Abstract
The mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) senses and relays environmental signals from growth factors and nutrients to metabolic networks and adaptive cellular systems to control the synthesis and breakdown of macromolecules. However, beyond inducing de novo lipid synthesis, the role of mTORC1 in controlling cellular lipid content remains poorly understood. Here we show that inhibition of mTORC1 via small molecule inhibitors or nutrient deprivation leads to the accumulation of intracellular triglycerides in both cultured cells and a mouse tumor model. The elevated triglyceride pool following mTORC1 inhibition stems from the lysosome-dependent, but autophagy independent, hydrolysis of phospholipid fatty acids. The liberated fatty acids are available for either triglyceride synthesis or beta-oxidation. Distinct from the established role of mTORC1 activation in promoting de novo lipid synthesis, our data indicate that mTORC1 inhibition triggers membrane phospholipid trafficking to the lysosome for catabolism and an adaptive shift in the use of constituent fatty acids for storage or energy production.
Editor summary:
Hosios et al. demonstrate that inhibition of mechanistic target of rapamycin complex 1 (mTORC1) in cells and in tumors in mice leads to a lysosome-dependent but autophagy-independent shift in membrane lipid metabolism, resulting in increased intracellular triglyceride pools.
Introduction
The coordinated regulation of anabolic and catabolic metabolism allows cells to adjust to their nutrient environment, promoting macromolecular synthesis when conditions are favorable to growth and engaging in adaptive metabolic pathways when nutrients are limited. While our knowledge of cell-intrinsic mechanisms that balance protein synthesis, folding, and degradation (i.e. proteostasis) between nutrient-rich and nutrient-poor conditions has advanced1-4, far less is known about cellular mechanisms influencing distinct lipid species in cells. A similar regulated balance must exist between intracellular fatty acids, membrane phospholipids, and neutral lipid stores that is coupled to both the nutrient state of the cell and to other co-occurring metabolic adaptations.
Mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) is a protein kinase complex that couples growth signals to the control of anabolic and catabolic metabolism3,5,6. mTORC1 is activated by growth factors and hormones (such as insulin) in normal cells and frequently via oncogenic mutations affecting its upstream regulation in human cancers. The ubiquitous growth-factor responsive phosphoinositide-3-kinase (PI3K)-AKR thymoma (AKT) pathway inactivates the tuberous sclerosis complex (TSC) protein complex, composed of TSC1, TSC2, and TBC1 domain family member 7 (TBC1D7), which is an essential inhibitor of mTORC1. Once active, mTORC1 stimulates protein synthesis and other biosynthetic processes through direct downstream effectors, including the S6 kinases (S6K) and eIF4E-binding proteins (4E-BP)5, and various transcription factors3,7. Notably, mTORC1 activates the sterol regulatory element binding protein (SREBP) transcription factors to induce the de novo synthesis of fatty acids and sterols8-11.
Beyond its regulation by growth factor and oncogenic signaling, mTORC1 activity is also responsive to the cellular nutrient environment3,5,12. Best characterized of these nutrient inputs into mTORC1 regulation is amino acid availability, which is sensed via pathways regulating the Rag GTPases12,13. In addition, mTORC1 is inhibited upon depletion of intracellular energy (e.g., ATP), glucose, oxygen, or cholesterol3,5,12,14. Collectively, signals reflecting the sufficient presence of these nutrients converge with signals from exogenous growth factors to promote mTORC1 activity when the cellular nutrient environment is favorable for anabolic growth and to inhibit it when nutrients are scarce. When mTORC1 activity is reduced upon nutrient deprivation, adaptive processes promoting catabolism and nutrient and energy conservation are activated. Bulk autophagy is induced when mTORC1 is inhibited, promoting the engulfing of organelles and cytosolic macromolecules into autophagosomes that fuse with the lysosome for degradation15. Autophagy allows cells to recycle nutrients and macromolecular building blocks for cellular homeostasis when nutrients are scarce. mTORC1 may also suppress the catabolism of nutrients scavenged from the extracellular environment through macropinocytosis by inhibiting their delivery to the lysosome through an unknown mechanism16.
mTORC1 regulates cellular lipid metabolism through several distinct mechanisms17. As noted, mTORC1 stimulates de novo synthesis of fatty acids and cholesterol through the SREBP family of transcription factors, an effect observed in response to physiological signals in the liver and oncogenic signals in cancer8-11,18,19. SREBP1 induces expression of all the enzymes needed for the production of fatty acids20 and may also induce those required for phospholipid biosynthesis21, perhaps allowing the mTORC1-SREBP1 circuit to directly enhance membrane biogenesis as part of a broader cell growth program. Additional control of lipid metabolism by mTORC1 has been documented in hepatocytes and adipocytes, specialized cell types affecting systemic lipid homeostasis. In hepatocytes, for example, mTORC1 has been found to stimulate phosphatidylcholine synthesis, which subsequently promotes lipoprotein secretion22, and to suppress fatty acid β-oxidation and ketogenesis23. In adipocytes, mTORC1 activation appears to promote triglyceride accumulation by activating de novo fatty acid synthesis, as well as suppressing lipolysis24, the regulation of which has been hypothesized to underlie the hyperlipidemia widely observed in rodents or humans chronically administered the mTOR inhibitor rapamycin or its analogs25,26. Nevertheless, how growth signals affecting the activation state of mTORC1 influence cell-intrinsic changes in lipid species remains underexplored.
A cell’s nutrient environment can profoundly influence lipid metabolism. Recent work has shown that hypoxia inhibits de novo lipid synthesis, rendering cells dependent on an exogenous source of fatty acids to grow27. Growth in hypoxic or lipid-depleted environments can also alter cellular fatty acid saturation, and adapting lipid metabolism to such environments is important to enabling cell and tumor growth28-30. Hypoxic cells also accumulate triglyceride stores in lipid droplets, an adaptation that may support survival and proliferation in this context28,31. Finally, nutrient starvation can lead to an accumulation of lipid droplets in cells, and prior studies have suggested that this effect is dependent on autophagy induced upon inactivation of mTORC132,33.
Seeking to identify how cells coordinate the synthesis and interconversion of complex lipid species with other growth processes, we used an unbiased lipidomics approach to quantify changes in cellular lipid species when mTORC1 is activated and inhibited. We observed numerous lipid classes being influenced by the activation state of mTORC1, including triglycerides, which were robustly and reproducibly elevated in cells and tumors following mTORC1 inhibition. We demonstrate that mTORC1 inhibition, via small molecule inhibitors or amino acid deprivation, stimulates lysosomal hydrolysis of phospholipids resulting in availability of the liberated fatty acids for use in either triglyceride synthesis or fatty acid oxidation. This adaptive shift in intracellular lipid species upon mTORC1 inhibition occurs independently of autophagy but involves endosomal delivery to the lysosome. Thus, this study reveals a process of lipid remodeling under poor growth conditions, resulting in the release of fatty acids from membrane phospholipids, facilitating their use for energy storage or production under conditions where de novo fatty acid synthesis is suppressed.
Results
Cells accumulate triglycerides following mTORC1 inhibition
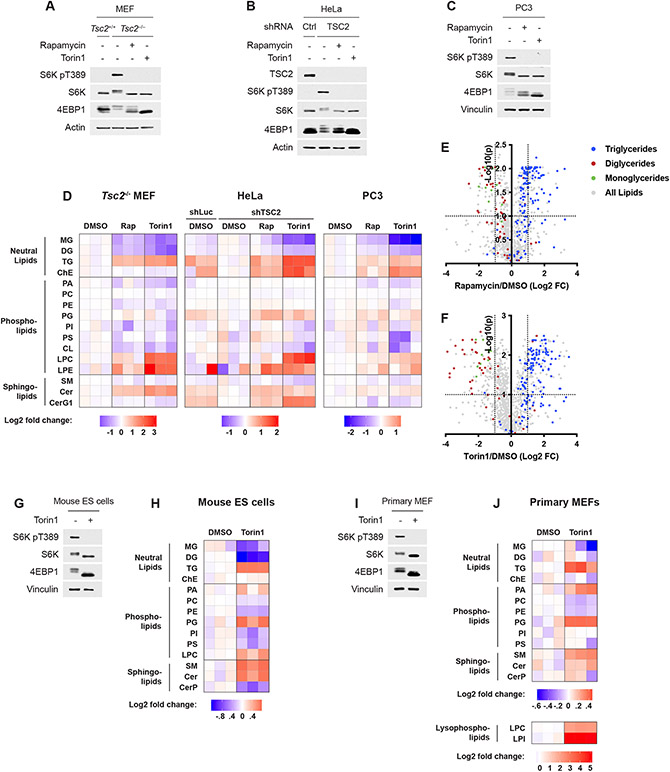
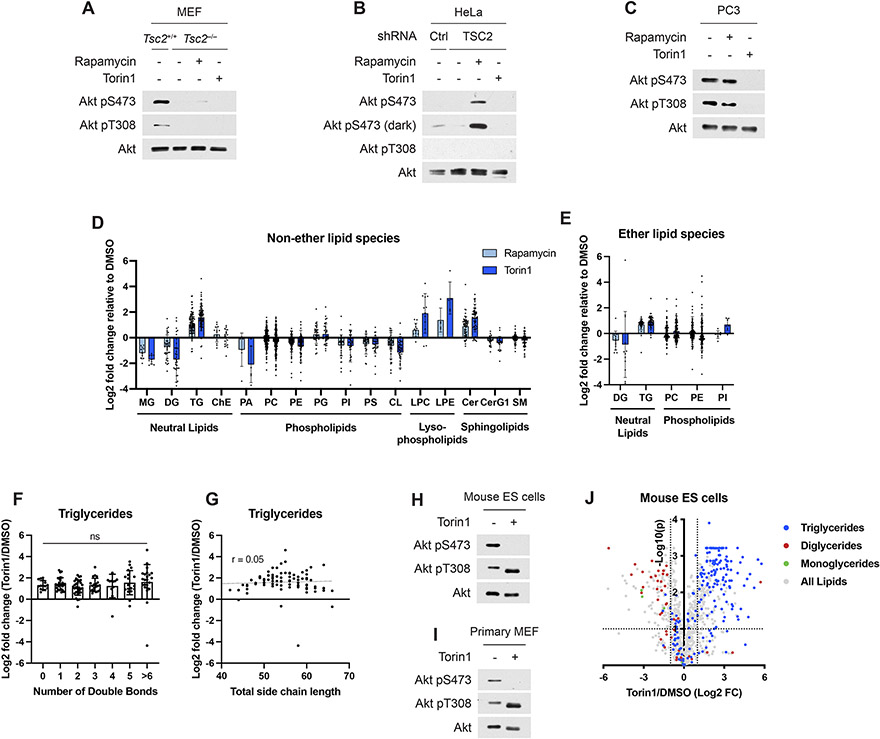
We profiled the lipidomes of three cell lines to identify lipid species whose abundance was affected by the activation state of mTORC1 (Fig. 1A-D). To isolate the PI3K-mTORC1 pathway from other growth factor-responsive pathways, we initially profiled three distinct cellular settings with genetic activation of the pathway. mTORC1 is activated in a growth factor-independent manner, as monitored by phosphorylation of S6K1 and a mobility shift in 4E-BP1, in Tsc2−/− mouse embryonic fibroblasts (MEFs), HeLa cells with shRNA-mediated knockdown of TSC2, and in PTEN null PC3 cells (Fig. 1A-C and Extended Data Fig. 1A-C). The effects of mTORC1 inhibition using both the partial, allosteric inhibitor rapamycin and the catalytic domain inhibitor torin1 on the abundance of different lipid classes (Fig. 1D) and individual lipid species (Extended Data Fig. 1D,E) was measured. Several lipid classes changed consistently with mTORC1 activity, most pronounced of which was an increase in triglyceride (TG) abundance when mTORC1 was inhibited. This was true for the summed ion counts for the whole class (Fig. 1D) and for the fold changes of each lipid individually (Fig. 1E,F and Extended Data Fig. 1D,E). While both inhibitors increased TGs, torin1, the more complete mTOR inhibitor, had a stronger effect. TG species were elevated following torin1 treatment regardless of their degree of saturation or total fatty-acid side chain length (Extended Data Fig. 1F,G). These lipidomic changes were also observed in primary cells in the presence of growth factors, including mouse embryonic stem cells and wild type, primary MEFs (Fig. 1G-J and Extended Data Fig. 1H-J), indicating that inhibiting mTORC1 leads to triglyceride accumulation in both genetically modified and normal cells. Other lipid classes also varied with mTORC1 activity across cellular settings, with mono- and diglycerides depleted and lysophospholipids and ceramides increased upon mTORC1 inhibition (Fig. 1D-F,H,J and Extended Data Fig. 1D,J, see below). Other classes, including cholesteryl esters, were influenced by mTORC1 activity in only a subset of the five cell lines analyzed via lipidomics.
Figure 1: Lipidomic analysis of cells treated with mTORC1 inhibitors.
(A-C) Immunoblots for Tsc2−/− MEFs (A), HeLa cells expressing shRNAs targeting luciferase (shLuc) or TSC2 (B), and PC3 cells (C) treated with vehicle (DMSO), rapamycin (20 nM), or torin1 (250 nM) for 16 h.
(D) Relative lipid class abundance determined by mass spectrometry lipidomics from cells treated in (A-C). Class abundance, determined from the summed ion counts of all lipids in that class, are shown normalized to vehicle-treated controls. MG, monoglyceride; DG, diglyceride; TG, triglyceride; ChE, cholesteryl ester; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; CL, cardiolipin; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; Cer, ceramide; SM, sphingomyelin; CerG1, glucosylceramide.
(E,F) Changes in the abundance of individual lipid species from the experiment in (D), with neutral lipids color coded, for Tsc2−/− MEFs treated with (E) rapamycin and (F) torin1. Dotted lines indicate a 2-fold change (FC) and an adjusted p-value of 0.1.
(G-J) Immunoblots (G,I) and relative lipid class abundance (H,J) determined by mass spectrometry lipidomics mouse embryonic stem (ES) cells (G,H) and wild type, primary MEFs (I,J) treated with vehicle (DMSO) or torin1 (250 nM) in the presence of serum for 16 h.
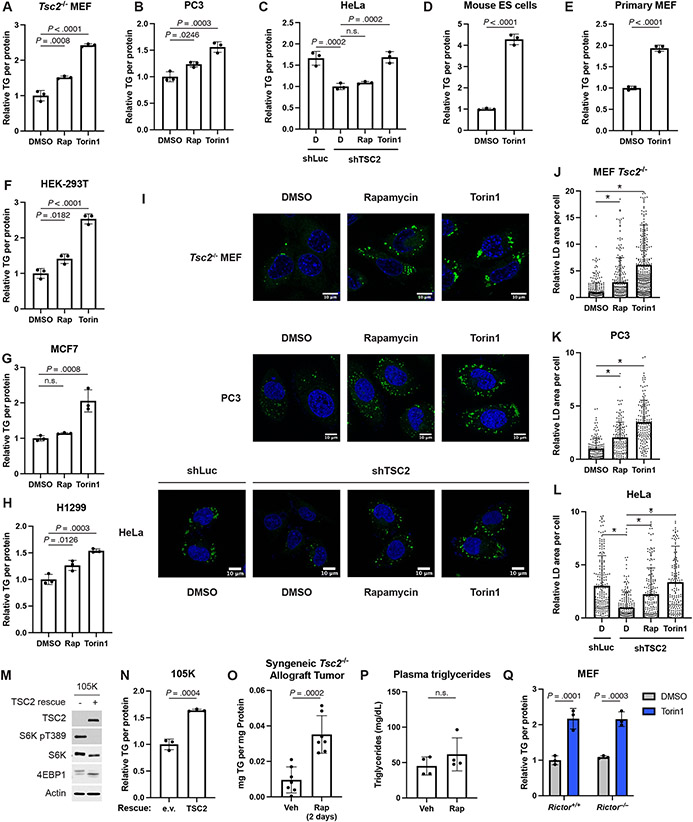
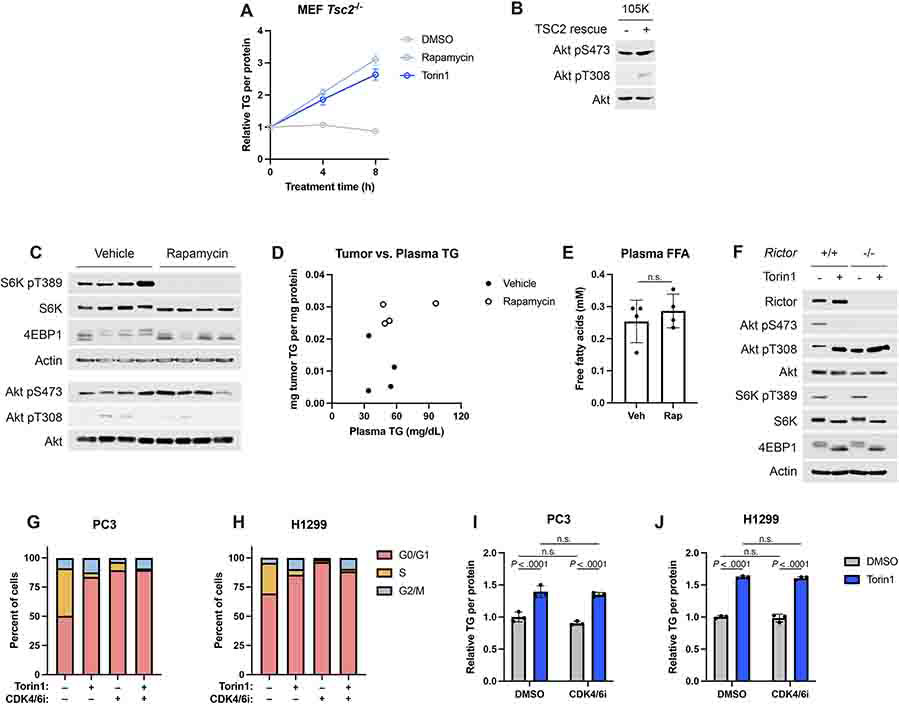
The elevation in total TG abundance following mTORC1 inhibition was validated in the Tsc2−/− MEFs, PC3, HeLa, and two sets of primary cells using an enzymatic assay (Fig. 2A-E), with TG levels beginning to rise within 4 hours of mTORC1 inhibition (Extended Data Fig. 2A). The effects of mTORC1 inhibitors on intracellular TGs was extended to three additional human cell lines (Fig. 2F-H), with torin1 again exerting a stronger effect than rapamycin on the increase in TGs. Cells store TGs and other neutral lipids in lipid droplets, and lipid droplet biogenesis can be induced when these lipid species are elevated. Consistent with this, treatment with rapamycin or torin1 was sufficient to increase lipid droplet area in Tsc2−/− MEFs, PC3, and HeLa cells (Fig. 2I-L). Rescuing expression of TSC2 in the murine Tsc2−/− renal-tumor-derived 105K cell line34 also suppressed mTORC1 signaling and increased TG levels (Fig. 2M,N and Extended Data Fig. 2B). Our findings were further extended to a syngeneic allograft tumor model in which mice bearing tumors derived from 105K cells were treated with vehicle or rapamycin for just two days to avoid confounding effects of tumors shrinking. Compared to vehicle, rapamycin treatment inhibited mTORC1 and significantly raised tumor TGs (Fig. 2O and Extended Data Fig. 2C). Although prolonged rapamycin treatment is known to raise plasma TGs in mice and humans26, neither plasma TGs nor free fatty acids changed significantly in these tumor-bearing mice with this short treatment (Fig. 2P and Extended Data Fig. 2D,E), suggesting that the change in tumor lipids could be tumor cell-autonomous. A caveat to this interpretation is that these measurements were made on ad libitum fed mice at the beginning of the light cycle, and if plasma TGs were elevated with rapamycin treatment at other times of day, this could influence tumor TG abundance. At least at this time point, there is no direct relationship between the abundance of tumor TGs and plasma TGs from the same mice (Extended Data Fig. 2D). We specifically used the Tsc2−/− tumor model in this experiment to uncouple tumor mTORC1 activity from extracellular hormones, such as insulin. Nonetheless, we cannot rule out a systemic effect of insulin on lipid metabolism influencing tumor TG content.
