Abstract
The causative agent of Chagas disease (CD), Trypanosoma cruzi, claims thousands of lives each year. Current diagnostic tools are insufficient to ensure parasitological detection in chronically infected patients has been achieved. A host-derived metabolic signature able to distinguish CD patients from uninfected individuals and assess antiparasitic treatment efficiency is introduced. Serum samples were collected from chronic CD patients, prior to and three years after treatment, and subjected to untargeted metabolomics analysis against demographically matched CD-negative controls. Five metabolites were confirmed by high-resolution tandem mass spectrometry. Several database matches for sex steroids were significantly altered in CD patients. A murine experiment corroborated sex steroid perturbation in T. cruzi-infected mice, particularly in male animals. Proteomics analysis also found increased steroidogenesis in the testes of infected mice. Metabolic alterations identified in this study shed light on the pathogenesis and provide the basis for developing novel assays for the diagnosis and screening of CD patients.
Keywords: Chagas disease, Trypanosoma cruzi, Biomarker discovery, Metabolomics, Proteomics, Mass spectrometry, Mouse model, Sex steroids, Testis, Testosterone, Taurine, Glutamine, Phenylalanyl-threonine, Pyroglutamyl-glycine
Chagas disease; Trypanosoma cruzi; Biomarker discovery; Metabolomics; Proteomics; Mass spectrometry; Mouse model; Sex steroids; Testis; Testosterone; Taurine; Glutamine; Phenylalanyl-threonine; Pyroglutamyl-glycine.
1. Introduction
American trypanosomiasis, also known as Chagas disease (CD), is caused by the protozoan parasite Trypanosoma cruzi and is mainly transmitted via the triatomine vector to the mammalian host [1]. Vector-borne transmission is largely limited to Latin America and US southern states, however, tourism and immigration from endemic countries have facilitated the spread of CD to other countries through non-vector routes including blood transfusion, organ transplantation, and congenital transmission [2]. Most CD patients remain infected for life after their initial exposure to T. cruzi. The acute phase of CD is often asymptomatic, and mild symptoms spontaneously resolve in most cases. The patient, however, remains chronically infected if untreated, and enters an indeterminate phase in which they remain seropositive for T. cruzi with no clinical signs and symptoms [3]. The infection can then progress into identifiable complications in the heart, gastrointestinal tract and central nervous system in the case of severe immunosuppression [4]. Chagas cardiomyopathy is a complication in 30% of chronically infected patients [5]. CD treatment relies on two nitroheterocyclic drugs: nifurtimox (Nfx) and benznidazole, effective during acute and early chronic phases of infection but much less so in people with long-standing infections. Moreover, their clinical effectiveness is limited by long treatment courses and potentially severe toxicity [6]. A major clinical challenge is the absence of cure biomarkers as serology results usually remain positive years to decades after treatment [1].
Disease biomarkers are a valuable tool for the diagnosis and follow-up screening of parasitic diseases, such as leishmaniasis, toxoplasmosis and malaria [7, 8, 9]. Several parasite-derived biomarkers have also been reported for CD, such as trypomastigote excreted-secreted antigens (TESA) and trypanosomal DNA [10, 11]. However, due to the high heterogeneity of parasitic burden and tissue distribution in CD, diagnosis using TESA and DNA-based methods are limited, often showing low sensitivity/specificity and poor reproducibility, respectively. Moreover, they are expensive and difficult to implement in endemic areas with limited resources and infrastructure. In contrast, methods based on host response or damage biomarkers are generally cost-efficient and do not require highly specialized equipment. Potential CD host biomarkers include immunological markers (cytokines and surface markers) elicited by the host cellular response to the infection [12]; inflammatory markers of cardiac damage [13]; hypercoagulability factors and other biochemical markers [14]. However, most host-derived approaches are in an early stage of development and further studies are required to validate their use in the diagnosis or prognosis of CD [14].
We have previously shown that various fragments of apolipoprotein A1 (ApoA1) and fibronectin have the potential to assess anti-trypanosomal treatment efficacy as CD biomarkers [15, 16]. ApoA1 is a major component of high-density lipoprotein (HDL) and plays a key role in lipid transport and metabolism. To understand the processes underlying ApoA1 degradation and lipid homoeostasis, we conducted metabolomics and lipidomics analyses on serum samples from South American patients with chronic CD before and after Nfx treatment. From these results, we describe a novel panel of metabolite biomarkers to potentially distinguish CD patients from CD-negative individuals and assess the efficiency of antiparasitic treatment. This work is the first untargeted metabolomics study of CD in humans and presents a novel CD host-parasite interaction. The metabolic panel described here could be used to develop novel assays for the diagnosis and follow-up screening of CD.
2. Methods
2.1. Sample collection and diagnosis
Serum samples from 24 chronic CD patients and 24 age-, sex- and ethnicity-matched CD-negative controls were collected at the Geneva University Hospitals (Geneva, Switzerland; protocol 11–162). Both cohorts were comprised of Latin American migrants originating from endemic areas for CD. Written informed consent was obtained from all participants. Patients received treatment with Nfx for 40 ± 24 days and were called back for a follow-up serum sample three years later. The seronegative status of the controls was confirmed by two serological tests as described elsewhere [17]. Five patients were also diagnosed with cardiomyopathy. Sociodemographic and clinical characteristics of the study participants are described in Table 1. Serum samples were frozen (−80 °C) within 1 h of collection and remained in freezer under identical storage conditions until defrosted at 4 °C for analysis. No internal standard was added to the serum samples.
Table 1.
Sociodemographic and clinical characteristics of the study participants.
| Number (%) or mean (range) |
|||
|---|---|---|---|
| Patients (N = 24) |
Controls (N = 24) |
||
| Sex | Female | 22 (92%) | 23 (96%) |
| Male | 2 (8%) | 1 (4%) | |
| Age (years) | 43 (28–56) | 39 (25–56) | |
| Origin | Bolivia | 21 | 23 |
| Brazil | 2 | 1 | |
| Argentina | 1 | 0 | |
| Staging | Indeterminate | 19 | 0 |
| Cardiomyopathy | 5 | 0 | |
| Duration of treatment (days) | 60 | 12 | 0 |
| 30–59 | 4 | 0 | |
| <30 | 10 | 0 | |
2.2. Metabolomics analysis
Serum (25 μL) was precipitated in methanol (1:3 v/v). The soluble fraction was dried under vacuum, reconstituted in 10% acetonitrile (ACN, 75 μL) and analyzed (12 μL) using a TripleTOF 5600 tandem mass spectrometer (Sciex, Concord, ON, Canada) equipped with a Shimadzu Nexera UHPLC and a 150 3 mm 3 μm (130 Å) Scherzo SM-C18 multi-mode HPLC column with water (A) and ACN (B), both containing 0.1% formic acid (FA), at a flow rate of 250 μL/min (40 °C). Liquid chromatography (LC) gradient started at 3% B, held for 3 min, increased linearly to 80% at 17.5 min, held for 2.5 min, increased to 95% at 20.5 min, and held for 3.5 min. Mass spectrometry (MS) analysis was performed in positive (5.5 kV) and negative (−4.5 kV) mode electrospray ionization (ESI) at 450 °C with a declustering potential of 60 V for m/z 80–975. MS/MS spectra were acquired at m/z 50–700 for the 5 most abundant ions using information-dependent acquisition and collision-induced dissociation (collision energy 30 ± 10 eV) for precursor ions >500 intensity units excluded for 20 min after 3 occurrences. Mass tolerances were set to 50 ppm and 50 mDa at the MS and MS/MS levels, and accumulation times were 0.3 s and 0.15 s for MS and MS/MS analyses, respectively. Total cycle time was 0.95 s.
