Abstract
One of the most studied defense mechanisms against invading pathogens, including viruses, are Toll-like receptors (TLRs). Among them, TLR3, TLR7, TLR8 and TLR9 detect different forms of viral nucleic acids in endosomal compartments, whereas TLR2 and TLR4 recognize viral structural and nonstructural proteins outside the cell. Although many different TLRs have been shown to be involved in SARS-CoV-2 infection and detection of different structural proteins, most studies have been performed in vitro and the results obtained are rather contradictory. In this study, we report using the unique advantages of the zebrafish model for in vivo imaging and gene editing that the S1 domain of the Spike protein from the Wuhan strain (S1WT) induced hyperinflammation in zebrafish larvae via a Tlr2/Myd88 signaling pathway and independently of interleukin-1β production. In addition, S1WT also triggered emergency myelopoiesis, but in this case through a Tlr2/Myd88-independent signaling pathway. These results shed light on the mechanisms involved in the fish host responses to viral proteins.
Keywords: Spike protein, Inflammation, Hematopoiesis, TLRs, Interleukin-1, Zebrafish
1. Introduction
Viral infections are very complex processes and require research at different interdisciplinary levels to obtain information on basic pathways leading to detailed explanations of the pathogenicity of the virus, host immune response and treatment development. Toll-like receptors (TLRs) are evolutionarily conserved pattern recognition receptors (PRRs) that discriminate between self and non-self by detecting pathogen-associated molecular patterns (PAMPs) and initiate the immune response activating signaling cascades that lead to the production of antimicrobial and proinflammatory molecules (Kawai and Akira, 2011; Lester and Li, 2014). All TLRs belong to type I transmembrane proteins, composed of an amino-terminal leucine-rich repeat-containing ectodomain (responsible for PAMP recognition), a transmembrane domain and cytoplasmic carboxy-terminal Toll-interleukin-1 receptor (IL-1R) homology (TIR) domain (responsible for activation of downstream signal transduction) (Akira and Takeda, 2004), which is also present in the interleukin-1 receptor (IL-1R). TLRs signals via the myeloid differentiation primary response 88 (MYD88) or TIR-domain containing adaptor inducing interferon-β (TRIF) (Akira, 2006; Kawai and Akira, 2010). Importantly, MYD88-dependent pathway can be activated by all TLRs except TLR3, which only signals through TRIF, while TLR4 activates both pathways (Takeda et al., 2003; Yamamoto et al., 2002).
To date there are many well characterized TLRs that have been linked to antiviral immunity. Among them, TLR3, TLR7, TLR8 and TLR9 detect different forms of viral nucleic acids in endosomal compartments, while TLR2 and TLR4 are able to recognize viral structural and nonstructural proteins outside the cell (Bieback et al., 2002; Kurt-Jones et al., 2000). TLR2 is located on the plasma membranes of immune, endothelial, and epithelial cells (Shuang Chen et al., 2007) to recognize mainly components of microbial cell walls and membranes, such as lipoproteins and peptidoglycans. As a heterodimeric receptor, it is paired with TLR1 or TLR6 to recognize different bacterial products such as triacylated lipopeptides (TLR1/TLR2) and diacylated lipopeptides (TLR1/TLR6). In case of viruses, it is assumed that TLR2 is able to recognize enveloped viral particles (Oliveira-Nascimento et al., 2012).
Both extracellular and intracellular TLR family receptors have been shown to play a role in SARS-CoV-2 viral detection. TLR2 recognizes the SARS-CoV-2 envelope protein, resulting in MYD88-dependent inflammation (Zheng et al., 2021). TLR3 is presumed to be critical in the recognition of double stranded RNA (dsRNA) from SARS-CoV-2, which is generated during viral replication, and stimulates endosomal TLR3 in addition to other intracellular receptors (Mabrey et al., 2021). TLR4 signaling by MYD88 and TRIF dependent pathways is proposed to detect viral structural proteins and glycolipids (Choudhury and Mukherjee, 2020; Zhao et al., 2021). Finally, TLR7 has been linked to COVID-19 severity in multiple studies, strongly suggesting a key role for TLR7 in COVID-19 pathogenesis (Mabrey et al., 2021).
Although TLRs are highly conserved during evolution, in fish they show different characteristics than those present in mammals (Zhang et al., 2014). In zebrafish, high expression of TLRs was detected in the skin, which may suggest their important role in the defense against pathogens. It is also worth mentioning that zebrafish has an almost complete set of 20 putative TLR variants, of which 10 have direct human orthologs, including TLR2 (Jault et al., 2004; Meijer et al., 2004). Tlr22 belongs to a fish-specific subfamily and recognizes double stranded RNA (Matsuo et al., 2008), whereas Tlr21 is present in birds, amphibians and fish with similar expression profiles and activity to Tlr9 (Yeh et al., 2013). Moreover, zebrafish show duplication of some mammalian TLRs including Tlr4ba/Tlr4bb for TLR4, andTlr5a/Tlr5b for TLR5, and Tlr8a/Tlr8b for TLR8 (Li et al., 2017). Interestingly, homologs of mammalian TLR6 and TLR10 are absent in fish, while other TLRs, such as Tlr14 and Tlr18, have also been identified. Strikingly, zebrafish Tlr4 paralogs do not recognized lipopolysaccharide and their ligands are still unknown (Sepulcre et al., 2009). In addition, ligands of some zebrafish Tlrs have already been identified, such as lipoproteins, lipopeptides or Pam3CSK4 that can be recognized by the heterodimers of Tlr2 (Yang et al., 2015) and flagellin by Tlr5 (Stockhammer et al., 2009; Yang et al., 2015). In addition, Tlr2-deficient larvae showed impaired neutrophil and macrophage migration to wound (Hu et al., 2021) and high susceptibility to Mycobacterium marinum infection (Hu et al., 2019). Notably, a rhabdoviruses glycoprotein-based mucosal vaccine was found to induce Tlr2-dependent immune responses and protection by activating antigen-presenting cells at local mucosal sites (Zhang et al., 2022). Furthermore, the TLR/IL-1R downstream signaling pathway is also highly conserved in zebrafish including the ortholog of Myd88 among others (Stein et al., 2007).