Figure 2: Cells accumulate TGs following mTORC1 inhibition.
(A-C) TGs measured by enzyme assay and normalized to protein content in Tsc2−/− MEFs (A), PC3 (B), and HeLa cells (C) treated as in Figure 1A-D graphed as mean ± SD relative to vehicle-treated cells, n=3.
(D,E) TGs measured and graphed as in (A-C) in mouse embryonic stem (ES) cells (D) and wild type, primary MEFs (E) treated as in Figure 1G-J.
(F-H) Triglyceride levels quantified by enzyme assay and normalized to protein content from the same sample in HEK-293T (F), MCF7 (G), and H1299 (H) cells following 16 hrs of vehicle, rapamycin (20 nM), or torin1 (250 nM) treatment, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(I) Representative images of lipid droplets (stained with Bodipy 493/503, green) in Tsc2−/− MEFs, PC3, and HeLa cells treated as in (A-C). Nuclei are shown in blue.
(J-L) Lipid droplet quantification for individual cells from (I) for Tsc2−/− MEFs (J), PC3 (K), and HeLa cells (L), graphed as mean ± SD relative to vehicle-treated cells, n=146–460 cells. * indicates P < .0001.
(M,N) Immunoblot (M) and triglycerides determined by enzyme assay (N) and normalized to protein content from the same sample in 105K cells re-expressing TSC2 or an empty vector, following 16 hrs of serum starvation, graphed as mean ± SD relative to empty-vector cells, n=3.
(O,P) TGs measured as in (A-C) in tumor (O) and plasma (P) from 105K tumor-bearing mice treated with vehicle or rapamycin (1 mg/kg) for two days, graphed as mean ± SD, n=7 tumors and n=4 plasma samples.
(Q) TGs in Rictor+/+ and −/− MEFs treated for 16 h with vehicle or Torin1 (250 nM), measured as in (A-C) and graphed as mean ± SD, n=3.
Statistical analysis by one-way ANOVA (A-C,F-H,J-L), two-tailed Student’s t-test (D,E), and two-way ANOVA (Q). n.s., not significant.
As a potent mTOR inhibitor, torin1 also inhibits mTORC2, a second mTOR kinase complex that is physically and functionally distinct from mTORC15 and phosphorylates Akt on S473 (Extended Data Fig. 2F). However, torin1 treatment increased TGs in MEFs lacking the essential mTORC2 component Rictor to a similar extent to wild-type MEFs, demonstrating that this effect is through mTORC1 inhibition (Fig. 2Q). Finally, mTORC1 inhibition, especially with mTOR catalytic domain inhibitors, such as torin1, can cause arrest or delay in the G1 phase of the cell cycle (Extended Data Fig. 2G,H), raising the possibility that cell cycle effects might contribute to the observed increase in TGs upon mTORC1 inhibition. PC3 and H1299 cells are sensitive to CDK4/6 inhibition for G1 arrest, so we chose these cell lines to compare the effects of torin1 to the CDK4/6 inhibitor palbociclib on cellular lipids. Despite the strong G1 arrest, treatment with the CDK4/6 inhibitor was not sufficient to raise TG levels nor did it alter the ability of torin1 to do so (Extended Data Fig. 2I,J).
Accumulated TGs derive from remodeled endogenous lipids
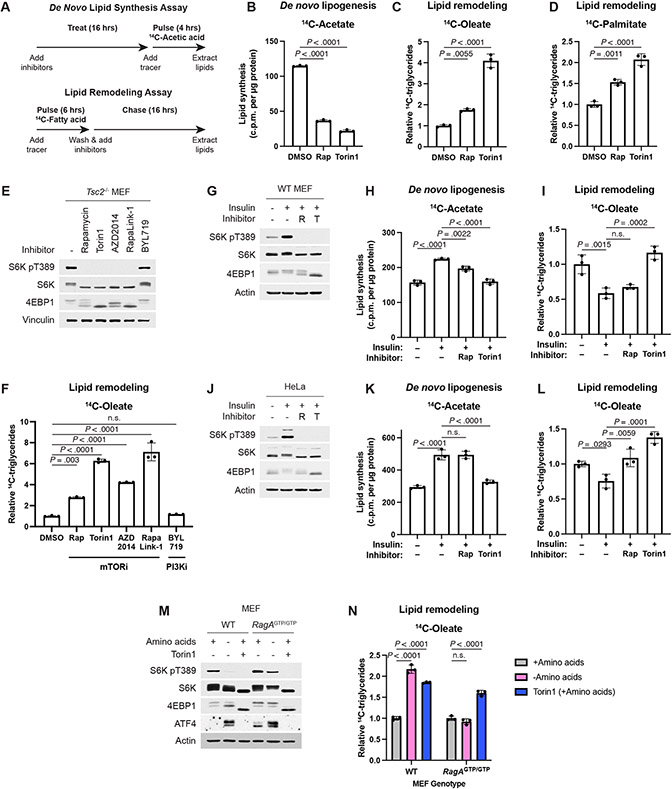
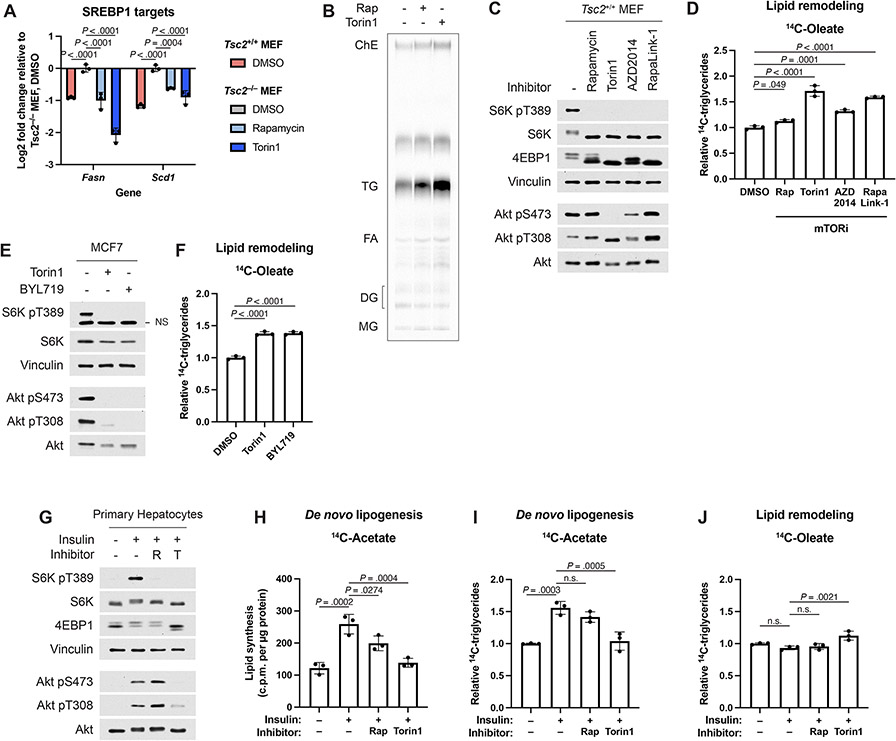
The fatty acids comprising the TGs increased by mTOR inhibitors could arise from de novo synthesis or from remodeling of fatty acid side chains from existing intracellular lipid species. We used two radioactive tracing assays to distinguish regulation of de novo lipogenesis and remodeling of endogenous lipids within the cell (Fig. 3A). De novo lipid synthesis was assayed by treating cells with mTORC1 inhibitors prior to 4 hours of labeling with 14C-acetate and measuring incorporation into a lipid extract. Consistent with mTORC1 stimulating de novo fatty acid synthesis via the SREBP transcription factors8-10, mTORC1 inhibitors reduced de novo lipid synthesis (Fig. 3B) as well as the expression of SREBP1 target genes required for fatty acid synthesis (Extended Data Fig. 3A). Thus, de novo fatty acid synthesis cannot account for the increase in TG levels upon mTORC1 inhibition.
Figure 3: mTORC1 suppresses endogenous lipid remodeling.
(A) Schematic for radioactive pulse-chase experiments conducted in this study. Lipid remodeling is assayed by a pulse-chase using 14C-fatty acids, and de novo lipid synthesis is assayed by labeling cells with 14C-acetic acid.
(B) De novo lipogenesis measured in Tsc2−/− MEFs treated vehicle, rapamycin (20 nM), or torin1 (250 nM) for 16 hrs, followed by 4 hrs labeling, as in (A), with [1-14C]-acetate, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(C,D) TGs measured in Tsc2−/− MEFs treated vehicle, rapamycin (20 nM), or torin1 (250 nM) for 16 hrs, after pulse-chase labeling, as in (A) with (C) [1-14C]-oleate or (D) [1-14C]-palmitate, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(E,F) Immunoblot (E) and TGs (F) assayed by pulse-chase with [1-14C]-oleate tracer (6 h) in Tsc2−/− MEFs followed by chase in cold medium for 16 h with vehicle, 20 nM rapamycin, 250 nM torin1, 500 nM AZD2014, 5 nM RapaLink-1, or 1 μM BYL719 treatment, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(G-L) Immunoblots (G,J), de novo lipogenesis (H,K) and lipid remodeling (I,L), assayed as in (B-D), from wild-type MEFs (G-I) and HeLa cells (J-L) serum starved for 16 h, pre-treated with vehicle, rapamycin (20 nM), or torin1 (250 nM) for 30 min, and stimulated with insulin (100 nM) for 8 h, graphed as mean ± SD relative to vehicle-treated unstimulated cells, n=3.
(M,N) Immunoblot (M) and 14C-TGs (N) in wild type and RagAGTP/GTP (RagAQ66L) MEFs following 16 h in amino acid-replete medium with vehicle or torin1 (250 nM) or amino acid-free medium with vehicle, measured and graphed as mean ± SD relative to amino acid replete, vehicle-treated cells, n=3.
Statistical analysis by one-way ANOVA (B-D,F,H,I,K,L) and two-way ANOVA (N). n.s., not significant.
We next used a radioactive pulse-chase assay to investigate the remodeling dynamics of existing intracellular lipid pools upon mTORC1 inhibition (Fig. 3A). Intracellular lipid species were pulse labeled (6 hours) with trace amounts of 14C-oleate or -palmitate, representing the two most common fatty acids in cells, and then treated with vehicle or mTORC1 inhibitors during the chase phase of the experiment. Neutral lipids were subsequently resolved by thin layer chromatography (TLC) (Example shown in Extended Data Fig. 3B). As seen in the steady state measurements above, labeled TGs accumulated from both oleate and palmitate tracers following treatment with rapamycin or torin1, indicating that they derive from an intracellular source of lipids affected by mTORC1 inhibition (Fig. 3C,D).
The structurally unrelated mTOR catalytic domain inhibitor AZD2014 and a hybrid compound of a rapamycin analog linked to an mTOR catalytic domain inhibitor, RapaLink-1, also increased TG accumulation from remodeled fatty acids under conditions of both genetic (Tsc2−/− MEFs) and serum-stimulated (Tsc2+/+) mTORC1 activation (Fig. 3E,F and Extended Data Fig. 3C,D). In the human breast cancer cell line MCF7, which carries a PI3K-p110α activating mutation (PIK3CAE545K) that drives growth factor-independent mTORC1 activation, both torin1 and the PI3K-p110α-selective inhibitor BYL719 likewise caused an accumulation of TGs derived from fatty acid remodeling (Extended Data Fig. 3E,F). As loss of TSC2 activates mTORC1 independent of PI3K activity, BYL719 did not alter mTORC1 signaling or TG levels in Tsc2−/− MEFs (Fig. 3E,F), also confirming that this inhibitor does not act directly on mTOR, as is common for many PI3K inhibitors.
Insulin and nutrient availability regulate lipid remodeling
We next assessed how physiological signals regulating mTORC1 influence lipid metabolism using cell lines that lack mutations in the PI3K-mTORC1 pathway and are, thus, growth-dependent for mTORC1 activation. Consistent with insulin stimulating de novo fatty acid synthesis, at least in part, through mTORC1-dependent mechanisms11,18, in both wild-type MEFs and HeLa cells, insulin promoted de novo lipid synthesis from acetate in a manner sensitive to mTOR inhibitors (Fig. 3G-L). However, in parallel pulse-chase assays, insulin decreased remodeling of endogenous fatty acids into TGs, and this suppression was blocked with mTOR inhibitors, especially torin1. Consistent with previous studies11,18, insulin-stimulated de novo lipid synthesis through mTORC1 in primary hepatocytes, including incorporation of acetate-derived fatty acids into TGs (Extended Data Fig. 3G-I). However, neither insulin nor mTOR inhibitors strongly influenced remodeling of endogenous fatty acids into TGs in hepatocytes (Extended Data Fig. 3J), indicating distinct cell-intrinsic mechanisms of regulation in this specialized setting that influences whole-body lipid metabolism. Unlike wild-type cells, cells expressing a constitutively GTP-bound RagA mutant (RagAGTP/GTP) sustain mTORC1 signaling upon amino acid withdrawal, while other amino acid-sensing mechanisms, such as the integrated stress response leading to the induction of ATF4, remain intact (Fig. 3M). Importantly, amino acid withdrawal inhibited mTORC1 and lead to an increase in TG levels in wild type MEFs, but not in the RagAGTP/GTP cells, whereas torin1 raises TG levels in both cell lines (Fig. 3N). These experiments confirm that nutrient and growth-factor inputs to mTORC1, in addition to pharmacological inhibitors, can similarly influence cellular TG content.
