Abstract
Prime editing (PE) is a precision gene editing technology that enables the programmable installation of substitutions, insertions, and deletions in cells and animals without requiring double-strand DNA breaks (DSBs). The mechanism of prime editing makes it less dependent on cellular replication and endogenous DNA repair than HDR-based approaches, and its ability to precisely install edits without creating DSBs minimizes indels and other undesired outcomes. The capabilities of prime editing have also expanded since its original publication. Enhanced prime editing systems, PE4 and PE5, manipulate DNA repair pathways to increase prime editing efficiency and reduce indels. Other advances that improve prime editing efficiency include engineered pegRNAs (epegRNAs), which include a structured RNA motif to stabilize and protect pegRNA 3′ ends, and the PEmax architecture, which improves editor expression and nuclear localization. New applications such as twin prime editing (twinPE) can precisely insert or delete hundreds of base pairs of DNA and can be used in tandem with recombinases to achieve gene-sized (>5 kb) insertions and inversions. Achieving optimal prime editing requires careful experimental design, and the large number of parameters that influence prime editing outcomes can be daunting. This protocol describes current best practices for conducting prime editing and twinPE experiments and describes the design and optimization of pegRNAs. We also offer guidelines for how to select the proper PE system (PE1 to PE5, and twinPE) for a given application. Finally, we provide detailed instructions on how to perform prime editing in mammalian cells. Compared to other procedures for editing human cells, prime editing offers greater precision and versatility, and can be completed within 2–4 weeks.
Keywords: Genome editing, CRISPR, Cas9, prime editing
Editorial Summary:
This protocol describes prime editing and twinPE experiments, and the design and optimization of pegRNAs. The Authors provide guidelines for selecting the proper PE system for a given application, and instructions on how to perform prime editing in mammalian cells.
Introduction
CRISPR-Cas systems enable manipulation of genes in living systems with unprecedented speed, convenience, and programmability1,2. CRISPR-derived editing agents for basic research have revolutionized our understanding of biological systems, and have also been used ex vivo and in vivo to treat patients with sickle cell disease, β-thalassemia, and transthyretin amyloidosis3,4. The reliance of early gene editing techniques on double-strand DNA breaks (DSBs), however, limits the types of edits that can be made with programmable nucleases such as CRISPR-Cas9 primarily to those that disrupt or delete genes. In addition, DSBs can also result in a variety of undesirable outcomes, such as unwanted mixtures of insertions and deletions (indels) at the target site, translocations5–8, large deletions9,10, aneuploidy11,12, chromothrypsis9,13, and p53 activation that can enrich oncogenic cells14. While homology-directed repair (HDR) using DSBs and donor DNA templates has been successfully used to correct, rather than disrupt, mutations in cell types including stem cells and T cells15–17, HDR-mediated correction has proven inefficient in most therapeutically relevant cell types due to the cell-cycle dependence of cellular machinery required for HDR.
The difficulties inherent in correcting genes using nucleases limits our ability to study and potentially treat genetic diseases, most of which require targeted gene correction, rather than gene disruption, for treatment. These considerations stimulated the development of precision programmable gene correction technologies that do not require cutting the DNA double helix. One such example of a DSB-free gene editing method that can mediate gene correction, in addition to gene disruption, is base editing. Cytosine base editors (CBEs) and adenine base editors (ABEs) can precisely install C•G-to-T•A mutations and A•T-to-G•C mutations, respectively, without requiring DSBs2,18–21. Base editors have been used both ex vivo and in vivo to rescue animal models of sickle cell disease22, Hutchinson-Gilford Progeria23, and several other genetic diseases24, but are limited to the installation of transition point mutations and, in some cases, C•G-to-G•C transversions25–29.
To further expand the scope of precise gene correction without requiring DSBs, we recently developed prime editing15. Prime editors (PEs) enable precise, highly versatile substitution, insertion, deletion, or combination edits without requiring DSBs15. The original prime editor, PE1, is composed of a Cas9(H840A) nickase fused to the M-MLV reverse transcriptase (RT) (Fig. 1) and uses a modified sgRNA called a prime editing guide RNA (pegRNA) (Fig. 2). A pegRNA possesses an additional 3′ extension containing a reverse transcription template (RTT), which encodes the desired edit, and a primer-binding site (PBS), which is complementary to the genomic target. Once delivered into a cell, the spacer of the pegRNA targets the prime editor protein to a specific target locus. The Cas9 domain then binds and nicks the target DNA, exposing a 3′ end. The PBS of the pegRNA then anneals to this 3′ end, and the RT domain of the prime editor uses the resulting DNA/RNA duplex as a substrate. The target DNA 3′ end serves as a primer, and the RT extends the flap, synthesizing the sequence encoded by the RTT of the pegRNA. The resulting newly synthesized DNA 3′ flap contains the desired edit (a substitution, insertion, deletion, or a combination thereof), followed by downstream homology. This downstream homology leads to flap equilibration and hybridization of the edited 3′ flap onto the unedited complementary target strand. Subsequent DNA repair, including the cell’s innate propensity to cleave 5′ DNA flaps, incorporates the edit into both target DNA strands (Fig. 1). The PE2 prime editor uses an engineered RT that contains five mutations that together strongly increase the efficiency of prime editing.
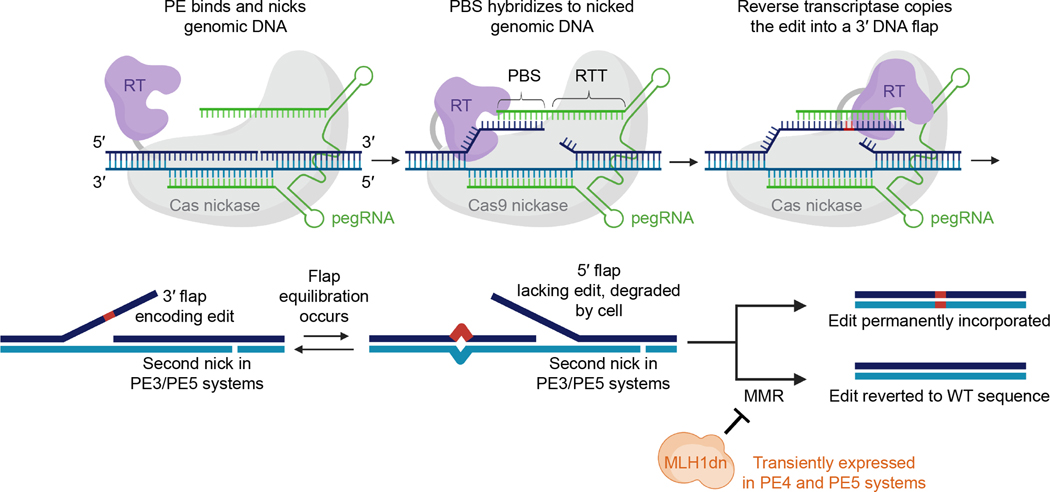
Figure 1. Mechanism of prime editing.
The steps shown above are the putative mechanism for prime editing using various editing systems and an unmodified pegRNA. Cas9 nickase (grey) is recruited to a target DNA site (blue) by a pegRNA (green) and nicks the target site to create a 3′ end of DNA. The primer binding site (PBS) of the pegRNA can then anneal to the genomic DNA flap. This duplex is recognized by a reverse transcriptase (purple), which reverse transcribes nucleotides extending from the target site 3′ end, copying the sequence encoded in the reverse transcription template (RTT) of the pegRNA. Reverse transcription produces a 3′ flap that contains the desired prime edit as well as downstream homology to the rest of the target DNA site. The 3′ flap equilibrates with the corresponding 5′ flap, which does not contain the desired edit. Cellular degradation of the 5′ flap, ligation of the edited 3′ flap into the genome, and repair of the complementary genomic DNA strand by DNA repair or replication results in stable installation of the edit. Prior to repair of the complementary strand, cellular mismatch repair (MMR) can revert the edit back to the unedited sequence. In the PE3 and PE5 systems, a second nick is installed in the complementary strand of DNA, ≥~50 bp away (and typically downstream) from the pegRNA-guided nick. This additional nick biases MMR in favor of editing. In the PE4 and PE5 systems, an engineered dominant-negative MLH1 mutant (MLH1dn, shown in orange) inhibits cellular mismatch repair and thus favors desired prime editing outcomes. This mechanism is based on data collected in previous publications15,30.
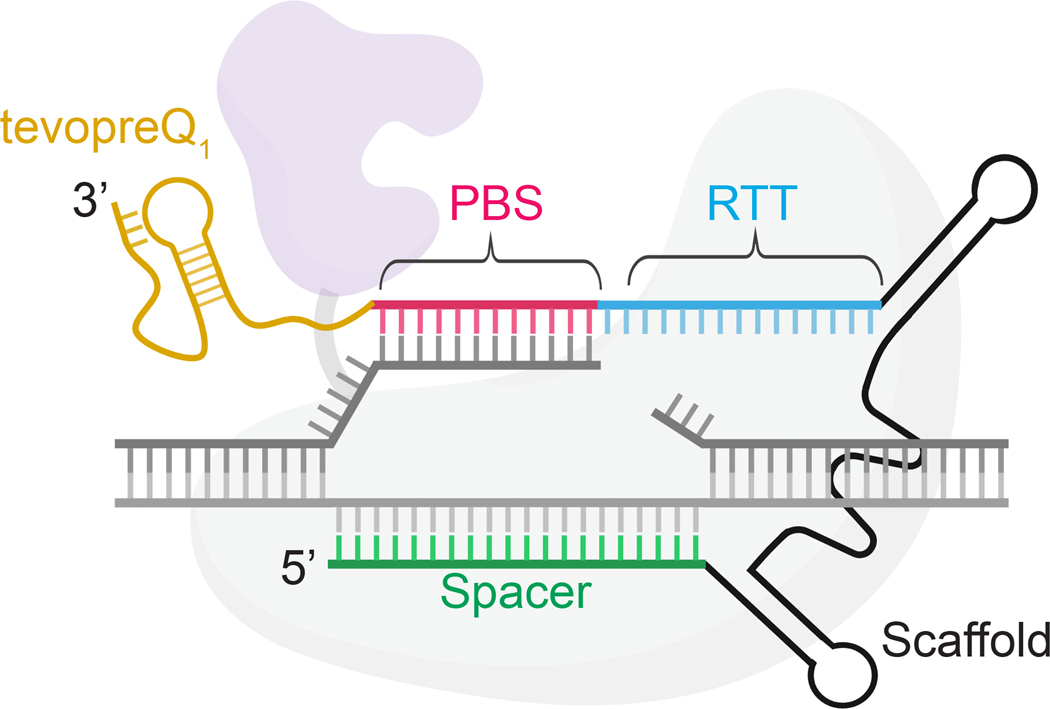
Figure 2. Architecture of an engineered prime editing guide RNA (epegRNA).
From 5′ to 3′, epegRNAs consist of a spacer (green), scaffold (black), RTT (reverse transcription template, shown in blue), PBS (primer binding site, shown in pink), and 3′ structural motif such as tevopreQ1 (shown in gold). The prime editor protein is shown in the background, with Cas9 in light grey and the reverse transcriptase shown in purple. The target genomic DNA is shown in grey, with the nicked and edited strand shown in dark grey and then complementary stand in light grey. The architecture of epegRNAs has been described in previous work31.
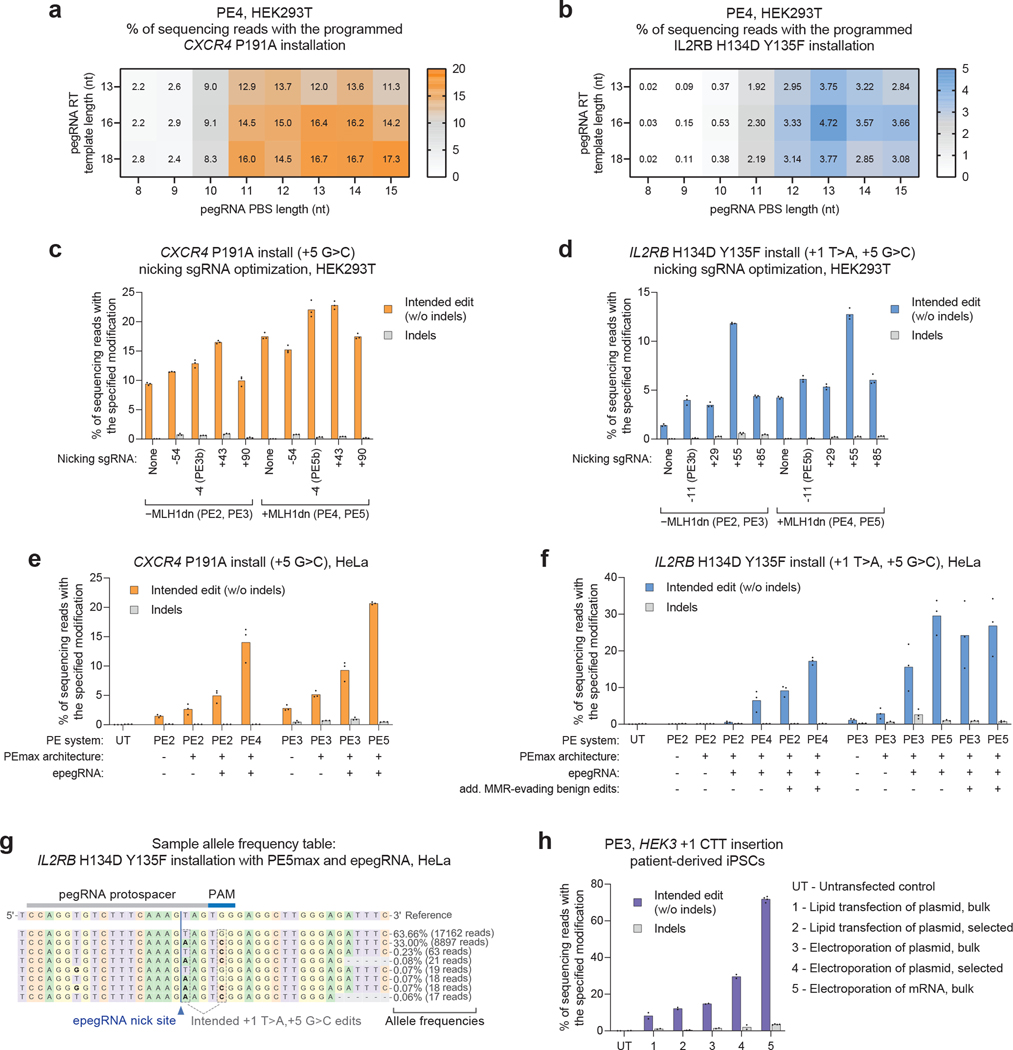
Prime editing intermediates are susceptible to cellular mismatch repair (MMR), which can reduce prime editing efficiency by reverting the edited DNA strand back to the starting sequence15,30. The PE3 system mitigates this possibility by adding an sgRNA that targets the editor to nick the non-edited strand of DNA. Because no 3′ extension is included on this additional sgRNA, a prime editor that engages this sgRNA only nicks the non-edited stand. Due to the nick-directed nature of eukaryotic MMR18, the additional nick biases outcomes towards replacement of the nicked non-edited strand using the edited strand as a template15. PE3 achieves higher editing efficiency than PE2, but typically results in more indel byproducts. Subsequent versions of prime editors, PE4 and PE5, transiently inhibit MMR to bias outcomes in favor of editing while also minimizing indels30 (described in the prime editing developments section below).
Compared to DSB-mediated genome editing techniques, prime editing offers a much higher editing:indel ratio and is less dependent on cellular repair pathways. Efficient prime editing has been demonstrated in many cell types, including primary cortical neurons, T cells, iPSCs, and patient-derived fibroblasts15,30,31. Additionally, because the desired edit is encoded in the pegRNA, delivery of an exogenous DNA template is not required, which simplifies basic research experiments and greatly facilitates in vivo delivery. Finally, off-target edits are minimized in prime editing. Cas9-dependent off target editing is much less frequent with prime editors than with Cas9 nuclease15,32–34, likely because prime editing requires three distinct DNA hybridization events with the spacer, PBS, and 3′ homology encoded by the pegRNA in order for productive editing to take place, and each event provides an opportunity to reject an off-target sequence. Additionally, three recent studies did not detect any Cas9-independent off targets from prime editing, as measured by clonal whole-genome sequencing of edited human stem cell-derived intestinal and liver organoids, embryonic stem cells, and rice plants33,35,36. Overall, prime editing offers versatile, efficient, and precise genome editing across many cell types with minimal off-target edits. This protocol details how to use prime editing in mammalian cells and how to choose a prime editing system that is well-matched for a given application.
Prime editing developments and comparisons with other methods
The mechanism of prime editing involves a complex series of events, each of which is influenced by the structure of the prime editor and pegRNA, as well as cellular factors. Since our initial report of prime editing, we and others have targeted several aspects of the PE system for improvement. When combined, these improvements are often additive, offering on average a 3.5-fold (in HEK293T cells) to 72-fold (in HeLa cells) increase in editing efficiency relative to originally published prime editing systems30,31. These improvements are particularly helpful when applying prime editing in vivo or in difficult-to-transfect cell types31,37. Various enhancements and their potential use cases are summarized below and in Table 1.
Table 1.
