Abstract
Rationale
Bacterial lung microbiota are correlated with lung inflammation and acute respiratory distress syndrome (ARDS) and altered in severe coronavirus disease (COVID-19). However, the association between lung microbiota (including fungi) and resolution of ARDS in COVID-19 remains unclear. We hypothesized that increased lung bacterial and fungal burdens are related to nonresolving ARDS and mortality in COVID-19.
Objectives
To determine the relation between lung microbiota and clinical outcomes of COVID-19–related ARDS.
Methods
This observational cohort study enrolled mechanically ventilated patients with COVID-19. All patients had ARDS and underwent bronchoscopy with BAL. Lung microbiota were profiled using 16S rRNA gene sequencing and quantitative PCR targeting the 16S and 18S rRNA genes. Key features of lung microbiota (bacterial and fungal burden, α-diversity, and community composition) served as predictors. Our primary outcome was successful extubation adjudicated 60 days after intubation, analyzed using a competing risk regression model with mortality as competing risk.
Measurements and Main Results
BAL samples of 114 unique patients with COVID-19 were analyzed. Patients with increased lung bacterial and fungal burden were less likely to be extubated (subdistribution hazard ratio, 0.64 [95% confidence interval, 0.42–0.97]; P = 0.034 and 0.59 [95% confidence interval, 0.42–0.83]; P = 0.0027 per log10 increase in bacterial and fungal burden, respectively) and had higher mortality (bacterial burden, P = 0.012; fungal burden, P = 0.0498). Lung microbiota composition was associated with successful extubation (P = 0.0045). Proinflammatory cytokines (e.g., tumor necrosis factor-α) were associated with the microbial burdens.
Conclusions
Bacterial and fungal lung microbiota are related to nonresolving ARDS in COVID-19 and represent an important contributor to heterogeneity in COVID-19–related ARDS.
Keywords: lung microbiome, host–microbial interactions, critical illness, artificial respiration
At a Glance Commentary
Scientific Knowledge on the Subject
Recent studies have shown that bacterial lung microbiota are correlated with lung inflammation and altered in acute respiratory distress syndrome (ARDS) and critically ill patients with coronavirus disease (COVID-19). Yet, the association between lung microbiota—including fungi—and resolution of ARDS in the setting of COVID-19 remains unclear.
What This Study Adds to the Field
In this cohort of 114 mechanically ventilated patients with COVID-19, we found that patients with an increased lung bacterial and fungal burden had a lower incidence of liberation from invasive mechanical ventilation and a higher mortality. The bacterial and fungal burden in BAL fluid were correlated with alveolar proinflammatory cytokines such as TNF-α and IL-1β, and lung microbiota community composition was associated with successful extubation. Our findings confirm the importance of the lung microbiome in ARDS and COVID-19 and highlight the significance of the—often overlooked—pulmonary fungal burden in critically ill patients. The lung microbiome has an important role in the heterogeneity in clinical outcomes of COVID-19–related ARDS.
In 15–30% of hospitalized patients with coronavirus disease (COVID-19), ICU admission is necessary. Almost all of these critically ill patients with COVID-19 fulfill the criteria for acute respiratory distress syndrome (ARDS) (1–3). The accompanying lung injury does not resolve quickly, frequently resulting in multiple weeks of invasive mechanical ventilation (4). Dynamic changes of biomarkers related to endothelial dysfunction, coagulopathy, epithelial injury, and innate immune responses in plasma are predictive of such adverse outcomes (5–8). Studies of the alveolar response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection are sparse but suggest that the alveolar space is a major contributor to a persistent proinflammatory environment (8–10). Yet, it remains unclear what biological processes prevent the resolution of lung injury in these patients.
Enrichment of pulmonary microbiota with gut-specific bacteria has been a consistent observation in patients with ARDS (11, 12). A higher bacterial burden in the lung has been associated with worse outcomes in idiopathic pulmonary fibrosis, after lung transplant and in mechanically ventilated patients without COVID-19 (12–14). Changes in the abundance and composition of lung microbiota have been linked to altered host immune responses, increased alveolar inflammation, and development of lung injury (15–17). Therefore, dysbiosis of the lung microbiome may contribute to nonresolving ARDS and increased mortality.
In COVID-19, several studies characterized the upper respiratory tract microbiome and showed an association between microbial composition and disease severity, likely because clinical practices (e.g., type of oxygen support and time in the ICU) drive variation within these communities (18–20). Changes in upper respiratory tract microbiota, however, do not reflect alveolar changes (21), and only a few studies—mainly small scale—described lower respiratory tract microbiota during COVID-19 (18, 19, 22). These studies found a distinct lung microbiota composition in patients with COVID-19 compared with critically ill patients without COVID-19 and healthy control subjects. In a larger cohort, microbial signatures (such as bacterial burden, SARS-CoV-2 viral load, and enrichment with an oral commensal) in BAL fluid of 118 patients with COVID-19 were associated with mortality (23). However, the association between lung microbiome dysbiosis and resolution of ARDS in the setting of COVID-19 remains to be determined.
We hypothesized that patients with COVID-19 with a higher bacterial and fungal burden were less likely to be liberated from invasive mechanical ventilation and have increased mortality, and that this could be linked to lung microbiome dysbiosis. Furthermore, we aimed to evaluate how dysbiosis relates to secondary infections, such as aspergillosis and ventilator-associated pneumonia, and to the alveolar host response. To this end, we characterized the microbiota in 163 BAL samples of 114 mechanically ventilated patients with COVID-19–related ARDS.
Methods
Study Cohort
We performed an observational cohort study in adult patients with COVID-19 admitted to the ICUs of the Amsterdam University Medical Centers, both location Academic Medical Center and VU Medical Center. Details on the study cohort and clinical data recording are provided in the online supplement. In brief, we used BAL fluid collected during routine clinical care of consecutive mechanically ventilated patients with COVID-19 included between September 2020 and July 2021. SARS-CoV-2 infection was confirmed by positive reverse transcription PCR result, and all patients had ARDS according to the Berlin criteria (24). A diagnostic bronchoscopy with BAL was performed routinely once weekly if there was no clinical improvement, or in case of deterioration. Samples were excluded if there was too little leftover BAL volume (⩽3 ml), inadequate sample storage (⩾24 h before storage at −80°C), or the inability to pass appropriate quality control (<200 high-quality sequencing reads) (15). Study procedures were approved by the review committee of the Amsterdam UMC Biobank (reference: 2020–065). Written informed consent was provided by all participants or their legal representatives.
Sample Collection and Processing
BAL samples were collected during video-assisted bronchoscopy by instilling 4 × 20 ml 0.9% NaCl into the lung at a subsegmental level (10). Fractions one, three, and four were sent for diagnostic microbiological evaluation as part of standard care. Fraction two was centrifuged (300 g, 10 min, 4°C), and supernatant was stored at −80°C until further analysis.
Details on sample processing have been earlier described by our group and are provided in the online supplement (7, 10, 25–27). In summary, bacterial microbiota were characterized by 16S rRNA gene sequencing targeting the V3-V4 region. Bacterial and fungal burden were quantified using quantitative PCR targeting the 16S and 18S rRNA gene, respectively. Buffers and solutions used during DNA isolation, sequencing, and quantification were used as negative experimental controls. Concentrations of cytokines were measured using a Luminex multiplex assay (R&D Systems) on a BioPlex200 (BioRad) (10).
Outcomes and Predictor Variables
Our primary clinical outcome was successful liberation from mechanical ventilation, adjudicated at Day 60 after initiation of mechanical ventilation. For time-to-event analyses, our primary approach was to calculate subdistribution hazard ratios (SHRs) using competing risk regression models, treating extubation as outcome and mortality as competing risk (28). For other analyses, patients were stratified as 1) those who reached the primary outcome and 2) those who died within 60 days or were still mechanically ventilated at Day 60. Sensitivity analyses included outcome assessment at 60 and 90 days after sample collection (rather than after intubation).
We prespecified that bacterial and fungal DNA burden, Shannon α-diversity, and community composition would serve as predictors of clinical outcomes, as measured in the first available BAL sample per patient. These key features of the lung microbiome, except fungal burden, have been previously reported to predict clinical outcomes in acute and chronic lung diseases (12–15). In addition, we examined the relation between markers of alveolar inflammation and the bacterial and fungal burden.