The zebrafish has been used to study SARS-CoV-2 infection (Tyrkalska et al., 2022a). For example, zebrafish larvae have been exposed to SARS-CoV-2 by bath immersion but, unfortunately, they were not infected and did not show signs of viral replication (Laghi et al., 2022). However, the microinjection into the swim bladder and coelomic cavity of 4 dpf larvae was successful and, although no signs of disease were observed, RNA replication was detected in the swim bladder but not the celomic cavity (Laghi et al., 2022). Other laboratories have injected recombinant domains or fragments of Spike protein of SARS-CoV-2 and found acquired and native immune response in zebrafish larvae and adults (Ventura Fernandes et al., 2022), and even adverse effects, such as olfactory damage and dysfunction.
In this study, we show using the zebrafish model that the hyperinflammation induced by the Spike protein of SARS-CoV-2 is mediated via a Tlr2/Myd88 signaling pathway and independently of Il1b production. In addition, Spike protein-induced emergency myelopoiesis, an inflammation-induced hematopoiesis process to replenish innate immune cells in the periphery, is independent of Tlr2/Myd88 in this model.
2. Methods
2.1. Animals
Zebrafish (Danio rerio H.) were obtained from the Zebrafish International Resource Center and mated, staged, raised and processed as described (Westerfield, 2000). The lines Tg(mpx:eGFP) i114 (Renshaw et al., 2006), Tg(lyz:DsRED2) nz50 (Hall et al., 2007), Tg(mfap4:mCherry-F) ump6 referred to as Tg(mfap4:mCherry) (Phan et al., 2018), Tg(NFkB-RE:eGFP) sh235 referred to as nfkb:eGFP (Kanther et al., 2011), myd88 hu3668/hu3568 mutant (van der Vaart et al., 2012), and casper (mitfa w2/w2 ; mpv17 a9/a9) (White et al., 2008) were previously described. The experiments performed comply with the Guidelines of the European Union Council (Directive 2010/63/EU) and the Spanish RD 53/2013. The experiments and procedures were performed as approved by the Bioethical Committee of the University of Murcia (approval number #669/2020).
2.2. Analysis of gene expression
Total RNA was extracted from whole head/tail part of the zebrafish body with TRIzol reagent (Invitrogen) following the manufacturer's instructions and treated with DNase I, amplification grade (1 U/mg RNA: Invitrogen). SuperScript IV RNase H Reverse Transcriptase (Invitrogen) was used to synthesize first-strand cDNA with random primer from 1 mg of total RNA at 50 °C for 50 min. Real-time PCR was performed with an ABIPRISM 7500 instrument (Applied Biosystems) using SYBR Green PCR Core Reagents (Applied Biosystems). The reaction mixtures were incubated for 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 1 min at 60 °C, and finally 15 s at 95 °C, 1 min 60 °C, and 15 s at 95 °C. For each mRNA, gene expression was normalized to the ribosomal protein S11 (rps11) content in each sample using the Pfaffl method (Pfaffl, 2001). The primers used are shown in Table S1. In all cases, each PCR was performed with samples in triplicate and repeated with at least two independent samples.
2.3. CRISPR and recombinant protein injections, and chemical treatments in zebrafish
crRNA for zebrafish il1b, tlr2 (Table S2) or negative control (Catalog #1072544), and tracrRNA were resuspended in Nuclease-Free Duplex Buffer to 100 μM. One μl of each was mixed and incubated for 5 min at 95 °C for duplexing. After removing from the heat and cooling to room temperature, 1.43 μl of Nuclease-Free Duplex Buffer was added to the duplex, giving a final concentration of 1000 ng/μl. Finally, the injection mixture was prepared by mixing 1 μl of duplex, 2.55 μl of Nuclease-Free Duplex Buffer, 0.25 μl Cas9 Nuclease V3 (IDT, 1081058) and 0.25 μl of phenol red, resulting in final concentrations of 250 ng/μl of gRNA duplex and 500 ng/μl of Cas9. The prepared mix was microinjected into the yolk sac of one-to eight-cell-stage embryos using a microinjector (Narishige) (0.5–1 nl per embryo). The same amounts of gRNA were used in all experimental groups. The efficiency of gRNA was checked by amplifying the target sequence with a specific pair of primers (Table S1) and the TIDE webtool (https://tide.nki.nl/). Embryos injected with crIl1b or crTlr2 were sorted at 2 hpf to choose the ones in the same developmental stage and raised at similar densities. At 24 hpf, the number of dead/alive embryos was determined and within the surviving group the number of embryos with any malformation was scored. At 26 hpf, the number of otic vesicle structures that could fit between the eye and otic vesicle in each larva were estimated. The higher the number of otic vesicles fitted, the lower the level of the larval development (Kimmel et al., 1995).
Recombinant His-tagged Spike S1 wild-type produced in baculovirus-insect cells and with <1.0 EU per μg protein as determined by the LAL method (#40591-V08B1, Sino Biological) or flagellin (Invivogen) at a concentration of 0.25 mg/ml supplemented with phenol red were injected into the hindbrain ventricle (1 nl) of 48 hpf zebrafish larvae.
In some experiments, 24 hpf embryos were treated with 0.3% N-Phenylthiourea (PTU) to inhibit melanogenesis.
2.4. Caspase-1 activity assays
Caspase-1 activity was determined with the fluorometric substrate Z-YVAD 7-Amido-4-trifluoromethylcoumarin (Z-YVAD-AFC, caspase-1 substrate VI, Calbiochem) as described previously (Angosto et al., 2012; Lopez-Castejon et al., 2008). A representative graph of caspase-1 activity of three repeats is shown in the figures.
