Lothian birth cohort of 1936, has assessed prospective associations. The authors found evidence that extrinsic AgeAccel was associated with CRP and IL-6 cross-sectionally, but no evidence that baseline extrinsic AgeAccel was associated with either marker after 9 years of follow-up. Our findings, together with those of Stevenson et al., suggest that inflammation, at least insofar as it is captured by several well-established markers, may not be meaningfully longitudinally associated with epigenetic aging. There are, however, other plausible interpretations of this finding. First, there may exist a clear relationship between inflammation and later AgeAccel, although confined to inflammatory mediators not measured in this study. Second, it may be that the association between baseline inflammation and follow-up AgeAccel is more apparent after a shorter follow-up duration (eg, 5 years), and declines in magnitude with increasing follow-up time.
The effect sizes of the cross-sectional associations between inflammation and AgeAccel were generally modest. A SD increase in log IL-6, for instance, was associated with a 0.26 SD increase in GrimAgeAccel at follow-up (the largest coefficient observed). This is approximately equivalent to +1 year of GrimAgeAccel for each SD increase in log IL-6. An indication of the potential clinical importance of this effect size can be evaluated through comparison with previous literature. One recent study, using data from a nationally representative cohort of American adults aged over 50 years, found that class II obesity (BMI ≥ 35) was associated with +1.1 years of GrimAgeAccel, relative to those with an underweight or normal BMI (30). A recent study of non-Hispanic White women found that lifetime alcohol use had a relatively smaller association with GrimAgeAccel (+0.3 years per 135 drinks/year; approximately 1 SD) (42). These associations, however, are considerably smaller than those which have been reported for male sex (+3 years GrimAgeAccel) or current versus never smoking (+7.3 years GrimAgeAccel) (30). Thus, the magnitude of the cross-sectional association between inflammatory markers, particularly CRP and IL-6, and AgeAccel may be similar to or greater than that of some well-established risk factors of disease (ie, obesity and alcohol intake), though clearly less important than characteristics such as sex and smoking.
In the mortality analyses, a measure of biological aging based on inflammation (the age-adjusted inflammaging signature) and AgeAccel measures were independently associated with mortality. The addition of inflammaging to a model containing all AgeAccel variables improved the prediction of mortality (c-index increase from 0.61 to 0.64), indicating that the 2 sets of markers are highly complementary. This finding is consistent with the results of a recent investigation using data from the Framingham Offspring Study. The authors constructed a measure of “inflammation age” (distinct from the inflammaging signature included in this study) and found that this measure, extrinsic AgeAccel, and an additional measure of biological age based on clinical variables, were independently associated with mortality (43).
Strengths and novelties of this study include the use of a wide range of inflammation-related markers, including those of the TK pathway, the inclusion of the more recent “second-generation” epigenetic aging measures, and its longitudinal design. A primary limitation of the study relates to the potential for selection bias. Only participants who were alive at follow-up were eligible for inclusion in the sample. As mortality may be a common effect of inflammation and epigenetic age, conditioning on survival at follow-up could introduce selection bias. Additionally, 32% of participants did not attend the follow-up visit at wave 2 of the MCCS and could not be included in this study. If participation at follow-up was related to ill-health, associated with inflammation and epigenetic aging (3,4), this could introduce further selection bias. Each of these selection biases may induce a likely negative spurious association between AgeAccel and inflammation, potentially leading to an underestimation of the association between the 2 measures.
An additional issue is the circularity in some of the hypotheses. For instance, PhenoAge and DunedinPoAm were developed using an algorithm that included CRP as one of its inputs, and cystatin C is an input of GrimAge. It is therefore unsurprising that these inflammatory markers would be associated with the respective epigenetic age measures of which they form part. Nevertheless, it is interesting to note that associations for those pairs (eg, CRP and PhenoAge) were generally not larger than associations observed for other inflammation or TK markers. Another limitation is measurement error. Although the reliability of AgeAccel and the inflammatory markers was not assessed in this study, previous research indicates that epigenetic age variables have high reliability (44) while inflammatory marker variables are measured with more error (ICCs ~ 0.6) (10). Such measurement error was likely nondifferential and would likely have introduced bias towards the null, leading to an underestimation of the association between inflammation and epigenetic aging. We previously found in the MCCS that dried blood spots had lower reliability than peripheral blood mononuclear cells (24) which could have added to measurement error; nevertheless in, for example, the mortality analysis, the HRs were very similar to those obtained in other studies for inflammatory markers (45), PhenoAge (11) and GrimAge (12). Future studies may benefit from performing repeated assays of the inflammatory markers in order to explicitly capture measurement and biological variation and produce more reliable results. Recent iterations of the epigenetic age measures for DunedinPoAm (46), GrimAge, and PhenoAge (47) have substantially improved their test–retest reliability and should be considered in future studies. Another limitation relates to the use of only 2 time points. Two time-point studies are unable to assess the rate or trajectory of change over time, which may have offered greater insight into the longitudinal relationships between these measures (48). Additional time points (eg, in 5-year intervals) would allow assessment of temporal patterns in the prospective relationship between baseline inflammation and AgeAccel. Although our study included a comprehensive set of aging-related markers, many more exist that should be considered in future studies. For example, white blood cell proportions estimated from the methylation data were considered but not included in the current study, in order to focus on well-characterized biological markers of inflammation-related and epigenetic aging. It is nevertheless likely that the immune system is another putative marker of aging that may connect inflammation and epigenetic pathways and could contribute to further improvements in the prediction of mortality. Finally, we did not assess effect modification of the relationship between inflammation and epigenetic aging, nor of the relationship between inflammaging, epigenetic age, and mortality. It is plausible that these relationships may have differed according to characteristics such as sex, age, or genetic predisposition, and future studies should consider such a possibility.