Use cases for various PE systems and modifications
| PE system | Uses |
|
PE1 Cas9 (H840A)–WT RT |
Not recommended; PE1 was the prototype prime editor from which PE2 was developed. |
|
PE2 Cas9(H840A)–engineered RT |
PE2 yields lower editing than PE3-5. However, PE2 may be preferred if: • Secondary nicks from PE3/PE5 generate an unacceptable frequency of indels, and long-term MLH1dn expression in the PE4/PE5 systems is not desired • If the application does not require optimized editing levels (i.e., creating a cell line), PE2 is the simplest and fastest method, as a nicking guide does not need to be optimized • If high editing efficiency is achieved without PE3-5 systems, for example due to the MMR-evading nature of the edit, or the addition of silent nearby mutations |
|
PE3 / PE3b PE2 + Additional nicking sgRNA (PE3b if nicking sgRNA protospacer overlaps with edit) |
PE3 and PE4 offer similar editing efficiencies; if PE3 does not generate substantial indels at the target locus and yields high editing efficiency, then it can serve as a good choice. Importantly, the relative editing of PE3 and PE4 varies by cell type. PE3 also provides the highest editing efficiency without inhibiting cellular MMR. Note: Several nicking sgRNAs (positioned both upstream and downstream of the edit) should be screened for optimal editing efficiency and a high editing:indel ratio. If an appropriate PAM exists, PE3b nicking sgRNAs should be screened as well and will usually provide the highest efficiencies and fewest indel byproducts. |
|
PE4 PE2 + MLH1dn |
PE4 is most useful when indels at the target site must be minimized or in applications that cannot use nicking sgRNAs; it yields improved editing relative to PE2, but its efficiency relative to PE3 varies depending on cell type. Note: cellular effects of long-term MLH1dn expression (>5 days) have not been assessed. If MLH1dn expression could interfere with downstream experiments, do not use. Note: of less benefit compared to PE2 in MMR-deficient cell types. |
|
PE5 / PE5b PE2 + Additional nicking sgRNA + MLH1dn (PE5b if nicking sgRNA protospacer overlaps with edit) |
PE5 typically yields the highest editing efficiency out of all PE systems, and offers substantially reduced indels compared to PE3. Note: cellular effects of long-term MLH1dn expression (>5 days) have not been assessed. If MLH1dn expression could interfere with downstream experiments, do not use. See PE3 information for notes on nicking sgRNA design. Note: of less benefit compared to PE3 in MMR-deficient cell types. |
| Protein Architecture | Uses |
| Original architecture15 | Not recommended; no longer state-of-the-art. |
| Max architecture30 (Addgene #174820) | Use for all applications. The max architecture is always the same as or better in editing efficiency than the original architecture across all edits and cell types tested. |
| pegRNA | Uses |
| pegRNA | Not recommended unless practical limitations such as chemical synthesis limitations prevent the use of epegRNAs. |
| epegRNA | Recommended for all applications: epegRNAs almost always offer high editing efficiencies than pegRNAs across all edits and cell types tested. |
| Silent mutations | Uses |
| None | If a given application does not allow silent mutations to be incorporated (efficient prime editing can still be achieved without them). |
| PAM (or seed)-disrupting mutations | Recommended if possible. Disruption of the PAM or seed region reduces re-binding and nicking of the edited product. Note: check a codon usage table to ensure that the mutations are silent and that the silent changes do not create a highly disfavored codon. |
| MMR-evading mutations | Installing multiple contiguous or tightly clustered mutations can help increase editing efficiency, especially if the PE2 system is being used. Different silent or benign mutations, in addition to the desired edit alone, should be tested whenever possible. Note: check a codon usage table to ensure that the mutations do not use highly disfavored codons. |
For a given prime editing experiment, one option from each category above is selected. When selecting PE systems and the incorporation of silent mutations, though, the optimal version will depend on the edit, cell type, and application. For these decisions, empirical testing for each site and mutation is needed to ensure optimal editing.
pegRNA Improvements
The pegRNA is responsible for both targeting the editor and encoding the desired edit. Because the elements of the pegRNA that encode the edit are located at the 3′ end for commonly used 3′-extended pegRNAs, exonucleolytic degradation is a concern. Indeed, we recently discovered that cellular degradation of pegRNAs can result in truncated, editing-incompetent pegRNAs that poison prime editing in cells by occupying target DNA sites and prime editor proteins without the possibility of productive editing. To address this issue, we developed engineered pegRNAs (epegRNAs). epegRNAs contain a structured 3′ motif that enhances stability and prevents 3′ degradation, which in turn results in an average improvement in editing efficiency of 1.5-fold to 4-fold over traditional pegRNAs31. Given the ease of incorporating the epegRNA modification and the large editing improvements that it provides, we strongly suggest the use of epegRNAs for all prime editing applications. In our original report of epegRNAs, we described two different 3′ structural motifs: mpknot and tevopreQ1. Similar studies have demonstrated benefits from using pegRNAs with a 3′ Csy4 recognition sequence38 or a Zika exoribonuclease-resistant RNA motif39. While all of these motifs can substantially enhance prime editing, we recommend the use of tevopreQ1 throughout this protocol, simply to decrease the number of epegRNAs that must be tested.
Similarly, we and others40 have also found that the “flip and extension” (F+E) sgRNA scaffold modification, which was previously shown to enhance Cas9 activity31,41, can also improve prime editing in some circumstances. This sgRNA scaffold modification, which extends one of the scaffold hairpins and disrupts a spacer-proximal UUUU sequence that may act as a Poll III transcriptional terminator, significantly increased editing at a subset of the sites tested31. Because this improvement is less generalizable across sites, we recommend using an unmodified scaffold for initial epegRNA screening. However, testing an F+E-modified version of the eventual optimized epegRNA could further increase editing efficiency. To summarize, we recommend using an epegRNA harboring the tevopreQ1 motif, including during PBS and RTT screening. After optimized PBS and RTT lengths have been achieved, changing the 3′ modification to the mpknot motif or changing the scaffold to the F+E sequence could further enhance editing.
Manipulating the cellular determinants of prime editing
The PE3 system uses an additional sgRNA to nick the unedited strand of the genome, which directs nick-directed eukaryotic MMR to favor an edited outcome. Due to the importance of DNA repair events during prime editing, we applied the Repair-seq CRISPRi screening platform42 to identify the cellular determinants of prime editing outcomes30. Strikingly, knockdown of MMR proteins led to substantial increases in prime editing efficiencies and decreases in indel frequencies, even when the PE3 system is used.
Based on this observation, we engineered MLH1dn, a dominant-negative variant of the MMR protein MLH1. When transiently co-expressed with prime editing machinery, MLH1dn temporarily inhibits MMR, which greatly enhances prime editing efficiency and minimizes indels across several cell types. When the PE2 or PE3 systems are used with MLH1dn, they are referred to as PE4 and PE5, respectively30. We also demonstrated that careful design of pegRNAs can cause prime editing intermediates to evade MMR, without requiring a secondary nick or MLH1dn, by installing silent or benign mutations near the target edit30. Larger distortions of the DNA double helix are less efficiently recognized by MMR proteins, so introducing additional mutations adjacent to the desired edit impedes engagement of prime editing heteroduplex intermediates by MMR, thereby increasing prime editing efficiencies. Guidelines on when and how to use various MMR manipulation tools are provided in the experimental design section, in Fig. 3, and in Table 1.
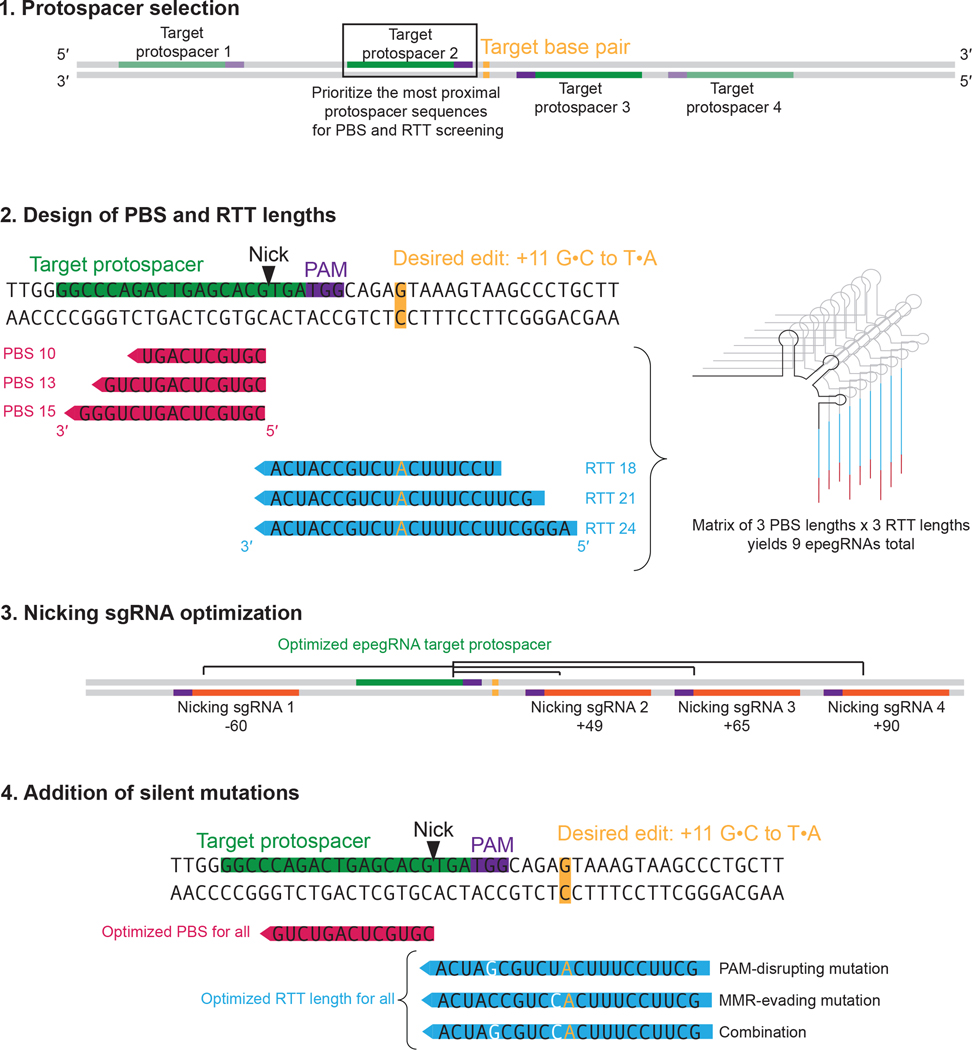
Figure 3. Experimental design of epegRNAs.
Protospacers (shown in green) should first be identified based on available PAM sequences (shown in purple). Of these protospacer candidates, the ones closest to the desired edit (shown in gold) should be tried first. Second, for a minimal initial screen, PBS (shown in pink) lengths of 10, 13, and 15 nt and RTT (shown in blue) lengths that extend at least 7 nt beyond the desired edit are designed. Note: the epegRNA modification is not shown here for simplicity, but it should be included in all pegRNA designs by default. Third, nicking sgRNAs (shown in orange) are designed to target the opposite strand, typically downstream of the initial nick. Finally, PAM-disrupting or silent mutations are identified and added to the RTT of the epegRNAs. This approach combines insights gained from several publications15,30,31.
Improvements to the prime editor protein architecture
Engineering the architecture of the editor protein has also improved prime editing efficiency. Our lab recently developed the PEmax architecture, which contains four improvements relative to the original editor: optimization of the nuclear localization signals (NLSs), codon usage, and linkers, as well as two Cas9 mutations that were previously shown to increase Cas9 nuclease activity30,43. Other laboratories have also manipulated the original prime editor architecture to create systems such as PE2*37, CMP-PE44, and hyPE245. Based on our comparison of various PE systems reported as of late 2021 (ref. 30), we recommend the PEmax architecture for all prime editing applications.
Larger genomic changes with twinPE, PEDAR, PRIME-Del, dual-pegRNA, HOPE, and GRAND
Traditional prime editing can mediate the efficient insertion or deletion of several dozen base pairs. To increase the size of insertions and deletions that are possible with prime editing, we recently developed twin prime editing (twinPE). In twinPE, two prime editing events occur on opposite strands of DNA, such that the newly synthesized genomic flaps are complementary to each other (Fig. 4). This method directly installs the edit on both DNA strands instead of requiring the cell to synthesize the non-reverse-transcribed strand. TwinPE is capable of making larger edits (for example, ≥780 bp deletions and ≥108 bp insertions) more efficiently than traditional prime editing methods46.
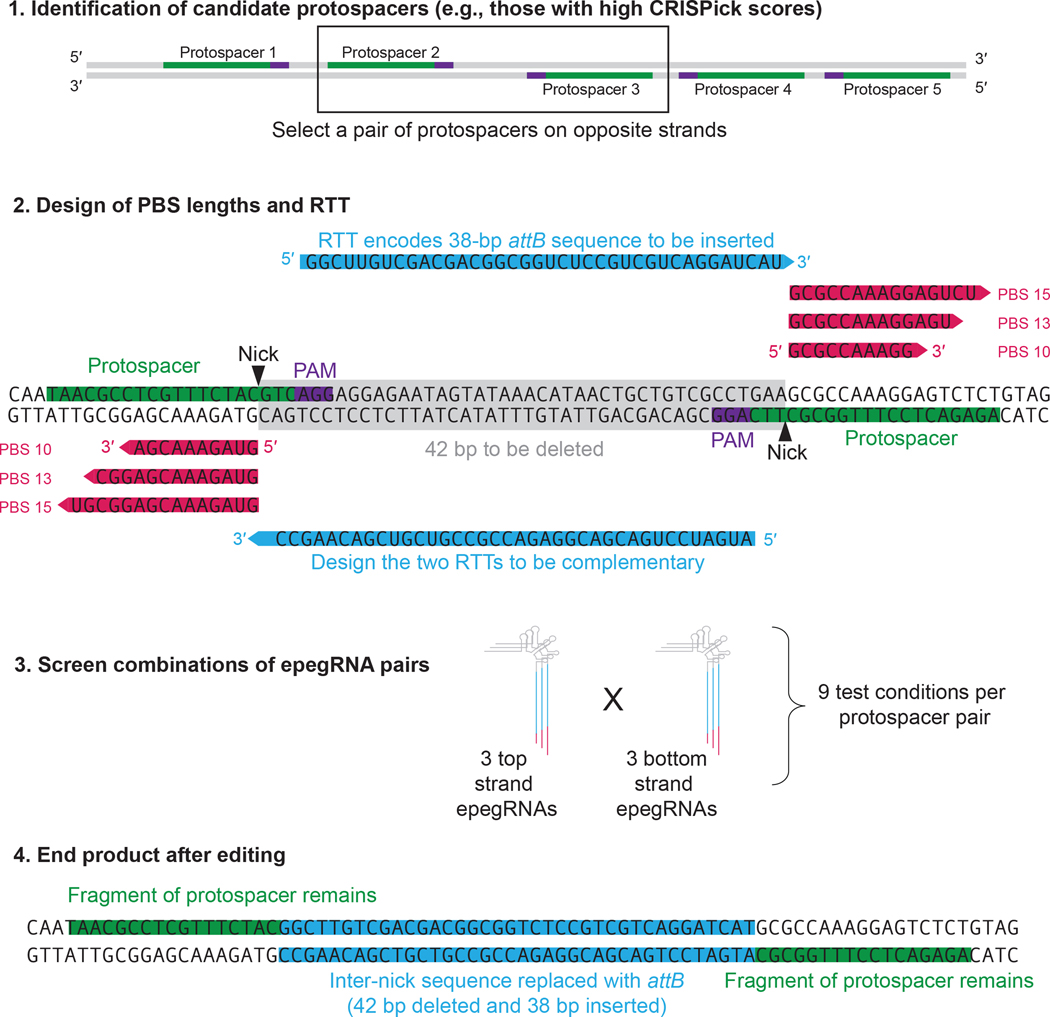
Figure 4. Experimental design for twinPE.
First, high-efficiency protospacers as predicted by CRISPick should be identified. Protospacer pairs should then be selected (minimum inter-nick distance of 30 nt). Second, PBS lengths of 10, 13, and 15 nt should be tried for each protospacer. For RTT design, the desired insertion should be encoded on one epegRNA, and its reverse complement should be encoded on the other. Third, for twinPE, epegRNA screening is not a matrix of PBS lengths × RTT lengths, but is instead a matrix of top and bottom strand epegRNAs, each of which will have three possible PBS lengths. Note: the epegRNA modification is not shown here for sake of simplicity, but should be included in all pegRNA designs by default. An example is shown of a twinPE product, in which the sequence between the two nicks is replaced with the sequence encoded in the RTTs of the epegRNAs. This approach combines insights gained from several previously published works15,31,46.
Several additional dual pegRNA prime editing approaches have been described by others including PRIME-Del47, PEDAR48, dual-pegRNAs49, HOPE50, and GRAND51. These systems differ in the extent and location of complementarity between the two new DNA strands, and in how the starting DNA sequence between the two nicks is manipulated. In twinPE and GRAND, the inter-nick sequence is deleted and replaced with the new sequence encoded by pegRNAs (Fig. 4). These newly synthesized strands are complementary to each other and can thus spontaneously anneal following reverse transcription. PRIME-Del is similar to twinPE, except the newly-synthesized DNA strands are not only complementary to each other but are also complementary to the genomic sequence upstream of the nick on the opposite DNA strand. PEDAR is similar as well, but instead of using a Cas9 nickase, a Cas9 nuclease is used in the prime editor protein. Finally, the dual-pegRNA method and HOPE differ from the other three methods in that they do not delete any sequence in between the two nicks. In this protocol, we refer to twinPE based on our more extensive experience with this method, but we anticipate that many of the strategies and procedures may also apply to prime editing with PRIME-Del, PEDAR, paired pegRNAs, HOPE, and GRAND.
Prime editing and site-specific recombinases to mediate gene insertion and inversion
Our group has also shown that PE and twinPE can install recombinase recognition sequences, and following the installation of these sequences, recombinases can mediate kb-scale changes46. In a sequential-transfection strategy, we first used twinPE to generate cells with a homozygous attB site at CCR5 and then used this site as a substrate in a second transfection of BxbI recombinase and an attP 5.6-kb donor plasmid, achieving up to 17% donor knock-in efficiency. In a single-transfection strategy, we treated unedited cells with prime editor, twinPE pegRNAs encoding the attB recombinase site, the corresponding BxbI recombinase, and a 5.6-kb attP donor plasmid to achieve up to 5.5% donor plasmid knock-in efficiency. We used a similar single-transfection strategy to insert factor IX cDNA into the human albumin locus and detected editing-dependent production of human factor IX protein in culture media. We also used two simultaneous twinPE editing events to install both the attB and attP sequences into the HEK293T genome, flanking a 39-kb inversion at the IDS locus that has been shown to cause Hunter syndrome. In a single RNA nucleofection of all PE and recombinase elements, we achieved 2.1–2.6% correction of this 39-kb pathogenic inversion. Independently, Ioannidi and coworkers have also used prime editing to incorporate recombinase sites to support gene-sized targeted insertion in a system they called PASTE52.
Alternate Cas9 and reverse transcriptase homologs
In principle, different Cas9 homologs can be used for prime editing, but in practice, non-SpCas9 prime editors have thus far mediated less efficient editing30. For other genome editing tools, the primary motivation for using alternate Cas9 domains is to access a wider array of PAM sequences. However, PAM flexibility is not critical for PE, as it offers a much wider range of distances between the PAM and the desired edit than base editing, and either DNA strand can be targeted to achieve a desired edit. Due to this inherent flexibility, we currently recommend using SpCas9 for all prime editing applications. If an NGG PAM is not present, alternate Cas9 domains can be tested, but editing efficiency may be lower. Instead, we recommend using twinPE to install the target mutation from two distal NGG PAMs. Similarly, different RT domains such as the cauliflower mosaic virus RT (RT-CaMV) and the E. coli BL21 retron RT (RT-retron) have been used for prime editing53. However, these reverse transcriptases yielded lower editing efficiencies than the engineered M-MLV RT used in PE2. While alternate reverse transcriptase domains could eventually prove useful, their prime editing properties may need to be improved before they should be chosen over PE2’s engineered M-MLV.
