Abstract
Kartogenin (KGN), a novel small-molecule compound, has been considered a promising chondrogenic promoter in cartilage regeneration. However, whether KGN also participates in osteogenesis and bone regeneration remains unclear. This research was designed to explore the roles of KGN on osteogenic differentiation in bone marrow mesenchymal stem cells (BMMSCs) as well as determine the possible mechanism of osteogenesis. We revealed that KGN enhanced the osteogenic differentiation capacity of BMMSCs without affecting cell proliferation, during which autophagic activities and the expression of autophagy-related genes were promoted. Moreover, KGN upregulated the phosphorylation level of the Smad1/5/9 signaling, and inhibition and activation of Smad signaling were also applied to validate the involvement of Smad in BMMSCs during KGN treatment. In summary, this study shows that KGN promotes osteogenic differentiation of BMMSCs through enhancing autophagic levels and upregulating Smad1/5/9 signaling mechanically.
1. Introduction
Bone defects can occur due to various reasons, such as trauma, infections, and congenital malformations. Although bone tissue has the capacity for self-healing, many bone defects or fractures remain a major challenge in regeneration medicine owing to certain disease states or the defect size. Research on bone marrow mesenchymal stem cells (BMMSCs) has provided new treatment strategies for complex bone defect diseases [1, 2]. However, most of the current therapies for bone defect repair using BMMSCs suffer from weaknesses such as poor osteogenic differentiation ratio, low level of bone fusion, and unsatisfied osteogenesis stability; therefore, there is an urgent need for further exploration of the BMMSC differentiation mechanism and optimization of their directional differentiation regulation in bone tissue engineering to promote stem cell-based bone defect repair and reconstruction. Kartogenin (KGN) is a novel small molecular compound which was studied by Johnson et al. [3] first in 2012. It was demonstrated to be capable of exerting chondroprotective effects in vitro by inducing chondrogenic differentiation of mesenchymal stem cells (MSCs) and proved efficacious in osteoarthritis animal models. Benefitting from the high stability, KGN has been broadly explored for tissue regeneration, especially for cartilage repair. To date, it has been reported that KGN can induce the differentiation of MSCs into chondrocytes both in vivo and vitro [4, 5]. Moreover, it has been studied in the fields of tendon-bone healing, wound healing, and limb development [6]. Nevertheless, there is a relative paucity of research on the effects of KGN on bone regeneration. According to Wang et al. [7], KGN promoted intracellular antioxidant activity by activating AMPK-SIRT1 signaling pathways, thereby promoting osteogenesis. Therefore, it is interesting to determine whether KGN can promote the osteogenic activity of BMMSCs and the specific mechanisms of KGN in stem cells.
MSCs have a multidifferentiation capacity and are able to maintain bone homeostasis, which has been broadly studied in the field of bone regeneration [8, 9]. Decker al. [10] revealed that KGN activates transforming growth factor-β/Drosophila mothers against decapentaplegic protein (TGF-β/Smad) signaling pathway, upregulates the phosphorylation level of Smad2/3, and stimulates mouse embryonic limb development. As an important signaling pathway, TGF-β/Smad is indispensable for mediating the osteogenic differentiation commitment of BMMSCs [11, 12]. Phosphorylation of Smad1/5/9 mediated by the bone morphogenetic protein- (BMP-) 2 receptor causes activated isoforms to associate with Smad4, followed by their translocation into cell nucleus, inducing osteogenesis-related gene transcription, making Smad1/5/9 signaling essential for MSC osteogenic signaling [12]. Comprising a subset of TGF-β superfamily, BMPs modulate numerous biological processes, such as formation of bone and cartilage during embryogenesis [13], and can result in Smad-specific transcription factor phosphorylation [14]. Among the Smad proteins, Smad1, Smad5, and Smad9 are closely associated with osteogenic differentiation [15]. Following the binding of BMPs with the specific receptor, phosphorylation of BMP-responsive Smad 1, 5, and 9 results in complex formation of R-Smads, accumulating with the common-mediator Smad4 in the nucleus, which is dedicated to osteogenesis-related gene expression regulation [16, 17], such as the transcription of Runt-related transcription factor 2 (Runx2), an essential regulatory factor in osteogenesis, leading to the initiation of the expression of other osteogenic genes [12, 18]. On this basis, whether KGN can upregulate the expression of Smad1/5/9 to promote BMMSC osteogenic differentiation needs to be confirmed.
Autophagy is a highly conserved fundamental cellular biological process that helps maintain homeostasis and enhances cell survival under cellular stress conditions [19]. Autophagy is also indispensable in cell trilineage differentiation [20], such as osteogenic differentiation [21]. In recent years, there have been studies concentrating on autophagy and osteogenic effects of MSCs. It has been reported that autophagy can enhance osteogenesis by mediating osteoblast differentiation and mineralization, especially affecting autophagosomes in extracellular calcium transportation [22]. Autophagic activity is also closely concerned with MSC differentiation potential by regulating either adipogenic or osteoblastic lineages, and autophagic level could influence bone-related cell survival rate under unfavorable situations [19]. Several markers have been proposed for monitoring autophagy, such as LC3, as well as autophagy-related genes such as LC3, Atg7, and Beclin1. LC3 generally participates in the elongation-closure of autophagosome formation and is present in two forms [23]: LC3-I, which is distributed in the cytoplasm and unconjugated to lipids, and LC3-II, located within the autophagosome membrane and conjugated to the lipid phosphatidylethanolamine, is widely used as a specific autophagosome marker to access autophagic flux.
The purpose of this research was to investigate the effects of KGN in promoting MSC osteogenesis and explore the possible mechanisms. Our findings are expected to provide new insights into small molecular compound-induced BMMSC osteogenesis in bone tissue engineering as well as new direction for clinical complex bone defect repair and reconstruction.
2. Materials and Methods
2.1. Isolation and Culture of BMMSCs
C57BL/6J mice (6-8 wk; female) were purchased from SiPeiFu Biotechnology Co., Ltd. (SPF; Beijing, China). All nucleated cells were obtained from mouse femurs and tibia bone marrow, and cells (1.5 × 107 cells per 100 mm culture dish) (Corning, NY, USA) were seeded at 37°C, 5% CO2. After 2 d, nonattached cells were discarded, and adherent cells were cultured for 14 d in complete alpha minimum essential medium (Biological Industries, Beit Haemek, Israel) adding fetal bovine serum (20%; Gibco), L-glutamine (2 mM; Gibco), 2-mercaptoethanol (55 μM; Gibco), and penicillin/streptomycin (1%; Gibco). The isolation and identification of MSCs were described in our previous studies [24–26]. The medium was refreshed per three days, and then, colony-forming attached cells were passaged once for further experimental use.
