Abstract
Circulating sex steroid hormones are critical for neural function and development of neuroplasticity in many regions of the central nervous system. In the spinal cord, our knowledge of steroid hormone influence mostly derives from mechanistic studies of pain processing in dorsal spinal cord circuits; less is known regarding hormonal influence of ventral spinal motor function. Gonadectomy (surgical removal of the testes in males and ovaries in females) rapidly and persistently reduces circulating sex steroids in both females and males, providing a means to interrogate the role of hormones on neural function. Here we provide a next-generation RNA sequencing (RNA-seq) data set to evaluate the impact of gonadectomy on the transcriptome of ventral spinal cord tissue of adult female and male rats.
Subject terms: Neurology, Neurophysiology
| Measurement(s) | Ventral spinal cord gene expression |
| Technology Type(s) | RNA Sequencing |
| Factor Type(s) | Ventral spinal cord gene expression |
| Sample Characteristic - Organism | Rattus norvegicus |
| Sample Characteristic - Location | United States |
Background & Summary
Our understanding of how sex steroid hormones influence central nervous system function across the lifespan has rapidly advanced over the past 30 years. The principle steroid hormones estrogen and testosterone are now recognized to exert pleiotropic effects on virtually all areas of the brain and spinal cord1. Primarily synthesized in the gonads and adrenal cortex, steroid hormones are released into the blood circulation, readily cross the blood brain barrier, and influence neuronal signaling through nuclear and non-nuclear receptor mechanisms1–3. In the spinal cord, steroid hormones directly modulate neurotransmitter release and synaptic neurotransmission4, facilitate myelination5,6, provide neuroprotection7–10, support axonal regeneration11 following spinal cord injury, and mediate the induction of neuroplasticity2,12–18. This knowledge comes primarily from studies of pain processing in the dorsal spinal cord, hippocampal learning and memory, and from rodent models of neuroprotection following spinal cord injury. Less understood is the direct role steroid hormones play in ventral spinal cord function and spinal motor output.
Gonadectomy, the surgical removal of the primary sex organs (ovaries in females; testes in males), induces a rapid and sustained reduction in circulating steroid hormones2,17,19. This relatively simple procedure is routinely used in basic science studies to ascertain the role of steroid hormones on function, particularly in the CNS. However, gonadectomy-induced reductions in steroid hormones are likely to alter the fundamental properties of the neural system being studied, complicating comparisons with gonadally-intact groups. This study used next-generation RNA sequencing (RNA-seq) to evaluate the impact of gonadectomy on the transcriptome of ventral spinal cord tissue of adult female and male rats. These data provide valuable, sex-specific gene expression profiles for the ventral cervical spinal cords of female and male rats and suggest unique, sexually dimorphic transcriptomic changes resulting from gonadectomy.
Methods
All experimental procedures were approved by the University of Minnesota Institutional Animal Care and Use Committee and conformed to guidelines detailed in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Pairs of young-adult (3–4 months) Sprague-Dawley rats (Envigo; Indianapolis, IN, USA) were housed in a mixed-sex rodent room within facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. Rats were acclimatized to the rodent facilities for at least 1 week prior to the start of experimentation and were maintained on a 12 hr light-dark cycle (6am – 6 pm) with food and water available ad libitum. A total of 22 rats were used for this study: 10 male rats and 12 virgin female rats.
Gonadectomy
Half of the male rats (n = 5) and half of the female rats (n = 6) underwent gonadectomy surgeries to reduce levels of circulating sex hormones. This is a common procedure to study the impact of circulating sex hormones on neural function and consists of removing the ovaries bilaterally in female rats (ovariectomy) or removing the bilateral testes (castration) in males. The castrated male rats in this study are referred to as gonadectomized (GDX) and females as ovariectomized (OVX).
At least 2 hours prior to surgery, all rats were provided Buprenorphine SR-LAB (1 mg/kg; s.q. ZooPharm; Fort Collins, CO, USA) for long-lasting pain control. For OVX surgeries, female rats were anesthetized with isoflurane in a closed chamber and then maintained via nose cone (2–3% in 50% O2) for the duration of surgery. Adequacy of anesthesia was confirmed by the lack of response to toe-pinch and eye-blink reflexes. Following removal of fur and skin cleansing with chlorhexidine (Prevantics; PDI Inc. Orangeburg, NY, USA), bilateral incisions were made through the dorsolateral skin and muscle layers to expose the ovarian fat pads. The ovaries were exteriorized and separated from the uterine horns using electrocautery to minimize bleeding. Uterine horns were then replaced into the abdominal cavity. Muscle layers were approximated and sutured with 4-0 absorbable suture (Covidien, Mansfield, MA, USA), and the skin incision was closed with wound clips (9 mm autoclips, MikRon Precision Inc. Gardena, CA, USA). Control rats (n = 6) were ovarian-intact and age-matched to OVX rats.