2.3. Metabolite identification and confirmation
Peak finding was performed in MarkerView software (Sciex) for 1.5–18.5 min retention time (RT) with predefined minimum spectral width (0.01 Da) and LC peak width (2 scans). RT tolerance and mass tolerance were 0.2 min and 0.01 Da, respectively. Molecular features with p > 0.01 (Student's t-test) and fold-change (patient/control) < ±1.5 were excluded. Isotopologues, adduct ions, and those resulting from in-source fragmentation of common neutral losses were also excluded. The mass spectra associated with the remaining features were visually inspected in PeakView software (Sciex) to confirm their exact mass and charge state. Confirmed molecular features were searched against the Human Metabolome Database (HMDB) [18], the Metabolite and Chemical Entity Database (METLIN) [19], and the Accurate Mass Metabolite Spectral Library (AMMSL v1.0; Sciex). LC-MS metabolite search was performed with a mass tolerance of 5 ppm for proton transfer adducts. Metabolite identification was performed based on the four following criteria: (1) Only protonated or deprotonated ions were accepted. (2) Features with arguable chromatographic behaviour were rejected; for example, hydrophilic compounds found at a late retention time were dismissed. (3) Physiologically irrelevant compounds such as xenobiotics and odd-chain fatty acids were not retained. (4) Structures with questionable chemical stability such as protonated esters or epoxides were also excluded. SciexOS software was used for the MS/MS spectral library searching using the “candidate search” algorithm with a precursor and fragment mass tolerance of 0.2 Da. Selected metabolites were confirmed by LC-HRMS/MS using RT and MS/MS spectral matching with synthetic standards. Phe-Thr standard was synthesized and purified at the Center of Excellence in Orphan Disease Research–Courtois Foundation (CERMO-FC) at Université du Québec à Montréal (Montreal, QC, Canada). pGlu-Gly and Asn-Gly-Phe-Lys were purchased from CanPeptide (Saint Laurent, QC, Canada). Steroid standards were purchased from Steraloids (Newport, RI, USA) and authorized for import by Health Canada Office of Controlled Substances (authorization # 49322.10.19 & 49323.10.19). Other standards were purchased in analytical grade from Sigma-Aldrich (Saint Louis, MO, USA). Standard solutions (1.0 mg/mL) were made in 50% MS-grade ACN (Sigma-Aldrich). LC-HRMS/MS analysis was conducted at Université du Québec à Montréal. Univariate and multivariate (with respect to metadata confounders) analyses were performed using Microsoft Excel and MetaboAnalyst web application [20], respectively.
2.4. Synthesis and purification of phenylalanyl-threonine
Phe-Thr standard was produced by Fmoc chemistry solid-phase peptide synthesis. Fmoc-L-Thr (tBu)-OH (795 mg, 2 mmol) was linked to 2-chlorotrityl chloride resin (1.176 g, 1 mmol, 100–200 mesh, 1% divinylbenzene, Matrix Innovation) by overnight incubation in N,N-diisopropylethylamine (DIEA, 523 μL, 3 mmol) and dichloromethane (DCM, 20 mL) with agitation by nitrogen sparging. After multiple washes of the resin with DCM, a small aliquot of resin was dried, weighted (10 mg) and the loading efficiency was quantified by measuring dibenzofulvene absorbance at 290 nm after Fmoc removal by 20% piperidine in dimethylformamide (DMF, 5 min), yielding 0.46 mmol/g (54%). The resin was capped (9 mL DCM, 1 mL methanol, 0.5 mL DIEA) for 1 h, washed (DCM twice, then DMF), Fmoc was removed (20% piperidine in DMF, 15 min), and the resin was washed again (DMF, DCM, DMF sequentially, 2 min each). Fmoc-L-Phe-OH (628 mg, 1.62 mmol) was coupled to resin-bound Thr using O-(1H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HCTU, 670 mg, 1.62 mmol) and DIEA (280 μL, 1.62 mmol) for 30 min. The resin was washed, the Fmoc group was removed, the resin was washed with diethyl ether and air-dried. The peptide was cleaved off from the resin (20 mL of 95% trifluoroacetic acid/2.5% water/2.5% triisopropylsilane, 2 h) and the resulting solution was concentrated. The crude peptide was precipitated in cold diethyl ether, air-dried, dissolved in deionized water, and lyophilized. The product was then dissolved in 0.5 mL water and purified by size-exclusion chromatography (20 mL packed Sephadex G-10, water elution, 1 mL fractions). Pure fractions were pooled and lyophilized, yielding Phe-Thr (102 mg, 38% total yield). HR-MS (LC-TOF): calcd. for [C13H18N2O4+H]+ 267.1339; found 267.1338. 1H NMR (300 MHz, D2O): δ 7.41–7.26 (m, 5H), 4.40 (d, J = 3.8 Hz, 1H), 4.36 (t, J = 7.0 Hz, 1H), 4.28 (m, 1H), 3.28 (dd, J = 14.2, 6.7 Hz, 1H), 3.18 (dd, J = 14.2, 7.4 Hz, 1H), 2.68 (s, 1H), 1.17 (d, J = 6.5 Hz, 3H). 13C NMR (75.44 MHz, D2O): δ 173.1, 169.4, 133.5, 129.4, 129.1, 128.0, 67.1, 58.4, 54.2, 36.7, 18.8.
2.5. Free fatty acid and cholesterol measurement
Serum (20 μL) was precipitated in acetone (1:10 v/v). The soluble fraction was transferred into a clean tube and evaporated (56 °C, 1 h). Serum lipids were hydrolyzed with potassium hydroxide (1 M in 70% ethanol, 80 °C, 4 h). The solution was then acidified with hydrochloric acid and extracted twice with pentane. Pentane was evaporated (40 °C, 1 h), sample was additionally heated (80 °C, 1 h) to remove moisture and reconstituted in ACN (25 μL). A non-endogenous fatty acid (pentadecanoic acid, 15:0) was added (50 μg/mL) as internal standard. Sample was then derivatized with N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA, 25 μL, 80 °C, 45 min). Two microliters of the BSTFA derivative was injected into an Agilent 7890-5975C GC-MS instrument (Agilent Technologies, Santa Clara, CA, USA) with a split ratio of 5:1 and inlet temperature of 280 °C. Gas chromatography (GC) separation was done at a starting temperature of 160 °C, then increased to 180 °C (13 °C/min, 0 min), to 225 °C (2 °C/min, 1 min), and to 310 °C (17 °C/min, 2 min) on a HP-5MS 30 m × 250 μm × 0.25 μm capillary column (Agilent) at a carrier gas flow rate of 1.2 mL/min. Selected-ion monitoring (SIM) was conducted in positive ionization mode for m/z 314 (C15:0), 326 (C16:1), 328 (C16:0), 352 (C18:2), 354 (C18:1), 356 (C18:0), 376/150 (C20:4) and 458 (cholesterol) with a dwell time of 0.01 s. To minimize statistical bias, samples were randomized and divided into 5 groups of ∼15 samples. Reference standards were purchased from Sigma-Aldrich and used to characterize each analyte based on m/z value and RT. Quantification was performed using the peak area ratio of each analyte to the internal standard. All analytes were detected in all the samples analysed. GC-MS analysis was conducted at Université de Montréal (Montreal, QC, Canada).
2.6. Parasite culturing and transformation
Logarithmic-phase epimastigotes of the Brazil strain [21] were washed three times in EMEM complete medium supplemented with 5% FBS and gentamycin (Wisent, Saint Bruno, QC, Canada) and added to Vero cell monolayers (ATCC CCL-81). Infected cells were washed on the third and fifth days post-infection. Subsequently, the supernatant containing trypomastigotes was collected, centrifuged (1900 g, 20 min), washed in PBS (Wisent) and counted in a Neubauer haemocytometer (Hausser Scientific, Horsham, PA, USA).
2.7. Murine infection and hormone assessment
Six- to eight-week-old CD-1 mice (Charles River Laboratories, Senneville, QC, Canada) were assigned to four study groups of equal number of males and females each (eight subgroups total). Mice were intraperitoneally injected with 10,000 culture-derived T. cruzi trypomastigotes and their weights were monitored weekly. Blood was collected from the lateral saphenous vein weekly in heparinized microcapillaries for immediate detection of parasites in the buffy coat, followed by serum collection and storage at –80 °C. One group was euthanized (isoflurane/CO2) on day 22 post-infection (PI) (acute phase), underwent cardiac puncture for blood and serum collection. One group was treated with Nfx (100 mg/kg body weight) on day 74 PI (chronic phase) for 20 days q. d. by gavage. The study was terminated on day 110 PI, and blood, serum, testes, and ovaries were collected. Samples were flash frozen in liquid nitrogen immediately after acquisition and stored at –80 °C until analysis. All procedures were carried out in accordance with the guidelines of the Canadian Council on Animal Care, as approved by the Animal Care Committee of McGill University (Animal Use Protocol 7631). The width of the testes was measured weekly with a digital caliper (General Tools, Secaucus, NJ, USA) before, throughout and after the treatment during the chronic phase of infection from day 70 to day 105 PI. Measurements were made to the nearest 0.1 mm. Serum testosterone, progesterone and estradiol levels were assessed by commercially available enzyme-linked immunosorbent assay (ELISA) kits at the Ligand Assay and Analysis Core Facility of the University of Virginia Center for Research in Reproduction (Charlottesville, VA, USA). Nfx was donated by Dr. Eric Chatelain (Drugs for Neglected Diseases initiative, Geneva, Switzerland).