Conclusion
In conclusion, cross-sectionally, but not longitudinally, concentrations of several inflammation-related markers were positively associated with epigenetic aging, but most of the variation in AgeAccel cannot be accounted for by consideration of a wide-ranging panel of inflammation-related markers. Epigenetic aging and inflammaging were independently associated with mortality in a large sample of older adults. These 2 sets of markers therefore offer complementary information that may be valuably used in combination, potentially improving upon existing models of biological aging. Future research should consider the combined use of markers of inflammaging and epigenetic age, alongside other candidate markers of biological age such as transcriptomic age (49) or metabolomic age (50) to better reflect the complexity of aging. Ultimately, the integration and consideration of markers representing the many facets of biological aging may contribute to improved prediction and understanding of age-related diseases.
Supplementary Material
Acknowledgments
Vital statuses were ascertained through the Victorian Cancer Registry and the Australian Institute of Health and Welfare, including the National Death Index and the Australian Cancer Database.
Contributor Information
Lachlan Cribb, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Parkville, Victoria, Australia.
Allison M Hodge, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Parkville, Victoria, Australia; Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, Victoria, Australia.
Chenglong Yu, Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, Victoria, Australia.
Sherly X Li, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Parkville, Victoria, Australia; Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, Victoria, Australia; Medical Research Council Epidemiology Unit, University of Cambridge, Cambridge, UK.
Dallas R English, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Parkville, Victoria, Australia; Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, Victoria, Australia.
Enes Makalic, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Parkville, Victoria, Australia.
Melissa C Southey, Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, Victoria, Australia; Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, Victoria, Australia; Department of Clinical Pathology, The University of Melbourne, Parkville, Victoria, Australia.
Roger L Milne, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Parkville, Victoria, Australia; Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, Victoria, Australia; Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, Victoria, Australia.
Graham G Giles, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Parkville, Victoria, Australia; Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, Victoria, Australia; Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, Victoria, Australia.
Pierre-Antoine Dugué, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Parkville, Victoria, Australia; Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, Victoria, Australia; Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, Victoria, Australia.
Funding
Melbourne Collaborative Cohort Study (MCCS) cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further augmented by Australian National Health and Medical Research Council grants 209057, 396414, and 1074383 and by infrastructure provided by Cancer Council Victoria. The nested case–control methylation studies were supported by the National Health and Medical Research Council grants 1011618, 1026892, 1027505, 1050198, 1043616, and 1074383. This work was further supported by National Health and Medical Research Council grants 1106016, 1088405, and 1164455. MCS is a National Health and Medical Research Council Senior Research Fellow (GNT1155163).
Conflict of Interest
None declared.
