For patients with coronavirus disease (COVID-19) that require management in an ICU, mortality varies between 30–70% and keystone treatment still relies on supportive measures such as invasive mechanical ventilation, vasopressor administration, and renal replacement therapy (1–6). Among these critically ill patients, there is significant heterogeneity in the natural history of the disease process varying from patients requiring transient ventilatory support, others developing thrombosis, cardiovascular complications and, frequently, prolonged mechanical ventilation, and death. Despite a growing understanding of the pathophysiological derangements that occur during a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, the reasons for the heterogeneous evolution among the most severe cases are not well understood. Multiple investigations have utilized a case-control design comparing patients infected with SARS-CoV-2 with differing degrees of severity (e.g., hospitalized patients versus not hospitalized or those admitted to an ICU versus those in the general wards) or with noninfected individuals. Although significant knowledge has been gained from these investigations, they do not help in our understanding of the heterogeneous evolution among critically ill patients with COVID-19. Although there is increased interest in molecular profiling, or endotyping, to uncover biomarkers that may help us understand patients’ heterogeneity, few studies have focused on applying this to a cohort with similar disease severity (e.g., exclusively critically ill patients) and with longitudinal follow-up. Furthermore, most investigations have focused on noninvasive assessments, such as blood, to develop biomarkers that may predict disease outcome. Although that for sure would be quite convenient, the samples we really need to study are those collected from the primary site of the disease: the lung (7).
In this issue of the Journal, Kullberg and colleagues (pp. 846–856) studied how the lower airway microbiome on 114 critically ill, mechanically ventilated patients infected with SARS-CoV-2 can be associated with poor clinical outcome (8). In these patients, BAL samples were obtained after days or weeks from initial intubation (median time from intubation to sample collection was 9 d) and in 32 patients they were able to analyze follow-up samples. The authors evaluated for microbial signatures associated with successful liberation from mechanical ventilation by day 60 after intubation (versus deceased or intubated >60 d). To do that, the lower airway microbiota was characterized by 16S rRNA gene sequencing and bacterial and fungal load were measured by qPCR targeting the 16S and 18S rRNA genes, respectively. In line with a prior study of >140 critically ill patients with COVID-19 (9), poor clinical outcome was associated with higher bacterial load. Importantly, the authors showed that increased fungal load was also associated with poor clinical outcome. The authors also found that some inflammatory markers measured in these samples correlated with bacteria/fungal load, such as tumor necrosis factor α. Moreover, the study aimed to evaluate if secondary bacterial pneumonia, defined here as BAL culture positivity (which occurred in 22% of cases), was associated with changes in the microbiota. They found that BAL culture positivity was indeed associated with increased bacterial load and there was some concordance between the isolated bacterial strain and the increased relative abundance in the 16S rRNA gene sequencing data.
In the context of other investigations on lower airway samples from critically ill patients with COVID-19, these results solidify the notion that it is not just uncontrollable SARS-CoV-2 infection that kills patients with COVID-19, though that’s likely one big part of it. In a different study using metatranscriptomic and metagenomic approaches on bronchoalveolar samples from 142 critically ill patients, mortality was associated with increased lower airway viral load and a decrease in anti–SARS-CoV-2 specific IgG levels (9). However, the association of poor outcome with increased microbial load, other than SARS-CoV-2, was found in both studies supporting that other microbes may be contributing to the disease process. Alternatively, the dynamic change in microbes other than SARS-CoV-2 may uncover alterations in the host immune response and serve as biomarkers that may allow us to risk-stratify patients (Figure 1A).
Figure 1.