TG synthesis is associated with phospholipid catabolism
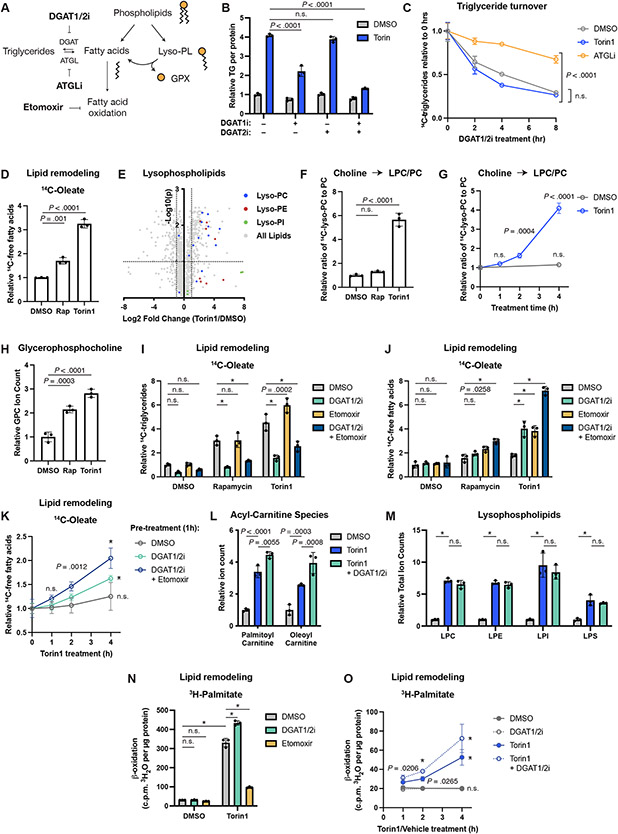
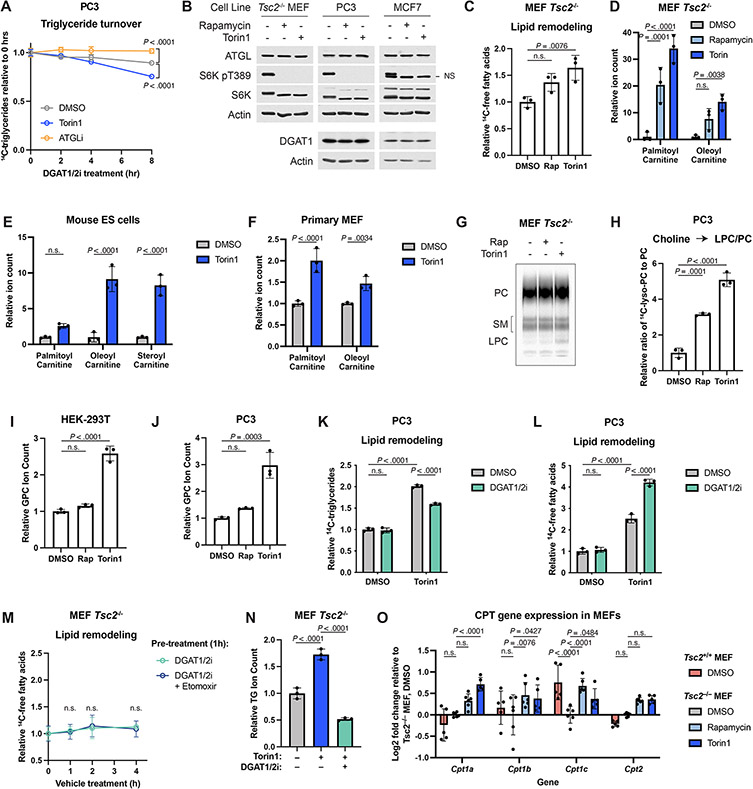
Diacylglycerol acyltransferase 1 and 2 (DGAT1 and 2) catalyze the final step in TG biosynthesis from DG and a fatty acyl-CoA (Fig. 4A). Inhibiting DGAT1 significantly reduced the torin1-mediated increase in TG levels, while DGAT2 inhibition alone had no effect (Fig. 4B). Combined DGAT1 and 2 inhibition further reduced TG abundance, indicating that cells synthesize TGs when mTORC1 is inhibited, with DGAT1 playing a predominant role. In contrast, TG turnover, as assayed by the disappearance of radiolabeled TGs following treatment with DGAT inhibitors, was not inhibited by torin1 (Fig. 4C and Extended Data Fig. 4A). Rates of TG turnover vary widely between the two cell systems examined (Tsc2−/− MEFs and PC3 cells), but distinct from inhibition of adipose TG lipase (ATGL), which significantly slowed TG turnover in both lines, torin1 failed to do so. mTORC1 inhibitors also did not change ATGL or DGAT1 protein levels (Extended Data Fig. 4B). Thus, intracellular TG levels rise upon mTORC1 inhibition via increased synthesis, not decreased turnover.
Figure 4: TG accumulation is linked to other changes in lipid metabolism.
(A) Relationship between phospholipids and neutral lipids. Lyso-PL, lysophospholipid; GPX, glycerophosphodiester.
(B) TGs measured enzymatically in Tsc2−/− MEFs treated with vehicle, torin1 (250 nM), and DGAT1 and/or DGAT2 inhibitor (each at 3 μM) for 16 hrs. Mean ± SD relative to vehicle, n=3.
(C) TG turnover in Tsc2−/− MEFs pulse-labeled with [1-14C]-oleate. Following 1 hr pre-treatment with Torin1 or ATGLi (20 μM), TG synthesis was inhibited with DGAT inhibitors (3 μM each). 14C-TGs graphed as mean ± SD relative to 0 hrs, n=3.
(D) 14C-Fatty acids in Tsc2−/− MEFs treated with vehicle, rapamycin (20 nM), or torin1 (250 nM) for 16 hrs, measured as in Figure 3C. Mean ± SD, n=3.
(E) Individual lysophospholipid species from the experiment in Tsc2−/− MEFs in Figure 1D. Dotted lines indicate a 2-fold change and an adjusted p-value of 0.1.
(F,G) Ratio of lysophosphatidylcholine to phosphatidylcholine in Tsc2−/− MEFs pulse-chased [methyl-14C]-choline, then treated with inhibitors for 16 hrs (F) or 0-4 hrs (G). Mean ± SD relative to vehicle (F) or relative to 0 hrs (G), n=3.
(H) Glycerophosphocholine levels following 16 hrs of inhibitor treatment in Tsc2−/− MEFs; mean ± SD relative to vehicle-treated cells, n=3.
(I,J) 14C-TG (I) and 14C-fatty acids (J) in Tsc2−/− MEFs treated for 16 hrs with vehicle, rapamycin, or torin1 in the presence of vehicle, DGAT inhibitors (3 μM each), and/or etomoxir (20 μM); mean ± SD, n=3.
(K) 14C-Fatty acids in Tsc2−/− MEFs following torin1 treatment after 1 hr pre-treatment with vehicle, DGAT inhibitors, and etomoxir; mean ± SD, n=3.
(L,M) Acyl-carnitines and lysophospholipids detected by mass spectrometry in Tsc2−/− MEFs treated for 16 hrs with torin1 and DGAT inhibitors; mean ± SD relative to vehicle, n=3.
(N) β-oxidation of [9,10-3H]-palmitate in Tsc2−/− MEFs following pulse-chase and 16 hr treatment with vehicle, torin1, DGAT inhibitors, or etomoxir. Mean ± SD, n=3.
(O) β-oxidation in Tsc2−/− MEFs following a time course of vehicle or torin1 after 1 hr pre-treatment with vehicle or DGAT inhibitors. Mean ± SD, n=3.
Statistics: one-way ANOVA (F,H), two-way ANOVA (B,G,I-O), extra sum-of-squares F-test (C). n.s., not significant. * P < .0001.
Given these findings, we hypothesized that the TGs synthesized following mTORC1 inhibition might be derived from fatty acids released upon phospholipid degradation (Fig. 4A) and that these liberated fatty acids might also be available for mitochondrial fatty acid oxidation. Consistent with this idea, radiolabeled free fatty acids accumulated in cells treated with rapamycin or torin1 (Fig. 4D and Extended Data Fig. 4C), and acyl-carnitine species were elevated, including in primary mouse ES cells and MEFs, indicative of a pool of fatty acids prepared for fatty acid oxidation (Extended Data Fig. 4D-F). Fatty acids can also be incorporated into cholesteryl esters, which were found to be increased when mTORC1 was inhibited in HeLa and PC3 cells but not in Tsc2−/− MEFs via lipidomics (Fig. 1D). An increase in cholesteryl esters was evident in Tsc2−/− MEFs upon mTORC1 inhibition in pulse-chase experiments (Extended Data Fig. 3B). Importantly, hydrolysis of fatty acid side chains from phospholipid catabolism produces lysophospholipids, and both the total abundance and individual levels of these lipid species were elevated following mTORC1 inhibition across the cell lines profiled via lipidomics (Fig. 1D,H,J and 4E). A pulse-chase using radiolabeled choline in both Tsc2−/− MEFs and PC3 cells followed by TLC for separation of polar lipid classes confirmed that lysophosphatidylcholine (LPC) increased after both overnight and acute torin1 treatment during the chase phase (Fig. 4F,G and Extended Data Fig. 4G,H). Lysophospholipid hydrolysis yields a second fatty acid and a glycerophosphodiester head group (GPX in Fig. 4A), such as glycerophosphocholine (GPC) from LPC. Indeed, GPC levels increased upon mTORC1 inhibition (Fig. 4H, Extended Data Fig. 4I,J), suggesting that both fatty acids of the phospholipids were being hydrolyzed.
These data suggest an interrelationship between the accumulated lipid species measured upon mTORC1 inhibition (Fig. 4A). Indeed, DGAT inhibitors prevented the accumulation of TGs following mTORC1 inhibition with a concomitant increase in free fatty acid levels in both Tsc2−/− MEFs and PC3 cells (Fig. 4I,J and Extended Data Fig. 4K,L). Likewise, the fatty acid oxidation inhibitor etomoxir also raised fatty acid levels in the presence of mTORC1 inhibitors, an effect that was additive with DGAT inhibitors. Importantly, inhibiting these two fates of free fatty acids had no effect on fatty acid abundance when mTORC1 was active, indicating that fatty acid accumulation triggered by mTORC1 inhibition occurs upstream of TG synthesis and fatty acid oxidation. Etomoxir further raised TG levels in the presence of torin1, consistent with these two processes drawing from the same pool of free fatty acids under such conditions. Temporally, torin1 treatment led to a rapid increase in free fatty acids in cells pre-treated with DGAT inhibitors and etomoxir, which had no effect on their own (Fig. 4K and Extended Data Fig. 4M).
Lipidomic profiling confirmed that the torin1-induced increase in steady state TG species was blocked by the DGAT inhibitors (Extended Data Fig. 4N). In this analysis, torin1 treatment also raised acyl-carnitine levels, which were further increased by the DGAT inhibitors (Fig. 4L), again indicating that fatty acid oxidation and TG synthesis are drawing from the same pool of fatty acids made available upon mTORC1 inhibition. In contrast, the increase in lysophospholipids stimulated by torin1 was not altered by DGAT inhibition, consistent with the changes in these lipids being upstream of fatty acid use for TG synthesis (Fig. 4M).
Consistent with the changes in acyl-carnitines, inhibiting mTORC1 increased β-oxidation of endogenous fatty acids, as measured by the production of 3H2O from [9,10-3H]-palmitate delivered in the pulse-chase assay of lipid remodeling, and DGAT inhibition further enhanced this effect (Fig. 4N). Etomoxir blocked this induction as expected. mTORC1 inhibition rapidly stimulated fatty acid oxidation, which was more pronounced when also inhibiting TG synthesis (Fig. 4O). Over the longer term, transcriptional regulation of carnitine palmitoyltransferase (CPT) isoforms might also contribute to the regulation of fatty acid oxidation by mTORC1 (Extended Data Fig. 4O), as observed in the mouse liver23.
Lysosomes generate phospholipid degradation products
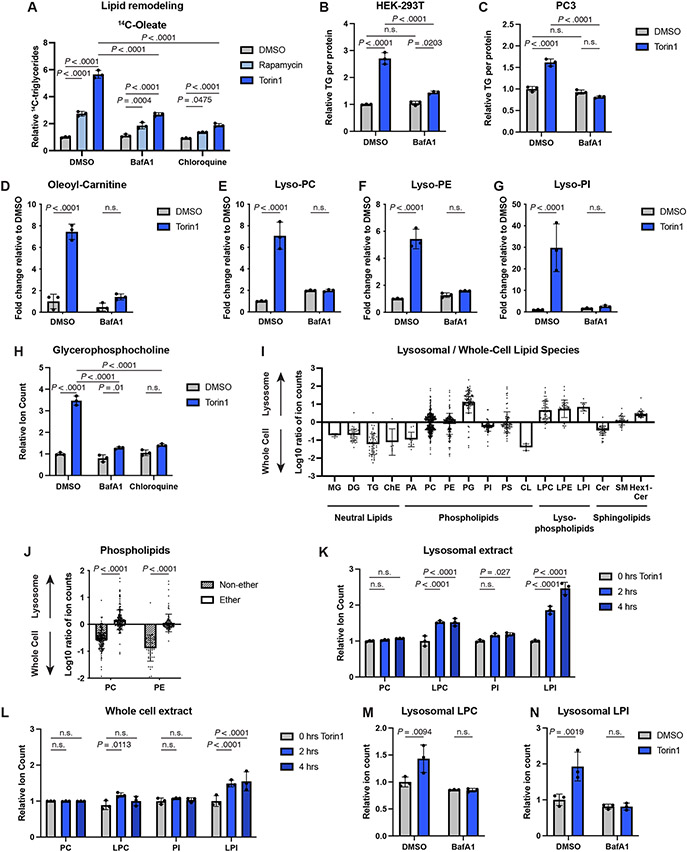
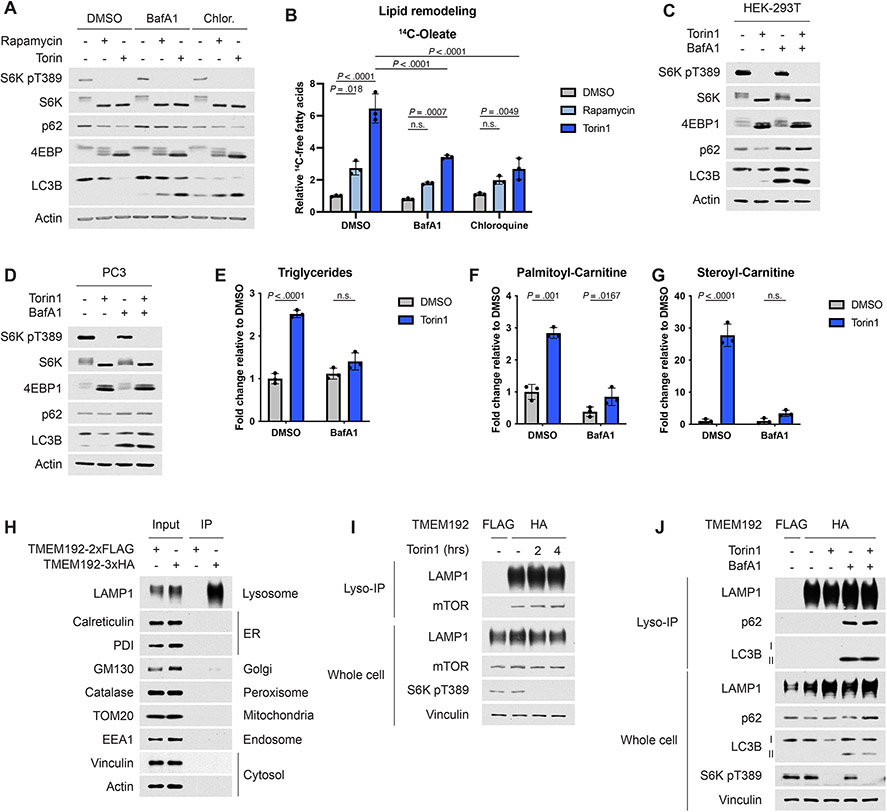
Having observed products of phospholipid breakdown increase following mTORC1 inhibition, we investigated if lysosomal activity was required for the shift in cellular lipid content documented, as was previously reported in starved cells32,33. Two lysosome inhibitors, bafilomycin A1 (BafA1) and chloroquine, significantly diminished TG accumulation induced by mTORC1 inhibition in Tsc2−/− MEFs (Fig. 5A), with inhibition of lysosomal processes confirmed via immunoblots of p62 and LC3B (Extended Data Fig. 5A). The increase in free fatty acids observed in the lipid remodeling assay upon mTORC1 inhibition was also blunted by these inhibitors (Extended Data Fig. 5B). BafA1 had a similar effect in HEK-293T and PC3 cells treated with torin1 (Fig. 5B,C and Extended Data Fig. 5C,D).
Figure 5: Changes in intracellular lipid species following mTORC1 inhibition require lysosomal function.
(A) TGs assayed by [1-14C]-oleate tracer in Tsc2−/− MEFs treated with vehicle, rapamycin (20 nM), or torin1 (250 nM), in the presence of vehicle (DMSO), bafilomycin A1 (BafA1, 250 nM) or chloroquine (100 μM) for 16 hrs, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(B,C) TGs measured by enzyme assay and normalized to protein content in HEK-293T (B) and PC3 (C) cells treated for 16 hrs with torin1 and/or BafA1, as in (A), graphed as mean ± SD relative to vehicle-treated cells, n=3.
(D-G) Oleoyl-carnitine (D) and lysophospholipid class sums (E-G) quantified by lipidomics in Tsc2−/− MEFs treated as in (B,C), graphed as mean ± SD relative to vehicle-treated cells, n=3.
(H) Relative glycerophosphocholine levels, measured by mass spectrometry, in MEFs treated as in (A), graphed as mean ± SD relative to vehicle-treated cells, n=3.
(I) Individual lipid species detected in HEK293T lipid extracts from lysosomal immunopurification or whole cells, grouped by class. The Log10 average ratio of ion counts in the fractions is shown. Bars represent the mean fold enrichment for individual lipid species in that class ± SD for n=3 replicates.
(J) Individual phosphatidylcholine (PC) and phosphatidyethanolamine (PE) species detected in the experiment in (I) and graphed by whether they contain an ether linkage or not. Plotted as group mean ± SD for n=3 replicates.
(K,L) Relative lysophospholipid and phospholipid class sums for species detected in lysosomal (K) and whole cell (L) lipid extracts from HEK-293T cells treated with torin1 (250 nM) for 0, 2, or 4 hrs, graphed as mean ± SD relative to the 0 hr timepoint, n=3.
