Abstract
Many modifiable and non-modifiable risk factors have been associated with hypertension. However, current screening programs are still failing in identifying individuals at higher risk of hypertension. Given the major impact of high blood pressure on cardiovascular events and mortality, there is an urgent need to find new strategies to improve hypertension detection. We aimed to explore whether a machine learning (ML) algorithm can help identifying individuals predictors of hypertension. We analysed the data set generated by the questionnaires administered during the World Hypertension Day from 2015 to 2019. A total of 20206 individuals have been included for analysis. We tested five ML algorithms, exploiting different balancing techniques. Moreover, we computed the performance of the medical protocol currently adopted in the screening programs. Results show that a gain of sensitivity reflects in a loss of specificity, bringing to a scenario where there is not an algorithm and a configuration which properly outperforms against the others. However, Random Forest provides interesting performances (0.818 sensitivity – 0.629 specificity) compared with medical protocols (0.906 sensitivity – 0.230 specificity). Detection of hypertension at a population level still remains challenging and a machine learning approach could help in making screening programs more precise and cost effective, when based on accurate data collection. More studies are needed to identify new features to be acquired and to further improve the performances of ML models.
Keywords: Hypertension, Data analysis, Prevention
Introduction
Arterial hypertension still remains the most important modifiable risk factor for cardiovascular disease worldwide. Despite extensive knowledge about ways to prevent and treat hypertension, the global incidence and prevalence of hypertension and its cardiovascular complications are still elevated mainly due to inadequacies in prevention, detection and control [1, 2]. The high variability characterising blood pressure (BP) values, together with the lack of specific symptoms of this condition, make the detection of hypertension still challenging.
Since 2005, the World Hypertension League has been leading a global campaign to raise awareness of the importance of hypertension through annual screening programs. During the 2018 survey, among 502079 participants found to have hypertension, only 59.5% were aware of having such condition [3]. This evidence confirms that current screening programs conducted by the National Health Services are still failing in detecting appropriately hypertension. Therefore, there is an urgent need to find new strategies to improve hypertension detection at a population level, given that identifying risk factors for hypertension may facilitate earlier interventions, aimed at preventing future development of hypertension, early detecting its appearance and reducing the incidence of its long-term consequences.
In the recent years, artificial intelligence (AI), which includes all computer systems able to perform tasks normally requiring human intelligence, has been successfully applied to healthcare and shown to be a valid tool in managing different clinical conditions [4, 5].
The Italian Society of Hypertension (SIIA) conducts every year a national campaign to increase awareness of the importance of high BP detection. Over the years, several questionnaires related to hypertension have been administered, generating a large dataset. In particular, 37110 individuals participated in the World Hypertension Day campaigns from 2015 to 2019. From the initial dataset, 20206 subjects have been selected for the present study, after removing those with high BP already diagnosed, out of which 4192 (20.75%) with newly discovered hypertension. Data include demographics, risk factor information, questions about general knowledge on hypertension and three measures of systolic and diastolic BP and heart rate. The aim was to apply supervised machine learning (ML) algorithms to such a large dataset in order to find a model capable of detecting unknown hypertension, and comparing their performances with the current screening protocols. Data were split into a training set with 14144 records and a validation set with 6062 samples. We used five ML models – logistic regression, decision tree, random forest, support vector machine and XGBoost – trained performing a 10-fold cross-validation process on the original training set in the first place, and then applying both oversampling and undersampling techniques for managing the imbalanced nature of data.
The performance of the models was evaluated estimating the sensitivity, specificity, accuracy and precision of the different trained models. Results show that, among different ML algorithms and different balancing techniques exploited, there is not an algorithm and a configuration which properly outperforms against the others, even though Random Forest is the most promising one in all the three schemes. The undersampling experiments are those that provide highest sensitivity scores, but a gain in sensitivity often reflects in poor sensitivity and overall accuracy. In particular, the model that best performs in terms of sensitivity was obtained with XGBoost under undersampling scheme, which however showed poor specificity and, consequently, overall low accuracy. The best compromise is given by Random Forest in the undersampling experiment, that provides sensitivity of 0.818, specificity of 0.629, an overall accuracy of 0.681 and AUC of 0.816.
Medical protocols result in a high sensitivity of 0.906 but have a poor specificity of 0.230, thus including in the screening program subjects that likely will not develop hypertension. This result highlights that the current protocols are not optimised and that novel strategies are needed to avoid unnecessary expanses and to reach a larger population.
Future studies should develop and apply new techniques and algorithms with the goal to improve the model performances, possibly evaluating how accurate data acquisition could be enriched with additional new information that may support the automatic prediction of hypertension occurrence.
Related work
In recent years, AI has been successfully applied to healthcare as a valid medical tool in different clinical conditions [4, 5]. ML in particular is designed for performing high accuracy predictions on individuals’ outcome without explicit programming but based on learning patterns from acquired data.
Different ML algorithms have been applied in the field of hypertension with very heterogeneous results [6–10]. The review by Martinez-Ríos et al. [11] provides a comprehensive analysis of the literature in the field. The conclusion was that ML has proven to be useful in classifying hypertensive subjects, even though there is not a general agreement on which algorithms perform better, which metrics must be measured to evaluate the model and, mostly, which type of data and features must be acquired to replicate the studies and possibly train predictors that outperform the clinical protocols currently adopted.