2.2. Cell Proliferation Assay
The survival rates of BMMSCs were performed using Cell Counting Kit-8 (CCK-8) assay (Solarbio). BMMSCs were cultured with medium (100 μL per well) in the 96-well plates at a density of 1.0 × 103 per well. CCK-8 reagent was diluted with culture medium to prepare the working solution after 24 h cultivation and incubated for extra 1.5 h as the instructions suggested. Subsequently, optical density value measurement was performed by a microplate reader (Bio-Rad, Virginia, USA) at the 450 nm wavelength. We performed this assay at 0 h, 12 h, and 24 h. Proliferation of the MSC population was also examined using a Ki67 incorporation assay. Mouse BMMSCs were seeded and cultured on φ20 mm glass chamber slide (Corning). The cultures were then incubated with medium containing Ki67 solution (1 : 100; Invitrogen, CA, USA) for another 20 h and stained using a Ki67 Staining Kit (Invitrogen). The positive Ki67 cell and overall cell counts were taken, respectively, and the proportions were calculated.
2.3. Transmission Electron Microscopy
Transmission electron microscope (TEM) was applied to observe autophagosomes. Cells were fixed in 4% formaldehyde and washed in PBS, followed by postfixation in 1% OsO4 at 4°C overnight. The samples were then dehydrated stepwisely in ascending ethanol, washed with propylene oxide, and then embedded with epoxy resin as previous study described [27]. Sections were stained with lead citrate as well as uranyl acetate.
2.4. Osteogenic Differentiation Assay
BMMSCs at passage 2 were prepared for osteogenic induction, and the osteogenic medium was used for BMMSC culture which contained complete culture medium plus ascorbic acid (50 mg/L; Sigma-Aldrich, MO, USA), β-glycerophosphate (10 mM; Sigma-Aldrich), and dexamethasone (10 nM; Sigma-Aldrich), with fresh OM replaced every three days. After two weeks of induction, 4% paraformaldehyde (Solarbio, Beijing, China) was used for cell fixing. The formation of mineralized nodule was detected using Alizarin Red Staining (ARS) kit (Sigma-Aldrich). Quantification analysis of positively staining areas was performed by Image Pro Plus 6.0v (Media Cybernetics, CA, USA) and presented by ratio.
2.5. Western Blotting Analysis
Total proteins of BMMSCs were obtained by RIPA buffer (Solarbio) adding the inhibitor cocktail of protease/phosphatase (Thermo Fisher Scientific, MA, USA). BCA protein assay kit (Beyotime, Shanghai, China) was used for concentration quantification. Equal amounts of protein suspension were applied on a 4-12% SDS-polyacrylamide gel (Beyotime) and then electroblotted onto polyvinylidene difluoride membrane (Thermo). Membranes were then blocked with 5% nonfat milk in TBST. Primary antibodies mouse-monoclonal anti-Runx2 (1 : 1,000; Santa Cruz, CA, USA; sc-101145), mouse-monoclonal anti-alkaline phosphatase (Alp) (1 : 1,000; Santa Cruz, CA, USA; sc-365765), rabbit-polyclonal anti-autophagy-related gene 7 (Atg7; 1 : 1,000; Proteintech, NJ, USA; 10088-2-AP), rabbit-polyclonal anti-microtubule-associated protein light chain 3 (LC3; 1 : 1,000; Cell Signaling Technology, MA, USA; 4108S), rabbit-polyclonal anti-Smad1/5/9 (1 : 1,000; Affinity Biosciences, CA, USA; AF0614), and rabbit-polyclonal anti-phosphorylated-Smad1/5/9 (pSmad1/5/9; 1 : 1,000; Affinity Biosciences; AF8313) were performed overnight in order to the membranes' molecular probing. Then, membrane incubation was implemented using horseradish peroxidase-conjugated mouse or rabbit secondary antibodies (1 : 15,000; Beyotime), after which SuperSignal West Pico Substrate (Thermo) was applied for band enhancement. Quantification analysis was conducted by ImageJ 16.0v software and relatively normalized to β-actin (1 : 5,000; Zhongshan Golden Bridge Biotechnology, China).
2.6. Immunofluorescent Staining
BMMSCs were harvested and treated with 4% paraformaldehyde and 0.1% Triton X-100 solution (Beyotime) for fixation and permeabilization, respectively, and afterwards incubated with 5% goat serum (Beyotime). Then BMMSCSs were probed with primary antibodies against LC3, Runx2, and ALP (1 : 100) overnight at 4 degree Celsius, and the secondary antibody (1 : 200, Beyotime) was then used for another an hour at room temperature. DAPI (Beyotime) was used for nucleuses counterstaining. The laser scanning confocal microscope was employed for staining visualization (LSM 510; Zeiss, Germany). The Image Pro Plus 6.0v software was performed for quantitative analytical purposes.
2.7. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total cellular RNA extraction was firstly performed with TRIzol reagent (Invitrogen), and then, complementary DNA was synthesised with a PrimeScript™ RT reagent Kit (TaKaRa, Tokyo, Japan) and gene-specific primers (Table 1). qRT-PCR was conducted by SYBR Green Q-PCR Master Mix (Thermo) on 7900HT Fast-Time PCR System. Relative expression levels were analyzed using 2−ΔΔCt calculating method with normalization to GAPDH.
Table 1.