Probable and proven COVID-19– associated pulmonary aspergillosis (CAPA) was defined according to the 2020 European Confederation of Medical Mycology and International Society for Human Animal Mycology definition (supplementary methods) (29). As earlier described, and in line with European Respiratory Society, European Society of Clinical Microbiology and Infectious Diseases, European Society of Intensive Care Medicine and Latin American Thoracic Association recommendations (30–32), a positive microbiologic (nondirected) BAL culture (cutoff of 104 CFU/ml) by the clinical laboratory confirmed bacterial ventilator-associated pneumonia. Cultures were considered nonpathogenic when only normal respiratory bacteria were detected. Negative cultures were those without growth (<104 CFU/ml). For patients with more than one BAL sample available, the presence of CAPA and bacterial ventilator-associated pneumonia were determined separately for each BAL.
Statistical Analysis
Details of our statistical methods are provided in the online supplement. We calculated α-diversity using the Shannon Diversity Index (phyloseq package). Our primary approach for time-to-event analyses was to calculate the SHR using Fine and Gray competing risk regression, with extubation as outcome and mortality as competing event (survival and survminer packages) (28). Time zero was defined as the day of intubation. Multivariable models were adjusted for age, sex, body mass index, BAL bacterial culture result, presence of proven or probable CAPA, severity of illness, and antifungal and antibiotic exposure. We quantified the severity of illness with the Sequential Organ Failure Assessment score at sample collection, and used an earlier-described detailed quantitative model to calculate an ICU antibiotic-exposure score that considers dosing duration, timing of administration, and antibiotic type (see the methods and Table E1 in the online supplement) (15, 33).
Differences in community composition (β-diversity) were visualized by principal coordinates analyses with the weighted Unifrac distance (which takes bacterial phylogeny into account). Statistical significance was determined by permutational multivariate ANOVA (vegan package, adonis2 function, 9,999 permutations). Bacterial taxa explaining differences in β-diversity were identified using biplot analysis, Wilcoxon rank sum tests, and random forest and DESeq2 models. Spearman correlations between cytokine concentrations and bacterial or fungal burden were calculated, with Benjamini-Hochberg correction for multiple comparisons. Continuous data are presented as median with interquartile range and analyzed using a Wilcoxon rank sum test. Two-tailed degree of significance between groups was set at (adjusted) P < 0.05.
Results
Study cohort
Lung microbiota of 159 mechanically ventilated patients with COVID-19 with ARDS were profiled by 16S rRNA gene sequencing of BAL samples. After exclusion of clinical samples of 45 patients (Table E2) owing to a very low number of high-quality reads (<200), 163 samples of 114 unique patients passed appropriate quality controls. A Consolidated Standards of Reporting Trials diagram of study inclusions is depicted in Figure E1. Table 1 shows the demographic and clinical characteristics of the 114 included patients. Of 114 patients, 94.7% received systemic antibiotics between ICU admission and first sampling (Figure E2), 53.5% died during ICU stay, and 60-day mortality was 52.6% (Table 1).
Table 1.
Clinical Characteristics of the Study Population
| Characteristic | Total (n = 114) | Extubated (n = 44) | Deceased or Intubated (n = 70) | P Value |
|---|---|---|---|---|
| Age, y | 64.0 (58.0–71.0) | 63.0 (58.0–68.0) | 65.5 (59.0–72.0) | 0.08 |
| Male sex | 84 (73.7) | 31 (70.5) | 53 (75.7) | 0.69 |
| BMI, kg/m2 | 27.8 (24.7–31.5) | 29.8 (25.8–32.7) | 27.5 (24.6–31.0) | 0.10 |
| Current or former smoker | 34 (29.8) | 15 (34.1) | 19 (27.1) | 0.38 |
| Transferred from another hospital | 68 (59.6) | 29 (65.9)) | 39 (55.7) | 0.38 |
| Comorbidities | ||||
| COPD | 7 (6.1) | 3 (6.8) | 4 (5.7) | 1.00 |
| Asthma | 14 (12.3) | 6 (13.6) | 8 (11.4) | 0.96 |
| Cardiovascular disease | 49 (43.0) | 21 (47.7) | 28 (40.0) | 0.54 |
| Diabetes | 31 (27.2) | 11 (25.0) | 20 (28.6) | 0.84 |
| Malignancy | 8 (7.0) | 3 (6.8) | 5 (7.1) | 1.00 |
| Kidney disease | 13 (11.4) | 3 (6.8) | 10 (14.3) | 0.36 |
| Hepatic disease | 1 (0.9) | 0 (0.0) | 1 (1.4) | 1.00 |
| Neurological disorder | 2 (1.8) | 2 (4.5) | 0 (0.0) | 0.29 |
| Treatment and sample collection | ||||
| Systemic steroids between ICU admission and sampling | 101 (88.6) | 36 (81.8) | 65 (92.9) | 0.13 |
| IL-6 receptor antagonists | 8 (7.0) | 4 (9.1) | 4 (5.7) | 0.76 |
| Systemic antibiotic exposure before ICU admission* | 87 (76.3) | 32 (72.7) | 55 (78.6) | 0.63 |
| Systemic antibiotic exposure between ICU admission and sampling | 108 (94.7) | 41 (93.2) | 67 (95.7) | 0.87 |
| Systemic antifungal exposure between ICU admission and sampling | 16 (14.0) | 7 (15.9) | 9 (12.9) | 0.86 |
| ICU length of stay before sample collection, d | 10.5 (6.0–16.0) | 11.5 (7.5–20.0) | 10.0 (6.0–14.8) | 0.45 |
| Mechanical ventilation before sample collection, d | 9.0 (5.0–14.8) | 9.0 (5.0–16.8) | 9.0 (5.0–14.0) | 0.64 |
| Severity of disease at day of sample collection | ||||
| SOFA score | 8.0 (7.0–10.0) | 8.0 (6.8–10.0) | 8.0 (7.0–10.8) | 0.63 |
| PaO2/FiO2 | 101.5 (79.1–124.2) | 120.7 (92.1–133.4) | 95.8 (77.8–117.2) | 0.03 |
| Follow-up samples | ||||
| Second BAL sample available | 32 (28.1) | 14 (28.6) | 18 (27.7) | — |
| Third BAL sample available | 14 (12.3) | 3 (6.1) | 11 (16.9) | — |
| Fourth BAL sample available | 3 (2.6) | 1 (2.0) | 2 (3.1) | — |
| Total samples available | 163 | 67 | 96 | — |
| Outcomes | ||||
| Probable COVID-19– associated pulmonary aspergillosis† | 30 (26.3) | 11 (25.0) | 19 (27.1) | 0.97 |
| Positive bacterial microbiologic (nondirected) BAL culture | 25 (21.9) | 10 (22.7) | 15 (21.4) | 1.00 |
| ICU length of stay, d | 30.0 (18.0–46.0) | 39.0 (22.0–49.5) | 24.5 (18.0–38.0) | 0.32 |
| Length of mechanical ventilation, d | 27.0 (16.3–42.0) | 32.0 (15.5–45.0) | 23.0 (16.3–35.0) | 0.97 |
| ICU mortality | 61 (53.5) | 0 (0.0) | 61 (87.1) | — |
| 60-d mortality‡ | 60 (52.6) | 0 (0.0) | 60 (85.7) | — |
| 90-d mortality‡ | 62 (54.4) | 0 (0.0) | 62 (88.6) | — |
Definition of abbreviations: BMI = body mass index; COPD = chronic obstructive pulmonary disease; COVID-19 = coronavirus disease; SOFA = sequential organ failure assessment.
Clinical characteristics of the study population, stratified by clinical outcome at Day 60 after the start of mechanical ventilation. Values are n (%) or median (interquartile range).
Systemic antibiotic exposure in the 30 d before ICU admission at the study center.
Defined according to the 2020 European Confederation of Medical Mycology and the International Society for Human and Animal Mycology consensus definition.
Calculated with the day of intubation as Day 0.
Lung Microbiota of BAL Samples Are Distinct from Negative Experimental Controls
Owing to the low bacterial mass, lung microbiome studies are vulnerable to contamination during DNA extraction and sequencing (34). Therefore, we compared the microbial signal of our BAL samples with negative experimental control samples (n = 32 in 16S rRNA gene sequencing and n = 46 in quantitative PCR). We found large spread in bacterial and fungal DNA burden, ranging from comparable to negative controls to 1,000-fold greater than background, and confirmed that BAL samples contained significantly more bacterial and fungal DNA than negative experimental controls (Figure E3). Although some overlap in community composition was found, there was a detectable and statistically significant difference between BAL samples and negative controls (P = 0.0001) (Figure E3). Consequently, although several taxa were detected in both sample types (e.g., Massilia, Ralstonia, and Pseudarthrobacter spp.), the bacteria identified in BAL samples collectively differed from those in negative controls (Figure E3F). BAL samples contained a greater microbial signal and a microbiome distinct from negative experimental controls, despite some overlap and potential contamination.
