Abstract
Patient-derived xenograft (PDX) models are more faithful in maintaining the characteristics of human tumors than cell lines and are widely used in drug development, although they have some disadvantages, including their relative low success rate, long turn-around time, and high costs. The collagen gel droplet embedded culture drug sensitivity test (CD-DST) has been used as an in-vitro drug sensitivity test for patients with cancer because of its high success rate of primary cell culture, high sensitivity, and good clinical relevance, but it is based on an in-vitro cell culture and may not simulate the tumor microenvironment accurately. This study aims to combine a PDX model with CD-DST to evaluate the efficiency of antitumor agents. KRpep-2d, a small peptide targeting KRAS (G12D), and oxaliplatin were used to verify the feasibility of this approach. Whole-exome sequencing and Sanger sequencing were first applied to test and validate the KRAS mutation status of a panel of colorectal cancer PDX tissues. One PDX model was verified to carry KRAS (G12D) mutation and was used for in-vivo and the CD-DST drug tests. We then established the PDX mouse model from the patient with the KRAS (G12D) mutation and obtained viable cancer cells derived from the same PDX model. Next, the antitumor abilities of KRpep-2d and oxaliplatin were estimated in the PDX model and the CD-DST. We found that KRpep-2d showed no significant antitumor effect on the xenograft model or on cancer cells derived from the same PDX model. In contrast, oxaliplatin showed significant inhibitory effects in both tests. In conclusion, the PDX model in combination with the CD-DST assay is a comprehensive and feasible method of evaluating the antitumor properties of compounds and could be applied for new drug discovery.
Keywords: KRAS (G12D), Colorectal cancer, KRpep-2d, Oxaliplatin, PDX model, CD-DST, Drug sensitivity
KRAS (G12D); Colorectal cancer; KRpep-2d; Oxaliplatin; PDX model; CD-DST; Drug sensitivity.
1. Introduction
Colorectal cancer (CRC) is one of the most common malignancies in the world, ranking third in incidence among cancers. In 2020, approximately 1.8 million patients were newly diagnosed with CRC worldwide, and there were more than 900,000 CRC-related deaths globally, ranking second in the world in terms of the mortality rate for all cancer. The treatment regimen for CRC includes surgical resection, chemotherapy, and radiotherapy [1]. However, chemotherapy resistance and postoperative recurrence remain the main causes of death. In recent years, the development and application of molecular targeted therapy has brought the treatment of colorectal cancer into a new stage. In-depth study of the molecular mechanisms of targeted drugs has also promoted individualized treatment feasibility. At present, targeted therapy of CRC mainly focuses on the epidermal growth factor receptor (EGFR) signaling pathway and vascular endothelial growth factor (VEGF), using bevacizumab, cetuximab, and panitumumab as the target drugs accordingly. Although targeted therapy has greatly improved the survival of patients with cancer, the response to treatment remains largely unpredictable. Recently, some novel approaches, such as patient-derived xenograft (PDX) models and patient-derived organoids, for predicting drug sensitivity based on individual tumor samples have emerged. These powerful tools help identify drug efficacy and understand the biological characteristics of the tumor.
PDX models are more faithful in maintaining the characteristics of human tumor than cell lines. The PDX model can retain the architecture, microenvironment, and heterogeneity of human tumors [2, 3, 4]. It can also mimic the tumorigenesis process and drug resistance route of the original tumors. Therefore, the PDX model is widely used in drug development, including for the evaluation of pharmacodynamics [5], cancer drug screening [6], drug combination tests and drug resistance studies [7]. However, PDX models have some disadvantages, including their relative low success rate, long turn-around time, and high costs. The collagen gel droplet embedded culture drug sensitivity test (CD-DST) is a drug sensitivity test based on a three-dimensional (3D) culture of primary tumor cells. The advantages of CD-DST include its high success rate of primary cell culture, requirement of little starting material, high sensitivity, and good clinical relevance [8]. It has been applied in drug screening in non-small cell lung cancer [9, 10], gastric cancer [11, 12], colorectal cancer [13, 14], breast cancer [15, 16], and ovarian cancer [17, 18].
As an oncogene in many human tumors, KRAS is a critical downstream effector in the EGFR signaling pathway and is commonly found to have constitutively activating mutation, including the most well-known G12D mutation. The mutation rate of KRAS in patients with colorectal cancer is 30%–35% [19, 20]. EGFR inhibitors, including cetuximab and panitumumab, are used to treat colorectal cancer with wild type KRAS. No survival benefits were observed in patients with KRAS mutations [21, 22]. Therefore, the development of drugs targeting KRAS mutations is urgently needed. Kotaro Sakamoto et al. [23] discovered the cyclic peptide, KRpep-2d, which has a molecular weight of 2561.01 Da and a chemical formula of C108H182N44O25S2. An enzymatic test showed that the IC50 of KRpep-2d against KRAS (G12D) mutant was 1.6 nM. Krpep-2d had a significant selective inhibitory effect on the lung cancer cell line A427 (KRAS (G12D) mutation), but it had no inhibitory effect on A549 cells (KRAS (G12C) mutation). KRpep-2d also selectively inhibits the phosphorylation of the KRAS downstream signaling protein ERK1/2 in A427 cells. Based on these data, KRpep-2d is considered a highly active and potential KRAS (G12D) inhibitor, but the efficiency of KRpep-2d seems to be cancer cell-type dependent.
In a previous study, we successfully established 20 cases of colorectal cancer PDX models. These PDX models were thoroughly characterized and biobanked [24]. In this study, the KRAS mutation status was tested and one case with KRAS (G12D) mutation was identified. We used the PDX model combined with a CD-DST assay to evaluate the antitumor effects of KRpep-2d and oxaliplatin in the KRAS (G12D) mutant colorectal cancer in vivo and in vitro. These approaches have referential significance for new drug discovery.
2. Materials and methods
2.1. Patients and tumor tissue
Patients enrolled in the study provided informed consent for using tumor tissue for in-vitro and in-vivo studies. This study was approved by institutional ethics committee. All patients were diagnosed with colorectal cancer. Samples were collected after surgical removal of the tumor tissue. The fresh tumor tissue was then used to establish the PDX model.
