Abstract
The periosteum on the skeletal surface creates a unique micro-environment for cortical bone homeostasis, but how this micro-environment is formed remains a mystery. In our study, we observed the cells in the periosteum presented elongated spindle-like morphology within the aligned collagen fibers, which is in accordance with the differentiated osteoblasts lining on the cortical surface. We planted the bone marrow stromal cells(BMSCs), the regular shaped progenitor cells, on collagen-coated aligned fibers, presenting similar cell morphology as observed in the natural periosteum. The aligned collagen topology induced the elongation of BMSCs, whichfacilitated the osteogenic process. Transcriptome analysis suggested the aligned collagen induced the regular shaped cells to present part of the periosteum derived stromal cells(PDSCs) characteristics by showing close correlation of the two cell populations. In addition, the elevated expression of PDSCs markers in the cells grown on the aligned collagen-coated fibers further indicated the function of periosteal topology in manipulating cells’ behavior. Enrichment analysis revealed cell-extracellular matrix interaction was the major pathway initiating this process, which created an osteo-friendly micro-environment as well. At last, we found the aligned topology of collagen induced mechano-growth factor expression as the result of Igf1 alternative splicing, guiding the progenitor cells behavior and osteogenic process in the periosteum. This study uncovers the key role of the aligned topology of collagen in the periosteum and explains the mechanism in creating the periosteal micro-environment, which gives the inspiration for artificial periosteum design.
Keywords: Periosteum, Aligned collagen, Osteogenic micro-environment, Igf1 alternative splicing
Graphical abstract

1. Introduction
The natural periosteum is a thin layer of connective tissue covers the outer surface of bone and connects to bone by strong collagenous fibers. The periosteum extends to the outer circumferential and interstitial lamellae of bone [1]. Previous studies have attributed the regenerative capacity of autografts to periosteum, which accounts for about 70% of bone and cartilage formation at the fracture healing site [2,3].
It has been observed that collagen orientation in periosteum is aligned with preferential directions of tissue growth [4]. Meanwhile, emerging evidence has confirmed the role of topology to enhance osteo-integration and bone formation no matter in vitro or in vivo [5,6]. Interestingly, the influence of topology on osteo-specific function seems to be most effectively translated to an in vivo response when applied in combination with microscale features, indicating a possible synergy of the cellular and extra-cellular mechanisms [[7], [8], [9], [10]]. As reported, macrophage lineage cells recruit periosteum-maintained cells, like Nestin+ and LepR+ cells, for cortical bone formation. No matter which, the Nestin+ or LepR+ cells were observed to travel through the aligned collagen layer and showed higher osteogenic capacity for cortical bone development or homeostasis [11]. This phenomenon takes us the question that if the oriented topology established a unique micro-environment for cells maintenance in the periosteum.
Previous studies have revealed that changes to the tissue hierarchy may have significant effects on cell adhesion, integrin-triggered signaling pathways and cellular function [12]. While in skeletal system, topology has also been shown to regulate over multiple aspects of skeleton derived cells behavior including migration [13], proliferation [14], differentiation [15,16] and genetic expression [17]. It has been proved the aligned topology facilitates osteogenic differentiation of mesenchymal stromal cells derived from bone marrow or adipose tissue [18,19], indicating the specific role of periosteal structure in skeletal construction and remodeling. However, it still lacks comprehensive analysis of the micro-environment that established by the aligned collagen in the periosteum.
To elucidate this enigma, we fabricated the randomized and aligned electrospun fibers coated with type I collagen to mimic the topographic structure of the natural periosteum and analyzed the behavior changes of the cells grown on it. We not only demonstrated the aligned topology induced bone marrow stromal cells(BMSCs) to present part of the periosteum derived stromal cells (PDSCs) characters, but also identified the increased IGF-1 expression and its alternative splicing exert the key regulatory function in this process. This study will deepen our understanding of the biological function of the periosteal topology, which is also helpful in facilitating the construction of the artificial periosteum for clinical applications.
2. Results
2.1. Topological analysis of the natural periosteum from rat femur
To understand the topological characteristics of the periosteum, we first observed the periosteal structure with scanning electron microscope(SEM). The femur samples were collected from 8-week male rat. SEM showed the collagen fibers in the periosteum were loose and well oriented, closely connecting to the out surface of the cortical bone. Within the aligned fibers, we also observed the embedded cells, showing the spindle like shape(Fig. 1A). Similar morphological characteristics of the collagen topology and cells in the periosteum were presented on tissue sections with hematoxylin-eosin (HE) staining of the rat femur(Fig. 1B).
Fig. 1.
The aligned structure of the periosteum. A. 8-week rat femur was subjected to SEM observation of the periosteum in metaphysis and diaphysis. B. H&E staining of the periosteum on 8-week rat femur. Scale bar: 200um and40μm. C. The LepR staining of the periosteum and bone marrow area in the metaphysis and diaphysis of the rat femur. Scale bar: 50um. D. The OCN and DMP1 staining of the periosteum of the rat femur. Scale bar: 50um.
To further analyze the cell morphology in the periosteum collagen fibers, we performed immunofluorescent staining of the rat femur with the antibodies of LepR, which is a subtype of stromal cells for skeletal development and remodeling in the periosteum. We see clear that LepR positive cells were embedded in the collagen fibers, showing spindle-like shape. Meanwhile, we also analyzed the cell morphology of the LepR cells in the bone marrow. The LepR+ cells were found to appear both in the metaphyseal and diaphyseal parts of the rat femur. All the cells were in random shape with a spread pattern(Fig. 1C). Interestingly, the matured osteoblasts, shown by osteocalcin(OCN), in the periosteum were also in spindle-like shape, which are in line with the morphological characteristics of the stromal cells we observed(Fig. 1D).
The results above give us a hint of the relationship between periosteal topology induced morphological change and cell behavior. Therefore, in the periosteum, if the oriented collagen fibers would change the cells maintenance and fate in the periosteum is worth to be investigated.