(M,N) Relative lysophospholipid class sums for species detected in lysosomal lipid extracts from HEK-293T cells pre-treated for 1 hr with vehicle or BafA1 (250 nM) and then treated with vehicle or torin1 (250 nM) for 4 hrs, graphed as mean ± SD relative to vehicle-treated cells, n=3.
Statistical analysis by two-way ANOVA (A-H,J-L). n.s., not significant.
Lipidomic profiling confirmed that torin1 treatment failed to increase TGs in the presence of BafA1 (Extended Data Fig. 5E). Furthermore, the torin1-stimulated increase in acyl-carnitine species (Fig. 5D and Extended Data Fig. 5F,G) and lysophospholipids (Fig. 5E-G) was blocked by BafA1. Finally, accumulation of the phospholipid catabolism product GPC upon mTORC1 inhibition was also blunted in the presence of lysosome inhibitors (Fig. 5H). Thus, phospholipid catabolism in the lysosome gives rise to fatty acids, lysophospholipids, and hydrophilic head groups when mTORC1 is inhibited (Fig. 4A), with the freed fatty acids subsequently used for extralysosomal TG synthesis or mitochondrial fatty acid oxidation.
To specifically monitor changes in lysosomal lipids, we adapted a rapid lysosome immunopurification method (Lyso-IP)35 for lipidomic profiling. This method was previously established in HEK-293T cells, which enable sufficient recovery of low abundance lysosomes for biochemical analyses35. Importantly, inhibiting mTORC1 leads to a lysosome-dependent increase in triglycerides in HEK-293T cells (Fig. 5B). Lyso-IPs from HEK-293T cells expressing FLAG- or HA-TMEM192, a lysosomal membrane protein, were performed using anti-HA beads, with the FLAG-TMEM192 cell sample serving as a negative control. This approach efficiently enriched for membranous compartments containing lysosomal markers but not that of other cellular organelles (Extended Data Fig. 5H). A comparison of the lysosomal and whole cell lipidomes in untreated cells confirmed that neutral lipids, which predominantly reside in lipid droplets, and cardiolipin, a mitochondrial lipid, were enriched in the whole cell relative to the lysosome (Fig. 5I). Interestingly, lysophospholipid species showed enrichment in the lysosome. In contrast to other phospholipids, phosphatidylglycerol (PG) also appeared to be enriched in the lysosomal extract; however, this is likely due to the fact that PG is isomeric with the endo-lysosomal lipid bis(monoacylglycerol)phosphate (BMP)36. Notably, phospholipids containing only ester-linked fatty acids (i.e. non-ether) were significantly depleted in the lysosome relative to the whole cell (Fig. 5J), suggesting that lysosomal membranes contain a higher proportion of ether phospholipids.
Lysosomal and whole-cell lipids were analyzed in this assay following acute inhibition of mTORC1 (Extended Data Fig. 5I). Torin1 modestly enhanced mTORC1 binding to the lysosome, consistent with prior reports37. Lysophosphatidylcholine and lysophosphatidylinositol levels rose significantly in the lysosome after 2 hours of torin1 treatment (Fig. 5K) but were more modestly increased in the whole cell extract (Fig. 5L). In contrast, lysosomal phosphatidylcholine and phosphatidylinositol were not substantially altered by torin1 treatment, indicating that the lysosomal lipidome was not grossly affected. Finally, BafA1 prevented the torin1-induced increase in lysosomal lysophospholipids (Fig. 5M,N) and led to lysosomal accumulation of the autophagy markers p62 and LC3B-II (Extended Data Fig. 5J). Collectively, these results further indicate that mTORC1 inhibition promotes phospholipid catabolism in the lysosome.
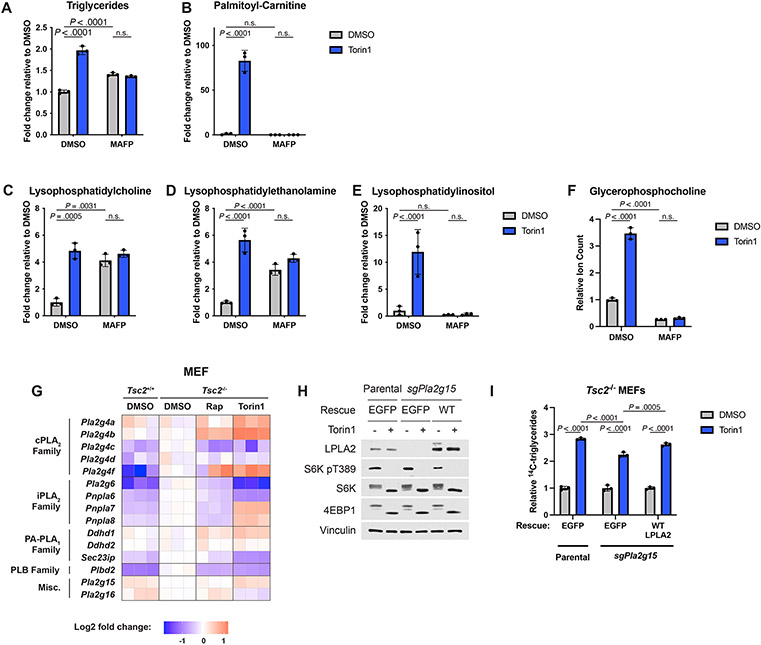
Phospholipase A1/A2 and lysophospholipases that release free fatty acids are serine hydrolases, and the pan-serine hydrolase inhibitor methoxy arachidonyl fluorophosphonate (MAFP) rendered many lipid classes and GPC insensitive to torin1 treatment (Extended Data Fig. 6A-F). It is worth noting that MAFP treatment, while completely blocking the ability of torin1 to increase TGs, partially elevated TG levels on its own, likely due to inhibition of ATGL, which is also a member of the serine hydrolase family (Extended Data Fig. 6A). Likewise MAFP treatment completely blocked the effects of torin1 on lysophospholipids but raised the levels of some lysophospholipid species on its own, consistent with MAFP inhibiting both phospholipase and lysophospholipase, activities that both generate and consume lysophospholipids, respectively (Extended Data Fig. 6C,D).
Expression of several intracellular phospholipase A1/A2 enzymes increased upon mTORC1 inhibition in MEFs (Extended Data Fig. 6G), but among these enzymes only lysosomal phospholipase A2 (LPLA2, encoded by Pla2g15) is known to localize to the lysosome38. However, unlike lysosome inhibitors or MAFP, CRISPR/Cas9 deletion of Pla2g15 did not strongly block the increase in TG following torin1 treatment. A modest but significant decrease in torin1-induced TG accumulation that could be partially reversed by restoring expression of this enzyme was noted, suggesting that it may be partially involved in this regulation but that other lysosomal lipases must also exist (Extended Data Fig. 6H,I).
Autophagy is dispensable for lipid remodeling
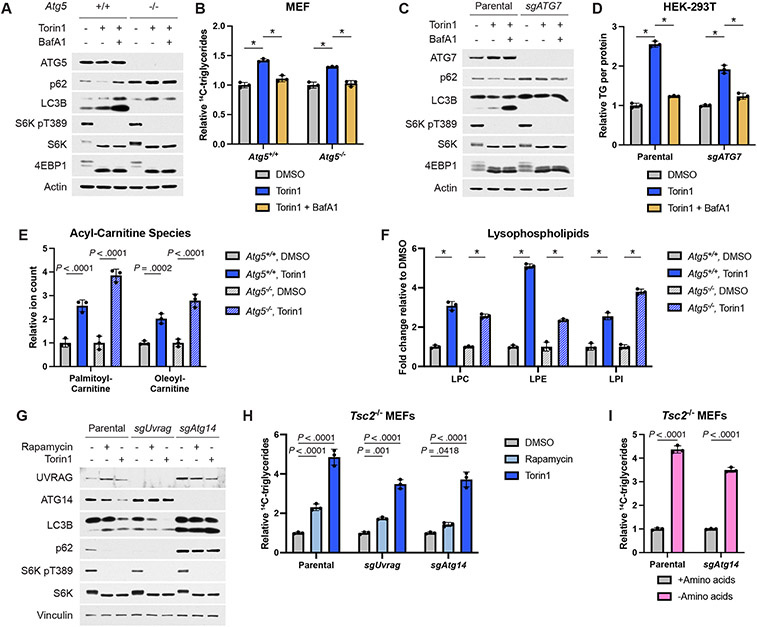
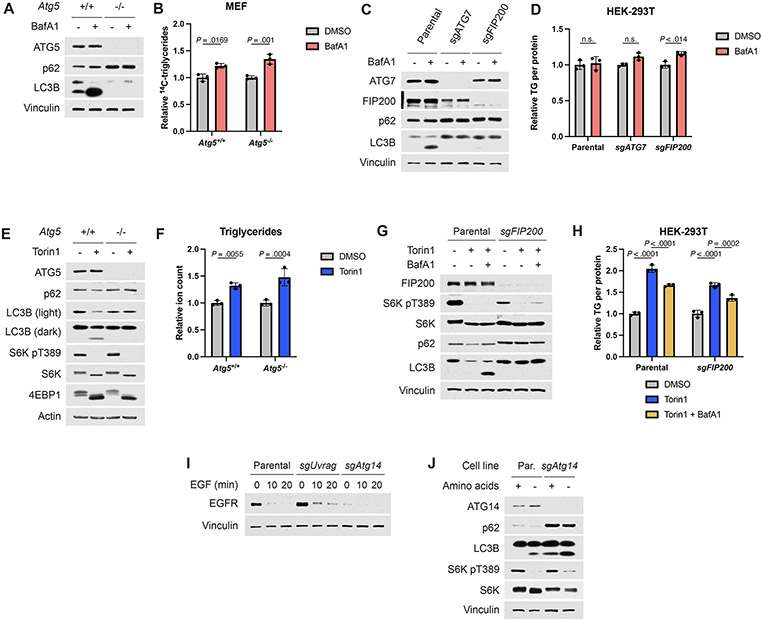
Autophagy has been implicated in the accumulation of lipid droplets in starved cells33 and serves as a major macromolecule recycling pathway when mTORC1 is inhibited. However, autophagy-defective Atg5−/− MEFs accumulated TGs produced from endogenous lipid species to a similar extent as their wild-type counterparts when treated with torin1, with the defect in torin1-induced autophagy confirmed by sustained p62 levels and a failure to lipidate LC3B, which is detected as a faster migrating form on immunoblots (Fig. 6A,B). However, the torin1-induced increase in TG levels remained sensitive to BafA1, further indicating that this ATG5-independent process downstream of mTORC1 inhibition is dependent on lysosomal function. Autophagy-deficient HEK-293T cells with CRISPR/Cas9-mediated deletion of ATG7 showed similar results (Fig. 6C,D). Importantly, BafA1 treatment alone did not reduce TG levels in these cells, confirming that its ability to suppress TG accumulation is specific to contexts of mTORC1 inhibition (Extended Data Fig. 7A-D). Lipidomic profiling confirmed steady state accumulation of TGs in Atg5−/− cells following torin1 treatment (Extended Data Fig. 7E,F), and both acyl-carnitine and lysophospholipid species also increased independently of autophagy upon mTORC1 inhibition (Fig. 6E,F).
Figure 6: TG and other lipid species accumulate independently of autophagy following mTORC1 inhibition.
(A,B) Immunoblot (A) and TGs measured by pulse-chase with [1-14C]-oleate tracer (B) in Atg5+/+ and −/− MEFs treated for 16 hrs with vehicle, torin1 (250 nM), and/or BafA1 (250 nM), graphed as mean ± SD relative to vehicle-treated cells, n=3.
(C,D) Immunoblot (C) and TGs (D) measured by enzyme assay and normalized to protein content in parental and ATG7 KO HEK-293T cells treated as in (A,B), graphed as mean ± SD relative to vehicle-treated cells, n=3.
(E,F) Acyl-carnitine (E) and lysophospholipid (F) species measured by mass spectrometry in Atg5+/+ and −/− MEFs treated for 16 hrs with vehicle or torin1 (250 nM), graphed as mean ± SD relative to vehicle-treated cells, n=3.
(G) Immunoblot for autophagy markers in parental Tsc2−/− MEFs and sgUvrag and sgAtg14 clones following 4 hrs treatment with vehicle, rapamycin (20 nM), or torin1 (250 nM).
(H) TG accumulation measured by pulse-chase with [1-14C]-oleate tracer in the cells treated as in (G) for 16 hrs, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(I) TG accumulation in parental Tsc2−/− MEFs and sgAtg14 clone following 16 hrs in amino acid-replete or free medium, measured and graphed as in (H) relative to amino acid-replete cells, n=3.
Statistical analysis by two-way ANOVA (B,D,E,F,H,I). n.s., not significant. * indicates P < .0001.
Studies suggest that autophagosome formation and delivery of some cellular components to the lysosome can still occur in the absence of ATG5 or ATG7, albeit at a diminished rate39. However, loss of FIP200, resulting in impaired activation of the autophagy initiating kinases ULK1 and 2, results in complete inhibition of autophagy39. Indeed, HEK-293T cells with CRISPR/Cas9-mediated deletion of FIP200 were unable to carry out torin1-induced autophagy (Extended Data Fig. 7G). However, these cells still accumulated TGs in a BafA1-sensitive fashion when mTORC1 was inhibited (Extended Data Fig. 7H).
The class-3 PI3K VPS34 regulates autophagy and vesicle trafficking, at least in part, via complexes containing ATG14 and UVRAG, both of which have been described to be influenced by mTORC1 signaling40,41. Like FIP200, ATG14 is essential for the initiation of autophagy39. However, CRISPR/Cas9-mediated deletion of neither Atg14 nor Uvrag in Tsc2−/− MEFs blocked the induction of TGs produced from lipid remodeling upon mTORC1 inhibition (Fig. 6G,H). Unlike ATG14, knockout of UVRAG did not affect autophagy induced by mTOR inhibitors, but its loss did modestly reduce EGF-stimulated EGFR degradation (Fig. 6G and Extended Data Fig. 7I), a process involving endocytic delivery to the lysosome40. Like pharmacological inhibition of mTORC1, amino acid deprivation induces autophagy in an ATG14-dependent manner (Extended Data Fig. 7J). However, amino acid withdrawal increased TGs similarly in wild-type and Atg14-null cells (Fig. 6I), further confirming that this response trigged by mTORC1 inhibition is independent of autophagy.
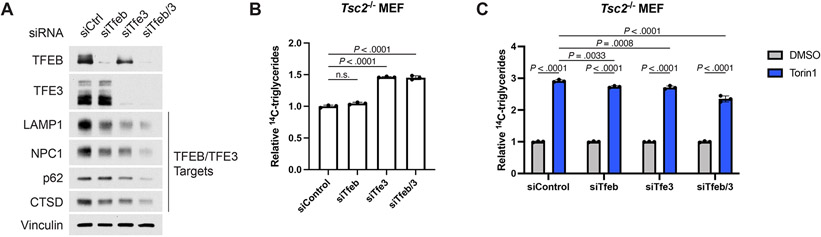
mTORC1 also regulates autophagy and lysosome biogenesis through the MiT-TFE family transcription factors TFEB and TFE342. siRNA-mediated knockdown of TFEB, TFE3, and especially the two combined greatly reduced expression of proteins encoded by their target genes (Extended Data Fig. 8A). While, TFEB/TFE3 knockdown modestly reduced TG accumulation following torin1 treatment, TG induction was largely intact (Extended Data Fig. 8B,C). As the canonical targets of these transcription factors are lysosomal components, it seems likely that decreased lysosome function following TFEB/TFE3 knockdown might underlie the small but significant reduction in torin1-induced TG accumulation observed. The rather acute effects of mTOR inhibitors on lipid remodeling revealed above (Fig. 4G,K,O and Extended Data Fig. 2A) suggest involvement of additional, non-transcriptional mechanisms.