2.2. Animals
Female Balb/c-nu mice (4–6 weeks old; body weight, 18–20 g) were purchased from Gempharmatech Co., Ltd. (Nanjing, China) and housed in a specific pathogen-free (SPF) environment with an individually ventilated caging system (IVC). All mice received humane care in compliance with the guidelines on animal welfare of the National Committee for Animal Experiments. This study was approved by the Ethics Committee for Clinical Research and Laboratory Animal of the First Affiliated Hospital of Sun Yat-sen University (lunshen [2018] NO.48).
2.3. KRAS (G12D) inhibitor and oxaliplatin
The KRAS (G12D) inhibitor KRpep-2d (Selleck, Shanghai, China) was dissolved in sterile water for injection (Shijiazhuang No.4 Pharmaceutical Co., Ltd., Shijiazhuang, China) to a concentration of 100 mg/mL and further diluted to 10 mg/mL immediately prior to use. Oxaliplatin (CENEXI-Laboratoires THISSEN S.A., Braine-L' Alleud, Belgium) was dissolved in sterile water for injection to a concentration of 5 mg/mL and further diluted to 1 mg/mL and 0.5 mg/mL immediately prior to use.
2.4. Screening for KRAS (G12D) mutation
Whole-exome sequencing (WES) was performed on a library of PDX models of colorectal cancer established earlier by our group [24]. DNA of PDX tumor tissue stored at -80 °C was extracted using the MagPure Tissue & Blood DNA LQ Kit (Magen, Peking, China). The DNA was then quantified using the Qubit dsDNA HS Assay Kit (Life Technologies, California, USA), and 200 ng of DNA was sheared using Biorupter (Diagenode, Seraing, Belgium) to acquire 150–200 bp fragments. The ends of the DNA fragment were repaired, and Illumina Adaptor was added (Fast Library Prep Kit, iGeneTech, Peking, China). After the sequencing library was constructed, the whole exons were captured with AIExomeV1-COSMIC (iGeneTech, Peking, China) and sequenced on the Illumina NovaSeq 6000 platform (Illumina, San Diego, California, USA) with 150 base paired-end reads. Raw data were filtered to remove low quality reads using FastQC. Then clean reads were mapped to the reference genome UCSC hg19 using BWA. After removing duplications, SNV and InDel were called and annotated using GATK.
2.5. Validation of KRAS (G12D) mutation by Sanger sequencing
PCR primers flanking codon 12 of KRAS were used to amplify the region. The sequences of the primers were KRAS-F: 5′-TACTGGTGGAGTATTTGATAGTG-3′ and KRAS-R: 5′-CTGTATCAAAGAATGGTCCTG-3’. The PCR products were analyzed using 2.0% agarose gel electrophoresis to determine the band size. Monodirectional sequencing was performed using the ABI 3730 automated sequencer (Applied Biosystems, Förster City, California, USA).
2.6. Effects of KRpep-2d and oxaliplatin in PDX model of KRAS (G12D) mutant colorectal cancer
The tumor-bearing mouse of the third-generation PDX model of KRAS (G12D) mutant colorectal cancer was sacrificed. The tumors were removed under sterile conditions. Tumor tissues were washed three times with a phosphate buffered saline (PBS) solution containing 2% penicillin/streptomycin, and necrotic tissue was removed. The tumor tissue was divided into two parts, one for the passaging of PDX and the other for the CD-DST assay. For PDX passaging, the tumor tissue was cut into 3 mm × 2 mm × 1 mm pieces and inoculated subcutaneously on the right side of the dorsal front of mice (n = 20). When the tumor grew to approximately 100 mm3, the mice were divided into a control group (n = 5), an oxaliplatin group (5 mg/kg, n = 5; 10 mg/kg, n = 5) and a KRpep-2d group (100 mg/kg, n = 5). KRpep-2d or water were administered by gavage once daily for 21 days in the KRpep-2d and control groups. Oxaliplatin was administered by intraperitoneal injection twice every week for 21 days in the oxaliplatin group. The tumor size was measured two or three times a week, and the mice were weighed every day. At the end of the experiment, mice were sacrificed, and tumors were dissected, weighed, and photographed. Tumor volume was calculated using the formula (tumor volume = length × width2/2), and the tumor growth curves were plotted. The tumor growth inhibition rate was calculated using the following formula: tumor growth inhibition rate = (average tumor weight of control group − average tumor weight of administration group)/average tumor weight of control group × 100%.
2.7. Hematoxylin and eosin staining
Tissues were harvested and fixed in 10% neutral-buffered formalin solution after tumor resection. The tissue was dehydrated and immersed in the wax prior to paraffin embedding. Tissue blocks were cut into 4 μm-thick slices on a microtome (Leica Biosystems, Deer Park, New York, USA). Finally, the sections were stained with hematoxylin and eosin and reviewed by a pathologist.
2.8. Immunohistochemistry
Following de-paraffinization of tissue sections, antigen retrieval was performed using a citrate buffer solution (pH 7.4) in an automatic dewaxing antigen repair instrument (GeneTech, Shanghai, China). Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in PBS. Non-specific staining was blocked using bovine serum albumin (BSA) for 40 min at room temperature. Anti-Ki67 primary antibody working solution (LBP, Guangzhou, China) was applied on tissue sections and incubated overnight at 4 °C in a humidified chamber. The next day, biotin conjugated goat anti-rabbit IgG (LBP, Guangzhou, China) secondary antibody, followed by Poly-HRP (LBP, Guangzhou, China) incubation was performed for 30 min each at room temperature. Staining was visualized using a DAB detection system (LBP, Guangzhou, China). Images were obtained via a Leica DM2000 microscope with a Mshot MS60 digital camera. For quantification of staining, representative images were obtained from the stained slides at 40X objective magnification. For each treatment condition, ten representative fields of view from five individual tumors were analyzed. The images were analyzed with the ImageJ software.