References
- 1. Kennedy BK, Berger SL, Brunet A, et al. Geroscience: linking aging to chronic disease. Cell. 2014;159(4):709–713. doi: 10.1016/j.cell.2014.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. Inflammaging: a new immune–metabolic viewpoint for age-related diseases. Nat Rev Endocrinol. 2018;14(10):576–590. doi: 10.1038/s41574-018-0059-4 [DOI] [PubMed] [Google Scholar]
- 4. Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- 5. Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine. 2017;21:29–36. doi: 10.1016/j.ebiom.2017.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dugué P-A, Li S, Hopper JL, Milne RL. Chapter 3—DNA methylation–based measures of biological aging. In: Tollefsbol TO, ed. Epigenetics in Human Disease. 2nd ed. Vol. 6. Academic Press; 2018:39–64. doi: 10.1016/B978-0-12-812215-0.00003-0 [DOI] [Google Scholar]
- 7. Dugué P-A, Bassett JK, Wong EM, et al. Biological aging measures based on blood DNA methylation and risk of cancer: a prospective study. JNCI Cancer Spectr. 2021;5(1). doi: 10.1093/jncics/pkaa109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4–S9. doi: 10.1093/gerona/glu057 [DOI] [PubMed] [Google Scholar]
- 9. Cervenka I, Agudelo LZ, Ruas JL. Kynurenines: tryptophan’s metabolites in exercise, inflammation, and mental health. Science. 2017;357(6349):eaaf9794. doi: 10.1126/science.aaf9794 [DOI] [PubMed] [Google Scholar]
- 10. Dugué P-A, Hodge AM, Ulvik A, et al. Association of markers of inflammation, the kynurenine pathway and B vitamins with age and mortality, and a signature of inflammaging. J Gerontol A Biol Sci Med Sci. 2022;77(4):826–836. doi: 10.1093/gerona/glab163 [DOI] [PubMed] [Google Scholar]
- 11. Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–591. doi: 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019;11(2):303–327. doi: 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y, Wilson R, Heiss J, et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat Commun. 2017;8(1):14617. doi: 10.1038/ncomms14617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Belsky DW, Caspi A, Arseneault L, et al. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. Elife. 2020;9. doi: 10.7554/eLife.54870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Irvin MR, Aslibekyan S, Do A, et al. Metabolic and inflammatory biomarkers are associated with epigenetic aging acceleration estimates in the GOLDN study. Clin Epigenetics. 2018;10(1):56. doi: 10.1186/s13148-018-0481-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stevenson AJ, McCartney DL, Harris SE, et al. Trajectories of inflammatory biomarkers over the eighth decade and their associations with immune cell profiles and epigenetic ageing. Clin Epigenetics. 2018;10(1):159. doi: 10.1186/s13148-018-0585-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Milne RL, Fletcher AS, MacInnis RJ, et al. Cohort profile: The Melbourne Collaborative Cohort Study (Health 2020). Int J Epidemiol. 2017;46(6):1757–1757i. doi: 10.1093/ije/dyx085 [DOI] [PubMed] [Google Scholar]
- 19. Dugué P-A, Wilson R, Lehne B, et al. Alcohol consumption is associated with widespread changes in blood DNA methylation: analysis of cross-sectional and longitudinal data. Addict Biol. 2021;26(1):e12855. doi: 10.1111/adb.12855 [DOI] [PubMed] [Google Scholar]
- 20. Dugué P-A, Bassett JK, Joo JE, et al. DNA methylation-based biological aging and cancer risk and survival: pooled analysis of seven prospective studies. Int J Cancer. 2018;142(8):1611–1619. doi: 10.1002/ijc.31189 [DOI] [PubMed] [Google Scholar]
- 21. Dugué P-A, Hodge AM, Wong EM, et al. Methylation marks of prenatal exposure to maternal smoking and risk of cancer in adulthood. Int J Epidemiol. 2021;50(1):105–115. doi: 10.1093/ije/dyaa210 [DOI] [PubMed] [Google Scholar]
- 22. Jayasekara H, English DR, Room R, MacInnis RJ. Alcohol consumption over time and risk of death: a systematic review and meta-analysis. Am J Epidemiol. 2014;179(9):1049–1059. doi: 10.1093/aje/kwu028 [DOI] [PubMed] [Google Scholar]
- 23. Joo JE, Wong EM, Baglietto L, et al. The use of DNA from archival dried blood spots with the Infinium HumanMethylation450 array. BMC Biotechnol. 2013;13:23. doi: 10.1186/1472-6750-13-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dugué P-A, English DR, MacInnis RJ, et al. Reliability of DNA methylation measures from dried blood spots and mononuclear cells using the HumanMethylation450k BeadArray. Sci Rep. 2016;6(1):30317. doi: 10.1038/srep30317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R1153156. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Midttun O, Hustad S, Ueland PM. Quantitative profiling of biomarkers related to B-vitamin status, tryptophan metabolism and inflammation in human plasma by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2009;23(9):1371–1379. doi: 10.1002/rcm.4013 [DOI] [PubMed] [Google Scholar]
- 27. Schröcksnadel K, Wirleitner B, Winkler C, Fuchs D. Monitoring tryptophan metabolism in chronic immune activation. Clin Chim Acta. 2006;364(1-2):82–90. doi: 10.1016/j.cca.2005.06.013 [DOI] [PubMed] [Google Scholar]