Endosomal trafficking enables lipid remodeling
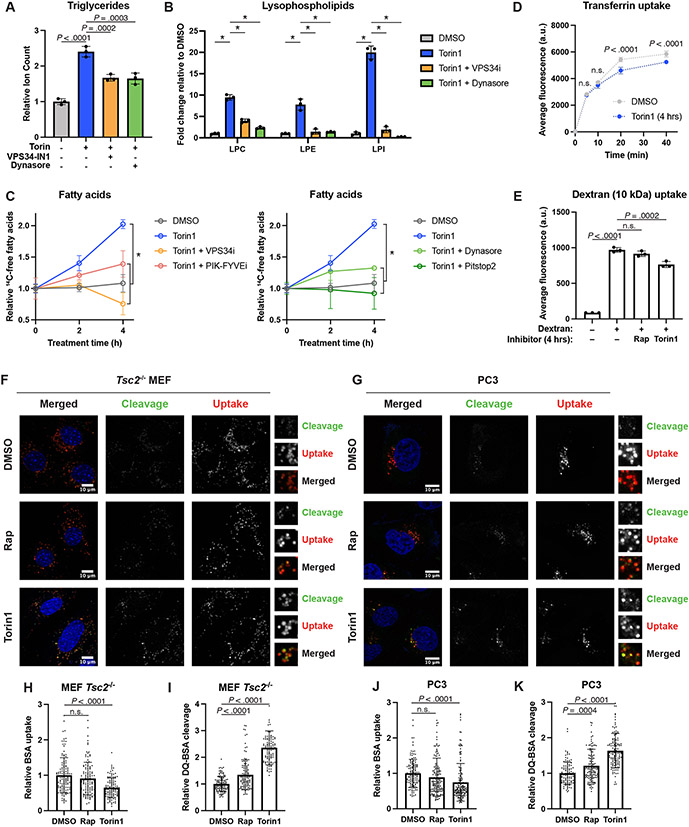
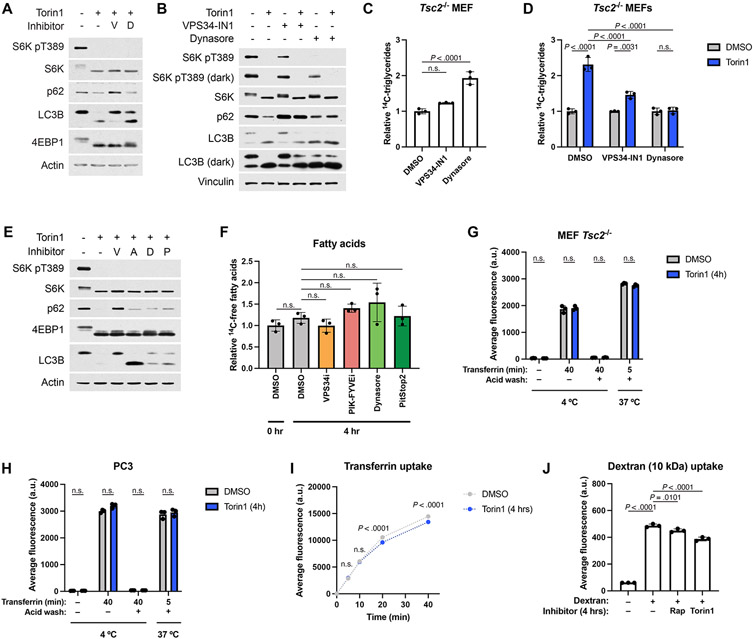
Beyond autophagy, cellular material is also transported to the lysosome via the endosomal system, and endosomes might serve as an important means of delivering phospholipids to the lysosome for degradation. Treating Tsc2−/− MEFs with inhibitors of VPS34 (VPS34-IN1) or dynamin (Dynasore), both of which can inhibit endocytosis43,44, reduced TG accumulation caused by inhibition of mTORC1, as measured by lipidomic profiling (Fig. 7A). It is worth noting that the VPS34 inhibitor also perturbed autophagy (Extended Data Fig. 9A), which was found through the above genetic analyses to be dispensable for the mTORC1-regulated shift in cellular lipid species. Neither inhibitor reduced TG accumulation in the absence of torin1 (Extended Data Fig. 9B-D). Consistent with reduced delivery of phospholipids to the lysosome, both inhibitors also blocked the torin1-induced increase in lysophospholipid species (Fig. 7B). Importantly, treatment with VPS34-IN1, Dynasore, or additional inhibitors of endocytosis, including the PIK-FYVE inhibitor apilimod and the dynamin inhibitor Pitstop2 also resulted in a significant reduction in torin1-induced fatty acid accumulation (Fig. 7C and Extended Data Fig. 9E), without reducing fatty acid levels in the absence of torin1 (Extended Data Fig. 9F). Together, these data indicate that endosomal trafficking is required for the lysosome-dependent shift in cellular lipid content upon inhibition of mTORC1.
Figure 7: Induction of endosomal delivery to the lysosome is required for the changes in cellular lipid species following mTORC1 inhibition.
(A,B) Relative TG (A) and lysophospholipid (B) class sums measured by mass spectrometry in Tsc2−/− MEFs treated for 16 hrs with vehicle, torin1 (250 nM), VPS34-IN1 (5 μM), and/or Dynasore (80 μM), graphed as mean ± SD relative to vehicle-treated cells, n=3.
(C) Fatty acid accumulation measured by pulse-chase with [1-14C]-oleate tracer in the presence of DGAT inhibitors (3 μM each) and etomoxir (20 μM) in Tsc2−/− MEFs over a time course of vehicle or torin1 (250 nM) treatment, following 1 hr pre-treatment with endocytosis inhibitors (5 μM VPS34-IN1, 1 μM PIK-FYVEi, 80 μM Dynasore, or 20 μM PitStop 2). Graphed as mean ± SD relative to vehicle-treated cells at time 0, n=3.
(D,E) Endocytosis of transferrin (time course in D) and 10-kDa dextran (E) following 4 hrs of vehicle, rapamycin (20 nM), or torin1 (250 nM) treatment in Tsc2−/− MEFs, graphed as mean ± SD, n=3.
(F,G) mTORC1 inhibition enhances hydrolysis of endocytosed BSA in Tsc2−/− MEFs (F) and PC3 cells (G). Following 1 hr pretreatment with vehicle, rapamycin (20 nM) or torin1 (250 μM), cells were co-labeled for 3 hrs with BSA Alexa Fluor 647 (uptake, red in merged) and DQ-Green BSA (cleavage, green in merged) (10 μg/mL each). Nuclei are shown in blue.
(H-K) Quantification of the results from the experiments shown in (F) and (G). (H,J) Relative BSA Alexa Fluor 647 uptake into individual cells. (I,K) Relative DQ-Green-BSA fluorescence (cleavage) normalized to BSA Alexa Fluor 647 (uptake) on a per-cell basis. Graphed as mean ± SD relative to vehicle-treated cells, n=108-133 cells.
a.u., Arbitrary Units. Statistical analysis by two-way ANOVA (A-E), and one-way ANOVA (H-K). n.s., not significant. In (B) and (C) * indicates P < 0.001.
To determine whether mTORC1 inhibition influenced endocytosis, we assayed the uptake of transferrin (Tf), representative of clathrin-mediated endocytosis, and dextran, representative of fluid-phase endocytosis45. Tf uptake was not increased, but modestly and significantly reduced upon torin1 treatment in both Tsc2−/− MEFs and PC3 cells, without altering the baseline amount of Tf receptor at the plasma membrane (Fig. 7D and Extended Data Fig. 9G-I). Likewise 10-kDa dextran uptake was modestly reduced when mTORC1 was inhibited (Fig. 7E and Extended Data Fig. 9J), possibly due to decreased macropinocytosis, one form of fluid phase endocytosis suggested to be influenced by mTORC146. Collectively, these results suggest that mTORC1 does not suppress, but rather positively contributes to, two major forms of endocytosis, thus making it unlikely that mTORC1 inhibition triggers lipid remodeling via enhanced plasma membrane endocytosis.
Previous studies have suggested that, after internalization, the delivery of endosomal material to the lysosome may be enhanced when mTORC1 is inhibited16,47. DQ-Green bovine serum albumin (BSA) is an endocytosis tracer whose fluorescence is dependent on delivery and hydrolysis in the lysosome16. BSA-AlexaFluor-647 is constitutively fluorescent, allowing BSA uptake to be measured as well. As controls for these two tracers, ethyl-isopropyl-amiloride (EIPA), which blocks macropinocytosis, reduced the uptake of BSA-AlexaFluor-647, and lysosomal protease inhibitors reduced the hydrolysis of DQ-Green-BSA (Extended Data Fig. 10A-D), which co-localized with LysoTracker (Extended Data Fig. 10E). To assay potential effects of mTORC1 inhibition on endolysosomal trafficking, Tsc2−/− MEFs and PC3 cells were co-labeled with both BSA conjugates for 3 hours in the presence of vehicle, rapamycin, or torin1 (Fig. 7F-K). Consistent with the data above on dextran uptake, inhibiting mTORC1 partially reduced BSA uptake (Fig. 7H,J). In contrast, DQ-BSA cleavage was significantly increased by both rapamycin and torin1 treatment (Fig. 7I,K), indicating a stimulated increase in delivery of endosomal cargo to the lysosome upon mTORC1 inhibition. Although DQ-BSA is a protein cargo, this finding is consistent with the possibility that more endosomal material, including membrane lipids, are delivered to the lysosome when mTORC1 is inactivated.
Discussion
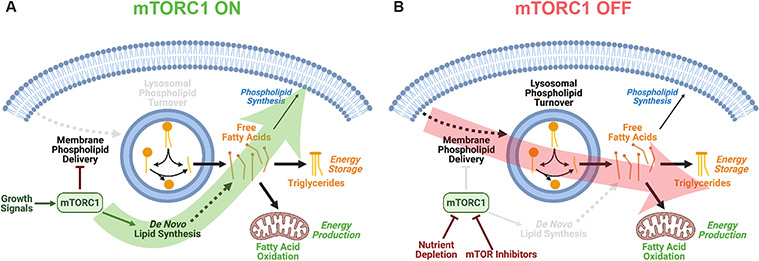
mTORC1 coordinately regulates many metabolic processes to allow cells to adapt to dynamic signals from exogenous growth factors and intracellular nutrients. In this study, we have identified an adaptive process by which cells catabolize membrane lipids when mTORC1 is inhibited, such as during nutrient depletion. Our findings indicate that mTORC1 suppresses the lysosome-dependent degradation of phospholipids, manifested in fatty acids becoming available for energy storage as TGs or energy production via fatty acid oxidation upon mTORC1 inhibition (Fig. 8). Under pro-growth conditions, activated mTORC1 induces de novo synthesis of fatty acids, an anabolic process that consumes nutrients, energy, and cellular reducing power, via the SREBP transcription factors. Distinct cellular states likely govern the fate of newly synthesized fatty acids when mTORC1 is active. However, mTORC1 and SREBP have been shown to stimulate synthesis of the most abundant membrane phospholipid, phosphatidylcholine22, and this might channel de novo synthesized fatty acids toward new membrane biogenesis when mTORC1 is active (Fig 8A).
Figure 8: Model of the differential effects of mTORC1 activation and inhibition on cellular lipid metabolism.
(A) mTORC1 activation by pro-growth signals induces de novo fatty acid synthesis as part of its growth-promoting activity.
(B) mTORC1 inhibition leads to membrane phospholipid delivery and turnover in the lysososome, with the released fatty acids being available for energy storage as triglycerides or energy production via beta-oxidation in the mitochondria.
Accumulation of TGs upon mTORC1 inhibition has also been reported in yeast and fruit flies48,49 and in nutrient starved mammalian cells32,33. Collectively, our data from numerous mouse and human cell lines and types indicate that this process may originate from stimulated membrane phospholipid delivery and turnover in the lysosome. Phospholipid catabolism may provide an advantage to cells when mTORC1 is inhibited, as would occur under poor growth conditions when nutrients or energy are scarce. Phospholipid hydrolysis releases two fatty acids that can fuel fatty acid oxidation for energy or be incorporated into TGs to be stored for future use (Fig 8B). Our results indicate that TG synthesis from the fatty acids liberated upon mTORC1 inhibition limits the rate of fatty acid oxidation. Consistent with this, Nguyen et al.32 proposed that TG synthesis is important to prevent mitochondrial dysfunction resulting from fatty acid overload in starved cells. Others have suggested that in some contexts TG synthesis prevents overflow of fatty acids into ceramides and other stress-inducing lipid species28. This may explain why DGAT inhibitors have a deleterious effect on hypoxic cells, which normally accumulate TGs in lipid droplets28,50. TG synthesis can also prevent lipotoxicity and ER stress in response to treatment with exogenous fatty acids or during lipolysis of intracellular lipid stores51,52.
Recent reports have indicated that mTORC1 inhibition causes TG accumulation in the liver and primary hepatocytes due to impaired lipoprotein secretion22. Our experiments specifically analyzed the cell-intrinsic redistribution of intracellular lipids, rather than the packaging of extracellular lipids, which likely explains why we did not observe the same TG accumulation upon mTORC1 inhibition in isolated hepatocytes. Hepatocytes play a specialized role in controlling whole body lipid metabolism, thus their mechanisms of regulating intracellular lipid pools likely differ from most other cells, which do not appreciably secrete lipoprotein.
Mono- and diglycerides (MGs and DGs) consistently decreased when mTORC1 was inhibited across the cell types analyzed. Although inhibited TG lipolysis could lead to lower cellular MGs and DGs, TG turnover was not impaired following mTORC1 inhibition. Hormone-sensitive lipase (HSL), which preferentially hydrolyzes DGs but can also act on other lipids including MGs and TGs53, has been proposed to be more active when mTORC1 is inhibited in adipocytes24. HSL was below the limit of detection in the cell types analyzed in this study, but if present its activation could influence MG and DG abundance. A more likely explanation is that these neutral lipids are depleted by the enhanced incorporation of fatty acids for TG synthesis observed upon mTORC1 inhibition.
The increase in lysophospholipid species upon mTORC1 inhibition was particularly enlightening in understanding the origins of the lipid changes observed in this study. Intracellular lysophospholipids can be generated as part of membrane remodeling via the Lands’ cycle, through which cytosolic phospholipase A2 enzymes cleave a fatty acid from membrane phospholipids to create lysophospholipids that are then re-esterified to regenerate phospholipids. While disrupting the re-esterification step of the Lands’ cycle could raise lysophospholipid levels, our evidence suggests that the mTORC1-regulated effects occur independently of this mechanism. The Lands’ cycle interconverts phospholipids and lysophospholipids, without additional hydrolysis of the lysophospholipid. However, we find that mTORC1 inhibition leads to accumulation of both free fatty acids and the polar phospholipid headgroup GPC, indicative of complete hydrolysis of abundant phospholipids such as phosphatidylcholine. Furthermore, the Lands’ cycle occurs in the cytosol, and the increase in lysophospholipids following mTORC1 inhibition occurs within the lysosome and depends upon the pH of the lysosomal lumen. Thus, the shift in intracellular lipids that we document appears to be initiated by lysosomal delivery and catabolism of phospholipids, rather than changes in specific Land’s cycle reactions.
Priolo et al.54 previously linked mTORC1 activity to changes in lysophospholipids in lymphangioleiomyomatosis (LAM), a proliferative lung disorder driven my aberrant activation of mTORC1 most commonly resulting from loss of TSC2. While its origin is unknown, plasma lysophospholipids were found to be modestly elevated in LAM patients. However, treatment of a LAM-derived cell line with rapamycin or torin1 also increased lysophospholipid levels, consistent with the observations here. Notably, the effects on lysophospholipids in this previous study were attributed to regulated expression of several phospholipases, including adipocyte phospholipase A2 (AdPLA, encoded by Pla2g16), which may be upregulated in LAM55 but is not thought to be a lysosomal lipase. As part of this current study, deletion of the one established lysosomal phospholipase A2, LPLA238, did not strongly block the effects of mTORC1 inhibitors on lipid metabolism, suggesting that additional phospholipases also likely act in the lysosomal lumen. Although we chose to focus on the lysosome because of the significant ability of lysosome inhibitors to block lipid remodeling upon mTORC1 inhibition, several cytosolic phospholipases were transcriptionally upregulated following mTORC1 inhibition, rendering it possible that these enzymes might also liberate fatty acids from phospholipids to contribute to TG synthesis. In some experiments BafA1 was not able to completely reverse the TG accumulation caused by torin1 treatment, perhaps suggestive of an additional role for extra-lysosomal mechanisms.
While not linked previously to changes in cellular lipid content, elevated endosomal delivery to the lysosome when mTORC1 is inhibited has been suggested by other studies. Palm et al.16 proposed that macropinocytosis and subsequent lysosomal catabolism of extracellular protein promote the survival of KrasG12D MEFs under amino-depleted conditions. Dauner et al.47 observed that rapamycin-treated cells diverted plasma membrane lipids and receptors, including the Tf receptor (TfR) and low density lipoprotein (LDL) receptor (LDLR), to the lysosome and concluded that mTORC1 activity might promote efficient recycling of these components back to the plasma membrane. While we did not investigate the fate of endocytosed TfR, we consistently observed a modest reduction in Tf endocytosis in cells treated with torin1, perhaps consistent with less recycling of endocytosed TfR back to the plasma membrane. The same study also observed increased lysosomal degradation of LDLR47, which, if coupled to LDL endocytosis, could provide an exogenous source of lipids for use by cells when mTORC1 is inhibited. A role for the endolysosomal system in adaptation to nutrient deprivation is also supported by data from starved yeast, which use the endosomal sorting complexes required for transport (ESCRT) pathway to traffic membrane proteins to the vacuole to be catabolized4. Together with autophagy, this pathway plays a critical role in maintaining cellular amino acid supply following starvation, and our findings suggest that endocytic trafficking of phospholipids to the lysosome may play an analogous role in the maintenance and repurposing of cellular fatty acid pools.
Although the molecular mechanism(s) linking mTORC1 inhibition to enhanced endosomal flux to the lysosome has yet to be defined, endosome dynamics could be regulated by mTORC1 at various steps. A number of protein complexes are involved in the recycling of endosomal components back to the plasma membrane or in directing vesicles to the lysosome or other intracellular locations56. While the mechanisms governing the fate of specific transmembrane receptors and protein cargo within the endomembrane system has been carefully studied, less is known about the factors governing the fate of lipids carried on endosomal vesicles. The ESCRT-mediated transfer of endosomal membrane components into intralumenal vesicles within late endosomes and lysosomes exposes phospholipids to hydrolases and represents an additional potential point of regulation36,57. A recent study proposed that mTORC1 regulates endocytosis transcriptionally, through its inhibition of TFEB58. Although we observed that knockdown of both TFEB and the related transcription factor TFE3 modestly decreased the torin1-induced accumulation of TG, this may be a consequence of reduced lysosome abundance following two days of siRNA treatment, as these transcription factors regulate many proteins required for lysosome biogenesis42. Although these observations do not rule out the involvement of direct regulation of TFEB/TFE3 by mTORC1, the effects reported in this study are likely TFEB-independent. While mTORC1 inhibition activates TFEB in most cell types, it paradoxically inhibits TFEB in TSC2-deficient settings59, and both TSC2-expressing and -deficient cells showed similar consistent changes in lipid species and enhanced lysosomal delivery of cargo in response to mTORC1 inhibitors in our study. Furthermore, activation of TFEB in the mouse liver and of its homologue in C. elegans have both been linked to decreased neutral lipid stores60, contrary to what would be expected if TFEB were driving the effects of mTORC1 inhibition described here.