2.9. CD-DST assay for drug screening
The PDX tumor tissue was processed following the instructions of the Cell Pre-culture Kit (Guangzhou Darui Biotechnology Co., Ltd., Guangzhou, China) to obtain viable cancer cells. Then cancer cells were mixed with type I collagen solution (Guangzhou Darui Biotechnology Co., Ltd., Guangzhou, China) to obtain a final cell density of 1 × 105/mL. Three drops of the collagen/cell mixture (30 μL per droplet) were seeded in each well of a six-well plate on ice, and gel was formed by incubating the plate at 37 °C in a CO2 incubator for 30 min. Four groups were designed: the 0-time group, control group, oxaliplatin group, and KRpep-2d group. The tumor cells of the 0-time group were cultured in DMEM/F12 medium (GIBCO, Grand Island, New York, USA) containing 10% fetal bovine serum (Bovogen Biologicals Pty Ltd., East Keilor VIC, Australia) at 37 °C in 5% CO2 for 24 h. Then, neutral red was added to each well at a final concentration of 50 μg/mL, and the colonies were fixed in 10% neutral-buffered formalin, washed with water, air-dried, and quantified by optical density image analysis. The tumor cells of the oxaliplatin and KRpep-2d groups were exposed to oxaliplatin or KRpep-2d at concentrations of 100, 50, 10, 1, 0.1 μM and were incubated for 24 h. After removal of the medium containing oxaliplatin or KRpep-2d, each well was rinsed twice with 4 mL of DMEM/F12 medium. Next, cells were overlaid with 3 mL of PCM-2 serum-free medium (Guangzhou Darui Biotechnology Co., Ltd., Guangzhou, China), and incubated for five days. The tumor cells of the control group did not receive drug treatment and were processed as the oxaliplatin group and the KRpep-2d group. The growth rate (GR) of tumor cells was calculated using the following formula: growth rate (GR) = mean value of OD540 in the control group/mean value of OD540 in the 0-time group; a GR value ≥0.8 represented a successful test. In-vitro sensitivity was expressed as the T/C ratio, where T is the optical density of the treatment group, and C is the optical density of control group. T/C > 50% was defined as low sensitivity (resistant in vitro), and T/C ≤ 50% was defined as high sensitivity (effective in vitro).
2.10. Statistical analysis
Qualitative data are expressed as the mean ± standard deviation. Differences between groups were analyzed by one-way analysis of variance or the Mann–Whitney test. All statistical analyses were conducted using SPSS 19.0 software (SPSS Inc., Chicago, Illinois, USA). P-values < 0.05 were considered statistically significant.
3. Results
3.1. Screening and validation of KRAS mutation in CRC PDX models
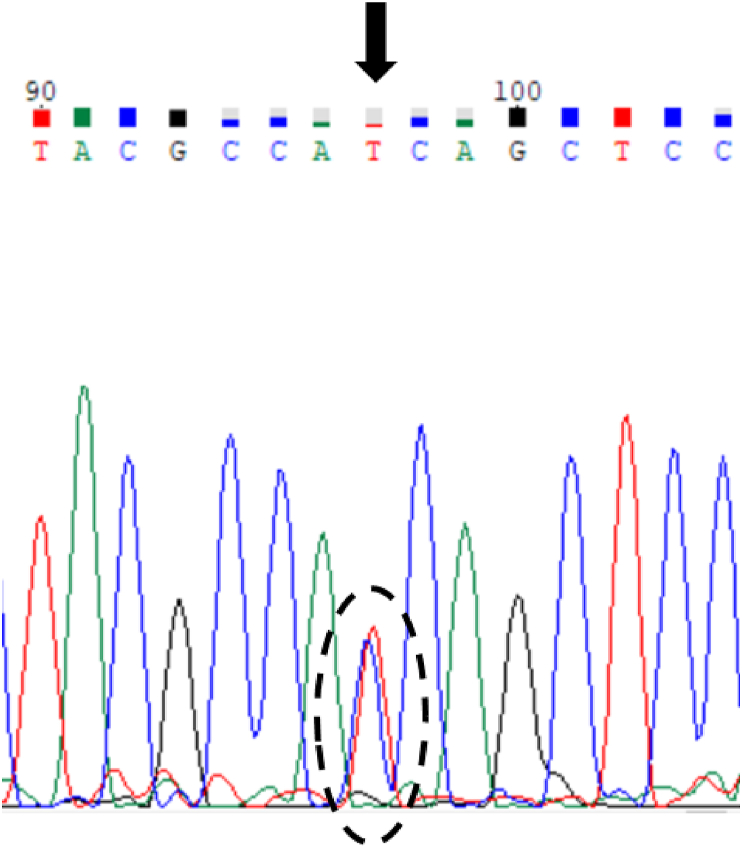
To screen for common cancer gene mutations, WES was performed on fifteen cases of CRC PDX models previously established by our group [24]. The results showed that five cases of CRC PDX models carried KRAS mutations, and one case (CRC19) carried KRAS (G12D) mutation (Table 1). Sanger sequencing confirmed the presence of the G12D mutation in KRAS in case, CRC19 (Figure 1).
Table 1.
KRAS (G12D) mutation detected by WES in a PDX model of colorectal cancer.
| Sample | Gene | Chromosome | Exon | cDNA | Protein | Ref | Alter | Mutation rate (%) |
|---|---|---|---|---|---|---|---|---|
| CRC19 | KRAS | 12 | 2 | c.G35A | p.G12D | C | T | 23.8 |
Figure 1.
Sanger sequencing of KRAS in colorectal cancer case CRC19 (c.G35A C>T; p.G12D).
