Abstract
The vast majority of patients possess one or more pharmacogenetic variants that can influence optimal medication use. When pharmacogenetic data are used to guide drug choice and dosing, evidence points to improved disease outcomes, fewer adverse effects, and lower healthcare spending. Although its science is well established, clinical use of pharmacogenetic data to guide drug therapy is still in its infancy. Pharmacogenetics essentially involves the intersection of an individual’s genetic data with their medications, which makes pharmacists uniquely qualified to provide clinical support and education in this field. In fact, most pharmacogenetics implementations, to date, have been led by pharmacists as leaders or members of a multidisciplinary team or as individual practitioners. A successful large-scale pharmacogenetics implementation requires coordination and synergy among administrators, clinicians, informatics teams, laboratories, and patients. Because clinical implementation of pharmacogenetics is in its early stages, there is an urgent need for guidance and dissemination of shared experiences to provide a framework for clinicians. Many early adopters of pharmacogenetics have explored various strategies among diverse practice settings. This article relies on the experiences of early adopters to provide guidance for critical steps along the pathway to implementation, including strategies to engage stakeholders; evaluate pharmacogenetic evidence; coordinate laboratory testing, results interpretation and their integration into the electronic health record; identify reimbursement avenues; educate providers and patients; and maintain a successful program. Learning from early adopters’ published experiences and strategies can allow clinicians leading a new pharmacogenetics implementation to avoid pitfalls and adapt and apply lessons learned by others to their own practice.
Keywords: Pharmacogenetics, Pharmacogenomics, Pharmacy, Implementation
BACKGROUND
Under the umbrella of precision medicine lies pharmacogenetics, the science of using an individual’s deoxyribonucleic acid (DNA) to predict their response to medications.1 While pharmacogenetics is well established in the research field, its clinical use to inform drug therapy changes is still in its early stages.2 Genetic variability has consistently been shown to affect many medications’ pharmacokinetic and pharmacodynamic endpoints. Evidence suggests utilizing pharmacogenetics to guide drug therapy selection and dosing can improve disease outcomes,3–5 decrease adverse effects,6,7 and decrease healthcare spending.8 More than 90% of people carry at least one clinically-actionable pharmacogenetic variant9 and between 20–50%10,11 of Americans are taking a medication subject to a Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline. A study of nearly 700 individuals found 16% had an actionable result for at least one of their current medications.12
This paper combines lessons learned from the authors’ practice within University of Florida (UF) Health and information available in the published literature from other early adopters of pharmacogenetics to discuss various steps and best practices for implementing a clinical pharmacogenetics service.13–16 The purpose of this paper is to provide a foundation and guidance for pharmacists wishing to initiate a clinical pharmacogenetics service at their institution.
GUIDING PRINCIPLES
While pharmacy often leads the initiation of a clinical pharmacogenetics service, successful implementation requires a multidisciplinary effort to make key decisions about the targeted patient population, implementation strategy, key prescribers, pharmacogenetic testing process and content, and support for interpretation of genotype results. Establishing a strong leadership team and guiding principles that will shape the implementation is the starting point in this process for many institutions. When developing guiding principles, individuals should consider specific needs and resources of their institution. These principles will be essential to ensure the long-term success and sustainability of any implementation effort.
In the authors’ experience, institutions wishing to implement clinical pharmacogenetic testing should consider the following decision points when developing guiding principles: 1) stakeholders and engagement strategies; 2) evidence evaluation strategy; 3) pharmacogenetic testing logistics; 4) electronic health record integration and test interpretation; 5) reimbursement strategies; 6) educational strategies; and 7) program maintenance strategies. Table 1 provides a use-case scenario of one institution’s application of these principles.17–19
Table 1.
Use Case Scenario: Implementation of Guiding Principles in University of Florida (UF) Health Precision Medicine Program (PMP)
| Program Overview: The UF Health PMP, a pharmacist-led multi-disciplinary clinical implementation initiative established in 2011 has genotyped more than 10,000 patients to date. Seven unique gene-drug pairs have been implemented across inpatient and outpatient settings, including an established pharmacogenetics consult clinic. Currently, testing is available in all patient care areas, supported by 2 clinical pharmacogenetics specialists and trainees. | |
| Stakeholders and Engagement | • Pharmacist-led multidisciplinary clinical implementation initiative engaging patients, pharmacists, providers, health system clinical and administrative leaders, and pathology. |
| Evidence Evaluation Strategy | • Evidence evaluated and assessed by an internal committee, similar to a Pharmacy and Therapeutics Committee, with representation from UF College of Pharmacy and UF Health clinicians. |
| Pharmacogenetic Testing Logistics | • Majority of testing performed by UF Health Pathology Laboratories. • Launched initially with a preemptive genotyping focus, using chip-based genotyping that generated multiple genotypes, some of which were stored in a research database with a plan to migrate to the electronic health record (EHR) as sufficient clinical data emerge.17 Then transitioned to single-gene tests: cytochrome P450 (CYP)2C19, followed by thiopurine S-methyltransferase (TPMT) in 2014, CYP2D6 in 2015, and NUDT15 in 2019. In 2019, UF Health developed and launched a clinically focused multi-gene panel while still offering several single-gene tests. |
| Electronic Health Record Integration and Test Interpretation | • Laboratory results section in Epic EHR houses discrete genotype and phenotype results that trigger clinical decision support (CDS) alerts. • The majority of alerts at UF Health are active, pop-up alerts that disrupt workflow appropriately (e.g., only fire for actionable gene-drug pair) • Consultation notes are placed in the EHR by pharmacists with providers notified by Epic in-basket message.18 • Currently transitioning to the Epic Genomics to allow for consolidation of all genetics information into one profile, creation of multiple forms of CDS, and improved pharmacogenetic result readability within the patient profile. |
| Reimbursement Strategies | • Non-research clinical tests for ambulatory care patients are billed to insurance with pre-test patient education on potential out-of-pocket costs. Inpatient tests within UF Health are billed through Diagnostic Related Grouping (DRG). A portion of tests are covered by research funding. |
| Educational Strategies | • Patient education in face-to-face encounters, through print materials, and via a video sent to patients prior to their clinic appointment (available at https://youtu.be/b9FyOAEXzBw). • Provider education includes traditional presentations, printed tip sheets to facilitate referrals and point-of-care decision making, and monthly patient case presentations of actual clinic patients to providers. |
| Program Maintenance Strategies | • A Research Electronic Data Capture (REDCap) database was created to track prospective data for quality improvement. The database tracks volume of patients with pharmacogenetic tests ordered, volume of patients seen in the consult clinic and associated billing information, pharmacogenetic test results, medication data, pharmacist recommendations made, medication changes in response to recommendations, and CDS alerts fired. • The clinical workbench in Epic is used to monitor functionality of CDS alerts on a monthly basis, including alert frequency and provider’s actions after viewing the alert. • A human-factor interaction approach was used to conduct a usability evaluation to evaluate and enhance the design of pharmacogenetics alerts.19 |
STAKEHOLDER ENGAGEMENT
Nearly all clinical pharmacogenetic implementations include a pharmacist in a leadership or clinical role. Other stakeholders are varied and may include institutional and practice leadership, prescribers, and patients.13,20 Institutional leadership buy-in is necessary to ensure that resources and personnel are sufficiently available to implement and sustain the service. In the majority of institutions, prescribers are ultimately responsible for acting upon pharmacogenetic test results to change pharmacotherapy. Therefore, physicians and other prescribers must support use of pharmacogenetic data in their daily clinical workflow. Having a “physician champion” or strong support of a prescriber from the outset of a new service also will help shape the type of pharmacogenetic interpretation and results provided. If the pharmacy service can ensure that their support is focused on concise, meaningful, and useful information that benefits prescribers, the service will have met an important goal. Early engagement with institutional leadership and prescribers is often one of the most important predictors of long-term success of an implementation.
Patients are the end-users of their pharmacogenetic test results, and results are ultimately their data. As such, patients often act as their own advocates to inform prescribers that they have pharmacogenetic test results available. In many cases, patients can be the drivers who are seeking out pharmacogenetic testing because of their own failed trials with previous medications. Even when prescribers encourage patients to undergo testing, the patient must still be agreeable and willing. The pharmacist can play a key role in educating patients to ensure they understand the benefits and limitations of pharmacogenetics.
EVIDENCE EVALUATION
Evaluating the evidence is an essential step in determining which gene-drug pairs will be included in the implementation. To date, most clinical programs have employed a governance structure for this process that includes one or more committees and/or subcommittees to oversee evidence evaluation. This may resemble a Pharmacy and Therapeutics structure and should include multidisciplinary expertise and include individual(s) trained in pharmacogenetic evidence evaluation and interpretation. In a scoping review of 18 pharmacogenomic programs, Luczak and colleagues reported diversity in committee members, including physicians, pharmacists, genetic counselors, Information Technology (IT) support, and finance/reimbursement specialists.13
Scientific evidence to support clinical implementation of pharmacogenetics is available through online databases (e.g., Pharmacogenomics Knowledgebase [PharmGKB]), guidelines (e.g., Clinical Pharmacogenetics Implementation Consortium [CPIC]), and United States Food and Drug Administration (FDA)-approved labeling information or guidance. When choosing gene-drug pairs the institution must decide on the criteria they believe proves the gene-drug pair is of high-quality evidence and ready for implementation. Resources are readily available to evaluate the evidence (Table 2).21 Institution-specific drug utilization data can help implementers prioritize gene-drug pairs.
Table 2.