Lung Bacterial and Fungal Burden Are Associated with Clinical Outcomes of COVID-19–related ARDS
We first asked whether earlier-described key features of the lung microbiome (bacterial and fungal burden and α-diversity) were associated with nonresolving ARDS in COVID-19. In competing risk analysis, using the first available BAL sample per patient (n = 114), an increasing bacterial burden was associated with a lower incidence of extubation (per log10 increase in bacterial burden, subdistribution hazard ratio [SHR], 0.64 [95% confidence interval (CI), 0.42–0.97]; P = 0.034) (Figure 1A). The cause-specific hazard ratio for mortality was 1.36 (95% CI, 1.06–1.75; P = 0.016) (Table E3) and for extubation, 0.79 (95% CI, 0.51–1.23; P = 0.30), suggesting an indirect effect of the strong association between the bacterial burden and the competing event (mortality) on the SHR for extubation. In other words, patients with an increased bacterial burden might have a lower hazard of extubation because deceased patients cannot be successfully extubated and hence have a “lower chance” of extubation.
Figure 1.
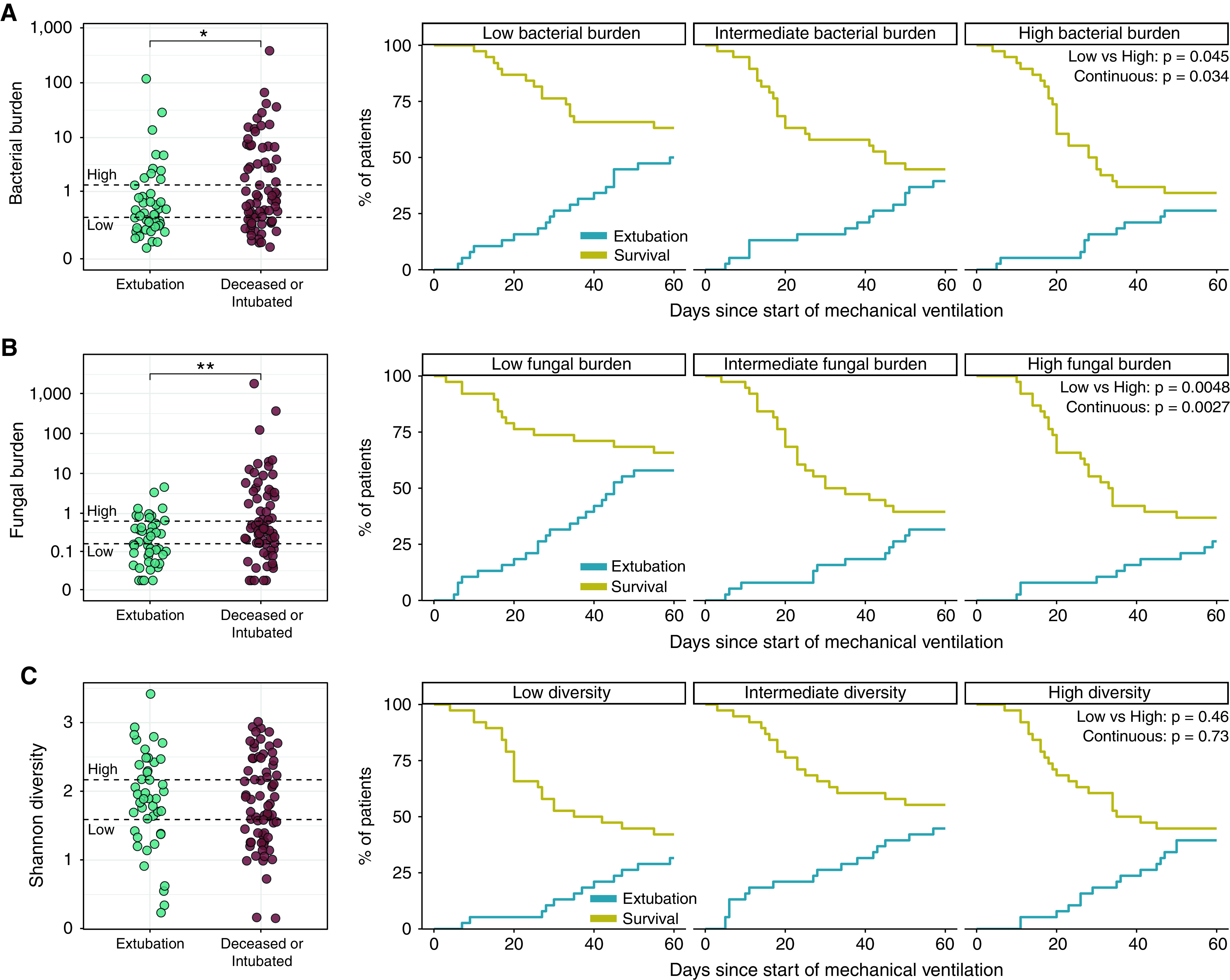
Lung bacterial and fungal burden are correlated with clinical outcomes of coronavirus disease (COVID-19)-related acute respiratory distress syndrome. (A) Lung bacterial DNA burden (quantified using a 16S rRNA gene quantitative PCR assay) was lower in patients with COVID-19 successfully extubated at Day 60 after initiation of mechanical ventilation (n = 44 of 114 unique patients with COVID-19). Patients with the lowest bacterial burden had a higher incidence of successful extubation than those with high bacterial burdens. (B) Likewise, lung fungal DNA burden (quantified by quantitative PCR assay targeting the 18S rRNA gene) was lower in patients who were successfully extubated, and patients with high lung burdens of fungal DNA were less likely to be extubated. (C) Shannon α-diversity did not correlate with the liberation of mechanical ventilation. At Day 60 after initiation of mechanical ventilation, 10 patients were still intubated (and thus did not achieve extubation or death). Hypothesis testing was performed using a Wilcoxon rank sum test (left panels) and Fine and Gray competing risk regression models, treating extubation as outcome and mortality as competing risk. *P < 0.05 and **P < 0.01.
To align with previous studies (12–14), we split bacterial burden into tertiles (low, intermediate, and high) and compared the incidence of extubation and mortality. Patients in the highest bacterial burden tertile were significantly less likely to be extubated and alive than with patients in the lowest tertile (SHR, 0.46 [95% CI, 0.21–0.98]; P = 0.045) (Figure 1A).
Likewise, patients with a higher fungal DNA burden were less likely to be liberated from mechanical ventilation, either when analyzed continuously (SHR, 0.59 [95% CI, 0.42–0.83]; P = 0.0027) or when comparing tertiles of fungal DNA burden (SHR, 0.34 [95% CI, 0.16–0.72]; P = 0.0048 for lowest vs. highest burden) (Figure 1B). The cause-specific hazard ratio for extubation was 0.64 (95% CI, 0.45–0.90; P = 0.012) (Table E3) and for mortality, 1.14 (95% CI, 0.92–1.41; P = 0.24). We found no significant association between lung bacterial diversity and our primary clinical outcome (Figure 1C).
After adjustment for potential confounders (age, sex, body mass index, BAL bacterial culture result, presence of proven or probable CAPA, severity of illness, and antifungal and antibiotic exposure) in multivariable analysis, higher bacterial and fungal burden (per log10 increase) remained independently associated with a lower incidence of extubation (SHR, 0.56 [95% CI, 0.36–0.88]; P = 0.011 for bacterial burden; SHR, 0.56 [95% CI, 0.38–0.84]; P = 0.0051 for fungal burden) (Table E4).
Two sensitivity analyses were performed to test particular assumptions of above-described models. First, adjudication of clinical outcomes at 60 days after sample collection (instead of 60 days after intubation) to account for immortal time bias yielded similar associations between higher bacterial and fungal burden and a lower chance of extubation (SHR, 0.64 [95% CI, 0.43–0.95]; P = 0.026 for bacterial burden; SHR, 0.74 [95% CI, 0.57–0.96]; P = 0.024 for fungal burden) (Figure E4). Second, when we extended our follow-up and assessed clinical outcomes at 90 days after sample collection, similar effect estimates were found (SHR, 0.63 [95% CI, 0.42–0.93]; P = 0.021 for bacterial burden; SHR, 0.72 [95% CI, 0.52–0.98]; P = 0.036 for fungal burden) (Figure E5).