Primer sequences for qRT-PCR.
| Gene name | Primer sequence |
|---|---|
| Runx2-F | CCGGGAACCAAGAAGGCACA |
| Runx2-R | AGGCGGGACACCTACTCTCA |
| Alp-F | GTGACTACCACTCGGGTGAAC |
| Alp-R | CTCTGGTGGCATCTCGTTATC |
| Beclin1-F | AGGCGAAACCAGGAGAGAC |
| Beclin1-R | CCTCCCCGATCAGAGTGAA |
| Atg12-F | TCCCCGGAACGAGGAACTC |
| Atg12-R | TTCGCTCCACAGCCCATTTC |
| Dram1-F | TCATCTCCTACGTGGTCGC |
| Dram1-R | CTGCGCCAAGAAATGCAGAG |
| Atg3-F | ACACGGTGAAGGGAAAGGC |
| Atg3-R | TGGTGGACTAAGTGATCTCCAG |
| GAPDH-F | TGGCCTTCCGTGTTCCTAC |
| GAPDH-R | GAGTTGCTGTTGAAGTCGCA |
2.8. Reagents and Chemicals
KGN (Selleck Chemicals, TX, USA) was dissolved with dimethylsulfoxide (DMSO) as the stock solution. Cells treated with 0.005% DMSO were used as controls. Other reagents applied in the study are listed as follows: 3-methyladenine (3-MA; Selleck Chemicals, TX, USA) (5 mM for 24 h) was used as autophagy inhibitor. Both the signal inhibitor LDN-193189 (LDN; 0.5 μM for 12 h) and the activator SB4 (0.1 μM for 24 h) were purchased from MedChemExpress (MCE, NJ, USA).
2.9. Statistical Analysis
Data processing and analysis were conducted with SPSS 23.0 v (IBM, NY, USA) as well as GraphPad Prism 8.1v for Mac (GraphPad Software, CA, USA). All values calculated are shown as the mean and standard deviation (mean ± S.D.), representing at least 3 independent experiments. Comparison of two groups was executed by independent sample t test analysis or Mann–Whitney rank-sum test. For difference analysis among more than two groups, one-way ANOVA with Tukey's test was conducted. Criteria for statistical significance were set at p < 0.05.
3. Results
3.1. KGN Promotes the Osteogenic Capacity of BMMSCs
The concentration of KGN has been suggested in previous studies [28–32], and the optimal concentration of KGN for osteogenesis in BMMSCs was 10 μM in this study (Figure S1). To determine whether proliferation capacity was affected by KGN, CCK-8 was performed, and the result suggested that single KGN treatment of 10 μM for 72 h did not influence cell growth (Figure 1(a)). Ki67, a marker of cell proliferation activity [33], was also measured by immunofluorescence staining, and it showed no statistical difference (Figures 1(b) and 1(c)). Next, we tested whether KGN promotes the osteogenic ability of BMMSCs. ARS staining demonstrated that matrix mineralization was visibly enhanced by KGN treatment for 14 days compared with the control group (Figures 1(d) and 1(e)). Western blotting analysis also indicated that KGN upregulated ALP protein levels in BMMSCs (Figures 1(f)–1(h)). Additionally, relative expression levels of ALP and Runx2 also showed an upward trend after KGN induction (Figures 1(i) and 1(j)). These data indicate that KGN could enhance BMMSC osteogenic differentiation without affecting cell proliferation activity.
Figure 1.

KGN enhances the osteogenic ability of BMMSCs without affecting cell proliferation. Cell proliferation capacity of BMMSCs was measured by CCK-8 (a), immunofluorescent staining (b) of Ki67, and semiquantification (c) of (b) between the control and KGN-treated BMMSCs. p > 0.05, independent Student's t test. ARS staining detection (d) and semiquantification (e) of mineralized nodules. KGN promoted osteogenesis of BMMSCs. Scale bar = 500 μm. ∗∗∗p < 0.001, independent Student's t test. Western blotting (f) and semiquantification analysis (g, h) and qRT-PCR (i, j) of osteogenic markers ALP and Runx2. KGN upregulated the expression of osteogenic genes. ∗∗p < 0.01 and ∗∗∗p < 0.001, independent Student's t test.
3.2. KGN Upregulates Autophagic Activity of BMMSCs
To evaluate whether KGN affected the autophagic activity of BMMSC during osteogenic differentiation, TEM was conducted to determine the formation of autophagosomes in BMMSCs morphologically, which is considered as the standard for detecting autophagy [34] (Figure 2(a)). LC3, as a marker of autophagy, was also analyzed by immunofluorescence staining. The results suggested that the expression level of LC3 increased distinctly after KGN induction (Figures 2(b) and 2(c)). In addition, a panel of autophagic markers was used to detect autophagic activity. ATG7 and cleaved LC3 product, LC3II, all presented an elevated level after KGN treatment, proving that KGN is closely associated with autophagy (Figures 2(d)–2(f)). Consistently, results from qRT-PCR also revealed a remarkable increase in the expression of the autophagy-related markers of Beclin1, Atg12, Dram1, and Atg3 (Figures 2(g)–2(j)). Taken together, these results suggest reinforcement of autophagic activity in KGN-treated BMMSCs. To confirm this result, an early autophagy inhibitor 3-MA was used and effectively suppressed autophagic level in BMMSCs, as assessed by western blotting (Figures 2(k)–2(m)).
Figure 2.

Autophagy is involved in the process of KGN-induced BMMSC osteogenesis. (a) The ultrastructural features of autophagic vacuoles by TEM. Black arrows suggest autophagosomes or autophagolysosomes in BMMSCs. Scale bar = 500 nm. Detection of LC3 (red) and DAPI (blue) by immunofluorescent staining (b) in the control and KGN-treated BMMSCs and semiquantification analysis (c). KGN upregulated the expression of autophagic marker LC3. Scale bar = 50 μm. ∗∗p < 0.01, independent Student's t test. (d) The effects of KGN on autophagy-associated markers were evaluated by western blotting, (e, f) semiquantification of (d), and qRT-PCR of autophagy markers (g–j). KGN promoted the expression of ATG7 and downregulated the LC3-I/LC3-II ratio. ∗∗p < 0.01 and ∗∗∗p < 0.001, independent Student's t test. (k) The effects of inhibitor 3-MA on autophagy-associated markers were evaluated by western blotting. (l, m) Semiquantification of (k). 3-MA inhibited the expression of ATG7 and upregulated the LC3-I/LC3-II ratio. ∗∗p < 0.01, independent Student's t test.