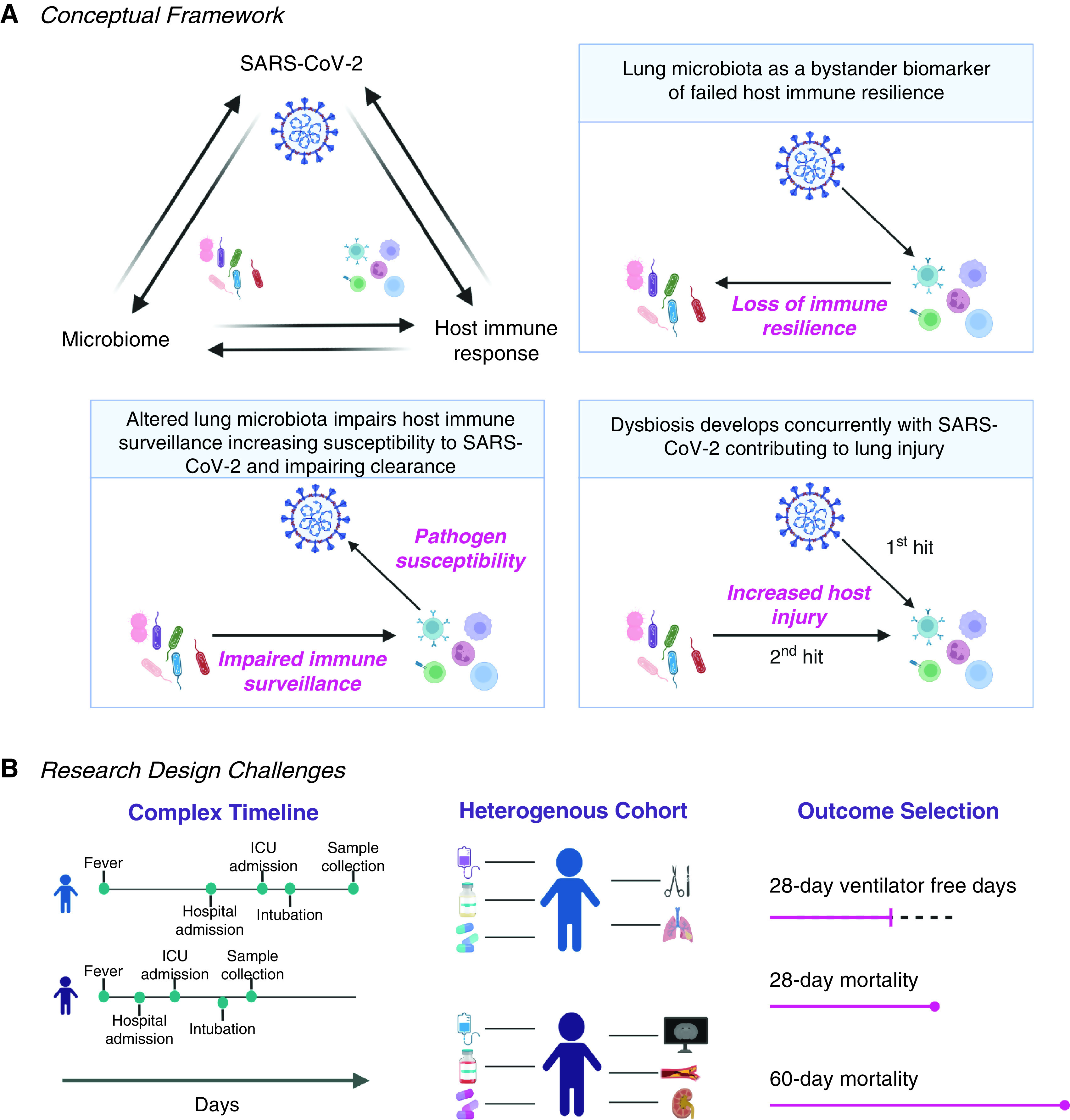
(A) Schematic representation of possible causal relationships between the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the lower airway microbiome, and the host immune system in the setting of coronavirus disease (COVID-19) infection. (B) Important considerations and challenges for the design of studies evaluating biomarkers for critically ill patients with COVID-19 infection.
The results of the paper now published in the Blue Journal highlights several challenges of conducting research in critically ill patients (Figure 1B). First, ideal biomarkers should be developed at early time points where interventions are more likely to affect the course of disease and should be able help us understand temporal changes in a patient’s conditions. In the current study, for the most part, samples were obtained on the second week of mechanical ventilation or after. Considering that, at the beginning of the pandemic, many professional societies recommended against performing bronchoscopic procedures in patients with COVID-19 under the concern for transmission to healthcare workers (10), having collected this many samples is already commendable. However, we need to do better. To develop clinically usable biomarkers among critically ill patients, samples will need to be obtained at early time points, e.g., upon admission to the ICU or soon after intubation, and longitudinally. In the study of critically ill patients with SARS-CoV-2, another challenge is the correct selection of time-dependent outcomes as traditional “critical care outcomes” such as 28-day mortality or 28-day ventilator free days may not appropriately capture the complexity of clinical trajectories in this disease. As we learned in this pandemic, critically ill patients with COVID-19 commonly require lengthy mechanical ventilation and a large proportion of these patients will succumb way after the first month. This has led to the recommendation of 60 days from hospitalization as a landmark for evaluating mortality (11). To complicate things further, the time from a patient’s onset of symptoms to hospitalization, or from hospitalization to ICU admission (or intubation), can vary greatly. As shown in the current paper and others, individuals can spend only a few days to multiple weeks in the hospital before being admitted to the ICU (11). This discrepancy becomes particularly relevant when defining the outcome of interest, as one would hope that biomarkers predict outcome among patients with similar degree of severity (in this case, when respiratory failure is declared and intubation is needed) and with the time to outcome adjusted for when measurements are made. Here, the authors evaluated their biomarkers’ accuracy to predict 60-day extubation success (versus extubation failure or mortality) from intubation and, in secondary analyses, from sample collection. This highlights how establishing proper clinical outcomes for group comparisons remains a significant challenge. Another major challenge is being able to properly capture time-dependent confounders in these complex patients. One example is the use of antibiotics and immunosuppression, broadly used in this cohort and of particular interest given the effects on the lower airway microbiome. Although there has been some standardization in care provided to these patients since the start of the pandemic (take, for example, how steroid use has changed), the use of many forms of immunosuppression and antibiotics differ widely and require careful adjustment and, ideally, longitudinal investigations. For example, here the authors included antibiotic exposure as part of their model but we cannot evaluate the individual effect of different antibiotics. Finally, it is particularly challenging to identify secondary bacterial pneumonias in these critically ill patients in the setting of viral acute respiratory distress syndrome (12, 13). Although the authors here used BAL culture positivity to identify those with potential VAPs, an important next step is to use other clinical data available (e.g., worsening oxygenation, imaging) as well.