2.2. Construction of the type I collagen-modified aligned electrospun fibers
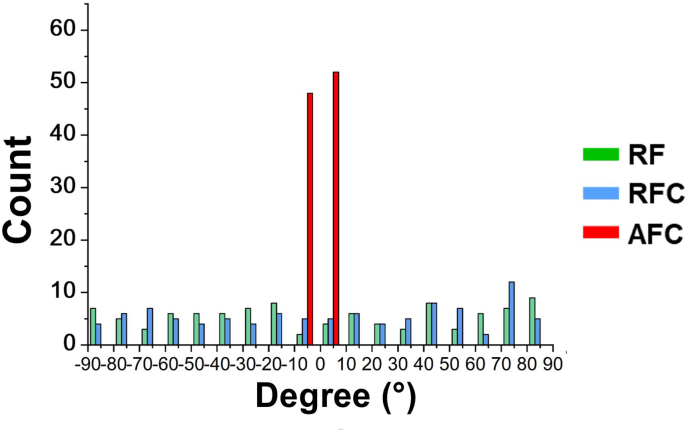
To imitate the aligned topology of the periosteum, we constructed the random and aligned poly lactic acid (PLA) electrospun fibers that assembled with or without rat tail type I collagen. We divided the three groups as RF(random fibers), RFC(random fibers coated with type I collagen) and AFC(aligned fibers coated with type I collagen). Electrospun fibers in RF and RFC exhibited inter-crossed alignment, in which the angles between fibers ranged from 0 to 90°. While most of the fibers in the AFC group showed uniformed fiber alignment, forming the angles only ranged from 0 to 10° between the fibers within the scaffold (Fig. S1). SEM scanning of the three groups showed the fibers in the RF and RFC groups were intermeshed, while the fibers in the AFC group was orientally arranged in one direction. Then, we measured the diameters of the electrospun fibers in the three groups. The data showed the averaged diameter of the fibers from the RF, RFC and AFC groups were 668.88 ± 140.88 nm, 1057.63 ± 228.99 nm and 1042.13 ± 183.74 nm, respectively (Fig. S2. A). We further analyzed several parameters to reflect the mechanical properties of the fibers in the three groups. First, we evaluated the Young's modulus as 12.92 ± 2.06 MPa, 18.52 ± 1.36 MPa and 32.85 ± 1.03 MPa in the RF, RFC and AFC groups (Fig. S2. B). Furthermore, we measured the water contact angle (WCA) and showed that the WCA was lower in the collagen modified two groups, indicating ideal contact ability of the collagen modified materials (Fig. S2. C). These data demonstrated that the type I collagen was evenly coated on the outer surface of the fibers in the RFC and AFC groups and the orientation was aligned in the AFC group, creating an ideal morphological environment to investigate the cells behavior grown on them(Fig. 2A).
Figure 2.
Construction of the type I collagen modified aligned electrospun fibers. The constructed electrospun fibers were divide into four groups, including random fibers(RF), random fibers with type I collagen modification(RFC) and aligned fibers with type I collagen modification(AFC). A. The Three groups of fibers were observed under SEM. Scale bar: 10um. B. F-actin staining of the BMSCs grown on the fibers of the three groups. Scale bar: 200um. C. SEM observation of the BMSCs grown on the fibers of the three groups. The upper panel of pictures are original SEM images and the bottom panel of pictures are artificially colored with the scaffolds and BMSCs. Scale bar: 50um.
To observe the cells morphological change in each group, we seeded the rat bone marrow stromal cells on the scaffolds in the three groups. Then, we stained the cells with F-actin to show the cytoskeleton of the cells on random or aligned fibers. The cells on the AFC group were arranged in one direction as compared to the RF and RFC groups. In addition, the cell bodies were evenly stretched in the RFC and AFC group as compared to the RF group(Fig. 2B). We further observed the cells morphology with SEM directly. Similar to the morphological changes observed in the cytoskeleton staining, SEM images showed the cells in the RF group were spread out on all sides, while the cells in the RFC group were spread more even and larger. On the other hand, the cells grown in the AFC group were elongated with spindle-like morphology, which are similar like the embedded cells in the natural periosteum(Fig. 2C). The results above suggest the constructed scaffold with the aligned topology and collagen modification is suitable for further investigations of the specific periosteal topology.
2.3. The aligned collagen topology facilitates osteogenesis in vitro
As the osteogenic micro-environment in the periosteum, the periosteum derived stromal cells presented higher osteo-differentiation ability for cortical bone formation and fracture healing [20]. Hence, we tried to test if the structured fibers that mimic the periosteum topology would change the osteo-differentiation ability of the bone marrow stromal cells(BMSCs). After growing for 7 days in osteogenic induction free medium, ALP staining was performed. It showed little positive staining in the RF group, while the positive staining was significantly increased in the RFC, which was even stronger in the AFC group(Fig. 3A). Alizarin Red S(ARS) staining of the cells with 14 days culture also showed the similar trend as that in the ALP staining results(Fig. 3B).
Fig. 3.
The aligned collagen topology facilitated osteogenesis in vitro. BMSCs were isolated from 8-week rat femur and planted on RF, RFC and AFC group of scaffold for 7 days and 14 days with osteogenic induction free medium. A. ALP staining of the BMSCs cultured for 7 days. B. ARS staining of the BMSCs cultured for 14 days. Osteogenic related genes expression on day 7 and day 14 was evaluated. C-E. Alp, Ocn and Opn expression on the 7th culture day. F–H. Alp, Ocn and Opn expression on the 14th culture day. (∗P<0.05, ∗∗P<0.01).
The osteo-differentiation ability in the three groups was further evaluated by quantitative polymerase chain reaction(qPCR) of the osteogenic related genes, including alkaline phosphatase(Alp), osteocalcin(Ocn) and osteopontin(Opn). We see from the results that the three osteogenic genes expression increased significantly in the RFC group and went higher in the AFC group on both 7 and 14 days culture(Fig. 3C–H). The data confirmed the two key elements of collagen and topology in the natural periosteum facilitate osteogenic differentiation in vitro, reflecting the possibility for using the constructed scaffold to further analyze the topology related functions of the periosteum.
2.4. The aligned collagen topology enhances bone regeneration in vivo
To further investigate the osteogenic attribution of the type I collagen coated aligned fibers in vivo, we employed rat cranial defect model. The constructed films with different topology or coated with assembled collagen were evenly covered on the surface of the cranial defect sites. The cranial tissue was collected after 4 weeks and 8 weeks, respectively. Micro-CT(uCT) analysis showed in 4 weeks, the cranial defect was partly filled with regenerated bone tissue in all groups. The volume of new bone formation was significantly larger in the RFC group and AFC group than that in the RF group, in which the AFC group was even greater. The trend was similar when it came to the time point at 8 weeks. Impressively, the defect was almost fully regenerated with new bone in the group of collagen-coated aligned electrospun fibers group(Fig. 4A&4 B).