2.8. Statistical analysis
Widths of testes were evaluated using one-way analysis of variance (ANOVA) followed by Tukey's multiple comparisons test. Sex hormone levels and weight change were evaluated using two-way ANOVA for the following two factors: sex (male vs female) and disease phase (uninfected vs acute, uninfected vs chronic, uninfected vs treated, acute vs chronic, acute vs treated, chronic vs treated) followed by Bonferroni post-hoc test for significant interactions when appropriate. When no interaction was found, unpaired t-tests were performed on the factor where significance was observed. The results are presented as the mean ± standard error of the mean (SEM). Differences were statistically significant at a p-value of < 0.05. Statistical analyses were performed using GraphPad Prism 5.00 (GraphPad Software, San Diego, CA, USA).
2.9. Proteomics analysis of mouse testes
Testes were thawed on ice, surgically separated from epididymis and ductus deferens, rinsed with PBS and dissected transversely. One section (∼25 mg) was used for DNA extraction and T. cruzi PCR. The remainder (25–120 mg) was thoroughly ground in 8 M urea, 0.1 M NaCl, 25 mM Tris HCl pH 8.2 with Pierce Protease Inhibitor (Thermo Scientific, Waltham, MA, USA) to minimize proteolytic degradation. The suspension was further homogenized (10 min, 4 °C) using a 400 W/20 kHz Branson Digital Sonifier (Shanghai, China) for 15 s at 50% maximum power and vigorously mixed for 10 s. The sonication/mixing cycle was repeated three more times. The insoluble fraction was separated by centrifugation (18,000 g, 4 °C, 10 min). The protein concentration of the supernatant was determined by Pierce BCA Protein Assay Kit (Thermo Scientific). An equivalent of 0.5 mg total protein was then diluted 10-fold in 0.1 M ammonium bicarbonate pH 8.0, reduced with dithiothreitol (10 μL, 0.1 M, 65 °C, 30 min) and alkylated with 2-iodoacetamide (10 μL, 0.25 M, 25 °C, 30 min) in the dark. Protein digestion was performed with porcine pancreas trypsin (Sigma-Aldrich; 5 μL, 2.0 mg/mL) overnight (37 °C, 16 h) at a 1:50 enzyme-to-protein ratio. Digestion was stopped by decreasing pH to 4.0 with 10% acetic acid. Sample was centrifuged (18,000 g, 4 °C, 10 min) and the supernatant was cleaned up using an Oasis HLB solid-phase extraction cartridge (Waters, Milford, MA, USA) following manufacturer's instructions. Peptides were eluted in methanol (0.1% FA), evaporated under vacuum (Labconco, Kansas City, MO, USA; 40 °C, 3 h) and reconstituted in buffer A (120 μL, see below). Strong cation exchange fractionation was performed using a published method [22]. Briefly, the peptide mixture was injected (100 μL) on to a Zorbax 300-SCX 150 mm × 2.1 mm column with 5 μm (300 Å) particles (Agilent) using a Beckman System Gold HPLC (Beckman Coulter, Brea, CA, USA) at a flow rate of 0.25 mL/min with a gradient of 0–50% B in 15 min, up to 100% B at 25 min, then held for an additional 5 min at 100% B, where buffers A and B were 10 mM potassium dihydrogen phosphate in 25% ACN (pH 2.75), and 1 M potassium chloride in buffer A (pH 2.75), respectively. UV absorbance was monitored at 220 and 280 nm. For each sample, 3-min (0.75-mL) fractions were aliquoted into 1.5-ml tubes from 2 to 32 min. Fractions were evaporated to dryness under vacuum and reconstituted (60 μL) in MS-grade water (0.1% FA, Sigma-Aldrich). LC-HRMS/MS analysis was performed (20 μL) on a Maxis II quadrupole-time-of-flight mass spectrometer (Bruker, Billerica, MA, USA) equipped with a Dionex UltiMate 3000 UHPLC system (Thermo Scientific) and an Aeris Peptide XB-C18 1.7 μm 100 Å 150 × 2.1 mm HPLC column (Phenomenex, Torrance, CA, USA) with water (A) and ACN (B), both containing 0.1% FA, at a flow rate of 125 μL/min (50 °C). LC gradient started at 5% B, held for 3 min, was increased to 45% at 25 min, and to 80% at 32 min. Apollo ESI source (Bruker) operated in positive mode with endplate offset and capillary voltage of 0.5 and 4.5 kV, respectively. A continuous flow of 99.5% pure N2 was provided (Parker, Mayfield Heights, OH, USA) as dry gas (200 °C) at a flow rate of 6 L/min. MS and MS/MS spectra were acquired at m/z 300–2200 using data-dependent acquisition and collision-induced dissociation with a collision energy of 21–55 eV depending on the precursor ion m/z value and charge state. Precursor ions with a charge state of 2–5 were preferred and singly charged ions were excluded. Redundant ions were also excluded for 3 min. Acquisition time was 0.25 s for MS and 0.06–0.25 s for MS/MS scan, depending on precursor ion signal intensity. MS instrument was internally calibrated via post-column infusion of ESI-L Low-Concentration Tuning Mix (Agilent). Samples were analyzed in random order in the sequence to avoid biases due to the injection order. Moreover, at least one blank (5 μL, mobile phase A) was inserted between each two analyses to minimize carryover. Proteomics analysis was conducted at Research Institute of the McGill University Health Centre (Montreal, QC, Canada).
2.10. Proteomics data analysis
LC-HRMS/MS data were imported into the MaxQuant software [23] and searched at 1% false-discovery rate (FDR) using the Andromeda search algorithm against the mouse subset of the UniProt/SwissProt protein database (17,023 sequences, downloaded in January 2020) for tryptic peptides allowing up to 2 missed cleavages. Methionine oxidation and N-terminal acetylation were defined as variable modifications while cysteine carbamidomethylation was set as fixed modification. Mass tolerances were 70 and 6 mDa for the first and main peptide searches, respectively. To account for LC retention shifts, the “match between runs” option was enabled with a match time window of 0.7 min and an alignment time window of 20 min. Only database searches with an FDR-adjusted p < 0.01 were accepted as protein hits. Proteome-wide label-free quantification (LFQ) was performed based on 2 razor unique peptides per protein [24]. Statistical testing was performed by Perseus software [25] at the 2-tailed α level of 0.05 (p < 0.05) to identify significantly differentially abundant proteins (fold-change >1.2) across the two cohorts. Permutation-based FDR [26] was used to correct p-values for multiple hypothesis testing. Protein accession numbers were uploaded to PANTHER [27] for gene ontology classification. Pathway over-representation analysis (ORA) was performed by InnateDB [28] using a hypergeometric search algorithm, Benjamini-Hochberg correction method, and pathway annotations from Reactome, INOH, NETPATH, KEGG, Biocarta and NCI databases. Only over-represented pathways with a corrected p-value < 0.05 were accepted. LC-HRMS/MS raw data from this analysis have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [29] with the dataset identifier PXD017802.
2.11. DNA extraction and PCR analysis
DNA extraction was carried out using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following manufacturer's instructions. PCR reactions were performed using a Bio-Rad T100 Thermal Cycler (Hercules, CA, USA; donated by Lucy Riddell via McGill University Health Centre Foundation) with 30 μL of 1× GoTaq Hot Start Polymerase Master Mix (Promega, Madison, WI, USA), containing 1.7 μM of each primer [30], and 20 μL of template DNA (final volume 50 μL). PCR amplification was conducted by 30 cycles of 94 °C (30 s), 50 °C (2 min), 72 °C (2 min) followed by final extension at 72 °C (5 min) and 37 °C (10 min). Control negative and positive DNA samples were included in the analysis to verify that carryover DNA contamination had not occurred. Detection of amplified DNA was accomplished by electrophoresis of PCR product (10 μL) on 2% agarose gel previously stained with SYBR Safe (0.01% v/w) (Life Technologies, Carlsbad, CA, USA). Fluorescent bands were visualized by an Omega Lum C imaging system (Aplegen, San Francisco, CA, USA). PCR analysis was conducted at Research Institute of the McGill University Health Centre.