Applications of prime editing
Despite being published just over two years ago, prime editing has already been used in a wide variety of studies. These applications have included editing in many workhorse cell lines such as HEK293T, HeLa, U2OS, and K562 cells15,24,30,31, as well as more therapeutically-relevant cells including patient-derived fibroblasts, iPSCs, and T cells30,31 and in animals34,37,54–59. Using PE4 and PE5, up to 40% editing has been achieved in patient-derived iPSCs, and up to 60% editing has been achieved in primary human T cells30. Prime editing has also been used for basic research applications such as lineage tracing60 and saturation mutagenesis in human cells61 and plants62. Many model organisms have also been created using prime editors; prime editing in rabbit embryos yielded an animal model of Tay-Sachs disease63, and PE has been used to install edits in mouse zygotes34,56,57. RNP-mediated delivery of the prime editor into zebrafish embryos has also generated up to 30% editing40. Finally, in vivo prime editing has been shown using hydrodynamic injection, adenovirus, and adeno-associated virus (AAV) delivery methods37,54,55,58,64. In general, in vivo prime editing efficiencies have been modest. However, it is important to note that virtually all published in vivo editing studies used the original PE2 or PE3 prime editors with unmodified pegRNAs. We anticipate that using epegRNAs and PE4max or PE5max will result in marked improvements in in vivo prime editing efficiencies.
Limitations
A logistical barrier to the use of prime editing is that editing efficiencies are strongly dependent on the PBS and RTT of the pegRNA, and the optimal choices for each component are not evident for most sites or edits. Our lab has developed general guidelines for pegRNA design (see experimental design section and Figs. 2–4); but within these guidelines, typically dozens of potential pegRNAs could be used for a given edit. A recent study by Kim et al. attempted to use libraries of edits and corresponding pegRNAs to identify additional design principles65. Their data suggests that for a +5 G to C edit, a 13-nt PBS and a 12-nt RTT are ideal starting points; this recommendation may be helpful in situations where pegRNA screening is not possible. However, we have also encountered many sites such as the RNF2 and HEK4 loci where a PBS of 13 is not optimal, and we frequently find that a 12-nt RTT is not desirable, especially for edits that are distal from the nick or mutate more than one base. Thus, when it is essential to achieve the highest editing possible, empirical screening of PBS and RTT lengths is required, even when using current-generation prime editing systems. This process is resource-intensive, as many pegRNA constructs must be generated and evaluated. To facilitate this screening, we recommend performing a pegRNA screen in an easy-to-transfect cell line such as HEK293T cells for human mutations or N2A cells for mouse mutations. If an easy-to-transfect cell line with a given mutation is not available, cell lines can be made to facilitate screening.
Finally, prime editing precision and in vivo prime editing efficiency can be improved. In vivo delivery of a prime editor, particularly using AAV, is more challenging than delivery of Cas9 nuclease or a base editor due to the prime editor’s large size. Removing the RNaseH domain of the RT has enabled AAV delivery, but in vivo editing efficiencies reported to date have been modest37,54,55,58. In addition, while prime editing is very precise overall, it can produce undesired byproducts. Like other genome-editing methods, prime editing can produce indels at the target site. Prime editing generally results in substantially fewer indels than nuclease-based approaches such as Cas9-mediated HDR, but indels can still occur, especially for the PE3 and PE5 systems. Comparatively, the PE2 and PE4 systems typically minimize indel frequencies, though they may be less efficient. Another type of prime editing byproduct results from reverse transcription into the pegRNA scaffold. Fortunately, the frequency of these scaffold insertions is typically low (1.7% on average)15, likely because the cell usually excises flaps that are unable hybridize to the unedited DNA strand due to their mismatched 3′ termini. Finally, while MLH1dn is extremely useful for short-term editing, long-term MMR inhibition could lead to adverse cellular effects or mutagenesis. Therefore, optimization of in vivo editing efficiency, improved editor size and precision, and analysis of off-target PE4/PE5 effects will further expand the application scope of prime editing.
Prime editing experimental design
There are four main decisions to make when designing a prime editing experiment: (1) pegRNA design, (2) selection of the prime editing system, (3) selection of prime editor architecture, and (4) installation of silent mutations. While some aspects of these decisions are relatively straightforward (for example, we currently recommend that the PEmax architecture and the epegRNA modification always be used), other decisions are dependent on the edit, target cell type, and delivery method. Guidelines for making these decisions are explained below and in Table 1.
Designing candidate epegRNAs
When considering pegRNA design, epegRNAs should be used over unmodified pegRNAs whenever possible due to their increased efficiency. A standard epegRNA has five components: the spacer, scaffold, RTT, PBS, and tevopreQ1 motif (Fig. 2). The scaffold and tevopreQ1 portions are constant, but the spacer, PBS, and RTT should be optimized for each new edit. The first step of epegRNA optimization is to scan the target locus for candidate protospacer sequences that are immediately 5′ of an appropriate PAM sequence (NGG for SpCas9). Only bases 3′ of the nick induced by the Cas9 domain of the editor can be edited. Therefore, as a frame of reference, we consider the first base 3′ of the epegRNA-directed nick—the first editable base—to be the +1 position. While the mechanism of PE enables a broad editing window, we find that targeting protospacers more proximal to the desired editing site generally yields higher editing efficiencies. Ideal candidate protospacer sequences should therefore be as close to the desired editing site as possible while keeping the target site in the editable region of PE (i.e., 3′ of the nick, see Fig. 3). Importantly, if the epegRNA will be expressed from a plasmid via the U6 RNA polymerase III promoter, a 5′ G at the start of the spacer is necessary to initiate transcription efficiently and should be incorporated into the epegRNA design.
After identifying candidate protospacers, PBS and RTT lengths must be optimized. The rules governing the best PBS and RTT lengths for a given locus and edit are not completely understood, but optimizing these lengths empirically for a specific edit is important to maximize editing efficiency. The number of PBS and RTT lengths that should be screened for a given application depends on the editing efficiency needed and resources available. The number of possible combinations can be large. In our experiences, optimal PBS lengths have ranged from 8 to 15 nt, and the optimal RTT range is even larger (10 to 74 nt). While screening this entire matrix for a given edit would maximize the likelihood of identifying the optimal epegRNA, it is not practical for most applications. Sufficiently active epegRNAs can often be determined with a less intensive screening campaign. For a typical epegRNA screen, we recommend examining a small matrix of PBS and RTT lengths for each protospacer. PBS lengths of 10, 13, and 15 are promising candidates for most sites.
Unlike the PBS, the RTT design is dictated by the edit to be installed15. For small changes such as SNPs, the shortest RTT length tested should encode at least 7 nt of homology downstream of the edit to promote hybridization to the complementary genomic strand. For larger edits such as the insertion of epitope tags, a longer stretch of downstream homology (~20 nt minimum) is recommended. In addition to this edit-dependent minimum RTT length, we recommend trying two longer RTT lengths (~4–10 nt longer than the minimum) as well. This creates a 3 PBS × 3 RTT matrix, representing 9 epegRNAs total for a first-pass assessment. This process is summarized in Fig. 3. Screening should be performed in a workhorse cell line such as HEK293T cells for human targets and N2A cells for murine targets. Additionally, we also strongly recommend screening epegRNAs on the exact target sequence for editing (this may require creating a cell line that harbors the target mutation—which can often be created), as small changes in the target sequence or epegRNA sequence can lead to large changes in editing outcomes.
Several potential pitfalls should be avoided when designing epegRNAs. For epegRNAs expressed from a plasmid using the U6 RNA polymerase III promoter, four or more consecutive uridines in the pegRNA sequence may act as a transcriptional terminator and prematurely truncate the epegRNA 66. Therefore, the sequences of the spacer, PBS, and RTT should avoid such poly(U) tracts if possible. Additionally, we (but not others65) have observed that beginning the RTT sequence with a cytosine lowers editing, likely because it disturbs the structure of the epegRNA scaffold15. Therefore, we recommend designing the 3′ extension to not begin with cytosine and omitting designs that would do so when screening for optimal RTTs. Online tools such as PrimeDesign67 and other similar resources68,69,70 have also been developed to aid in pegRNA sequence generation.
Choice of prime editing system (PE1-PE5) and prime editor architecture
We have reported five prime editing systems as of this writing. PE1 lacks the substantial benefits of reverse transcriptase engineering and other improvements, and is rarely preferred over other systems. PE2, PE3, PE4, and PE5 can each be favored for different applications. See Table 1 for a summary of each editing system and detailed guidelines for when to use each one. Importantly, when performing the epegRNA screen described above, PE2 or PE4 should be used to simplify the screening process, as they do not require simultaneous nicking sgRNA optimization.
When using the PE3 or PE5 system, a secondary nicking guide will need to be designed. Several nicking guide protospacers should be tried to maximize editing efficiency while minimizing the incorporation of indels. Generally, the optimal secondary nick is 50–90 nt upstream or downstream of the epegRNA-guided nick. However, if a PAM is positioned near the desired edit, a PE3b/PE5b nicking sgRNA, which only directs nicking of the unedited strand after editing of reverse-transcribed strand occurs, can be used and typically minimizes indel byproducts. To design a PE3b/PE5b nicking sgRNA, we recommend positioning the protospacer of the nicking sgRNA such that it overlaps with the edited base(s) on the other strand, as shown in Fig. 5. Because the PE3b/PE5b systems tend to generate fewer indels than PE3/PE5, we recommend trying PE3b or PE5b whenever possible—that is, whenever a properly positioned PAM exists on the unedited strand. For the PE3, PE3b, PE5, and PE5b systems, the U6 RNA polymerase III promoter may be used for nicking sgRNA expression; if this is the case, a 5′ G at the start of the spacer is required for transcription initiation. A final consideration for design of the nicking sgRNA is that differences in DNA repair between cell types may require re-optimization of the nicking sgRNA after transitioning between different cell lines, even for the same edit.
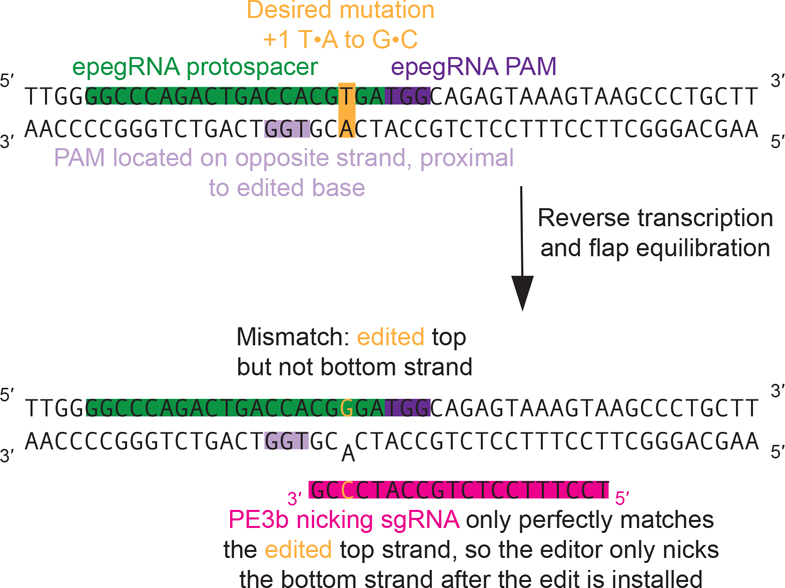
Figure 5. Design of a PE3b/PE5b nicking sgRNA.
To use the PE3b or PE5b systems, a PAM needs to be present on the non-edited strand close to the edit. A nicking sgRNA can then be designed such that it can only bind and direct nicking of the non-edited strand after reverse transcription and flap equilibration have occurred. Such a PE3b/PE5b nicking sgRNA has a spacer that is perfectly complementary to the edited DNA sequence, but contains mismatches with the unedited sequence.
Converting PE2 to PE4, or PE3 to PE5, is simple experimentally; an extra plasmid or other construct providing MLH1dn is added to the transfection mixture. Importantly, while the addition of MLH1dn may not be as helpful for some edits in MMR-deficient cells such as HEK293T cells, it can drastically improve editing efficiency for the same edit in a more MMR-competent cell type. Therefore, even if using PE4 or PE5 in initial screening in HEK293T cells shows only modest benefits, we recommend testing these PE systems again later on in the target cell type. Short-term expression of MLH1dn has been shown to be minimally perturbative to cells, but long-term expression effects have not been evaluated30. Therefore, delivery methods in which PE machinery would be constitutively expressed for a long period of time may warrant selecting PE2 and PE3 over PE4 and PE5, especially if the phenomenon being investigated is sensitive to MMR. Finally, regarding the architecture of the protein component of the prime editor, we strongly recommend using the PEmax improvements. Compared to the originally described prime editor, PEmax has improved nuclear localization, codons, and linkers, in addition to mutations in the Cas9 domain that increase activity30.
Introduction of silent mutations
Two categories of silent mutations can be installed to achieve higher editing efficiencies. The first class is mutations that disrupt either the PAM (positions +5–6) or the seed region (positions +1–3) of the target site. PAM or seed-disrupting edits partially prevent Cas9 from re-binding and re-nicking the target strand, which otherwise could result in indels or the reversion of a desired edit back to the wild-type sequence15. To include PAM or seed-disrupting mutations, simply encode them in the RTT of the epegRNA along with the original target edit (Fig. 3). PAM-disrupting and seed-disrupting mutations are almost always beneficial, and we recommend including them if possible.
The second class of silent mutations is MMR-evading target-adjacent mutations. Because the inclusion of additional mutations adjacent to the target mutation results in more significant helix distortion, these regions are less likely to be recognized by cellular MMR proteins. This strategy is particularly useful for desired edits that are point mutations and insertions and deletions of fewer than 13 nt30. To include MMR-evading mutations, encode them in the RTT of the epegRNA along with the desired edit (Fig. 3). Silent mismatches (particularly C•C mismatches), within 5 nt of the desired edit are typically the most beneficial. Notably, the effects of MMR-evading mutations are less consistent than those of PAM-disrupting mutations, and certain mismatch types are more effective than others. For this reason, we recommend first optimizing the epegRNA without any MMR-evading silent mutations and then adding these mutations afterward. For both MMR-evading mutations and PAM- or seed-disrupting mutations in coding regions, a codon usage table should be checked to ensure that the additional mutations do not create a highly disfavored codon.
Iteration to maximize editing efficiency
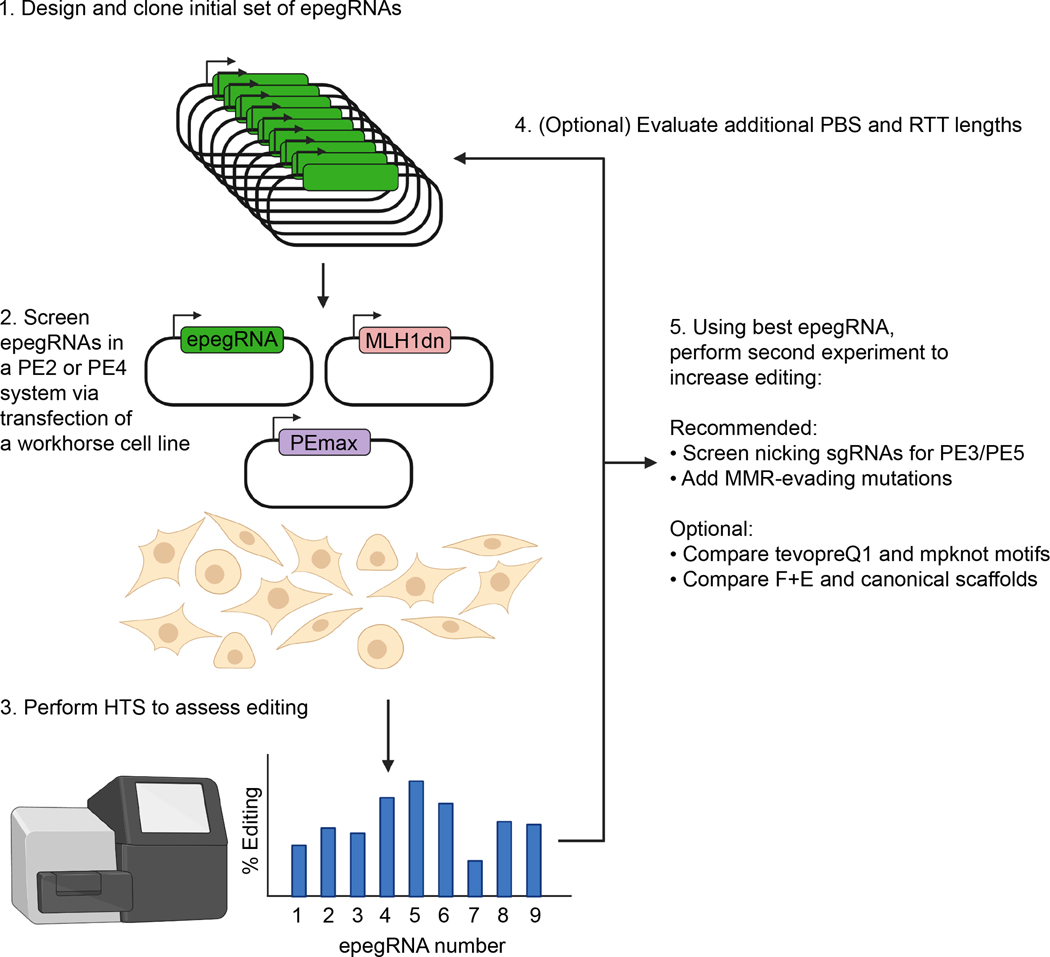
For applications in which editing efficiency must be maximized, we recommend several iterative rounds of optimization. Initially, one should screen for PBS and RTT lengths using the PE2 or PE4 systems, which do not require a nicking sgRNA. Typically, this initial panel will reveal an optimal PBS and/or RTT length; these optimal lengths can then be carried forward in a more refined screen. For instance, if the optimal PBS length is found to be 10 nt in the initial screen, PBS lengths of 9 and 11 nt can be tried, or many different RTT lengths can be screened with the 10-bp PBS. Using optimized PBS and RTT lengths, other aspects of the epegRNA can then be tested. For instance, PAM-disrupting mutations and/or MMR-evading mutations can be encoded in the RTT, and the mpknot motif and F+E scaffold can be evaluated. Finally, nicking sgRNAs and the PE system (PE2-PE5) can be optimized. Even after editing has been optimized in a workhorse cell line, it is beneficial to re-optimize some aspects such as PE system and nicking sgRNAs, due to the specific cell type effects of these changes. This cycle of iterative improvements, summarized in Fig. 6, can be repeated until editing efficiencies plateau.