Fig. 4.
The aligned collagen topology enhanced bone regeneration in vivo. The scaffold from RF, RFC and AFC groups were planted and covered the defect of rat calvaria. A. micro-CT analysis of the defect repair of the rat calvaria after 4 weeks and 8 weeks. B. Quantitative analysis of the micro-CT data of the bone defect in rat calvaria. C. OCN staining of the rat calvaria defect sections from RF, RFC and AFC groups. Scale bar: 1 mm and 100um.
The results were further confirmed by OCN, one of the osteoblastic cell markers, staining with the collected samples. It showed the positive staining of OCN was significantly increased in the RFC group and even more in the AFC group, confirming the enhanced osteoblastic differentiation with collagen coated aligned fibers(Fig. 4C). The data above suggest the collagen coated aligned fibers are promising scaffold to mimic the structure of the natural periosteum and used to analyze the micro-environment established within the periosteum.
2.5. Periosteal topology reshapes the cells to show periosteum derived stromal cells(PDSCs) characteristics
To evaluate the comprehensive characteristics of the cells grown on the collagen coated aligned fibers, we planted regularly shaped stromal cells on the scaffolds in the three groups. The transcriptome sequencing was performed and the osteogenic related genes expression was analyzed in each group, with the PDSCs as positive control. After sequencing, the differentially expressed genes(DEGs) with more than 1.5-fold change(P value < 0.05) were obtained. The DEGs between every two of the RF, RFC and AFC groups were extracted. Meanwhile, the unsupervised hierarchical clustering based on genes related to osteogenesis(Gene ontology(GO) term contains “osteo-” or “bone”) were applied to the following analysis to eliminate the background noise [21].
The heatmap of the collected DEGs were constructed with all the groups analyzed. We can see that, generally, nearly half of the genes expression was gradually decreased from the RF group to the RFC group and to the AFC group, in which, most of them were bone remodeling activating genes, such as pthlh, sfrp4 and Ccr1, and bone catabolic genes, like Il17 and Pdk4. The rest genes expression was gradually increased from the RF group to the RFC group and to the AFC group, and most of which were genes for bone anabolism, including Tnfrsf11b(OPG), Col1a1, Igf1 and Ptn. The sample-to-sample cluster analysis showed RF and RFC groups were firstly linked, and then linked to the AFC group, which was finally linked to the PDSCs group(Fig. 5A). The similar result was further confirmed by correlation analysis exhibiting the gradual change of the characteristics of stromal cells to PDSCs from RF to AFC group(Fig. 5B). Furthermore, Principal component analysis(PCA) analysis showed all the samples were well clustered within their own groups. The RFC group became close to the PDSCs group in PC1 and PC2 dimensions as compared to the RF group, and the AFC group was even more close to the PDSCs group both in PC1 and PC2 dimensions(Fig. 5C).
Fig. 5.
Periosteal topology induced transformation of BMSCs to PDSCs-like cells. The BMSCs grown on RF, RFC and AFC scaffold for 3 days were subjected to transcriptome analysis. The DEGs(fold change>1.5, P value<0.05) with GO term contains “osteo-” or “bone” were subjected to the following analysis. A. Heatmap analysis of the DEGs selected. Correlation analysis(B) and PCA analysis(C) of the samples from RF, RFC and AFC groups compared with PDSCs. D. qPCR evaluation of the PDSCs marker genes(Mx1, Ctsk and Postn) and osteogenic related genes(Col1a1 and BMP2), as well as stemness related genes (Klf4 and Sox2). (∗P<0.05, ∗∗P<0.01) E and F. Immunofluorescent staining of Mx1(E) and Postn(F) of the BMSCs grown on RF, RFC and AFC groups. Scale bar: 50um.
To further confirm the PDSCs-like characteristics induced by the aligned collagen topology, we performed qPCR of the PDSCs marker genes, including Mx1, Ctsk, Postn and osteogenic related genes, including Col1a1 and Bmp2. The data showed the PDSCs marker genes expression was dramatically increased in the AFC group when compared with the RF group, with similar change of the osteogenic related genes. Meanwhile, to confirm stemness that maintained within the stromal cells grown on the scaffold, Klf4 and Sox2 genes expression was also quantified. The data showed no matter in which group, the cells maintained their stemness for further diffrentiation(Fig. 5D). Furthermore, immuno-staining of MX-1 and POSTN was performed, both of which were significantly increased in the AFC group as compared to the RF and RFC groups, indicating the stromal cells show PDSCs characteristics when grown on the collagen coated aligned fibers. The above results indicate the aligned topology of collagen in the natural periosteum creates a specific micro-environment for cortical bone homeostasis.
2.6. The osteogenic micro-environment established by the aligned topology of collagen in the periosteum
Periosteal topology creates a specific micro-environment for cortical bone homeostasis. As analyzed with the transcriptome sequencing data, we found totally 891 DEGs between the AFC and RFC groups, 690 DEGs between the AFC and RF groups, as well as 679 DEGs between the RFC and RF groups. Between the AFC and RF groups, 382 genes were up-regulated and 308 were down-regulated. GO and kyoto encyclopedia of genes and genomes(KEGG) were analyzed based on the up- and down-regulated DEGs, respectively. GO analysis revealed the enrichment of abundant terms, in which the top enriched terms were mostly related with extracellular regulations, including extracellular space, extracellular matrix etc.(Fig. 6A–C). The KEGG analysis also hits the extracellular matrix receptors interaction signaling. Hence, combine with the GO analysis, the results indicate the aligned topology of collagen transforms the cells characteristics through extracellular interactions.
Fig. 6.
The osteogenic micro-environment created by periosteal topology. All the up-regulated(A) and down-regulated(B) DEGs(fold change>1.5, P value<0.05) were subjected to GO analysis. The up-(C) and down-(D) regulated DEGs also went through KEGG analysis. E. Part of the enriched KEGG signaling pathways were further analyzed by GSEA analysis, including Cell Surface and Extracellular Matrix from GO analysis and Wnt, ECM-receptor interaction, Osteoclast differentiation and NF-Kappa B signaling pathways from KEGG analysis.