3. Results
3.1. Chronic CD patients have an altered metabolomic profile
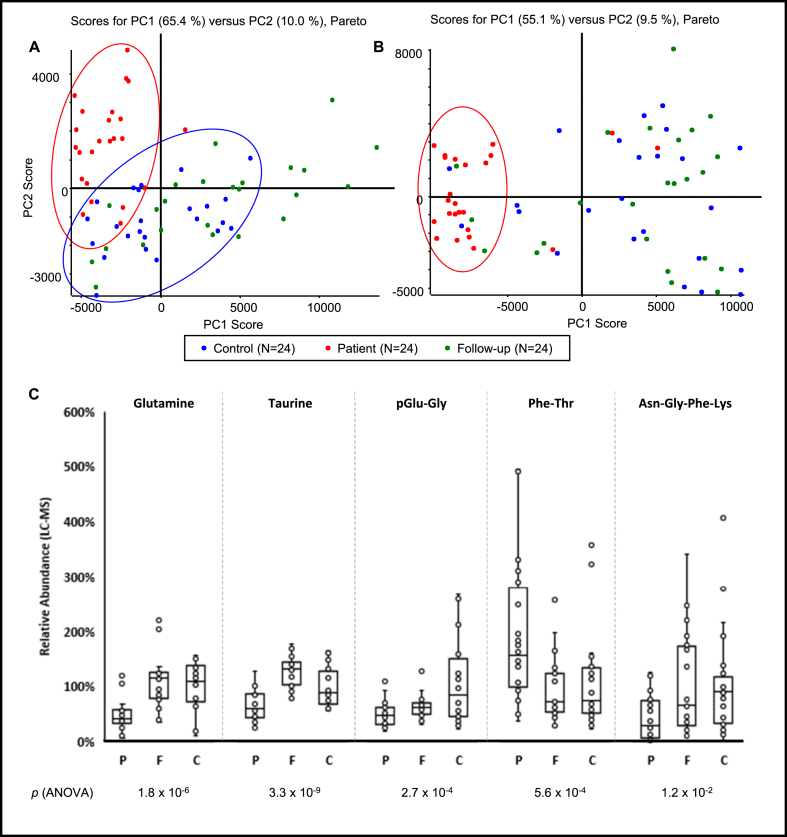
To identify metabolic changes associated with chronic CD, liquid chromatography combined with positive/negative ionization mass spectrometry (MS) was performed on the serum of confirmed CD patients before and after Nfx treatment, as well as healthy controls. A total of 9,127 and 9,317 features were detected in positive and negative ionization modes, respectively, from these samples. Using a non-supervised principal component analysis (PCA) with Pareto scaling and Student's t-test, the number of positively charged and negatively charged features with significantly (p < 0.01) altered abundance were reduced to 1,053 and 1,822, respectively. Using this subset of features, sample grouping within PCA scoring plots demonstrated that untreated CD patients and uninfected controls were distinct from both positively and negatively-charged metabolites. However, the same distinct groupings between groups was not as obvious in PCA plots from negative mode data. Sera from Nfx-treated patients did not show any clear grouping in either ionization mode (Figure 1A, B).
Figure 1.
Chronic CD patients have a significantly altered metabolomic profile. Scoring plots from principal component analysis (PCA) of positive (A) and negative (B) ionization mass spectrometry of sera from Chagas disease (CD) patients (red), post-treatment follow-up samples (green) and demographically matched CD-negative controls (blue). Samples clustered against the principal components (PC1 & PC2) are comprised of constituents (i.e., metabolites) with significantly different abundances. Five metabolites with significantly different levels in patients (fold-change ≥ ±1.5, patients/controls) were identified by liquid chromatography - high resolution tandem mass spectrometry (LC-HRMS/MS) (C). Significance levels were calculated using ANOVA and F-test. Error bars represent mean ± standard error of mean. % Abundances were normalized based on average control levels. P, F and C on the x-axis represent patient, follow-up, and control groups, respectively. Circles on the PCA plots were drawn to demonstrate sample grouping and do not represent mathematical confidence intervals.
Comparing the untreated CD patients with those of uninfected controls, the relative abundance of 371 positively charged ions and 775 negatively charged ions were increased/decreased by more than 50%, respectively. To identify unreliable metabolite candidates, the MS spectra associated with each feature were visually inspected to ensure proper peak shape and monoisotopic mass. Thirty percent of the candidates were retained (344/1,146) leading to 42 acceptable MS/MS database matches for putative metabolite identifications. Of these, 11 were increased and 31 were decreased in the CD patients relative to uninfected controls (Table S1).
Five metabolites were confirmed using synthetic standards by exact mass measurements of precursor and fragment ions in MS/MS spectra and chromatographic retention time, including taurine, glutamine and three small peptides: pGlu-Gly, Phe-Thr, Asn-Gly-Phe-Lys. Phe-Thr was more abundant in CD patients, while other compounds were decreased in those patients. Following Nfx treatment, four of these metabolites returned to levels matching those of healthy controls, with pGlu-Gly being the exception (Figure 1C). No strong correlation was found between the identified metabolites, age, cardiomyopathy, or treatment duration (Pearson's r < 0.4). However, taurine and Asn-Gly-Phe-Lys moderately correlated (| r | = 0.4–0.8) with patient's sex (Table S2). Taurine was more abundant in male patients (male/female levels = 1.2) before the treatment and less abundant in them (male/female levels = 0.7) after the treatment. Asn-Gly-Phe-Lys was more abundant in male patients before (male/female levels = 2.9) and after (male/female levels = 2.0) the treatment. Multivariate analysis confirmed that the above metabolites were not confounded by metadata variables, including age, country of origin, diagnosis, or sex (Figure S1). Two database matches for sex steroids had increased levels in CD patients. The MS/MS spectra of these compounds matched with 9 progestogens/androgens. However, we could not confirm the structure of these metabolites with synthetic standards. LC-HRMS/MS data of confirmed metabolites and reference standards are included in Figure S2.
3.2. Anti-trypanosomal treatment restores serum lipid levels in CD patients
Several major serum fatty acids, including palmitic acid (C16:0), linoleic acid (C18:2), stearic acid (C18:0), arachidonic acid (C20:4), and cholesterol (C27:1) were significantly increased (p < 0.03; ANOVA) in CD patients relative to healthy controls. Palmitoleic acid (C16:1) and oleic acid (C18:1) showed a trend towards increased levels in CD patients compared to healthy controls. They all returned to control levels after treatment, except palmitoleic acid that was unchanged (Figure S3). Several tentative lipid oxidation products, such as oxocaprylic acid (C8:0 + O), nonanedione (C9:0), hydroxycapric acid (C10:0) and hydroxylauric acid (C12:0) had decreased levels in CD patients. Fourteen unconfirmed metabolites were associated with phospholipids (Table S1). Of these, 2 increased and 12 decreased in the patients. All lipid metabolites had levels comparable to CD-negative controls after Nfx treatment.
3.3. T. cruzi infection in mice leads to altered sex steroid metabolism and testes morphology
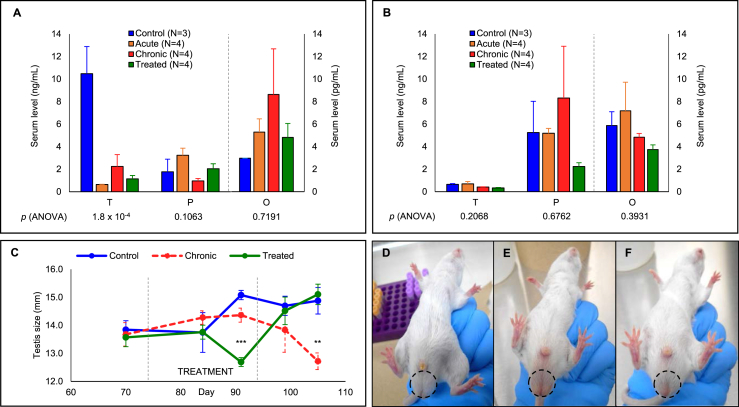
Our metabolomics analysis of human sera suggested that several steroid metabolites were changing in CD patients. To assess the effects of chronic T. cruzi infection on sex steroid metabolism in vivo, mice were infected with T. cruzi trypomastigotes and their major steroid hormones monitored during both acute and chronic infection phases and following Nfx treatment. Trypomastigotes were detected in the buffy coat of all infected mice until day 25 post-infection (PI), but at day 42, PI parasites were no longer detected, indicating a transition from acute to chronic phase CD [31]. To control for sex-specific differences in the response to infection, male and female groups were analyzed independently. At the experimental endpoint (day 110 PI), the mean serum testosterone level of uninfected male mice was 10.48 ± 2.40 ng/mL, while the testosterone levels of T. cruzi infected animals were drastically reduced (0.64 ± 0.02 ng/mL, 2.25 ± 1.05 ng/mL and 1.15 ± 0.30 ng/mL for acute, chronic and chronic-treated groups, respectively) (Figure 2A). As expected, uninfected female mice had substantially lower levels of testosterone at baseline (0.65 ± 0.07 ng/mL), which did not differ significantly in any of the other experimental groups (Figure 2B). In contrast to our findings with human CD patients, neither progesterone nor estradiol were significantly altered in mice infected with T. cruzi.
Figure 2.