2.5. In vivo imaging
To study immune cell recruitment to the injection site and Nfkb activation, 2 dpf mpx:eGFP, mfap4:mcherry or nfkb:egfp larvae were anaesthetized in embryonic medium with 0.16 mg/ml buffered tricaine. Images of the hindbrain, head or whole-body area were taken 3, 6, 12 and 24 h post-injection (hpi) using a Leica MZ16F fluorescence stereomicroscope. The number of neutrophils or macrophages was determined by visual counting and the fluorescence intensity was obtained and analyzed with ImageJ (FIJI) software (Schindelin et al., 2012).
Neutral red stains zebrafish macrophage granules and the procedure was performed as originally reported (Herbomel et al., 2001). Briefly, macrophage staining was performed on live 3 dpf larvae and was obtained by incubating the embryos in 2.5 g/ml of neutral red (in embryonic medium) at 25–30 °C in the dark for 5–8 h. The larvae were anaesthetized in 0.16 mg/ml buffered tricaine and imaged using a Leica MZ16F fluorescence stereo microscope.
In all experiments, images were pooled from at least 3 independent experiments performed by two people and using blinded samples.
2.6. Statistical analysis
Data are shown as mean ± s.e.m. and were analyzed by analysis of variance and a Tukey multiple range test to determine differences between groups. The differences between two samples were analyzed by Student's t-test.
3. Results
3.1. Myd88 is required for hyperinflammation but dispensable for emergency myelopoiesis induced by S1WT
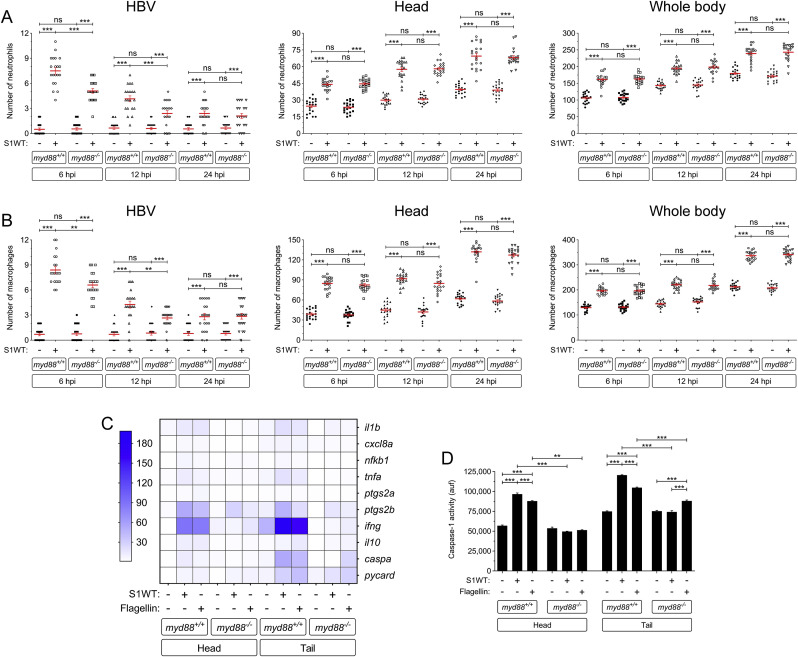
As the specific TLRs activated by the spike protein of SARS-CoV-2 are controversial, either recombinant S1 from the Wuhan strain (S1WT) or flagellin (positive control that activates Tlr5) were injected into the hindbrain of Myd88-deficient zebrafish and the recruitment to the injection site and the total number of neutrophils and macrophages in the head and the whole body were analyzed at 6, 12 and 24 hpi . Although robust recruitment of both neutrophils and macrophages was observed in wild type larvae, a significantly lower recruitment of both immune cells was observed in Myd88-deficient larvae at 6 and 12 hpi (Fig. 1 A and B). In contrast, no differences in total number of neutrophils and macrophages in the head or whole body were observed between mutant and wild type larvae at any timepoint (Fig. 1A and B). These results suggest that S1WT-induced emergency myelopoiesis is Myd88-independent and, therefore, depends exclusively on the inflammasome (Tyrkalska et al., 2022b).
Fig. 1.
Myd88 is required for hyperinflammation but dispensable for emergency myelopoiesis induced by S1WT. Recombinant S1WT (+) or vehicle (−) were injected in the hindbrain ventricle (HBV) of 2 dpf Myd88-deficient Tg(mpx:eGFP) (A) or wild type (B–D) larvae. Neutrophil (A) and macrophage (neutral red positive cells) (B) recruitment and number were analyzed at 6, 12 and 24 hpi by fluorescence (A) or brightfield (B) microscopy, the transcript levels of the indicated genes were analyzed at 12 hpi by RT-qPCR in larval head and tail (C), and caspase-1 activity was determined at 24 hpi using a fluorogenic substrate (D). Each dot represents one individual and the mean ± S.E.M. for each group is also shown. P values were calculated using one-way ANOVA and Tukey multiple range test. RT-qPCR data are depicted as a heat map in C with higher expression shown in darker color. ns, not significant, **p ≤ 0.01, ***p ≤ 0.001. auf, arbitrary units of fluorescence.
To further analyze the impact of Myd88 in the local and systemic inflammation induced by S1WT, samples were collected at 12 hpi from heads and the rest of the body for RT-qPCR analysis. Although flagellin and S1WT induced similar gene expression patterns, Myd88 deficiency impaired the induction of transcript levels of genes encoding the inflammatory mediators Il1b, Cxcl8a, Nfkb1, Tnfa, Ptgs2a, Ptgs2b, Infg and Il10 by both S1WT and flagellin (Fig. 1C and Fig. S1A-S1H). Curiously, mRNA levels of genes encoding the canonical inflammasome effector Caspa also appear to be Myd88 dependent, as S1WT failed to induce their expression in Myd88-deficient larvae, while transcript levels of the gene encoding the inflammasome adaptor Asc (pycard gene) were largely unaffected (Fig. 1C, Fig. S1I and S1J). Similarly, Myd88 deficiency impaired the induction of caspase-1 activity by S1WT and flagellin locally and systemically. These results suggest that Myd88 plays a key role in the hyperinflammation induced by S1WT in zebrafish.