Our results suggest that cells rewire lipid metabolism following mTORC1 inhibition and shift towards a program of phospholipid catabolism to recycle or repurpose fatty acids. Recent work has demonstrated that upregulating fatty acid oxidation is a critical adaptation allowing cancer cells to survive therapeutic inhibition of the mitogen-activate protein kinase (BRAF-MEK-ERK) pathway61. Targeting this pathway in settings of its oncogenic activation should also inhibit mTORC1, suggesting that the shift in lipid metabolism that our study has revealed could contribute to cell survival in such contexts. Future studies will seek to elucidate the precise mechanism by which mTORC1 regulates the autophagy-independent delivery of membrane components to the lysosome and to determine if that process could be exploited for therapeutic benefit.
Materials and Methods
Cell culture
Cells were maintained in DMEM without pyruvate, or RPMI 1640 for MCF7 cells, with 10% fetal bovine serum (FBS) and 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C with 5% CO2. Littermate-derived Tsc2+/+ (referred to as WT) and Tsc2−/− mouse embryonic fibroblasts (MEFs) immortalized by Trp53 deletion were a gift from David Kwiatkowski (Brigham and Women’s Hospital). Littermate-derived Atg5+/+ and Atg5−/− MEFs were a gift from Gökhan Hotamisligil (Harvard T.H. Chan School of Public Health). 105K (Tsc2−/−) renal cystadenoma cells with pLXIN-IRES-hygromycin empty vector or TSC2 were described previously34 and were a gift from Elizabeth Henske (Brigham and Women’s Hospital). Rictor+/+ and Rictor−/− MEFs, RagAGTP/GTP MEFs, and sgATG7 HEK-293T cells were gifts from David Sabatini, and sgFIP200 HEK-293T cells were a gift from Wade Harper (Harvard Medical School). HeLa cells stably expressing pMSCV-PM mir30-shLuciferase (shLuc) and -shTSC2 were described previously62. Primary MEFs (Lonza M-FB-481) were propagated for fewer than 4 passages. Mouse embryonic stem cells were cultured on plates coated with 0.1% gelatin (Sigma G1890) and maintained in DMEM with pyruvate plus 15% FBS and 1x MEM NEAA (Thermo 11140050). Before use, this medium was freshly supplemented with 100 μM β-mercaptoethanol and 100 U/mL mLIF (Sigma ESG1106).
For all experiments, Tsc2−/− MEFs, HeLa, 105K, PC3, MCF7, and H1299 cells and derivatives were washed twice with phosphate-buffered saline (PBS) and changed to medium lacking FBS for 16 hrs. For insulin stimulation, cells were pre-treated for 30 min with vehicle, rapamycin, or torin1 prior to addition of 100 nM insulin (Alpha Diagnostic INSL16-N-5) for 8 hrs. For epidermal growth factor (EGF) stimulation, cells were stimulated with 100 ng/mL EGF (Thermo Scientific, PHG0311). For experiments with ≤4 hr treatment with torin1, cells were pre-treated with other inhibitors for 1 hr prior to torin1. Amino acid free medium was made from powder (US Biological D9800-13) with glucose supplemented to 4.5 g/L. Amino acid replete medium was made from this by supplementing amino acids to their DMEM concentrations. 10% dialyzed FBS (Thermo) was added to these media for experiments with WT and RagAGTP/GTP MEFs, and serum-free were used for the Tsc2−/− parental and sgAtg14 MEFs.
Animal care
Animal studies were conducted with the approval of the Harvard Medical Area Standing Committee on Animals IACUC at an AAALAC International and USDA-accredited facility. All protocols concerning animal care and animal studies were consistent with the Guide for the Care and Use of Laboratory Animals. Maximum tumor size permitted on this protocol is 2-cm in the longest diameter, and this was not exceeded in any experiment. Allograft tumor studies were initiated in 6-week old female C57BL/6J mice, and primary hepatocytes were isolated from 8-week old female C57BL/6J mice, all acquired from Jackson Laboratories (000664). Mice were group housed (3-4 per cage), with autoclaved food (PicoLab Diet #5053) and water provided ad libitum in a room temperature, pathogen-free facility with a 12:12 hr light:dark cycle in standard static microisolator top cages.
Chemical Inhibitors
Inhibitor product information and concentrations used are listed in Supplementary Table 1.
Plasmids
pSpCas9n(BB)-2A-GFP (PX461) PX461 was a gift from Feng Zhang (Addgene 48140). psPAX2 (Addgene 12260) and pMD2.G (Addgene 12259) were gifts from Didier Trono. pLJC5-Tmem192-2xFlag (Addgene 102929), pLJC5-Tmem192-3xHA (Addgene 102930), and pLJM1-EGFP (Addgene 19319) were gifts from David Sabatini. Mouse Pla2g15 cDNA (Horizon Discovery MMM1013-202763537) was cloned into the pLJM1 backbone derived from pLJM1-EGFP digested with AgeI-HF and EcoRI-HF (both from NEB) using the In-Fusion HD Cloning System (Takara 638910) according to the manufacturer’s instructions. Primers were designed using the In-Fusion Cloning Primer Design Tool (Takara).
Immunoblots
Cells were washed twice in ice cold PBS and scraped into NP-40 lysis buffer with phosphatase inhibitors (40 mM HEPES pH 7.4, 120 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 50 mM NaF, and 0.5 mM sodium orthovanadate) with protease inhibitor cocktail (Sigma P8340) and 1 μg/mL microcystin-LR (Enzo ALX-350-012). After centrifuging for 10 min at maximum speed at 4 °C, the supernatant was reserved, and protein concentration was determined using the Bio-Rad Protein Assay Dye Reagent. Protein concentrations were normalized, and samples were heated to 70 °C for 10 min with Laemmli Sample Buffer. To detect DGAT1, cells were lysed in 1% SDS lysis buffer with phosphatase inhibitors (10 mM Tris-HCl pH 7.4, 1 mM EDTA, 1% SDS, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 50 mM NaF, and 0.5 mM sodium orthovanadate) with protease inhibitor cocktail and microcystin-LR. Samples were passed through a 27-G needle five times, centrifuged for 10 min at 1000 x g at room temperature, and processed as above.
Proteins were resolved by Laemmli SDS-PAGE. Protein was transferred to 0.2 μm nitrocellulose membranes, unless probing for LC3B, DGAT1, UVRAG, and ATG14, in which case PVDF membranes were used. Membranes were blocked in Tris-buffered saline with 0.1% Tween-20 (TBS-T) plus 5% non-fat dry milk and probed with the primary antibody overnight at 4 °C in TBS-T plus 5% BSA. Antibodies used are listed in Supplementary Table 2.
Membranes were then washed with TBS-T and probed for 1 hr at room temperature with secondary antibodies. Proteins were visualized with SuperSignal West Pico PLUS or Femto Chemiluminescent Substrates (Thermo 34577 and 34096) and film.
Lipidomic profiling
Cells were trypsinized, counted, and an equal number of cells were pelleted for 5 min at 1000 x g at 4 °C. Cells were resuspended in 200 μL HPLC-grade water and vortexed with 2.5 mL HPLC-grade methanol in glass tubes. 5 mL methyl tert-butyl ether (MTBE) were added, and samples were rocked for 1 hr at room temperature. 1.5 mL water were added, and samples were vortexed and centrifuged for 10 min at 1000 x g at room temperature. The upper phase was dried under nitrogen gas stream and stored at −80 °C.
Samples were resuspended in 35 μL 1:1 LCMS-grade isopropanol:methanol and analyzed by liquid chromatography-mass spectrometry as described previously63, using a high-resolution hybrid QExactive HF Orbitrap mass spectrometer (Thermo) run in data-dependent acquisition mode (Top 8) with positive/negative ion polarity switching. Additional details can be found in the Supplementary Methods. Lipid species were identified and quantified using LipidSearch 4.1.30 software (Thermo) using an internal database of ≥20 main lipid classes and ≥80 subclasses.
To confirm signal linearity, a pooled sample was generated by mixing 5 μL of each sample. This was diluted with 1:1 isopropanol:methanol to produce 0.3x- and 0.1x samples. These dilutions and a blank were analyzed. For each lipid species in this dilution series, the Pearson correlation constant for ion count and sample concentration was calculated, and only lipids with r>.9 were included in the final analysis. Individual lipid species abundance was normalized to the sample’s total ion count. Lipids were sorted by class, and the summed ion intensity for each class in each sample was computed in R.
Metabolomic profiling
Cells were washed twice with ice-cold PBS and scraped into ice-cold 80% HPLC-grade methanol. Samples were vortexed and centrifuged for 10 min at maximum speed at 4 °C. Supernatants were dried under nitrogen gas stream and stored at −80 °C. Samples were resuspended in 20 μL LCMS-grade water and analyzed by liquid chromatography-mass spectrometry via selected reaction monitoring (SRM) as described previously64. Additional details can be found in the Supplementary Methods. Ion counts for each metabolite were quantified by integrating the peak area of the total ion current for each SRM transition using MultiQuant v3.0 software (AB/SCIEX). Ion counts were normalized to protein measured from parallel plates lysed in NP-40 lysis buffer.
Triglyceride enzyme assay
Cells were washed twice with ice-cold PBS, scraped into 1 mL PBS, and pelleted for 5 min at 1000 x g at 4 °C. Cell pellets were resuspended in 30-50 μL 1x NP-40 Substitute Assay Reagent (Cayman Chemical). 5 μL of this resuspension were diluted into NP-40 lysis buffer and pelleted for 5 min at maximum speed at 4 °C, and the supernatant was used for protein quantification. The remaining sample was heated to 80 °C for 5 min, vortexed, and heated again. Samples were clarified for 10 min at 10 000 x g at room temperature. Triglyceride concentration in the supernatant was quantified using the Triglyceride Colorimetric Assay (Cayman Chemical 10010303). Triglycerides were normalized to protein for each sample. Tumor samples were homogenized into 160 μL water in 1.5 mL NAVY Bead Lysis Kit tubes in a Bullet Blender Tissue Homogenizer (both from Next Advance). The sample was then mixed with 40 μL of 5x NP-40 Substitute Assay Reagent and triglycerides and protein were assayed as above.
Microscopy
Slides and plates were imaged using a Nikon Ti-E inverted microscope (Nikon Instruments) with a 60x oil-immersion objective lens. Images were recorded with a Zyla cMOS camera, and NIS Elements software was used for acquisition parameters, shutters, filter positions and focus control.
Lipid droplet quantification
Cells plated in glass bottom dishes (MatTek P35G-1.5-14-C) were labeled for 15 min at 37 °C in DMEM containing 1 μM Bodipy 493/503 and 1 μM Hoechst 33342 (Thermo D3922 and H3570) and subsequently washed. Total lipid droplet area in each cell was quantified using Cell Profiler.
Allograft tumor studies
One million 105K cells in PBS mixed with an equal volume of Matrigel (Corning 356237) were injected subcutaneously into the flank of seven female 6-week-old C57BL/6J mice. Tumor size was measured by caliper. When tumors reached an average size of 500 mm3, mice were randomly assigned to vehicle or control groups, ensuring that average weight and tumor size did not differ between groups. Mice were injected intraperitoneally with vehicle (PBS, 5% PEG-400, 5% Tween-80, 5% ethanol) or 1 mg/kg rapamycin in vehicle on two consecutive days at 9 am. Mice were fed ad libitum for the duration of the experiment. At 9 am, 24 hours after the final injection, blood was collected by retro-orbital bleed following anaesthesia with isoflurane, transferred to EDTA-coated tubes (Sarstedt 20.1288.100), and centrifuged 20 min at 2000 x g at 4 °C to collect plasma. Mice were sacrificed by CO2 asphyxiation and tumors were resected and flash frozen.
Analysis of mouse plasma lipids
Plasma triglycerides were quantified using the Infinity Triglycerides Reagent (Thermo TR22421), and plasma free fatty acids were quantified using the Wako NEFA-HR(2) Reagent (FujiFilm LABNEFA-M1) according to the manufacturers’ protocols.
Cell cycle analysis
Cells were labeled with 10 μM 5-ethynyl-2'-deoxyuridine for 2 hrs at 37 °C. Cells were trypsinized and labeled using the Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay Kit (Thermo C10425) and FxCycle Violet (Thermo F10347) according to the manufacturer’s instructions. Cells were quantified by flow cytometry using FACS Diva on an LSR II flow cytometer (BD). Data was analyzed using FlowJo.
Carbon-14 lipid measurements
To assay neutral lipids, cells were labeled medium containing 0.5 μCi/mL [1-14C]-oleic acid or [1-14C]-palmitic acid (Perkin Elmer NEC317 or NEC075H) for 6 hrs. To assay lipids containing choline, cells were labeled in DMEM lacking choline (US Biological D9800-14 supplemented with 0.4 mM glycine and 0.4 mM serine) plus 10% dialyzed FBS and 4 μCi/mL [methyl-14C]-choline (American Radiolabeled Chemicals 0208). [methyl-14C]-choline was dried under nitrogen gas stream and resuspended in medium.
Following 6 hrs, cells were washed three times in PBS and chased in unlabeled medium with inhibitors for 16 hrs. Alternatively, cells were chased for 16 hrs in unlabeled medium, then pretreated for 1 hr with inhibitors prior to torin1 treatment. Cells were washed twice with ice-cold PBS, and lipids were extracted twice with 300 μL 3:2 hexane:isopropanol by rocking the plate for 10 min at room temperature. Extracts were pooled and dried under nitrogen gas stream.
Samples resuspended in 50 μL 2:1 chloroform:methanol and spotted on Silica Gel HL 250um 20x20 cm Channeled TLC plates (Miles Scientific P43911). Neutral lipids were resolved with 80:20:1 hexane:diethyl ether:glacial acetic acid, or phospholipids were resolved with 25:15:4:2 chloroform:methanol:glacial acetic acid:water. TLC plates were exposed to BAS-IP MS Phosphor Storage Screens for up to 2 days, and screens were scanned using a GE Typhoon FLA 7000. Lipid abundance was quantified in Fiji. Bands corresponding to specific lipid classes were identified by co-migration with 20 μg of lipid standards visualized with iodine vapors. Standards: trioleoyl-glycerol (Sigma T7140), dioleoyl-glycerol (Sigma 8894), oleic acid (Sigma O1383), phosphatidylcholine (Avanti 840051C), lysophosphatidylcholine (Avanti 845874C), cholesteryl-oleate (Sigma C9253), and Mono-, Di-, & Triglyceride Mix (Sigma 1787).
After extraction, tissue culture plates were air-dried. Protein was extracted by adding 300 μL 0.3 M NaOH plus 0.1% SDS to each well and shaking 4 hrs at room temperature. Protein was assayed by BCA Assay (Thermo 23225) to normalize lipid content of each sample.
De novo lipogenesis
Following 16 hours of inhibitor treatment (Tsc2−/− MEFs) or 4 hours of insulin stimulation (WT MEFs and HeLa cells), [1-14C]-acetic acid (Perkin Elmer NEC084H001MC) was spiked into the medium at 4 μCi/mL (final). After 4 hours at 37 °C, cells were washed three times with ice cold PBS and scraped into 250 μL cold PBS plus 0.5% Triton-X100 with protease inhibitors. 150 μL crude lysate were mixed with 500 μL 2:1 chloroform:methanol and vortexed. Following addition of 125 μL water, the samples were vortexed and centrifuged for 15 min at 1000 x g at room temperature. 250 μL of the lower/organic phase were mixed with Emulsifier Safe scintillation cocktail, and carbon-14 was quantified on a Hidex 300 SL Counter. Remaining crude lysate was clarified for 10 min at maximum speed, and protein was quantified in the supernatant to normalize carbon-14 incorporated into the organic phase.
Hepatocyte isolation and labeling
Hepatocytes were isolated from 8-week old female C57BL/6J mice (Jackson Laboratories 000664) according to previously published protocols64. Briefly, livers were perfused with Liberase TM (Sigma LIBTM-RO) via the hepatic portal vein. Hepatocytes were pelleted, washed, and resuspended in DMEM plus 2.5% FBS. Viable cell count was determined by Trypan Blue exclusion using a hemocytometer. Hepatocytes were pelleted 5 min at 1000 x g at room temperature, resuspended in DMEM plus 1% FBS, and plated on collagen I-coated plates (Corning 62405): for immunoblot, 1.5 x 106 viable cells per well in a 6-well plate; for radioactive assays, 7.5 x 105 viable cells per well in a 12-well plate (for pulse-chase, medium included 0.5 μCi/mL [1-14C]-oleic acid). Five hours later cells were washed three times with PBS and incubated in serum-free medium M199 with 100 nM dexamethasone, 100 nM triiodothyronine, and 1 nM insulin for 16 hrs. Cells were washed and the medium was changed to M199 + 100 nM dexamethasone + 100 nM triiodothyronine without insulin for 30 min. Hepatocytes were then stimulated with 100 nM insulin for 6 hrs.