Key Resources for Evaluating Evidence of Pharmacogenetic Associations
| Resource | Key information to be gained by the individual |
URL |
|---|---|---|
| PharmGKB | • Review annotated literature (e.g., prescribing information, drug label, international pharmacogenetics (pgx) guidelines, clinical and variants) • Review drug pathway • Download allele definition, allele functionality, frequency, diplotype-phenotype tables |
https://www.pharmgkb.org/ |
| CPIC | • Download CPIC guidelines and supplements • Obtain key updates on guidelines since publication • Clinical decision support resources |
https://cpicpgx.org/ |
| DPWG | • Download DPWG guidelines • Read background on enzymes |
https://www.knmp.nl/patientenzorg/medicatiebewaking/farmacogenetica/pharmacogenetics-1/pharmacogenetics |
| Association for Molecular Pathology with College of American Pathologists | • Review published Guidelines on Genotyping Allele Selection for various pharmacogenes | https://www.amp.org/clinical-practice/practice-guidelines/ |
| PharmVar | • Review variants of genes; nomenclature, evidence level, clinical function |
https://www.pharmvar.org/ |
PharmGKB: Pharmagogenomics Knowledege Base; CPIC: Clinical Pharmacogenetics Implementation Consortium; DPWG: Dutch Pharmacogenetics Working Group; PharmVar: Pharmacogene Variation
Selection of the Gene(s) for Testing
CPIC is an international consortium of pharmacogenetic experts that evaluates pharmacogenetic evidence and develops consensus guidelines to assist clinicians with applying pharmacogenetic test results to prescribing decisions.22 As of late 2021, 26 CPIC guidelines have been published, with gene-drug pairs prioritized for guideline development based largely on the strength of evidence supporting genetic associations with reduced drug effectiveness or poor tolerability and the availability of alternative therapeutic strategies when such an association exists. Guideline development includes rigorous review and grading of the evidence for genetic contributions to drug response. When data exist for multiple genes related to a specific drug or drug class (e.g. cytochrome P450 [CYP]2D6, OPRM1, and COMT for opioids), the writing committee reviews the evidence for each gene, and grades the evidence as weak, moderate, or high. They similarly grade therapeutic recommendations based on genotype results as optional, moderate, or strong. No recommendation is provided when the data are considered to be insufficient to support genotype-guided prescribing.
The CPIC grading system can guide clinicians in choosing which genes to target for testing, with priority given to those with moderate to strong evidence and recommendations. For example, CPIC ranked most of the evidence for OPRM1 and COMT associations with opioid response as weak to moderate whereas much of the evidence with CYP2D6 associations with response to codeine and tramadol was ranked as moderate to high.23 Thus, strong recommendations are provided for avoiding codeine and tramadol in patients with certain CYP2D6 genotypes, whereas no recommendations are provided for other genes. Consistent with CPIC evidence grades and recommendations, pharmacogenetic testing to guide opioid prescribing at major medical institutions is limited to the CYP2D6 gene.4 While other genotypes may be provided by some commercial testing companies, based on CPIC guidance, they should not be used to guide opioid selection until stronger data become available supporting their use. CYP2C19 and CYP2D6 together account for nine of the CPIC guidelines published, and consequently are two of the most common genes implemented in various institutions.
Once the gene(s) is/are decided, a decision on implementing (multiple) single gene test(s) or a panel-based test must be made. Factoring in whether the program wants to support preemptive or reactive testing, along with prescriber preference, may aid this decision.24 Turnaround time of results and its importance for the gene-drug pair being implemented are other factors to consider. For example, for CYP2C19-clopidogrel, it is ideal to have the results returned quickly, which may not be possible if CYP2C19 is a part of a panel.
A thorough review of the resources highlighted above and in Table 2 can inform both the choice of gene-drug pairs and institutional criteria (or internal threshold) for testing. An example of potential institutional criteria for testing is displayed in Table 3. Many health systems have adopted a governing or regulatory body with expertise in pharmacogenetics, pathology, and genomic medicine to provide guidance with these choices initially and over time as new evidence emerges.
Table 3.
Sample Institutional Criteria for Pharmacogenetic Testing
| The following criteria should be met for any gene-drug pair implemented clinically at this institution: |
| 1. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines are available for the gene-drug pair and clinical recommendations align with CPIC guidance; or |
| 2. Pharmacogenetic testing is required or suggested in the FDA-approved drug labeling; or |
| 3. The gene-drug pair is included in the FDA Table of Pharmacogenetic Associations; or |
| 4. Pharmacogenetic testing is recommended in disease-specific treatment guidelines. |
PHARMACOGENETIC TESTING LOGISTICS
Once the decision is made to implement pharmacogenetic testing in practice and the evidence for specific gene-drug pairs have been evaluated, decisions are needed regarding the genotyping process. These decisions include which gene alleles and how to obtain genotype results. Specific resources, including CPIC guidelines22, Association for Molecular Pathology (AMP) recommendations for clinical genotyping allele selection, and the National Institutes of Health Genetic Testing Registry (GTR), can inform these decisions.
Selection of Gene Allele(s) for Testing
To assist clinical laboratories with designing pharmacogenetic testing assays and promote testing standardization across laboratories, the Pharmacogenomic Working Group of the Association for Molecular Pathology (AMP) Clinical Practice Committee defines which variants should, at a minimum, be included on clinical pharmacogenetic genotyping assays.25,26 The writing committee consists of experts in pharmacogenetic clinical testing and research and includes representatives from CPIC, the Dutch Pharmacogenetics Working Group, and the College of American Pathologists. The committee determines which alleles to designate as Tier 1 or “must-test” based on their functional impact, frequency in multiethnic populations, availability of reference materials for establishing clinical assays, and feasibility of variant detection in a clinical laboratory. The working group has published guidelines addressing four genes as of late 2021: CYP2C19, CYP2C9, CYP2D6, and Vitamin K epOxide Reductase Complex subunit 1 (VKORC1), which are among the most commonly tested pharmacogenes in clinical practice.25–28 Recommendations for additional genes are expected. In the event that there are no AMP guidelines available for the gene of interest, other resources to consider in allele selection are CPIC guidelines, PharmGKB, and consultation with experts at other institutions who are implementing testing into practice.
When testing for variants within a gene that influence drug response, and none are detected, patients are reported to have a normal genotype. For example, if a laboratory tests only for the CYP2C9*2 and *3 alleles, which are associated with reduced to absent enzyme function, and neither are detected, then the patient is reported to have a the *1/*1 genotype and the normal metabolizer phenotype. In other words, the *1 allele is reported by default when no variant allele is detected. This does not rule out the presence of other variants not captured on the testing platform yet with important effects on protein function or gene expression. In fact, while testing for the CYP2C9*2 and *3 alleles will detect the majority of variants associated with reduced or absence enzyme activity in European and Asian populations, it detects a small portion of important variants present in African ancestry patients. Specifically, approximately 6% of African ancestry patients have a *2 or *3 allele, while approximately 18% have a *5, *6, *8, or *11 allele, which also result in reduced CYP2C9 activity.29 In the case of genotype-guided warfarin dosing, failure to account for these other alleles can lead to significant overestimation of warfarin dose requirements and supratherapeutic anticoagulation in those of African ancestry.30,31 This led to CPIC recommendations to avoid dosing warfarin based on genotype unless the *5, *6, *8, and *11 alleles are tested for and illustrates the importance of carefully evaluating alleles to include on genotyping assays.29 If using an outside laboratory rather than designing an assay to be used in-house, it is critical to ensure that the laboratory is testing for all variants impacting drug response that occur at an appreciable frequency across ancestry groups.
Selection of Laboratory
Luczak and colleagues reported that 15 of the 18 programs assessed in their review used an in-house laboratory for pharmacogenetic testing.13 Notably, the majority of programs included in this review had an academic affiliation. The percentage of programs with technical expertise and genomic testing capability to support in-house testing may differ from this in a real-world setting. Barriers to in-house testing may include the cost of obtaining genotyping equipment and/or setting up and validating testing assays and the lack of certified personnel to conduct testing. The voluntary Genetic Testing Registry can serve as a resource to identify laboratories that offer testing, recognizing that given its voluntary nature, it may not be all inclusive. When assessing commercial laboratories for pharmacogenetic testing, there are several important factors to consider, including which alleles are tested as discussed. Other factors have been extensively reviewed elsewhere32 and are summarized in Table 4.
Table 4.