Besides, the KEGG analysis with the up-regulated DEGs reflects the enrichment of Wnt signaling, known as the key signaling to facilitate osteogenesis. Meanwhile, in the KEGG analysis with the down-regulated DEGs, osteoclast differentiation and NF-κB signaling pathways were enriched as the tops(Fig. 6C and D). The key signaling pathways that enriched were further confirmed by gene set enrichment analysis(GSEA) analysis, which also confirmed the corresponding change that observed in the GO and KEGG analysis of the cells grown in the three groups(Fig. 6E). These data showed clearly the aligned collagen topology established a suitable micro-environment for osteogenesis within the periosteum.
2.7. The periosteal topology initiates Igf1 alternative splicing
Based on the transcriptome data, we performed Venn-analysis with the DEGs from every two of the RF, RFC and AFC groups. The results showed totally 44 differentially expressed genes were included in the intersection sets of the RF, RFC and AFC groups(Fig. 7A). Then, the protein-protein interaction analysis of the 44 genes revealed 8 key node genes, including Igf1, Cxcl12, Agtr1a and Ptprz1 etc.(Fig. 7B). Among the 8 genes, Igf1 was found to be in the leading position and closely related with osteogenic micro-environment coordination. Meanwhile, the Igf1 gene hits the most enriched terms either in the GO or KEGG analysis between the AFC and RF groups(Fig. 7C and D). The results indicate that Igf1 is potentially orchestrating the topology induced micro-environment in the periosteum.
Fig. 7.
The alternative splicing of Igf1 orchestrates the periosteal topology established osteogenic micro-environment. A. Venn analysis of RFC vs RF, AFC vs RFC and AFC vs RF groups with 44 shared DEGs selected. B. Protein-protein interaction analysis of the selected 44 genes above, among which Igf1 is in the core position. C, D. The top 15 DEGs that hits the most GO(C) or KEGG(D) terms were listed, in which Igf1 is the top 1 in either GO and KEGG terms. E. Immunofluorescent staining of LepR and IGF-1 of the rat femur periosteum. Red color represents LepR and green color represents IGF-1. Scale bar: 50um. F, G. The schematic diagram of previously reported Igf1 alternative splicing and the identified variants in BMSCs cultured on the scaffolds we constructed. H. qPCR analysis of the detected exons expression on the scaffolds from RF, RFC and AFC groups. (∗P<0.05, ∗∗P<0.01).
Subsequently, we evaluated Insulin-like growth factor type 1 (IGF-1) protein expression by co-staining of LepR and IGF-1 in the rat femur periosteum. The results showed clear that either in the metaphyseal or diaphyseal area of the periosteum, the positive IGF-1 staining was largely co-localized with LepR. As expected, the IGF-1 expression level was relatively higher in the metaphyseal periosteum compared to the diaphyseal periosteum(Fig. 7E).
Previous studies have indicated Igf1 alternative splicing leads to the production of different proteins, including mature (IGF-1 and mechano-growth factor(MGF). There are 6 exons in Igf1 gene. All mRNA splice variants contain exons 3 and 4, which encode the mature 70-amino-acid IGF-I peptide. mRNAs containing exon 4 spliced to exon 6 are designated as IGF-IEa [[22], [23], [24], [25]], whereas those containing exon 4 spliced to exon 5 and exon 6 are designated IGF-IEb(MGF) in rodents(Fig. 7F). In our study, we first analyzed which alternative splicing was initiated in the specific collagen topology. Transcriptome data revealed no exon 2 in either of the samples. Exon 5 was only detected in a low level in 2 of the 3 samples from the RF group, while its expression was abundantly detected in both RFC and AFC groups, which was significantly higher in the AFC group(Fig. 7G). Meanwhile, qPCR data identified two types of variable splicing with exon 2 deletion(Δ2) and exon 2 + 5 deletion(Δ2+5) among all the three groups. No exon 1 deletion(Δ1) or exon 1 + 5 deletion(Δ1+5) was observed(Fig. 7G). qPCR data also showed exon 5 + 6 expression was gradually increased in RFC and AFC groups as compared to the RF group(Fig. 7H), suggesting the aligned topology of collagen activates Igf1 gene through regulation of its variable splicing.
2.8. Periosteal topology induced MGF expression promoted BMSCs transformation
To understand the function of Igf1 alternative splicing in mediating periosteum micro-environment, we constructed the corresponding plasmid with exon 2 (Δ2) or exon 2 + 5(Δ2+5) deletion. Then, the plasmids were transfected into the BMSCs in the cell culture plates and subsequently substituted with osteogenic induction medium. After 14 days culture, the cells were performed with ARS staining. The BMSCs transfected with Δ2 plasmid showed the most wide and deepest ARS staining as compared to the Δ2+5 and the control groups(Fig. 8A&8 B). qPCR analysis also revealed that BMSCs transfected with Δ2 plasmid exhibited higher expression of the osteogenic genes, including Alp, Col1a1 and Ocn(Fig. 8C–E). We further performed this test with BMSCs that planted on the random fibers scaffold with osteogenic-induction free medium. The results clearly showed the BMSCs transfected with the Δ2 plasmid were more easily to transform into osteoblasts reflected by ARS staining(Fig. 8F and G). Then, the qPCR data also confirmed the results as that in the ARS staining(Fig. 8H–J). All the above results confirmed the Igf1 alternative splicing with exon 5 + 6(encoding MGF) is the orchestrator in mediating the aligned collagen topology induced osteogenic micro-environment within the periosteum.
Fig. 8.
MGF expression promotes BMSCs transformation. A. BMSCs transfected with control, △2 + 5 and △2 plasmids were cultured with osteogenic induction medium in dish. On day 14, the cells were stained with ARS to evaluate the osteogenic differentiation of BMSCs. Scale bar: 500um. B. Quantitative analysis of ARS staining in A. C-E. qPCR evaluation of the osteogenic genes, including ALP, Col1a1 and OCN, expression in A. F. BMSCs transfected with control, △2 + 5 and △2 plasmids were cultured on RF scaffold with osteogenic induction free medium. On day 14, the cells were stained with ARS. Scale bar: 500um. G. Quantitative analysis of ARS staining in F. H-J. qPCR evaluation of the osteogenic genes, including ALP, Col1a1 and OCN, expression in F. (∗P<0.05, ∗∗P<0.01).