To be effective, predictions should be based on data that are accurately, easily and massively collected to screen a population whose size is as large as possible. For instance, predictive models based on genetic data make a mass screening not feasible [7]. Promising results are presented in [10] where four ML models were evaluated on 11 easy-to-collect variables, risk factors (such as smoking, drinking, family history) and anthropometric data. However, data were all acquired in the same local hospital, making results possibly not generalisable to individuals living in other areas.
Materials and methods
Study subjects
Data were collected from questionnaires administered during the World Hypertension Day from 2015 to 2019. Individuals willing to participate were asked to fill in an anonymous questionnaire and had their BP measured according to the European Society of Hypertension (ESH) standards (3 consecutive BP readings were performed by trained health personnel with validated automated devices after 5 min rest) [3].
Demographic information (age, sex, BMI), self-reported information on cardio- vascular risk factors (hypertension, diabetes, smoking, high cholesterol, kidney disease, family history of cardiovascular diseases), sleep complaints (snoring, witnessed apneas, daytime sleepiness) and prior cardiovascular diseases (previous cardio and cerebrovascular events, previous myocardial infarction) as well as information about the awareness of hypertension and its health consequences were collected through the questionnaire. Questionnaires were administered in medical check-up points in different cities streets and squares, thus involving a very diverse population.
Preprocessing
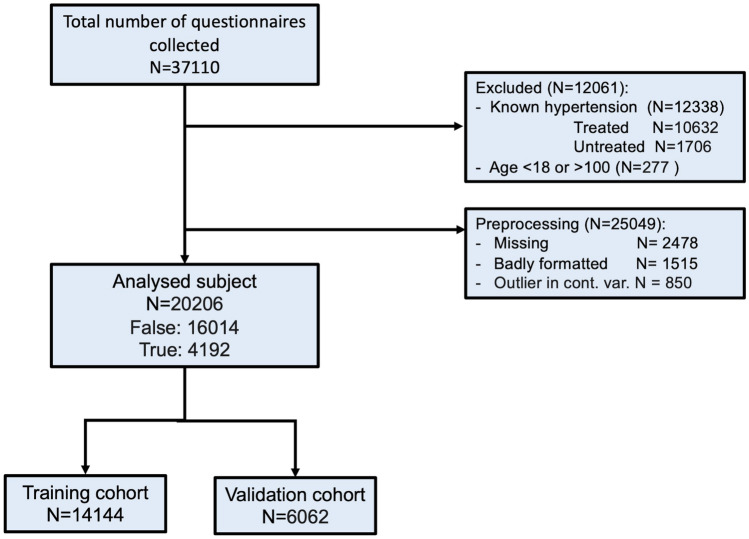
In order to analyse the raw data, a set of preprocessing techniques have been applied to the dataset. We included data of adult subjects only (age 18 and 100 years) with new-onset hypertension. Patients with known hypertension or already treated for hypertension were excluded. Participants were classified as newly detected hypertension if, computed the mean value of the last two BP measurements, at least one between systolic and diastolic BP was equal or greater than 140 or 90 mmHg respectively [12]. Records with missing values have been removed since some of the algorithms adopted in the present study do not support missing values. Outliers for normally distributed data (age, height, weight), defined as those values deviating from the mean more than three times the standard deviation (outside the range ) were also excluded from the analysis because were classified as errors in data acquisition. The whole preprocessing procedure is shown in Fig. 1.
Fig. 1.
Preprocessing flowchart
Features on top of which our analysis grounds, are listed in Tables 1 and 2. All the features containing categorical values have been converted into a set of dummy variables, each one modelling one of the categories, for a total of 28 dummy variables and 2 continuous ones. For each feature in the table, a statistic description is available, mean/std for continuous variable, or % for dummy variables. Features include the information collected from questionnaires, namely individual’s medical history, subject demographic, clinical characteristics, lifestyle factors, anthropometric measurements, and general knowledge on hypertension.
Table 1.
Descriptive statistics for continuous variables (subjects n = 20206)
| Feature | Mean | SD | Median |
|---|---|---|---|
| Age | 50.89 | 17.3 | 52 |
| BMI | 24.74 | 3.86 | 24.38 |
Table 2.
Descriptive statistics for dummy variables. Percentage are of true values
| Feature | Percentage |
|---|---|
| Female | 51.65 |
| Cardiovascular risk factors | |
| smoker | 20.11 |
| chronic kidney disease | 2.83 |
| diabetes mellitus | 4.27 |
| previous heart ischemic events | 3.52 |
| high cholesterol | 19.23 |
| previous brain ischemic events | 1.63 |
| family history of hypertension | 20.18 |
| Previous hypertensive emergency | 2.37 |
| Sleep complaints | |
| daytime somnolence | 28.12 |
| snoring | 29.23 |
| sleep apnea | 9.11 |
| Awareness of hypertension health consequences | |
| heart ischemia | 66.54 |
| brain ischemia | 54.25 |
| renal insufficiency | 76.79 |
| liver insufficiency | 87.25 |
| blindness | 23.10 |
| diabetes mellitus | 15.37 |
| Awareness of habits to prevent hypertension | |
| low Kcal diet | 20.20 |
| low fat and salt diet | 65.47 |
| low alcohol | 50.06 |
| drink one glass of wine per day | 19.23 |
| 30 min fitness | 62.29 |
| intensive fitness | 9.66 |
| no coffee | 71.98 |
| no smoking | 61.61 |
| periodic medical check | 44.76 |
| after symptoms medical check | 9.73 |
Accordingly, our dataset is composed of 37110 individuals before applying exclusion criteria, which led to 20206 subjects being included in the analyses. This cohort includes middle aged subjects almost equally distributed in term of sex (51.65% females), with a normal BMI (24.74 kg/m2). Most of the included subjects were in primary prevention as only 3.52% reported a previous cardiovascular event.