Trypanosoma cruzi infection in mice leads to altered sex steroid metabolism and testes morphology. Serum levels of testosterone (T), progesterone (P) (in ng/mL) and estradiol (O) (in pg/mL) in wild-type CD-1 male (A) and female (B) mice ± acute or chronic T. cruzi infection ±20-day nifurtimox treatment, at day 105 post-infection. Infected male mice had significantly decreased testosterone and increased estradiol levels in both acute and chronic phases whereas female mice had increased estradiol and progesterone levels specifically in acute phase and chronic phase, respectively. Testes width was measured before, during and after the treatment (C) for uninfected (D), chronically infected (E) and treated (F) males. The testes shrank in size in the infected group; however, they presented similar width outcome to those of the uninfected group post-treatment. Significance levels were calculated using ANOVA and F-test: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Error bars represent mean ± standard error of mean.
The testes width of male mice was measured during the chronic phase of T. cruzi infection, and during and after treatment (Figure 2C). Prior to treatment, the testes widths were comparable for all experimental groups (13.8 ± 0.3 mm, 13.7 ± 0.4 mm, and 13.6 ± 0.3 mm for uninfected, chronically infected, and treated groups, respectively). As expected, the testes width of uninfected mice gradually increased throughout the experiment. In contrast, the testes width of chronically infected untreated mice showed an initial increase which slowed by day 90 PI and decreased rapidly thereafter. During Nfx treatment, the testes width of treated mice was reduced at day 91 PI, but following treatment completion, this trend reversed and testes width rapidly increased. By the experimental endpoint, the testes of chronically infected Nfx-treated mice were indistinguishable in width and appearance from those of the uninfected mice, while the testes of untreated mice were significantly smaller than the other experimental groups (Figure 2D−F). No significant change in total body weight was observed for either male or female mice during the study.
3.4. Proteomics reveal increased energy metabolism and steroidogenesis within the testes of chronic T. cruzi-infected mice
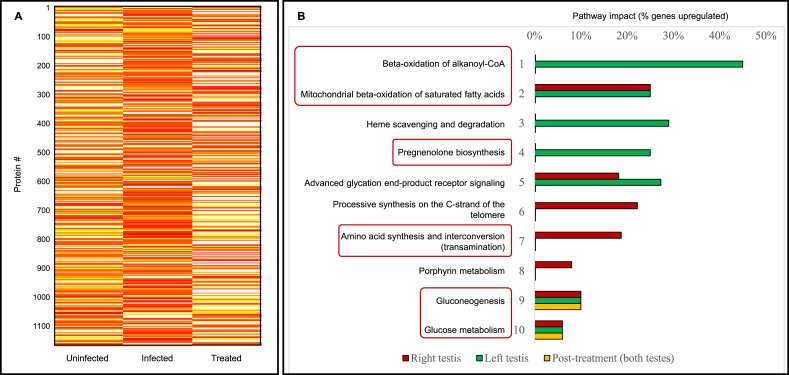
To gain deeper insight into the effects of chronic T. cruzi infection on host metabolism, proteomic analysis was performed on the testes of mice chronically infected with T. cruzi. Using a cut-off of 1% FDR, 1,130 proteins were identified within all testes samples of infected and uninfected mice. Two-hundred and sixteen of these proteins were over-expressed (fold-change > 1.2), 695 were under-expressed (fold-change ≤ 1.2), and 219 did not change in chronically infected mice relative to uninfected controls. Of the proteins that were increased in chronically infected mouse testes, less than one-fourth (45/216) were over-expressed following Nfx treatment (Figure 3A).
Figure 3.
An altered proteomic profile leads to increased energy metabolism and steroidogenesis within the testes of chronic T. cruzi-infected mice. (A) Abundance levels of proteins identified in uninfected, T. cruzi-infected, and nifurtimox (Nfx)-treated mice. Yellow and red indicate decreased and increased proteins, respectively. Most proteins are significantly altered in infected mice but return to those of uninfected controls after antiparasitic treatment. (B) Pathways significantly over-represented in the testes of T. cruzi-infected mice relative to uninfected controls before and after Nfx treatment. Pathway alteration in the right and left testes were displayed separately. While beta-oxidation of alkanoyl-coenzyme A, heme scavenging and degradation and pregnenolone biosynthesis were specifically upregulated in the left testis, processive synthesis on the C-strand of the telomere, amino acid transamination and porphyrin metabolism were more drastically altered in the right testis. Gluconeogenesis and glucose metabolism pathways remained perturbed after the treatment.
Independent analysis of the left and right testes showed a greater number of over-expressed proteins in the right testes compared to the left (215 and 181, respectively). Fifty-six proteins were over-expressed in both testes of infected mice. On average, testes from infected animals had a lower protein content (–16%, ns). However, while compared to the control group, left testes had lost more of their protein content (–27%, ns) compared to the right testes (–4%, ns). A total of 51 over-represented pathways (corrected p-value <0.05) were identified in the testes of T. cruzi-infected mice. Most highly affected pathways included: beta-oxidation of alkanoyl-coenzyme A, beta-oxidation of saturated fatty acids, heme scavenging and degradation, pregnenolone biosynthesis, advanced glycation end-product (AGE) receptor signaling and amino acid synthesis and interconversion (transamination). Highly affected over-represented pathways differed in the right and left testes of the infected animals. Within the right testis, pathways that were most highly affected included the calnexin/calreticulin cycle, beta-oxidation of saturated fatty acids, amino acid synthesis and interconversion (transamination) and AGE receptor signaling. In contrast, the most highly affected pathways identified in the left testis were breakdown of the nuclear lamina, beta-oxidation of alkanoyl coenzyme A, pregnenolone biosynthesis, lysine catabolism and beta-oxidation of saturated fatty acids. The most altered pathways in the testes of treated mice were gluconeogenesis, glyoxylate and dicarboxylate metabolism, citrate cycle, and glucose metabolism. Gene ontology analysis revealed that the exosome, cytoplasm, and mitochondrion were the upregulated cellular components in the testes of infected mice. Likewise, oxidation-reduction, cell redox and protein folding were the top three upregulated biological processes, whereas malate dehydrogenase activity, NEDD8-activating enzyme activity and 15-hydroxyprostaglandin dehydrogenase activity were the most upregulated molecular functions in the testes of infected animals. Figure 3B displays most highly affected pathways in the testes of T. cruzi-infected mice before and after Nfx treatment relative to uninfected controls. For the complete list of mouse testis proteins and the list of over-represented pathways refer to Table S3.
4. Discussion
In this study, we conducted untargeted metabolomics analysis on sera from patients with chronic CD and demographically matched CD-negative controls using both positive and negative ion modes. Metabolite features associated with CD and selected lipid components were measured before and after anti-trypanosomal treatment. Sex steroids were among the most significantly altered metabolites in CD patients. The impact of T. cruzi infection on sex steroid levels was later assessed in a mouse model. Mice were selected as their immune response and disease progression following T. cruzi infection is shown to closely mimic human CD [32].
Sex steroids play critical roles in the reproduction and survival of many parasites, such as Toxoplasma gondii [33], Schistosoma mansoni [34], Plasmodium falciparum [35] and Trichinella spiralis [36]. Protozoan parasites synthesize steroid hormones from cholesterol, ergosterol or other 24-alkylated sterols [37]. In mammals, steroid hormones have more intricate functions, such as regulating osmotic pressure and the stress response, as well as initiating and maintaining sexual differentiation and reproduction. Mammals produce steroid hormones via de novo steroidogenesis in the adrenal cortex, the gonads, the placenta, and the brain. Steroidogenic tissues utilize cholesterol as a precursor for mitochondrial biosynthesis of pregnenolone, from which all other steroid hormones are synthesized. Sex steroids are primarily produced in the gonads. Progesterone and testosterone are synthesized in testicular Leydig cells and ovarian theca cells, whereas estrone and estradiol are mainly products of the ovarian granulosa cells [38]. Trypanosomal infection has been associated with altered sex steroid metabolism in mammals. Dromedary bulls infected with Trypanosoma evansi were shown to have perturbed serum sex steroid levels [39] while in female rats T. evansi infection resulted in altered levels of estradiol and progesterone [40]. In both rats and humans, Trypanosoma brucei infection causes gonadosteroidal disorders [41, 42]. Our data suggest that T. cruzi infection alters sex steroids in humans and mice.