3.3. KGN Improves Osteogenesis of BMMSCs through Autophagy Activation
Autophagy plays an important role during BMMSC differentiation into primary functional cells in bone formation, regulating their physiological functions to maintain bone homeostasis [35]. Although KGN has been shown to elevate autophagic activity in BMMSCs and promote osteogenic differentiation, the underlying mechanism remains elusive. Therefore, 3-MA was applied prior to the induction by KGN to identify its autophagic role during BMMSC osteogenic differentiation. On day 14 after treatment, 3-MA was found to have attenuated the number of calcium nodules induced by KGN (Figures 3(a) and 3(b)). Compared with the control group, immunofluorescence staining also indicated that the KGN-induced expression of both ALP and Runx2 was suppressed by 3-MA (Figures 3(c)–3(f)). Consistent with this, the protein and mRNA levels of ALP and Runx2 showed an appreciable decline when 3-MA was involved (Figures 3(g)–3(k)). This suggested autophagy performs important functions in osteogenic differentiation caused by KGN, and suppression of autophagy leads to the inhibition of osteogenic differentiation.
Figure 3.

KGN increases the osteogenic capability of BMMSCs via upregulating autophagy. ARS (a) and semiquantification (b) of mineralized nodules. 3-MA-mediated autophagy suppression inhibited KGN-induced BMMSC osteogenesis. Scale bar = 500 μm. ∗p < 0.05 and ∗∗∗p < 0.001, one-way ANOVA test. (c, d) Detection of ALP (green), Runx2 (green), and DAPI (blue) by immunofluorescent staining in BMMSCs. (e, f) Semiquantification of (c) and (d). 3-MA-mediated autophagy suppression inhibited KGN-induced expression of osteogenic genes. Scale bar = 50 μm. ∗∗p < 0.01 and ∗∗∗p < 0.001, one-way ANOVA test. (g) Western blotting, (h, i) semiquantification of (g), and qRT-PCR assay (j, k) of ALP and Runx2. 3-MA-mediated autophagy suppression inhibited KGN-induced expression of osteogenic genes. ∗∗p < 0.01 and ∗∗∗p < 0.001, one-way ANOVA test.
3.4. KGN Promotes Osteogenesis of BMMSCs through Smad1/5/9 Signal after Autophagy Activation
Considering the crucial role of Smad signaling in MSC osteogenic differentiation [12], we investigated whether the Smad1/5/9 pathway participates in KGN-induced osteogenic differentiation. To test whether autophagy and the Smad1/5/9 pathways mediated osteogenic differentiation, 3-MA and LDN were applied for the above two inhibitions, respectively. As assessed by western blotting, KGN upregulated the phosphorylation of Smad1/5/9, and 3-MA attenuated this effect (Figures 4(a) and 4(b)), whereas Smad1/5/9 remained stable. Moreover, the phosphorylation level of Smad1/5/9 was compressed by LDN (Figures 4(c) and 4(d)). After treatment of LDN, the calcium nodules were significantly decreased compared with the KGN group by ARS, suggesting the osteogenesis effect was greatly weakened by LDN (Figures 4(e) and 4(f)). Consistently, LDN also suppressed both the protein and mRNA levels of ALP and Runx2 induced by KGN (Figures 4(g)–4(k)). Also, similar findings were also presented in immunofluorescence staining (Figures 4(l)–4(o)), suggesting LDN blocked the KGN-induced increase in ALP and Runx2 expression levels.
Figure 4.
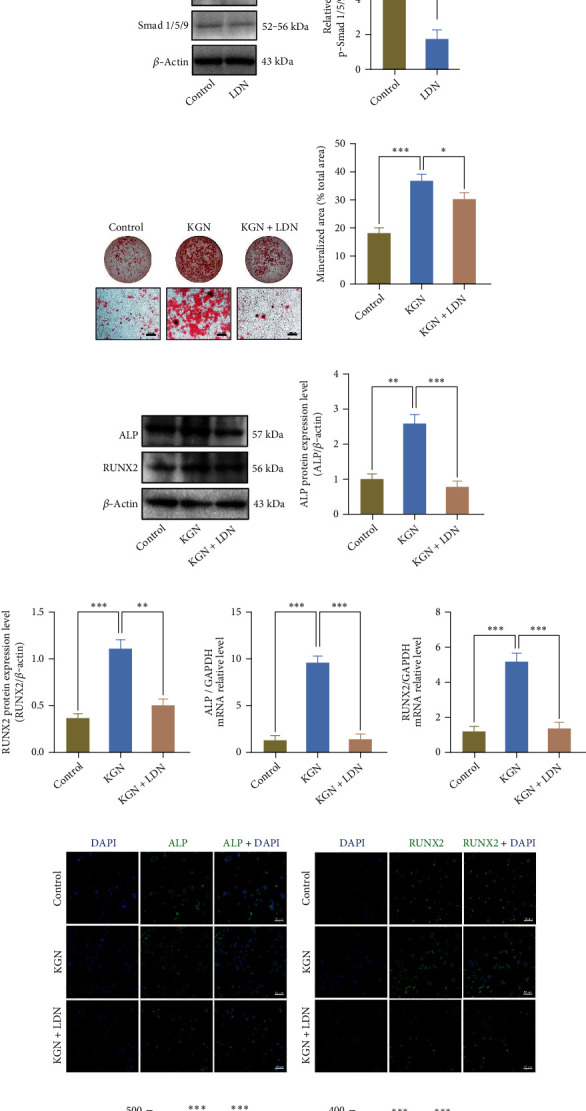
KGN elevates autophagic activities in BMMSCs and prompts osteogenesis through Smad1/5/9 signal pathway. Western blotting (a) and semiquantification (b) of Smad and phosphorylation level of Smad1/5/9. 3-MA-mediated autophagy suppression inhibited KGN-induced Smad1/5/9 phosphorylation increase. ∗∗∗p < 0.001, one-way ANOVA test. Western blotting (c) and semiquantification (d) of the phosphorylation level of Smad1/5/9 in the control and LDN-treated BMMSCs. LDN effectively inhibited phosphorylation level of Smad 1/5/9. ∗∗p < 0.01, independent Student's t test. ARS detection (e) and semiquantification (f) of mineralized nodules. LDN inhibited KGN-induced osteogenesis. Scale bar = 500 μm. ∗p < 0.05 and ∗∗∗p < 0.001, one-way ANOVA test. (g) Western blotting, (h, i) semiquantification of (g), and (j, k) qRT-PCR assay of ALP and Runx2. LDN inhibited KGN-induced expression of osteogenic genes. ∗∗p < 0.01 and ∗∗∗p < 0.001, one-way ANOVA test. (l, m) Detection of ALP (green), Runx2 (green), and DAPI (blue) by immunofluorescent staining in BMMSCs. (n, o) Semiquantification of (l) and (m). LDN inhibited KGN-induced expression of osteogenic genes. Scale bar = 50 μm. ∗∗∗p < 0.001, one-way ANOVA test.