3.2. The in-vivo effects of KRpep-2d and oxaliplatin in a CRC PDX model
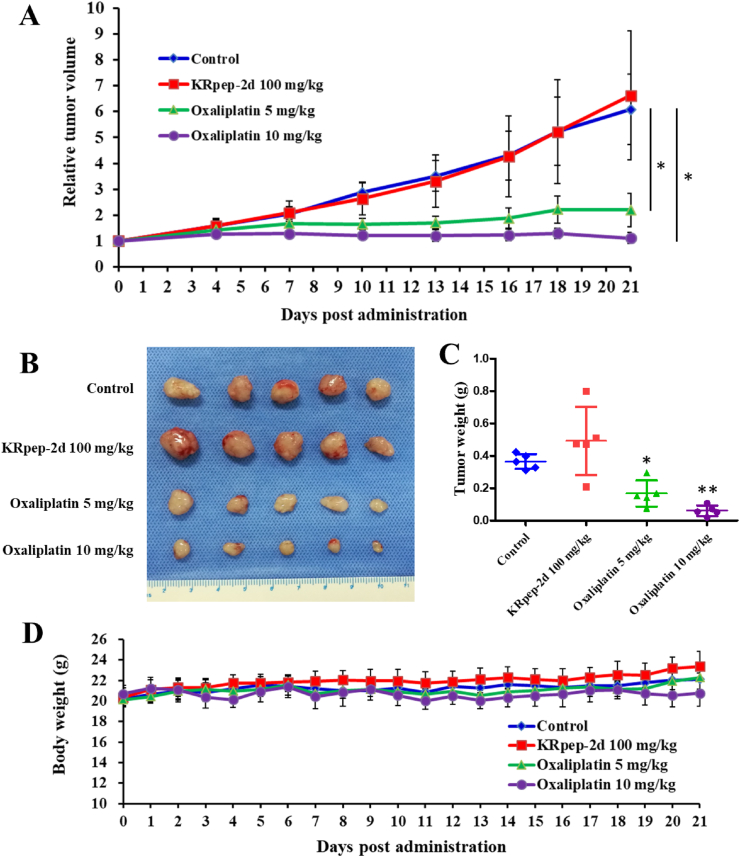
The in-vivo effects of KRpep-2d and oxaliplatin were tested in the PDX model. Tumors in the control and KRpep-2d groups grew quickly (Figure 2A), with no significant difference in tumor sizes between them (P > 0.05). Compared to that in the control group, tumors in the oxaliplatin treatment group were smaller at both doses of 5 and 10 mg/kg (P < 0.05). At the end of the experiment, mice were sacrificed, and tumors were dissected. The tumor weights between the control and KRpep-2d groups were not significantly different (Figure 2B and 2C) (P = 0.102). Compared to that in the control group, the tumor weight in the oxaliplatin group at doses of 5 and 10 mg/kg were significantly lower (P = 0.016 and 0.001, respectively). The tumor growth inhibition rates of KRpep-2d (100 mg/kg) and oxaliplatin (5 and 10 mg/kg) were −34.8%, 54.3% and 83.3%, respectively. The oral administration of KRpep-2d was generally tolerable. The body weights in the KRpep-2d treatment and oxaliplatin groups were comparable to that in the control group (Figure 2D). In addition, gross examination of the internal organs of KRpep-2d- or oxaliplatin-treated mice revealed no apparent changes. This experiment indicated that KRpep-2d had no significant antitumor effects in the CRC PDX model with KRAS (G12D) mutation.
Figure 2.
Antitumor effects of KRpep-2d and oxaliplatin in a CRC PDX model. (A) Growth curves of PDX tumors in each group. No significant difference was observed in tumor volumes between the control and KRpep-2d groups (P > 0.05). Compared to that in the control group, oxaliplatin could significantly inhibit the growth of the xenograft at doses of 5 and 10 mg/kg (P < 0.05). (B) Photo of the PDX tumors harvested from the control, KRpep-2d, and oxaliplatin groups. (C) Tumor weight of all groups. The tumor weights between the control and KRpep-2d groups were not significantly different (P = 0.102). Compared to that in the control group, the tumor weight of the oxaliplatin group at 5 and 10 mg/kg were significantly lower (P = 0.016 and 0.001, respectively). (D) The body weight of mice in each group. The weights of the mice in all groups did not change significantly during the observation period.
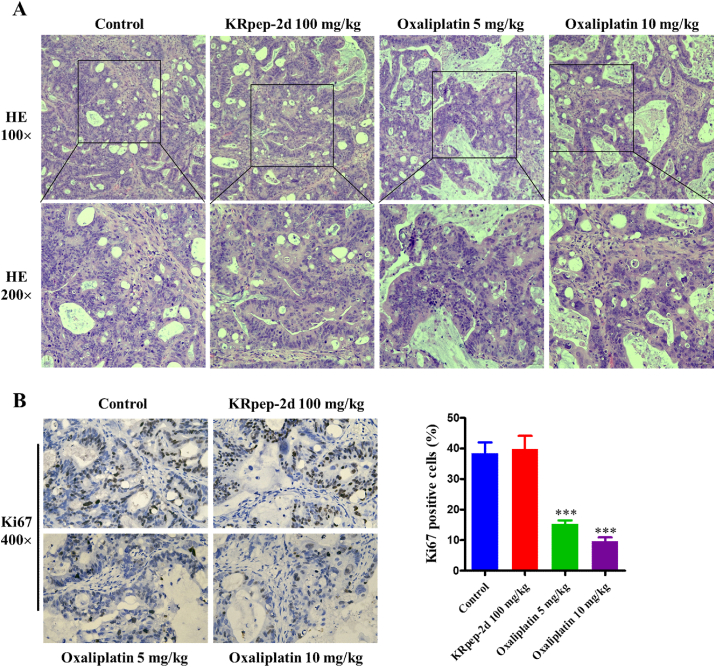
We then performed hematoxylin and eosin staining and immunohistochemistry on the harvested PDX tissues from both treatment and control groups. The tumor cells in the control and KRpep-2d groups were irregularly arranged with obvious atypia. The tumor cells were mainly round or oval, with different cell sizes, dark staining of chromatin, large cell nuclei, and many nuclear division phases. No obvious treatment effects, such as necrosis or fibrosis, were revealed in the KRpep-2d group. In the oxaliplatin groups (5 and 10 mg/kg), cancer cells were uniform in size and had hydropic degeneration, cytoplasmic hyalinization, nuclear pyknosis, and lysis. Part of the tumor tissue displayed degeneration and severe necrosis. Some scattered cancer cells showed apoptosis, forming apoptotic bodies in a dose-dependent manner (Figure 3A). Ki67 expression rates of the control, KRpep-2d, and oxaliplatin groups (5 and 10 mg/kg) were (38.4 ± 3.6)%, (39.8 ± 4.3)%, (15.3 ± 1.2)%, and (9.6 ± 1.4)% respectively. The Ki67 expression rates between the control and KRpep-2d groups were not significantly different (P = 0.958). Compared to that in the control group, the Ki67 expression rates of the oxaliplatin group at 5 and 10 mg/kg were much lower (P = 5∗10−9 and P = 2∗10−10). Oxaliplatin could significantly inhibit the proliferation of tumor cells in a PDX model in a dose-dependent manner (Figure 3B).