Bottom line, we need more studies including early time points, longitudinal samples, and detailed consideration of all possible confounders. Why? Disentangling this web of microbial, pathogen, and host dynamics is the only way we will be able to risk-stratify patients and identify treatable traits (14).
Footnotes
This work was supported in part by R37 CA244775 (L.N.S., NCI/NIH), and Stony Wold Herbert Inc. Foundation (C.R.B.).
Originally Published in Press as DOI: 10.1164/rccm.202206-1074ED on June 13, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Arentz M, Yim E, Klaff L, Lokhandwala S, Riedo FX, Chong M, et al. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA . 2020;323:1612–1614. doi: 10.1001/jama.2020.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhatraju PK, Ghassemieh BJ, Nichols M, Kim R, Jerome KR, Nalla AK, et al. Covid-19 in critically ill patients in the Seattle region — case series. N Engl J Med . 2020;382:2012–2022. doi: 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cummings MJ, Baldwin MR, Abrams D, Jacobson SD, Meyer BJ, Balough EM, et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet . 2020;395:1763–1770. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, et al. COVID-19 Lombardy ICU Network Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. JAMA . 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, et al. the Northwell COVID-19 Research Consortium Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA . 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med . 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dickson RP. Lung microbiota and COVID-19 severity. Nat Microbiol . 2021;6:1217–1218. doi: 10.1038/s41564-021-00969-x. [DOI] [PubMed] [Google Scholar]
- 8. Kullberg RFJ, de Brabander J, Boers LS, Biemond JJ, Nossent EJ, Heunks LMA, et al. ArtDECO consortium and the Amsterdam UMC COVID-19 Biobank Study Group Lung microbiota of critically ill COVID-19 patients are associated with non-resolving acute respiratory distress syndrome. Am J Respir Crit Care Med . 2022;206:846–856. doi: 10.1164/rccm.202202-0274OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sulaiman I, Chung M, Angel L, Tsay JJ, Wu BG, Yeung ST, et al. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat Microbiol . 2021;6:1245–1258. doi: 10.1038/s41564-021-00961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wahidi MM, Lamb C, Murgu S, Musani A, Shojaee S, Sachdeva A, et al. American Association for Bronchology and Interventional Pulmonology (AABIP) statement on the use of bronchoscopy and respiratory specimen collection in patients with suspected or confirmed COVID-19 infection. J Bronchology Interv Pulmonol . 2020;27:e52–e54. doi: 10.1097/LBR.0000000000000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis . 2020;20:e192–e197. doi: 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bell RC, Coalson JJ, Smith JD, Johanson WG., Jr Multiple organ system failure and infection in adult respiratory distress syndrome. Ann Intern Med . 1983;99:293–298. doi: 10.7326/0003-4819-99-3-293. [DOI] [PubMed] [Google Scholar]
- 13. Koenig SM, Truwit JD. Ventilator-associated pneumonia: diagnosis, treatment, and prevention. Clin Microbiol Rev . 2006;19:637–657. doi: 10.1128/CMR.00051-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bos LDJ, Laffey JG, Ware LB, Heijnen NFL, Sinha P, Patel B, et al. Towards a biological definition of ARDS: are treatable traits the solution? Intensive Care Med Exp . 2022;10:8. doi: 10.1186/s40635-022-00435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]