The GDX procedure in males was performed using similar surgical preparations. Male rats were anesthetized with isoflurane in a closed chamber and then maintained via nose cone (2–3% in 50% O2) for the duration of surgery. Adequacy of anesthesia was confirmed by the lack of response to toe-pinch and eye-blink reflexes. From a supine position, fur was removed over the bilateral testes and skin cleansed with chlorhexidine. Testes were exteriorized via small, bilateral scrotal incisions. The testes were removed using electrocautery to minimize bleeding and vas deferens were replaced back into the scrotal sac. The skin incisions were closed using veterinary surgical glue (Vetbond, 3 M, St. Paul, MN, USA) with special care taken to avoid the penile or anal openings. Control rats (n = 5) had intact testes and were age-matched to GDX rats. Following OVX and GDX surgeries, rats were recovered in temperature-controlled chambers until mobilizing independently, and then were returned to their home cages.
Ventral cervical spinal cord tissue isolation
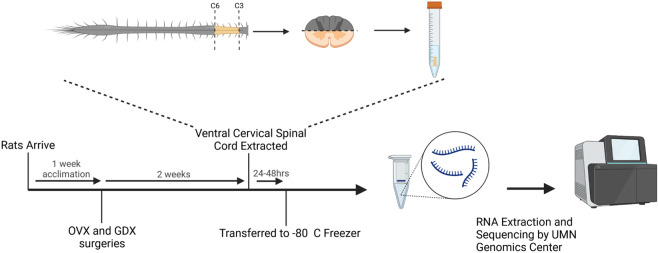
Rats were deeply anesthetized with isoflurane in a closed container, transcardially perfused with ice-cold saline (pH 7.4), and the spinal cords immediately extracted from C3-C6 (Fig. 1). The dorsal half of the tissue segments were discarded to isolate only the ventral spinal cord. Samples were quickly transferred to ice-cold RNAlater solution (Invitrogen) and refrigerated at 4 °C for 48 hours. Cords were subsequently removed from RNAlater solution, placed into fresh 1.5 mL conical tubes and stored at −80 °C until processed for RNA extraction (Fig. 1). The ventral cervical spinal cord in this region contains upper motor neurons of UE musculature and the phrenic motor pool, a column of motor neurons innervating the diaphragm muscle via the phrenic nerve. Spinal cord tissue from rats receiving castration or ovariectomy was processed 2 weeks post-surgery (Fig. 1), to align with recent studies indicating that respiratory function and the expression of phrenic neuroplasticity are impacted by gonadectomy in rats2,16,17.
Fig. 1.
Experimental workflow. Following transcardial perfusion, spinal cord segments from C3-C6 were extracted and the dorsal half of the tissue segments discarded to isolate only the ventral spinal cord. Samples were quickly transferred to ice-cold RNAlater solution and refrigerated at 4 °C for up to 48 hours. The tissue was subsequently removed from RNAlater solution, placed into fresh 1.5mL conical tubes and stored at -80 °C until processed for RNA extraction and sequencing.
RNA isolation and sequencing
All samples were sent on dry ice to the University of Minnesota Genomic Center for RNA isolation, library preparation and sequencing. RNA was isolated using the RNeasy mini kit (Qiagen Corp., Santa Clarita, CA) according to manufacturer’s instructions. The purity of RNA was checked with Nanodrop (Thermo Fisher Scientific). All RNAs showed a 260:280 ratio of >2.00 (Table 1)
Table 1.
RNA quantification and quality controls including 260/280 ratios and RNA integrity numbers (RIN#).
| Sample | Tissue | RNA Concentration (ng/µL) | 260/280 Ratio | RIN # |
|---|---|---|---|---|
| Intact Female 1 | Ventral Spinal Cord - IF1 | 226 | 2.09 | 8.4 |
| Intact Female 2 | Ventral Spinal Cord - IF2 | 102 | 2.05 | 8.4 |
| Intact Female 3 | Ventral Spinal Cord - IF3 | 212 | 2.07 | 7.8 |
| Intact Female 4 | Ventral Spinal Cord - IF4 | 251 | 2.07 | 8.4 |
| Intact Female 5 | Ventral Spinal Cord - IF5 | 141 | 2.06 | 7.9 |
| Intact Female 6 | Ventral Spinal Cord - IF6 | 262 | 2.07 | 8.6 |
| OVX Female 1 | Ventral Spinal Cord - XF1 | 144 | 2.08 | 8.0 |
| OVX Female 2 | Ventral Spinal Cord - XF2 | 177 | 2.06 | 8.8 |
| OVX Female 3 | Ventral Spinal Cord - XF3 | 192 | 2.1 | 8.1 |
| OVX Female 4 | Ventral Spinal Cord - XF4 | 177 | 2.07 | 8.0 |
| OVX Female 5 | Ventral Spinal Cord - XF5 | 230 | 2.09 | 8.2 |
| OVX Female 6 | Ventral Spinal Cord - XF6 | 160 | 2.06 | 7.9 |
| Intact Male 1 | Ventral Spinal Cord - IM1 | 59 | 2.08 | 8.5 |
| Intact Male 2 | Ventral Spinal Cord - IM2 | 176 | 2.09 | 8.2 |
| Intact Male 3 | Ventral Spinal Cord - IM3 | 120 | 2.07 | 8.4 |
| Intact Male 4 | Ventral Spinal Cord - IM5 | 142 | 2.05 | 8.4 |
| Intact Male 5 | Ventral Spinal Cord - IM6 | 225 | 2.06 | 8.1 |