Lung Microbiota Community Composition Is Associated with Successful Extubation in COVID-19–related ARDS
We next asked if community composition of lung bacterial microbiota differed between patients who were extubated and those deceased or still intubated by Day 60. When visualizing microbial composition of the first available BAL sample per patient (n = 114) by principal coordinate analysis, we observed wide variation in composition and some overlap between outcome groups. However, there was a small difference in community composition between outcome groups, which was confirmed statistically via permutation (P = 0.0045) (Figure 2A and Figure E6A) and remained significant when adjusted for potential confounders (P = 0.033). Outcome assessment at 60 and 90 days after sample collection did not meaningfully impact these results (Figures E6B and E6C).
Figure 2.
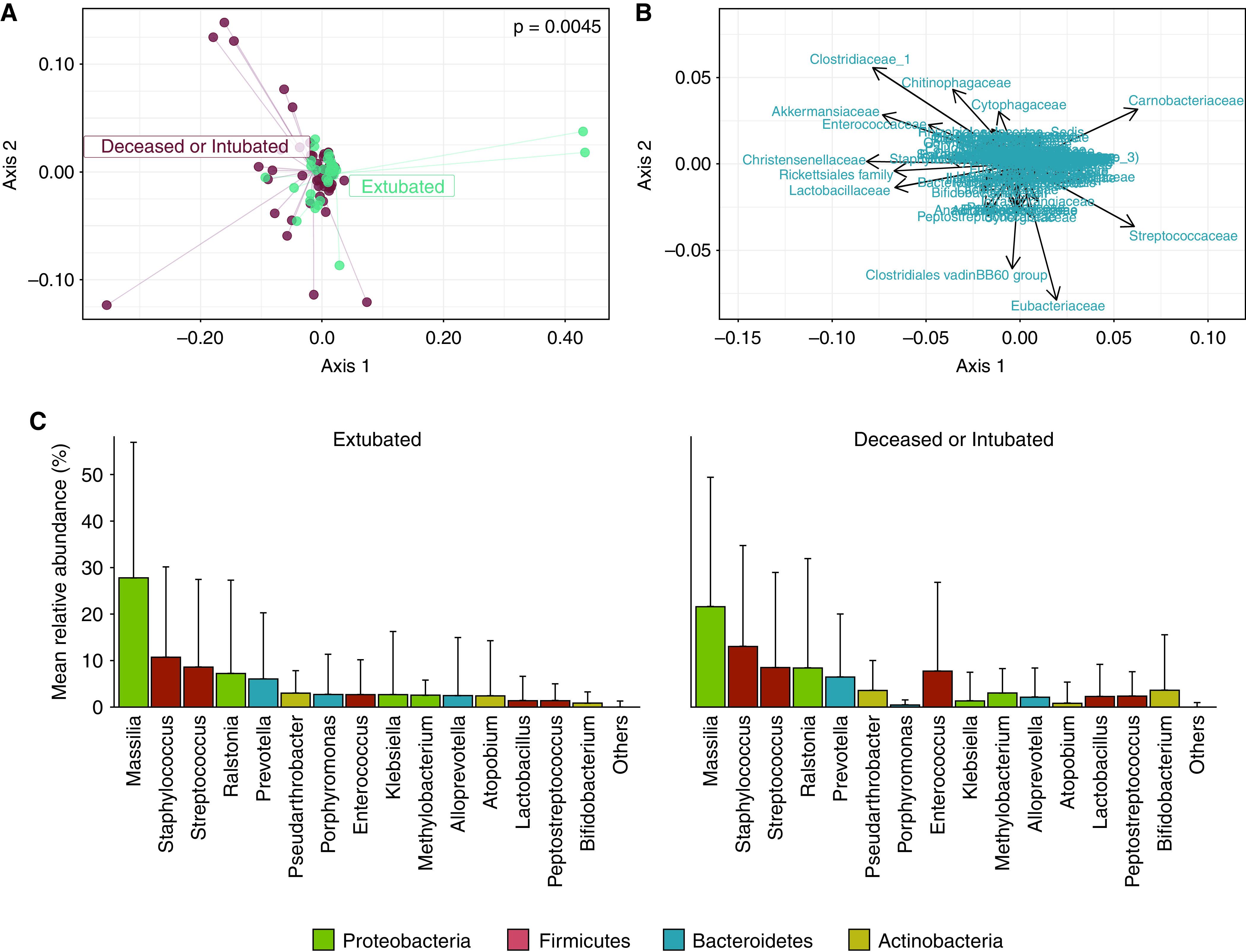
Lung microbiota community composition is associated with successful extubation in coronavirus disease (COVID-19)-related acute respiratory distress syndrome but not driven by specific bacterial genera. (A) Community composition of lung bacterial microbiota differed between patients who were extubated (n = 44) and those who were deceased or still intubated (n = 70) at 60 days after initiation of mechanical ventilation. Significance of differences in community composition are determined using permutational multivariate ANOVA with the weighted UniFrac distance. Principal coordinates analysis focusing on the center of the figure (leaving out outlying samples with distinct microbiota) is depicted in Figure E6 in the online supplement. (B and C) Biplot analysis (B) and rank abundance analysis (C) revealed no earlier reported bacterial taxa (family and genus level, respectively) driving the differences in community composition. Contaminant taxa (e.g., Massilia and Ralstonia spp.) were abundant in both groups. Rank abundance analysis included the 15 most abundant genera in decreasing order of relative abundance (mean ± SD), and no genera breached Benjamini-Hochberg adjusted significance (α < 0.05; Wilcoxon rank sum test).
Notably, a significant association between the bacterial burden and β-diversity (P = 0.003) was found, which indicates that variation in community composition is linked to differences in total bacterial burden. However, the relation between community composition and successful extubation remained significant when adjusted for the bacterial burden (P = 0.043). We thus concluded that lung microbiota community composition was associated with successful extubation in COVID-19–related ARDS, although sequencing contamination could have influenced these differences.
To identify individual bacterial taxa associated with successful extubation, biplot and rank abundance analyses were used. In contrast to earlier studies in critically ill patients in the ICU with and without COVID-19 (11, 12, 15, 23), differences in community composition were not driven by pathogenic or gut- or oral-associated bacteria (Figures 2B and 2C). Bacterial genera that discriminated between outcome groups (identified by random forest analyses) differed from the bacteria observed in biplot analysis (Figures E7A and E7B). Moreover, a DESeq2 model detected only Porphyromonas as a differentially abundant genus between outcome groups that was also identified by random forest analyses (Figure E7C). Because no other bacterial taxa were consistently identified by these different approaches and the detection of Porphyromonas did not correlate with the liberation of mechanical ventilation (Figure E7D), we concluded that no individual bacterial genera were definitively associated with successful extubation.
Lung Microbiota Alterations Correspond with Secondary Pulmonary Infections
The pathogenesis of pneumonia involves lung microbiome dysbiosis, characterized by increased microbial burden, pathogen overgrowth, and inflammation (35, 36). Therefore, we aimed to verify if secondary bacterial and fungal infections were associated with a higher bacterial and fungal burden, respectively, and asked whether bacterial pneumonia was associated with elevated relative abundances of the causative pathogens in BAL fluid (n = 163 samples). An overview of bacterial culture results is depicted in Figure E8.
Secondary bacterial pneumonia was associated with a higher lung bacterial burden than in patients with a negative BAL culture (Figure 3A). The bacterial burden of patients with a culture positive for (nonpathogenic) oral bacteria was increased as well. In other words, bacterial growth (⩾104 CFU/ml) in conventional BAL cultures reflected higher bacterial counts, irrespective of the type of bacteria present (either pathogenic or normal respiratory bacteria). Although we observed overlap in fungal burden between patients with and without proven or probable CAPA, fungal burden of patients with CAPA was significantly increased (Figure 3B).
Figure 3.