Factors to consider when selecting a reference laboratory for pharmacogenetic testing32
| Factor | Questions to consider |
|---|---|
| Laboratory certification | Can results be used clinically? In the U.S., this requires that the laboratory be College of American Pathologists (CAP) accredited and Clinical Laboratory Improvement Amendments (CLIA) certified. |
| Alleles tested | Does the laboratory test for the minimum set of alleles (i.e. Tier 1 alleles) recommended by the Association for Molecular Pathology (AMP) Pharmacogenomic Working Group? If no AMP recommendations are available for the gene of interest, are the alleles described in CPIC guidelines included? Are there other alleles that affect drug response and occur at an appreciable frequency across ancestry groups? What alleles are other sites testing for clinically? |
| Financial factors | Is any financial assistance with the cost of testing available for patients from the company? |
| Sample collection | What kind of sample is required (e.g. blood, buccal cell, saliva)? Is the sample type feasible to collect at your site (e.g. is phlebotomy easily accessible for patients if blood is required)? Does the company provide sample collection materials or do they need to be supplied internally? |
| Test turnaround time | Will the time needed to get test results returned fit the clinical workflow (e.g. will results be available in time to inform prescribing decisions)? |
| Return of results | Can results be integrated into the electronic health record as discrete data to allow for automated clinical decision support? If results are returned as Portable Document Format (PDF) reports, how will these be stored in the electronic health record so that they are accessible to clinicians? |
| Interpretation of results | Does the laboratory provide any interpretation of the results (e.g. assign phenotype based on genotype results)? If so, are interpretations consistent with CPIC guidelines? |
Test Consent Consideration
A sometimes-debated topic is whether specific patient consent is necessary prior to pharmacogenetic testing for clinical purposes, outside of a research study. In many institutions, pharmacogenetic testing is treated in the same vein as a routine laboratory test, with consent covered under the general consent for treatment provided by the patient.33 This may differ from testing for genetic risk for disease, in which positive test results could have a significant psychological impact on the patient and his or her family. In this case, patient consent may be necessary to ensure that potential risks have been disclosed to the patient and that the patient is willing to go forward with testing. The consequences of pharmacogenetic testing are usually less concerning as alternative dosing or therapy is generally available in the event that the patient is genetically predisposed to having a poor response to adverse effects from standard therapy. Nonetheless, data show that patients expect to be involved in decisions about pharmacogenetic testing, and thus, obtaining verbal consent at a minimum would seem warranted under most circumstances.33,34 Some institutions, such as programs at the University of Colorado obtain patient consent to release test results into the patient portal.35
ELECTRONIC HEALTH RECORD INTEGRATION
The integration of pharmacogenetic test results into the local EHR is an essential step in implementing pharmacogenetics clinically.36,37 Successful EHR integration can be defined as the ability to order the pharmacogenetic test electronically, return of test results as discrete variables, and availability of clear and concise result interpretation. Discrete variables allow pharmacogenetic clinical decision support (CDS) to operate seamlessly and logically. Nearly all published descriptions of clinical pharmacogenetic implementations report inclusion of CDS components ranging from active or passive clinical decision alerts to clinical consultation notes delivered via email or EHR inbasket.13
Pharmacogenetics Laboratory Order Within the EHR System
The ability for prescribers to order a pharmacogenetic laboratory test within their EHR system is the first important component of EHR integration. Ideally, ordering a pharmacogenetic laboratory test should be integrated seamlessly to allow prescribers to incorporate pharmacogenetic information into patient care. However, this process can be particularly confusing in clinical practice and can have negative ramifications (e.g., wrong or repeat test ordered) for prescribers and/or patients. Specifically, incorrect pharmacogenetic test orders can potentially delay the care process or cause unnecessary patient costs. Therefore, the EHR system should clearly communicate to prescribers how to order pharmacogenetic laboratory tests and provide instructions when possible on use of internal or external laboratories, single-gene testing versus panel-based testing, and other considerations. CDS tools, including alerts and order sets are common approaches that healthcare systems use to assist prescribers with these choices.38 For example, at University of Alabama at Birmingham the CYP2C19 order option appears in the post-percutaneous coronary intervention (PCI) order set.39
Furthermore, the ability for prescribers to order external labs within the EHR is crucial for institutions that do not support internal pharmacogenetics labs. Available surveys on prescribers’ perception with pharmacogenetics indicate that if external pharmacogenetic labs are not available for prescribers to order pharmacogenomic testing within the EHR, prescribers are less likely to adopt pharmacogenetics clinically.40,41 Additionally, this functionality significantly affects the institution’s ability to integrate pharmacogenetic test results into the continuity of care for patients, especially for result interpretation and CDS development.38
Discrete pharmacogenetic test results, defined as health data stored at the lowest level of granularity, are an essential component of EHR integration. Discrete pharmacogenetic test results, usually in the form of genotypes and/or phenotypes, allow data to be measurable and reportable within the EHR system. As a result, pharmacogenetic test results can be interpreted, displayed in the patient profile, utilized for CDS, or incorporated with active medications for appropriate phenoconversion interpretations.42 Phenoconversion occurs when a drug inhibits the CYP450 enzyme and results in a different phenotype, e.g., a patient who is a CYP2D6 Normal Metabolizer taking bupropion will phenoconvert to a CYP2D6 Poor Metabolizer. For external lab results, the process of aligning lab result components using common standards (such as HL7) are essential to generate discrete results. Non-discrete results (e.g., scanned Portable Document Format [PDF] file) can create a burden for providers to locate and extract information since they are not stored in a standard location.42 Some EHRs do allow for free text entry of discrete variables, but this is susceptible to human error. Having laboratory results entered as discrete variables allows for reinterpretation or revision of the genotype-based phenotype if new evidence emerges, which would be challenging with non-discrete results.
Term Standardization for Communicating Pharmacogenetic Test Results
Variable terminology has been and is still used to report pharmacogenetic test results among various laboratories, pathology reports, and in the EHR, which can cause significant confusion43 among laboratory personnel, researchers, and clinicians, and may hinder portability of patients’ genetic test results. For example, at UF Health, the internal pathology laboratory reports CYP2C19 *1/*1 as a “normal metabolizer,” and a CYP2C19 *1/*2 result as an “intermediate metabolizer.” In contrast, one external laboratory resulting CYP2C19 tests directly into the EHR reported a CYP2C19 *1/*1 as “negative,” and a CYP2C19 *1/*2 as only “*2.” This led to inconsistencies in the EHR and the CDS alerts firing within this system, since Best Practice Advisory alerts in Epic were built to fire off of diplotype. In this case pharmacists worked directly with internal systems and the external laboratory to standardize reporting.
To avoid confusion, it is recommended that implementers adopt the standardized terminology for pharmacogenetic allele function and inferred phenotypes proposed by CPIC and adopted in CPIC guidelines43 in pathology laboratory reporting, EHR displays of test results, and CDS rules whenever possible. In addition to CPIC guidance, an HL7 standard is available to reporting of these values. Use of established standardized terminology among laboratories and researchers and within the EHR can help decrease confusion and support successful implementation and adoption of pharmacogenetic testing. Additionally, it supports sharing of pharmacogenetic data among diverse healthcare systems that may have varying levels of knowledge and/or experience with pharmacogenetic test reporting and application.
Pharmacogenetic Test Result Interpretation
The EHR is an optimal tool to assist prescribers with interpreting pharmacogenetic test results because it can display phenotype results in multiple locations and workflows. For example, the patient profile can house the individuals’ pharmacogenetic test results (genotype and phenotype), and CDS alerts can appear at various stages of the prescriber’s workflow (Figure 1). When possible, all genomic information (somatic and germline) should be available within the same location to improve access. Within Epic’s Genomic Module, a genomics profile can be created to store and display all genomic information for the provider and/or the patient (Figure 2). This Genomic Module is relatively new, but several institutions have already implemented it with pharmacogenetic data.42,44,45 Due to the historical lack of a standard repository for pharmacogenetic data, many implementation sites have used workaround solutions to display test results, such as the problem list field as with Boston Children’s Hospital or the allergy list field.46 However, this is not an ideal solution since laboratory results are not easily found and have sometimes been misplaced in random locations. Linking pharmacogenetic test results directly with an associated medication is challenging in the EHR. While Epic does not currently have this functionality, it is possible in Cerner through use of in-house custom web-based applications or by manually pulling pharmacogenetic test results into the drug order field or a laboratory results field.47
Figure 1.
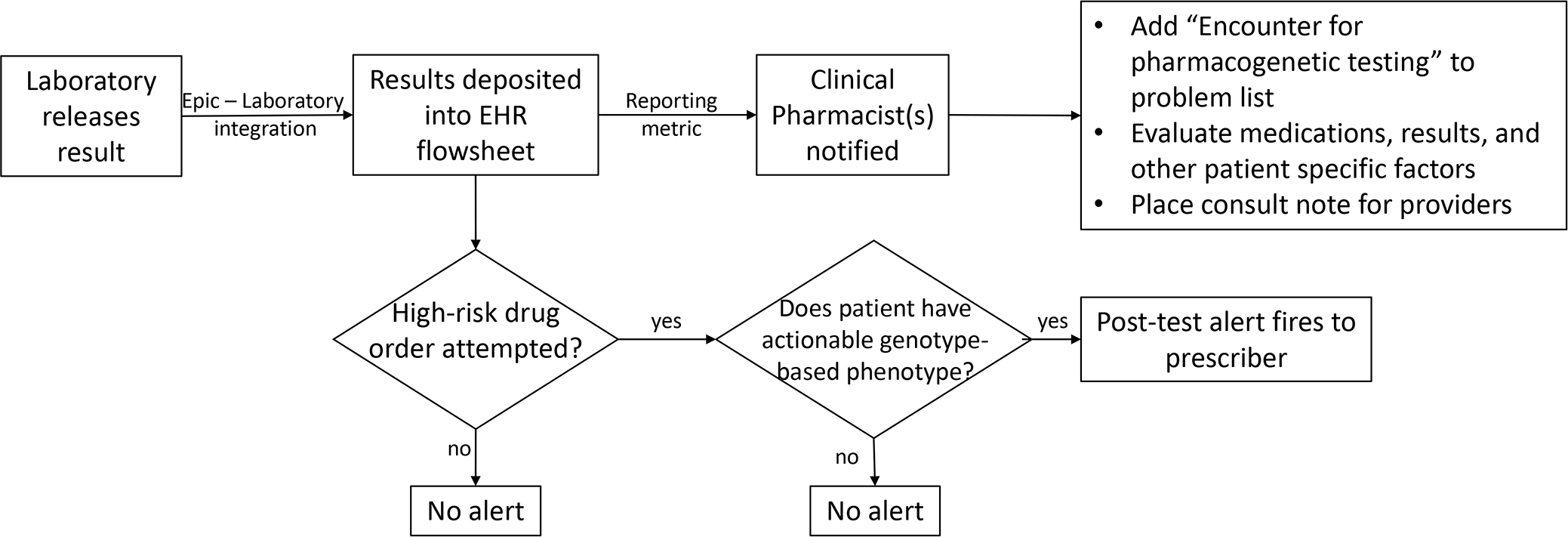
Interpretation of Pharmacogenetic Test Result in the Electronic Health Record The electronic health record (EHR) is an optimal tool to assist prescribers in interpreting the pharmacogenetic test result. If results are deposited as discrete results, clinical decision support alerts can fire at appropriate times.
This figure was adapted from Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol. 2015;55:89–106.
Figure 2.