- 28. Ulvik A, Midttun O, Pedersen ER, Eussen SJ, Nygård O, Ueland PM. Evidence for increased catabolism of vitamin B-6 during systemic inflammation. Am J Clin Nutr. 2014;100(1):250–255. doi: 10.3945/ajcn.114.083196 [DOI] [PubMed] [Google Scholar]
- 29. Ueland PM, Ulvik A, Rios-Avila L, Midttun O, Gregory JF. Direct and functional biomarkers of vitamin B6 status. Annu Rev Nutr. 2015;35:33–70. doi: 10.1146/annurev-nutr-071714-034330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crimmins EM, Thyagarajan B, Levine ME, Weir DR, Faul J. Associations of age, sex, race/ethnicity, and education with 13 epigenetic clocks in a nationally representative U.S. sample: The Health and Retirement Study. J Gerontol A Biol Sci Med Sci. 2021;76(6):1117–1123. doi: 10.1093/gerona/glab016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dugué P-A, Bassett JK, Joo JE, et al. Association of DNA methylation-based biological age with health risk factors and overall and cause-specific mortality. Am J Epidemiol. 2018;187(3):529–538. doi: 10.1093/aje/kwx291 [DOI] [PubMed] [Google Scholar]
- 32. Yu C, Hodge AM, Wong EM, et al. Does genetic predisposition modify the effect of lifestyle-related factors on DNA methylation? Epigenetics. 2022:1–10. doi: 10.1080/15592294.2022.2088038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fiorito G, McCrory C, Robinson O, et al. Socioeconomic position, lifestyle habits and biomarkers of epigenetic aging: a multi-cohort analysis. Aging (Albany NY). 2019;11(7):2045–2070. doi: 10.18632/aging.101900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stekhoven DJ, Bühlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28(1):112–118. doi: 10.1093/bioinformatics/btr597 [DOI] [PubMed] [Google Scholar]
- 35. Carvalho CM, Polson NG, Scott JG. The horseshoe estimator for sparse signals. Biometrika. 2010;97(2):465–480. doi: 10.1093/biomet/asq017 [DOI] [Google Scholar]
- 36. Piironen J, Vehtari A. Sparsity information and regularization in the horseshoe and other shrinkage priors. Electron J Stat. 2017;11(2):5018–5051. doi: 10.1214/17-EJS1337SI [DOI] [Google Scholar]
- 37. Bürkner P-C. brms: an R package for Bayesian multilevel models using Stan. J Stat Soft. 2017;80(1):1–28. doi: 10.18637/jss.v080.i01 [DOI] [Google Scholar]
- 38. Gelman A, Goodrich B, Gabry J, Vehtari A. R-squared for Bayesian regression models. Null. 2019;73(3):307–309. doi: 10.1080/00031305.2018.1549100 [DOI] [Google Scholar]
- 39. Harrell FEJ, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247(18):2543–2546. [PubMed] [Google Scholar]
- 40. Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21(1):128–138. doi: 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang R-C, Lillycrop KA, Beilin LJ, et al. Epigenetic age acceleration in adolescence associates with BMI, inflammation, and risk score for middle age cardiovascular disease. J Clin Endocrinol Metab. 2019;104(7):3012–3024. doi: 10.1210/jc.2018-02076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kresovich JK, Martinez Lopez AM, Garval EL, et al. Alcohol consumption and methylation-based measures of biological age. J Gerontol A Biol Sci Med Sci. 2021;76(12):2107–2111. doi: 10.1093/gerona/glab149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murabito JM, Zhao Q, Larson MG, et al. Measures of biologic age in a community sample predict mortality and age-related disease: the Framingham Offspring Study. J Gerontol A Biol Sci Med Sci. 2018;73(6):757–762. doi: 10.1093/gerona/glx144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ori APS, Lu AT, Horvath S, Ophoff RA. A systematic evaluation of 41 DNA methylation predictors across 101 data preprocessing and normalization strategies highlights considerable variation in algorithm performance. bioRxiv, doi: 10.1101/2021.09.29.462387 [DOI] [Google Scholar]
- 45. Zuo H, Ueland PM, Ulvik A, et al. Plasma biomarkers of inflammation, the kynurenine pathway, and risks of all-cause, cancer, and cardiovascular disease mortality: the Hordaland Health Study. Am J Epidemiol. 2016;183(4):249–258. doi: 10.1093/aje/kwv242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Belsky DW, Caspi A, Corcoran DL, et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife. 2022;11:e73420. doi: 10.7554/eLife.73420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Higgins-Chen A, Thrush K, Hu-Seliger T, Wang Y, Hagg S, Levine M. A Computational solution to bolster epigenetic clock reliability for clinical trials and longitudinal tracking. Innov Aging. 2021;5(Suppl 1):5. doi: 10.1093/geroni/igab046.015 [DOI] [Google Scholar]
- 48. Singer JD, Willett JB.. Applied Longitudinal Data Analysis: Modeling Change and Event Occurrence. New York: Oxford Academic; 2003. doi: 10.1093/acprof:oso/9780195152968.001.0001 [DOI] [Google Scholar]
- 49. Peters MJ, Joehanes R, Pilling LC, et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6(1):8570. doi: 10.1038/ncomms9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robinson O, Chadeau Hyam M, Karaman I, et al. Determinants of accelerated metabolomic and epigenetic aging in a UK cohort. Aging Cell. 2020;19(6):e13149. doi: 10.1111/acel.13149 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.