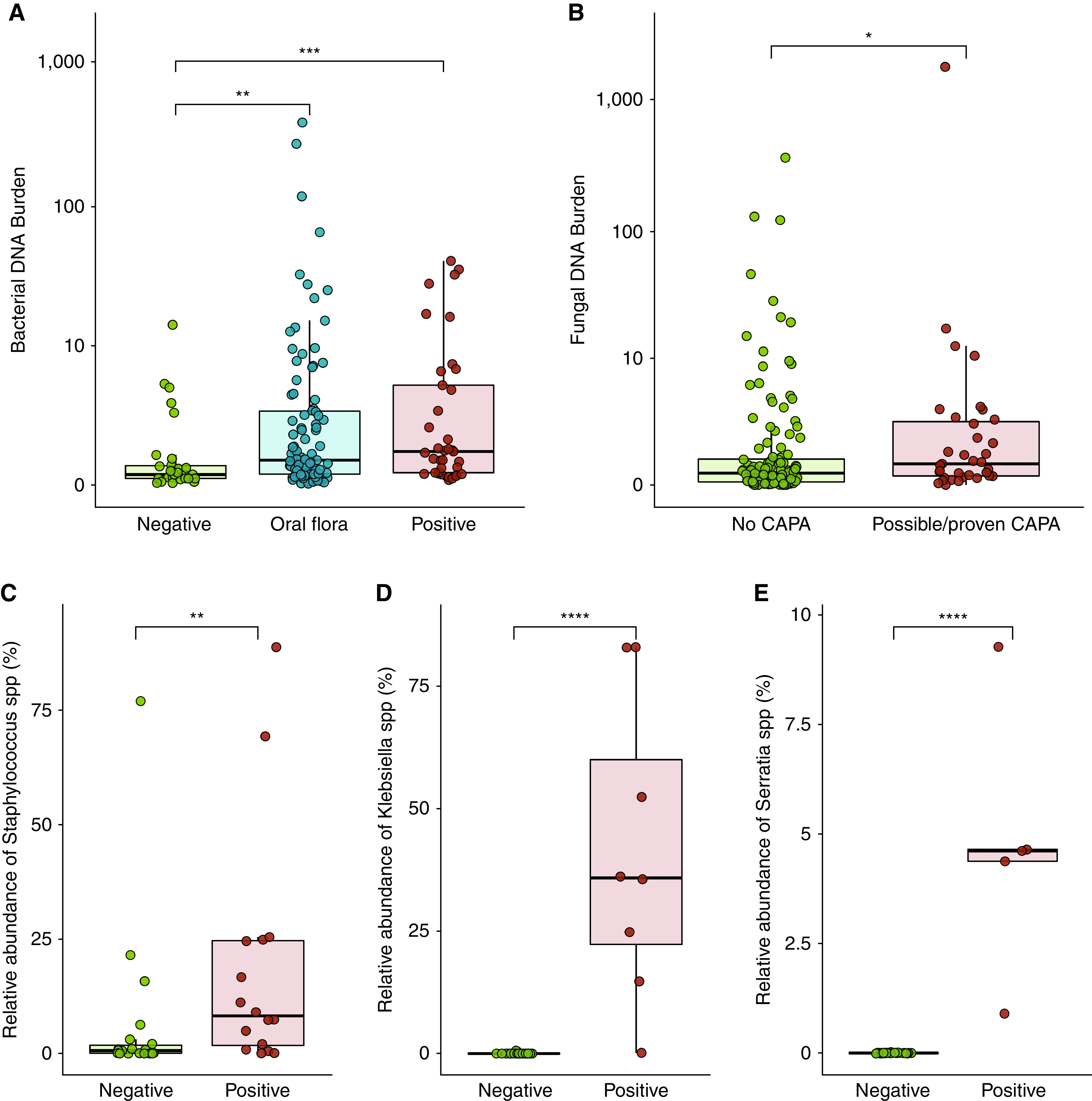
Lung microbiota alterations correspond with secondary pulmonary infections in severe coronavirus disease (COVID-19). (A) Increased lung bacterial DNA burden in patients with a positive microbiologic culture (n = 39 BAL samples) and in those for whom oral bacteria were cultured (n = 95), compared with those with negative cultures (n = 29). (B) The lung fungal DNA burden was higher in patients with proven or probable COVID-19–associated pulmonary aspergillosis (CAPA) (n = 35 BAL samples of 30 unique patients with COVID-19), although considerable overlap did occur. (C–E) High relative abundances (proportion of total 16S rRNA reads) of Staphylococcus species (C), Klebsiella spp. (D), and Serratia spp. (E) in BAL samples of patients with microbiological diagnosis of these pathogens as obtained via nondirected culture. In the boxplots, the rectangle spans the interquartile range with a line at the median. P values are calculated using a Wilcoxon rank sum test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Next, we evaluated if causative pathogens in BAL could be identified using 16S rRNA gene sequencing. For the three most common pathogens in our cohort (Staphylococcus aureus, Klebsiella, and Serratia spp. [Figure E8]), we compared their relative abundance in BAL between patients with microbiological diagnosis of these pathogens and patients negative for these pathogens. Patients with a confirmed secondary pneumonia caused by S. aureus, Klebsiella, or Serratia spp. harbored higher lung relative abundances of Staphylococcus (P = 0.005), Klebsiella (P < 0.0001), and Serratia spp. (P < 0.0001), respectively, than those with none or other causative pathogens (Figure 3C). These findings indicate that lung microbiota alterations correspond with secondary pulmonary infections.
Lung Bacterial and Fungal Burden Are Correlated with Alveolar Inflammatory Cytokine Responses
Having determined that lung bacterial and fungal burden are associated with clinical outcomes of COVID-19–related ARDS, we sought to determine whether these burdens are correlated with alveolar cytokine responses, as measured in both initial and follow-up samples. Twenty-five out of 63 (39.7%) measured biomarkers were associated with lung bacterial burden, and 35 out of 63 (55.6%) with fungal burden (Figure E9). Next, we focused on cytokines that serve as markers of (alveolar) inflammation in ARDS or COVID-19 (10, 37–40). A higher bacterial DNA burden in BAL samples was correlated with increased alveolar concentrations of the proinflammatory cytokines TNF-α (tumor necrosis factor-α), IL-6, IL-1β and TGF-α (transforming growth factor-α) (Figure 4A). Moreover, the fungal burden was positively correlated with alveolar concentrations of proinflammatory cytokines (TNF-α and TGF-α) and ILs involved in the antifungal response (IL-12p70 and IL-17A) (Figure 4B).
Figure 4.