3.2. Il1b signaling is not involved in S1WT-induced hyperinflammation in zebrafish
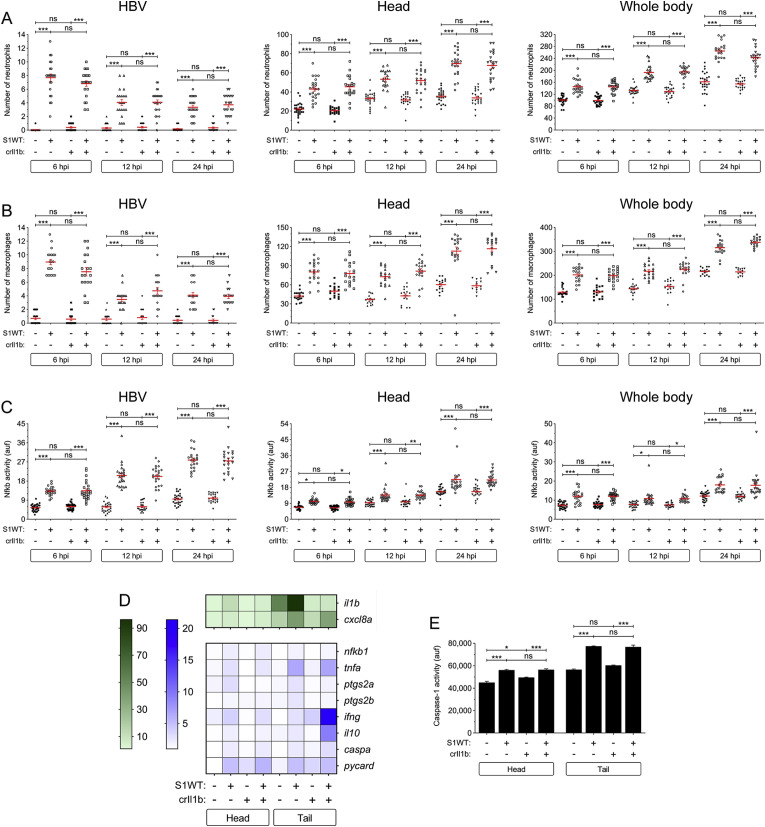
To learn if the hyperinflammation induced by S1WT was dependent of Tlr signaling, we knocked down Il1b, as its receptor also signals through Myd88. For this purpose, we used a specific guide RNA that provided 70% of efficiency and showed neither toxicity nor malformations in embryos (Fig. S2A and S3A-S3C). The results showed that Il1b-deficiency failed to affect S1WT-induced neutrophil and macrophage recruitment, neutrophilia and monocytosis (Fig. 2 A and B), and Nfkb activity (Fig. 2C). In addition, the transcript levels of genes encoding major inflammatory molecules were similarly induced by S1WT in wild type and Il1b-deficient larvae, apart from those of il1b itself which were drastically decreased and those of ptgs2a and ptgs2b which were weakly decreased (Fig. 2D and S4A-S4F). Curiously, i110 and infg mRNA levels were systemically higher in Il1b-deficient larvae than in their wild type siblings (Fig. 2D, S4G and S4H), while those of genes encoding the inflammasome components Caspa and Pycard, and caspase-1 activity were rather similar in Il1b-deficient and wild type larvae (Fig. 2D and E, S4I and S4J). These results taken together confirmed the impact of the genetic inhibition of il1b gene and that S1WT-induced hyperinflammation is largely Il1b-independent.
Fig. 2.
Il1b signaling is not involved in S1WT-induced hyperinflammation in zebrafish. One-cell stage zebrafish eggs of Tg(lyz:dsRED2) (A), Tg(mfap4:mCherry) (B), Tg(NFkB-RE:eGFP) (C) and wild type (D, E) were microinjected with control or il1b crRNA/Cas9 complexes. At 2 dpf, recombinant S1WT (+) or vehicle (−) were injected in the hindbrain ventricle (HBV) of control and Il1b-deficient larvae. Neutrophil (A) and macrophage (B) recruitment and number, and Nfkb activation (C) were analyzed at 6, 12 and 24 hpi by fluorescence microscopy, the transcript levels of the indicated genes were analyzed at 12 hpi by RT-qPCR (D), and caspase-1 activity was determined at 24 hpi using a fluorogenic substrate (E). Each dot represents one individual and the mean ± S.E.M. for each group is also shown. RT-qPCR data are depicted as a heat map in D with higher expression shown in darker color. P values were calculated using one-way ANOVA and Tukey multiple range test. ns, not significant, *≤p0.05, **p ≤ 0.01, ***p ≤ 0.001. auf, arbitrary units of fluorescence.