Figure 3.
Hematoxylin and eosin (HE) staining and Ki67 expression of the xenografts. (A) Results of HE staining in each group. No obvious treatment effects, such as necrosis or fibrosis, were revealed in the KRpep-2d group. In the oxaliplatin groups (5 and 10 mg/kg), the cancer cells showed changes related to treatment. (B) Results of Ki67 expression in each group. Ki67 expression rates of the control, KRpep-2d, and oxaliplatin groups (5 and 10 mg/kg) were (38.4 ± 3.6)%, (39.8 ± 4.3)%, (15.3 ± 1.2)% and (9.6 ± 1.4)% respectively. The Ki67 expression rates between the control and KRpep-2d groups were not significantly different (P = 0.958). Compared to that in the control group, the Ki67 expression rates of oxaliplatin group at 5 and 10 mg/kg were much lower (P = 5∗10−9 and P = 2∗10−10).
3.3. The in-vitro effects of KRpep-2d and oxaliplatin in CD-DST assay
We evaluated the antitumor effects of KRpep-2d and oxaliplatin with CD-DST assay using the same tissue as that used for the PDX model. Firstly, the cell GR was calculated to evaluate the validity of the assay. The cell viabilities indicated by OD540 were significantly higher in control group as compared to those in the 0-time group (GR = 4.61 and 4.33, respectively; Table 2), indicating the cells were proliferating well and the CD-DST assay was valid.
Table 2.
Values of OD540 in the 0-time and control groups and GR analysis of this CD-DST assay.
| Samples | Groups | Values of OD540 | Values of GR |
|---|---|---|---|
| KRpep-2d | 0-time | 17.61 ± 1.32 | 4.61 |
| Control | 81.17 ± 20.62 | ||
| Oxaliplatin | 0-time | 17.72 ± 0.51 | 4.33 |
| Control | 76.65 ± 13.14 |
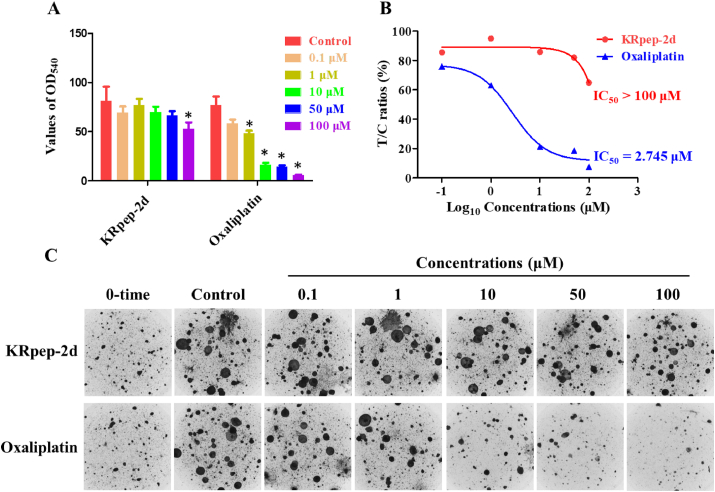
Next, the OD540 values of the KRpep-2d and oxaliplatin treatment groups were compared to that of the control group to derive the treatment effects. The T/C value of KRpep-2d was larger than 50% even at the highest concentration, indicating the value of IC50 was more than 100 μΜ. Oxaliplatin could significantly inhibit the growth of tumor cells with IC50 at 2.745 μM (Figure 4A and 4B). A visual check of cell colonies from the control and treatment groups confirmed the quantification results (Figure 4C). Therefore, KRpep-2d was ineffective in the CD-DST assay using KRAS (G12D) mutant colorectal cancer.
Figure 4.
In-vitro effects of KRpep-2d and oxaliplatin in the CD-DST assay. (A) Values of OD540 in the control, KRpep-2d-, and oxaliplatin-treatment groups. Compared to that in the control group, the values of OD540 in the KRpep-2d group at 100 μM and in the oxaliplatin group at 100, 50, 10, and 1 μM were significantly different (P < 0.05). (B) T/C ratios and IC50 values of KRpep-2d and oxaliplatin in tumor cells. T/C ratios of KRpep-2d were all >50%, indicating IC50 > 100 μM. T/C ratios of oxaliplatin were <50% at 100, 50 and 10 μM, with an IC50 = 2.745 μM. (C) The images of tumor cell colonies in collagen gel droplets from different treatment groups.
4. Discussion
The first PDX model was reported by Jørgen Rygaard and Carl O. Poulsen in 1969 [25]. After more than a half century, PDX has become one of the standard models in pre-clinical studies. PDX models can represent human cancer cells while maintaining the tumor microenvironments. PDX models have many applications, including in the development of anticancer drugs, study of cancer biology, personalized medicine, and immunotherapy [26]. Our facility center has established a large biobank of PDX models, including more than 200 cases derived from 30 cancer types [24, 27, 28, 29, 30, 31, 32, 33, 34, 35]. This is a good resource for pre-clinical drug evaluation. However, the PDX model is hindered by its long turn-around time and high costs. In addition, the stromal cells in the PDX tumor are gradually replaced by mouse stromal cells, making it unclear whether it can truly represent the human tumor microenvironment. CD-DST is an in-vitro anticancer drug sensitivity test. CD-DST requires few cells input and can maintain the characteristics of the original tumor. It has been used to guide treatment for several cancers, including colorectal cancer [36]. The disadvantage of the CD-DST assay is that it is based on an in-vitro cell culture and may not simulate the tumor microenvironment accurately. Nevertheless, PDX and CD-DST are better for pre-clinical drug evaluation than traditional 2D culture of cancer cell lines, and a combination of the two may be a superior approach.