Fatty acid oxidation
Cells were labeled with 20 μCi/mL [9,10-3H]-palmitic acid (Perkin Elmer NET043) for 8 hrs at 37 °C. Tracer was removed by washing cells three times in PBS, and cells were serum starved in unlabeled medium for 16 hrs. For longer-term treatment, inhibitors were added for the entire 16-hr duration of the chase period. For shorter-term treatment, following the chase period, cells were pretreated for 1 hr with DGAT1 and 2 inhibitors or etomoxir prior to addition of torin1 for the indicated durations. Medium was refreshed for the final hour of the experiment to quantify production of 3H2O, indicative of fatty acid β-oxidation. Medium was pelleted for 10 min at maximum speed at room temperature, and 350 μL were applied to 350 mg 1x8 Dowex Chloride Resin (Sigma 44340) that had been prewashed with water and dispensed into Spin-X Centrifuge Tube Filters (Costar). Tubes were centrifuged 3 min at 4000 x g at room temperature. 3H2O in the flow through was quantified using Emulsifier Safe scintillation cocktail on a Hidex 300 SL counter. Cells were lysed in NP-40 lysis buffer and protein was quantified to normalize 3H2O produced.
Gene expression analysis
RNA was purified from cells using the TRIzol Reagent (Thermo 15596026), and cDNA was synthesized from 1 μg of RNA using the SuperScript III First-Strand Synthesis System (Thermo 18080051), according to the manufacturer’s instructions. Samples were assayed via quantitative RT-PCR using iTaq Universal SYBR Green Supermix (Bio-Rad 1725125) and a CFX Connect Realtime PCR Detection System (Bio-Rad). Relative transcript abundance was quantified using the ΔΔCT method, normalizing to mouse β-actin. qPCR primers were synthesized by Integrated DNA Technologies and are listed in Supplementary Table 3.
Lentivirus
Lentivirus was generated in HEK-293T cells transfected with a transfer plasmid, psPAX2, and pMD2.G using polyethylenimine. Target cells were infected two days later with viral media filtered through a 0.45 μM PES filter (Millipore) with 8 μg/mL (final) polybrene (Santa Cruz). Cells were selected with 1-4 μg/mL puromycin.
Lysosome immunoprecipitation (Lyso-IP)
Lyso-IP experiments were carried out according to previously published protocols 35 with modifications made for lipid extraction. HEK-293T cells infected with pLJC5-Tmem192-3xHA (Addgene 102930) or 2xFLAG (Addgene 102929) were grown to confluence in a 15 cm plate for each sample. After inhibitor treatment, cells were washed twice with ice cold PBS, scraped into 1 mL of ice-cold PBS containing potassium (KPBS: 136 mM KCl, 10 mM potassium phosphate pH 7.4), and pelleted for 2 min at 1000 x g at 4 C. Subsequent steps were carried out in the cold room.
Pellets were resuspended in 1 mL KPBS. 25 μL were removed and immediately mixed with NP-40 lysis buffer containing protease inhibitors for a whole cell protein sample or 225 μL 80% HPLC-grade methanol for whole cell lipidomics. Remaining cell suspension was lysed with 20 strokes in a tight-fitting dounce homogenizer. Nuclei and debris were removed by centrifuging for 2 min at 1000 x g at 4 °C. Supernatant was mixed with 100 μL resuspended Pierce Anti-HA Magnetic Beads (Thermo 88837) that was washed three times with KPBS. The beads were rocked with the lysate for 10 min at 4 °C and washed four times with KPBS using a DynaMag magnet.
Following the final wash, beads were suspended in 100 μL NP-40 lysis buffer with protease inhibitors for a lysosomal protein sample or in 225 μL 80% methanol for lysosomal lipid extraction. For lipid extraction, samples were stored at −80 °C at this point until all cells were processed. Then 600 μL MTBE were added and samples were mixed for 1 hr at room temperature. 105 μL HPLC-grade water were added, and phases were separated by centrifugation for 10 min at 1000 x g at room temperature. The upper phase was collected and dried under nitrogen gas stream. Lipids were analyzed as described above. Lipid species whose abundance in the Lyso-IP from HA-tagged cells was ≤4-fold higher than the average abundance in the negative control Lyso-IP from FLAG-tagged cells were excluded from the final analysis.
CRISPR-Cas9 mediated gene deletion
MEFs were transfected with pairs of PX461 plasmids containing small guide RNAs (sgRNA) and Cas9-D10A nickase mutant. The targeted sequences are listed in Supplementary Table 4, and corresponding oligonucleotides were synthesized by Integrated DNA Technologies and cloned into PX461 (Addgene 48140). Resulting plasmids were verified by Sanger sequencing (Genewiz). Nucleofector kit (Lonza, VAPL-1004) and Nucleofector II Device (Lonza) were used according to the manufacturer’s protocols. Two days after transfection, cells were trypsinized, and stained with DAPI to mark dead cells. Single DAPI-negative, GFP-positive cells were sorted into individual wells in a 96-well plate containing DMEM + 10% FBS using a Sony SH800Z Cell Sorter. The resulting colonies were screened to identify clones with successful deletions. Mutant MEF lines newly generated in this study are available upon reasonable request.
siRNA mediated knockdown
Cells were transfected with 50 nM of each siRNA using Lipofectamine RNAiMAX (Thermo) according to the manufacturer’s protocol. 48 hours post-transfection, cells were labeled by pulse-chase and treated with inhibitors as described above. siRNAs from Horizon Discovery: siControl (ON-TARGETplus Non-targeting pool, D-001810-10), siTfe3 (ON-TARGETplus Mouse Tfe3 siRNA Smart Pool, L-054750-00), and siTfeb (ON-TARGETplus Mouse Tfe3 siRNA Smart Pool, L-054750-00).
Transferrin uptake assay
Cells were treated with torin1 in DMEM + 10% FBS for 4 hrs prior to the transferrin (Tf) uptake assay. Cells were washed twice with PBS and incubated in DMEM + 25 mM HEPES pH 7.4 with 5 μg/mL Tf-AlexaFluor-647 (Thermo T23366) at 37 °C. Cells were transferred to ice and washed once with ice-cold PBS, twice with acid wash buffer (150 mM NaCl, 1 mM MgCl2, 0.125 mM CaCl2, 0.1 M glycine, pH 2.5) to remove membrane-bound extracellular Tf, and twice with PBS. Cells were detached by in cold PBS + 10 mM EDTA, pelleted for 5 min at 1000 x g at 4 °C, and resuspended in PBS + 1% BSA. Tf uptake was quantified from sample mean fluorescence for >2000 cells using a Fortessa (BD Biosciences). Where indicated, the acid wash was replaced with PBS and/or cells were incubated with Tf at 4 °C.
Dextran uptake assays
Cells were labeled with 20 μg/mL 10 kDa Dextran-AlexaFluor-647 (Thermo D22914) for 30 min at 37 °C. Cells were washed twice with PBS, trypsinized, pelleted for 5 min at 1000 x g at 4 °C, and resuspended in PBS plus 1% BSA. Dextran uptake was quantified from sample mean fluorescence for >2000 cells using a Fortessa (BD Biosciences).
BSA Cleavage Assay
Following 1 hr inhibitor treatment, cells in glass bottom dishes (MatTek, P35G-1.5-14-C) were labeled with 10 μg/mL DQ-Green BSA and 10 μg/mL BSA AlexaFluor-647 (Thermo D12050 and A34785) for 3 hrs at 37 °C. 1 μM Hoechst 33324 was added to the medium for 20 min before imaging. To image lysosomes, BSA AlexaFluor-647 was omitted and cells were stained with 50 nM LysoTracker Deep Red (Thermo Scientific L12492) for 20 min prior to imaging. BSA uptake (AlexaFluor 647 fluorescence) and cleavage (DQ-Green BSA fluorescence) were quantified for each cell in Fiji, and cleavage was normalized to uptake.
Statistics & Reproducibility
Statistical analysis was carried out in Microsoft Excel and GraphPad Prism 9, and statistical details for each experiment can be found in the corresponding figure legend. Data are presented as the mean ± standard deviation (SD).
An unpaired two-tailed Student’s t-test was used for comparisons between two groups. A one-way ANOVA with Tukey’s multiple comparison test was used for comparisons between three or more groups. A two-way ANOVA with Sidak's multiple comparison test was used for experiments with combinations of inhibitors/genotypes. An extra sum-of-squares F-test was used to compare triglyceride turnover data fit by non-linear regression to an exponential decay. Alpha was set at 0.05 for significance and p-values are reported for significant results.
For lipidomic data, the total abundance of a given lipid class reported is the sum of ion counts for individual lipids in that class. Changes in the abundances of individual lipid species are reported where indicated. To generate volcano plots, the abundances of individual lipid species in treated cells were compared to control cells, and a p-value was determined using an unpaired two-tailed Student’s t-test. These p-values were adjusted for multiple comparisons in R using the Benjamini-Hochberg method, and the −log10 transform of these adjusted p-values is reported.
Experiments reported in this study were repeated at least two times, and in many cases more than three times, with similar results obtained each time to confirm reproducibility. Immunoblots were conducted once per experiment shown as positive controls to validate the on-target efficacy of the inhibitors, growth factors, and genetic perturbations used. TLC scans shown in Extended Data Figures 3B and 4G are representative of at least three independent replicates and are provided here to demonstrate effective chromatographic resolution of lipid classes.
Extended Data
Extended Data Fig. 1.
(A-C) Immunoblots for AKT and pAKT corresponding to Figures 1A-C.
(D,E) Fold changes of individual lipids grouped by class from the lipidomics experiment shown in Figure 1D. (D) Non-ether and (E) ether lipids are separated. Dots represent the fold change of each individual lipid species relative its abundance in vehicle-treated cells. Bars represent the mean fold change for that class, graphed ± SD for n=3 replicates.
(F,G) Triglyceride species from the results in Figure 1C stratified by the total number of double bonds (F) or carbon atoms (G) in their side chains. Each point represents an individual triglyceride. Group mean ± SD is shown in (F). Pearson’s r and line of best fit are shown in (G). Data represent n=3 replicates.
(H,I) Immunoblots for AKT and pAKT corresponding to Figures 1G-J.
(J) Changes in the abundance of individual lipid species from the experiment in Figure 1H, with neutral lipids color coded, for mouse ES cells treated with torin1. Dotted lines indicate a 2-fold change (FC) and an adjusted p-value of 0.1.
Statistical analysis by one-way ANOVA (F); n.s., not significant.
Extended Data Fig. 2.
(A) Triglyceride accumulation measured by enzyme assay and normalized to protein content from the same sample in Tsc2−/− MEFs serum starved for 16 hrs, followed by 0-8 hrs of vehicle, rapamycin (20 nM), or torin1 (250 nM) treatment, graphed as mean ± SD relative to the 0 hr timepoint, n=3.
(B) Immunoblot for AKT and pAKT corresponding to Figure 2M.
(C) Immunoblot for the tumors shown in Figure 2O.
(D) Tumor TG versus plasma TG for individual mice shown in Figures 2O,P.
(E) Plasma non-esterified fatty acids (NEFA) measured for the mice shown in Figure 2O, graphed as mean ± SD, n=5 mice.
(F) Immunoblot corresponding to the experiment shown in Figure 2Q.
(G,H) Cell cycle profile for PC3 (G) and H1299 (H) cells treated for 16 hrs with vehicle, palbociclib (CDK4/6i, 100 nM), and/or torin1 (250 nM), n=1.
(I,J) Triglyceride levels quantified by enzyme assay and normalized to protein content from the same sample for cells treated as in (G,H), graphed as mean ± SD relative to vehicle-treated cells, n=3.
Statistical analysis by two-way ANOVA (I,J) and two-tailed Student’s t-test (E); n.s., not significant.
Extended Data Fig. 3.
(A) SREBP1 target gene expression in Tsc2+/+ and Tsc2−/− MEFs treated for 16 hrs with vehicle, rapamycin (20 nM), or torin1 (250 nM), graphed as mean ± SD relative to vehicle-treated cells, n=3.
(B) Representative thin layer chromatogram for Tsc2−/− MEFs labeled with [1-14C]-oleate and treated with vehicle, rapamycin (20 nM), or torin1 (250 nM) for 16 hrs, developed to separate neutral lipids. ChE, cholesteryl-esters; TG, triglycerides; FA, fatty acids; DG, diglycerides; MG, monoglycerides.
(C,D) Immunoblot (C) and TGs (D) assayed by pulse-chase with [1-14C]-oleate tracer (6 h) in Tsc2+/+ MEFs followed by chase in cold medium for 16 h with vehicle, 20 nM rapamycin, 250 nM torin1, 500 nM AZD2014, or 5 nM RapaLink-1, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(E,F) Immunoblot (E) and TGs (F) assayed by pulse-chase with [1-14C]-oleate tracer (6 h) in MCF7 cells followed by chase in cold medium for 16 h with vehicle, 250 nM torin1, or 1 μM BYL719 treatment, graphed as mean ± SD relative to vehicle-treated cells, n=3. NS denotes a non-specific band.
(G-J) Immunoblot (G), de novo lipogenesis of all lipids (H), TG labeled from de novo lipogenesis (I), and lipid remodeling (J), assayed as in Figures 3H,I in primary mouse hepatocytes serum starved for 16 hrs, pretreated for 30 min with vehicle, rapamycin (20 nM), or torin1 (250 nM), and stimulated with insulin (100 nM) for 6 hrs, graphed as mean ± SD relative to vehicle-treated unstimulated cells, n=3.
Statistical analysis by one-way ANOVA (A,D,F,H-J); n.s., not significant.
Extended Data Fig. 4.
(A) 14C-TG turnover in PC3 cells treated with vehicle, torin1, or ATGLi, measured as in Figure 4C. 14C-TG abundance is graphed over time as mean ± SD relative to 0 hrs, n=3.
(B) Immunoblots for ATGL and DGAT1 levels from two separate experiments in cell lines treated with vehicle, rapamycin, or torin1 for 16 hrs. NS denotes a non-specific band.
(C) 14C-Fatty acid levels in Tsc2−/− MEFs labeled with [1-14C]-palmitate, treated with vehicle, rapamycin, or torin1 for 16 hrs, graphed as mean ± SD relative to vehicle, n=3.
(D) Acyl-carnitine species in Tsc2−/− MEFs in the experiment in Figure 1D, graphed as mean ± SD relative to vehicle, n=3.
(E,F) Acyl-carnitine species detected in mouse embryonic stem (ES) cells (E) and wild type, primary MEFs (F) in the experiments in Figures 1H,J, graphed as mean ± SD relative to vehicle, n=3.
(G) Representative thin layer chromatogram for Tsc2−/− MEFs traced with [methyl-14C]-choline and treated with vehicle, rapamycin, or torin1 for 16 hrs, developed to separate phospholipids. PC, phosphatidylcholine; SM, sphingomyelin; LPC, lysophosphatidylcholine.
(H) Ratio of lysophosphatidylcholine to phosphatidylcholine in PC3 cells pulse-labeled with [methyl-14C]-choline as in (G); mean ± SD relative to vehicle, n=3.
(I,J) Glycerophosphocholine levels in HEK-293T (I) and PC3 (J) cells treated for 16 hrs with vehicle, rapamycin, or torin1, graphed as mean ± SD relative to vehicle, n=3.
(K,L) 14C-TG (K) and 14C-fatty acids (L) in PC3 cells measured as in Figures 4I,J and graphed as mean ± SD, n=3.
(M) Fatty acid accumulation in Tsc2−/− MEFs labeled with [1-14C]-oleate following vehicle (DMSO) treatment with 1 hr pre-treatment with DGAT inhibitors and/or etomoxir; mean ± SD relative to 0 hrs, n=3.
(N) Triglyceride ion counts in the experiment shown in Figures 4L,M; mean ± SD relative to vehicle, n=3.
(O) Carnitine palmitoyl-transferase (CPT) gene expression in MEFs treated for 16 hrs with inhibitors, graphed as mean ± SD relative to vehicle, n=6.
Statistics: by one-way ANOVA (C,H-J,N), two-way ANOVA (D-F,K-M,O), and extra sum-of-squares F-test (A). n.s., not significant.
Extended Data Fig. 5.
(A,B) Immunoblot (A) and fatty acids (B) corresponding from the experiment shown in Figure 5A. Fatty acids quantification is graphed as mean ± SD relative to vehicle-treated cells, n=3.
(C,D) Immunoblots corresponding to the experiments shown in Figures 5B,C.
(E-G) Triglyceride and acyl-carnitine levels from the experiment shown in Figures 5D-G.
(H) Immunoblot for organelle markers in whole cell lysates or in HA-tag immunopurified lysosomes (IP) from HEK-293T cells expressing TMEM192-2xFLAG or 3xHA.
(I) Immunoblot corresponding to the experiment shown in Figures 5K,L.
(J) Immunoblot corresponding to the experiment shown in Figures 5M,N. LC3B is indicated in its unmodified (LC3B-I) and lipidated (LC3B-II) forms.
Statistical analysis by two-way ANOVA (B,E-G). n.s., not significant.
Extended Data Fig. 6.