3. Discussion
The periosteum is simply divided into two layers by the contains. The outer fibrous layer and the inner cambial layer. The inner layer contains multiple osteoprogenitor cells, which participate in the growth, development and regeneration process of bone. The outer layer is thicker and consists of a matrix layer and a fibroblastic/collagenous layer. This layer consists of longitudinally oriented cells and well-organized collagen fibers are aligned in the direction of bone growth, reflecting the growth trend.
The importance of the periosteum in bone reconstruction is becoming well known. Due to collagen is the main component of the periosteum microenvironment, numerous recent studies have constructed tissue engineered periosteum to mimic the microenvironment by using collagen and hydrogels [26,27]. Periosteal unique oriented micro-topological makes periosteum tough and has also been found to contribute to osteogenesis [28]. At present, numerous studies have shown the importance of PDSCs in osteogenesis, which inspired us to explore tissue engineering with the stimulation from periosteum structure [29]. Shi et al. used grooved micropatterns on the surface of collagen membrane inherited from the periosteum effectively directed cell alignment as the way of natural periosteum, which significantly enhanced osteogenic capacity [30]. Yang et al. found that micropattern on hydroxyapatite nanoparticles can sustainably provide a periosteum-mimetic microenvironment, which could significantly enhance vascularized ossification and accelerate new bone formation [31]. We believe that a synergistic effect of collagen and oriented structure may be responsible for inducing the regularly shaped stromal cells to gain PDSCs characteristics. Therefore, we used oriented PLA electrospinning and type I collagen (the most abundant ECM in the periosteum) to simulate the periosteum micro-environment.
IGF-1 is one of the most abundant substances in the bone matrix, which has long been known to play a role in bone growth [32], because of its effects in regulating osteoblastic and osteoclastic cell populations [33]. Previous research showed that low level of IGF-1 in serum resultsjiu in impaired subperiosteal expansion combined with impaired endosteal apposition and lack of compensatory changes in mineralization throughout growth and aging [34]. MGF is a splice variant of IGF-1 and Deng et al. found that injecting MGF into the bone defect gap results in accelerated bone healing [35], but the exact mechanism is unclear. It has been demonstrated that MGF enhances hBMSCs differentiation by lengthening G1 phase of cell cycle [36]. Through our study, it was found that oriented topology and collagen may induce the stromal cells to show PDSCs characteristics by activating MGF, which may become a new target for periosteum tissue engineering to repair bone defects.
In this study, we tried to explore the topological function of the periosteum in influencing the cells behavior by growing the stromal cells on the scaffolds with different orientations. However, based on study design, there still exists several flaws. First, the scaffolds we constructed are not fully originated from or replicated the natural periosteum, so the cells may show other characters in the natural conditions. Second, the cells we used are stromal cells from the bone marrow, which may not originate from the lineage of periosteal progenitor cells, so it only partly reflected the periosteal topology induced cells behavioral change. Therefore, more studies are in need to further explore the mystery within the periosteum.
In conclusion, our study revealed the aligned collagen topology in the periosteum induces regularly shaped stromal cells to show PDSCs characteristics, which are inclined to differentiate into osteo-linage cells and contribute to cortical bone homeostasis. The underlined mechanism is through activation of Igf1 alternative splicing and production of MGF.
4. Materials and methods
4.1. Micro-anatomy analysis of the periosteum
The femur of male Sprague-Dawley (SD) rats at 8w was used to observe the periosteal structure. After fixed with formalin for 24 h, the femur was decalcified with 14% EDTA for 30 days, and then cut along the sagittal plane of the femur with a thickness of 40um. Select slices containing intact parts of the bone marrow cavity and periosteum tissue. The slices were pasted directly onto the sample platform to which conductive adhesive had been applied. The apparatus was placed in a cool ventilated area and the slices were dried for 24 h. After metal spraying, the slices were observed under SEM as described above.
4.2. Isolation of rat bone marrow stromal cells (BMSCs) and periosteum derived stromal cells (PDSCs)
BMSCs were obtained from Sprague-Dawley rats femoral bone marrow. Both ends of the femurs were cut-off and flushed with PBS (HyClone, Logan, UT). PSCs were collected from rat femurs too. Muscle tissue was removed from the femurs and rinsed out the bone marrow. Then, the femurs were cut up and placed directly on the bottom of the cell culture dish. All cells were cultured at 37 °C in a humidified 5% CO2 incubator with Alpha's modified Eagle's medium (α-MEM, Hyclone Co. Ltd., Logan, UT). After 3 days, nonadherent cells were removed by repeated washing with prewarmed PBS and adherent cells were further cultured. The medium was changed every 3 days.
4.3. Fabrication of electrospun scaffolds
The electrospun scaffold was synthesized using PLLA (Daigang Co. Ltd., Shandong, China), dichloromethane (DCM, Qiangsheng Functional Chemistry Co. Ltd., Jiangsu, China), and N,N-dimethylformamide (DMF, Qiangshun Chemical Reagent Co. Ltd., Shanghai, China). Normally, 1 g of PLLA was dissolved in 4.5 g DCM completely. After that, 2.25 g DMF was added and stirred for half an hour. The solution was placed into a 10 mL syringe, equipped with a 0.41 mm-diameter needle. The electrospinning voltage was set to 10 kV and solution feed rate was 0.3 mL/h. For non-oriented scaffold, the obtainer is a round metal collector covered by aluminum foil. For oriented scaffold, the electrospun scaffold can be obtained by collecting the paralled fiber between a pair of parallel electrode rods. The distance between the needle and the collector is 15 cm.
4.4. Preparation of collagen modified scaffolds
Specifically, 0.15 mL sodium hydroxide (NaOH, 0.1 M) and 0.18 mL phosphate buffer saline (PBS, 10x, HyClone Co. Ltd., Logan, UT) were first mixed. And 0.67 mL collagen type I solution (Rat Tail, Corning Co. Ltd., MA, USA) was mixed into it. All of above are done on ice. Then drip it on the electrospinning surface and incubated at 37 °C for 30 min. After collagen self-assembly, use PBS 1x to wash the scaffold for 3 times.