Figure 6. Experimental workflow for PE optimization.
To optimize prime editing at a new locus, first design and clone an initial set of epegRNAs. These epegRNAs are then screened via transfection in workhorse cell lines, such as HEK293T cells or N2A cells. PE2 or PE4 should be used for this initial screen to avoid screening nicking sgRNAs in tandem. Based on sequencing results from this initial screen, additional optimization can be performed. We recommend screening additional PBS and RTT lengths if low editing efficiency is observed. Once optimal PBS and RTT lengths are found, additional improvements, such as nicking sgRNAs and MMR-evading mutations, can be tested using the optimized epegRNA. This approach combines insights gained from several previously published works15,30,31.
Experimental design for twin prime editing
We recommend using epegRNAs and the PEmax architecture for twinPE. The only exception to this rule may be if the additional sequence length from a 3′ motif could make impractical the chemical synthesis of an unusually long epegRNA or its expression from the U6 promoter. Second, unlike other PE schemes, twin prime editing does not require the design of nicking sgRNAs or the use of MLH1dn. The only aspect that should be optimized is a pair of epegRNAs, which have the same architecture as epegRNAs used for typical prime editing. The first step is to identify protospacer combinations to use. However, many possible protospacers typically exist due to the flexibility of the twinPE system. To prioritize protospacers that are likely to yield high editing efficiency, we recommend using the CRISPick design tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design), which can predict the Cas9 nuclease cutting efficiency at a particular protospacer71. Because Cas9 nuclease efficiency is the strongest predictor of prime editing efficiency65, it makes sense that we have observed a loose correlation between a protospacer’s CRISPick score and the PE efficiency at that protospacer.
Out of the list of promising protospacers, appropriately spaced pairs of protospacers on opposite DNA strands should be selected. The distance between the two nicks should be at least 30 bp, as inter-nick distances smaller than this can lead to steric clashes between the two editor proteins. The upper limit of the inter-nick distance is dependent on the desired edit; we have used protospacers as far as 800 bp apart, although most high-efficiency inter-nick distances are between 40 and 150 bp46. We recommend trying ~5 protospacer combinations in total. For each protospacer, PBS lengths should be optimized, following the same general guidelines used for traditional epegRNA design (10, 13, and 15 bp to start). Conversely, in twinPE, the RTT does not require extensive optimization or screening. The RTTs for a pair of twinPE epegRNAs are typically each other’s reverse complement, as shown in Fig. 4. Due to these guidelines, experimenters will need to screen 9 epegRNA combinations for each pair of protospacers (3 PBS lengths for the top protospacer × 3 PBS lengths for the bottom protospacer). Finally, one important aspect of twinPE experimental design is that, if the desired edit is a deletion, editing efficiency can be overestimated due to bias during sample preparation and sequencing. While we found this bias to be relatively small (<10%) for deletions 50 bp or less in length, bias increases as deletion size increases. Therefore, when performing large deletions, or when quantification must be highly accurate, we recommend using unique molecular identifiers (UMIs)46. UMIs, which barcode individual molecules during the first step of high-throughput sequencing (HTS) sample preparation, allow for PCR duplicates to be detected during downstream analysis. De-duplication mitigates the bias that arises during sample preparation and enables more accurate quantification.
Choice of delivery method
Efficient delivery of prime editing components is necessary to achieve efficient editing. During pegRNA optimization, we strongly recommend using an easily transfected cell line, such as HEK293T cells for human genome editing or N2A cells for mouse genome editing. In these cells, the efficiency and high-throughput nature of lipid transfection greatly expedites initial rounds of pegRNA screening and prime editor optimization. For other cell types, the most efficient method for delivery will vary, and many therapeutically relevant cell types are not easily transfected. One way to improve editing efficiency in such cell types is to instead deliver plasmids encoding editing systems by electroporation and include a selectable or screenable marker on the prime editor plasmid. Following electroporation, cells harboring the prime editor can be enriched using the marker to increase editing levels among the selected or screened cells. More promisingly, we have found that in vitro-transcribed mRNA encoding the prime editor protein, co-electroporated with chemically modified synthetic epegRNAs and (if needed) nicking sgRNAs, can support efficient editing in cell types such as patient-derived iPSCs, primary human T cells, and patient-derived fibroblasts30,31. In this protocol, we describe procedures for plasmid transfection into HEK293T cells and electroporation of mRNA into patient-derived fibroblasts. These methods are promising starting points, but some parameters will need to be re-optimized for other cell types. RNP delivery of prime editors has also been reported, but will not be covered in this protocol40.
Experimental controls
In all prime editing experiments, an unedited negative control should be included. This control allows experimenters to be confident that desired editing or other observed mutations at the target locus are PE-dependent. This control is particularly important when attempting to edit a mutation for which cells are heterozygous or contain genetic variability before treatment. Irregularities such as SNPs or indels that endogenously occur at the target locus can be identified using this control. It is also important to note that plasmid quality, transfection efficiency, and the health of the edited cells can affect editing efficiency. For this reason, it is important to include internal controls when comparing two different editing approaches. For example, when comparing two pegRNAs designed to make the same edit, the two should ideally be tested side-by-side in the same experiment. Finally, if attempting to edit a new target locus for the first time, it is helpful to include a positive control using a previously validated pegRNA to edit a well-characterized site (such as the HEK3 locus in human cells or the DNMT1 locus in mouse cells). The editing efficiency achieved for this positive control should be compared to previously published values to ensure that experimental techniques and analyses are being performed correctly.
Materials
Reagents
Prime editor, epegRNA, and sgRNA preparation
Plasmids: PEmax (pCMV-PEmax, Addgene ID: 174820), tevopreq1 epegRNA cloning vector (pU6-tevopreq1-GG-acceptor, Addgene ID: 174038), sgRNA cloning vector (pU6-pegRNA-GG-acceptor, Addgene ID: 132777), PEmax mRNA IVT template plasmid (pT7-PEmax, Addgene ID: 178113), hMLH1dn (pEF1a-MLH1dn, Addgene ID: 174824), hMLH1dn mRNA IVT template plasmid (pT7-hMLH1dn, Addgene ID: 178114).
Oligos for sgRNA, pegRNA, and epegRNA Golden Gate cloning, can be designed as shown in Table 2. Alternatively, eBlocks from IDT or similar gene fragment products from other vendors can be used for a simple isothermal assembly reaction with the gene fragment overhangs and PCR primers listed in Table 2. Custom chemically modified sgRNAs and epegRNAs can also be ordered from Agilent, IDT, or other vendors.
PCR primers for sequencing edited DNA and amplifying template DNA for mRNA transcription can also be designed as shown in Table 2.
Nuclease-free water (Qiagen, cat. no. 129115)
Phusion U Green Multiplex PCR Master Mix, 2x (Thermo Fisher Scientific, cat no. F564L) or any other high-fidelity polymerase.
SeaKem LE Agarose (Lonza, cat. no. 50004)
Ethidium bromide solution, 10 mg/ml (Millipore Sigma, cat. no. E1510-10ML)
UltraPure TAE Buffer, 10× (Thermo Fisher Scientific, cat. no. 15558026)
TriDye 1 kb Plus DNA Ladder (NEB, cat no. N3270S)
Gel Loading Dye, Purple (6X) (NEB, cat no. B7024S)
QIAquick PCR purification kit (Qiagen, cat. no. 28104)
S.O.C. medium (Thermo Fisher Scientific, cat. no. 15544034)
LB medium (United States Biological, cat. no. L1505)
LB agar medium (Millipore Sigma, cat. no. L2897)
Carbenicillin, 50 mg/ml, sterile filtered (Gold Biotechnology, cat. no. C-103)
Illustra TempliPhi 100 Amplification Kit (Cytiva, cat. no. 25640010)
Qiagen Plasmid Plus Midi Kit (Qiagen, cat. no. 12945)
PureYield Plasmid Miniprep System (Promega, cat no. A1222)
TE Buffer, 1× (Thermo Fisher Scientific, cat. no. 12090015)
Table 2.
Example oligonucleotide sequences for prime editing procedure
| Step | Oligo Name | Sequence (5’ -3’) | Purpose | Usage |
|---|---|---|---|---|
| 3A(i) | Golden Gate Part 1, top oligo | CACC(N20–21)GTTTT | Top oligo with cloning overhangs to insert the desired spacer (target) sequence | Replace the (N20–21) with the desired epegRNA or sgRNA spacer sequence. |
| 3A(i) | Golden Gate Part 1, bottom oligo | CTCTAAAAC(N20–21) | Bottom oligo with cloning overhangs to insert the desired spacer (target) sequence | Replace the (N20–21) with the reverse complement of the desired epegRNA or sgRNA spacer sequence. |
| 3A(i) | Golden Gate Part 2, top oligo | /5Phos/AGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCG | Top oligo with cloning overhangs to insert a standard SpCas9 sgRNA scaffold sequence in an epegRNA cloning reaction | This oligo must be 5′ phosphorylated for the epegRNA Golden Gate assembly to work. The position of the 5′ phosphorylation is indicated with the bolded /5Phos/ notation. If cloning sgRNAs, alternate Golden Gate Part 2 oligos must be used (see below). |
| 3A(i) | Golden Gate Part 2, bottom oligo | /5Phos/GCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAG | Bottom oligo with cloning overhangs to insert a standard SpCas9 sgRNA scaffold sequence in an epegRNA cloning reaction | This oligo must be 5′ phosphorylated for the epegRNA Golden Gate assembly to work. The position of the 5′ phosphorylation is indicated with the bolded /5Phos/ notation. If cloning sgRNAs, alternate Golden Gate Part 2 oligos must be used (see below). |
| 3A(i) | Golden Gate Part 2, top oligo [sgRNA alternate] | /5Phos/AGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGC | Top oligo with cloning overhangs to insert a standard SpCas9 sgRNA scaffold sequence in a sgRNA cloning reaction | This oligo must be 5′ phosphorylated for the sgRNA Golden Gate assembly to work. The position of the 5′ phosphorylation is indicated with the bolded /5Phos/ notation. If cloning epegRNAs, alternate Golden Gate Part 2 oligos must be used (see above). |
| 3A(i) | Golden Gate Part 2, bottom oligo [sgRNA alternate] | /5Phos/AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAG | Bottom oligo with cloning overhangs to insert a standard SpCas9 sgRNA scaffold sequence in an sgRNA cloning reaction | This oligo must be 5′ phosphorylated for the sgRNA Golden Gate assembly to work. The position of the 5′ phosphorylation is indicated with the bolded /5Phos/ notation. If cloning epegRNAs, alternate Golden Gate Part 2 oligos must be used (see above). |
| 3A(i) | Golden Gate Part 3, top oligo | GTGC(Nextenison) | Top oligo with cloning overhangs to insert the desired epegRNA RTT/PBS 3′ extension | Replace the (Nextenison) with the desired epegRNA RTT/PBS 3′ extension. |
| 3A(i) | Golden Gate Part 3, bottom oligo | CGCG(Nextension) | Bottom oligo with cloning overhangs to insert the desired epegRNA RTT/PBS 3′ extension | Replace the (Nextenison) with the reverse complement of the desired epegRNA rTt/PBS 3′ extension. |
| 3B(i) | Isothermal gene fragment | CTTGGCTTTATATATCTTGTGGAAAGGACGAAACACC(NepegRNA)TTTTTTTAAGCTTGGGCCGCTCGAGGTACCTCTCTACATATGACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTC | Gene fragment to insert a complete epegRNA or sgRNA with isothermal assembly overhangs. (Note: unlike the other sequences listed in this table, the isothermal gene fragment should be a double-stranded DNA piece, not a single-stranded oligonucleotide.) |
Replace the (NepegRNA) with the sequence of the desired epegRNA or sgRNA. epegRNA sequences should include a spacer, sgRNA scaffold, RTT, PBS and 3′ structural motif. sgRNA sequences should include a spacer and sgRNA scaffold. The underlined sequence is an isothermal assembly cloning overhang that overlaps the human U6 promoter from the 3′ end of the PCR amplified product from Step 3B(iii). The italicized sequence is an isothermal assembly cloning overhang that overlaps to the 5′ end of the PCR amplified product from Step 3B(iii). |
| 3B(ii) | Isothermal assembly forward primer | CAAAAATCGACGCTCAAGTC | Forward primer to PCR amplify pU6-tevopreq1-GG-acceptor plasmid for isothermal assembly | Anneals to the pU6-tevopreq1-GG-acceptor plasmid origin of replication to amplify the full-length plasmid for an isothermal assembly. |
| 3B(ii) | Isothermal assembly reverse primer | ACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAG | Reverse primer to PCR amplify pU6-tevopreq1-GG-acceptor plasmid for isothermal assembly | Anneals to the pU6-tevopreq1-GG-acceptor plasmid human U6 promoter sequence to amplify the full-length plasmid for an isothermal assembly. |
| 4 | in vitro transcription forward primer | TCGAGCTCGGTACCTAATACGACTCACTATAAGG | Forward primer for PCR amplification of a template for in vitro transcription of editor mRNA | Amplification with this primer installs a functional T7 promoter sequence into the generated PCR amplicon. |
| 4 | in vitro transcription reverse primer | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCTTCCTACTCAGGCTTTATTCAAAGACCA | Reverse primer for PCR amplification of a template for in vitro transcription of editor mRNA | Amplification with this primer installs a templated poly(A) tail. |
| 27 | PCR1 Forward | ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNN (NAnneal) | Forward primer for amplifying gDNA in preparation for HTS | Replace NAnneal with a sequence matching the 5′ end of the targeted genomic site (for a 300 bp amplicon, this primer should typically anneal to the genomic region ~150 nt 5′ of the target prime edit). The Illumina PCR1 forward adapter is shown in italics. |
| 27 | PCR1 Reverse | TGGAGTTCAGACGTGTGCTCTTCCGATCT(NAnneal) | Reverse primer for amplifying gDNA in preparation for HTS | Replace NAnneal with a sequence matching the reverse complement of the 3′ end of the targeted genomic site (for a 300 bp amplicon, this primer should typically anneal to the genomic region ~150 nt 3′ of the target prime edit). The Illumina PCR1 reverse adapter is shown in italics. |
Golden Gate cloning of epegRNAs and sgRNAs
BsaI-HFv2 (NEB, cat. no. R3733S)
NcoI-HF (NEB, cat. no. R3193S)
PvuII-HF (NEB, cat. no. R3151S)
rCutsmart Buffer, 10× (NEB, cat. no. B6004S or provided with restriction enzymes)
Tris-HCl, pH 8.0, 1 M solution (Thermo Fisher Scientific, cat. no. 15568025)
NaCl, 5M solution (Thermo Fisher Scientific, cat. no. AM9760G)
T4 DNA Ligase (NEB, cat. no. M0202S)
T4 DNA Ligase Reaction Buffer, 10x provided with the T4 DNA ligase, but can also be ordered separately (NEB, cat. no. B0202S).
T4 Polynucleotide Kinase, necessary if sgRNA scaffold oligos for Golden Gate method will be manually phosphorylated (NEB, cat. no. M0201S)
QIAquick Gel Extraction Kit (Qiagen, cat. no. 28704)
Isothermal assembly of epegRNAs and sgRNAs
NEBuilder HiFi DNA Assembly Master Mix (NEB, cat. no. E2621S) or other preferred isothermal assembly mastermix
DpnI (NEB, cat. no. R0176S)
rCutsmart Buffer, 10× is provided with the restriction enzyme, but can also be ordered separately (NEB, cat. no. B6004S).
Phusion High-Fidelity PCR Master Mix with HF Buffer (NEB, cat. no. M0531S) or any other high-fidelity polymerase with a DpnI-compatible reaction buffer.
in vitro transcription of prime editor mRNA
HiScribe T7 High Yield RNA Synthesis Kit (NEB cat. no. E2040S)
CleanCap Reagent AG (Trilink, cat. no. N-7113)
N1-Methylpseudouridine-5′-Triphosphate (Trilink, cat. no. N-1081)
7.5M LiCl Precipitation Solution (Thermo Fisher Scientific, cat. no. AM9480).
RNaseZap RNase Decontamination Solution (Thermo Fisher Scientific, cat. no. AM9782) or equivalent product.
DNase I, RNase-free (NEB cat. no. M0303S)
Gel Loading Buffer II, Denaturing PAGE (Thermo Fisher, cat. no. AM8546G)
Millennium RNA Markers (Thermo Fisher, cat. no. AM7150)
SYBR Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific, cat. no. S11494)
Mammalian cell culture
MycoAlert Plus (Lonza, cat. no. LT07-710)
DMEM, high glucose, GlutaMAX Supplement (Thermo Fisher Scientific, cat. no. 10566016; phenol-red free: 21063029)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, cat. no. 16000044) FBS should be divided into aliquots and frozen at −20 °C if not in use for culture medium.
PBS, pH 7.4 (1×) (Thermo Fisher Scientific, cat. no. 10010023)
TrypLE Express Enzyme (1×), phenol red (Thermo Fisher Scientific, cat. no. 12605036; phenol-red free: 12604021)
Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, cat. no. 11668019)
Opti-MEM Reduced Serum Medium (Thermo Fisher Scientific, cat. no. 31985070)
GFP transfection marker: pmaxGFP (provided in Lonza Nucleofector kits such as SE Cell Line 4D-Nucleofector X Kit S; Lonza, cat. no. V4XC-1032)
Proteinase K (NEB, cat. no. P8107S)
Tris-HCl, pH 8.0, 1 M solution (Thermo Fisher Scientific, cat. no. 15568025)
SDS, 10% (wt/vol) solution (Thermo Fisher Scientific, cat. no. 15553027)
SE Cell Line 4D-Nucleofector X Kit S, for electroporation of editor mRNA (Lonza, cat. no. V4XC-1032)
Biological materials
One Shot Mach1 T1 Phage-Resistant Chemically Competent Escherichia coli (Thermo Fisher, cat. no. C862003) or preferred cloning strain
HEK293T cell line (ATCC, cat. no. CRL-3216; RRID: CVCL_0063) CAUTION: All cell lines should be regularly tested for mycoplasma
Primary human fibroblasts can be purchased from a biobank such as the Coriell Institute. Primary Tay–Sachs disease patient fibroblast cells were previously obtained from the Coriell Institute (cat. no. GM00221).
High-throughput sequencing analysis
Phusion U Green Multiplex PCR Master Mix, 2x (Thermo Fisher Scientific, cat. no. F564L) or any other high-fidelity polymerase.