Sex steroid levels affect sexual organ morphology in mammals [43]. Serum testosterone and estradiol levels are closely linked with total testicular volume [44, 45]. The drastic alterations in serum testosterone and estradiol levels in T. cruzi-infected male mice were consistent with their decreased testicle size. Testicle size is also associated with male reproductive features such as sperm count and motility [46]. Decreased reproductive efficiency in male animals with trypanosomiasis has been demonstrated. For example, Trypanosoma vivax infection was associated with reduced libido in Zebu bulls [47], while Arabian camels infected with T. evansi were less fertile [39]. Both studies reported decreased sperm count and/or serum testosterone levels in the infected animals. T. brucei accumulates in mouse testis and may be the cause of reduced fertility in infected mice [48]. T. brucei amastigotes have been observed in mouse testis epididymal adipose tissue which plays an important role in spermatogenesis and male reproductive performance [49]. It also provides a lipid-rich environment for the protozoan parasite. Furthermore, testes are physiologically maintained at ∼35 °C, which compared to internal organs, is closer to the optimal temperature for trypomastigote proliferation, i.e. 29–35 °C [50]. Testes could therefore act as reservoirs for the chronic phase of trypanosomiasis. Histopathological analysis has shown that T. cruzi colonizes both male and female reproductive systems in mice suggesting sexual intercourse as a potential route of transmission [51]. Though we did not identify any parasite-derived DNA or protein in the testes of T. cruzi-infected mice (Figure S4), proteomics analysis showed that the pregnenolone biosynthesis pathway was over-represented in the testes of the infected mice. Specifically, two major enzymes that convert cholesterol to pregnenolone in steroidogenic tissues, cytochrome P450 11A1 (CP11A) and adrenodoxin reductase (ADRO), were both increased (1.4- and 1.6-fold, respectively) in the infected group.
The left and right testes differ in size and in their production of endocrine factors including sex steroid hormones [52, 53]. The left testis is usually smaller and more involved in spermatogenesis [52] while the right testis is larger and appears more involved in steroidogenesis [53]. Our pathway analysis revealed that while right testes of the infected mice had more upregulated pathways associated with energy metabolism, the left testes were subject to increased pregnenolone biosynthesis and mineralocorticoid production (Table S3). In fact, both CP11A and ADRO were more abundant in the left testes of infected mice (2.2- and 2.5-fold increase, respectively) and not in the right testes (unchanged and 0.7-fold decrease, respectively) when compared to uninfected controls. Moreover, Nfx treatment did not decrease testicular expression of ADRO or CP11A. Particularly, CP11A almost doubled in the right testes and only slightly decreased in the left testes after treatment. Since infected mice had very low serum testosterone compared to the controls, the increased testicular synthesis of pregnenolone could potentially explain serum progesterone and estradiol over-expression in male infected animals. To elucidate these processes, future studies will compare the sexual reproducibility and mating behaviour of T. cruzi-infected male mice to an uninfected group. The libido, fertility, and gonad morphology of CD patients will also be compared to uninfected individuals and to CD patients after treatment.
Increased steroid levels deplete plasma glutamine in human [54]. Glutamine is the most abundant amino acid in the human body. It plays an important role in maintaining cell functions as a fuel and a precursor for the synthesis of purines, pyrimidines, glutathione, and a potential regulator of gluconeogenesis and protein biosynthesis. Glutamine is also essential to T. cruzi development [55], and its uptake is tightly regulated by the parasite during its lifecycle [56]. We found that glutamine was significantly decreased (2.1-fold) in CD patients, but whether this is a direct consequence of T. cruzi infection or a systemic response to steroid overabundance remains to be determined. Pathway analysis showed that the amino acid synthesis and interconversion (transamination) pathway was significantly upregulated in both right and left testes of T. cruzi-infected mice. Two important enzymes involved in glutamate metabolism, glutamate dehydrogenase 1 (DHE3; 1.2-fold increase) and aspartate aminotransferase (AATC; 1.4-fold increase) were more abundant in the infected group. Following Nfx treatment, LFQ intensity of DHE3 and AATC returned to a similar level to the uninfected group, suggesting that Gln/Glu metabolism could be linked to treatment efficiency.
Taurine is one of the most abundant amino acids throughout the body. It plays a role in Ca2+ homeostasis and serves a wide variety of functions in the central nervous system [57]. Taurine deficiency is associated with cardiomyopathy potentially via the renin angiotensin system [58]. It has also been seen in rarer CD manifestation, such as renal, hepatic, and gastrointestinal complications [60]. Moreover, a previous study has shown that taurine enhances the sexual response and mating ability in male rats [59]. We observed that serum taurine levels were significantly decreased (1.5-fold) in CD patients; however, there was no significant difference between those levels in the patients with and without cardiomyopathy. Same as glutamine, taurine levels increased to levels comparable to those of CD-negative controls following Nfx treatment.
The dipeptide Phe-Thr was significantly increased (1.9-fold) in the CD patients. Phe-Thr oligopeptides enhance αs1 casein gene expression and are believed to be important in milk protein synthesis in mammary epithelial cells [61]. Bioinformatics analysis demonstrated that of the 26,199 proteins included in the Swiss-Prot human protein database, 14,457 proteins contained at least one Phe-Thr dipeptide in their sequence. Pathway enrichment analysis revealed that the top four pathways altered by these proteins were GPCR downstream signaling, fertilization, reproduction, and interaction with the zona pellucida. The zona pellucida is a glycoprotein layer surrounding the plasma membrane of mammalian oocytes and ovarian follicles and plays an important role in sperm-oocyst interaction and fertilization [62]. Increased serum levels of Phe-Thr are a putative indication to excessive degradation of proteins involved in fertilization and reproduction in CD patients. Proteome dynamics assessment of Phe-Thr-containing proteins in T. cruzi-infected subjects at the chronic stage of infection can provide insight on the potential links between trypanosomiasis and sexual reproduction.
Two other short peptides were confirmed as potential biomarkers of chronic CD in humans. pGlu-Gly was previously associated with colorectal cancer [63] and patented as a serum biomarker for atherosclerosis [64]. pGlu-Gly was significantly decreased (2.0-fold) in the CD patients and unlike the other biomarkers described in this manuscript, did not return to control levels after the treatment. The p Glu-Gly dipeptide was found in 18,459 confirmed human proteins. The most significantly over-represented pathways associated with these proteins were arrhythmogenic right ventricular cardiomyopathy, extracellular matrix-receptor interaction, axon guidance and ATP-binding cassette transporters. Asn-Gly-Phe-Lys has not been previously reported as a disease biomarker. BLAST analysis showed that this tetrapeptide is only present in one human protein: armadillo-like helical domain-containing protein 4 (ARMD4). ARMD4 is not a well-studied protein, however, it has been associated with osmoregulation [65] and heart tissue development [66]. Asn-Gly-Phe-Lys was significantly decreased (2.4-fold) in the CD patients, although its levels were comparable to CD-negative controls after the treatment.
In conclusion, we identified five potential serum metabolite biomarkers that can distinguish chronic CD patients from uninfected and cured individuals. While the interplay between the identified metabolites remains to be investigated, this study suggests that steroidogenesis, fertilization/reproduction, and Gln/Glu metabolism may be part of a larger picture that can potentially explain the physiopathology of CD at the chronic phase of infection. It is imperative that the findings described here be validated in a larger study population of CD patients. Hence, our future direction will be the development of a method for high-throughput screening of serum samples from potentially T. cruzi-infected or treated individuals using a robust and sensitive method, such as a multiplex immunoassay, or a rapid mass spectrometry-based method for clinical validation of target metabolite biomarkers. In contrast to protein biomarkers, the detection and quantitation of metabolite biomarkers are simpler, faster, and more cost-efficient. The diagnostic tests based on these metabolic panels would be amenable to the point-of-care sites in CD endemic areas as well as clinics and blood screening centres in countries with a positive migration rate from regions with a high risk of T. cruzi infection.
Declarations
Author contribution statement
Makan Golizeh: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
John Nam; Fabio Vasquez Camargo; Alexandra Furtos: Analyzed and interpreted the data.
Eric Chatelain; Asieh Rasoolizadeh: Contributed reagents, materials, analysis tools or data.
Yves Jackson: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Leanne B. Ohlund: Performed the experiments; Analyzed and interpreted the data.
Louiza Mahrouche: Performed the experiments.
Lekha Sleno; Momar Ndao: Conceived and designed the experiments.
Funding statement
This work was supported by Drugs for Neglected Diseases initiative (DNDi) (UK Aid (UK), Swiss Agency for Development and Cooperation (SDC; Switzerland), and Médecins Sans Frontières International) [grant number 4759].
Momar Ndao was supported by the Public Health Agency of Canada/National Microbiology Laboratory (contract number 6D063-193271/001), the Foundation of the McGill University Health Centre, the Foundation of the Montreal General Hospital, the Research Institute of the McGill University Health Centre, and the R. Howard Webster Foundation.
Lekha Sleno was supported through the UQAM Strategic Research Chair program. (The LC-MS metabolomics platform at UQAM is supported by the CERMO-FC, through the Courtois Foundation).
Data availability statement
Data associated with this study has been deposited at the ProteomeXchange Consortium via the PRIDE partner repository under the accession number PXD017802.
Declaration of interest's statement
The authors declare no conflict of interest.
Additional information
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2022.e12380.