Besides, to further confirm the involvement of Smad1/5/9 pathways in KGN-induced osteogenesis, we employed its activator SB4 (Figures 5(a) and 5(b)) after KGN treatment over the course of 24 h. As shown above, 3-MA inhibited the phosphorylation of Smad1/5/9, whereas this effect was rescued by SB4 (Figures 5(c) and 5(d)). Consistent with this, ARS staining suggested that calcium salt deposits of KGN were reduced by 3-MA, whereas SB4 reduced the suppression effect (Figures 5(e) and 5(f)). These results were verified using western blotting (Figures 5(g)–5(i)), qRT-PCR (Figures 5(j) and 5(k)), and immunofluorescence staining (Figures 5(l)–5(o)). These results confirmed that KGN enhanced BMMSC osteogenic differentiation through the Smad1/5/9 pathways after autophagy activation.
Figure 5.
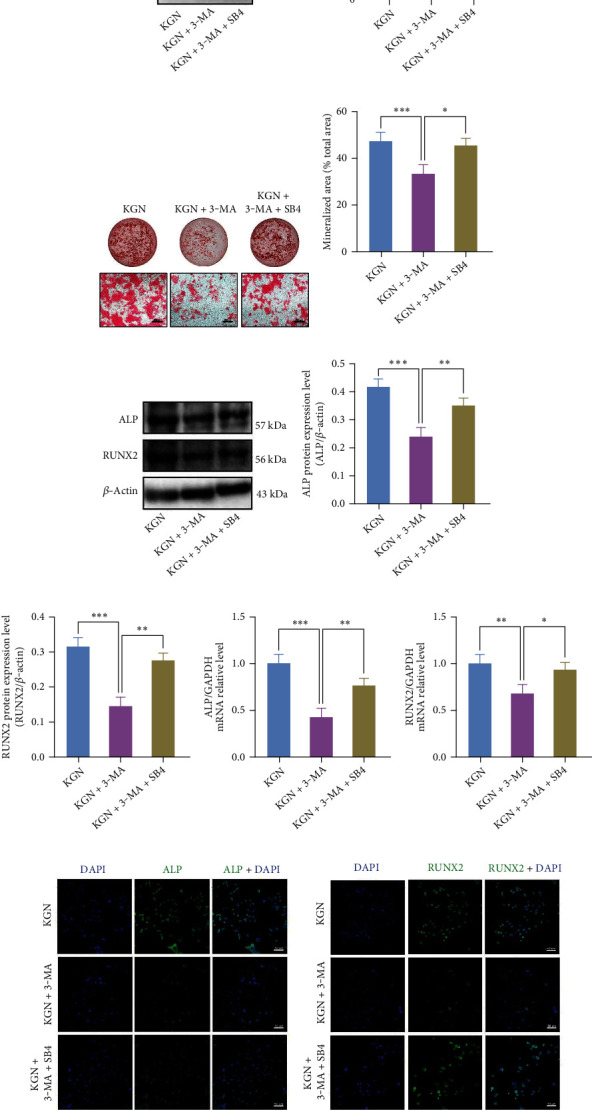
BMMSC osteogenic level promoted by KGN is suppressed by 3-MA, whereas rescued by SB4. Western blotting assay (a) and semiquantification (b) of Smad1/5/9 and phosphorylation level of Smad1/5/9 in the control and SB4-treated BMMSCs. SB4 promoted phosphorylation level of Smad1/5/9. ∗∗∗p < 0.001, independent Student's t test. Western blotting assay (c) and semiquantification (d) on Smad1/5/9 and phosphorylation level of Smad1/5/9. 3-MA suppressed KGN-induced increased phosphorylation level of Smad1/5/9, whereas SB4 reversed this effect. ∗p < 0.05 and ∗∗∗p < 0.001, one-way ANOVA test. ARS (e) and semiquantification (f) of mineralized nodules. 3-MA restrained KGN-induced BMMSC osteogenesis, whereas SB4 reversed this effect. Scale bar = 500 μm. ∗p < 0.05 and ∗∗∗p < 0.001, one-way ANOVA test. (g) Western blotting, (h, i) semiquantification of (g), and (j, k) qRT-PCR of ALP and Runx2. 3-MA suppressed KGN-induced expression of osteogenic genes whereas SB4 reversed this effect. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, one-way ANOVA test. (l, m) Detection of ALP (green), Runx2 (green), and DAPI (blue) by immunofluorescent staining in BMMSCs. (n, o) Semiquantification of (l) and (m). Scale bar = 50 μm. ∗p < 0.05 and ∗∗∗p < 0.001, one-way ANOVA test.
4. Discussion
As an essential part of bone tissue engineering, BMMSCs have a multilineage differentiation profile, migration and paracrine effects, and low immunogenicity risk, making them widely researched for osteoporosis and bone defect repair [36]. In recent years, studies of KGN have mostly focused on regenerative medicine. Most researches have concentrated on KGN roles for cartilage repair and osteoarthritis treatment. Johnson et al. [3] first demonstrated that KGN could induce MSC chondrogenic differentiation by enhancing the nuclear translocation of CBFβ in order to develop complexes with RUNX1 as well as facilitating to express COL II. So far, the chondrogenic promotion and protection functions of KGN have also been proved [37, 38]. Wang et al. [7] revealed that KGN attenuated reactive oxygen species in intracellular environments and promoted osteogenic differentiation of human BMMSCs. However, the relationship between KGN and osteogenesis improvement is not yet fully understood. In this study, we confirmed the significant inducing role of KGN in promoting BMMSC osteogenesis by upregulating autophagy activity and the Smad1/5/9 pathway. Several studies have revealed an obvious increase in the expression of early osteogenesis-associated markers, such as ALP and Runx2, during BMMSC osteogenic differentiation [39–41]. In our study, KGN elevated the expression level of ALP and Runx2 dominantly. Meanwhile, the proliferation activity of BMMSCs was not affected, indicating that KGN promoted BMMSC osteogenic differentiation capacity without affecting their proliferation ability.