QIAquick Gel Extraction Kit (Qiagen, cat. no. 28704)
Qubit double-stranded DNA High-Sensitivity Assay Kit (Thermo Fisher Scientific, cat. no. Q32854)
MiSeq Reagent Kit v2 (300-cycles) (Illumina, cat. no. MS-103-1002—Micro kit, ~4 million reads; MS-102-2002—standard kit, ~15 million reads)
Equipment
Filtered sterile pipette tips (e.g., Biotix, Rainin, VWR)
Serological pipettes, assorted (Corning)
Standard microcentrifuge tubes, 1.5 ml (Neptune Scientific, cat. no. 4445.X)
Standard PCR tube strips, 8 tubes/strip, 0.2 ml (Corning, cat. no. PCR-0208-C)
Standard PCR 1 × 8 strip caps, for 0.2-ml PCR tubes (Corning, cat. no. PCR-2CP-RT-C)
Falcon centrifuge tubes, polypropylene, 15 ml (VWR, cat. no. 62406-200)
Falcon centrifuge tubes, polypropylene, 50 ml (VWR, cat. no. 21008-940)
Corning 50-ml Mini Bioreactor (Corning, cat. no. 431720)
VWR 96-Well Deep-Well Plates with Automation Notches (VWR, cat. no. 76329-998)
Corning vacuum filter/storage bottle system, 0.22-μm pore, 33.2 cm2 polyethersulfone (PES) membrane (Corning, cat. no. 431097)
8-tube PCR strips, white for qPCR (Bio-rad, cat. no. TLS0851)
Flat PCR tube 8-cap strips, optical, ultraclear (Bio-rad, cat. no. TCS0803)
VWR 96-well PCR plate (VWR, cat. no. 89218-296)
Hard-shell 96-well PCR plates, for qPCR (Bio-rad, cat. no. HSP9655)
Microseal ‘F’ PCR plate seal, foil (Bio-rad, cat. no. MSF1001)
PCR plate heat seal, clear, optical, for qPCR reactions (Bio-rad, cat. no. 1814030)
Plastic inoculating loops, 10 μl (Copan, cat. no. COP-S10)
Non-tissue culture–treated bacteriological Petri dish, 100 × 15 mm (VWR, cat. no. 470210-568)
96-well clear flat-bottom TC-treated microplates with lids (Corning, cat. no. 353075)
Falcon TC-treated cell culture flask with vented cap, 75 cm2 (Corning, cat. no. 353136)
Light microscope with filters for fluorescence (Zeiss Axio Observer or comparable system)
Gel casting system (Bio-rad, cat. no. 1704412—caster; and Bio-rad, cat. no. 1704416—gel tray)
Gel electrophoresis system (Bio-rad, cat. no. 1704401)
Power supply for gel electrophoresis (Bio-rad, cat. no. 1645070)
PCR thermocycler with 48- and/or 96-well heating blocks (Bio-rad, cat. no. 1851197)
Real-time PCR detection system (e.g., Bio-rad CFX96 or comparable system)
Benchtop microcentrifuge (Eppendorf, cat. no. 5405000441)
Tabletop centrifuge with rotor fitting 50-ml conical tubes (Eppendorf, cat. no. 022623508 or comparable system)
Qubit 4 fluorometer (Thermo Fisher Scientific, cat. no. Q33238)
Nucleocounter NC-300 (Chemometec) or other cell counter
Lonza 4D Nucleofector with X unit (Lonza, cat. no. AAF-1002X and AAF-1002B)
37 °C, humidity- and CO2-regulated incubator (Thermo Fisher Scientific, cat. no. 51030284 or comparable system)
Benchtop vortexer (Fisher Scientific, cat. no. 02-215-414 or comparable system)
Blue-light transilluminator for gel cutting (VWR, cat. no. 76151-834 or comparable system)
Gel-imaging system (Bio-rad ChemiDoc or comparable system)
NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Fisher cat. no. ND-ONE-W)
MiSeq system (Illumina, cat. no. SY-410-1003)
Software
CRISPResso2 (https://github.com/pinellolab/CRISPResso2)
Geneious or preferred comparable software (https://www.geneious.com/)
Reagent setup
Oligonucleotide annealing buffer for Golden Gate cloning
To prepare 50 ml of annealing buffer, combine 500 μl 1 M Tris-HCl, pH 8.0 with 500 μl 5 M NaCl. Add nuclease-free water to a final volume of 50 ml. This solution can be stored at room temperature (25 °C) indefinitely.
Mammalian cell lysis buffer for gDNA extraction from HEK293Ts and primary fibroblasts
Mix 10 ml of 1 M pH 8.0 Tris-HCl, 5 ml of 10% (wt/vol) SDS solution, and nuclease-free water to a total volume of 1 liter. Store this incomplete buffer at room temperature (25 °C) for <6 months. Immediately before lysis, make a small aliquot of complete mammalian cell lysis buffer by adding a 1:1,000 (vol/vol) dilution of proteinase K (NEB).
DMEM culture medium with FBS for culturing HEK293T cells and primary human fibroblasts
Refer to final FBS concentration suggested for growth media by cell line vendors, especially when growing primary fibroblasts.
For HEK293T cells, prepare a 500 mL volume of 10% FBS-supplemented culture medium by adding 50 ml FBS to 450 ml DMEM and sterile filtering.
For primary human fibroblasts, prepare a 500 mL volume of 20% FBS-supplemented culture medium by adding 100 ml FBS to 400 ml DMEM and sterile filtering.
After supplementing with FBS, DMEM should be stored for a maximum of 3 weeks at 4 °C.
Procedure
Design of epegRNAs and nicking sgRNAs (Timing 1 day)
-
1
Follow the process outlined in the “Designing candidate epegRNAs” section to create a list of epegRNA spacer and RTT/PBS 3′ extension sequences
-
2
Follow the process outlined in “Choice of prime editing system (PE1-PE5) and editor architecture” to design nicking sgRNAs, if necessary.
Preparation of epegRNA or sgRNA constructs
-
3
When delivering epegRNAs and nicking sgRNAs as plasmids, either Golden Gate cloning (option A) or isothermal assembly (option B) can be used to generate constructs. If pegRNAs, epegRNAs, or nicking sgRNAs will instead be delivered as RNA, they should be purchased with chemical modifications that enhance editing (option C).
A. Generation of epegRNAs or sgRNAs by Golden Gate cloning (Timing 3 days)
[CRITICAL] This method is most useful for altering spacers and RTT/PBS 3′ extensions while keeping the scaffold and tevopreQ1 motif constant. The modified version of this procedure noted throughout is also useful for cloning nicking sgRNAs.
-
Design Golden Gate cloning oligonucleotides, following the examples listed in Table 2. Briefly, these oligos include:
Top and bottom oligos with cloning overhangs to insert the spacer sequence (Golden Gate part 1).
Top and bottom oligos with cloning overhangs to insert the SpCas9 sgRNA scaffold sequence (Golden Gate part 2). These can either be ordered with 5′ phosphorylation or they can be phosphorylated by the experimenter. Note: Golden Gate part 2 will be different between epegRNAs and nicking sgRNAs to account for the absence of an epegRNA RTT/PBS 3′ extension in nicking sgRNAs.
Top and bottom oligos with cloning overhangs to insert the desired epegRNA RTT/PBS 3′ extension (Golden Gate part 3). This is not required if cloning a nicking sgRNA.
-
In separate reactions for Golden Gate parts 1, 2, and 3, anneal ssDNA oligonucleotides to create the dsDNA parts necessary for Golden Gate assembly. Prepare the annealing reactions as follows:
Component Amount (μL) Final concentration Top oligonucleotide, 100 μM 1 μl 4 μM Bottom oligonucleotide, 100 μM 1 μl 4 μM Annealing buffer 23 μl - Total reaction volume 25 μl - [CRITICAL STEP] If cloning an sgRNA, only two annealing reactions (Part 1 for the spacer and the modified Part 2 for the scaffold) are necessary.
- Perform the annealing reactions under the following conditions in a thermocycler:
Step number Step description Duration 1 95 °C 3 m 2 Cool to 22 °C at 0.1 °C / s - -
Dilute annealed oligonucleotides by adding 75 μL H2O. The final concentration of each dsDNA Golden Gate part will now be 1 μM.
[CRITICAL STEP] Do not dilute the sgRNA scaffold (Golden Gate part 2) if oligos were purchased without 5′ phosphorylation.
[Pause Point] Golden Gate parts can be stored at −20 °C indefinitely.
- (Optional) If Golden Gate Part 2 oligos were purchased without 5′ phosphorylation, phosphorylate the annealed scaffold oligos (Golden Gate part 2) from Step 3A(iii). This step is not necessary if top and bottom oligos were purchased with 5′ phosphorylation. Assemble the T4 Polynucleotide Kinase in a reaction as follows:
Component Amount (μL) Final concentration 4 μM dsDNA Golden Gate part 2 (not phosphorylated) 25 μl 1 μM 10× T4 DNA ligase buffer 10 μl 1× T4 Polynucleotide Kinase (10U / μl) 2 μl 0.2 U / μl Nuclease-free H20 63 μl - Total reaction volume 100 μl - -
In a thermocycler, incubate at 37 °C for 60 minutes. Following this phosphorylation reaction, annealed scaffold oligonucleotides are now at a concentration of 1 μM.
[Pause Point] Phosphorylated and annealed oligonucleotides can be stored at −20 °C and reused indefinitely for future reactions.
-
Predigestion and agarose gel extraction of the epegRNA expression vector. We recommend cloning epegRNAs using the plasmid pU6-tevopreq1-GG-acceptor (Addgene ID: 174038) which already contains the tevopreQ1 3′ structural motif and a human U6 promoter.
[CRITICAL STEP] If cloning a nicking sgRNA, use the plasmid pU6-pegRNA-GG-acceptor (Addgene ID: 132777), which is a U6 promoter mammalian expression vector without the tevopreQ1 3′ structural motif.
- Prepare a triple restriction enzyme digest of 5μg of pU6-tevopreq1-GG-acceptor as follows:
Component Amount (μL) Final concentration pU6-tevopreq1-GG-acceptor, 5μg x μl 125 ng/μl BsaI-HFv2 (20U / μl) 1 μl 0.5 U/μl NcoI-HF (20U / μl) 1 μl 0.5 U/μl PvuII-HF (20U / μl) 1 μl 0.5 U/μl 10× rCutsmart Buffer 4 μl 1× Nuclease-free H20 to 40 μl - Total volume 40 μl - Incubate the reaction for 4–16 hours at 37 °C.
After the restriction digest, use agarose gel electrophoresis to verify successful digestion and gel extract the linearized cloning vector. Make a 1% agarose gel supplemented with 1:10,000 (vol/vol) ethidium bromide or preferred nucleic acid staining reagent. Mix the 40 μl restriction digest with 8 μl 6x purple loading dye (1x final concentration) and load all 48 μl into the gel along with a DNA ladder in a separate lane. Run the gel in a 1× TAE buffer at 140 V/cm for 20 min. Successfully digested plasmids will yield a prominent 2.2 kb restriction fragment corresponding to the desired backbone and an 825bp RFP dropout cassette.
Selectively excise and purify the 2.2 kb restriction fragment products using the QIAquick Gel Extraction kit (Qiagen) according to the manufacturer’s protocols. This 2.2 kb restriction fragment is Golden Gate part 4.
-
Elute in EB buffer (provided in the kit) or water and normalize the concentration to 30 ng/μl.
[Pause Point] Purified restriction fragments can be stored at −20 °C for several months.
-
Set up the Golden Gate reaction to assemble an epegRNA as follows:
[CRITICAL STEP] If cloning a nicking sgRNA, there will be no Golden Gate part 3, a different part 2 (as detailed in Table 2) than shown below, and a different part 4 (as detailed in Step 3Avii) than shown below.Component Amount (μL) Final concentration Annealed spacer oligonucleotides, 1μM
Golden Gate part 1 (from Step 3Aiv)1 μl 0.1 μM Annealed and phosphorylated sgRNA scaffold oligonucleotides, 1μM
Golden Gate part 2 (from Step 3Aiv or 3Avi)1 μl 0.1 μM Annealed epegRNA RTT/PBS 3′ extension oligonucleotides, 1μM
Golden Gate part 3 (from Step 3Aiv)1 μl 0.1 μM pU6-tevopreq1-GG-acceptor 2.2 kb fragment, 30ng / μl
Golden Gate part 4 (from Step 3Axii)1 μl 3 ng / μl BsaI-HFv2 (20U / μl) 0.2 μl 0.4 U/μl NcoI-HF (20U / μl) 0.2 μl 0.4 U/μl PvuII-HF (20U / μl) 0.2 μl 0.4 U/μl T4 DNA ligase (40U / μl) 0.50 μl 2 U/μl 10× T4 DNA ligase buffer 1 μl 1× Nuclease-free H20 3.9 μl - Total reaction volume 10 μl - - Perform the assembly reaction under the following conditions in a thermocycler:
Cycle number Step description Duration 1 20 °C 10m 2 37 °C 5m 3 80 °C 5m Following the completion of the Golden Gate assembly reaction, place the reactions on ice.
To transform the Golden Gate assembly into chemi-competent E. coli, combine 1 μl of each reaction and 10-μl of chemi-competent E. coli Mach1 cells or another chemi-competent strain.
Incubate the assembly/cell mix on ice for 10 min, heat-shock the mix at 42 °C for 30 s, and then immediately return the mix to ice for 1 min.
-
Add 100 μl of S.O.C. media to the mix and plate the entire volume on an LB-agar plate containing 50 μg/ml carbenicillin. Incubate overnight at 37 °C.
[CRITICAL STEP] Additional outgrowth after heat shock is not required.
[Pause Point] Transformed E. coli can be stored at 4 °C for 1 week.
-
Perform a rolling circle amplification (RCA) according to manufacturer (Cytiva) instructions. Briefly, pick individual RFP-negative colonies into 5 μl of sample buffer. Heat the mixture to 95 °C for 5 minutes in a thermocycler, and then add 5 μl of reaction buffer and 0.2 μl of enzyme. Incubate at 30 °C for at least 5 hours.
[CRITICAL STEP] Do not pick red colonies. These are colonies with undigested or reassembled pU6-tevopreq1-GG-acceptor plasmids.
[? Troubleshooting]
-
Sequencing of epegRNA or sgRNA expression plasmid. Using a preferred Sanger sequencing vendor, submit completed RCA reactions for sequence validation.
[CRITICAL STEP] Be sure to use a sequencing primer that will provide coverage of the epegRNA spacer, sgRNA scaffold, and RTT/PBS 3′ extension. Sequencing verification of the entire cloned epegRNA (or nicking sgRNA) sequence is necessary to avoid junction mutations or mutations from impure oligos.
In single wells of a 96-well deep-well plate, inoculate 1 ml cultures of sequence-verified colonies. LB media with 50 μg/ml carbenicillin should be used. Incubate at 37 °C with shaking for 20 h.
-
Use a Promega PureYield Plasmid Miniprep kit or another endotoxin-free plasmid preparation kit to isolate plasmid DNA from each 1 ml culture, according to the manufacturer’s instructions.
[Pause Point] Purified plasmids can be stored at −20 °C indefinitely.
B. Generation of epegRNAs or sgRNAs by isothermal assembly (Timing 3 days)
[CRITICAL] This method is recommended when one prefers a simpler two-component assembly and to have complete control over the entire epegRNA or nicking sgRNA sequence. Isothermal assembly for epegRNAs and sgRNAs is the same; only the gene fragments differ.
Design and order epegRNA or sgRNA isothermal assembly gene fragments following the examples listed in Table 2. These fragments should include all epegRNA elements (spacer, sgRNA scaffold, RTT, PBS, and 3′ structural motif) or sgRNA elements (spacer and sgRNA scaffold) between the two adapter sequences.
-
Perform a PCR using the isothermal assembly primers listed in Table 2 and the template pU6-tevopreq1-GG-acceptor (Addgene ID: 174038). The reaction is assembled as follows:
Component Amount (μL) Final concentration Isothermal assm. Forward primer, 100 μM 0.25 μl 0.5 μM Isothermal assm. Reverse primer, 100 μM 0.25 μl 0.5 μM pU6-tevopreq1-GG-acceptor, 200 ng/μl 0.05 μl 0.2 ng / μl Phusion HF 2× Master Mix 25 μl 1× Nuclease-free H20 24.5 μl - Total reaction volume 50 μl - [CRITICAL STEP] Phusion High-Fidelity PCR Master Mix with HF Buffer is specifically used because its buffer is compatible with a later DpnI digestion in Step 3B(v).
- Perform PCR using the following program:
Cycle number Denature Anneal Extend 1 98 °C, 3 m - - 2–36 98 °C, 15 s 61 °C, 15 s 72 °C, 1 m 37 - - 72 °C, 5 m Make a 1% agarose gel supplemented with 1:10,000 (vol/vol) ethidium bromide (or other DNA gel stain). Mix 1 μl of the PCR reaction with 4 μl water and 1 μl of 6x purple loading dye. Load this mix into the gel along with a ladder and run the gel in a 1× TAE buffer at 140 V/cm for 20 min. The correct PCR product will yield a prominent 2 kb band.
-
Digest the PCR reaction with DpnI (NEB), which removes the template plasmid input. This digestion is essential to minimize re-transformation of the PCR template. Add 1 μl of DpnI (20 U / μl) to the unpurified PCR and incubate at 37 °C for 15 minutes on a thermocycler.
[CRITICAL STEP] DpnI can be added directly to this reaction as it is active in the HF buffer supplied with the Phusion HF 2x Mastermix.
-
Purify the PCR products using the QIAquick PCR purification kit (Qiagen) according to the manufacturer’s instruction. Elute in water and dilute the PCR products to a concentration of 70 ng/μl.
[Pause Point] Purified amplicons can be stored at −20 °C indefinitely and reused for different cloning projects.
- Set up the isothermal reaction as follows:
Component Amount (μL) Final concentration NEBuilder HiFi DNA Assembly 2× Master Mix 6 μl 1× Gene fragment (from Step 3Bi), 10 ng/μl 5 μl 4.2 ng/μl 2 kb PCR amplicon (from Step 3B(vi), 70 ng/μl 1 μl 5.8 ng/μl Total reaction volume 12 μl - Incubate the isothermal assembly at 50 °C for 15–60 minutes on a thermocycler.
Following the completion of the isothermal assembly, place the reactions on ice.
-
For transformation and sequence verification, follow the same procedure used for the Golden Gate Assembly (Steps 3A (xvi-xxii)).
[CRITICAL STEP] In this method, the entire pU6-tevopreq1-GG-acceptor plasmid is amplified using PCR, which risks generating mutations throughout the entire plasmid. Therefore, when validating the sequence, be sure to use a sequencing primer or primers that will provide coverage of the vector’s entire U6 promoter, all epegRNA/sgRNA elements, and terminator. Mutations in any of these could yield ineffective constructs.