Acknowledgements
We thank Drs. Brian J. Ward, Robert Kiss, Makoto Nagano, Bernard Robaire, Daniel Bernard for helpful discussions; Milli Nath-Chowdhury, Maya Scott-Lourenço, Colton Strong, Elizabeth Ruiz-Lancheros, Amirhossein Daryaei for helping with the animal study and laboratory work; Dr. Kebba Sabally, Patrick Sabourin, Rami Tohme, Wilson Carreiro for technical support; Angela J. Brewer, Louis Cyr, Kelly Marshall, Nathalie Martel for logistics and administrative support; and James I. P. Stewart for reviewing the manuscript. Makan Golizeh thanks Dr. Momar Ndao for his exceptional support during this project.
Contributor Information
Lekha Sleno, Email: sleno.lekha@uqam.ca.
Momar Ndao, Email: momar.ndao@mcgill.ca.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Bern C. Chagas’ disease. N. Engl. J. Med. 2015;373:456–466. doi: 10.1056/NEJMra1410150. [DOI] [PubMed] [Google Scholar]
- 2.Echeverria L.E., Morillo C.A. American trypanosomiasis (Chagas disease) Infect. Dis. Clin. 2019;33:119–134. doi: 10.1016/j.idc.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Pérez-Molina J.A., Molina I. Chagas disease. Lancet. 2018;391:82–94. doi: 10.1016/S0140-6736(17)31612-4. [DOI] [PubMed] [Google Scholar]
- 4.Perez C.J., Lymbery A.J., Thompson R.C.A. Reactivation of Chagas disease: implications for global Health. Trends Parasitol. 2015;31:595–603. doi: 10.1016/j.pt.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. Chagas Disease (American Trypanosomiasis) Key Facts 2019. https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis).
- 6.Assíria Fontes Martins T., de Figueiredo Diniz L., Mazzeti A.L., da Silva do Nascimento Á.F., Caldas S., Caldas I.S., et al. Benznidazole/itraconazole combination treatment enhances anti-trypanosoma cruzi activity in experimental Chagas disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibarra-Meneses A.V., Moreno J., Carrillo E. New strategies and biomarkers for the control of visceral leishmaniasis. Trends Parasitol. 2020;36:29–38. doi: 10.1016/j.pt.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Isenberg J., Golizeh M., Belfort R.N., da Silva A.J., Burnier M.N., Ndao M. Peptidyl-prolyl cis-trans isomerase A – a novel biomarker of multi-episodic (recurrent) ocular toxoplasmosis. Exp. Eye Res. 2018;177:104–111. doi: 10.1016/j.exer.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 9.Krampa F.D., Aniweh Y., Awandare G.A., Kanyong P. Recent Progress in the Development of Diagnostic Tests for Malaria. Diagnostics. 2017;7 doi: 10.3390/diagnostics7030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balouz V., Agüero F., Buscaglia C.A. Chagas disease diagnostic applications: present knowledge and future steps. Adv. Parasitol. 2017;97:1–45. doi: 10.1016/bs.apar.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schijman A.G. Molecular diagnosis of Trypanosoma cruzi. Acta Trop. 2018;184:59–66. doi: 10.1016/j.actatropica.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Egüez K.E., Alonso-Padilla J., Terán C., Chipana Z., García W., Torrico F., et al. Rapid diagnostic tests duo as alternative to conventional serological assays for conclusive Chagas disease diagnosis. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bocchi E.A., Bestetti R.B., Scanavacca M.I., Cunha Neto E., Issa V.S. Chronic Chagas heart disease management. J. Am. Coll. Cardiol. 2017;70:1510–1524. doi: 10.1016/j.jacc.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Pinazo M.-J., Thomas M.-C., Bustamante J., Almeida IC de, Lopez M.-C., Gascon J. Biomarkers of therapeutic responses in chronic Chagas disease: state of the art and future perspectives. Mem. Inst. Oswaldo Cruz. 2015;110:422–432. doi: 10.1590/0074-02760140435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santamaria C., Chatelain E., Jackson Y., Miao Q., Ward B.J., Chappuis F., et al. Serum biomarkers predictive of cure in Chagas disease patients after nifurtimox treatment. BMC Infect. Dis. 2014;14:302. doi: 10.1186/1471-2334-14-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz-Lancheros E., Rasoolizadeh A., Chatelain E., Garcia-Bournissen F., Moroni S., Moscatelli G., et al. Validation of apolipoprotein A-1 and fibronectin fragments as markers of parasitological cure for congenital Chagas disease in children treated with benznidazole. Open Forum Infect. Dis. 2018;5:ofy236. doi: 10.1093/ofid/ofy236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson Y., Chatelain E., Mauris A., Holst M., Miao Q., Chappuis F., et al. Serological and parasitological response in chronic Chagas patients 3 years after nifurtimox treatment. BMC Infect. Dis. 2013;13:85. doi: 10.1186/1471-2334-13-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vázquez-Fresno R., et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guijas C., Montenegro-Burke J.R., Domingo-Almenara X., Palermo A., Warth B., Hermann G., et al. METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 2018;90:3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang Z., Zhou G., Ewald J., Chang L., Hacariz O., Basu N., et al. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022;17:1735–1761. doi: 10.1038/s41596-022-00710-w. [DOI] [PubMed] [Google Scholar]
- 21.Baum S.G., Wittner M., Nadler J.P., Horwitz S.B., Dennis J.E., Schiff P.B., et al. Taxol, a microtubule stabilizing agent, blocks the replication of Trypanosoma cruzi. Proc. Natl. Acad. Sci. U. S. A. 1981;78:4571–4575. doi: 10.1073/pnas.78.7.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golizeh M., Schneider C., Ohlund L.B., Sleno L. Multidimensional LC–MS/MS analysis of liver proteins in rat, mouse and human microsomal and S9 fractions. EuPA Open Proteomics. 2015;6:16–27. [Google Scholar]
- 23.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 24.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyanova S., Cox J. 2018. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research; pp. 133–148. [DOI] [PubMed] [Google Scholar]
- 26.Lage-Castellanos A., Martínez-Montes E., Hernández-Cabrera J.A., Galán L. False discovery rate and permutation test: an evaluation in ERP data analysis. Stat. Med. 2010;29:63–74. doi: 10.1002/sim.3784. [DOI] [PubMed] [Google Scholar]
- 27.Thomas P.D., Kejariwal A., Campbell M.J., Mi H., Diemer K., Guo N., et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynn D.J., Winsor G.L., Chan C., Richard N., Laird M.R., Barsky A., et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vizcaíno J.A., Côté R.G., Csordas A., Dianes J.A., Fabregat A., Foster J.M., et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ndao M., Kelly N., Normandin D., Maclean J.D., Whiteman A., Kokoskin E., et al. Trypanosoma cruzi infection of squirrel monkeys: comparison of blood smear examination, commercial enzyme-linked immunosorbent assay, and polymerase chain reaction analysis as screening tests for evaluation of monkey-related injuries. Comp. Med. 2000;50:658–665. [PubMed] [Google Scholar]
- 31.Levine R.A., Wardlaw S.C., Patton C.L. Detection of haematoparasites using quantitative buffy coat analysis tubes. Parasitol. Today. 1989;5:132–134. doi: 10.1016/0169-4758(89)90056-2. [DOI] [PubMed] [Google Scholar]
- 32.Costa S.C. Mouse as a model for Chagas disease: does mouse represent a good model for Chagas disease? Mem. Inst. Oswaldo Cruz. 1999;94(Suppl 1):269–272. doi: 10.1590/s0074-02761999000700045. [DOI] [PubMed] [Google Scholar]
- 33.Lim A., Kumar V., Hari Dass S.A., Vyas A. Toxoplasma gondii infection enhances testicular steroidogenesis in rats. Mol. Ecol. 2013;22:102–110. doi: 10.1111/mec.12042. [DOI] [PubMed] [Google Scholar]
- 34.Morrison D.D., Vande Waa E.A., Bennett J.L. Effects of steroids and steroid synthesis inhibitors on fecundity ofSchistosoma mansoni in vitro. J. Chem. Ecol. 1986;12:1901–1908. doi: 10.1007/BF01022391. [DOI] [PubMed] [Google Scholar]
- 35.Lingnau A., Margos G., Maier W.A., Seitz H.M. The effects of hormones on the gametocytogenesis of Plasmodium falciparum in vitro. Appl. Parasitol. 1993;34:153–160. [PubMed] [Google Scholar]