Autophagy, a homeostatic mechanism under various conditions, is required to eliminate cellular waste produced during the quiescent stage of stem cells [42]. The modulation of autophagy in MSCs could affect their differentiation capacity, as well as the fate of proliferation, activation, and function [42]. Past research has shown that autophagy plays an indispensable part in bone remodeling regulation [21]. In our study, the protein levels of autophagy-related genes of Atg7 as well as LC3-II/LC3-I ratio were both elevated after KGN treatment, and the expressions of Beclin1, Atg12, Dram1, and Atg3 were enhanced likewise. These results reflect the role of KGN in promoting autophagic activities of BMMSCs. Additionally, 3-MA, an autophagy inhibitor, was used to confirm the involvement of autophagy during KGN treatment. As a result of 3-MA-induced autophagy suppression, the protein and mRNA levels of ALP and Runx2 in KGN-treated BMMSCs were sharply downregulated, and osteogenic differentiation activity was attenuated. The finding demonstrated that autophagy is essential in KGN-induced osteogenic differentiation.
Studies have shown that autophagy is involved in TGF-β/Smad pathway [14, 43]. Zhang et al. demonstrated that insulin inhibits the osteogenic level of BMMSCs by suppressing autophagic activities and enhancing premature senescence via the TGF-β1 signaling pathway [44]. The phosphorylation of Smad2/3 is also affected by autophagy, which affects a range of functions [45], such as epithelial-to-mesenchymal transition and cell migration [46]. However, the impact of autophagy on Smad1/5/9 protein remains unclear, and current studies on the correlation between autophagy and Smad proteins mostly focus on the functions of cells such as the fibroblasts and endothelial cells. In this study, KGN was found to induce phosphorylation level of Smad1/5/9 in BMMSCs. A study by Decker et al. also proposed that KGN could prompt mouse embryo limb development mainly by upregulating the phosphorylation of Smad2/3 of E11.5 limb bud mesenchymal cells [10]. This phenomenon might be attributed to stemness properties and degrees of differentiation, and the effects of KGN on various Smad phosphorylation levels in embryonic or adult stem cells could impact the osteogenic capacity. Additionally, our results further suggested that LDN [47], a selective inhibitor of the Smad signaling pathway, could suppress the differentiation ability of BMMSCs, whereas the activator SB4 [48] could rescue the inhibiting impacts of 3-MA on the osteogenic differentiation in BMMSCs. These results indicate that KGN induced autophagic activity of BMMSCs and enhanced osteogenic differentiation ability by upregulating the phosphorylation level of Smad1/5/9, which was not found in previous studies. Therefore, our findings suggest that autophagy and Smad1/5/9 play important roles not only in fibroblasts or endothelial cells but also in BMMSCs for osteogenic differentiation. However, although the roles of KGN in BMMSC osteogenic differentiation have been shown here in vitro, more in vivo studies should be further conducted. Therefore, research on the specific functions and mechanisms of KGN would be indispensable, for example, the effects of KGN on bone repair and reconstruction in different cell niches (different age stages, various types of bone defects, osteoporosis model, etc.) [49, 50], and the relationship between KGN and osteogenesis/chondrogenesis under different conditions should also be elucidated. In addition, the application of KGN in vivo is worthy of further exploration, such as combining KGN with biological scaffold materials, seed cells and growth factors to meet the needs of bone tissue engineering, and even suitable drug delivery systems (DDS) to facilitate controlled delivery of KGN [51–53]. More in-depth research to identify the mechanisms of KGN including autophagy-related gene knockout and Smad pathway validation in vivo is also required so as to better understand the role of KGN in bone regeneration.
5. Conclusions
In summary, our study has revealed that KGN plays a key role in promoting osteogenic differentiation of BMMSCs via the modulation of autophagy and phosphorylation level of Smad1/5/9 (Figure 6). The research also improves the understanding of the properties of MSCs and the roles of Smad isoforms as well as the autophagy/Smad1/5/9 signaling pathway, thereby facilitating further studies to explore osteogenic capacity of MSCs in bone regeneration and clinical treatment of bone tissue engineering.
Figure 6.

Schematic diagram of the study. KGN promotes autophagic activity of BMMSCs and upregulates the phosphorylation level of Smad1/5/9, thereby enhancing osteogenesis of BMMSCs.
Acknowledgments
This work was supported by the National Science Foundation of China (grant numbers 81970909 and 51903003), Young Scientists Fund of PKUSS (grant numbers 20110203 and 20170109), Open Foundation of the State Key Laboratory of Military Stomatology (grant number 2020KA01), Key R&D Plan of Ningxia Hui Autonomous Region (grant number 2020BCG01001-4), China Oral Disease Foundation (grant number A2021057), National Multidisciplinary Cooperative Diagnosis and Treatment Capacity Building Project of PKUSS (grant number PKUSSNMP202020), Young Elite Scientist Sponsorship Program by CAST (grant number 2021QNRC001), and Natural Science Foundation of Beijing Municipality (grant number 7192290).
Contributor Information
Yanheng Zhou, Email: yanhengzhou@vip.163.com.
Dawei Liu, Email: liudawei@bjmu.edu.cn.
Data Availability
The authors confirm that all data underlying the findings are fully available. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare that they have no competing interests.
Authors' Contributions
Huichun Yan and Tingting Yu have contributed equally to this work as co-first authors.
Supplementary Materials
Supplementary Figure 1: the titration data of KGN for BMMSC osteogenesis. (A) ARS and (B) semiquantification of mineralized nodules of KGN-induced BMMSC osteogenesis at different concentrations from 0.1 μM to 100 μM. Scale bar = 500 μm. (C, D) qRT-PCR showed the expression levels of ALP and Runx2 at different concentrations of KGN. ∗p < 0.05 and ∗∗∗p < 0.001, one-way ANOVA test.