[? Troubleshooting]
C. Acquiring purified, chemically modified, synthetic epegRNAs, pegRNAs, or sgRNAs (Timing 1–6 weeks)
[CRITICAL] In general, researchers can deliver epegRNAs, pegRNAs, and nicking sgRNAs either as plasmids (e.g. Step 20A) or as chemically modified synthetic RNAs (e.g. Step 20B). Delivery of chemically modified synthetic RNAs is preferred if the PE protein components will be delivered as in vitro transcribed mRNAs (produced in Steps 4–19). The use of in vitro transcribed mRNA and synthetic guide RNAs can enable higher editing than plasmid delivery in certain cell types.
[CRITICAL] When ordering synthetic epegRNAs from Agilent, IDT, or other vendors, it is important that the ends of the RNA are chemically modified to prevent degradation in cells. Include 2′O-methyl groups on the first three and last three nucleotides and replace the first three and last three phosphodiester bonds with phosphorothioate bonds. We recommend ordering enough synthetic RNA to use 90 pmol of epegRNA and 60 pmol of nicking sgRNA per sample, but these amounts may need optimization for each different electroporation system and cell type.
Dissolve lyophilized synthetic epegRNAs and/or sgRNAs in TE buffer. Resuspend RNAs to a concentration of 100–300 μM and store at −20 °C for ≤1 year.
Preparation of in vitro transcribed PEmax mRNA (Optional) (Timing 1–2 Days)
[CRITICAL] These steps are only necessary when delivering prime editors as mRNA transcripts (e.g. Step 20B). mRNA delivery can greatly enhance editing in some cell types, as shown in Fig. 7h.
Figure 7. Example results.
(a, b) Heat map showing a PE4 system screen of PBS lengths and RTT lengths for (a) CXCR4 P191A installation and (b) IL2RB H134D + Y135F installation. Note that the optimal PBS and RTT lengths are different between the two sites shown in a and b. Values shown in the heat map cells reflect the mean of n=3 independent replicates. (c, d) Application of nicking sgRNAs at the CXCR4 locus (c) and the IL2RB locus (d). Nicking sgRNAs improve editing in both the PE4 and PE5 system, and MLH1dn improves editing with and without a nicking sgRNA. All values from n=3 independent replicates are shown. (e, f) Editing of the CXCR4 locus (e) and the IL2RB locus (f) in HeLa cells, which are less amenable to prime editing; here, the use of epegRNAs, the PEmax architecture, and MLH1dn dramatically improves editing over the original conditions (PE2 and PE3 with an unmodified pegRNA). All values from n=3 independent replicates are shown. (g) Example allele table generated by CRISPResso2. (h) Example of delivery optimization in patient-derived iPSC cells. Relative to lipid transfection and plasmid electroporation, mRNA electroporation generated a large improvement in editing efficiency. All values from n=3 independent replicates are shown. Data shown in a-h was uniquely collected for this protocol, and is deposited at the NCBI Sequence Read Archive database under PRJNA817825, but experimental techniques are identical to previously reported work15,30,31.
-
4DNA templates for in vitro transcription must be linear, not circular. To generate a linear in vitro transcription template, PCR amplify PEmax and/or MLH1dn from mRNA transcription template plasmids (Addgene ID: 178113 and 178114, respectively) using the primers listed in Table 2. Set up the following reaction:
Component Amount (μL) Final concentration in vitro transcription forward primer, 100 μM 0.75 μl 0.25 μM in vitro transcription reverse primer, 100 μM 0.75 μl 0.25 μM mRNA transcription template plasmid, 40 ng/μl 6 μl 0.8 ng/μl Phusion U Green Multiplex Master Mix, 2× 150 μl 1× Nuclease-free H20 142.5 μl - Total reaction volume 300 μl - [CRITICAL STEP] This reaction is a scaled-up version of a standard 50 μL PCR. We find that total DNA yields from this PCR can be relatively low and that pooling multiple 50 μL PCRs into a single PCR purification column (Step 6) provides enough template for subsequent in vitro transcription (Step 8). Using typical equipment, this 300 μL mastermix will need to be divided into six individual 50 μL reactions on a thermocycler.
-
5Perform the PCR under the following conditions:
Cycle number Denature Anneal Extend 1 98 °C, 2 m - - 2–36 98 °C, 15 s 71.4 °C, 30 s 72 °C, 3:30 m (PEmax); 72 °C, 1:15 m (MLH1dn) 37 - - 72 °C, 5 m -
6
Purify the PCR products from the 300 μL mastermix using a single silica column from the QIAquick PCR purification kit (Qiagen) according to the manufacturer’s protocols. Elute in EB (provided with the kit) and quantify purified product concentration by UV-Vis spectrophotometry (NanoDrop) or equivalent method.
[CRITICAL STEP] The mRNA transcription template plasmid contains a T7 promoter disabled by a single nucleotide mutation. PCR amplification with the in vitro transcription forward primer generates an amplicon with a repaired T7 promoter. The disabled T7 promoter on the template plasmid prevents transcription initiation and obviates the need to remove the template plasmid via DpnI digest or gel purification. Instead, a simple silica column cleanup can be used in this step.
-
7
After PCR purification, verify amplification via agarose gel (0.7%, supplemented with 1:10,000 (vol/vol) ethidium bromide or other nucleic acid stain) electrophoresis. Dilute 100ng of purified PCR product in 5 μl of nuclease-free water and mix with 1 μL of 6x purple loading dye. Load this mix into the gel along with ladder in a separate lane. Run the gel in a 1× TAE buffer at 140 V/cm for 20 min. Successfully amplified in vitro transcription templates will yield a distinct 6.5 kb amplicon.
[? Troubleshooting]
-
8
Using the HiScribe T7 High Yield RNA Synthesis Kit (NEB), set up an in vitro transcription reaction as follows, scaling the reaction up or down as needed:
[CRITICAL STEP] This reaction follows the manufacturer-suggested protocol for HiScribe T7 High Yield RNA Synthesis Kit when using Trilink’s CleanCap Reagent AG to enable co-transcriptional capping. However, we additionally replace the kit’s 100 mM UTP with Trilink’s 100mM N1-Methylpseudouridine-5′-Triphosphate.
[CRITICAL STEP] RNAse-free technique is essential during this step and all subsequent in vitro transcription steps. RNAse contamination will compromise mRNA integrity and produce sub-optimal results. Before starting an in vitro transcription reaction setup, decontaminate all work surfaces, pipettes, and other materials with an RNase decontamination solution, such as RNaseZap (Thermo Fisher) and ensure that tubes, pipette tips, and other disposables are RNAse free.Component Amount (μL) Final concentration Nuclease-free H20 24.4 - x μl - 10X Reaction Buffer 2 μl 0.5× ATP, 100 mM 2 μl 5 mM CTP, 100 mM 2 μl 5 mM GTP, 100 mM 2 μl 5 mM N1-Methylpseudouridine-5′-Triphosphate, 100 mM 2 μl 5 mM CleanCap Reagent AG, 100mM 1.6 μl 4 mM Purified linear Template DNA from Step 6 x μl 1 μg total T7 RNA polymerase mix 4 μl - Total reaction volume 40 μl - -
9
Incubate the in vitro transcription reaction at 37 °C for 2 hours in a thermocycler or a dry air incubator.
-
10Remove template DNA by DNase (NEB) treatment. Set up DNAse digest as listed below:
Component Amount (μL) Final concentration Step 9 reaction mix 40 μl - Nuclease-free H20 136 μl - DNAse I reaction buffer, 10× 20 μl 1× DNAse I, RNAse free 2 U/μl 4 μl 0.04 U/μl Total reaction volume 200 μl - -
11
Incubate the DNAse I treatment at 37 °C for 15 minutes in a thermocycler.
-
12
Purify the synthesized RNA by lithium chloride precipitation: mix the 200 μl reaction from Step 10 with 100 μl 7.5 M LiCl.
-
13
Incubate the mixture at −20 °C for 30 minutes.
-
14
Centrifuge at 21,000 × g in a microcentrifuge for 15 minutes. A temperature-controlled microcentrifuge set to 4 °C is preferred, if available.
-
15
A white pellet of precipitated RNA will form in the tube. Pipette off the supernatant and wash the pellet with ice-cold 70% ethanol. Do not remove the 70% ethanol.
-
16
Centrifuge again at top speed in a microcentrifuge for 5 minutes.
-
17
Remove all the 70% ethanol without disturbing the pellet. Resuspend the pellet in nuclease-free water or 10 mM Tris, 1 mM EDTA. Quantify purified mRNA concentration by UV-Vis spectrophotometry (NanoDrop) or equivalent method.
-
18
Verify successful and precise transcription via agarose gel electrophoresis (2.0%, supplemented with 1:10,000 (vol/vol) SYBR Gold nucleic acid staining reagent, Thermo Fisher Scientific): dilute 300ng of purified Step 17 product in 5 μl of nuclease-free water and mix with 5 μL 2x Gel Loading Buffer II (Thermo Fisher). Also dilute 2.5 μl of Millennium RNA Markers (Thermo Fisher) in 2.5 μl nuclease-free water and mix with 5 μL 2x Gel Loading Buffer II. Heat both 10 μl mixtures on a thermocycler for 10 minutes at 70 °C. Load both mixtures into separate lanes of the 2% gel and perform electrophoresis in a 1× TAE buffer at 140 V/cm for 20–30 min. Successfully transcribed mRNAs will yield a distinct 6.5 kb (PEmax) or 2.4 kb (MLH1dn) mRNA transcript.
[? Troubleshooting]
-
19
If gel electrophoresis confirms that the transcribed mRNA is high quality, distribute the purified mRNA into working aliquots of 5 – 20 μl.
[CRITICAL STEP] Multiple freeze-thaw cycles will result in mRNA degradation and should be avoided whenever possible. Preparing multiple aliquots is essential to maximizing the shelf life of in vitro transcribed mRNAs.
[Pause Point] Purified mRNA transcripts can be stored at −80 °C for several months if not subjected to multiple freeze-thaw cycles.
Verification of prime editing in HEK293T cells or primary human fibroblasts
-
20
Prime editing can be verified in a variety of mammalian cell types, including HEK293T cells (option A) or primary human fibroblasts (option B). We recommend HEK293T cells as a workhorse cell line for prime editing epegRNA optimization. Primary cells, such as primary human fibroblasts, can be used to verify prime editing correction of pathogenic mutations in patient cells.
A. Prime editing in HEK293T cells via plasmid transfection (Timing 4–5 Days)
[CRITICAL] In this example transfection protocol, we describe a PE5 transfection, which typically yields the highest editing efficiency out of all PE systems and drastically reduces indels relative to PE3. PE5 requires expression plasmids for four PE components: (1) PEmax (2) an epegRNA (3) a nicking sgRNA, and (4) MLH1dn. In systems such as PE2, PE3, PE3b, and PE4, the nicking sgRNA and/or MLH1dn are not included and would be excluded from this protocol. For twinPE transfections, two epegRNAs are used instead of an epegRNA and a nicking sgRNA. (See Table 3 for plasmid amounts to be used for each PE system.)
Table 3.
DNA amounts for lipid transfection, based on prime editor system
| PE System | Amounts of Transfection Components |
|---|---|
| PE2 | 200 ng PEmax plasmid 66 ng epegRNA plasmid |
| PE3/PE3b | 200 ng PEmax plasmid 66 ng epegRNA plasmid 22 ng nicking sgRNA plasmid |
| PE4 | 200 ng PEmax plasmid 66 ng epegRNA plasmid 100 ng MLHIdn plasmid |
| PE5/PE5b | 200 ng PEmax plasmid 66 ng epegRNA plasmid 22 ng nicking sgRNA plasmid 100 ng MLHIdn plasmid |
| TwinPE | 200 ng PEmax plasmid 33 ng epegRNA 1 plasmid 33 ng epegRNA 2 plasmid |
| TwinPE + recombinase single-transfection targeted donor integration *Optimized for 48-well plate |
500 ng PEmax plasmid 50 ng epegRNA 1 plasmid 50 ng epegRNA 2 plasmid 200 ng Bxbl plasmid 200 ng recombination donor plasmid |
All of these amounts, except for those associated with single-transfection integration, have been optimized for 96-well plate transfections of HEK293T cells using 0.5 μl per well of Lipofectamine 2000.The single-transfection integration amounts have been optimized for 48-well plate transfections of HEK293T cells using 1 μl per well of Lipofectamine 2000.
Plasmid preparation. Order or clone expression plasmids for all desired prime editing components: prime editor (PEmax architecture, Addgene #174820), epegRNA, nicking sgRNA, and MLH1dn (Addgene #174824). See Steps 3A or 3B for epegRNA and nicking sgRNA cloning instructions.
Generate transfection-grade preparations of expression vectors using endotoxin-free plasmid isolation kits such as Qiagen Plasmid Plus Midi Kit (Qiagen) or PureYield Plasmid Miniprep System (Promega) according to the manufacturer’s protocol.
-
HEK293T cell culture. Follow the vendor-specified (ATCC) protocol to culture HEK293T cells. Briefly, use DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (vol/vol) and grow HEK293Ts in T75 tissue culture flasks maintained at 37 °C and 5% CO2.
[CRITICAL STEP] Penicillin and streptomycin can be included during the culture of HEK293Ts. However, they should be avoided when plating cells for transfection: using antibiotics during transfections can affect both transfection efficiency and cell viability.
Culture HEK293T cells until 70% confluent. When 70% confluent, passage cells by removing growth medium, washing the cell monolayer with 1x PBS, and then removing the PBS wash, being careful to not detach the monolayer from the surface of the flask.
Add 2 ml of TrypLE (Thermo Fisher Scientific) and incubate at 37 °C and 5% CO2 for 5 minutes to dissociate the adherent cells.
After incubation, add 10mL of pre-warmed media to the flask. Pipette up and down to detach cells from the flask’s growth surface and to disperse clumps of cells.
-
Continue to subculture the cells by reseeding into a new T75 flask and/or preparing 96-well plates for plasmid transfection as detailed in Step 20A viii-ix.
[CRITICAL STEP] Do not grow HEK293T cell cultures beyond 80% confluency and dispose of cells after passage 20. We generally passage HEK293T cell cultures at a ratio between 1:5 and 1:10 every 2–3 days.
We perform experiments in 96-well plates, using 1.6–1.8 × 104 cells in 100 μl of FBS-supplemented DMEM per well. To plate HEK293T cells for transfection, firstly count the dissociated cells (from Step 20A(vi)) using a Nucleocounter NC-3000 (Chemometec) or other cell counter according to manufacturer instructions. Dilute the cells to a concentration of 1.6–1.8 × 105 cells/mL in FBS-supplemented DMEM.
-
Plate 100 μl of the diluted cell mix (from Step 20A(viii)) into each well of a 96 well plate. This will result in 1.6–1.8 × 104 cells per well.
[CRITICAL STEP] Cell viability and transfection efficiency are affected by the density at which cells are plated. Plating too many cells will reduce transfection efficiency and plating too few cells will result in excessive cell death.
Perform transfection 18–24h after plating (Step 20A(ix)), at which point cells should be approximately 70–80% confluent.
-
Transfection Mix Preparation. For the transfection of each well, mix the desired combinations of prime editor, epegRNA, nicking sgRNA, and MLH1dn expression plasmids from Step 20Aii following the transfection setup below:
[CRITICAL STEP] Every well of a PE5 editing experiment will receive a plasmid dose of each PE5 editing component: prime editor, epegRNA, nicking sgRNA, MLH1dn. When screening epegRNAs, we recommend normalizing the concentration of all epegRNA plasmids and making a mastermix of the other PE5 components to simplify the experimental workflow. For example, if screening 15 epegRNAs in a PE5 experiment, make a mastermix of 15 equivalents (plus overage) of prime editor plasmid, MLH1dn plasmid, sgRNA plasmid, and Opti-MEM.
[CRITICAL STEP] Including an unedited negative control at this stage is crucial. To do so, one can either omit the pegRNA and nicking sgRNA, or include a non-targeting pegRNA and nicking sgRNA pair.Component Amount per well Volume (μl) per well Prime editor vector (Step 20A(ii)) 200 ng variable epegRNA vector (Step 3a or 3b) 66 ng variable Nicking sgRNA vector (Step 3a or 3b) 22 ng variable MLH1dn vector (Step 20A(ii)) 100 ng variable Opti-MEM Reduced Serum Medium (Thermo Fisher Scientific) - to 5 μl Total reaction DNA and volume 388 ng 5 μl -
Prepare a lipid solution of 0.5 μl of Lipofectamine 2000 (Thermo Fisher Scientific) per well diluted into 4.5 μl of Opti-MEM per well, following the manufacturer’s instructions.
[CRITICAL STEP] In this protocol we describe using lipofectamine 2000 in HEK293T cells. Amounts of lipid and DNA will vary based on the transfection reagent and target cell type.
Add 5 μl of the separately prepared lipid mixture (from Step 20Axii) to each well of the plasmid mixture (from Step 20A(xi)) to a total volume of 10 μl and incubate for 10 minutes.
-
Transfer all 10 μL of the mix from Step 20A(xiii) to each well of the previously prepared 96-well tissue culture plate (Step 20A(ix)). Return the plate to the incubator at 37 °C and 5% CO2 when all wells have been treated.
[CRITICAL STEP] Take care to gently add the DNA and lipid mixture to the culture well. Forcefully ejecting liquid against the plated cell monolayer may dislodge cells from the growth surface or lead to toxicity.
B. Prime editing in primary human fibroblasts via RNA electroporation (Timing 4–5 Days)
[CRITICAL STEP] In this procedure, PEmax and MLH1dn are delivered as in vitro transcribed mRNAs (from Step 19), and the epegRNA and nicking sgRNA are delivered as chemically modified synthetic RNAs (from Step 3C).
[CRITICAL STEP] We have observed that prime editing efficiency is highest in primary human fibroblasts when mRNA and synthetic RNA prime editing reagents are delivered by electroporation. In this example, we describe a PE5 electroporation, which typically yields the highest editing efficiency out of all PE systems and reduces indels relative to PE3. A PE5 editing experiment requires four PE components: (1) PEmax (2) an epegRNA (3) a nicking sgRNA (4) MLH1dn. In systems such as PE2, PE3, PE3b, and PE4 the nicking sgRNA and/or MLH1dn are not included.
[CRITICAL STEP] Here, electroporation is conducted using the Lonza 4D Nucleofector with X unit (Lonza) but can be completed with an alternative electroporation system. The conditions described here were optimized for primary human fibroblasts: considerable optimization of electroporation conditions for other cell types should be expected. Protocols for optimization are available from electroporation equipment manufacturers.