3.3. Tlr2 mediates the S1WT-induced hyperinflammation in zebrafish
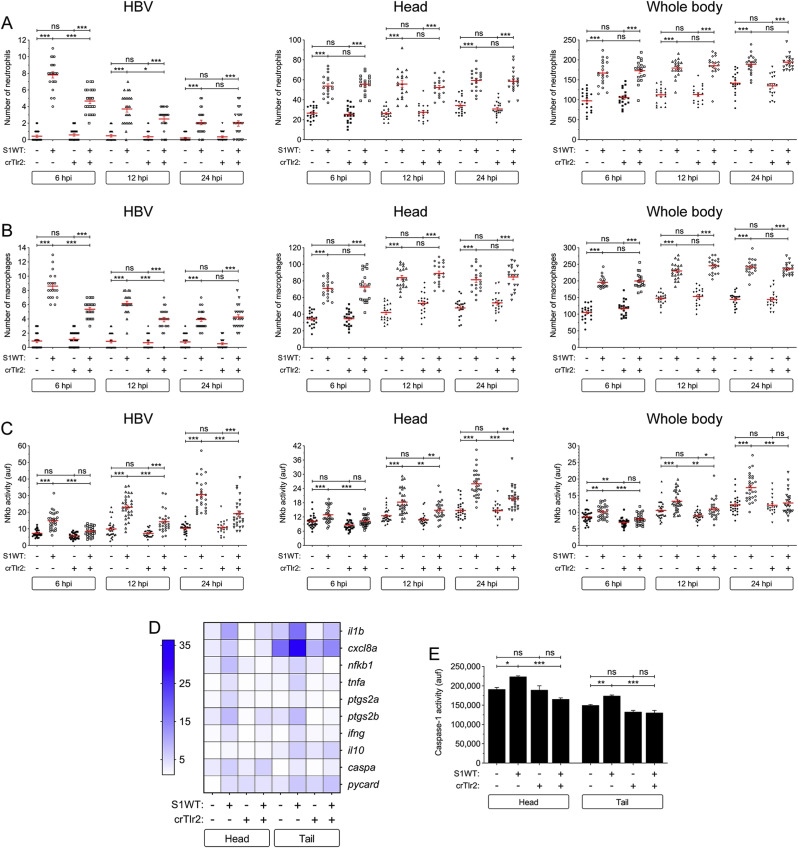
Since Myd88 acts downstream of almost all TLRs apart from TLR3 (Takeda et al., 2003; Yamamoto et al., 2002), we decided to check the expression levels of the orthologs of the TLRs shown to be involved in the responses to SARS-CoV-2, namely tlr2, tlr4ba, tlr4bb and tlr7, and tlr3 as a negative control, upon S1WT hindbrain injection. Notably, only the transcript levels of tlr2 increased after S1WT injection (Figs. S5A–E), suggesting S1WT signals via Tlr2 in zebrafish. We then knocked down Tlr2 using a specific guide RNA that resulted in approximately 70% efficiency (Fig. S2B) and showed no detrimental effect in larval development (Figs. S3D–S3F). We found that Tlr2 deficiency did not affect S1WT-induced neutrophilia and monocytosis at any of the times tested (Figs. S3A and 3B), further confirming that S1WT-induced emergency myelopoiesis is Tlr2/Myd88 independent. However, neutrophil and macrophage recruitment at the S1WT injection site was partially impaired in Tlr2-deficient larvae at 6 and 12 hpi (Fig. 3 A and B). Moreover, Tlr2-deficient larvae also showed lower Nfkb activity than wild type larvae not only at the site of the injection but also in the head and the whole body at all analyzed timepoints (Fig. 3C).
Fig. 3.
Tlr2 mediates the S1WT-induced hyperinflammation in zebrafish. One-cell stage zebrafish eggs of Tg(lyz:dsRED2) (A), Tg(mfap4:mCherry) (B), Tg(NFkB-RE:eGFP) (C) and wild type (D, E) were microinjected with control or tlr2 crRNA/Cas9 complexes. At 2 dpf, recombinant S1WT (+) or vehicle (−) were injected in the hindbrain ventricle (HBV) of control and Tlr2-deficient larvae. Neutrophil (A) and macrophage (B) recruitment and number, and Nfkb activation (C) were analyzed at 6, 12 and 24 hpi by fluorescence microscopy, the transcript levels of the indicated genes were analyzed at 12 hpi by RT-qPCR (D), and caspase-1 activity was determined at 24 hpi using a fluorogenic substrate (E). Each dot represents one individual and the mean ± S.E.M. for each group is also shown. RT-qPCR data are depicted as a heat map in D with higher expression shown in darker color. P values were calculated using one-way ANOVA and Tukey multiple range test. ns, not significant, *≤p0.05, **p ≤ 0.01, ***p ≤ 0.001. auf, arbitrary units of fluorescence.
The above results were then confirmed by RT-qPCR. Thus, the transcript levels of tlr2 significantly decreased in Tlr2-deficient animals, further confirming the high efficiency of the crRNA used (Fig. S5A). Furthermore, the transcript levels of il1b, cxcl8a, nfkb1, tnfa, ptgs2a, ptgs2b and ifng were lower locally and systemically in the S1WT injected Tlr2-deficient larvae than in their wild type siblings (Fig. 3D and S6A-S6G). However, no significant differences were observed in the mRNA levels of genes encoding anti-inflammatory I110 and the inflammasome components Caspa and Pycard (Fig. 3H-J). Surprisingly, the induction of caspase-1 by S1WT was also attenuated in Tlr2-deficient larvae locally and systemically (Fig. 3E).
4. Discussion
Since the beginning of the COVID-19 pandemic, scientists are trying to find the molecular basis of SARS-CoV-2 virulence and host immune responses to find targeted treatments to moderate the severe symptoms of the disease and save people's lives. Although both TLRs and the inflammasome pathways have been found to be involved in COVID-19, a mechanistic understanding of their involvement in COVID-19 progression is still unclear (Khanmohammadi and Rezaei, 2021; Vora et al., 2021). It has been suggested that the imbalance between the generation of excessive inflammation through TLR/MyD88 pathway and IFN-β/TRIF pathway plays a key role in COVID-19 severity (Mabrey et al., 2021). Computer-based modelling has found that the S protein of SARS-CoV-2 is predicted to bind to TLR4 (Choudhury and Mukherjee, 2020) and, more interestingly, SARS-CoV-2 S1 protein engaged TLR4 and strongly activated the inflammatory response leading to the production of pro-inflammatory mediators through nuclear factor κB (NF-κB) and stress-activated mitogen-activated protein kinase (MAPK) signaling pathways (Shirato and Kizaki, 2021; Zhao et al., 2021). Moreover, it was suggested that SARS-CoV-2 S glycoprotein binds and activates TLR4, leading to increased cell surface expression of ACE2 which, in turn, would facilitate viral entry and cause the COVID-19-associated cytokine storm syndrome (CSS) (Aboudounya and Heads, 2021). Similarly, it has been proposed that the TLR2 signaling pathway is activated following SARS-CoV-2 infection, resulting in strong production of proinflammatory cytokines, suggesting that it may contribute to the severity of COVID-19 (Sariol and Perlman, 2021). TLR2 is known to form heterodimers with TLR1 and TLR6, which increases its ligand diversity and allows detection of different kinds of pathogens, including viruses (Oliveira-Nascimento et al., 2012). Recently, the envelope protein (E) of SARS-CoV-2 has been found to be a ligand of human and mouse TLR2 and, surprisingly, plays a critical role in COVID-19-associated CSS in the K18-hACE2 transgenic mice model (Zheng et al., 2021). However, this study has reported that S1+S2 of SARS-CoV-2 failed to activate mouse macrophages and human PBMCs (Zheng et al., 2021). In stark contrast, another study found that TLR2 recognizes the SARS-CoV-2 S protein and then dimerizes with TLR1 or TLR6 to activate the NF-κB pathway and promote CSS (Khan et al., 2021).