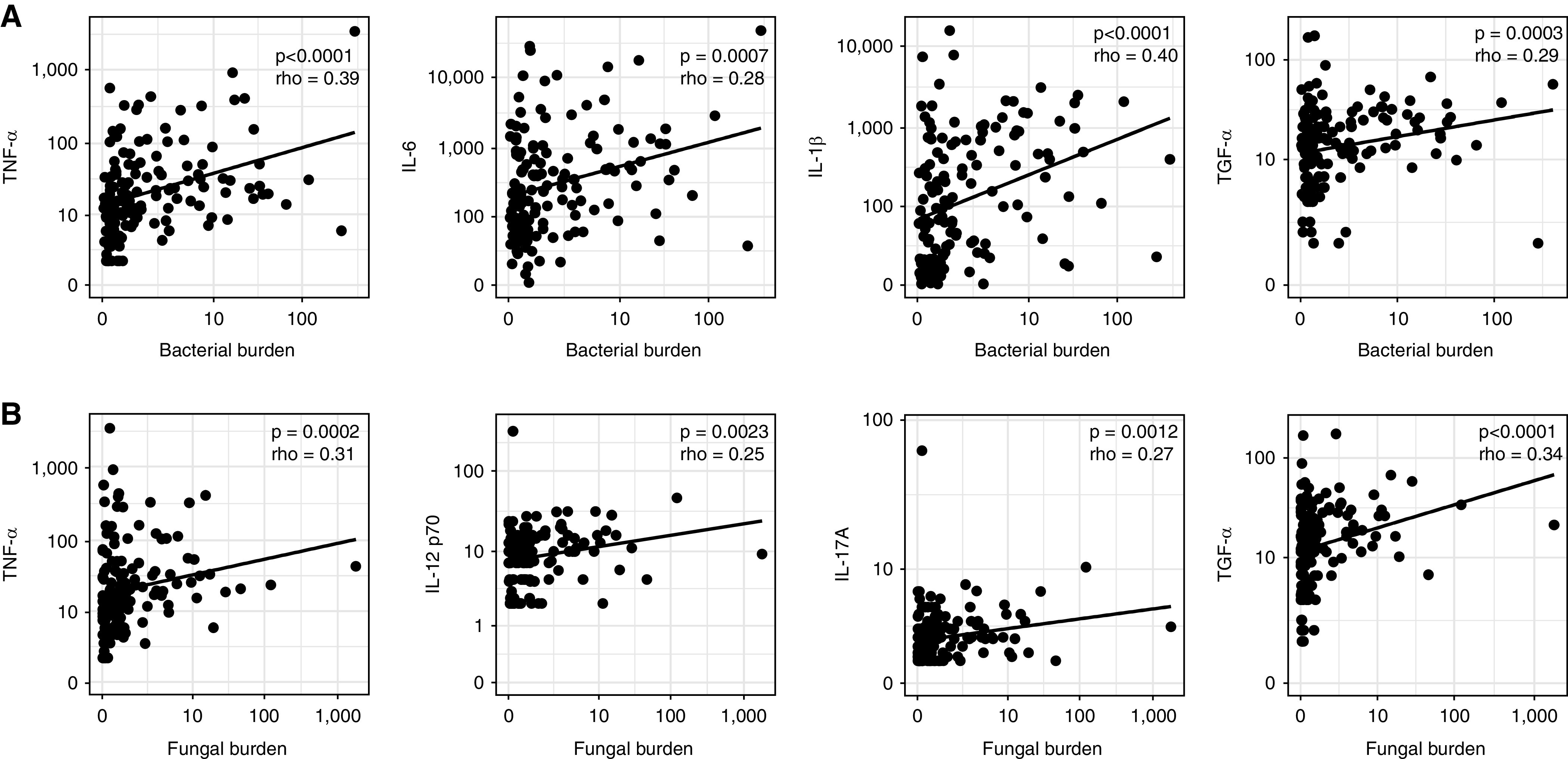
Lung bacterial and fungal burden are correlated with alveolar inflammatory cytokine responses in coronavirus disease (COVID-19)-related acute respiratory distress syndrome. (A) The lung bacterial DNA burden of patients with COVID-19 (n = 163 BAL samples of 114 unique patients) was positively correlated with alveolar concentrations of the proinflammatory cytokines TNF-α (tumor necrosis factor-α), IL-6, IL-1β and TGF-α (tumor growth factor-α. (B) Fungal DNA burden was positively correlated with the alveolar concentrations of proinflammatory cytokines (TNF-α and TGF-α) and ILs involved in the antifungal response (IL-12p70 and IL-17A). Statistical significance was determined using Spearman correlations between cytokine measurements and DNA burdens.
Discussion
In this observational cohort study of mechanically ventilated patients with COVID-19, we found that patients with an increased lung bacterial and fungal burden had a lower incidence of liberation from invasive mechanical ventilation and higher mortality. The bacterial and fungal burdens were correlated with alveolar proinflammatory cytokines and lung microbiota community composition was associated with successful extubation. Our findings suggest that lung microbiota are related to nonresolving ARDS in COVID-19, and represent an important contributor to heterogeneity in clinical outcomes of COVID-19–related ARDS.
The present study validates earlier studies in ARDS and COVID-19, as an increased pulmonary bacterial burden has been associated with fewer ventilator-free days and higher mortality (12, 17, 23). Although the importance of pulmonary bacterial burden has now been demonstrated across cohorts irrespective of regional and temporal differences, this is the first study to demonstrate that a higher pulmonary fungal burden is also associated with alveolar inflammation and nonresolving ARDS, highlighting the clinical relevance of fungal lung microbiota.
Lung microbiota alterations might affect host immune responses and increase alveolar inflammation, independent of acute infections (15–17). For example, lung microbiota composition is linked with variation in TNF-α, and microbial factors may activate inflammasomes resulting in IL-1β release (11, 39, 41). Supporting this immunomodulatory role of lung microbiota, we indeed found that lung bacterial and fungal burden correlated with cytokines involved in inflammasome activation and markers of alveolar inflammation (e.g., TNF-α, IL-6, and IL-1β) that have been implicated in ARDS development and are important features of severe COVID-19 (6, 37–40). Although our findings suggest that changes in the abundance of lung microbiota are linked to alveolar inflammation, lung injury in turn alters the lung microbiome (42). Lung microbiota could thus also be an innocent bystander in the pathogenesis of COVID-19–related ARDS. Moreover, the lower incidence (i.e., SHR) of extubation in patients with an increased bacterial burden in this study could be influenced by their higher risk of mortality because the SHR includes both patients without any event and those who had the competing event (43, 44). Nevertheless, our study is the first to reveal that the pulmonary fungal burden is associated with clinical outcomes of COVID-19–associated ARDS, and we confirm the role of bacterial lung microbiota dysbiosis in ARDS and COVID-19 (11, 12, 15, 17, 23).
We found that lung microbiota alterations correspond with secondary bacterial pulmonary infections (defined as a positive microbiologic BAL culture). Yet, the association between lung bacterial burden and clinical outcomes remained significant when controlled for several potential confounders, including the presence of secondary bacterial pneumonia and antibiotic exposure. Similarly, the fungal burden was associated with clinical outcomes independent of the presence of CAPA, which accentuates the difficulties associated with classifying aspergillosis (45, 46). Furthermore, these findings suggest that culture-independent techniques can be used to identify clinically relevant bacteria in BAL samples. Conventional microbiologic cultures take up to 48–72 hours and often fail to identify causative pathogens. Novel sequencing-based tests could help overcome these challenges (36, 47).
This study has several shortcomings. First, although clinical outcomes in our cohort were roughly comparable to others (48), the extremes of disease severity could have been underrepresented; the most critically ill patients conceivably could not undergo bronchoscopy safely, and patients with mild disease were extubated before sampling. This is reflected in the mortality rate of this cohort, which is higher than anticipated for an unselected ICU population (49). Consequently, our findings provide insights into host-microbiota interactions in COVID-19–associated ARDS, rather than offering a model to predict clinical outcomes. Second, we used cell-free BAL fluid, thereby selectively excluding cell-associated bacteria and reducing the microbial burden (50). Although the microbial signal in our samples was distinct from negative control specimens, we found some overlap with these negative controls, suggesting potential contamination. Third, 16S rRNA gene sequencing provides limited taxonomic resolution at species level and no insight in nonbacterial communities. Using metagenomic sequencing could reveal which fungi are responsible for differences in fungal burden and provide improved characterization of bacteria involved in secondary pulmonary infections. Finally, although the relation between clinical outcomes and lung microbiota remained significant in multivariable models that controlled for confounders, residual confounding cannot be excluded. Moreover, owing to the observational design of our study, we could not establish a causal relation. It thus remains to be determined by which mechanism lung microbiota affect clinical outcomes and how this knowledge could be used as a therapeutic target in COVID-19–related ARDS.
In conclusion, critically ill patients with COVID-19 with increased lung bacterial and fungal burden had higher mortality and lower incidences of liberation from invasive mechanical ventilation. Lung microbiota are related to nonresolving ARDS in COVID-19 and represent an important contributor to heterogeneity in COVID-19–related ARDS.
Acknowledgments
Acknowledgment
The authors thank all the patients and their families who participated in this study. They also thank Jorn Hartman and Theodorus B.M. Hakvoort for their expert technical assistance with the workup of the microbiota samples.
ArtDECO Consortium Members: Amsterdam University Medical Centers, Amsterdam, the Netherlands: Esther J. Nossent, JanWillem Duitman, Anno Saris, Heder De Vries, Lilian J. Meijboom, Lieuwe D.J. Bos, Siebe G. Blok, Alex R. Schuurman, Tom D.Y. Reijnders, Juan J. Garcia Vallejo, Hetty Bontkes, Alexander P.J. Vlaar, W. Joost Wiersinga, René Lutter, Tom van der Poll, Harm Jan Bogaard, and Leo Heunks.