References
- 1.Xia B., Deng Y., Lv Y., Chen G. Stem cell recruitment based on scaffold features for bone tissue engineering. Biomaterials Science . 2021;9(4):1189–1203. doi: 10.1039/D0BM01591A. [DOI] [PubMed] [Google Scholar]
- 2.Wang X., Wang Y., Gou W., Lu Q., Peng J., Lu S. Role of mesenchymal stem cells in bone regeneration and fracture repair: a review. International Orthopaedics . 2013;37(12):2491–2498. doi: 10.1007/s00264-013-2059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson K., Zhu S., Tremblay M. S., et al. A stem Cell–Based approach to cartilage repair. Science . 2012;336(6082):717–721. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]
- 4.Xu X., Liang Y., Li X., et al. Exosome-mediated delivery of kartogenin for chondrogenesis of synovial fluid- derived mesenchymal stem cells and cartilage regeneration. Biomaterials . 2021;269, article 120539 doi: 10.1016/j.biomaterials.2020.120539. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W., Chen R., Xu X., et al. Construction of biocompatible hydrogel scaffolds with a long-term drug release for facilitating cartilage repair. Frontiers in Pharmacology . 2022 doi: 10.3389/fphar.2022.922032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai J. Y., Zhang L., Chen J., Chen S. Y. Kartogenin and its application in regenerative medicine. Current Medical Science . 2019;39(1):16–20. doi: 10.1007/s11596-019-1994-6. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Chen G., Yan J., et al. Upregulation of SIRT1 by kartogenin enhances antioxidant functions and promotes osteogenesis in human mesenchymal stem cells. Oxidative Medicine and Cellular Longevity . 2018;2018:15. doi: 10.1155/2018/1368142.1368142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caplan A. I., Correa D. The MSC: an injury drugstore. Cell Stem Cell . 2011;9(1):11–15. doi: 10.1016/j.stem.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grayson W. L., Bunnell B. A., Martin E., Frazier T., Hung B. P., Gimble J. M. Stromal cells and stem cells in clinical bone regeneration. Nature Reviews Endocrinology . 2015;11(3):140–150. doi: 10.1038/nrendo.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decker R. S., Koyama E., Enomoto-Iwamoto M., et al. Mouse limb skeletal growth and synovial joint development are coordinately enhanced by kartogenin. Developmental Biology . 2014;395(2):255–267. doi: 10.1016/j.ydbio.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grier W. K., Tiffany A. S., Ramsey M. D., Harley B. A. C. Incorporating β-cyclodextrin into collagen scaffolds to sequester growth factors and modulate mesenchymal stem cell activity. Acta Biomaterialia . 2018;76:116–125. doi: 10.1016/j.actbio.2018.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M., Chen G., Li Y. P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res . 2016;4(1):p. 16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salazar V. S., Gamer L. W., Rosen V. BMP signalling in skeletal development, disease and repair. Nature Reviews. Endocrinology . 2016;12(4):203–221. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- 14.Newman A. C., Kemp A. J., Drabsch Y., Behrends C., Wilkinson S. Autophagy acts through TRAF3 and RELB to regulate gene expression via antagonism of SMAD proteins. Nature Communications . 2017;8(1):p. 1537. doi: 10.1038/s41467-017-00859-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J. H., Kim S. C., Kim K. M., et al. Isorhamnetin attenuates liver fibrosis by inhibiting TGF-β/Smad signaling and relieving oxidative stress. European Journal of Pharmacology . 2016;783:92–102. doi: 10.1016/j.ejphar.2016.04.042. [DOI] [PubMed] [Google Scholar]
- 16.Kopf J., Paarmann P., Hiepen C., Horbelt D., Knaus P. BMP growth factor signaling in a biomechanical context. BioFactors . 2014;40(2):171–187. doi: 10.1002/biof.1137. [DOI] [PubMed] [Google Scholar]
- 17.Higashi K., Matsuzaki E., Hashimoto Y., et al. Sphingosine-1-phosphate/S1PR2-mediated signaling triggers Smad1/5/8 phosphorylation and thereby induces Runx2 expression in osteoblasts. Bone . 2016;93:1–11. doi: 10.1016/j.bone.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Zou M.-L., Chen Z.-H., Teng Y.-Y., et al. The Smad dependent TGF-β and BMP signaling pathway in bone remodeling and therapies. Frontiers in Molecular Biosciences . 2021;8 doi: 10.3389/fmolb.2021.593310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakovljevic J., Harrell C. R., Fellabaum C., Arsenijevic A., Jovicic N., Volarevic V. Modulation of autophagy as new approach in mesenchymal stem cell-based therapy. Biomedicine & Pharmacotherapy . 2018;104:404–410. doi: 10.1016/j.biopha.2018.05.061. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima N., Levine B. Autophagy in mammalian development and differentiation. Nature Cell Biology . 2010;12(9):823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao L., Xiao Y. The autophagy in osteoimmonology: self-eating, maintenance, and beyond. Frontiers in Endocrinology . 2019;10 doi: 10.3389/fendo.2019.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin X., Zhou C., Li J., et al. Autophagy in bone homeostasis and the onset of osteoporosis. Bone Res . 2019;7(1):p. 28. doi: 10.1038/s41413-019-0058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nuschke A., Rodrigues M., Stolz D. B., Chu C. T., Griffith L., Wells A. Human mesenchymal stem cells/multipotent stromal cells consume accumulated autophagosomes early in differentiation. Stem Cell Research & Therapy . 2014;5(6):p. 140. doi: 10.1186/scrt530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D., Kou X., Chen C., et al. Circulating apoptotic bodies maintain mesenchymal stem cell homeostasis and ameliorate osteopenia via transferring multiple cellular factors. Cell Research . 2018;28(9):918–933. doi: 10.1038/s41422-018-0070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang R., Yu T., Kou X., et al. Tet1 and Tet2 maintain mesenchymal stem cell homeostasis via demethylation of the P2rX7 promoter. Nature Communications . 2018;9(1):p. 2143. doi: 10.1038/s41467-018-04464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C., Akiyama K., Wang D., et al. mTOR inhibition rescues osteopenia in mice with systemic sclerosis. The Journal of Experimental Medicine . 2015;212(1):73–91. doi: 10.1084/jem.20140643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang D., Kang R., Livesey K. M., et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metabolism . 2011;13(6):701–711. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon Y., Patel M., Um S., et al. Folic acid pretreatment and its sustained delivery for chondrogenic differentiation of MSCs. Journal of Controlled Release . 2022;343:118–130. doi: 10.1016/j.jconrel.2022.01.018. [DOI] [PubMed] [Google Scholar]