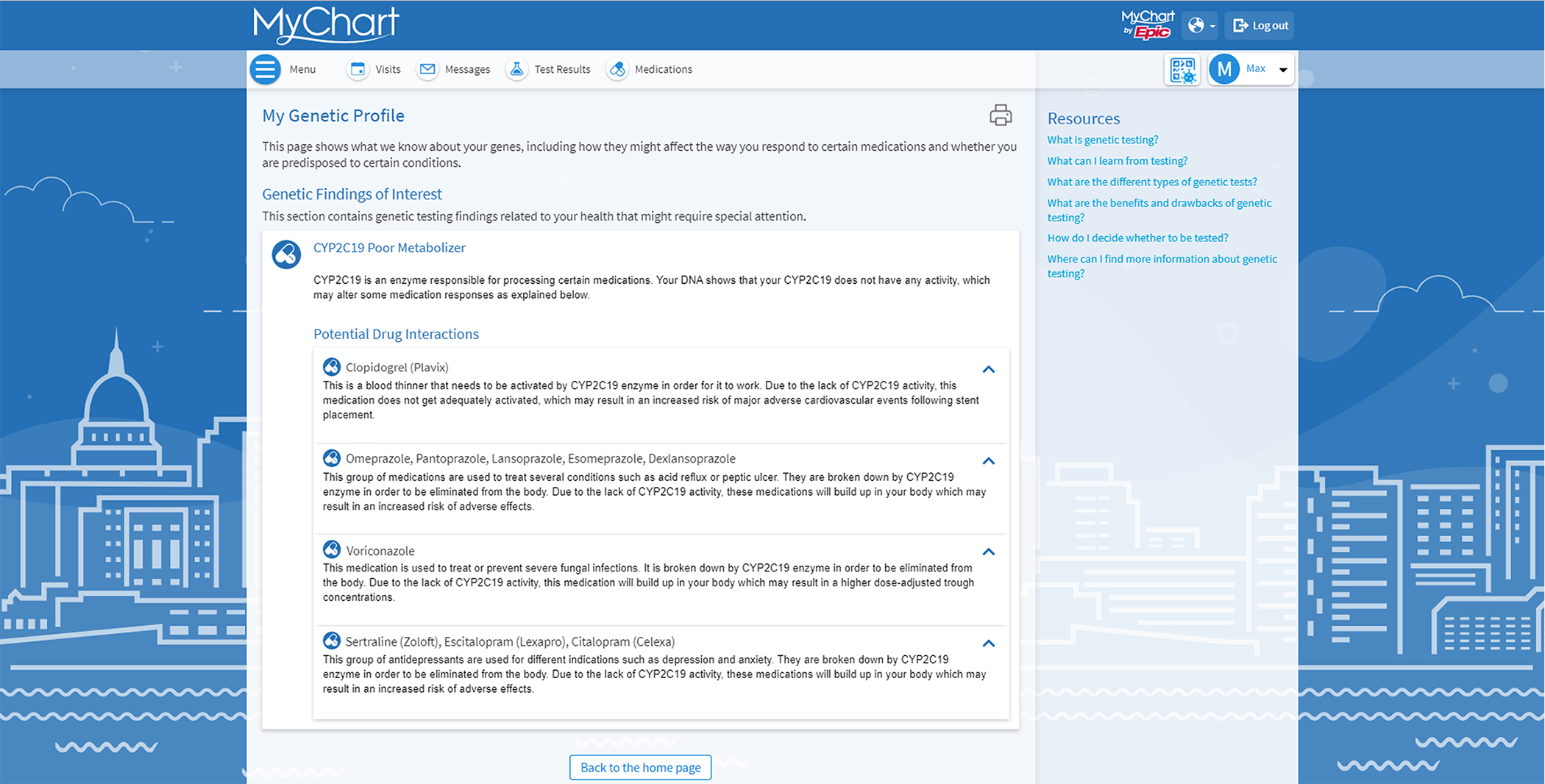
Patient Facing Genetic Profile Within EPIC’s Genomic Module, a genomics profile can be created to display all genomic information for the provider and/or the patient. Above is an example of what the profile looks like in the patient view.
Clinical Decision Support
CDS, arguably, is the best available option to assist prescribers in utilizing pharmacogenetic test results or in prompting prescribers to order a test within the normal patient care process.42 If provided at the appropriate time, CDS alerts prescribers that pharmacogenetic test results are available and provides guidance on their use. Alerts may also appear as “pre-test” and notify the prescriber that results are not available but are recommended to have before prescribing a drug (e.g., TPMT-thiopurines) Detailed tutorials on creating internal CDS or utilizing an external platform for CDS exist.42,48,49 CDS alerts can be interruptive and active (e.g., pop-up alerts that require action) or passive alerts that appear in the background (e.g., informational, require no action, Figure 3). Some institutions, including Duke University, rely on pharmacist’s consultation notes placed manually in the EHR.13 This method has been referred to as a form of “static” CDS.
Figure 3.
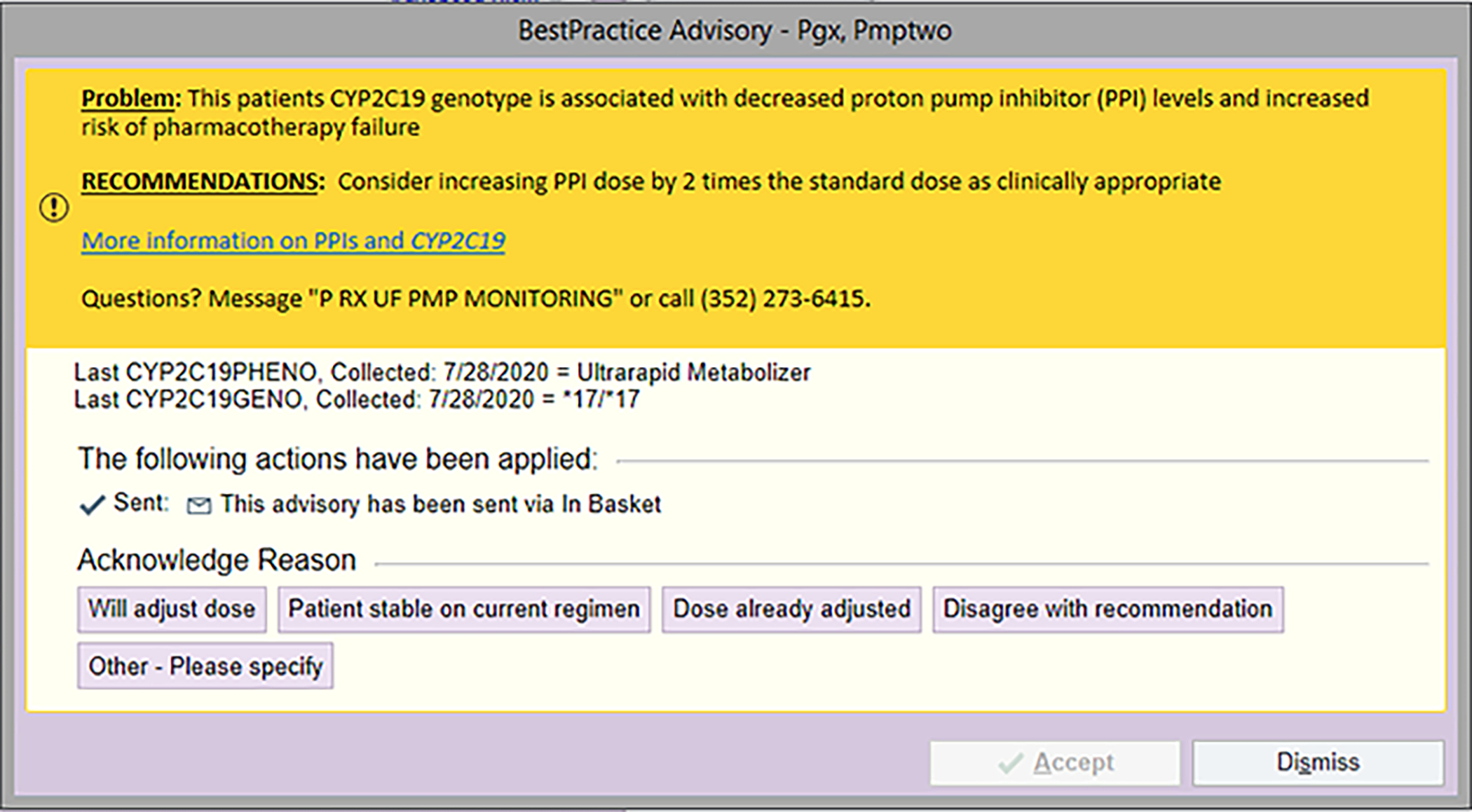
Sample Health Best Practice Advisory This is an example of an alert that fires when certain proton pump inhibitors are ordered (e.g., omeprazole) and the individual is a CYP2C19 Ultrarapid Metabolizer.
This figure is used with permission from Epic.’
REIMBURSEMENT
When establishing a new clinical pharmacogenetics service, consideration should be given to reimbursement for the test itself and reimbursement for the pharmacist’s time, as with any other pharmacist-led service.
Reimbursement for the Pharmacogenetic Test
Reimbursement for pharmacogenetic testing is determined by each payer. Individual payer decisions are generally based on an internal determination of whether a pharmacogenetic test changes the clinical management and/or the outcome of the patient. Coverage policies vary among payers and may need to be reviewed on a case-by-case basis. 58 Although reimbursement for pharmacogenetic testing is inconsistent, the landscape for reimbursement is becoming more favorable.50 In 2012, the American Medical Association created current procedural terminology (CPT) codes to support documentation and reimbursement of pharmacogenetic testing (Table 5). In recent years, the Center for Medicare and Medicaid Services (CMS) has issued policies on reimbursement for pharmacogenetic testing through Medicare Administrative Contractors (MACs), which can pave the way for policy changes with other insurers. In June 2020, Palmetto GBA, issued a final local coverage determination (LCD) for pharmacogenetic testing and ten of the twelve MACs in the US have adopted similar policies for coverage of pharmacogenetic testing to date (Figure 4). 59 In some cases, private payers such as United HealthCare have adopted transparent coverage policies for pharmacogenetic testing. 60 Pharmacists within our outside of the program can verify coverage of pharmacogenetic testing with patients’ payers and/or facilitate prior authorization prior to submitting a test for reimbursement. For example, Cleveland Clinic Health System partners with an external laboratory to provide prior authorization services.13
Table 5.