(A-F) Lipid class sums (A-E) and glycerophosphocholine levels (F) detected in Tsc2−/− MEFs treated with vehicle, MAFP (a non-specific serine hydrolase inhibitor, 10 μM), and/or torin1 (250 nM) for 16 hrs, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(G) Phospholipase A1 (PLA1) and A2 (PLA2) gene expression in Tsc2+/+ and Tsc2−/− MEFs treated for 16 hrs with vehicle, rapamycin (20 nM), or torin1 (250 nM), calculated relative to vehicle-treated Tsc2−/− MEFs, n=3. cPLA2, cytosolic PLA2; iPLA2, calcium-independent PLA2; PA-PLA1, phosphatidic acid-preferring PLA1; PLB, phospholipase B.
(H,I) Immunoblot (H) and TGs measured by pulse-chase with [1-14C]-oleate tracer (I) in parental and Pla2g15 knockout Tsc2−/− MEFs expressing enhanced green fluorescent protein (EGFP) or wildtype LPLA2 (PLA2G15) treated for 16 hrs with vehicle or torin1 (250 nM), graphed as mean ± SD relative to vehicle-treated cells, n=3.
Statistical analysis by two-way ANOVA (A-F,I). n.s., not significant.
Extended Data Fig. 7.
(A,B) Immunoblot (A) and TGs measured by pulse-chase with [1-14C]-oleate tracer (B) in Atg5+/+ and −/− MEFs treated for 16 hrs with vehicle or BafA1 (250 nM), graphed as mean ± SD relative to vehicle-treated cells, n=3.
(C,D) Immunoblot (C) and triglyceride levels measured by enzyme assay (D) and normalized to protein content from the same sample for parental, ATG7 knockout, and FIP200 knockout HEK-293T cells treated with vehicle or bafilomycin A1 (250 nM) for 16 hrs, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(E,F) Immunoblot (E) and triglyceride levels (F) corresponding to the results shown in Figures 6E,F. Triglyceride levels are graphed as mean ± SD relative to vehicle-treated cells, n=3.
(G,H) Immunoblot (G) and triglyceride levels measured by enzyme assay (H) and normalized to protein content from the same sample for parental and FIP200 knockout HEK-293T cells treated with vehicle, torin1 (250 nM), and/or bafilomycin A1 (250 nM) for 16 hrs, graphed as mean ± SD relative to vehicle-treated cells, n=3.
(I) EGFR degradation assay in parental Tsc2−/− MEFs and sgUvrag or sgAtg14 clones. Cells were serum starved for 16 hrs and stimulated with EGF (10 ng/mL) for the indicated times.
(J) Immunoblot for autophagy markers in parental Tsc2−/− MEFs and sgAtg14 clone after 4 hrs in amino acid replete or free medium.
Statistical analysis by two-way ANOVA (B,D,F,H). n.s., not significant.
Extended Data Fig. 8.
(A) Immunoblot for TFEB, TFE3, and proteins corresponding to their transcriptional target genes in Tsc2−/− MEFs treated for two days with control siRNA, siTfeb, or siTfe3.
(B,C) 14C-TGs assayed by pulse-chase with [1-14C]-oleate tracer (6 h) in Tsc2−/− MEFs treated as in (A) followed by chase in cold medium for 16 hrs with vehicle or 250 nM torin1, graphed as mean ± SD relative to control siRNA-treated cells (B) and relative to vehicle-treated cells (C), n=3.
Statistical analysis by one-way ANOVA (B) and two-way ANOVA (C). n.s., not significant.
Extended Data Fig. 9.
(A) Immunoblot corresponding to the experiment shown in Figures 7A,B.
(B-D) Immunoblots (B) and 14C-TGs (C,D) assayed by pulse-chase with [1-14C]-oleate tracer (6 h) in Tsc2−/− MEFs followed by chase in cold medium for 16 hrs with vehicle, 250 nM torin1, 5 μM VPS34-IN1, or 80 μM Dynasore, graphed as mean ± SD relative to vehicle-treated cells (C) and relative to no-torin1 controls (D), n=3.
(E) Immunoblot corresponding to the experiment shown in Figure 7C.
(F) Fatty acid accumulation measured by pulse-chase with [1-14C]-oleate tracer in the presence of DGAT inhibitors (3 μM each) and etomoxir (20 μM) in Tsc2−/− MEFs following a 4 hr treatment with endocytosis inhibitors (5 μM VPS34-IN1, 1 μM PIK-FYVEi, 80 μM Dynasore, or 20 μM PitStop 2). Graphed as mean ± SD relative to vehicle-treated cells at time 0, n=3.
(G,H) Binding of transferrin to Tsc2−/− MEFs (G) and PC3 (H) cells treated for 4 hrs with vehicle or torin1 (250 nM) and kept on ice while labeling with transferrin, confirming that the amount of plasma membrane transferrin receptor is not regulated by mTOR and that surface-bound transferrin is eliminated following an acid wash. Graphed as mean ± SD, n=3.
(I,J) Endocytosis assays in PC3 cells, measuring the uptake of transferrin (I) or 10 kDa dextran (J), following 4 hrs of vehicle, rapamycin (20 nM), or torin1 (250 nM) treatment, graphed as mean ± SD, n=3.
a.u., Arbitrary Units. Statistical analysis by one-way ANOVA (C,F,J), and two-way ANOVA (D,G-I). n.s., not significant.
Extended Data Fig. 10.
(A) BSA uptake in Tsc2−/− MEFs treated with torin1 (250 nM) and vehicle or EIPA (20 μM) for 1 hr prior to labeling with 10 μg/mL BSA Alexa Fluor 647 (uptake, red) for 3 hrs. Nuclei are shown in blue.
(B) BSA cleavage in Tsc2−/− MEFs treated with torin1 (250 nM) and vehicle or protease inhibitors (2 μM E64d, 2 μM Pepstatin A, and 10 μM Leupeptin) for 1 hr prior to labeling with 10 μg/mL each of BSA Alexa Fluor 647 (red, uptake) and DG-Green BSA (green, cleavage) for 3 hrs. Nuclei are shown in blue.
(C,D) Quantification of the effect of EIPA on BSA uptake (C) and protease inhibitors on BSA cleavage (D) for cells treated as in (A,B), graphed as mean ± SD relative to vehicle-treated cells, n=57-84 cells.
(E) Co-localization of hydrolyzed DQ-Green BSA (green) and LysoTracker Deep Red (red) in Tsc2−/− MEFs pre-treated with torin1 (250 nM) for 1 hr prior to labeling with 10 μg/mL DQ-Green BSA for 3 hrs. Nuclei are shown in blue. These images are representative of two replicate experiments in which similar results were obtained.
Supplementary Material
Acknowledgments
We thank members of the Manning lab for helpful input, in particular Sandra Schrötter for her guidance in microscopy studies. We thank David Kwiatkowski (Brigham and Women’s Hospital), Elizabeth Henske (Brigham and Women’s Hospital), David Sabatini, and Wade Harper (Harvard Medical School) for providing cell lines and other reagents. We thank Alex Kreutzberger, Anwesha Sanyal, and Tomas Kirchausen for helpful conversations and initial guidance in studying endocytosis, and Zon Weng Lai and Yohannes Ambaw from the Harvard-Chan Advanced Multi-omics Platform for guidance in piloting the lyso-IP lipidomics experiments. Cell sorting was performed with the Harvard Medical School Systems Biology Flow Cytometry Core. A.M.H. was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. This study was funded by NIH grants R35-CA197459 (B.D.M) and P01-CA120964 (B.D.M. and J.M.A.).
Footnotes
Declaration of interests
B.D.M. is a member of the scientific advisory board and a shareholder of Navitor Pharmaceuticals. All other authors declare no competing financial interests.
Editor recognition statement:
Primary handling editor: Yanina-Yasmin Pesch, in collaboration with the Nature Metabolism team.
Reviewer recognition statement:
Nature Metabolism thanks Martin Giera, Mathieu Laplante and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data Availability
All data generated or analysed during this study are included in this published article (and its supplementary information files).
References:
- 1.Rousseau A & Bertolotti A An evolutionarily conserved pathway controls proteasome homeostasis. Nature 536, 184–189, doi: 10.1038/nature18943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ottens F, Franz A & Hoppe T Build-UPS and break-downs: metabolism impacts on proteostasis and aging. Cell Death Differ 28, 505–521, doi: 10.1038/s41418-020-00682-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Sahra I & Manning BD mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol 45, 72–82, doi: 10.1016/j.ceb.2017.02.012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller M et al. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. Elife 4, e07736, doi: 10.7554/eLife.07736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saxton RA & Sabatini DM mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976, doi: 10.1016/j.cell.2017.02.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J & Guan KL mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol 21, 63–71, doi: 10.1038/s41556-018-0205-1 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Torrence ME & Manning BD Nutrient Sensing in Cancer. Annual Review of Cancer Biology 2, 251–269, doi: 10.1146/annurev-cancerbio-030617-050329 (2018). [DOI] [Google Scholar]
- 8.Duvel K et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39, 171–183, doi: 10.1016/j.molcel.2010.06.022 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson TR et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420, doi: 10.1016/j.cell.2011.06.034 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porstmann T et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab 8, 224–236, doi: 10.1016/j.cmet.2008.07.007 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Brown MS & Goldstein JL Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A 107, 3441–3446, doi: 10.1073/pnas.0914798107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Condon KJ & Sabatini DM Nutrient regulation of mTORC1 at a glance. J Cell Sci 132, doi: 10.1242/jcs.222570 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E, Goraksha-Hicks P, Li L, Neufeld TP & Guan KL Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 10, 935–945, doi: 10.1038/ncb1753 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castellano BM et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 355, 1306–1311, doi: 10.1126/science.aag1417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dikic I & Elazar Z Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19, 349–364, doi: 10.1038/s41580-018-0003-4 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Palm W et al. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell 162, 259–270, doi: 10.1016/j.cell.2015.06.017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caron A, Richard D & Laplante M The Roles of mTOR Complexes in Lipid Metabolism. Annual Review of Nutrition 35, 321–348, doi: 10.1146/annurev-nutr-071714-034355 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Yecies JL et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab 14, 21–32, doi: 10.1016/j.cmet.2011.06.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricoult SJ, Yecies JL, Ben-Sahra I & Manning BD Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene 35, 1250–1260, doi: 10.1038/onc.2015.179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton JD, Goldstein JL & Brown MS SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109, 1125–1131, doi: 10.1172/JCI15593 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagen RM, Rodriguez-Cuenca S & Vidal-Puig A An allostatic control of membrane lipid composition by SREBP1. FEBS Lett 584, 2689–2698, doi: 10.1016/j.febslet.2010.04.004 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Quinn WJ 3rd et al. mTORC1 stimulates phosphatidylcholine synthesis to promote triglyceride secretion. J Clin Invest 127, 4207–4215, doi: 10.1172/JCI96036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sengupta S, Peterson TR, Laplante M, Oh S & Sabatini DM mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104, doi: 10.1038/nature09584 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Soliman GA, Acosta-Jaquez HA & Fingar DC mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids 45, 1089–1100, doi: 10.1007/s11745-010-3488-y (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paolella LM et al. mTORC1 restrains adipocyte lipolysis to prevent systemic hyperlipidemia. Mol Metab 32, 136–147, doi: 10.1016/j.molmet.2019.12.003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrisett JD et al. Effects of sirolimus on plasma lipids, lipoprotein levels, and fatty acid metabolism in renal transplant patients. J Lipid Res 43, 1170–1180 (2002). [PubMed] [Google Scholar]
- 27.Li Z et al. Cancer cells depend on environmental lipids for proliferation when electron acceptors are limited. bioRxiv, 2020.2006.2008.134890, doi: 10.1101/2020.06.08.134890 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ackerman D et al. Triglycerides Promote Lipid Homeostasis during Hypoxic Stress by Balancing Fatty Acid Saturation. Cell Rep 24, 2596–2605 e2595, doi: 10.1016/j.celrep.2018.08.015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lien EC et al. Low glycaemic diets alter lipid metabolism to influence tumour growth. Nature 599, 302–307, doi: 10.1038/s41586-021-04049-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young RM et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev 27, 1115–1131, doi: 10.1101/gad.198630.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu B et al. HIF2alpha-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov 5, 652–667, doi: 10.1158/2159-8290.CD-14-1507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen TB et al. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev Cell 42, 9–21 e25, doi: 10.1016/j.devcel.2017.06.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rambold AS, Cohen S & Lippincott-Schwartz J Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell 32, 678–692, doi: 10.1016/j.devcel.2015.01.029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filippakis H et al. Lysosomal regulation of cholesterol homeostasis in tuberous sclerosis complex is mediated via NPC1 and LDL-R. Oncotarget 8, 38099–38112, doi: 10.18632/oncotarget.17485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu-Remaileh M et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813, doi: 10.1126/science.aan6298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Showalter MR et al. The Emerging and Diverse Roles of Bis(monoacylglycero) Phosphate Lipids in Cellular Physiology and Disease. Int J Mol Sci 21, doi: 10.3390/ijms21218067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohsaki Y, Suzuki M, Shinohara Y & Fujimoto T Lysosomal accumulation of mTOR is enhanced by rapamycin. Histochem Cell Biol 134, 537–544, doi: 10.1007/s00418-010-0759-x (2010). [DOI] [PubMed] [Google Scholar]
- 38.Shayman JA & Tesmer JJG Lysosomal phospholipase A2. Biochim Biophys Acta Mol Cell Biol Lipids 1864, 932–940, doi: 10.1016/j.bbalip.2018.07.012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuboyama K et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036–1041, doi: 10.1126/science.aaf6136 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Kim YM et al. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell 57, 207–218, doi: 10.1016/j.molcel.2014.11.013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan HX, Russell RC & Guan KL Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 9, 1983–1995, doi: 10.4161/auto.26058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Napolitano G & Ballabio A TFEB at a glance. J Cell Sci 129, 2475–2481, doi: 10.1242/jcs.146365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macia E et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 10, 839–850, doi: 10.1016/j.devcel.2006.04.002 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Bago R et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J 463, 413–427, doi: 10.1042/BJ20140889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rennick JJ, Johnston APR & Parton RG Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat Nanotechnol 16, 266–276, doi: 10.1038/s41565-021-00858-8 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Filippakis H et al. Vps34-mediated macropinocytosis in Tuberous Sclerosis Complex 2-deficient cells supports tumorigenesis. Sci Rep 8, 14161, doi: 10.1038/s41598-018-32256-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dauner K, Eid W, Raghupathy R, Presley JF & Zha X mTOR complex 1 activity is required to maintain the canonical endocytic recycling pathway against lysosomal delivery. J Biol Chem 292, 5737–5747, doi: 10.1074/jbc.M116.771451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madeira JB et al. TORC1 inhibition induces lipid droplet replenishment in yeast. Mol Cell Biol 35, 737–746, doi: 10.1128/MCB.01314-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bjedov I et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab 11, 35–46, doi: 10.1016/j.cmet.2009.11.010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilcock DJ et al. DGAT1 is a lipid metabolism oncoprotein that enables cancer cells to accumulate fatty acid while avoiding lipotoxicity. bioRxiv, 2020.2006.2023.166603, doi: 10.1101/2020.06.23.166603 (2020). [DOI] [Google Scholar]
- 51.Chitraju C et al. Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell Metab 26, 407–418 e403, doi: 10.1016/j.cmet.2017.07.012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piccolis M et al. Probing the Global Cellular Responses to Lipotoxicity Caused by Saturated Fatty Acids. Mol Cell 74, 32–44 e38, doi: 10.1016/j.molcel.2019.01.036 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fredrikson G, Stralfors P, Nilsson NO & Belfrage P Hormone-sensitive lipase of rat adipose tissue. Purification and some properties. J Biol Chem 256, 6311–6320 (1981). [PubMed] [Google Scholar]
- 54.Priolo C et al. Tuberous sclerosis complex 2 loss increases lysophosphatidylcholine synthesis in lymphangioleiomyomatosis. Am J Respir Cell Mol Biol 53, 33–41, doi: 10.1165/rcmb.2014-0379RC (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li C et al. Rapamycin-insensitive up-regulation of adipocyte phospholipase A2 in tuberous sclerosis and lymphangioleiomyomatosis. PLoS One 9, e104809, doi: 10.1371/journal.pone.0104809 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naslavsky N & Caplan S The enigmatic endosome - sorting the ins and outs of endocytic trafficking. J Cell Sci 131, doi: 10.1242/jcs.216499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bissig C & Gruenberg J Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb Perspect Biol 5, a016816, doi: 10.1101/cshperspect.a016816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nnah IC et al. TFEB-driven endocytosis coordinates MTORC1 signaling and autophagy. Autophagy 15, 151–164, doi: 10.1080/15548627.2018.1511504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alesi N et al. TSC2 regulates lysosome biogenesis via a non-canonical RAGC and TFEB-dependent mechanism. Nat Commun 12, 4245, doi: 10.1038/s41467-021-24499-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Settembre C et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 15, 647–658, doi: 10.1038/ncb2718 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oren Y et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature 596, 576–582, doi: 10.1038/s41586-021-03796-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang J, Dibble CC, Matsuzaki M & Manning BD The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol 28, 4104–4115, doi: 10.1128/MCB.00289-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Breitkopf SB et al. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 13, doi: 10.1007/s11306-016-1157-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Byles V et al. Hepatic mTORC1 signaling activates ATF4 as part of its metabolic response to feeding and insulin. Mol Metab 53, 101309, doi: 10.1016/j.molmet.2021.101309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its supplementary information files).