Amsterdam UMC COVID-19 Biobank Study Group Members: Sanne de Bruin, Alex R. Schuurman, Rutger Koning, Michiel A. van Agtmael, Anne G. Algera, Frank E.H.P. van Baarle, Diane J.C. Bax, Martijn Beudel, Harm Jan Bogaard, Marije Bomers, Lieuwe D.J. Bos, Michela Botta, Justin de Brabander, Godelieve J. de Bree, Marianne Bugiani, Esther B. Bulle, Osoul Chouchane, Alex P.M. Cloherty, Paul E. Elbers, Lucas M. Fleuren, Suzanne E. Geerlings, Bart F. Geerts, Theo B.H. Geijtenbeek, Armand R.J. Girbes, Bram Goorhuis, Martin P. Grobusch, Florianne M.J. Hafkamp, Laura A. Hagens, Jörg Hamann, Vanessa C. Harris, Robert Hemke, Sabine M. Hermans, Leo M.A. Heunks, Markus W. Hollmann, Janneke Horn, Joppe W. Hovius, Menno D. de Jong, Niels van Mourik, Jeannine F Nellen, Esther J. Nossent, Frederique Paulus, Tom D.Y. Reijnders, Edgar Peters, Tom van der Poll, Bennedikt Preckel, S. Jorinde Raasveld, Michiel Schinkel, Marcus J. Schultz, Kim Sigaloff, Marleen A. Slim, Marry R. Smit, Cornelis S. Stijnis, Willemke Stilma, Charlotte E. Teunissen, Patrick Thoral, Anissa M. Tsonas, Marc van der Valk, Denise P. Veelo, Heder de Vries, Michèle van Vugt, Dorien Wouters, A.H. Zwinderman, Matthijs C. Brouwer, W. Joost Wiersinga, Alexander P.J. Vlaar, Diederik van de Beek.
Footnotes
A complete list of ArtDECO Consortium and Amsterdam UMC COVID-19 Biobank Study Group members may be found before the beginning of the References.
Supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organization for Scientific Research) under VIDI grant 91716475 to W.J.W. and VENI grant 016.1860.046 to J.D.; and by an Amsterdam University Medical Centers Ph.D. scholarship to R.F.J.K. and fellowship to L.D.J.B.
Author Contributions: Conception and design: R.F.J.K., J.d.B., L.S.B., E.J.N., L.M.A.H., A.P.J.V., P.I.B., T.v.d.P., J.D., L.D.J.B., and W.J.W. Processing of specimens and generation of data: R.F.J.K., J.d.B., L.S.B., and J.J.B. Analysis and interpretation of data: R.F.J.K., L.D.J.B., and W.J.W. Drafting and revision of manuscript: R.F.J.K., J.d.B., L.S.B., J.J.B., E.J.N., L.M.A.H., A.P.J.V., P.I.B., T.v.d.P., J.D., L.D.J.B., and W.J.W. Final approval of the manuscript: R.F.J.K., J.d.B., L.S.B., J.J.B., E.J.N., L.M.A.H., A.P.J.V., P.I.B., T.v.d.P., J.D., L.D.J.B., and W.J.W.
Data sharing statement: Sequence data have been deposited in the European Nucleotide Archive (accession number PRJEB50057). Original code used for analysis is publicly available at github.com/rfjkullberg/COVID_lung_microbiome.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202202-0274OC May 26, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
Contributor Information
on behalf of the ArtDECO Consortium and the Amsterdam UMC COVID-19 Biobank Study Group:
Anno Saris, Heder De Vries, Lilian J. Meijboom, Siebe G. Blok, Alex R. Schuurman, Tom D.Y. Reijnders, Juan J. Garcia Vallejo, Hetty Bontkes, René Lutter, Harm Jan Bogaard, S. de Bruin, R. Koning, M.A. van Agtmael, A.G. Algera, F.E.H.P. van Baarle, D.J.C. Bax, M. Beudel, M. Bomers, M. Botta, G.J. de Bree, M. Bugiani, E.B. Bulle, O. Chouchane, A.P.M. Cloherty, P.E. Elbers, L.M. Fleuren, S.E. Geerlings, B.F. Geerts, T.B.H. Geijtenbeek, A.R.J. Girbes, A. Goorhuis, M.P. Grobusch, F.M.J. Hafkamp, L.A. Hagens, J. Hamann, V.C. Harris, R. Hemke, S.M. Hermans, M.W. Hollmann, J. Horn, J.W. Hovius, M.D. de Jong, N. van Mourik, J.F Nellen, F. Paulus, E. Peters, B. Preckel, S.J. Raasveld, M. Schinkel, M.J. Schultz, K. Sigaloff, M.R. Smit, C. Stijnis, W. Stilma, C.E. Teunissen, P. Thoral, A.M. Tsonas, M. van der Valk, D.P. Veelo, M. van Vugt, D. Wouters, A.H. Zwinderman, M.C. Brouwer, and D. van de Beek
References
- 1. Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA . 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 2. Karagiannidis C, Windisch W, McAuley DF, Welte T, Busse R. Major differences in ICU admissions during the first and second COVID-19 wave in Germany. Lancet Respir Med . 2021;9:e47–e48. doi: 10.1016/S2213-2600(21)00101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Torres Acosta MA, Singer BD. Pathogenesis of COVID-19-induced ARDS: implications for an ageing population. Eur Respir J . 2020;56:2002049. doi: 10.1183/13993003.02049-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Botta M, Tsonas AM, Pillay J, Boers LS, Algera AG, Bos LDJ, et al. PRoVENT-COVID Collaborative Group Ventilation management and clinical outcomes in invasively ventilated patients with COVID-19 (PRoVENT-COVID): a national, multicentre, observational cohort study. Lancet Respir Med . 2021;9:139–148. doi: 10.1016/S2213-2600(20)30459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leisman DE, Mehta A, Thompson BT, Charland NC, Gonye ALK, Gushterova I, et al. Alveolar, endothelial, and organ injury marker dynamics in severe COVID-19. Am J Respir Crit Care Med . 2022;205:507–519. doi: 10.1164/rccm.202106-1514OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Osuchowski MF, Winkler MS, Skirecki T, Cajander S, Shankar-Hari M, Lachmann G, et al. The COVID-19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir Med . 2021;9:622–642. doi: 10.1016/S2213-2600(21)00218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Bruin S, Bos LD, van Roon MA, Tuip-de Boer AM, Schuurman AR, Koel-Simmelinck MJA, et al. Amsterdam UMC COVID-19 Biobank Investigators Clinical features and prognostic factors in Covid-19: a prospective cohort study. EBioMedicine . 2021;67:103378. doi: 10.1016/j.ebiom.2021.103378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grant RA, Morales-Nebreda L, Markov NS, Swaminathan S, Querrey M, Guzman ER, et al. NU SCRIPT Study Investigators Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature . 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D’Agnillo F, Walters KA, Xiao Y, Sheng ZM, Scherler K, Park J, et al. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci Transl Med . 2021;13:eabj7790. doi: 10.1126/scitranslmed.abj7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saris A, Reijnders TDY, Nossent EJ, Schuurman AR, Verhoeff J, Asten SV, et al. ArtDECO consortium and the Amsterdam UMC COVID study group Distinct cellular immune profiles in the airways and blood of critically ill patients with COVID-19. Thorax . 2021;76:1010–1019. doi: 10.1136/thoraxjnl-2020-216256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol . 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dickson RP, Schultz MJ, van der Poll T, Schouten LR, Falkowski NR, Luth JE, et al. Biomarker Analysis in Septic ICU Patients (BASIC) Consortium Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med . 2020;201:555–563. doi: 10.1164/rccm.201907-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O’Dwyer DN, Ashley SL, Gurczynski SJ, Xia M, Wilke C, Falkowski NR, et al. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am J Respir Crit Care Med . 2019;199:1127–1138. doi: 10.1164/rccm.201809-1650OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Combs MP, Wheeler DS, Luth JE, Falkowski NR, Walker NM, Erb-Downward JR, et al. Lung microbiota predict chronic rejection in healthy lung transplant recipients: a prospective cohort study. Lancet Respir Med . 2021;9:601–612. doi: 10.1016/S2213-2600(20)30405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kitsios GD, Yang H, Yang L, Qin S, Fitch A, Wang XH, et al. Respiratory tract dysbiosis is associated with worse outcomes in mechanically ventilated patients. Am J Respir Crit Care Med . 2020;202:1666–1677. doi: 10.1164/rccm.201912-2441OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol . 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kyo M, Nishioka K, Nakaya T, Kida Y, Tanabe Y, Ohshimo S, et al. Unique patterns of lower respiratory tract microbiota are associated with inflammation and hospital mortality in acute respiratory distress syndrome. Respir Res . 2019;20:246. doi: 10.1186/s12931-019-1203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Merenstein C, Liang G, Whiteside SA, Cobián-Güemes AG, Merlino MS, Taylor LJ, et al. Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. MBio . 2021;12:e0177721. doi: 10.1128/mBio.01777-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lloréns-Rico V, Gregory AC, Van Weyenbergh J, Jansen S, Van Buyten T, Qian J, et al. CONTAGIOUS collaborators Clinical practices underlie COVID-19 patient respiratory microbiome composition and its interactions with the host. Nat Commun . 