4.5. Physical and chemical characterization
The morphology and topology of different scaffolds were analyzed using scanning electron microscopy (SEM; SU8010, Hitachi Co. Ltd., Tokyo, Japan) The surface hydrophilicity of the scaffolds was evaluated by measuring the water contact angles (WCAs) with a sessile drop shape method and a drop analysis system (DSA25, KRUSS Co. Ltd., Hamburg, Germany).
To evaluate the mechanical properties, dry fibrous scaffolds were punched into dumbbell-shaped specimens (15.0 × 3.0 × 0.13 mm3) before testing. The uniaxial tensile test was carried out by a mechanical testing machine (E10000, Instron Co. Ltd., Shanghai, China) to obtain a strain-stress curve. At least 10 indentations were conducted for each sample.
4.6. Cell morphology on scaffolds
To observe the cell morphology on different scaffolds, rat BMSCs were cultured on the surface of scaffolds. After 48 h, the culture medium was removed and cell were washed 3 times with PBS. Then the cells were fixed in 4% paraformaldehyde (PFA) for 40 min. For cytoskeletal staining, the samples were permeabilized with 0.3% Triton X-100 for 10 min and incubated with phalloidin for 30 min at room temperature. The nuclei were counterstained with DAPI. Images were obtained using a Zeiss Axiovert 200 fluorescence microscope (Carl Zeiss Inc., Thornwood, NY). For direct observation, the fixed samples were treated with gradient ethanol. SEM was performed before critical point drying and sputter-coated with gold.
4.7. Cell viability assay
The proliferation of BMSCs was measured by Cell Counting Kit-8 (CCK-8, Dojindo, Japan). Briefly, scaffolds were set at the bottom of the 96-well plates. The BMSCs (1 × 104/mL) were seed subsequently with α-MEM (200 μL/well) at 37 °C with 5% CO2. After incubation, the cell medium was removed and replaced with 100 μL of culture medium and 10 μL of CCK-8 reagent for another 2 h for color development. The optical density (OD) value was collected by a microplate reader (BioTek, VT) at 450 nm.
4.8. Osteogenic induction experiment
BMSCs were seeded on the scaffolds at a density of 2 × 105 cells/well in 24-well plates. After 7 or 14 days of culture, the culture medium was removed. At day 7, following fixation, the cells on different scaffolds were incubated with a BCIP/NBT ALP color development kit (Beyotime Co. Ltd., Shanghai, China) for 30 min at room temperature and then images were taken. Mineralization of BMSCs was analyzed on day 14 using alizarin red staining (ARS, Cyagen Biosciences Co. Ltd., Guangzhou, China). After being fixed, the cells were stained with 1% alizarin red for 30 min and then images were taken.
For real-time quantitative polymerase chain reaction (RT-qPCR), BMSCs were seeded on the scaffolds at a density of 5 × 105 cells/well in 6-well plates. After 7 or 14 days of culture, the culture medium was removed. TRIzol (Takara Co. Ltd., Tokyo, Japan) was used to extract the total RNA. Total RNA yields were measured using Nano Drop (Thermo Fisher Scientific Co. Ltd., MA, USA). One microgram of total RNA from each sample was reverse transcribed according to the manufacturer's instructions using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen, OR, USA). All RT-qPCR reactions were prepared using the iTaq Universal SYBR Green Master (Thermo Fisher Scientific Co. Ltd., MA, USA). The 10 μL volume reaction component included 5 μL of Master Mix (Green, 2X, Beyotime, Shanghai, China), 1 μL of forward and reverse primers, 1.5 μL nuclease free water and 2.5 μL of cDNA template. Col I, ALP, OCN and OPN were selected as target gene primers. Relative expressions were calculated and normalized to actin gene expression. Primer sequences are listed in Table 1.
Table 1.
PCR primer sequences.
| Target gene | Forward primer sequence (5′–3′) | Reverse primer sequence (5′–3′) |
|---|---|---|
| Collagen 1 (Col-1) | CAGGCTGGTGTGATGGGATT | CCAAGGTCTCCAGGAACACC |
| Alkaline phosphatase (ALP) | TATGTCTGGAACCGCACTGAAC | CACTAGCAAGAAGAAGCCTTTGG |
| Osteocalcin (OCN) | AACGGTGGTGCCATAGATGC | AGGACCCTCTCTCTGCTCAC |
| Osteopontin (OPN) | GCGGTTCACTTTGAGGACAC | TATGAGGCGGGGATAGTCTTT |
4.9. Animal models
SD male rats were provided by the Animal Facility of Soochow University. The animal handling and surgical procedures were conducted in accordance with protocols approved by the Ethics Committee at the First Affiliated Hospital of Soochow University. All SD rats were 6-8w and were designed to harvest at 4 and 8 weeks after surgery. Different scaffolds were covered on the skull defects (5 mm in diameter). The bone formation capability was evaluated by Micro-CT scanning (65 kV, 385 mA, 1 mm Al filter). Newly formed bone was quantified in five sections of at least five different defects per animal group as previously described. New bone formation on each section was expressed as a percentage of the total area of the defect. After this, samples were decalcified, embedded in paraffin, sliced, and stained.
4.10. Immunofluorescence staining
The expression of Nestin (Abcam), Leptin Receptor (Santa Cruz Biotechnology), OCN (Takara) and IGF1 (Abcam) was analyzed using immunofluorescence. Briefly, the prepared sections were rinsed with PBS three times, permeated with 0.3% Triton X-100 in PBS, and sequentially treated with 3% bovine serum albumin (BSA) to block nonspecific binding sites. The sections were stained against the primary antibodies (Nestin, 1:200, Leptin Receptor, 1:50, OCN, 1:200, IGF1, 1:200) at 4 °C overnight. Then, the sections were incubated in the corresponding secondary antibody (Abcam). The nuclei were further stained with DAPI. The samples were scanned using a fluorescence microscope (Zeiss Axiovert 200; Carl Zeiss Inc., Thornwood, NY).
4.11. Immunohistochemical staining
The expression of OCN in the defect area was analyzed using immunohistochemical staining. The previous steps are basically the same as immunofluorescence staining. The diluted concentration of primary antibody is 1:400. The sections were incubated with HRP labeled secondary antibody at room temperature for 1 h, then washed with PBS for three times, followed by DAB for 10 s. Finally, the nucleus was re-stained with hematoxylin. The samples were scanned using a microscope (Zeiss Axiovert 200; Carl Zeiss Inc., Thornwood, NY).