Data analyses with machine learning algorithms
Algorithms have been trained to predict, on top of the features of Tables 1 and 2, an individual’s hypertension risk. Since the goal it to detect hypertension, the database was split into two classes: 16014 subjects (79.25 %) with normal BP and 4192 subjects with newly discovered hypertension (20.75 %).
Due to the imbalance of the two groups, in order to avoid that the trained model overfits on the majority class, a combination of techniques was adopted: (i) accuracy was combined with precision and sensitivity to evaluate the model performance and a different coefficient was used to evaluate the errors of the classes (ii) the minority class was oversampled, (iii) the majority class undersampled. Accordingly, in this study, we performed different experiments in order to find the best predictors and achieve better performance.
Data were randomly split into a training set (70%, n = 14144), used for model construction and development, and a validation set (30%, n = 6062), used to test the performance of the derived model.
As a first experiment, we adopted the imbalanced dataset and we scaled errors with weights inversely proportional to the double of class frequencies in the input data. Given N as the samples number, the number of classes in the problem and the number of occurrences of class i, the weight to balance class i is
Then, we tested the performances of the algorithms exploiting resampling methods to transform the composition of the training dataset and balance the class distribution. Oversampling consists in creating synthetic samples of the under-represented class, thus generating a training set with two classes equally populated: we adopted to this purpose the SMOTE technique [13] and obtained a training set with 22420 samples (11210 positive samples and 11210 negative samples). Validation has been performed on the same test set as for the previous experiment. Conversely, undersampling consists in reducing the data by eliminating examples belonging to the majority class: we adopted a random strategy to this purpose.
For all the three schemes of experiments, we ran a Grid Search with 10- fold cross-validation to prevent overfitting on the training set and chose the hyperparameters of the prediction model. We chose to adopt sensitivity as the main measure of model to be maximised during the training phase, in order to find the model that optimises the number of positive individuals correctly classified (true positive). The best model built during the training phase has been selected and tested on the validation set. To evaluate the performance of the derived prediction model sensitivity, specificity, precision and accuracy have been computed. Moreover, the receiver operating characteristic (ROC) curve and validated area under the curve (AUC) value were derived.
This workflow has been adopted to evaluate different machine learning algorithms:
Logistic Regression: a binary statistical model that estimates the probability of an event occurring, based on a given dataset of independent variables. To map predictions and their probabilities, this method uses a sigmoid logistic function of the form [14];
Decision tree classifier: a scheme that uses a tree-like model of decisions to classify an input [15];
Random Forest classifier: an ensemble learning technique that uses a combination of decision trees as the base classifiers. Each tree is constructed from a sample from the original dataset and collected outputs are combined to obtain the final classification [15].
Support vector machine (SVM) classifier: a class of linear algorithms that generate a hyperplane to separates different classes of data with as wide a margin as possible [14].
XGBoost model: a scalable, distributed gradient-boosted decision tree that, similarly to random forests, combines multiple machine learning algorithms to obtain a better model [16].
Since the majority of features, available in the dataset, were categorical, we avoided those machine learning algorithms thought to work on numerical values, e.g., K-Nearest Neighbors (K-NN). All analyses were performed using the scikit-learn library [17].
Data analyses with medical protocols
In order to make our evaluation as complete as possible, we formalised the medical protocol extracted from the WHO list of risk factors1. The set of rules derived has been applied to the original dataset and a confusion matrix defined, basically reporting how many positive inidviduals were intercepted and how many were left out, as well as how many negative individuals were classified as positive, bringing to overscreening and unnecessary medical tests. In particular, dealing with the features reported in Table 2 if an individual was positive to one of the factors reported Table 3, he/she was classified as positive.
Table 3.
WHO risk factors
| Feature |
|---|
| Age > 65 years |
| BMI > 25 |
| Smoke |
| Renal insufficiency |
| Diabetes mellitus |
| Previous heart ischemic events |
| Previous brain ischemic events |
| High cholesterol |
| Family history of hypertension |
| Sleep complaints |
Results
The clinical goal of this study was to maximise the number of individuals included in the screening who will likely develop hypertension, while minimising included individuals who will not develop hypertension. As such, we needed to select the model that provides the best compromise between sensitivity and specificity.