As one of the most frequently mutated oncogenes in cancer, KRAS is very difficult to target. In 2021, the US Food and Drug Administration (FDA) approved sotorasib, the first KRAS inhibitor in history for the treatment of non-small cell lung cancer [37]. Sotorasib is a small molecular inhibitor of KRAS (G12C) that forms an irreversible covalent bond with the cysteine of KRAS (G12C), locking the protein in an inactive state that prevents downstream signaling, without affecting wild-type KRAS. Unfortunately, identifying compounds that can target KRAS mutants G12D and G12V is both difficult and time-consuming.
Cyclic peptides are polypeptide chains that contain a circular sequence of bonds. Cyclic peptides have a wide range of biological activities, such as anticancer, antiviral, antibacterial, and enzyme inhibition, as well as being used as drugs [38, 39, 40, 41]. KRAS mutation is very common in colorectal cancer and other tumor types. Patients with the KRAS mutation cannot benefit from EGFR inhibitors. As a selective inhibitory peptide to KRAS (G12D), KRpep-2d has great potential to be used in patients carrying the mutation. Currently, its inhibitory effects on KRAS (G12D) are only demonstrated at the molecular and cellular levels, and the in-vivo effects of KRpep-2d have not been reported. In this study, we report the in-vivo antitumor property of KRpep-2d in KRAS (G12D) mutant colorectal cancer. To test the feasibility of using KRpep-2d for in-vivo study, we selected a high dose (100 mg/kg) of KRpep-2d to evaluate the safety of different drug delivery routes. Our data showed that when a single 100 mg/kg dose of KRpep-2d was administered, subcutaneous injection and intraperitoneal injection showed different degrees of toxic reactions, and intravenous injection even resulted in the death of the mice. Only oral administration showed no apparent toxic reaction. Therefore, KRpep-2d was delivered by gavage for the in-vivo experiment.
Unfortunately, KRpep-2d administrated at dose of 100 mg/kg showed no tumor growth inhibition on the xenograft of the PDX model of KRAS (G12D) mutant colorectal cancer. Multiple reasons may account for this observation. The drug delivery route may be a key issue. To balance safety and effectiveness, we chose to administrate KRpep-2d by gavage for the in-vivo experiment. Most peptides are easily degraded after oral administration, and have difficulty penetrating the intestinal mucosa [42]. Thus, oral administration may not be effective in delivering KRpep-2d. Actually, we tried to give the tumor-bearing mice 500 mg/kg of KRpep-2d orally for seven days. Even at this high dosage, KRpep-2d did not show a tendency to inhibit the tumor growth, and no toxic reaction was observed. Thus, in further studies, the bioavailability and stability of the peptide when administrated orally need to be assessed carefully and thoroughly. In addition, the blood concentration and pharmacokinetics of KRpep-2d in mice after oral administration should be studied. Next, the safety and efficacy of an intratumor injection of low-dose KRpep-2d may be worth attempting. In fact, some researchers reported that certain peptides could significantly inhibit the tumor growth by intratumor injection [43, 44]. Finally, future studies could combine KRpep-2d with chemotherapy, such as oxaliplatin, to enhance the antitumor effects.
Similarly, KRpep-2d was not sensitive to the CRC19 tumor cells according to the evaluation rule, though its inhibitory rate against CRC19 tumor cells was 36.16% at 100 μM. The delivery of KRpep-2d in tumor cells may have been an issue when the in-vitro effect of KRpep-2d was evaluated by CD-DST assay. It may be challenging for KRpep-2d to pass through the collagen gel given its relatively large shape. In contrast, oxaliplatin has a smaller molecular weight, at 397.29 Da, making it easy to pass through the collagen gel and act on the tumor cells, where it exhibited a significant inhibitory effect on the CRC19 tumor cells, with an IC50 value of 2.745 μM. Therefore, the delivery of KRpep-2d in tumor cells should be evaluated in the CD-DST assay in further studies.
In this study, we tested the efficacy of KRpep-2d in only one model because only one case (CRC19) carried the KRAS (G12D) mutation from among fifteen cases of CRC PDX models tested by WES. This is a limitation for this study. In further studies, more cases with the KRAS (G12D) mutation are needed to study the potential therapeutical effect of KRpep-2d to colorectal cancer. However, the results of the in-vitro and in-vivo of the two compounds were consistent for each drug.
In conclusion, the antitumor effects of KRpep-2d and oxaliplatin on KRAS (G12D) mutant colorectal cancer were successfully evaluated using a PDX model combined with CD-DST assay. The PDX model in combination with the CD-DST assay is a comprehensive and feasible approach for the pre-clinical evaluation of antitumor drugs.
Declarations
Author contribution statement
Wu-guo Li: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Wei Chen: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Jia-lin Wang; Guang-yin Zhao; Lian-zhou Chen; Yong Wan; Qian-xin Luo; Wen-wen Li; Hao-ji Huang; Wen-ying Li; Wu Li; Yu-tong Yang: Performed the experiments; Analyzed and interpreted the data.
Dai-ci Chen: Conceived and designed the experiments; Wrote the paper.
Qiao Su: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Funding statement
Dai-ci Chen was supported by National Natural Science Foundation of China [31970703], and Natural Science Foundation of Guangdong Province [2021A1515010544 and 2022A1515012472].
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest’s statement
The authors declare no competing interests.
Additional information
No additional information is available for this paper.
Contributor Information
Daici Chen, Email: chendc3@mail.sysu.edu.cn.