-
Primary human fibroblast cell culture. Follow the vendor-specified protocol to maintain fibroblasts (Coriell Institute) in cell culture. Briefly, grow fibroblasts in T75 tissue culture flasks in DMEM (Thermo Fisher Scientific) supplemented with 20% (vol/vol) FBS (Thermo Fisher Scientific) at 37 °C and 5% CO2.
[CRITICAL STEP] We have found that in general, DMEM supplemented with 20% FBS is suitable for most primary fibroblasts, but always reference vendor-recommended growth instructions.
Passage fibroblasts until 70% confluent. When 70% confluent, passage cells by removing growth medium, washing the cell monolayer with 1x PBS, and then removing the PBS wash.
Add 3 ml of TrypLE (Thermo Fisher Scientific) and incubate at 37 °C and 5% CO2 for 5 minutes to dissociate the adherent cells.
After incubation, add 10mL of FBS-supplemented DMEM to the flask. Pipette repeatedly to detach cells from the flask’s growth surface dissociate the cells.
-
Reseed dissociated cells into a fresh flask to continue subculture or use the cells immediately for an RNA electroporation.
[CRITICAL STEP] Common maintenance antibiotics such as penicillin and streptomycin can be included during the fibroblast culture, however they may affect cell physiology.
[CRITICAL STEP] Do not allow cells to reach a confluency higher than 80%. For most primary fibroblast cell lines we work with, passaging at a 1:5 ratio every 2–3 days is sufficient. However, growth characteristics will likely vary between cell lines and may need to be adjusted.
Cell preparation for Lonza electroporation. Count dissociated cells from Step 20B(v), using a Nucleocounter NC-3000 (Chemometec) or other cell counter to determine the density of the dissociated cells.
Calculate the total number of cells required, using 1.0 × 105 fibroblasts per electroporation well. Centrifuge the total number of required cells in an appropriately sized tube at 150g for 5 minutes at room temperature (25 °C).
A pellet of cells will form. Remove and discard supernatant by vacuum aspiration or pipetting and wash the pellet of cells with 1 ml of PBS. Resuspend the cells in the PBS.
Repeat the 5 min centrifugation (Step 20B(vii)) to pellet the cells again. Remove and discard the supernatant.
Prepare the electroporation buffer for the Lonza SE nucleofection kit (Lonza) during the centrifugation steps. For each electroporation, mix 16.4 μl of Lonza SE nucleofector solution with 3.6 μl of Lonza SE supplement solution, for a total of 20 μl prepared electroporation buffer per electroporation.
Resuspend the pelleted cells from Step 20B(ix) with the prepared nucleofection solution from Step 20B(x). For example, if one intended to prepare 5 electroporations, a washed pellet of 5 × 105 cells would be resuspended in 100 μl of prepared electroporation buffer.
-
Prepare the prime editor reagent mixture. Prepare the following final reagent mix for the electroporation reaction as follows:
Component Amount Volume (μl) Prime editor in vitro transcribed mRNA (2 μg/μl stock) (Step 19) 1 μg 0.5 μl epegRNA synthetic RNA (200μM stock) (Step 3C) 90 pmol 0.45 μl Nicking sgRNA synthetic RNA (100μM stock) (Step 3C) 60 pmol 0.6 μl MLH1dn in vitro transcribed mRNA (2 μg/μl stock) (Step 19) 1 μg 0.5 μl Fibroblasts in Lonza SE buffer (5000 cells/μl, from Step 20Bxi) 1 × 105 cells 20 μl Total reaction volume - 22.05 μl [CRITICAL STEP] Holding cells in the nucleofection buffer for extended periods of time reduces cell viability and electroporation efficiency. Work as quickly as possible once the washed pellet from Step 20B(ix) is resuspended in the nucleofection buffer from Step 20B(x). If preparing many electroporations, premix the RNA components from Step 20B(xii) and hold them on ice until Step 20B(xi) is complete.
[CRITICAL STEP] Including an unedited negative control at this stage is crucial. To do so, one can either omit the pegRNA and nicking sgRNA, or include a non-targeting pegRNA and nicking sgRNA pair.
-
Transfer the 22 μl reagent mix into the 20-μl nucleocuvette wells included in the Lonza SE kit.
[CRITICAL STEP] Air bubbles in the cuvette will disrupt the electroporation. Use a thin pipette tip (e.g., a common 10 μL tip) to disrupt bubbles or drag bubbles out of the cuvette.
Electroporate the reaction mix using program CM-130 on a Lonza 4D nucleofector.
After electroporation, add 80 μl of 37 °C FBS-supplemented DMEM growth media to each electroporation reaction and gently mix. Incubate for 10 min at room temperature (25 °C) to allow cells to recover.
Following the incubation at room temperature, gently mix and transfer 40 μL of the recovered cell mix to a 48-well tissue culture plate filled with 250 μl of 37 °C FBS-supplemented DMEM growth media and transfer it to an incubator at 37 °C and 5% CO2.
Lysis of mammalian cells for HTS (Timing 1 Day)
-
21
72 hours after lipid transfection of plasmids into HEK293T cells (Step 20A(xiv)) or electroporation of RNA into primary human fibroblasts (20B(xvi)), cells are lysed for gDNA harvesting and HTS analysis.
[CRITICAL STEP] Here, we describe a simple cell lysis method for harvesting gDNA without further purification steps. Many alternative methods for harvesting gDNA can be used.
-
22
Prepare a fresh aliquot of complete mammalian cell lysis buffer (See Reagent Setup) by adding a 1:1,000-fold (vol/vol) dilution of proteinase K (NEB) into stored incomplete cell lysis buffer.
-
23
Remove media from edited cells from Step 20A(iv) or Step 20B(xvi) and carefully wash with PBS. Do not disturb the plated monolayers. Remove any residual PBS.
-
24
Cell lysis. Add lysis buffer from Step 22 directly to PBS-washed plates from Step 23. For lysis of 96-well plates, use 50 μl lysis buffer per well.
[CRITICAL STEP] Lysis buffer volume may need to be adjusted for different cell types or different cell densities.
-
25
Incubate plates at 37 °C for 1 hour after adding lysis buffer.
[CRITICAL STEP] Adding fresh lysis buffer to cell monolayers will generate a viscous solution that is difficult to pipette. This incubation can be completed on a thermocycler but will be complicated by difficult liquid transfers. We recommend lysing cells directly in culture plates.
-
26
After incubation, transfer lysate into PCR plates or strips by pipetting. Inactivate proteinase K heating at 80 °C for 30 minutes on a thermocycler. Heat-inactivated lysis mix can be used as a PCR template in subsequent HTS analysis.
[Pause Point] Cell lysis mix can be stored at 4 °C for 1 week or −20 °C for several months.
HTS for prime editing analysis (Timing 1–2 Days)
-
27
Design and order PCR1 primers (see Table 2) to amplify the target genomic locus. We recommend using NCBI’s Primer-BLAST tool to aid with the design of PCR1 primers.
[CRITICAL STEP] Primers must amplify a region spanning at least from 25 bp upstream of the epegRNA-guided nick to 25 bp downstream of the 3′ flap generated by the RT or any secondary nick (whichever is longer). If PCR1 primers are too close to either nick site, accurate indel quantification with CRISPResso2 will not be possible (see Table 4).
[CRITICAL STEP] PCR1 primers require 5′ adaptor sequences (see Table 2) so that individual samples can be barcoded in a second PCR (PCR2; see Step 32). These barcodes enable the identification of individual samples during later HTS analysis.
-
28Prepare the PCR1 reaction as follows:
Component Amount (μL) Final concentration Phusion U Green Multiplex Master Mix, 2× 12.5 μl 1× PCR1 forward primer (Step 27, Table 2), 100 μM 0.125 μl 0.5 μM PCR1 reverse primer (Step 27, Table 2), 100 μM 0.125 μl 0.5 μM Lysis mix with harvested gDNA (Step 26) 1 μl - Nuclease-free H20 11.25 μl - Total reaction volume 25 μl - [CRITICAL STEP] We recommend starting with 1 μl of lysis mix as a PCR template, but optimization of this volume may be required. Post-transfection cell density, cell type, and lysis volume will influence gDNA yields from the lysis mix (Step 26) and may affect PCR performance. Assuming cells divide twice between seeding and lysis, there will be ~1,280 cells/μl of lysis buffer. Adding less than 1 μl of lysis mix to PCR1 risks bottlenecking downstream analysis by the number of cells analyzed, as opposed to the detection limit of the MiSeq.
[CRITICAL STEP] We use Phusion U Green Multiplex Mastermix for PCR1 and PCR2. It includes a density reagent and two electrophoresis tracking dyes for direct loading of PCR products into gels, which saves considerable time during the HTS library preparation. While convenient, these properties are not critical, and any other comparable high-fidelity DNA polymerase may be used.
-
29Perform PCR1 under the following conditions:
Cycle number Denature Anneal Extend 1 98 °C, 3 m - - 2–24 98 °C, 10 s 60 °C, 20 s 72 °C, 30 s 25 - - 72 °C, 5 m [CRITICAL STEP] Excessive cycles of amplification at this step and PCR2 (Step 32) can introduce amplification bias. Bias can be minimized (but not completely removed) by performing as few PCR cycles as possible. qPCR should be used to determine this minimum cycle number, which corresponds to the top of the linear range. 24–29 cycles are sufficient for most loci. The optimal number of cycles for PCR1 will vary between amplicons.
! CAUTION If the target edit is a large deletion, PCR bias is more likely to occur. We found that for deletions 50 bp or less, bias is typically in the single-digit percentage range, but for larger deletions, the amount of bias can increase to 30–40%(ref. 42).
-
30
Confirm efficient and precise amplification of PCR1 amplicons using gel electrophoresis. Run 5 μL of each PCR1 reaction on a 1% (wt/vol) agarose gel at 140 V/cm for 10 minutes. Amplicons should be the length of the amplified genomic locus plus approximately 70bp. The additional ~ 70 bp in length is from the included 5′ adaptors appended to the PCR1 primers (See Table 2).
[CRITICAL STEP] Unoptimized PCR1 primers can bind nonspecifically throughout the genome and produce multiple amplification bands after PCR1. We generally test 3–5 pairs of PCR1 primers for each new site to find a specific, high-efficiency pair. If a specific primer pair cannot be found, gel extraction of the desired band is possible following PCR2.
[? Troubleshooting]
-
31
Dilute PCR2 primers to 10 μM. Forward and reverse primer sequences for PCR2 are designated by Illumina: (https://support.illumina.com/downloads/illumina-adapter-sequences-document-1000000002694.html).
-
32Use PCR1 products (Step 30) as a PCR template for PCR2. This second amplification appends Illumina indices that uniquely barcode individual samples. The PCR2 primers bind to the 5′ adaptor sequences appended to the PCR1 primers (See Table 2). Prepare the PCR2 reaction as follows:
Component Amount (μL) Final concentration Phusion U Green Multiplex Master Mix, 2× 12.5 μl 1× PCR2 forward primer, 10 μM 1.25 μl 0.5 μM PCR2 reverse primer, 10 μM 1.25 μl 0.5 μM PCR1 unpurified product (Step 30) 1 μl - Nuclease-free H20 9 μl - Total reaction volume 25 μl - [CRITICAL STEP] Use a unique combination of PCR2-Forward and PCR2-Reverse Illumina indices for each sample. This will enable their identification for use in later HTS steps.
-
33Perform PCR2 under the following conditions:
Cycle number Denature Anneal Extend 1 98 °C, 3 m - - 2–7 98 °C, 10 s 60 °C, 20 s 72 °C, 30 s 8 - - 72 °C, 5 m [CRITICAL STEP] PCR2 is also susceptible to PCR bias. Optimize this PCR as directed in Step 29. In general, we find that 7–10 cycles are generally a good starting point.
-
34
Confirm efficient and precise amplification of PCR2 amplicons using gel electrophoresis. Run 5 μL of each PCR2 reaction on a 1% (wt/vol) agarose gel at 140 V/cm for 10 minutes. Amplicons should be the length of the amplified genomic locus plus approximately 130bp. The additional 130 bp in length is from the sum of included 5′ adaptors appended to the PCR1 primers (~ 70bp, See Table 2) and the length of the appended PCR2 Illumina indices (~ 60bp).
-
35
If all PCR2 products are approximately the same length (<100 bp difference), pool 2 μL of each PCR2 product into a single mastermix. This mastermix will be used for a subsequent gel extraction (Step 36) and should have a minimum volume of 40 μL to ensure enough PCR product is present for an efficient gel extraction. Increase the volume of each individual pooled PCR2 product as needed to reach the 40 μL minimum volume (e.g., 4 μl of each PCR2 product if there are only 10 PCR2 reactions). If PCR2 products have variable length (>100 bp difference), pool similarly-sized amplicons into separate mastermixes.
[CRITICAL STEP] Sequencing coverage for an individual PCR2 product will be directly related to the molar amount of that product pooled into the gel extraction mastermix (Step 36). PCR2 yields (evaluated via agarose gel band intensity) and desired sequencing coverage of each PCR2 sample should be considered jointly when pooling individual samples into the gel extraction mastermix. Volume inputs into the gel extraction mastermix can be varied to approximately achieve the desired level of sequencing coverage for each sample.
-
36
Load 40–60 μl of the gel extraction mastermix from Step 35 onto a 1% (wt/vol) agarose gel for gel extraction. Run the gel for 20–30 min at 140 V.
-
37
Excise the desired PCR2 band from the gel using a razor blade and purify the size-separated amplicon from the agarose using the QIAquick Gel Extraction Kit (Qiagen) or equivalent gel extraction kit, following manufacturer’s instructions. Elute the gel-extracted DNA in nuclease-free water.
[CRITICAL STEP] It is important to perform this gel extraction precisely. Shorter amplicons bind more efficiently to the MiSeq flow cell, so contamination with low-molecular weight primer dimer will cause the loss of many reads in the subsequent MiSeq run. Therefore, be careful to excise only the desired amplicon and exclude primer dimer. If PCR1 or PCR2 produced several bands, only the desired length band should be gel extracted. If a large insertion or deletion was performed, gel extract an inclusive range that would contain both the starting and ending amplicon lengths. For example, if an unedited target would produce a 350-bp band after PCR2 and a 50-bp insertion was edited into this target, an inclusive range of all amplicons between 350 bp and 400 bp should be excised from PCR2.
-
38
Quantify the concentration of the eluted DNA using a Qubit kit or similar technique, following manufacturer instructions.
! CAUTION Incorrectly determining the concentration of a library could result in a failed MiSeq run or insufficient sequencing coverage. Underestimating the concentration will cause overloading of the sequencer in downstream steps, which can cause the run to fail due to over-clustering. Overestimating the concentration will lead to too little sample being loaded onto the sequencer, yielding fewer sequencing reads per sample. It is essential to determine the library concentration accurately.
-
39
Dilute the library with nuclease-free water to precisely 4 nM using the concentration determined in Step 38.
-
40
40. Illumina MiSeq DNA sequencing. Follow the instructions in the Illumina user manual to complete the remaining library-preparation steps and load the sequencer.
Table 4. CRISPResso2 common batch parameters.
Careful analysis is required to ensure accurate assessment of editing and indels. As a starting point, we recommend the following parameters. Below the parameter descriptions, we have provided example setups for standard mode (to be used for SNPs) and HDR mode (to be used for insertions, deletions, or multiple base changes).
| Description of CRISPResso2 parameters important for prime editing analysis | |||||||
| r1 (fastq_r1) | Specifies the name of the fastq file to be analyzed (a second r2 entry is required for analyzing paired end reads). | ||||||
| a (amplicon_seq) | Specifies the nucleotide sequence of the unedited amplicon | ||||||
| n (name) | Specifies the desired output filename | ||||||
| g (guide_seq) | Specifies the nucleotide sequence of the protospacer targeted for editing | ||||||
| q (min_average_read_quality) | Specifies the minimum average phred quality score needed for a read to be included in the analysis. The recommended value is 30. | ||||||
| qwc (quantification_window_coordinates) | Specifies the region of the unedited reference amplicon that CRISPResso2 will analyze for indels. The specified range is inclusive and zero-indexed, meaning that the first nucleotide of the amplicon is position 0. We recommend setting a range spanning from 10 bp 5′ upstream of the pegRNA-guided nick to 10 bp 3′ downstream of the 3′ flap generated by the RT or any secondary nick, whichever is longer, such that the entire inter-nick distance, flanked by 10 bp on either side, is analyzed for indels. | ||||||
| e (expected_amplicon_seq) | Specifies the nucleotide sequence of the edited amplicon. Only include this parameter when running CRISPResso2 in HDR mode to quantify insertions, deletions, or multiple-base pair substitutions. | ||||||
| discard_indel_reads | When set to TRUE, CRISPResso2 will discard reads containing an indel and count the number of discarded reads with respect to the reference amplicon (and also the expected amplicon in HDR mode). Doing so streamlines quantification of PE indels, as discarded reads can be easily counted after analysis. | ||||||
| Example batch parameter file for CRISPResso2 standard mode | |||||||
| r1 | a | n | g | q | qwc | discard_indel_reads | |
| SampleX_filename | Unedited reference amplicon sequence | SampleX_outputname | Protospacer sequence | 30 | StartingBP-EndingBP | TRUE | |
| Example batch parameter file for CRISPResso2 HDR mode | |||||||
| r1 | a | n | g | q | qwc | Discard_indel_reads | e |
| SampleX_filename | Unedited reference amplicon sequence | SampleX_outputname | Protospacer sequence | 30 | StartingBP-EndingBP | TRUE | Edited reference amplicon sequence |
HTS analysis (Timing 1–4 Hours)
[CRITICAL] A variety of computational pipelines are suitable for analyzing sequencing data generated by genome editing experiments. Here we describe a typical workflow for batch quantification of prime editing efficiencies using CRISPResso2 that is commonly used in our laboratory. The following protocol assumes the user already has access to CRISPResso2 via Docker, Bioconda, or local installation. Additional details for using CRISPResso2 can be found in the public code repository (https://github.com/pinellolab/CRISPResso2) or original publication.
-
41
Generate individual standard mode or HDR mode tab-delimited batch parameter files for each target amplicon. Populate the files according to the guidelines in Table 4.
While CRISPResso2 can perform batch analysis on multiple amplicons in the same run, doing so will prevent the generation of certain summary tables and plots.
[CRITICAL STEP] The workflow for quantifying prime editing efficiency using CRISPResso2 differs slightly between quantifying single point mutations (requiring standard mode) versus insertions, deletions, or substitutions of multiple base pairs (requiring HDR mode).
-
42
Run CRISPResso2 using either standard mode or HDR mode for a specific amplicon by calling the appropriate batch parameter file from Step 41 (see Table 4). [CRITICAL STEP] If analyzing multiple samples that use the same pegRNA and nicking sgRNA, batch settings can be applied to either standard mode or HDR mode. Running CRISPResso2 using batch settings will generate summary files for each batch of samples. This greatly facilitates downstream analyses.