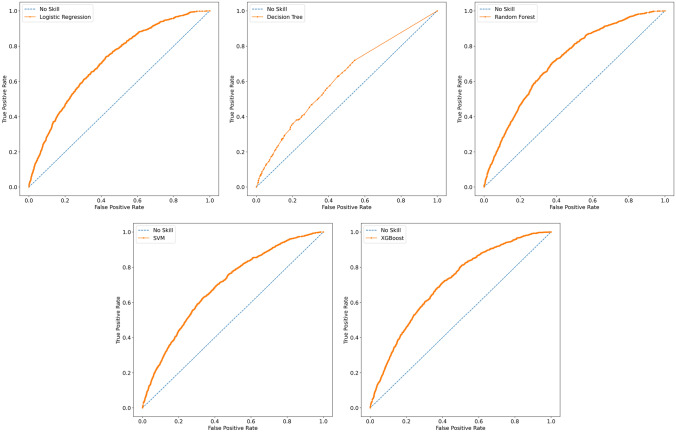
The evaluation metrics of the different models, for each of the three schemes evaluated, is presented in Table 4, highlighting the highest measure. Moreover, Fig. 2 shows the ROC curves associated with the best model found for each of the 5 classification algorithms from the original dataset. With the exception of decision tree, which provides the worse performances, from Fig. 2 we derive that, training algorithms on the original dataset, none of the ROC curves resulting from the other models outperforms and AUC remains low, thus requiring further experiments.
Table 4.
Performance in the test set of the three experimented schemes, original, oversampling and undersampling the training set, and of the medical protocol evaluation
| Original | |||||
|---|---|---|---|---|---|
| Model | Sensitivity | Specificity | Precision | Accuracy | AUC |
| Logistic Regression | 0.666 | 0.637 | 0.325 | 0.644 | 0.714 |
| Decision Tree | 0.524 | 0.656 | 0.285 | 0.629 | 0.616 |
| Random Forest | 0.582 | 0.723 | 0.351 | 0.694 | 0.716 |
| Support Vector Machines | 0.790 | 0.483 | 0.278 | 0.547 | 0.619 |
| XGBoost | 0.701 | 0.610 | 0.320 | 0.629 | 0.710 |
| Oversampling | |||||
|---|---|---|---|---|---|
| Model | Sensitivity | Specificity | Precision | Accuracy | AUC |
| Logistic Regression | 0.497 | 0.773 | 0.365 | 0.716 | 0.702 |
| Decision Tree | 0.352 | 0.743 | 0.304 | 0.698 | 0.590 |
| Random Forest | 0.368 | 0.846 | 0.386 | 0.747 | 0.712 |
| Support Vector Machines | 0.629 | 0.652 | 0.322 | 0.647 | 0.641 |
| XGBoost | 0.855 | 0.404 | 0.273 | 0.497 | 0.708 |
| Undersampling | |||||
|---|---|---|---|---|---|
| Model | Sensitivity | Specificity | Precision | Accuracy | AUC |
| Logistic Regression | 0.678 | 0.633 | 0.326 | 0.642 | 0.711 |
| Decision Tree | 0.739 | 0.610 | 0.332 | 0.637 | 0.739 |
| Random Forest | 0.818 | 0.629 | 0.377 | 0.681 | 0.816 |
| Support Vector Machines | 0.754 | 0.561 | 0.310 | 0.601 | 0.721 |
| XGBoost | 0.988 | 0.113 | 0.226 | 0.295 | 0.709 |
| Medical Protocol | |||||
|---|---|---|---|---|---|
| Model | Sensitivity | Specificity | Precision | Accuracy | AUC |
| Formal rules | 0.906 | 0.230 | 0.236 | 0.370 | 0.568 |
In each of the three experiments, and for each indicator, the highest value is marked in order to highlight the best performing model
Fig. 2.
ROC curves for the classification algorithms on the original dataset
Extending the study with resampling methods, we observe that oversampling significantly drops sensitivity and slightly improves specificity of the different algorithms, while undersampling affects positively both these indicators. Generally speaking, the model with the highest specificity, accuracy and precision was obtained with Random Forest in oversampling scheme. However, the sensitivity was 0.368 leading far more than 50% of positive individuals being classified as negative. XGBoost performs very well with a sensitivity of 0.855 in oversampling, and a very promising 0.988 in undersampling configuration, but specificity falls to 0.404 and 0.113 respectively, resulting in low overall accuracy. The best compromise was achieved with the Random Forest model, getting to a sensitivity of 0.818 and a specificity of 0.629, and an overall accuracy score of 0.681. The corresponding AUC is 0.816, the highest we obtained among the different experiments and models, and the corresponding ROC curve is reported in Fig. 3
Fig. 3.
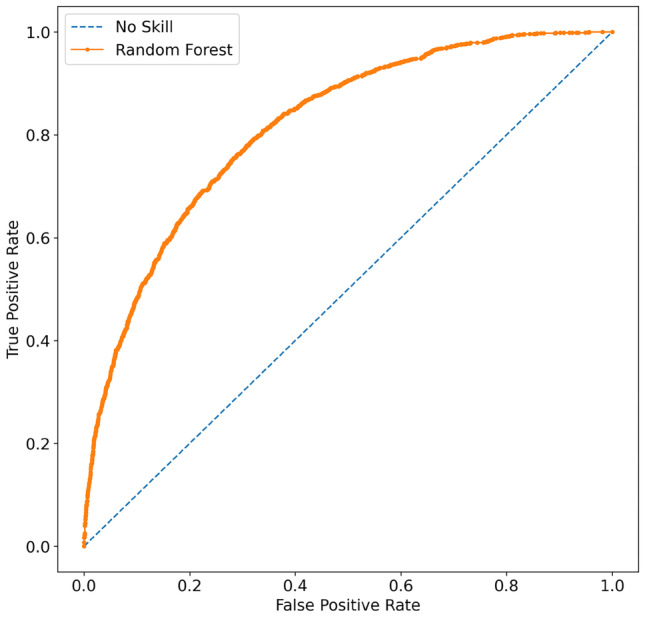
ROC curve for the best performing model, i.e., the Random-Forest in the undersampling experiment
When compared with ML algorithms, use of the known risk factors as performed in standard care yielded to a sensitivity of more than 90% but a specificity of 23%, leading to an accuracy of 37%.