Qiao Su, Email: suqiao@mail.sysu.edu.cn.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Stewart E.L., Mascaux C., Pham N.A., Sakashita S., Sykes J., et al. Clinical utility of patient-derived xenografts to determine biomarkers of prognosis and map resistance pathways in EGFR-mutant lung adenocarcinoma. J. Clin. Oncol. 2015;33(22):2472–2482. doi: 10.1200/JCO.2014.60.1492. [DOI] [PubMed] [Google Scholar]
- 3.Gu Q.Y., Zhang B., Sun H.Y., Xu Q., Tan Y.X., et al. Genomic characterization of a large panel of patient-derived hepatocellular carcinoma xenograft tumor models for preclinical development. Oncotarget. 2015;6(24):20160–20176. doi: 10.18632/oncotarget.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertotti A., Papp E., Jones S., Adleff V., Anagnostou V., et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526(7572):263–280. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kung P.P., Martinez R., Zhu Z., Zager M., Blasina A., et al. Chemogenetic evaluation of the mitotic Kinesin CENP-E reveals a critical role in triple-negative breast cancer. Mol. Cancer Therapeut. 2014;13(8):2104–2115. doi: 10.1158/1535-7163.MCT-14-0083-T. [DOI] [PubMed] [Google Scholar]
- 6.Fong E.L.S., Martinez M., Yang J., Mikos A.G., Navone N.M., et al. Hydrogel-based 3D model of patient-derived prostate xenograft tumors suitable for drug screening. Mol. Pharm. 2014;11(7):2040–2050. doi: 10.1021/mp500085p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monsma D.J., Cherba D.M., Eugster E.E., Dylewski D.L., Davidson P.T., et al. Melanoma patient derived xenografts acquire distinct Vemurafenib resistance mechanisms. Am. J. Cancer Res. 2015;5(4):1507–1518. PMID: 26101714. [PMC free article] [PubMed] [Google Scholar]
- 8.Wan Y. Southern Medical University; Guangzhou, China: 2018. Development and Clinical Application of the Kit for Anti-cancer Drug Sensitivity Test Based on 3D Cultured Tumor Cells. [Google Scholar]
- 9.Inoue M., Maeda H., Takeuchi Y., Fukuhara K., Shintani Y., et al. Collagen gel droplet-embedded culture drug sensitivity test for adjuvant chemotherapy after complete resection of non-small-cell lung cancer. Surg. Today. 2018;48(4):380–387. doi: 10.1007/s00595-017-1594-7. [DOI] [PubMed] [Google Scholar]
- 10.Higashiyama M., Okami J., Maeda J., Tokunaga T., Fujiwara A., et al. Differences in chemosensitivity between primary and paired metastatic lung cancer tissues: in vitro analysis based on the collagen gel droplet embedded culture drug test (CD-DST) J. Thorac. Dis. 2012;4(1):30–47. doi: 10.3978/j.issn.2072-1439.2011.05.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makino H., Nomura S., Kogo H., Wada N., Hayashi M., et al. Role of collagen gel droplet-embedded culture-drug sensitivity testing (CD-DST) for assessing the sensitivity of gastric cancer to chemotherapy drugs combined with other cancer therapeutic drugs. J. Nippon Med. Sch. 2022;89(4):412–421. doi: 10.1272/jnms.JNMS.2022_89-408. [DOI] [PubMed] [Google Scholar]
- 12.Tanigawa N., Yamaue H., Ohyama S., Sakuramoto S., Inada T., et al. Exploratory phase II trial in a multicenter setting to evaluate the clinical value of a chemosensitivity test in patients with gastric cancer (JACCRO-GC 04, Kubota memorial trial) Gastric Cancer. 2016;19(2):350–360. doi: 10.1007/s10120-015-0506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ochiai T., Nishimura K., Watanabe T., Kitajima M., Nakatani A., et al. Impact of primary tumor location as a predictive factor in patients suffering from colorectal cancer treated with cytotoxic anticancer agents based on the collagen gel droplet-embedded drug sensitivity test. Oncol. Lett. 2019;17(2):1842–1850. doi: 10.3892/ol.2018.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonoda H., Mekata E., Shimizu T., Miyake T., Ueki T., et al. Clinical predictive value of in vitro anticancer drug sensitivity test for the therapeutic effect of 5-FU based adjuvant chemotherapy in patients with stage II-III colorectal cancer: 10-year follow up results. J. Clin. Oncol. 2018;36(15) doi: 10.3892/mco.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhai L.L., Li S., Li X.Y., Li H.L., Gu F., et al. The nuclear expression of poly (ADP-ribose) polymerase-1 (PARP1) in invasive primary breast tumors is associated with chemotherapy sensitivity. Pathol. Res. Pract. 2015;211(2):130–137. doi: 10.1016/j.prp.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y., Lv F., Liu F.F., Guo X.J., Fan Y., et al. High expression of pyruvate Kinase M2 is associated with chemosensitivity to epirubicin and 5-fluorouracil in breast cancer. J. Cancer. 2015;6(11):1130–1139. doi: 10.7150/jca.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamagami W., Banno K., Kawaguchi M., Yanokura M., Kuwabara Y., et al. Use of the collagen gel droplet embedded drug sensitivity test to determine drug sensitivity against ovarian mature cystic teratoma with malignant transformation to adenocarcinoma: a case report. Chemotherapy. 2007;53(2):137–141. doi: 10.1159/000099985. [DOI] [PubMed] [Google Scholar]
- 18.Nagai N., Minamikawa K., Mukai K., Hirata E., Komatsu M., et al. Predicting the chemosensitivity of ovarian and uterine cancers with the collagen gel droplet culture drug-sensitivity test. Anti Cancer Drugs. 2005;16(5):525–531. doi: 10.1097/00001813-200506000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Akman T., Oztop I., Baskin Y., Unek I.T., Demir N., et al. The association of clinicopathological features and survival in colorectal cancer patients with kras mutation status. J. Cancer Res. Therapeut. 2016;12(1):96–102. doi: 10.4103/0973-1482.148684. [DOI] [PubMed] [Google Scholar]