2021;12:6243. doi: 10.1038/s41467-021-26500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ren L, Wang Y, Zhong J, Li X, Xiao Y, Li J, et al. Dynamics of the upper respiratory tract microbiota and its association with mortality in COVID-19. Am J Respir Crit Care Med . 2021;204:1379–1390. doi: 10.1164/rccm.202103-0814OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio . 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Pascale G, De Maio F, Carelli S, De Angelis G, Cacaci M, Montini L, et al. Staphylococcus aureus ventilator-associated pneumonia in patients with COVID-19: clinical features and potential inference with lung dysbiosis. Crit Care . 2021;25:197. doi: 10.1186/s13054-021-03623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sulaiman I, Chung M, Angel L, Tsay JJ, Wu BG, Yeung ST, et al. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat Microbiol . 2021;6:1245–1258. doi: 10.1038/s41564-021-00961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, et al. ARDS Definition Task Force Acute respiratory distress syndrome: the Berlin Definition. JAMA . 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 25. Nossent EJ, Schuurman AR, Reijnders TDY, Saris A, Jongerius I, Blok SG, et al. Pulmonary procoagulant and innate immune responses in critically ill COVID-19 patients. Front Immunol . 2021;12:664209. doi: 10.3389/fimmu.2021.664209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haak BW, Brands X, Davids M, Peters-Sengers H, Kullberg RFJ, van Houdt R, et al. Bacterial and viral respiratory tract microbiota and host characteristics in adults with lower respiratory tract infections: a case-control study. Clin Infect Dis . 2022;74:776–784. doi: 10.1093/cid/ciab568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haak BW, Argelaguet R, Kinsella CM, Kullberg RFJ, Lankelma JM, Deijs M, et al. Integrative transkingdom analysis of the gut microbiome in antibiotic perturbation and critical illness. mSystems . 2021;6:e01148-20. doi: 10.1128/mSystems.01148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yehya N, Harhay MO, Curley MAQ, Schoenfeld DA, Reeder RW. Reappraisal of ventilator-free days in critical care research. Am J Respir Crit Care Med . 2019;200:828–836. doi: 10.1164/rccm.201810-2050CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koehler P, Bassetti M, Chakrabarti A, Chen SCA, Colombo AL, Hoenigl M, et al. European Confederation of Medical Mycology; International Society for Human Animal Mycology; Asia Fungal Working Group; INFOCUS LATAM/ISHAM Working Group; ISHAM Pan Africa Mycology Working Group; European Society for Clinical Microbiology; Infectious Diseases Fungal Infection Study Group; ESCMID Study Group for Infections in Critically Ill Patients; Interregional Association of Clinical Microbiology and Antimicrobial Chemotherapy; Medical Mycology Society of Nigeria; Medical Mycology Society of China Medicine Education Association; Infectious Diseases Working Party of the German Society for Haematology and Medical Oncology; Association of Medical Microbiology; Infectious Disease Canada Defining and managing COVID-19-associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance Lancet Infect Dis 2021. 21 e149 e162 33333012 [Google Scholar]
- 30. van Oort PM, Nijsen TM, White IR, Knobel HH, Felton T, Rattray N, et al. BreathDx Consortium Untargeted molecular analysis of exhaled breath as a diagnostic test for ventilator-associated lower respiratory tract infections (BreathDx) Thorax . 2021;77:79–81. doi: 10.1136/thoraxjnl-2021-217362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Torres A, Niederman MS, Chastre J, Ewig S, Fernandez-Vandellos P, Hanberger H, et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: Guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociación Latinoamericana del Tórax (ALAT) Eur Respir J . 2017;50:1700582. doi: 10.1183/13993003.00582-2017. [DOI] [PubMed] [Google Scholar]
- 32. Pickens CO, Gao CA, Cuttica MJ, Smith SB, Pesce LL, Grant RA, et al. NU COVID Investigators Bacterial superinfection pneumonia in patients mechanically ventilated for COVID-19 pneumonia. Am J Respir Crit Care Med . 2021;204:921–932. doi: 10.1164/rccm.202106-1354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao J, Murray S, Lipuma JJ. Modeling the impact of antibiotic exposure on human microbiota. Sci Rep . 2014;4:4345. doi: 10.1038/srep04345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol . 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med. 2014;2:238–246. doi: 10.1016/S2213-2600(14)70028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langelier C, Kalantar KL, Moazed F, Wilson MR, Crawford ED, Deiss T, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci USA . 2018;115:E12353–E12362. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meduri GU, Kohler G, Headley S, Tolley E, Stentz F, Postlethwaite A. Inflammatory cytokines in the BAL of patients with ARDS. Persistent elevation over time predicts poor outcome. Chest . 1995;108:1303–1314. doi: 10.1378/chest.108.5.1303. [DOI] [PubMed] [Google Scholar]
- 38. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med . 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 39. Peukert K, Fox M, Schulz S, Feuerborn C, Frede S, Putensen C, et al. Inhibition of caspase-1 with tetracycline ameliorates acute lung injury. Am J Respir Crit Care Med . 2021;204:53–63. doi: 10.1164/rccm.202005-1916OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Madtes DK, Rubenfeld G, Klima LD, Milberg JA, Steinberg KP, Martin TR, et al. Elevated transforming growth factor-alpha levels in bronchoalveolar lavage fluid of patients with acute respiratory distress syndrome. Am J Respir Crit Care Med . 1998;158:424–430. doi: 10.1164/ajrccm.158.2.9711112. [DOI] [PubMed] [Google Scholar]
- 41. Ashley SL, Sjoding MW, Popova AP, Cui TX, Hoostal MJ, Schmidt TM, et al. Lung and gut microbiota are altered by hyperoxia and contribute to oxygen-induced lung injury in mice. Sci Transl Med . 2020;12:eaau9959. doi: 10.1126/scitranslmed.aau9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poroyko V, Meng F, Meliton A, Afonyushkin T, Ulanov A, Semenyuk E, et al. Alterations of lung microbiota in a mouse model of LPS-induced lung injury. Am J Physiol Lung Cell Mol Physiol . 2015;309:L76–L83. doi: 10.1152/ajplung.00061.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Austin PC, Lee DS, Fine JP. Introduction to the analysis of survival data in the presence of competing risks. Circulation . 2016;133:601–609. doi: 10.1161/CIRCULATIONAHA.115.017719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lau B, Cole SR, Gange SJ. Competing risk regression models for epidemiologic data. Am J Epidemiol . 2009;170:244–256. doi: 10.1093/aje/kwp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Arkel ALE, Rijpstra TA, Belderbos HNA, van Wijngaarden P, Verweij PE, Bentvelsen RG. COVID-19-associated pulmonary aspergillosis. Am J Respir Crit Care Med . 2020;202:132–135. doi: 10.1164/rccm.202004-1038LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fekkar A, Poignon C, Blaize M, Lampros A. Fungal infection during COVID-19: does Aspergillus mean secondary invasive aspergillosis? Am J Respir Crit Care Med . 2020;202:902–903. doi: 10.1164/rccm.202005-1945LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pendleton KM, Erb-Downward JR, Bao Y, Branton WR, Falkowski NR, Newton DW, et al. Rapid pathogen identification in bacterial pneumonia using real-time metagenomics. Am J Respir Crit Care Med . 2017;196:1610–1612. doi: 10.1164/rccm.201703-0537LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lim ZJ, Subramaniam A, Ponnapa Reddy M, Blecher G, Kadam U, Afroz A, et al. Case fatality rates for patients with COVID-19 requiring invasive mechanical ventilation. A meta-analysis. Am J Respir Crit Care Med . 2021;203:54–66. doi: 10.1164/rccm.202006-2405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carbonell R, Urgelés S, Rodríguez A, Bodí M, Martín-Loeches I, Solé-Violán J, et al. COVID-19 SEMICYUC Working Group Mortality comparison between the first and second/third waves among 3,795 critical COVID-19 patients with pneumonia admitted to the ICU: a multicentre retrospective cohort study. Lancet Reg Health Eur . 2021;11:100243. doi: 10.1016/j.lanepe.2021.100243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, et al. Cell-associated bacteria in the human lung microbiome. Microbiome . 2014;2:28. doi: 10.1186/2049-2618-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]