Current Procedural Terminology (CPT) Codes for Pharmacogenomic Testing for Common Gene-Drug Pairs
| Gene/Test | CPT Code | Intended use for Drug |
|---|---|---|
| CYP2C19 | 81225 | Clopidogrel, voriconazole, proton pump inhibitors, selected selective serotonin reuptake inhibitors (SSRIs) |
| CYP2C9 | 81227 | Phenytoin, warfarin, selected non-steroidal anti-inflammatory drugs |
| CYP4F2 | 81479 | Warfarin |
| VKORC1 | 81355 | Warfarin |
| CYP2D6 | 81226, 0070U, 0071U, 0072U, 0073U, 0074U, 0075U, 0076U | Atomoxetine, codeine, ondansetron, tropisetron, tamoxifen, selected SSRIs and tricyclic antidepressants |
| DPYD | 81232 | Fluoropyrimidines |
| G6PD | 81247 | Rasburicase |
| SLCO1B1 | 81328 | Simvastatin |
| NUDT15 | 81306 | Thiopurines |
| TPMT | 81335 | Thiopurines |
| UGT1A1 | 81350 | Atazanavir |
The complete list of CPT codes for pharmacogenomic testing is available at www.cms.gov.
Cytochrome P450 (CYP)
Vitamin K Epoxide Reductase Complex (VKORC)
Dihydropyrimidine dehydrogenase (DPYD)
glucose-6-phosphate-dehydrogenase (G6PD)
Solute carrier organic anion transporter (SLCO)
Nudix hydrolase (NUDT)
Thiopurine methyltransferase (TPMT)
UDP-glucuronosyltransferase (UGT)
Figure 4:
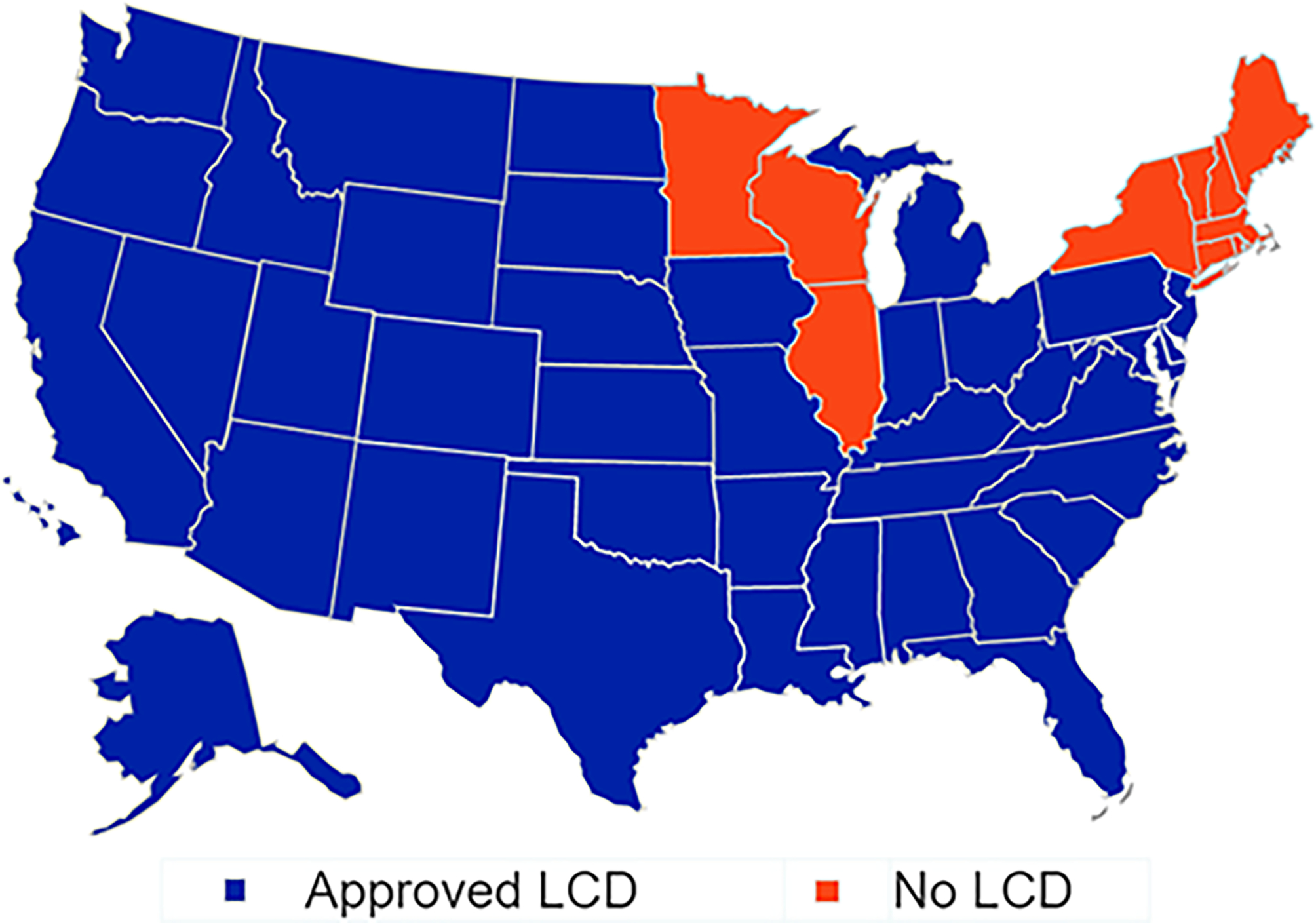
Pharmacogenetic Testing LCD Approval Status by State Local coverage determination (LCD) for pharmacogenetic testing and ten of the twelve MACs in the US have adopted similar policies for coverage of pharmacogenetic testing to date.
While third-party payers may cover all or part of the cost of pharmacogenetic tests, it is possible that any given provider may deny coverage altogether or cover only a portion of the cost so that the patient receives a bill to cover the total or remaining cost out-of-pocket. In addition, third party payers may not cover the cost of testing ordered preemptively in a patient without an immediate need for drug therapy. Given this, institutions may prefer to obtain written consent from the patient to acknowledge that there may be some out-of-pocket cost incurred with testing or, in the case of pre-emptive testing, that the patient is expected to cover the cost in full.
Reimbursement for Pharmacists’ Time
In addition to the cost of pharmacogenetic testing, reimbursement for clinicians’ time to interpret and apply test results, communicate with providers, and educate patients is a critical element of sustainability, as with any pharmacist-led service. It may be helpful to integrate clinical pharmacogenetics services into the existing billing structure for pharmacist-led comprehensive medication management services. However, additional challenges exist with regard to reimbursement for pharmacogenetic testing services. First, fewer patients are eligible for pharmacogenetic testing to guide drug therapy changes as compared with other pharmacist-led services such as anticoagulation management, which can impact financial sustainability. In addition, the results are lifetime results and as such majority of patients are not seen for repeat visits, thus to have a sustainable clinic there needs to a continual flow of new patients to test. Second, patient and provider education for pharmacogenetics may be more time consuming than for other areas of practice, which places an additional time and resource burden on pharmacists conducting clinical pharmacogenetics services. When possible, streamlining patient and provider education through print materials and/or other efforts (e.g., patient education videos) is recommended to maximize the pharmacist’s time.51
Established programs report using a variety of methods to pursue compensation for the pharmacist’s time in the outpatient setting, including use of a medication therapy management model.52 Increasingly, pharmacists are exploring established strategies for “incident-to” billing of the pharmacist’s time under a physician’s supervision, as with UF Health and Cleveland Clinic.13,53 A thorough review of pharmacist incident-to billing has been published and may be a useful guide for those exploring this option.53 In some cases, a collaborative practice agreement has been utilized between a pharmacist and a physician to facilitate clinical decision making and reimbursement, as with St. Jude Children’s Research Hospital.13 Other strategies include partnering with a billable provider, such as a genetic counselor, to deliver test results to patients, as employed by the NorthShore University Health System.54
EDUCATION
Education is a key intervention to increase clinician knowledge, skills, and confidence, which are essential for adoption and sustainability of a pharmacogenetics service.55,56 Education around the frequency of clinically-actionable phenotypes, prevalence of clinically actionable pharmacogenetic medications, and risks associated with prescribing medications incongruent with phenotype will increase prescriber’s awareness of genotype-informed prescribing and, in turn, may motivate them to adopt pharmacogenetics clinically. It is also important to review elements of an institution’s specific program and clinical workflow for pharmacogenetic testing, including who providers should contact if additional help is needed and what support is available, including CDS alert logistics.38
Education can be provided in multiple formats and usually encompasses small and large group settings (e.g., grand rounds), continuing education, and supplemental written materials.13 In the authors’ experience, recurring case-based educational events centered on actual clinical experiences within the institution have been key to changing prescribers’ behavior.