-
43
Quantify CRISPResso2 editing results using option A to quantify single point mutations from the standard mode output files, or option B to quantify insertions, deletions, or multiple-base pair substitutions from HDR mode output files:
A. Quantifying single point mutations from standard mode.
Open the “Nucleotide_percentage_summary.txt” file and, for each sample, collect the frequency of the desired edit.
Open the “CRISPRessoBatch_quantification_of_editing_frequency.txt” file and, for each sample, collect the values under “Reads aligned” and “Reads_aligned_all_amplicons”.
For each sample, derive the frequency of alleles containing only the desired edit (without indels) by dividing the “Reads aligned” value from Step 43A(ii) by the “Reads_aligned_all_amplicons” value from Step 43A(ii), and then multiplying that quotient by the edit frequency value from Step 43A(i).
B. Quantifying insertions, deletions, or multiple-base pair substitutions from HDR mode.
Open the “CRISPRessoBatch_quantification_of_editing_frequency.txt” file. When using HDR mode, two amplicons per sample are generated (HDR and reference). For each sample’s HDR amplicon, collect the values under “Reads aligned” and “Reads_aligned_all_amplicons.”
For each sample, derive the frequency of alleles containing only the desired edit (without indels) by dividing the “Reads aligned” value from Step 43B(i) by the “Reads_aligned_all_amplicons” value from Step 43B(i).
-
44
Quantify indels from standard mode or HDR mode. Open the “CRISPRessoBatch_quantification_of_editing_frequency.txt” and, for each sample, collect the values under “Discarded” and the value under “Reads_aligned_all_amplicons”.
[CRITICAL STEP] This step requires that the “discard_indel_reads” parameter was set to TRUE for the analysis (See Table 4).
[CRITICAL STEP] If running CRISPResso2 in HDR mode, sum the “Discarded” values from each sample’s reference amplicon and HDR amplicon and use this as the “Discarded” value in Step 45.
-
45
For each sample, derive the frequency of alleles containing an indel by dividing the “Discarded” value by “Reads_aligned_all_amplicons” value.
[? Troubleshooting]
-
46
Repeat Steps 41–45 as necessary for each amplicon to be analyzed.
Troubleshooting
Troubleshooting advice is summarized in Table 5.
Table 5.
Troubleshooting advice
| Step | Problem | Potential Causes | Solutions |
|---|---|---|---|
|
| |||
| 3A(xix)/3B(x) | epegRNA cloning fails: No colonies observed after cloning | For 3A(xix): The presence of many red colonies indicates backbone bleedthrough due to the incomplete digestion of the pU6-tevopreq1-GG-acceptor plasmid. | Repeat digestion with BsaI, PvuII, and NcoI and perform subsequent gel extraction |
| For 3A(xix): Oligos not properly phosphorylated | Check that PNK is being performed correctly if sgRNA scaffold oligos (Golden Gate part 2) were not purchased with 5′ phosphorylation | ||
| Overhangs incorrectly designed | Check overhang design, switch between 3A and 3B to try different methods | ||
| Incorrect antibiotic used | All epegRNA and pegRNA plasmids based on our designs yield carb/amp resistance | ||
|
| |||
| 7 | PCR amplification of mRNA transcription template plasmid fails | Non-specific amplification | Re-optimize PCR conditions to avoid aberrant primer binding: rerun the PCR(s) with different annealing temperatures and extension times. Ensure in vitro transcription forward and reverse primers are PAGE purified. Use gel electrophoresis to verify product purity. |
| Low-yield amplification | Scale up PCR beyond the suggested volume; then pool and concentrate products in a single silica column. Use gel electrophoresis to verify product purity. | ||
|
| |||
| 18 | mRNA gel electrophoresis shows wrong length transcript, smear, or no transcript | Incorrect length indicates sub-optimal input DNA quality | Check gel electrophoresis from Step 7. A high-quality DNA amplicon input is important. Refer to Step 7 troubleshooting. |
| Smear on mRNA gel indicates RNAse contamination | Determine the source of any RNAse contamination. Repeat mRNA prep from Step 8 with RNAse-free technique. | ||
| No Transcript: Sub-optimal input DNA quantity; precipitated RNA pellet lost during LiCl cleanup (Steps 12–17) | Ensure that IVT reaction is initiated with 1 μg of template DNA. Take care during the ethanol washes of the LiCl precipitation to not remove the pellet from the spin tube. Review IVT setup to ensure all reagents are included and in good condition. Ensure that technique is RNAse-free. | ||
|
| |||
| 41–46 | Observed editing rates are low or undetectable in workhorse cell line (HEK293T, N2A, etc.) | PBS and RTT are not optimized | Try more PBS and RTT lengths and combinations. |
| Inefficiently edited protospacer | Confirm Cas9 nuclease or base editing activity at that protospacer. Test more protospacers. | ||
| Lipid has oxidized and prevented efficient transfection | Repeat with fresh lipid and Opti-MEM. | ||
| Poor quality plasmids | Re-prep plasmid: run plasmid on a gel to ensure no RNA contamination, which manifests as a low-MW smear on EtBr gel. | ||
| Editor not being delivered | Use a western blot to test for editor expression; transfect easily monitored plasmid such as pmaxGFP to ensure transfection is working. | ||
| SNP in spacer relative to consensus HG38 sequence or other reference sequence | Sequence unedited cells from sample to check for this, adjust epegRNA components accordingly. | ||
| Not using optimal PE systems | Switch to epegRNA, use max architecture, or try PE4 or PE5. | ||
| epegRNA was incorrectly designed: edit not encoded in the 3′ epegRNA extension (causing the RT to synthesize the WT sequence), or the mutation was included in the spacer, preventing Cas9 from binding to the target locus | Check epegRNA design; use one of several web tools to re-design epegRNA and compare output with your epegRNA. | ||
| 5′ G not included, transcription from U6 promoter is inefficient (if using U6 promoter for epegRNA transcription) | Ensure that either the spacer sequence begins with a 5′ G, or if it does not, append an extra G at the 5′ end to extend the spacer length to 21 nt. | ||
| First nucleotide in epegRNA 3′ extension is a cytosine | We have observed that 3′ extensions starting with a cytosine generally result in lower prime editing. Redesign epegRNA RTT lengths to avoid starting the 3′ extension with a cytosine. | ||
| epegRNA contains a polyU stretch, which causes premature transcriptional termination of epegRNA | If the polyU stretch is in the RTT, consider adding a silent edit (if possible) to disrupt the polyU sequence. If the polyU stretch is in the spacer, consider targeting an alternative protospacer. | ||
| General technical issues | To parse apart epegRNA problems from experimental workflow errors, check that you can perform high-efficiency prime editing at a previously established site and with a validated edit. | ||
|
| |||
| 41–46 | Efficient editing in workhorse cell line, but inefficient editing in other cell types | Prime editor may not be expressing | Check editor expression with nuclease-mediated indel activity, base editor activity, or western blot. Re-optimize transfection or electroporation protocol, or change from plasmid to mRNA delivery. |
| Disconnect between cell line used for optimization and cell line of interest | Re-optimize nicking sgRNA in target cell line. Consider using epegRNAs and/or MLH1dn if these were excluded from initial optimizations, as these modifications tend to have a large impact in more challenging cell lines. | ||
|
| |||
| 30 | PCR1 amplification fails | Cell lysis is incomplete | If using complete mammalian cell lysis buffer (See Reagent Setup), confirm lysis buffer is pH 8. |
| PCR1 conditions may not be optimal | Try new combinations of PCR1 primers. Re-optimize thermal cycling steps, in particular the annealing temperature. Repeat the PCR1 with different gDNA template inputs, but keep gDNA input into each PCR consistent across reactions and make sure an adequate number of cells are analyzed. Run control PCRs of previously validated PCR1 primer sets (i.e., HEK3) to confirm that the lysis step worked properly. Use NCBI’s Primer-BLAST to verify that primer pairs do not bind undesired regions. | ||
|
| |||
| 41– 46 | High rates of indel incorporation | Nicking sgRNA is not optimal | Test more nicking sgRNAs, especially PE3b/PE5b nicking sgRNAs if possible. |
| MMR is inducing high indels | Switch to PE4 or PE5 system. | ||
Timing
| Steps 1–2, | Design of epegRNAs and nicking sgRNAs: 1 d |
| Step 3A, | Generation of epegRNAs or sgRNAs by Golden Gate cloning: 3 d |
| Step 3B, | Generation of epegRNAs or sgRNAs by isothermal assembly: 3 d |
| Step 3C, | Acquiring purified, chemically modified, synthetic epegRNAs, pegRNAs, or sgRNAs: 7–42 d |
| Steps 4–19, | Preparation of in vitro transcribed PEmax mRNA: 1–2 d |
| Step 20A, | Prime editing in HEK293T cells via plasmid transfection: 4–5 d |
| Step 20B, | Prime editing in primary human fibroblasts via RNA electroporation: 4–5 d |
| Steps 21–26, | Lysis of mammalian cells for HTS: 1 d |
| Steps 27–40, | HTS for prime editing analysis: 1–2 d |
| Steps 41–46, | HTS analysis: 1–4 h |
Anticipated results
With a few optimizations for the desired edit, prime editing can enable highly efficient and precise genome editing in mammalian cells. Here, we show the anticipated results from screening pegRNAs and nicking sgRNAs for prime editing an amenable cell line (HEK293T), which highlights the importance of optimizing pegRNA PBS and RTT length and sgRNA spacer sites (Fig. 7a–d). In a less amenable cell line (HeLa), we also demonstrate that the use of PEmax, epegRNAs, PE4/PE5 systems, and additional MMR-evading benign edits can substantially elevate editing efficiency compared to the original prime editing approaches (Fig. 7e–f). Analysis of high-throughput sequencing data with CRISPResso2 yields the allelic outcomes from editing, revealing the on-target purity of the intended genomic change (Fig. 7g). As shown in induced pluripotent stem cells72, the efficiency of prime editing can vary widely across delivery methods (plasmid DNA, mRNA; Fig. 7h) and should be optimized for the desired application.
Supplementary Material
Acknowledgements
We thank Daniel Gao and Tony Huang for helpful discussions and Anahita Vieira for feedback on this manuscript. We thank Andrew Anzalone and other Liu Laboratory members who advanced prime editing technology. Figures were created with BioRender (BioRender.com). This work was supported by US NIH U01AI142756, RM1HG009490, R01EB031172, and R35GM118062, the Howard Hughes Medical Institute, and the Bill & Melinda Gates Foundation. J.L.D., A.A.S., P.B.R., and P.J.C. are supported by the NSF Graduate Research Fellowship program. J.L.D is supported by a Fannie and John Hertz Foundation Fellowship.
Footnotes
Declaration of interests
J.L.D., A.A.S., P.B.R., P.J.C., and D.R.L. have filed patent applications on prime editing technologies and applications. D.R.L. is a consultant and equity holder of Prime Medicine, Beam Therapeutics, Pairwise Plants, and Chroma Medicine, companies that use genome editing or genome engineering.
Code availability
The code used for HTS processing and analysis are accessible at https://github.com/pinellolab/CRISPResso2.
Tweet: A protocol for designing and executing prime editing experiments in mammalian cells from David Liu’s lab. @AlexanderASousa @JordanDoman @liugroup
Related links
Key references using this protocol:
1. Anzalone, A.V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). https://doi.org/10.1038/s41586-019-1711-4
2. Nelson, J.W. et al. Engineered pegRNAs improve prime editing efficiency. Nature Biotechnology (2021). https://doi.org/10.1038/s41587-021-01039-7
3. Chen, P.J. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652 (2021). doi: 10.1016/j.cell.2021.09.018
4. Anzalone, A. V. et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nature Biotechnology (2021) doi:10.1038/s41587-021-01133-w
Data availability
Sequencing data used to generate Fig. 7 is deposited at the NCBI Sequence Read Archive database under PRJNA817825.
References
- 1.Jinek M et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzalone AV, Koblan LW & Liu DR Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Frangoul H et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N. Engl. J. Med. 384, 252–260 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Gillmore JD et al. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 385, 493–502 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Giannoukos G et al. UDiTaS™, a genome editing detection method for indels and genome rearrangements. BMC Genomics 19, 212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadtmauer EA et al. CRISPR-engineered T cells in patients with refractory cancer. Science 367, eaba7365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webber BR et al. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 10, 5222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turchiano G et al. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell 28, 1136–1147.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Kosicki M, Tomberg K & Bradley A Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Y et al. Large-Fragment Deletions Induced by Cas9 Cleavage while Not in the BEs System. Mol. Ther. - Nucleic Acids 21, 523–526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuccaro MV et al. Allele-Specific Chromosome Removal after Cas9 Cleavage in Human Embryos. Cell 183, 1650–1664.e15 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Alanis-Lobato G et al. Frequent loss of heterozygosity in CRISPR-Cas9–edited early human embryos. Proc. Natl. Acad. Sci. 118, e2004832117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leibowitz ML et al. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 53, 895–905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enache OM et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 52, 662–668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox DBT, Platt RJ & Zhang F Therapeutic genome editing: prospects and challenges. Nat. Med. 21, 121–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman JR, Taylor MRG & Boulton SJ Playing the End Game: DNA Double-Strand Break Repair Pathway Choice. Mol. Cell 47, 497–510 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida K et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter MF et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newby GA et al. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature 595, 295–302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koblan LW et al. In vivo base editing rescues Hutchinson–Gilford progeria syndrome in mice. Nature 589, 608–614 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newby GA & Liu DR In vivo somatic cell base editing and prime editing. Mol. Ther. 29, 3107–3124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koblan LW et al. Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat. Biotechnol. 39, 1414–1425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurt IC et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 39, 41–46 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L et al. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. Commun. 12, 1384 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan T et al. Optimization of C-to-G base editors with sequence context preference predictable by machine learning methods. Nat. Commun. 12, 4902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao D et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 39, 35–40 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Chen PJ et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson JW et al. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. (2021) doi: 10.1038/s41587-021-01039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim DY, Moon SB, Ko J-H, Kim Y-S & Kim D Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res. 48, 10576–10589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schene IF et al. Prime editing for functional repair in patient-derived disease models. Nat. Commun. 11, 5352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao P et al. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome Biol. 22, 83 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin S et al. Genome-wide specificity of prime editors in plants. Nat. Biotechnol. 39, 1292–1299 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Habib O, Habib G, Hwang G-H & Bae S Comprehensive analysis of prime editing outcomes in human embryonic stem cells. Nucleic Acids Res. 50, 1187–1197 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. 12, 2121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y et al. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res. 31, 1134–1136 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang G et al. Enhancement of prime editing via xrRNA motif-joined pegRNA. Nat. Commun. 13, 1856 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petri K et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. (2021) doi: 10.1038/s41587-021-00901-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen B et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hussmann JA et al. Mapping the genetic landscape of DNA double-strand break repair. Cell 184, 5653–5669.e25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spencer JM & Zhang X Deep mutational scanning of S. pyogenes Cas9 reveals important functional domains. Sci. Rep. 7, 16836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park S-J et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol. 22, 170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song M et al. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nat. Commun. 12, 5617 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anzalone AV et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 1–10 (2021) doi: 10.1038/s41587-021-01133-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi J et al. Precise genomic deletions using paired prime editing. Nat. Biotechnol. 1–9 (2021) doi: 10.1038/s41587-021-01025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang T, Zhang X-O, Weng Z & Xue W Deletion and replacement of long genomic sequences using prime editing. Nat. Biotechnol. 1–8 (2021) doi: 10.1038/s41587-021-01026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Q et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 39, 923–927 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Zhuang Y et al. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nat. Chem. Biol. 18, 29–37 (2022). [DOI] [PubMed] [Google Scholar]
- 51.Wang J Efficient targeted insertion of large DNA fragments without DNA donors. Nat. Methods 19, 25 (2022). [DOI] [PubMed] [Google Scholar]
- 52.Ioannidi EI et al. Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases. http://biorxiv.org/lookup/doi/10.1101/2021.11.01.466786 (2021) doi: 10.1101/2021.11.01.466786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin Q et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Zheng C et al. A flexible split prime editor using truncated reverse transcriptase improves dual-AAV delivery in mouse liver. Mol. Ther. S1525001622000053 (2022) doi: 10.1016/j.ymthe.2022.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhi S et al. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol. Ther. 30, 283–294 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y et al. Efficient generation of mouse models with the prime editing system. Cell Discov. 6, 1–4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin J et al. Modeling a cataract disorder in mice with prime editing. Mol. Ther. - Nucleic Acids 25, 494–501 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Böck D et al. In vivo prime editing of a metabolic liver disease in mice. Sci. Transl. Med. 14, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim Y et al. Adenine base editing and prime editing of chemically derived hepatic progenitors rescue genetic liver disease. Cell Stem Cell 28, 1614–1624.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Choi J et al. A temporally resolved, multiplex molecular recorder based on sequential genome editing. http://biorxiv.org/lookup/doi/10.1101/2021.11.05.467388 (2021) doi: 10.1101/2021.11.05.467388. [DOI] [Google Scholar]
- 61.Erwood S et al. Saturation variant interpretation using CRISPR prime editing. Nat. Biotechnol. 1–11 (2022) doi: 10.1038/s41587-021-01201-1. [DOI] [PubMed] [Google Scholar]
- 62.Xu R, Liu X, Li J, Qin R & Wei P Identification of herbicide resistance OsACC1 mutations via in planta prime-editing-library screening in rice. Nat. Plants 7, 888–892 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Qian Y et al. Efficient and precise generation of Tay–Sachs disease model in rabbit by prime editing system. Cell Discov. 7, 50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jang H et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 6, 181–194 (2022). [DOI] [PubMed] [Google Scholar]
- 65.Kim HK et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol. 39, 198–206 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Gao Z, Herrera-Carrillo E & Berkhout B Delineation of the Exact Transcription Termination Signal for Type 3 Polymerase III. Mol. Ther. - Nucleic Acids 10, 36–44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsu JY et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat. Commun. 12, 1034 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hwang G-H et al. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Res. 49, W499–W504 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson MV, Haldrup J, Thomsen EA, Wolff JH & Mikkelsen JG pegIT - a web-based design tool for prime editing. Nucleic Acids Res. 49, W505–W509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chow RD, Chen JS, Shen J & Chen S A web tool for the design of prime-editing guide RNAs. Nat. Biomed. Eng. 5, 190–194 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen P-F et al. Generation and characterization of human induced pluripotent stem cells (iPSCs) from three male and three female patients with CDKL5 Deficiency Disorder (CDD). Stem Cell Res. 53, 102276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data used to generate Fig. 7 is deposited at the NCBI Sequence Read Archive database under PRJNA817825.