- 20.Sakai K., Yoneshige A., Ito A., Ueda Y., Kondo S., et al. Performance of a novel KRAS mutation assay for formalin-fixed paraffin embedded tissues of colorectal cancer. SpringerPlus. 2015;4:7–12. doi: 10.1186/2193-1801-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L., Bhattacharya A., Li Y., Sexton S., Ling X., et al. Depleting receptor tyrosine kinases EGFR and HER2 overcomes resistance to EGFR inhibitors in colorectal cancer. J. Exp. Clin. Cancer Res. 2022;41(1) doi: 10.1186/s13046-022-02389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen J.B.E., Medema J.P., Gootjes E.C., Tauriello D.V.F., Verheul H.M.W. Mutant RAS and the tumor microenvironment as dual therapeutic targets for advanced colorectal cancer. Cancer Treat Rev. 2022;109 doi: 10.1016/j.ctrv.2022.102433. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto K., Kamada Y., Sameshima T., Yaguchi M., Niida A., et al. K-Ras(G12D)-selective inhibitory peptides generated by random peptide T7 phage display technology. Biochem. Biophys. Res. Commun. 2017;484(3):605–611. doi: 10.1016/j.bbrc.2017.01.147. [DOI] [PubMed] [Google Scholar]
- 24.Li W., Chen W., Ren H., Bi J., Li W., et al. Establishment of colorectal cancer patient-derived xenograft (PDX) models and its characterization with 18F-FDG PET/CT live imaging. J. Digest. Oncol. (Electronic Version) 2017;9(4):264–269. [Google Scholar]
- 25.Rygaard J., Povlsen C.O. Heterotransplantation of a human malignant tumour to “Nude” mice. Acta Pathol. Microbiol. Scand. 1969;77(4):758–760. doi: 10.1111/j.1699-0463.1969.tb04520.x. [DOI] [PubMed] [Google Scholar]
- 26.Abdolahi S., Ghazvinian Z., Muhammadnejad S., Saleh M., Aghdaei H.A., et al. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J. Transl. Med. 2022;20(1) doi: 10.1186/s12967-022-03405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Z.Y., He Q.T., Liang J.F., Li W.G., Su Q., et al. miR-31-5p is a potential circulating biomarker and therapeutic target for oral cancer. Mol. Ther. Nucleic Acids. 2019;16:471–480. doi: 10.1016/j.omtn.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou C., Su Q., Li W., Zhao J., Fan Z., et al. Isolation of primary human osteosarcoma cells and establishment of cell-derived xenograft model and patient-derived xenograft model. J. Clin. Orthoped. Res. 2019;4(5):286–290. [Google Scholar]
- 29.Ban L., Mei T., Su Q., Li W., Huang Z., et al. Anti-fungal drug itraconazole exerts anti-cancer effects in oral squamous cell carcinoma via suppressing Hedgehog pathway. Life Sci. 2020;254 doi: 10.1016/j.lfs.2020.117695. [DOI] [PubMed] [Google Scholar]
- 30.Hu Q.C., Peng J.M., Jiang L.B., Li W.G., Su Q., et al. Metformin as a senostatic drug enhances the anticancer efficacy of CDK4/6 inhibitor in head and neck squamous cell carcinoma. Cell Death Dis. 2020;11(10) doi: 10.1038/s41419-020-03126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu L.L., Chen Z.J., Lin X.Y., Tian L., Su Q., et al. Inhibition of BRD4 suppresses the malignancy of breast cancer cells via regulation of Snail. Cell Death Differ. 2020;27(1):255–268. doi: 10.1038/s41418-019-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Z.X., Su Q., Li W.G., Ren H., Huang H.Q., et al. Suppressed mitochondrial respiration via NOX5-mediated redox imbalance contributes to the antitumor activity of anlotinib in oral squamous cell carcinoma. J. Genet. Genom. 2021;48(7):582–594. doi: 10.1016/j.jgg.2021.06.014. [DOI] [PubMed] [Google Scholar]
- 33.Wang C.M., Li H.F., Wang X.K., Li W.G., Su Q., et al. Ailanthus altissima-derived ailanthone enhances gastric cancer cell apoptosis by inducing the repression of base excision repair by downregulating p23 expression. Int. J. Biol. Sci. 2021;17(11):2811–2825. doi: 10.7150/ijbs.60674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang M., Dong W., Xie R.H., Wu J.L., Su Q., et al. HSF1 facilitates the multistep process of lymphatic metastasis in bladder cancer via a novel PRMT5-WDR5-dependent transcriptional program. Cancer Commun. 2022;42(5):447–470. doi: 10.1002/cac2.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J.Y., Wang W.J., Zhang C.Y., Ling Y.Y., Hong X.J., et al. Ru(II)-modified TiO2 nanoparticles for hypoxia-adaptive photo-immunotherapy of oral squamous cell carcinoma. Biomaterials. 2022;289 doi: 10.1016/j.biomaterials.2022.121757. [DOI] [PubMed] [Google Scholar]
- 36.Prabhu A., Brandl A., Wakama S., Sako S., Ishibashi H., et al. Effect of oxaliplatin-based chemotherapy on chemosensitivity in patients with peritoneal metastasis from colorectal cancer treated with cytoreductive surgery and hyperthermic intraperitoneal chemotherapy: proof-of-concept study. BJS OPEN. 2021;5(2) doi: 10.1093/bjsopen/zraa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skoulidis F., Li B.T., Dy G.K., Price T.J., Falchook G.S., et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 2021;384(25):2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y., Phat C., Hong S.C. Structural diversity of marine cyclic peptides and their molecular mechanisms for anticancer, antibacterial, antifungal, and other clinical applications. Peptides. 2017;95:94–105. doi: 10.1016/j.peptides.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Zorzi A., Deyle K., Heinis C. Cyclic peptide therapeutics: past, present and future. Curr. Opin. Chem. Biol. 2017;38:24–29. doi: 10.1016/j.cbpa.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y.Y., Mao H., Chen L.X., Li L.F. Targeting signal pathways triggered by cyclic peptides in cancer: current trends and future challenges. Arch. Biochem. Biophys. 2021:701. doi: 10.1016/j.abb.2021.108776. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J.N., Xia Y.X., Zhang H.J. Natural cyclopeptides as anticancer agents in the last 20 years. Int. J. Mol. Sci. 2021;22(8) doi: 10.3390/ijms22083973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun L. Peptide-based drug development. Modern Chemistry & Applications. 2013;1(1):e103. [Google Scholar]
- 43.Varas M.A., Munoz-Montecinos C., Kallens V., Simon V., Allende M.L., et al. Exploiting Zebrafish xenografts for testing the in vivo antitumorigenic activity of microcin E492 against human colorectal cancer cells. Front. Microbiol. 2020:11. doi: 10.3389/fmicb.2020.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y.B., Ouyang M., Wang H.L., Zhang B.H., Guang W.H., et al. A cyclic peptide retards the proliferation of DU145 prostate cancer cells in vitro and in vivo through inhibition of FGFR2. Medcomm. 2020;1(3):362–375. doi: 10.1002/mco2.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.