In the present study, we used a newly established zebrafish model of COVID-19 (Tyrkalska et al., 2022b) based on the larval hindbrain injection of SARS-CoV-2 S1 protein to further understand the contribution of Tlr2 and Myd88 signaling pathway to the COVID-19-associated CSS. Genetic experiments demonstrated that Tlr2 sensed S1WT and induced hyperinflammation via Myd88 in zebrafish, as shown in mice (Khan et al., 2021). Furthermore, this model has also revealed: (i) S1WT-induced hyperinflammation is independent of the production of Il1b and (ii) S1WT-induced emergency myelopoiesis is Tlr2/Myd88-independent. On the one hand, the relevance of IL1B in the COVID-19-associated CSS is unclear, as a large controlled trial in hospitalized patients with COVID-19 found no therapeutic benefit of IL-1 blockade (Declercq et al., 2021), whereas another trial of early treatment of COVID-19 patients with anakinra, to block IL-1, found decreased patient severity and improved survival (Kyriazopoulou et al., 2021). On the other hand, although the relevance of emergency hematopoiesis in COVID-19 has been recognized, it has been less studied than the CSS. Thus, multi-omic single-cell immune profiling of COVID-19 patients has revealed that emergency myelopoiesis is a prominent feature of fatal COVID-19 (Wilk et al., 2021). In addition, COVID-19 patients in intensive care also show low levels of hemoglobin and circulating nucleated red cells, and erythroid progenitors can be infected by SARS-CoV-2 via ACE2 (Huerga Encabo et al., 2021). The zebrafish model is excellent for further understanding the role of altered hematopoiesis in COVID-19 and other viral diseases. Thus, although our results point to the relevance of the inflammasome in S1-driven emergency myelopoiesis and dispensability of Tlr, the decreased caspase-1 activity levels in Tlr2-and Myd88-deficient larvae injected with S1 suggest a crosstalk between these two pivotal inflammatory pathways.
In summary, the zebrafish model has demonstrated that while both Tlr2/Myd88 and inflammasome signaling pathways are required for the induction of hyperinflammation by the S protein of SARS-CoV-2, the Tlr2/Myd88 axis is dispensable for S protein-induced emergency myelopoiesis.
Funding
This work has been funded by Fundación Séneca, CARM, Spain (research grants 20793/PI/18 to VM and 00006/COVI/20 to VM and MLC), Saavedra Fajardo (postdoctoral contract to SC), the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie (grant agreement No.955576 – INFLANET), Spanish Ministry of Science and Innovation (Juan de la Cierva-Incorporación postdoctoral contract to SDT), co-funded with European Regional Development Funds and ZEBER funded by Consejería de Salud-CARM (postdoctoral contract to AM-L). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
SDT, VM and MLC conceived the study; SDT, AM-L, AP and SC performed the research; SDT, AM-L, AP, SC, MLC and VM analyzed the data; and SDT and VM wrote the manuscript with minor contributions from other authors.
Availability of data and material
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Declaration of competing interest
The authors declare no competing interest.
Acknowledgments
We thank I. Fuentes and P. Martinez for their excellent technical assistance, and Profs. Tobin, Crosier, Renshaw, Zon, Meijer and Lutfalla for the zebrafish lines.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dci.2022.104626.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- Aboudounya M.M., Heads R.J. COVID-19 and toll-like receptor 4 (TLR4): SARS-CoV-2 may bind and activate TLR4 to increase ACE2 expression, facilitating entry and causing hyperinflammation. Mediat. Inflamm. 2021;2021 doi: 10.1155/2021/8874339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S. TLR signaling. Curr. Top. Microbiol. Immunol. 2006;311:1–16. doi: 10.1007/3-540-32636-7_1. [DOI] [PubMed] [Google Scholar]
- Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Angosto D., López-Castejón G., López-Muñoz A., Sepulcre M.P., Arizcun M., Meseguer J., Mulero V. Evolution of inflammasome functions in vertebrates: inflammasome and caspase-1 trigger fish macrophage cell death but are dispensable for the processing of IL-1β. Innate Immun. 2012;18:815–824. doi: 10.1177/1753425912441956. [DOI] [PubMed] [Google Scholar]
- Bieback K., Lien E., Klagge I.M., Avota E., Schneider-Schaulies J., Duprex W.P., Wagner H., Kirschning C.J., Ter Meulen V., Schneider-Schaulies S. Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J. Virol. 2002;76:8729–8736. doi: 10.1128/JVI.76.17.8729-8736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Shuang, Wong M.H., Schulte D.J., Arditi M., Michelsen K.S. Differential expression of Toll-like receptor 2 (TLR2) and responses to TLR2 ligands between human and murine vascular endothelial cells. J. Endotoxin Res. 2007;13:281–296. doi: 10.1177/0968051907085096. [DOI] [PubMed] [Google Scholar]
- Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020;92:2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declercq J., Van Damme K.F.A., De Leeuw E., Maes B., Bosteels C., Tavernier S.J., De Buyser S., Colman R., Hites M., Verschelden G., Fivez T., Moerman F., Demedts I.K., Dauby N., De Schryver N., Govaerts E., Vandecasteele S.J., Van Laethem J., Anguille S., van der Hilst J., Misset B., Slabbynck H., Wittebole X., Lienart F., Legrand C., Buyse M., Stevens D., Bauters F., Seys L.J.M., Aegerter H., Smole U., Bosteels V., Hoste L., Naesens L., Haerynck F., Vandekerckhove L., Depuydt P., van Braeckel E., Rottey S., Peene I., Van Der Straeten C., Hulstaert F., Lambrecht B.N. Effect of anti-interleukin drugs in patients with COVID-19 and signs of cytokine release syndrome (COV-AID): a factorial, randomised, controlled trial. Lancet Respir. Med. 2021;9:1427–1438. doi: 10.1016/S2213-2600(21)00377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C., Flores M.V., Storm T., Crosier K., Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Thisse B., Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 2001;238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- Hu W., Yang S., Shimada Y., Munch M., Marin-Juez R., Meijer A.H., Spaink H.P. Infection and RNA-seq analysis of a zebrafish tlr2 mutant shows a broad function of this toll-like receptor in transcriptional and metabolic control and defense to Mycobacterium marinum infection. BMC Genom. 2019;20:878. doi: 10.1186/s12864-019-6265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., van Steijn L., Li C., Verbeek F.J., Cao L., Merks R.M.H., Spaink H.P. A novel function of TLR2 and MyD88 in the regulation of leukocyte cell migration behavior during wounding in zebrafish larvae. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.624571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerga Encabo H., Grey W., Garcia-Albornoz M., Wood H., Ulferts R., Aramburu I.V., Kulasekararaj A.G., Mufti G., Papayannopoulos V., Beale R., Bonnet D. Human erythroid progenitors are directly infected by SARS-CoV-2: implications for emerging erythropoiesis in severe COVID-19 patients. Stem Cell Rep. 2021;16:428–436. doi: 10.1016/j.stemcr.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jault C., Pichon L., Chluba J. Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Mol. Immunol. 2004;40:759–771. doi: 10.1016/j.molimm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Kanther M., Sun X., Mühlbauer M., Mackey L.C., Flynn E.J., Bagnat M., Jobin C., Rawls J.F. Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology. 2011;141:197–207. doi: 10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Khan S., Shafiei M.S., Longoria C., Schoggins J.W., Savani R.C., Zaki H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. Elife. 2021;10 doi: 10.7554/eLife.68563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanmohammadi S., Rezaei N. Role of Toll-like receptors in the pathogenesis of COVID-19. J. Med. Virol. 2021;93:2735–2739. doi: 10.1002/jmv.26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dynam. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kurt-Jones E.A., Popova L., Kwinn L., Haynes L.M., Jones L.P., Tripp R.A., Walsh E.E., Freeman M.W., Golenbock D.T., Anderson L.J., Finberg R.W. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- Kyriazopoulou E., Poulakou G., Milionis H., Metallidis S., Adamis G., Tsiakos K., Fragkou A., Rapti A., Damoulari C., Fantoni M., Kalomenidis I., Chrysos G., Angheben A., Kainis I., Alexiou Z., Castelli F., Serino F.S., Tsilika M., Bakakos P., Nicastri E., Tzavara V., Kostis E., Dagna L., Koufargyris P., Dimakou K., Savvanis S., Tzatzagou G., Chini M., Cavalli G., Bassetti M., Katrini K., Kotsis V., Tsoukalas G., Selmi C., Bliziotis I., Samarkos M., Doumas M., Ktena S., Masgala A., Papanikolaou I., Kosmidou M., Myrodia D.M., Argyraki A., Cardellino C.S., Koliakou K., Katsigianni E.I., Rapti V., Giannitsioti E., Cingolani A., Micha S., Akinosoglou K., Liatsis-Douvitsas O., Symbardi S., Gatselis N., Mouktaroudi M., Ippolito G., Florou E., Kotsaki A., Netea M.G., Eugen-Olsen J., Kyprianou M., Panagopoulos P., Dalekos G.N., Giamarellos-Bourboulis E.J. Early treatment of COVID-19 with anakinra guided by soluble urokinase plasminogen receptor plasma levels: a double-blind, randomized controlled phase 3 trial. Nat. Med. 2021;27:1752–1760. doi: 10.1038/s41591-021-01499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laghi V., Rezelj V., Boucontet L., Fretaud M., Da Costa B., Boudinot P., Salinas I., Lutfalla G., Vignuzzi M., Levraud J.P. Exploring zebrafish larvae as a COVID-19 model: probable abortive SARS-CoV-2 replication in the swim bladder. Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.790851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester S.N., Li K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014;426:1246–1264. doi: 10.1016/j.jmb.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cao X., Jin X., Jin T. Pattern recognition receptors in zebrafish provide functional and evolutionary insight into innate immune signaling pathways. Cell. Mol. Immunol. 2017;14:80–89. doi: 10.1038/cmi.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Castejon G., Sepulcre M.P., Mulero I., Pelegrin P., Meseguer J., Mulero V. Molecular and functional characterization of gilthead seabream Sparus aurata caspase-1: the first identification of an inflammatory caspase in fish. Mol. Immunol. 2008;45:49–57. doi: 10.1016/j.molimm.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Mabrey F.L., Morrell E.D., Wurfel M.M. TLRs in COVID-19: how they drive immunopathology and the rationale for modulation. Innate Immun. 2021;27:503–513. doi: 10.1177/17534259211051364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo A., Oshiumi H., Tsujita T., Mitani H., Kasai H., Yoshimizu M., Matsumoto M., Seya T. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J. Immunol. 2008;181:3474–3485. doi: 10.4049/jimmunol.181.5.3474. [DOI] [PubMed] [Google Scholar]