Discussion
Screening for hypertension still remains one of the most critical aspects in cardiovascular prevention. Appropriate screening programs could allow early detection of patients at risk and prevent the development of hypertension mediated organ damage. Big data analysis through AI algorithms could improve sensitivity and specificity of such screening programs.
In this study we evaluated and compared five supervised ML algorithms in predicting hypertension based on a large dataset acquired during a national campaign during the World Hypertension Day. Although the best performing model achieved with the Random Forest algorithm allowed identification of hypertensives and normotensives with acceptable accuracy, sensitivity appears to be suboptimal limiting its use in clinical practice. As such, the results of the present work highlight both the advantages and limitations of ML: if on one hand ML automates the entire data analysis, resulting in more comprehensive, deeper, and faster insights, it strongly depends on the quality of data. Despite the huge amount of scientific literature reporting ML applications in healthcare, suggesting that AI applications can improve accuracy and timeliness of diagnoses, results are not always convincing. Have data the sufficient quality to be adopted for ML analyses? Are data containing the information needed for a clinical evaluation? Accordingly, an appropriate and comprehensive data collection is of fundamental importance as predictive models are only as good as the the quality of data from which they are built.
However, adopting the standard approach that includes the evaluation of known risk factors identified by the WHO medical protocol, allowed acceptable sensitivity but a very low specificity, leading to an approach that causes high expanses for the health system and that is not sustainable from an economic point of view.
Nevertheless, integrating the best ML model with clinical algorithms could represent the best compromise that maximises the number of true positives and minimises the false negatives.
Conclusion
Supervised ML algorithms applied to a big dataset collected in Italy by the SIIA, allowed to identify hypertension with modest accuracy and suboptimal sensitivity. Such an approach, if further improved and tested in different cohorts, could help with hypertension screening and might represent a potentially cost-effective alternative to patients’ evaluation by physicians. Future research should consider the adoption of ML techniques to develop a clinical model which should combine in practice the accuracy in diagnosing hypertension with the need to reduce the costs particularly in low resource settings.
Acknowledgements
We thank all participating members of local SIIA centres listed in “Italian investigators” section.
A. Italian investigators
Francesco Cipollone - Centro per l’Aterosclerosi, l’Ipertensione Arteriosa AO Chieti
Claudio Ferri - Servizio Ambulatoriale per l’Ipertensione Arteriosa e la Prevenzione Cardiovascolare, Ospedale Regionale “San Salvatore” , L’Aquila
Francesca Mallamaci - Centro dell’Ipertensone, UO di Nefrologia Dialisi e Trapianto di Rene, Grande Ospedale Metropolitano “Bianchi Melacrino Morelli”, Reggio Calabria
Nicola De Luca - Centro Ipertensione, AOU “Federico II”, Napoli
Ferruccio Galletti - Medicina Interna, Ipertensione e Prevenzione Cardiovascolare, AOU “Federico II”, Napoli
Luciano Di Meo - ASL CE - Centro Ipertensione e Prevenzione Cardiovascolare - Distretto 14, Cellole (CE)
Teodoro Marotta - Poliambulatorio “Cesare Battisti” - Distretto 31 - ASL Napoli 1 Centro, Napoli
Aniello De Leo - Ambulatorio per l’Ipertensione Arteriosa e la Prevenzione del Rischio Cardiovascolare Ospedale “Fatebenefratelli”, Napoli
Giovanni Rosiello - ASL Napoli 1 Centro - UOSD di Patologia Cardiovascolare, Napoli
Michele Ciccarelli - AOU “San Giovanni di Dio e Ruggi d’Aragona”, Salerno
Claudio Borghi - Centro per la prevenzione e cura dell’Ipertensione Arteriosa, Policlinico “S. Orsola”, Bologna
Renzo Roncuzzi - Casa di Cura Villa Erbosa , Bologna
Vincenzo Mazzeo - Ambulatorio diagnosi e terapia dell’Ipertensione Arteriosa Presidio Ospedaliero “Pierantoni”, Forlì
Claudio Guadagni - Centro per la diagnosi e cura dell’Ipertensione Arteriosa (Polo Sanitario Ravenna 33), Ravenna