PROGRAM MAINTENANCE
Tracking Quality Improvement Metrics
As an emerging clinical science, pharmacogenetics is ideal for a learning health system approach, or one in which an institution commits to learning and continuously applying new insights for improvement purposes. Identifying and tracking clinical metrics of a new pharmacogenetics services serves multiple purposes, including but not limited to: ensuring CDS alerts are working properly, tracking program growth and sustainability to support personnel effort, identifying ordering trends in the health system that highlight success and may identify educational needs, and providing opportunities for dissemination and education on clinical implementation. When possible, tracking metrics should be initiated at program launch to prospectively measure the success of the implementation.13
It is common across multiple sites to track implementation outcomes (e.g., acceptability, adoption, costs, feasibility), service outcomes (e.g., effectiveness, safety, timeliness), and patient outcomes (e.g., symptomatology, satisfaction).57 Established programs have reported outcomes including patient and/or consult volume, referral information (e.g., department), turnaround time, number of actionable genotypes, interventions and acceptance rates, visit length, provider and patient satisfaction, costs, impacted medications, among others.13
In addition to collecting prospective data, retrospective data can be collected by utilizing various reporting functions within the medical record. For example, it is important to assess that CDS alerts continue to fire as expected, particularly after updates to the EHR, which may disrupt alert functionality. Tracking the alert frequency on a monthly basis allows us to closely monitor alert firing rates to detect a systemwide error. Specifically, within Epic the clinical workbench allows for collection of multiple CDS data elements such as alert names, alert date/time, locations, user names and specialties, patient names, encounter date, provider actions, and override reasons. Other EHRs should have similar reporting mechanisms.
Reinterpretation of Pharmacogenetic Test Results
Ongoing monitoring of emerging evidence will help to identify any changes needed in the genes and/or alleles tested for any given drug or drug class and should be an ongoing part of program maintenance. For example, FDA-cleared tests for CYP2C9 historically limited genotyping to the *2 and *3 alleles. However, the emergence of evidence on the importance of additional CYP2C9 alleles (e.g. *5, *6, *8, *11) in those of African ancestry prompted updates to CPIC guidelines and recent guidance from AMP recommending incorporating these additional alleles on clinical testing platforms.58 As another example, CPIC guidelines for genotype-informed thiopurine use were updated in 2018 to add the NUDT15 genotype in light of new data demonstrating the significant effect of this genotype on thiopurine.59 This revision required clinical programs to change laboratory testing procedures, clinical recommendations, CDS alerts, and provider education. The CPIC website serves as a resource for assessing for the need for changes in pharmacogenetic testing. CPIC continuously monitors emerging evidence and posts changes such as these to their website in between guideline updates. Institutions can benefit from seeking ongoing patient and provider feedback when revising CDS and educational strategies in light of new evidence. It is also useful to get patient’s perspective and feedback on materials developed to help improve the content.60
PUTTING IT ALL TOGETHER: ESTABLISHING A CLINICAL WORKFLOW FOR PHARMACOGENETIC SERVICES
Once strategies for each of the individual elements discussed above are determined, the clinical workflow should be defined. Ideally, EHR integration, pharmacist support, and patient care processes should be efficient and seamlessly integrated into the existing clinical workflow. In our experience, 4 elements should be addressed in this workflow to support a successful implementation: 1) patient identification for testing; 2) pharmacogenetic test ordering; 3) interpretation and application of test results (e.g., drug therapy changes); and 4) patient education (Figure 5).16 Factors that influence the optimal choice at each decision-making point are institution-specific and may be affected by the targeted patient population (e.g., specialty vs. primary care), patient status at the time of testing (e.g., inpatient vs. outpatient), practitioners involved in workflow and their respective role(s), and reimbursement avenues.
Figure 5.
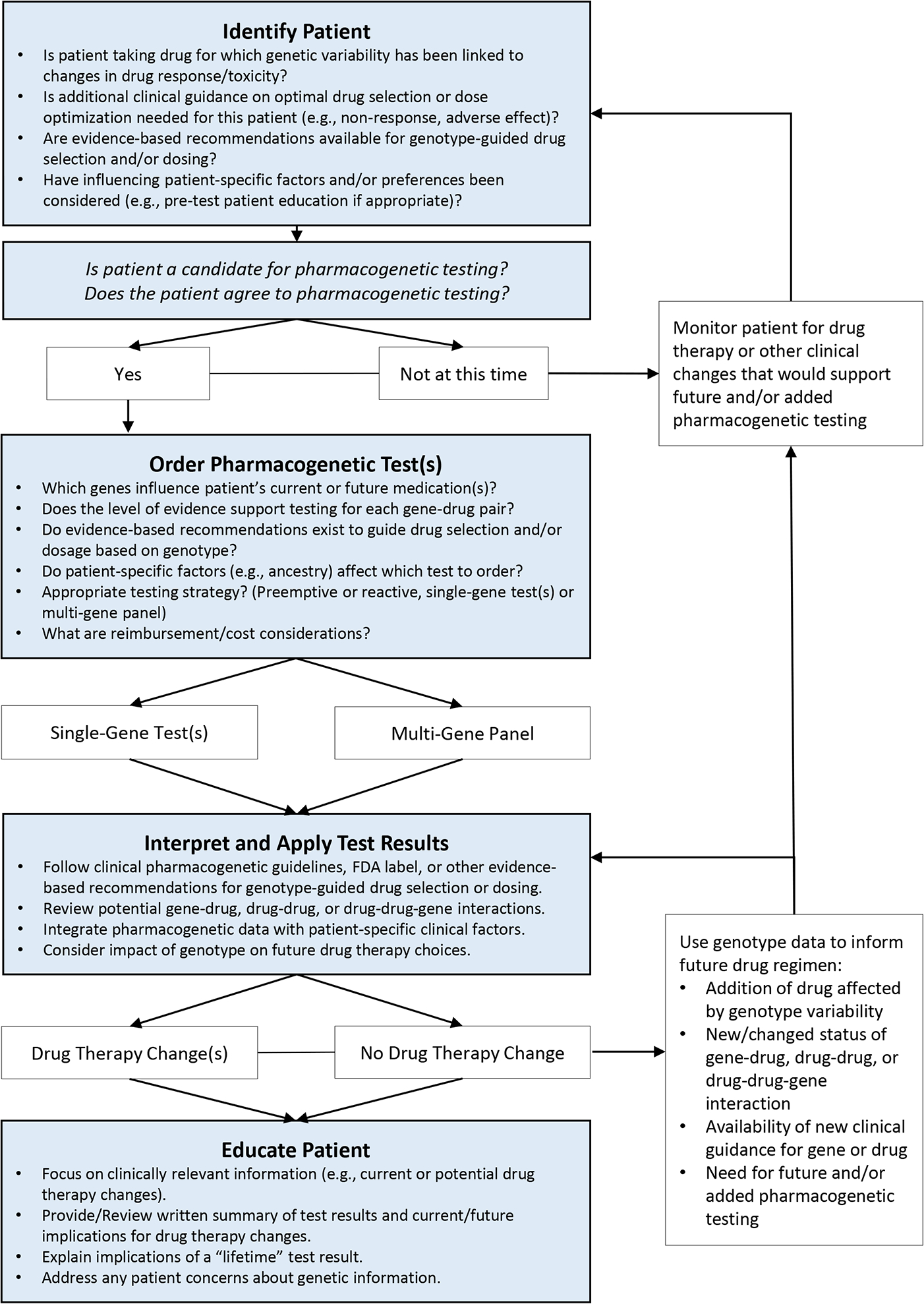
Clinical Decision Making Process for Pharmacogenetic Testing The clinical workflow should ideally integrate 4 elements to support a successful implementation: 1) patient identification; 2) pharmacogenetic test ordering; 3) interpretation and application of test results (e.g., drug therapy changes); and 4) patient education
This figure has been reprinted with permission from Weitzel KW, Duong BQ, Arwood MJ, Owusu-Obeng A, Abul-Husn NS, Bernhardt BA, Decker B, Denny JC, Dietrich E, Gums J, Madden EB, Pollin TI, Wu RR, Haga SB, Horowitz CR. A stepwise approach to implementing pharmacogenetic testing in the primary care setting. Pharmacogenomics. 2019 Oct;20(15):1103–1112.
Patient Identification
Patients who may benefit from pharmacogenetic testing are most often identified by a pharmacist, provider, or by the patients themselves. Factors such as patient status (inpatient vs. outpatient), medication use, and practitioner role(s) are important. In most cases, a test ordered for an outpatient will be reimbursed through the patient’s major medical insurance provider, while a test ordered for an inpatient will be covered through the health-system’s Diagnosis Related Group (DRG) scheme, which may be a key factor in long-term sustainability and require institutional buy-in. Optimal patient status for pharmacogenetic testing is also influenced by test turnaround time. In many cases, an inpatient will be discharged by the time the pharmacogenetic test result is received. In this case, a consult note can still be placed and routed to the primary care prescriber to follow up on. Medication use and payer mix characteristics in the targeted patient population are important, and ideally, should be analyzed in aggregate prior to starting a new service. Understanding which drugs are more commonly used, prevalence of diagnostic codes, and the type of payer(s) that will be billed for testing are essential. Since pharmacogenetic testing is relevant for defined medications, long term sustainability is more likely if enough patients are eligible to build momentum for the service. Finally, the method for patient identification and resources to support this are also important. In a very busy practice, for example, a regular patient screening process led by a pharmacist may be needed to ensure an adequate number of eligible patients are offered pharmacogenetic testing to maintain a sustainable service.
Pharmacogenetic Test Ordering
Once a patient is identified, the pharmacogenetic test will need to be ordered by an ordering provider. Again, with a busy clinical practice, a pharmacist may need to order the test. Alternatively, if the pharmacist does not have the authority to order a pharmacogenetic test, it may need to be done by the provider, in which case the process should be aligned with provider preferences and workflow. In some cases, an alternative provider such as a geneticist or pathologist may be established as the ordering provider for all pharmacogenetic tests. This can streamline workflow for pharmacists if they are not designated as ordering providers within their institution. Many commercial laboratories have providers on staff to serve as the ordering provider, which may also be an avenue if pharmacists are not authorized to order the test.