- 29.Zeng W. N., Zhang Y., Wang D., et al. Intra-articular injection of kartogenin-enhanced bone marrow-derived mesenchymal stem cells in the treatment of knee osteoarthritis in a rat model. The American Journal of Sports Medicine . 2021;49(10):2795–2809. doi: 10.1177/03635465211023183. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S., Hu P., Liu T., et al. Kartogenin hydrolysis product 4-aminobiphenyl distributes to cartilage and mediates cartilage regeneration. Theranostics . 2019;9(24):7108–7121. doi: 10.7150/thno.38182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baharlou Houreh A., Masaeli E., Nasr-Esfahani M. H. Chitosan/polycaprolactone multilayer hydrogel: a sustained kartogenin delivery model for cartilage regeneration. International Journal of Biological Macromolecules . 2021;177:589–600. doi: 10.1016/j.ijbiomac.2021.02.122. [DOI] [PubMed] [Google Scholar]
- 32.Hong Y., Liu N., Zhou R., et al. Combination therapy using kartogenin-based chondrogenesis and complex polymer scaffold for cartilage defect regeneration. ACS Biomaterials Science & Engineering . 2020;6(11):6276–6284. doi: 10.1021/acsbiomaterials.0c00724. [DOI] [PubMed] [Google Scholar]
- 33.Sobecki M., Mrouj K., Camasses A., et al. The cell proliferation antigen Ki-67 organises heterochromatin. eLife . 2016;5:e13722–e13722. doi: 10.7554/eLife.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klionsky D. J., Abdel-Aziz A. K., Abdelfatah S., et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1) Autophagy . 2021;17(1):1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Y.-F., Su T., Yang M., et al. The role of autophagy in bone homeostasis. Journal of Cellular Physiology . 2021;236(6):4152–4173. doi: 10.1002/jcp.30111. [DOI] [PubMed] [Google Scholar]
- 36.Andrzejewska A., Lukomska B., Janowski M. Concise review: mesenchymal stem cells: from roots to boost. Stem Cells . 2019;37(7):855–864. doi: 10.1002/stem.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X. Q., Shi D. Q., Shen Y. S., et al. Full-thickness cartilage defects are repaired via a microfracture technique and intraarticular injection of the small-molecule compound kartogenin. Arthritis Research & Therapy . 2015;17(1) doi: 10.1186/s13075-015-0537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Wang J. H. C. Kartogenin induces cartilage-like tissue formation in tendon-bone junction. Bone Research . 2014;2(1) doi: 10.1038/boneres.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vimalraj S. Alkaline phosphatase: structure, expression and its function in bone mineralization. Gene . 2020;754, article 144855 doi: 10.1016/j.gene.2020.144855. [DOI] [PubMed] [Google Scholar]
- 40.Bruderer M., Richards R. G., Alini M., Stoddart M. J. Role and regulation of RUNX2 in osteogenesis. European Cells & Materials . 2014;28:269–286. doi: 10.22203/eCM.v028a19. [DOI] [PubMed] [Google Scholar]
- 41.Komori T. Regulation of osteoblast differentiation by transcription factors. Journal of Cellular Biochemistry . 2006;99(5):1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 42.Ceccariglia S., Cargnoni A., Silini A. R., Parolini O. Autophagy: a potential key contributor to the therapeutic action of mesenchymal stem cells. Autophagy . 2020;16(1):28–37. doi: 10.1080/15548627.2019.1630223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki H. I., Kiyono K., Miyazono K. Regulation of autophagy by transforming growth factor-β (TGF-β) signaling. Autophagy . 2010;6(5):645–647. doi: 10.4161/auto.6.5.12046. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P., Zhang H., Lin J., et al. Insulin impedes osteogenesis of BMSCs by inhibiting autophagy and promoting premature senescence via the TGF-β1 pathway. Aging (Albany NY) . 2020;12(3):2084–2100. doi: 10.18632/aging.102723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y., Xiong L., Wang X., et al. Autophagy inhibition attenuates TGF-β2-induced epithelial-mesenchymal transition in lens epithelial cells. Life Sciences . 2021;265, article 118741 doi: 10.1016/j.lfs.2020.118741. [DOI] [PubMed] [Google Scholar]
- 46.Trelford C. B., Di Guglielmo G. M. Autophagy regulates transforming growth factor β signaling and receptor trafficking. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research . 2022;1869(9, article 119284) doi: 10.1016/j.bbamcr.2022.119284. [DOI] [PubMed] [Google Scholar]
- 47.Cuny G. D., Yu P. B., Laha J. K., et al. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorganic & Medicinal Chemistry Letters . 2008;18(15):4388–4392. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradford S. T. J., Ranghini E. J., Grimley E., Lee P. H., Dressler G. R. High-throughput screens for agonists of bone morphogenetic protein (BMP) signaling identify potent benzoxazole compounds. The Journal of Biological Chemistry . 2019;294(9):3125–3136. doi: 10.1074/jbc.RA118.006817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGovern J. A., Griffin M., Hutmacher D. W. Animal models for bone tissue engineering and modelling disease. Disease Models & Mechanisms . 2018;11(4) doi: 10.1242/dmm.033084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin H., Sohn J., Shen H., Langhans M. T., Tuan R. S. Bone marrow mesenchymal stem cells: aging and tissue engineering applications to enhance bone healing. Biomaterials . 2019;203:96–110. doi: 10.1016/j.biomaterials.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newman M. R., Benoit D. S. Local and targeted drug delivery for bone regeneration. Current Opinion in Biotechnology . 2016;40:125–132. doi: 10.1016/j.copbio.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., Newman M. R., Benoit D. S. W. Development of controlled drug delivery systems for bone fracture-targeted therapeutic delivery: a review. European Journal of Pharmaceutics and Biopharmaceutics . 2018;127:223–236. doi: 10.1016/j.ejpb.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng Y., Hoque J., Varghese S. Biomaterial-assisted local and systemic delivery of bioactive agents for bone repair. Acta Biomaterialia . 2019;93:152–168. doi: 10.1016/j.actbio.2019.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: the titration data of KGN for BMMSC osteogenesis. (A) ARS and (B) semiquantification of mineralized nodules of KGN-induced BMMSC osteogenesis at different concentrations from 0.1 μM to 100 μM. Scale bar = 500 μm. (C, D) qRT-PCR showed the expression levels of ALP and Runx2 at different concentrations of KGN. ∗p < 0.05 and ∗∗∗p < 0.001, one-way ANOVA test.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available. Further inquiries can be directed to the corresponding authors.