Leonardo Sechi - Centro Ipertensione, Clinica Medica Università di Udine
Bruno Fabris - Centro per lo Studio e la Cura dell’Ipertensione Arteriosa, Trieste
Massimo Volpe - Centro per la Diagnosi e Cura dell’Ipertensione Arteriosa, AO “Sant’Andrea”, Roma
Claudio Letizia - Centro dell’Ipertensione Secondaria, Centro di Riferimento della Regione Lazio dell’Ipertensione Secondaria ed Endocrinopatie di difficile diagnosi, Università di Roma “Sapienza”, AOU Policlinico “Umberto I” di Roma
Dario Manfellotto - Centro Ipertensione Arteriosa e Gestazionale, Ospedale “Fatebenefratelli” Roma
Marco Mettimano - Policlinico Universitario “Agostino Gemelli” di Roma
Roberto Pontremoli - Centro per la Diagnosi e Cura dell’Ipertensione Arteriosa, AOU “San Martino”, Genova
Roberto Ervo - Centro Dialisi Ventimiglia Ospedale Bordighera/Ventimiglia ASL 1 Imperiese, Bordighera
Aldo Pende - Centro per l’Ipertensione, AOU “San Martino”, Genova
Maria Lorenza Muiesan - Centro per la prevenzione e cura dell’Ipertensione Arteriosa, Università degli Studi di Brescia
Cristina Giannattasio - Ambulatorio Ipertensione, Ospedale Niguarda “Ca’ Granda” e Università di Milano-Bicocca, Milano
Gianfranco Parati - Istituto Scientifico Ospedale “San Luca” - IRCCS Istituto Auxologico Italiano, Università di Milano-Bicocca, Milano
Giuseppe Mancia - Centro Studi Ipertensione e Malattie Vascolari - Policlinico di Monza, Verano Brianza (MB)
Rosario Ariano - Ambulatorio per la diagnosi e terapia dell’ipertensione arteriosa, UO Nefrologia AO di Cremona
Guido Garavelli - Ambulatorio per Ipertensione Arteriosa, Istituto “Figlie di San Camillo”, Cremona
Fabio Albini - Ambulatorio Ipertensione e Protezione Cardiovascolare Milano Nord, Cusano Milanino (MI)
Massimo Crippa - Unità semplice di diagnosi e trattamento dell’Ipertensione arteriosa, PO Gardone Val Trompia, AO “Spedali Civili” di Brescia, Gardone Valtrompia (BS)
Flavio Acquistapace - GB Mangioni Hospital - GVM Care & Research, Lecco
Maria Teresa Lavazza - AO Ospedale di Legnano
Antonio Agrati, Giovanni Ferraro - Ambulatorio Medicina Generale Ipertensione Dipartimento Medico Polispecialistico Ospedale “Niguarda Cà Granda” Milano
Stefano Carugo - Asst - Santi Paolo e Carlo, Milano
Chiara Lonati - Centro Ipertensione Arteriosa, Ospedale Classificato “San Giuseppe”, Milano
Maurizio Turiel - Servizio di Cardiologia - IRCCS Istituto Galeazzi, Milano
Amedeo Mugellini - Centro per l’Ipertensione e la Fisiopatologia Cardiovascolare - Università degli Studi di Pavia - Sezione di Medicina Interna, Malattie Vascolari e Metaboliche IRCCS Policlinico “San Matteo”, Pavia
Claudio Pini - Azienda Socio Sanitaria Territoriale Lariana, San Fermo della Battaglia (CO)
Giuseppina Dognini - AO Treviglio, Ospedale Treviglio Caravaggio, SC Medicina Generale, Treviglio
Andrea Maresca - Centro per la diagnosi e Terapia dell’Ipertensione Arteriosa, UO Medicina1, Ospedale di Circolo e “Fondazione Macchi” ASST Sette Laghi, Varese
Riccardo Sarzani - Centro di Riferimento Regione Marche Ipertensione Arteriosa e Malattie Cardiovascolari Clinica di Medicina Interna e Geriatria Università Politecnica delle Marche e IRCCS-INRCA, Torrette di Ancona, Ancona
Giuseppe Lembo - IRCCS Neuromed - Polo Didattico Sede distaccata Molise, “Sapienza” Università di Roma, Pozzilli (IS)
Franco Veglio - Centro Ipertensione Arteriosa, Università di Torino, Torino
Aldo Ortensia - Ambulatorio per Ipertensione Nefrovascolare, AON “S.S. Antonio e Biagio e Cesare Arrigo” Alessandria
Alessandro Rossi - Servizio per Ipertensione Medicina Interna, Ospedale di Chieri, (TO)
Pietro Nazzaro - Centro di Prevenzione Cerebrovascolare ed Ipertensione Arteriosa “A.M.Pirrelli”, Bari
Giuseppe Ranieri - Centro Ipertensione Arteriosa, AOU Policlinico di Bari
Vito Vulpis - UOS Diagnosi e Cura dell’Ipertensione Arteriosa - Medicina Interna Ospedaliera, AUO Policlinico di Bari
Nicola Morelli - Presidio Territoriale Assistenziale “F. Jaia”, Conversano (BA)
Giuseppe Antonio De Giorgi - Clinica San Francesco s.r.l. - Fondatore Giovanni Tartaro , Galatina (LE)
Francesco Cocco - SC Cardiologia Utic - PO Manduria ASL TA, Manduria (TA)
Rachele Grifa - Ospedale “Casa Sollievo della Sofferenza” - IRCCS Dipartimento di Scienze Mediche - SC di Nefrologia e Dialisi, San Giovanni Rotondo (FG)