Interpreting and Applying Test Results
In a pharmacist-led clinical pharmacogenetics service, this step often mimics existing workflows for comprehensive medication management or clinical pharmacist services within an institution. Pharmacogenetics services are increasingly being integrated into comprehensive medication management because of commonalities with targeted diseases (e.g., depression, stroke, lipids) and the prevalence of polypharmacy. In these cases, recommendations for drug therapy changes based on pharmacogenetic test results can be approached in the same manner as existing pharmacist-led services for chronic disease management (e.g., diabetes). This strategy usually involves a pharmacist physically embedded in the clinical space for the patient visit, consultation with the provider for drug therapy recommendations, followed by patient education. Using this established approach may also be helpful from a reimbursement perspective if the pharmacogenetics service can be integrated into existing procedures to bill for pharmacists’ services within the institution.
In the outpatient setting, these services are often structured as a “one-visit” or “two-visit” model (Figure 6). In a single visit model, the pharmacogenetic test is ordered, most often by the prescriber at a routine clinic visit, and drug therapy changes are implemented in a follow-up visit with the pharmacist, along with patient education. In a two-visit model, an initial visit is conducted by the pharmacist to gather a comprehensive medication history and determine the appropriateness of testing. The second visit is focused on drug therapy changes and patient education. Whatever model is used, reimbursement of the pharmacist’s time should be considered. A single-visit model may be associated with increased efficiency and sustainability overall.54,61
Figure 6.
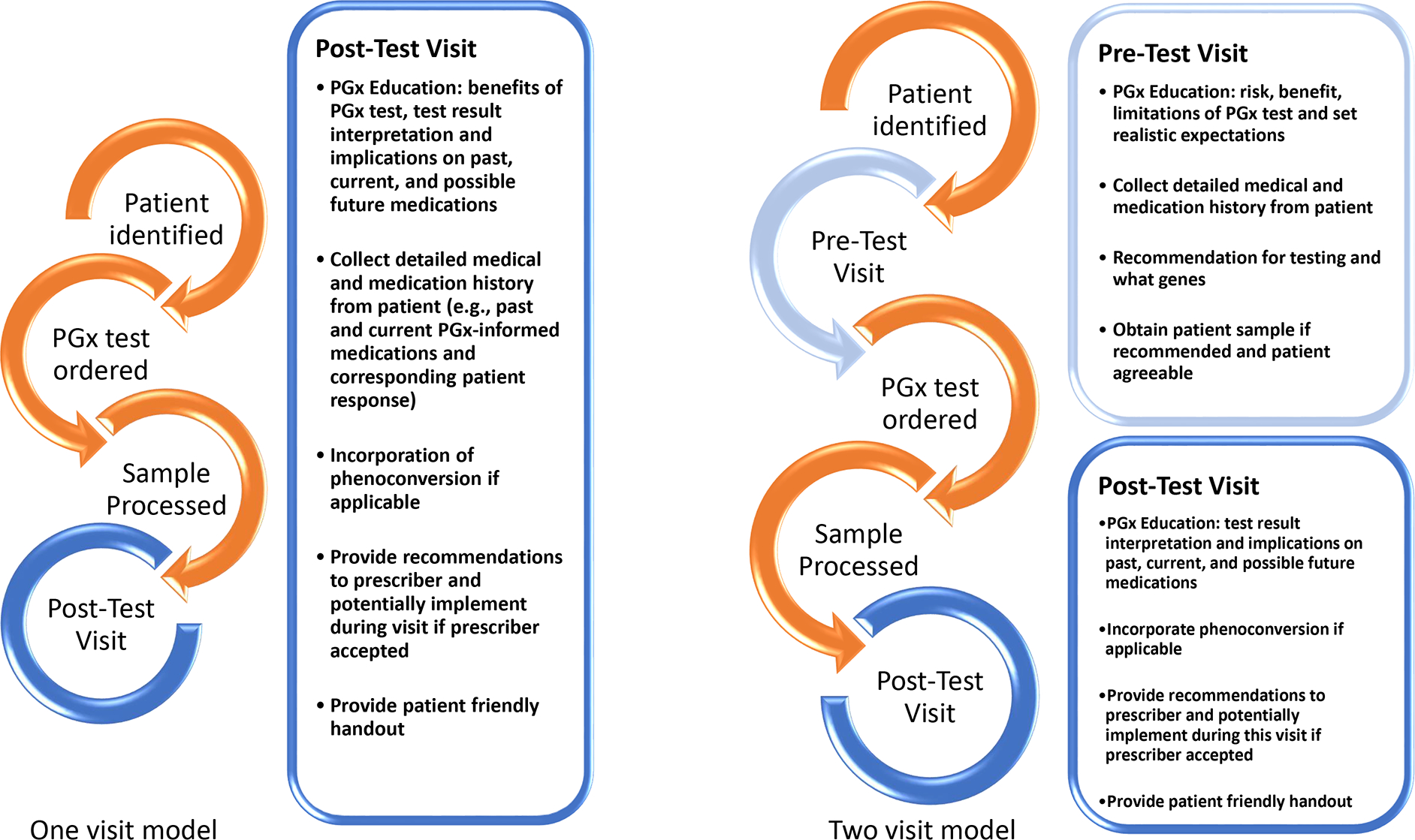
One- and Two-Visit Models for Pharmacogenomic Testing A pharmacogenetics consult clinic may follow a one- or two-visit model depending on the setting. Components of what makes up each visit are described in the figure above.
Alternatively, pharmacogenetic tests are ordered by the provider in the routine process of care with a pharmacogenetics-specialized pharmacist “covering” any test ordered within an institution. In this model, the clinical interpretation of the test results and drug therapy recommendations do not usually involve a face-to-face pharmacist visit. Instead, recommendations for drug therapy changes are communicated to prescribers via a clinical note placed remotely in the EHR.62 This approach is most successful when there is an existing mechanism to reimburse the pharmacist’s time for a clinical consult within an institution. It is unlikely that the pharmacist’s time can or will be reimbursed from the patient’s major medical service if no face-to-face visit takes place, even if the patient was seen as an outpatient at the time of the test order.
PATIENT EDUCATION
In most pharmacist-led pharmacogenetics services, patient education is conducted by the pharmacist. Other approaches have included joint education with pharmacist and a genetic counselor or geneticist, or education by the ordering provider. In some institutions, single-gene provider-initiated pharmacogenetic tests are considered part of the normal process of clinical care and patient education takes place as a part of the standard clinical workflow, as with any other non-genetic laboratory tests (e.g., serum creatinine). Whatever the mechanism, reimbursement for the pharmacist’s time should be considered when planning the workflow as with interpreting and applying test results. In addition, patient education should include guidance on the lifetime nature of test results and instruction to patients on making other health care providers aware of them in the future to avoid unnecessary duplicate testing.
Any of the above described pharmacist interactions may also be delivered via telehealth or “telepharmacy,” which adds another layer of complexity to reimbursement for pharmacists’ time. In general, Medicare does not currently reimburse pharmacists for telehealth because they are not recognized as prescribers. Instead, reimbursement for telehealth services may be accomplished through bundled payments or grants/contract services. Of note, when delivering services via telepharmacy, pharmacists must comply with the policies and regulations of the state in which the patient resides. Pharmacists should consult with their state board of pharmacy and the board of pharmacy for the state in which the patient resides, if different, before providing telehealth services.
CONCLUSION
Initiating a pharmacogenetics service is a complex undertaking with many potential barriers to overcome. Fortunately, many early adopters of clinical pharmacogenetics have documented their experiences and are helping to pave the way for others. By learning from and relying on published experiences and strategies from early adopter institutions, pharmacists implementing new clinical pharmacogenetics services can avoid common pitfalls and increase their likelihood of developing a successful and sustainable implementation.
Acknowledgments:
Supported by the National Human Genome Research Institute of the National Institutes of Health (NIH) under award number U01HG007269. Additional support was provided by the NIH National Institute of General Medical Sciences and the National Heart, Lung, and Blood Institute under award numbers NIH U01GM074492 and U01HL105198, respectively, as part of the Pharmacogenomics Research Network, and by institutional support from the University of Florida Clinical Translational Science Institute, which is supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflict of interest: All authors have no relevant conflicts of interest.