- Meijer A.H., Gabby Krens S.F., Medina Rodriguez I.A., He S., Bitter W., Ewa Snaar-Jagalska B., Spaink H.P. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol. Immunol. 2004;40:773–783. doi: 10.1016/j.molimm.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Oliveira-Nascimento L., Massari P., Wetzler L.M. The role of TLR2 in infection and immunity. Front. Immunol. 2012;3:79. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan Q.T., Sipka T., Gonzalez C., Levraud J.P., Lutfalla G., Nguyen-Chi M. Neutrophils use superoxide to control bacterial infection at a distance. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw S.A., Loynes C.A., Trushell D.M., Elworthy S., Ingham P.W., Whyte M.K. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- Sariol A., Perlman S. SARS-CoV-2 takes its Toll. Nat. Immunol. 2021;22:801–802. doi: 10.1038/s41590-021-00962-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre M.P., Alcaraz-Perez F., Lopez-Munoz A., Roca F.J., Meseguer J., Cayuela M.L., Mulero V. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J. Immunol. 2009;182:1836–1845. doi: 10.4049/jimmunol.0801755. [DOI] [PubMed] [Google Scholar]
- Shirato K., Kizaki T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein C., Caccamo M., Laird G., Leptin M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 2007;8:R251. doi: 10.1186/gb-2007-8-11-r251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockhammer O.W., Zakrzewska A., Hegedus Z., Spaink H.P., Meijer A.H. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J. Immunol. 2009;182:5641–5653. doi: 10.4049/jimmunol.0900082. [DOI] [PubMed] [Google Scholar]
- Takeda K., Kaisho T., Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- Tyrkalska S.D., Candel S., Pedoto A., Garcia-Moreno D., Alcaraz-Perez F., Sanchez-Ferrer A., Cayuela M.L., Mulero V. Vol. 2. FEMS Microbiol Rev; 2022. p. fuac042. (Zebrafish Models of COVID-19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrkalska S.D., Martinez-Lopez A., Arroyo A.B., Martinez-Morcillo F.J., Candel S., Garcia-Moreno D., Mesa-Del-Castillo P., Cayuela M.L., Mulero V. Differential proinflammatory activities of Spike proteins of SARS-CoV-2 variants of concern. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart M., Spaink H.P., Meijer A.H. Pathogen recognition and activation of the innate immune response in zebrafish. Adv Hematol. 2012;2012 doi: 10.1155/2012/159807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura Fernandes B.H., Feitosa N.M., Barbosa A.P., Bomfim C.G., Garnique A.M.B., Rosa I.F., Rodrigues M.S., Doretto L.B., Costa D.F., Camargo-Dos-Santos B., Franco G.A., Neto J.F., Lunardi J.S., Bellot M.S., Alves N.P.C., Costa C.C., Aracati M.F., Rodrigues L.F., Cirilo R.H., Colagrande R.M., Gomes F.I.F., Nakajima R.T., Belo M.A.A., Giaquinto P.C., de Oliveira S.L., Eto S.F., Fernandes D.C., Manrique W.G., Conde G., Rosales R.R.C., Todeschini I., Rivero I., Llontop E., Sgro G.G., Oka G.U., Bueno N.F., Ferraris F.K., de Magalhães M.T.Q., Medeiros R.J., Mendonça-Gomes J.M., Junqueira M.S., Conceição K., Pontes L.G., Condino-Neto A., Perez A.C., Barcellos L.J.G., Júnior J.D.C., Dorlass E.G., Camara N.O.S., Durigon E.L., Cunha F.Q., Nóbrega R.H., Machado-Santelli G.M., Farah C.S., Veras F.P., Galindo-Villegas J., Costa-Lotufo L.V., Cunha T.M., Chammas R., Carvalho L.R., Guzzo C.R., Malafaia G., Charlie-Silva I. Toxicity of spike fragments SARS-CoV-2 S protein for zebrafish: a tool to study its hazardous for human health? Sci. Total Environ. 2022;813 doi: 10.1016/j.scitotenv.2021.152345. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Vora S.M., Lieberman J., Wu H. Inflammasome activation at the crux of severe COVID-19. Nat. Rev. Immunol. 2021;21:694–703. doi: 10.1038/s41577-021-00588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. University of Oregon Press.; Eugene, OR: 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish Danio* (Brachydanio) Rerio. [Google Scholar]
- White R.M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E., Zon L.I. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Lee M.J., Wei B., Parks B., Pi R., Martinez-Colon G.J., Ranganath T., Zhao N.Q., Taylor S., Becker W., Stanford C.-B., Jimenez-Morales D., Blomkalns A.L., O'Hara R., Ashley E.A., Nadeau K.C., Yang S., Holmes S., Rabinovitch M., Rogers A.J., Greenleaf W.J., Blish C.A. Multi-omic profiling reveals widespread dysregulation of innate immunity and hematopoiesis in COVID-19. J. Exp. Med. 2021;218 doi: 10.1084/jem.20210582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., Akira S. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 2002;169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- Yang S., Marin-Juez R., Meijer A.H., Spaink H.P. Common and specific downstream signaling targets controlled by Tlr2 and Tlr5 innate immune signaling in zebrafish. BMC Genom. 2015;16:547. doi: 10.1186/s12864-015-1740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh D.W., Liu Y.L., Lo Y.C., Yuh C.H., Yu G.Y., Lo J.F., Luo Y., Xiang R., Chuang T.H. Toll-like receptor 9 and 21 have different ligand recognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20711–20716. doi: 10.1073/pnas.1305273110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kong X., Zhou C., Li L., Nie G., Li X. Toll-like receptor recognition of bacteria in fish: ligand specificity and signal pathways. Fish Shellfish Immunol. 2014;41:380–388. doi: 10.1016/j.fsi.2014.09.022. [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhao Z., Zhang P.Q., Guo S., Zhu B. TLR2-mediated mucosal immune priming boosts anti-rhabdoviral immunity in early vertebrates. Antivir. Res. 2022;203 doi: 10.1016/j.antiviral.2022.105346. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Kuang M., Li J., Zhu L., Jia Z., Guo X., Hu Y., Kong J., Yin H., Wang X., You F. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021;31:818–820. doi: 10.1038/s41422-021-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Karki R., Williams E.P., Yang D., Fitzpatrick E., Vogel P., Jonsson C.B., Kanneganti T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021;22:829–838. doi: 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Data will be made available on request.