Santina Cottone - UO Dipartimentale di Nefrologia ed Ipertensione, Palermo
Paola Belluardo - SC Cardiologia - UTIC Ospedale “Maggiore” di Modica, (RG)
Stefano Taddei - Centro per la cura e la diagnosi dell’Ipertensione Arteriosa, AOU Pisana, Pisa
Salvatore Lenti - Centro Ipertensione Arteriosa di II livello - UOSD Ipertensione, dislipidemia e rischio cardiovascolare - Dipartimento Medicina Interna - Ospedale “San Donato” Arezzo - USL sudest Toscana, Arezzo
Andrea Ungar - Centro di Riferimento Regionale per l’Ipertensione Arteriosa dell’anziano della Regione Toscana, Cardiologia e Medicina Geriatrica, AOU “Careggi” e Università di Firenze
Franco Cipollini - Ambulatorio Specialistico per l’Ipertensione Arteriosa, UO Medicina Interna, Ospedale “S. Jacopo”, Pistoia, Azienda USL Toscana Centro, Pistoia
Stefano Barolo - Medicina Interna Ospedale di Silandro, Silandro (BZ)
Giacomo Pucci - Centro Ipertensione Arteriosa - Dipartimento di Medicina, Università degli Studi di Perugia, SC di Medicina Interna, AO “S. Maria”, Terni
Maria Sabina Modesti - Ambulatorio per la Diagnosi e la Cura dell’Ipertensione Arteriosa - SC Medicina Interna - Ospedale Regionale “U. Parini” di Aosta
Pietro Minuz - UOC Medicina Generale per lo Studio ed il Trattamento della Malattia Ipertensiva, Dipartimento di Medicina Università di Verona ed AOUI Verona Policlinico “GB Rossi”, Verona
Francesca Saladini - ULSS 6 Euganea - UO Cardiologia, PO Cittadella-Camposampiero, (PD)
Francesco Fallo - Ambulatorio Divisionale e dell’Attività di Ricovero per il Settore dell’Ipertensione Arteriosa, A.O.U. Padova
Giancarlo Parisi - Ambulatorio Diagnostica vascolare e prevenzione cardiovascolare UO Medicina, Ospedale “Immacolata Concezione” - ULSS & Euganea, Piove di Sacco (PD)
Marcello Rattazzi - Università degli Studi di Padova, Dipartimento di Medicina-DIMED / ULSS 9 di Treviso, Dipartimento di Medicina Interna, SC di Medicina Interna 1, Treviso
Cristiana Leprotti - Centro per L’ipertensione Arteriosa, UOSD Venezia
Arrigo Cicero - Department of Medical and Surgical Sciences, Univ. Bologna
Guido Iaccarino - Department of Advanced Biomedical Sciences, Univ. Naples, Federico II
Giuseppe Mulè - Nephrology and Hypertension Unit of Palermo “Paolo Giaccone
Carmine Savoia - Department of Clinical and Molecular Medicine, Sapienza Univ., Roma
Carmine Arena - Centro Ospedaliero Militare di Milano
Susanna Cozzio - Ospedale di Rovereto, (TN)
Plinio Fabiani - Ospedale di Viareggio (LU)
Anna Fornasiero - Poliambulatori ULSS Berica, Lonigo (VI)
Chiara Grasselli - Azienda Ospedaliera S.M. Nuova, Reggio Emilia
Paola Mesiano - Ospedale San Giovanni Bosco, Torino
Claudio Pascale - Ospedale Cottolengo, Torino
Fabio Bracco - Poliambulatorio Cornareto , Carcare (SV)
Roberto Marini, Gorizia
Paolo Borgheresi - Poliambulatorio territoriale, Arezzo
Amalia Mariotti - Ospedale “Di Summa” - Brindisi
Antonio La Grotta - UNI-ASTI SS Polo Universitario, Asti
Grzegorz Bilo - Istituto Scientifico Ospedale “San Luca” - IRCCS Istituto Auxologico Italiano, Università di Milano-Bicocca, Milano
Author contribution
S.M., M.F.P and G.P. wrote the manuscript. S.M. and S.F. analysed data. M.F.P, C.B., C.F., G.G., G.P. was involved in data collection. All authors reviewed the manuscript.
Funding
This work was supported by the University of Urbino through the competitive call “Health and Food Safety Promotion”: grant BIT4Food: Blockchain and Internet of Things for Food Safety.
Data Availability
The participants of this study did not give written consent for their data to be shared publicly, so due to the sensitive nature of the research, supporting data is not available.
Declarations
Ethical approval
Questionnaires were collected as part of a nation-wide hypertension screening in Italy coordinated by the Italian Society of Hypertension. Informed consent was obtained for each participant onsite. This work was conducted in conformity with the Helsinki Declaration. No personal information was requested, as all forms were anonymous, making the identification of participants not possible.
Competing interests
The authors have no competing interests associated with this study, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sara Montagna and Martino Pengo contributed equally to this work.