4.12. Transcriptome analysis
The BMSCs and PSCs were analyzed. The BMSCs on scaffolds were used for transcriptome analysis too. The transcriptome sequencing was conducted by OE biotech Co. Ltd. (Shanghai, China) Cleaning reads were obtained using Trimmomatic and mapped to reference genome using hisat2. FPKM (fragments per kilobase of exon per million reads mapped) value of each gene was calculated using cufflinks. The DEGs, GO (gene ontology), and KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis were performed using R software. p < 0.05 and FoldChange >1.5 or FoldChange <0.5 was set as the threshold for significantly differential expression or differential enrichment.
The clustered heatmap in Fig. 5A were plotted using R package ‘pheatmap’ (ver. 1.0.12), and Fig. 5B (correlation matrix plot) was create using the ‘corrplot’ (ver. 0.92). The three-dimensional PCA Plots was analyzed by ‘pca3d’ (ver. 0.10.2) and visualized by Origin 2021b.
The Gene Ontology enrichment analysis and KEGG pathway enrichment analysis of DEGs were performed using the OE suite of online tools (https://cloud.oebiotech.cn/task/). The Gene Set Enrichment Analysis was performed using a gene list pre-ranked by fold change upon RF and RFC cells.
The network in Fig. 7B was created using the stringApp(ver.1.7.1) in Cytoscape(ver. 3.9.1).
4.13. Lentivirus construction and transfection
Lentiviruses are constructed primarily by Genomeditech Co. Ltd. BMSCs were seed in 6-well plates with a density of 5 × 105/well. After 10–12 h of culture, the old medium was removed. Replace the new medium with 1ml/well, adding 5ug/ml polybrene. Then the calculated dose of virus solution according to the concentration of MOI = 50 was added, infect the cells for 24–48 h.
4.14. Statistical analysis
For each experiment, five to ten cases were included in each group at each time point. All of the experiments were independently repeated at least three times. All data are provided as the mean ± standard deviation. Statistical analysis was performed with an unpaired two-tailed Student's t-test for single comparisons with GraphPad Prism 8 (GraphPad Software, CA). One-way analysis of variance (ANOVA) was used to compare data from more than two groups. Differences were considered significant at ∗p < 0.05 and ∗∗p < 0.01.
Credit author statement
Jun Pan, Hanwen Li, Kai Jin and Huaye Jiang performed most of the experiments. Ke Li, Yingchuang Tang, Yu Shi, Wenzheng Lin and Zixiang Liu analyzed the data. Kai Zhang, Kangwu Chen, Zhuobin Xu, Huihui Wang and Chun Pan made suggestions during the study. Huilin Yang, Qin Shi, Junjie Niu and Hao Chen designed the study. Hao Chen wrote the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81802132 & 8217090346),Natural Science Foundation of Jiangsu Province (SBK2021022619),Natural Science Fund for Colleges and Universities in Jiangsu Province (21KJB320009),Science and Technology Projects Fund of Yangzhou City (SSF2021000051).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100519.
Contributor Information
Huilin Yang, Email: suzhouspine@163.com.
Junjie Niu, Email: jjnewzzzx@sina.com.
Qin Shi, Email: shiqin@suda.edu.cn.
Hao Chen, Email: hchen2020@yzu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figure S1.
The alignment of the fibers in the scaffolds of the RF, RFC and AFC groups.
Figure S2.
The basic characteristics of the collagen-coated eletrospun fibers. Diameters(A), Young’s modulus (B) and the Water contact angle (C) of the fibers in the scaffolds from the RF, RFC and AFC groups.
Data availability
Data will be made available on request.
References
- 1.Augustin G., Antabak A., Davila S. The periosteum. Part 1: anatomy, histology and molecular biology. Injury. 2007;38:1115–1130. doi: 10.1016/j.injury.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X., Awad H.A., O'Keefe R.J., Guldberg R.E., Schwarz E.M. A perspective: engineering periosteum for structural bone graft healing. Clin. Orthop. Relat. Res. 2008;466:1777–1787. doi: 10.1007/s11999-008-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X., et al. Periosteal progenitor cell fate in segmental cortical bone graft transplantations: implications for functional tissue engineering. J. Bone Miner. Res. 2005;20:2124–2137. doi: 10.1359/JBMR.050806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foolen J., et al. Collagen orientation in periosteum and perichondrium is aligned with preferential directions of tissue growth. J. Orthop. Res. 2007;26:1263–1268. doi: 10.1002/jor.20586. [DOI] [PubMed] [Google Scholar]
- 5.Svensson S., et al. Osseointegration of titanium with an antimicrobial nanostructured noble metal coating. Nanomed. Nanotechnol. Biol. Med. 2013;9:1048–1056. doi: 10.1016/j.nano.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Ballo A., Agheli H., Lausmaa J., Thomsen P., Petronis S. Nanostructured model implants for in vivo studies: influence of well-defined nanotopology on de novo bone formation on titanium implants. Int. J. Nanomed. 2011;6:3415–3428. doi: 10.2147/IJN.S25867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J., et al. Multiscale patterned transplantable stem cell patches for bone tissue regeneration. Biomaterials. 2014;35:9058–9067. doi: 10.1016/j.biomaterials.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 8.Coelho P.G., et al. Nanometer-scale features on micrometer-scale surface texturing: a bone histological, gene expression, and nanomechanical study. Bone. 2014;65:25–32. doi: 10.1016/j.bone.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Koo S., Muhammad R., Peh G.S.L., Mehta J.S., Yim E.K.F. Micro- and nanotopology with extracellular matrix coating modulate human corneal endothelial cell behavior. Acta Biomater. 2014;10:1975–1984. doi: 10.1016/j.actbio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Wei Q., et al. Cellular modulation by the mechanical cues from biomaterials for tissue engineering. Biomaterials Translational. 2021;2:323–342. doi: 10.12336/biomatertransl.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao B., et al. Macrophage-lineage TRAP+ cells recruit periosteum-derived cells for periosteal osteogenesis and regeneration. J. Clin. Invest. 2019;129:2578–2594. doi: 10.1172/JCI98857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong T., et al. Nanomaterials and bone regeneration. Bone Res. 2015;3 doi: 10.1038/boneres.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamers E., et al. Dynamic cell adhesion and migration on nanoscale grooved substrates. European Cells. 2012;23:182. doi: 10.22203/ecm.v023a14. [DOI] [PubMed] [Google Scholar]