REFERENCES
- 1.Topić E 5. Pharmacogenomics and Personalized Medicine. EJIFCC. 2008;19(1):31–41. [PMC free article] [PubMed] [Google Scholar]
- 2.Owusu Obeng A, Fei K, Levy KD, Elsey AR, Pollin TI, Ramirez AH, Weitzel KW, Horowitz CR Physician-Reported Benefits and Barriers to Clinical Implementation of Genomic Medicine: A Multi-Site IGNITE-Network Survey. J Pers Med. 2018;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavallari LH, Lee CR, Beitelshees AL, et al. Multisite Investigation of Outcomes With Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. JACC Cardiovascular interventions. 2018;11(2):181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavallari LH, Van Driest SL, Prows CA, et al. Multi-site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genet Med. 2019;21(10):2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenblat JD, Lee Y, McIntyre RS. The effect of pharmacogenomic testing on response and remission rates in the acute treatment of major depressive disorder: A meta-analysis. J Affect Disord. 2018;241:484–491. [DOI] [PubMed] [Google Scholar]
- 6.Galli M, Benenati S, Franchi F, et al. Comparative effects of guided vs. potent P2Y12 inhibitor therapy in acute coronary syndrome: a network meta-analysis of 61 898 patients from 15 randomized trials. Eur Heart J. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang Q, Chen SQ, Ma LY, et al. Association between SLCO1B1 T521C polymorphism and risk of statin-induced myopathy: a meta-analysis. Pharmacogenomics J. 2018;18(6):721–729. [DOI] [PubMed] [Google Scholar]
- 8.Perlis RH, Mehta R, Edwards AM, Tiwari A, Imbens GW. Pharmacogenetic testing among patients with mood and anxiety disorders is associated with decreased utilization and cost: A propensity-score matched study. Depress Anxiety. 2018;35(10):946–952. [DOI] [PubMed] [Google Scholar]
- 9.Van Driest SL, Shi Y, Bowton EA, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clinical pharmacology and therapeutics. 2014;95(4):423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanfreau-Coffinier C, Hull LE, Lynch JA, et al. Projected Prevalence of Actionable Pharmacogenetic Variants and Level A Drugs Prescribed Among US Veterans Health Administration Pharmacy Users. JAMA Netw Open. 2019;2(6):e195345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicks JK, El Rouby N, Ong HH, et al. Opportunity for Genotype-Guided Prescribing Among Adult Patients in 11 US Health Systems. Clinical pharmacology and therapeutics. 2021;110(1):179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith DM, Peshkin BN, Springfield TB, et al. Pharmacogenetics in Practice: Estimating the Clinical Actionability of Pharmacogenetic Testing in Perioperative and Ambulatory Settings. Clin Transl Sci. 2020;13(3):618–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luczak T, Brown SJ, Armbruster D, Hundertmark M, Brown J, Stenehjem D. Strategies and settings of clinical pharmacogenetic implementation: a scoping review of pharmacogenetics programs. Pharmacogenomics. 2021;22(6):345–364. [DOI] [PubMed] [Google Scholar]
- 14.Arwood MJ, Chumnumwat S, Cavallari LH, Nutescu EA, Duarte JD. Implementing Pharmacogenomics at Your Institution: Establishment and Overcoming Implementation Challenges. Clin Transl Sci. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marrero RJ, Cicali EJ, Arwood MJ, et al. How to Transition from Single-Gene Pharmacogenetic Testing to Preemptive Panel-Based Testing: A Tutorial. Clinical pharmacology and therapeutics. 2020;108(3):557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weitzel KW, Duong BQ, Arwood MJ, et al. A stepwise approach to implementing pharmacogenetic testing in the primary care setting. Pharmacogenomics. 2019;20(15):1103–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavallari LH, Weitzel KW, Elsey AR, et al. Institutional profile: University of Florida Health Personalized Medicine Program. Pharmacogenomics. 2017;18(5):421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cicali EJ, Elchynski AL, Cook KJ, et al. How to Integrate CYP2D6 Phenoconversion Into Clinical Pharmacogenetics: A Tutorial. Clinical pharmacology and therapeutics. 2021;110(3):677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elchynski AL, Desai N, D’Silva D, et al. Utilizing a Human-Computer Interaction Approach to Evaluate the Design of Current Pharmacogenomics Clinical Decision Support. J Pers Med. 2021;11(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartzler A, McCarty CA, Rasmussen LV, et al. Stakeholder engagement: a key component of integrating genomic information into electronic health records. Genet Med. 2013;15(10):792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caudle KE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Relling MV, Klein TE. Evidence and resources to implement pharmacogenetic knowledge for precision medicine. Am J Health Syst Pharm. 2016;73(23):1977–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab. 2014;15(2):209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clinical pharmacology and therapeutics. 2021;110(4):888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weitzel KW, Cavallari LH, Lesko LJ. Preemptive Panel-Based Pharmacogenetic Testing: The Time is Now. Pharm Res. 2017;34(8):1551–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for Clinical CYP2D6 Genotyping Allele Selection: A Joint Consensus Recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J Mol Diagn. 2021;23(9):1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pratt VM, Del Tredici AL, Hachad H, et al. Recommendations for Clinical CYP2C19 Genotyping Allele Selection: A Report of the Association for Molecular Pathology. J Mol Diagn. 2018;20(3):269–276. [DOI] [PubMed] [Google Scholar]
- 27.Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for Clinical Warfarin Genotyping Allele Selection: A Report of the Association for Molecular Pathology and the College of American Pathologists. J Mol Diagn. 2020;22(7):847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for Clinical CYP2C9 Genotyping Allele Selection: A Joint Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2019;21(5):746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JA, Caudle KE, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clinical pharmacology and therapeutics. 2017;102(3):397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drozda K, Wong S, Patel SR, et al. Poor warfarin dose prediction with pharmacogenetic algorithms that exclude genotypes important for African Americans. Pharmacogenet Genomics. 2015;25(2):73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369(24):2283–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vo TT, Bell GC, Owusu Obeng A, Hicks JK, Dunnenberger HM. Pharmacogenomics Implementation: Considerations for Selecting a Reference Laboratory. Pharmacotherapy. 2017;37(9):1014–1022. [DOI] [PubMed] [Google Scholar]
- 33.Haga SB, Mills R. A review of consent practices and perspectives for pharmacogenetic testing. Pharmacogenomics. 2016;17(14):1595–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang SC, Bruce C, Hayden M, Rieder MJ. Public perceptions of pharmacogenetics. Pediatrics. 2014;133(5):e1258–1267. [DOI] [PubMed] [Google Scholar]
- 35.Aquilante CL, Kao DP, Trinkley KE, et al. Clinical implementation of pharmacogenomics via a health system-wide research biobank: the University of Colorado experience. Pharmacogenomics. 2020;21(6):375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caraballo PJ, Sutton JA, Giri J, et al. Integrating pharmacogenomics into the electronic health record by implementing genomic indicators. J Am Med Inform Assoc. 2020;27(1):154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottesman O, Scott SA, Ellis SB, et al. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clinical pharmacology and therapeutics. 2013;94(2):214–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caraballo PJ, Bielinski SJ, St Sauver JL, Weinshilboum RM. Electronic Medical Record-Integrated Pharmacogenomics and Related Clinical Decision Support Concepts. Clinical pharmacology and therapeutics. 2017;102(2):254–264. [DOI] [PubMed] [Google Scholar]
- 39.Harada S, Zhou Y, Duncan S, et al. Precision Medicine at the University of Alabama at Birmingham: Laying the foundational processes through implementation of Genotype-Guided Antiplatelet Therapy. Clinical pharmacology and therapeutics. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abou Diwan E, Zeitoun RI, Abou Haidar L, Cascorbi I, Khoueiry Zgheib N. Implementation and obstacles of pharmacogenetics in clinical practice: An international survey. Br J Clin Pharmacol. 2019;85(9):2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson JF, Field JR, Shi Y, et al. Attitudes of clinicians following large-scale pharmacogenomics implementation. Pharmacogenomics J. 2016;16(4):393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu M, Vnencak-Jones CL, Roland BP, et al. A Tutorial for Pharmacogenomics Implementation Through End-to-End Clinical Decision Support Based on Ten Years of Experience from PREDICT. Clinical pharmacology and therapeutics. 2021;109(1):101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med. 2017;19(2):215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau-Min KS, Asher SB, Chen J, et al. Real-world integration of genomic data into the electronic health record: the PennChart Genomics Initiative. Genet Med. 2021;23(4):603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gill PS, Yu FB, Porter-Gill PA, et al. Implementing Pharmacogenomics Testing: Single Center Experience at Arkansas Children’s Hospital. J Pers Med. 2021;11(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manzi SF, Fusaro VA, Chadwick L, et al. Creating a scalable clinical pharmacogenomics service with automated interpretation and medical record result integration - experience from a pediatric tertiary care facility. J Am Med Inform Assoc. 2017;24(1):74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;166C(1):45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wake DT, Smith DM, Kazi S, Dunnenberger HM. Pharmacogenomic Clinical Decision Support: A Review, How-to Guide, and Future Vision. Clinical pharmacology and therapeutics. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cook KJ, Duong BQ, Seligson ND, et al. Key Considerations for Selecting a Genomic Decision Support Platform for Implementing Pharmacogenomics. Clinical pharmacology and therapeutics. 2021;110(3):555–558. [DOI] [PubMed] [Google Scholar]
- 50.Empey PE, Pratt VM, Hoffman JM, Caudle KE, Klein TE. Expanding evidence leads to new pharmacogenomics payer coverage. Genet Med. 2021;23(5):830–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.S LR, Keeling NJ, Giri J, et al. PARC report: a health-systems focus on reimbursement and patient access to pharmacogenomics testing. Pharmacogenomics. 2020;21(11):785–796. [DOI] [PubMed] [Google Scholar]
- 52.Haga SB, Allen LaPointe NM, Moaddeb J. Challenges to integrating pharmacogenetic testing into medication therapy management. J Manag Care Spec Pharm. 2015;21(4):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dietrich E, Gums JG. Incident-to Billing for Pharmacists. J Manag Care Spec Pharm. 2018;24(12):1273–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunnenberger HM, Biszewski M, Bell GC, et al. Implementation of a multidisciplinary pharmacogenomics clinic in a community health system. Am J Health Syst Pharm. 2016;73(23):1956–1966. [DOI] [PubMed] [Google Scholar]
- 55.Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clinical pharmacology and therapeutics. 2012;91(3):450–458. [DOI] [PubMed] [Google Scholar]
- 56.Johansen Taber KA, Dickinson BD. Pharmacogenomic knowledge gaps and educational resource needs among physicians in selected specialties. Pharmgenomics Pers Med. 2014;7:145–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuteja S, Salloum RG, Elchynski AL, et al. Multisite evaluation of institutional processes and implementation determinants for pharmacogenetic testing to guide antidepressant therapy. Clin Transl Sci. 2022;15(2):371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theken KN, Lee CR, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clinical pharmacology and therapeutics. 2020;108(2):191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Relling MV, Schwab M, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2018 Update. Clinical pharmacology and therapeutics. 2019;105(5):1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Truong TM, Lipschultz E, Danahey K, Schierer E, Ratain MJ, O’Donnell PH. Assessment of Patient Knowledge and Perceptions of Pharmacogenomics Before and After Using a Mock Results Patient Web Portal. Clin Transl Sci. 2020;13(1):78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arwood MJ, Dietrich EA, Duong BQ, et al. Design and Early Implementation Successes and Challenges of a Pharmacogenetics Consult Clinic. J Clin Med. 2020;9(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hicks JK, Stowe D, Willner MA, et al. Implementation of Clinical Pharmacogenomics within a Large Health System: From Electronic Health Record Decision Support to Consultation Services. Pharmacotherapy. 2016;36(8):940–948. [DOI] [PubMed] [Google Scholar]