References
- 1.Collaborators GRF. Global burden of 87 risk factors in 204 countries and territories, 1990–2019: a systematic analysis for the global burden of disease study 2019. The Lancet. 2020;396(10258):1223–1249. doi: 10.1016/S0140-6736(20)30752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parati, G., Stergiou, G.S., Bilo, G., Kollias, A., Pengo, M., Ochoa, J.E., Agarwal, R., Asayama, K., Asmar, R., Burnier, M., De La Sierra, A., Giannattasio, C., Gosse, P., Head, G., Hoshide, S., Imai, Y., Kario, K., Li, Y., Manios, E., Mant, J., McManus, R.J., Mengden, T., Mihailidou, A.S., Muntner, P., Myers, M., Niiranen, T., Ntineri, A., O’Brien, E., Octavio, J., Ohkubo, T., Omboni, S., Padfield, P., Palatini, P., Pellegrini, D., Postel-Vinay, N., Ramirez, A.J., Sharman, J.E., Shennan, A., Silva, E., Topouchian, J., Torlasco, C., Wang, J.G., Weber, M.A., Whelton, P.K., White, W.B., Mancia, G.: Home blood pressure monitoring: methodology, clinical relevance and practical application: a 2021 position paper by the working group on blood pressure monitoring and cardiovascular variability of the european society of hypertension. Journal of Hypertension 39(9) (2021) [DOI] [PMC free article] [PubMed]
- 3.Beaney T, Burrell LM, Castillo RR, Charchar FJ, Cro S, Damasceno A, Kruger R, Nilsson PM, Prabhakaran D, Ramirez AJ, Schlaich MP, Schutte AE, Tomaszewski M, Touyz R, Wang JG, Weber MA, Poulter NR. the MMM Investigators: May Measurement Month 2018: a pragmatic global screening campaign to raise awareness of blood pressure by the International Society of Hypertension. European Heart Journal. 2019;40(25):2006–2017. doi: 10.1093/eurheartj/ehz300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nature Medicine. 2022;28(1):31–38. doi: 10.1038/s41591-021-01614-0. [DOI] [PubMed] [Google Scholar]
- 5.Topol E. High-performance medicine: the convergence of human and artificial intelligence. Nature Medicine. 2019;25(1):44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 6.Golino, H.F., Amaral, L.S.d.B., Duarte, S.F.P., Gomes, C.M.A., Soares, T.d.J., Reis, L.A.d., Santos, J.: Predicting increased blood pressure using machine learning. Journal of Obesity 2014, 637635 (2014). 10.1155/2014/637635 [DOI] [PMC free article] [PubMed]
- 7.Held E, Cape J, Tintle N. Comparing machine learning and logistic regression methods for predicting hypertension using a combination of gene expression and next-generation sequencing data. BMC Proceedings. 2016;10(7):34. doi: 10.1186/s12919-016-0020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanegae H, Suzuki K, Fukatani K, Ito T, Harada N, Kario K. Highly precise risk prediction model for new-onset hypertension using artificial intelligence techniques. The Journal of Clinical Hypertension. 2020;22(3):445–450. doi: 10.1111/jch.13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Yuan X, Wang W, Liu K, Qin Y, Sun X, Ma W, Zou Y, Zhang H, Zhou X, Wu H, Jiang X, Cai J, Chang W, Zhou S, Song L. Value of a machine learning approach for predicting clinical outcomes in young patients with hypertension. Hypertension. 2020;75(5):1271–1278. doi: 10.1161/HYPERTENSIONAHA.119.13404. [DOI] [PubMed] [Google Scholar]
- 10.Zhao, H., Zhang, X., Xu, Y., Gao, L., Ma, Z., Sun, Y., Wang, W.: Predicting the risk of hypertension based on several easy-to-collect risk factors: A machine learning method. Frontiers in Public Health 9 (2021). 10.3389/fpubh.2021.619429 [DOI] [PMC free article] [PubMed]
- 11.Martinez-Ríos, E., Montesinos, L., Alfaro-Ponce, M., Pecchia, L.: A review of machine learning in hypertension detection and blood pressure estimation based on clinical and physiological data. Biomedical Signal Processing and Control 68, 102813 (2021). 10.1016/j.bspc.2021.102813
- 12.Williams, B., Mancia, G., Spiering, W., Agabiti Rosei, E., Azizi, M., Burnier, M., Clement, D.L., Coca, A., de Simone, G., Dominiczak, A., Kahan, T., Mahfoud, F., Redon, J., Ruilope, L., Zanchetti, A., Kerins, M., Kjeldsen, S.E., Kreutz, R., Laurent, S., Lip, G.Y.H., McManus, R., Narkiewicz, K., Ruschitzka, F., Schmieder, R.E., Shlyakhto, E., Tsioufis, C., Aboyans, V., Desormais, I., Group, E.S.D.: 2018 ESC/ESH Guidelines for the management of arterial hypertension: The Task Force for the management of arterial hypertension of the European Society of Cardiology (ESC) and the European Society of Hypertension (ESH). European Heart Journal 39(33), 3021–3104 (2018). 10.1093/eurheartj/ehy339
- 13.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. Smote: Synthetic minority over-sampling technique. Journal of Artificial Intelligence Research. 2002;16(1):321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 14.Hastie, T., Tibshirani, R., Friedman, J.H., Friedman, J.H.: The elements of statistical learning: data mining, inference, and prediction, vol. 2. Springer (2009)
- 15.Breiman, L., Friedman, J.H., Olshen, R.A., Stone, C.J.: Classification and regression trees. Routledge (2017)
- 16.Chen, T., Guestrin, C.: XGBoost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD ’16, pp. 785–794. ACM, New York, NY, USA (2016). 10.1145/2939672.2939785 http://doi.acm.org/10.1145/2939672.2939785
- 17.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The participants of this study did not give written consent for their data to be shared publicly, so due to the sensitive nature of the research, supporting data is not available.