- 14.Hu J., Zhou Y., Huang L., Liu J., Lu H. Effect of nano-hydroxyapatite coating on the osteoinductivity of porous biphasic calcium phosphate ceramics. BMC Muscoskel. Disord. 2014;15:114. doi: 10.1186/1471-2474-15-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biggs M.J., et al. The use of nanoscale topology to modulate the dynamics of adhesion formation in primary osteoblasts and ERK/MAPK signalling in STRO-1+ enriched skeletal stem cells. Biomaterials. 2009;30:5094–5103. doi: 10.1016/j.biomaterials.2009.05.049. [DOI] [PubMed] [Google Scholar]
- 16.Metavarayuth K., Villarreal E., Wang H., Wang Q. Surface topology and free energy regulate osteogenesis of stem cells: effects of shape-controlled gold nanoparticles. Biomaterials Translational. 2021;2:165–173. doi: 10.12336/biomatertransl.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantawong F., et al. Whole proteome analysis of osteoprogenitor differentiation induced by disordered nanotopology and mediated by ERK signalling. Biomaterials. 2009;30:4723–4731. doi: 10.1016/j.biomaterials.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 18.Gao X., et al. Bioinspired design of polycaprolactone composite nanofibers as artificial bone extracellular matrix for bone regeneration application. ACS Appl. Mater. Interfaces. 2016;8:27594–27610. doi: 10.1021/acsami.6b10417. [DOI] [PubMed] [Google Scholar]
- 19.Yang L., et al. Topology induced stiffness alteration of stem cells influences osteogenic differentiation. Biomater. Sci. 2020;8:2638–2652. doi: 10.1039/d0bm00264j. [DOI] [PubMed] [Google Scholar]
- 20.Duchamp de Lageneste O., et al. Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin. Nat. Commun. 2018;9:773. doi: 10.1038/s41467-018-03124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Gene Ontology C. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rotwein P., Pollock K.M., Didier D.K., Krivi G.G. Organization and sequence of the human insulin-like growth factor I gene. Alternative RNA processing produces two insulin-like growth factor I precursor peptides. J. Biol. Chem. 1986;261:4828–4832. [PubMed] [Google Scholar]
- 23.Chew S.L., Lavender P., Clark A.J., Ross R.J. An alternatively spliced human insulin-like growth factor-I transcript with hepatic tissue expression that diverts away from the mitogenic IBE1 peptide. Endocrinology. 1995;136:1939–1944. doi: 10.1210/endo.136.5.7720641. [DOI] [PubMed] [Google Scholar]
- 24.Bell G.I., Stempien M.M., Fong N.M., Rall L.B. Sequences of liver cDNAs encoding two different mouse insulin-like growth factor I precursors. Nucleic Acids Res. 1986;14:7873–7882. doi: 10.1093/nar/14.20.7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts C.T., Jr., Lasky S.R., Lowe W.L., Jr., Seaman W.T., LeRoith D. Molecular cloning of rat insulin-like growth factor I complementary deoxyribonucleic acids: differential messenger ribonucleic acid processing and regulation by growth hormone in extrahepatic tissues. Mol. Endocrinol. 1987;1:243–248. doi: 10.1210/mend-1-3-243. [DOI] [PubMed] [Google Scholar]
- 26.Gupta S., et al. Periosteum-mimicking tissue-engineered composite for treating periosteum damage in critical-sized bone defects. Biomacromolecules. 2021;22:3237–3250. doi: 10.1021/acs.biomac.1c00319. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman M.D., Xie C., Zhang X., Benoit D.S. The effect of mesenchymal stem cells delivered via hydrogel-based tissue engineered periosteum on bone allograft healing. Biomaterials. 2013;34:8887–8898. doi: 10.1016/j.biomaterials.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi X., et al. Periosteum-mimetic structures made from freestanding microgrooved nanosheets. Adv. Mater. 2014;26:3290–3296. doi: 10.1002/adma.201305804. [DOI] [PubMed] [Google Scholar]
- 29.Li H., et al. Nanoscaled bionic periosteum orchestrating the osteogenic microenvironment for sequential bone regeneration. ACS Appl. Mater. Interfaces. 2020;12:36823–36836. doi: 10.1021/acsami.0c06906. [DOI] [PubMed] [Google Scholar]
- 30.Shi X., Chen S., Zhao Y., Lai C., Wu H. Enhanced osteogenesis by a biomimic pseudo-periosteum-involved tissue engineering strategy. Adv Healthc Mater. 2013;2:1229–1235. doi: 10.1002/adhm.201300012. [DOI] [PubMed] [Google Scholar]
- 31.Yang G., et al. Bioinspired membrane provides periosteum-mimetic microenvironment for accelerating vascularized bone regeneration. Biomaterials. 2021;268 doi: 10.1016/j.biomaterials.2020.120561. [DOI] [PubMed] [Google Scholar]
- 32.Langlois J.A., et al. Association between insulin-like growth factor I and bone mineral density in older women and men: the Framingham Heart Study. J. Clin. Endocrinol. Metab. 1998;83:4257–4262. doi: 10.1210/jcem.83.12.5308. [DOI] [PubMed] [Google Scholar]
- 33.Crane J.L., Cao X. Function of matrix IGF-1 in coupling bone resorption and formation. J. Mol. Med. (Berl.) 2014;92:107–115. doi: 10.1007/s00109-013-1084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakar S., et al. Serum IGF-1 determines skeletal strength by regulating subperiosteal expansion and trait interactions. J. Bone Miner. Res. 2009;24:1481–1492. doi: 10.1359/JBMR.090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng M., et al. Mechano growth factor E peptide promotes osteoblasts proliferation and bone-defect healing in rabbits. Int. Orthop. 2011;35:1099–1106. doi: 10.1007/s00264-010-1141-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., et al. Mechano-growth factor enhances differentiation of bone marrow-derived mesenchymal stem cells. Biotechnol. Lett. 2015;37:2341–2348. doi: 10.1007/s10529-015-1915-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.