- 36.Hernández-Bello R., Ramirez-Nieto R., Muñiz-Hernández S., Nava-Castro K., Pavón L., Sánchez-Acosta A.G., et al. Sex steroids effects on the molting process of the helminth human parasite Trichinella spiralis. J. Biomed. Biotechnol. 2011;2011:1–10. doi: 10.1155/2011/625380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano M.C., Jiménez P., Miranda-Brito C., Valdez R.A. Parasites and steroid hormones: corticosteroid and sex steroid synthesis, their role in the parasite physiology and development. Front. Neurosci. 2015;9:1–5. doi: 10.3389/fnins.2015.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiffer L., Barnard L., Baranowski E.S., Gilligan L.C., Taylor A.E., Arlt W., et al. Human steroid biosynthesis, metabolism and excretion are differentially reflected by serum and urine steroid metabolomes: a comprehensive review. J. Steroid Biochem. Mol. Biol. 2019;194 doi: 10.1016/j.jsbmb.2019.105439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Qarawi A.A. Infertility in the dromedary bull: a review of causes, relations and implications. Anim. Reprod. Sci. 2005;87:73–92. doi: 10.1016/j.anireprosci.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Faccio L., Da Silva A.S., Tonin A.A., França R.T., Gressler L.T., Copetti M.M., et al. Serum levels of LH, FSH, estradiol and progesterone in female rats experimentally infected by Trypanosoma evansi. Exp. Parasitol. 2013;135:110–115. doi: 10.1016/j.exppara.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Hublart M., Tetaert D., Croix D., Boutignon F., Degand P., Boersma A. Gonadotropic dysfunction produced by Trypanosoma brucei brucei in the rat. Acta Trop. 1990;47:177–184. doi: 10.1016/0001-706x(90)90024-t. [DOI] [PubMed] [Google Scholar]
- 42.Soudan B., Tetaert D., Racadot A., Degand P., Boersma A. Decrease of testosterone level during an experimental African trypanosomiasis: involvement of a testicular LH receptor desensitization. Acta Endocrinol. 1992;127:86–92. doi: 10.1530/acta.0.1270086. [DOI] [PubMed] [Google Scholar]
- 43.Preston B.T., Stevenson I.R., Lincoln G.A., Monfort S.L., Pilkington J.G., Wilson K. Testes size, testosterone production and reproductive behaviour in a natural mammalian mating system. J. Anim. Ecol. 2012;81:296–305. doi: 10.1111/j.1365-2656.2011.01907.x. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz-Olvera S.F., Rajmil O., Sanchez-Curbelo J.-R., Vinay J., Rodriguez-Espinosa J., Ruiz-Castañé E. Association of serum testosterone levels and testicular volume in adult patients. Andrologia. 2018;50 doi: 10.1111/and.12933. [DOI] [PubMed] [Google Scholar]
- 45.Leavy M., Trottmann M., Liedl B., Reese S., Stief C., Freitag B., et al. Effects of elevated β-estradiol levels on the functional morphology of the testis - new insights. Sci. Rep. 2017;7 doi: 10.1038/srep39931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bujan L., Mieusset R., Mansat A., Moatti J.P., Mondinat C., Pontonnier F. Testicular size in infertile men: relationship to semen characteristics and hormonal blood levels. Br. J. Urol. 1989;64:632–637. doi: 10.1111/j.1464-410x.1989.tb05325.x. [DOI] [PubMed] [Google Scholar]
- 47.Bittar J.F.F., Bassi P.B., Moura D.M., Garcia G.C., Martins-Filho O.A., Vasconcelos A.B., et al. Evaluation of parameters related to libido and semen quality in Zebu bulls naturally infected with Trypanosoma vivax. BMC Vet. Res. 2015;11:261. doi: 10.1186/s12917-015-0571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carvalho T., Trindade S., Pimenta S., Santos A.B., Rijo-Ferreira F., Figueiredo L.M. Trypanosoma brucei triggers a marked immune response in male reproductive organs. PLoS Neglected Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansel W. The essentiality of the epididymal fat pad for spermatogenesis. Endocrinology. 2010;151:5565–5567. doi: 10.1210/en.2010-1146. [DOI] [PubMed] [Google Scholar]
- 50.Dvorak J.A., Poore C.M. Trypanosoma cruzi: interaction with vertebrate cells in vitro. Exp. Parasitol. 1974;36:150–157. doi: 10.1016/0014-4894(74)90121-0. [DOI] [PubMed] [Google Scholar]
- 51.Lenzi H.L., Castelo-Branco M.T., Pelajo-Machado M., Oliveira D.N., Gattass C.R. Trypanosoma cruzi: compromise of reproductive system in acute murine infection. Acta Trop. 1998;71:117–129. doi: 10.1016/s0001-706x(98)00058-8. [DOI] [PubMed] [Google Scholar]
- 52.Ungerfeld R., Villagrán M., Lacuesta L., Vazquez N., Pérez W. Asymmetrical size and functionality of the pampas deer (Ozotoceros bezoarticus) testes: right testis is bigger but left testis is more efficient in spermatogenesis. Anat. Histol. Embryol. 2017;46:547–551. doi: 10.1111/ahe.12307. [DOI] [PubMed] [Google Scholar]
- 53.Gerendai I., Csaba Z., Csernus V. Lateralized effect of right- and left-sided vagotomy on testicular steroidogenesis and serum gonadotropin levels in hemicastrated peripubertal rats. Neuroendocrinol. Lett. 1995;17:193–198. [Google Scholar]
- 54.Thibault R., Welch S., Mauras N., Sager B., Altomare A., Haymond M., et al. Corticosteroids increase glutamine utilization in human splanchnic bed. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;294:G548–G553. doi: 10.1152/ajpgi.00461.2007. [DOI] [PubMed] [Google Scholar]
- 55.O’Daly J.A., Rodríguez M.B., Garlin G. Trypanosoma cruzi: growth requirements at different temperature in fetal bovine serum or peptide supplemented media. Exp. Parasitol. 1987;64:78–87. doi: 10.1016/0014-4894(87)90011-7. [DOI] [PubMed] [Google Scholar]
- 56.Damasceno F.S., Barisón M.J., Crispim M., Souza R.O.O., Marchese L., Silber A.M. L-Glutamine uptake is developmentally regulated and is involved in metacyclogenesis in Trypanosoma cruzi. Mol. Biochem. Parasitol. 2018;224:17–25. doi: 10.1016/j.molbiopara.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 57.Ripps H., Shen W. Review: taurine: a “very essential” amino acid. Mol. Vis. 2012;18:2673–2686. [PMC free article] [PubMed] [Google Scholar]
- 58.Qaradakhi T., Gadanec L.K., McSweeney K.R., Abraham J.R., Apostolopoulos V., Zulli A. The anti-inflammatory effect of taurine on cardiovascular disease. Nutrients. 2020;12 doi: 10.3390/nu12092847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J., Lin S., Feng Y., Wu G., Hu J. 2013. Taurine Enhances the Sexual Response and Mating Ability in Aged Male Rats; pp. 347–355. [DOI] [PubMed] [Google Scholar]
- 60.Bkaily G., Jazzar A., Normand A., Simon Y., Al-Khoury J., Jacques D. Taurine and cardiac disease: state of the art and perspectives. Can. J. Physiol. Pharmacol. 2020;98:67–73. doi: 10.1139/cjpp-2019-0313. [DOI] [PubMed] [Google Scholar]
- 61.Zhou M.M., Wu Y.M., Liu H.Y., Liu J.X. Effects of phenylalanine and threonine oligopeptides on milk protein synthesis in cultured bovine mammary epithelial cells. J. Anim. Physiol. Anim. Nutr. 2015;99:215–220. doi: 10.1111/jpn.12246. [DOI] [PubMed] [Google Scholar]
- 62.Gadella B.M. Interaction of sperm with the zona pellucida during fertilization. Soc. Reprod. Fertil. Suppl. 2010;67:267–287. doi: 10.7313/upo9781907284991.023. [DOI] [PubMed] [Google Scholar]
- 63.Goedert J.J., Sampson J.N., Moore S.C., Xiao Q., Xiong X., Hayes R.B., et al. Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014;35:2089–2096. doi: 10.1093/carcin/bgu131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voros S., Brown B.O., Marvasty I.B. 2019. Blood Based Biomarkers for Diagnosing Atherosclerotic Coronary Artery Disease. 20190391131. [Google Scholar]
- 65.Lemopoulos A., Uusi-Heikkilä S., Huusko A., Vasemägi A., Vainikka A. Comparison of migratory and resident populations of Brown trout reveals candidate genes for migration tendency. Genome Biol. Evol. 2018;10:1493–1503. doi: 10.1093/gbe/evy102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conway S.J., McConnell R., Simmons O., Snider P.L. Armadillo-like helical domain containing-4 is dynamically expressed in both the first and second heart fields. Gene Expr. Patterns. 2019;34 doi: 10.1016/j.gep.2019.119077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at the ProteomeXchange Consortium via the PRIDE partner repository under the accession number PXD017802.