| GDX Male 1 | Ventral Spinal Cord - XM1 | 181 | 2.09 | 8.2 |
| GDX Male 2 | Ventral Spinal Cord - XM2 | 190 | 2.04 | 8.3 |
| GDX Male 3 | Ventral Spinal Cord - XM4 | 295 | 2.06 | 8.4 |
| GDX Male 4 | Ventral Spinal Cord - XM5 | 262 | 2.08 | 8.0 |
| GDX Male 5 | Ventral Spinal Cord - XM6 | 218 | 2.07 | 8.0 |
RNA isolates were quantified using a fluorimetric RiboGreen assay and RNA integrity was assessed using capillary electrophoresis (Agilent BioAnalyzer 2100) to generate an RNA integrity number (RIN). All samples had RIN values ≥7.8 (avg. RIN 8.2; Table 1) and at least 1 ng total RNA. Sequencing libraries were generated using a SMARTer Stranded Total RNA Seq v2 – Pico Mammalian kit (Takara Bio). Briefly, >1 ng of total RNA were fragmented and then reverse transcribed into cDNA using random primers, with a template switching oligo incorporated during cDNA synthesis to allow for full length cDNA synthesis and retain strand specificity. Illumina sequencing adapters and barcodes were then added to the cDNA by PCR, followed by cleavage of ribosomal cDNA. Uncleaved fragments were then enriched by PCR for 12–16 cycles. Final library size distribution was again validated using capillary electrophoresis and quantified using fluorimetry (PicoGreen). Indexed libraries were normalized and pooled for sequencing. Libraries were then loaded onto a NextSeq. 550 (single read, Illumina) cartridge, where clustering occurred on-board the instrument. After clustering, sequencing was commenced using Illumina’s 2-color SBS chemistry. Following sequencing, Base call (.bcl) files for each cycle of sequencing were generated by Illumina Real Time Analysis software. Primary analysis and de-multiplexing were performed using Illumina bcl2fastq software. The end result of the bcl2fastq workflow was de-multiplexed FASTQ files for subsequent analysis. 151 bp fastq single-end reads (mean read depth ≥40 million per sample) were trimmed using Trimmomatic (version 0.33) enabled with the optional “-q” option; 3 bp sliding-window trimming from 3′ end required minimum Q30. Quality control of raw sequence data for each sample was performed using FastQC (Supplementary Figs. 1–22). Read mapping was performed via Hisat2 (v2.1.0) using the rat genome (Rnor_6.0.98) as reference. Mapping results are provided in Table 2. Average read-pairs per sample was 44.4 M with an average input fragment size of 188 bp (Table 2). Gene quantification was performed using Subread featureCounts for raw read counts (Fig. 2A). Sample variance was identified using principle component analysis (Fig. 2B). Clear separation between male and female samples indicate highly sex-specific expression profiles, and samples tended to cluster based on treatment (i.e. gonadally-intact vs. gonads removed; Fig. 2B). The impact of gonadectomy on gene expression is presented in Fig. 3. Differentially expressed genes (DEGs) were identified using the edgeR (negative binomial) feature in CLCGWB (Qiagen, Redwood City, CA) using raw read counts. The frequency distribution of DEGs relative to their fold-change are plotted at three levels of statistical stringency (p < 0.1; p < 0.01; p < 0.0001; Fig. 3). Removal of the gonads appeared to have a more robust effect on the spinal transcriptome of male rats as a total of 1642 genes were significantly up or down regulated at p < 0.1, while 273 genes remained significantly different at p < 0.001 (Fig. 3A). Of these, 191 genes were significantly upregulated and 82 downregulated. Female gene expression in the ventral cervical spinal cord was minimally affected by removal of the gonads. Only 114 genes were significantly changed following OVX at lower statistical stringency (p < 0.1), with 6 DEGs remaining significantly upregulated at p < 0.001 (Fig. 3B).
Table 2.
RNA-Seq mapping results and NCBI data accession codes for each sample.
| Sample | Tissue | Average input fragment size (bp) | Raw Read-pairs (per million) | % sequence alignment | NCBI SRA Data Accession |
|---|---|---|---|---|---|
| Intact Female 1 | Ventral Spinal Cord - IF1 | 162 | 51.8 | 98.03 | GSM6112668 |
| Intact Female 2 | Ventral Spinal Cord - IF2 | 243 | 41.7 | 97.97 | GSM6112669 |
| Intact Female 3 | Ventral Spinal Cord - IF3 | 245 | 47.6 | 97.75 | GSM6112670 |
| Intact Female 4 | Ventral Spinal Cord - IF4 | 162 | 43.6 | 97.51 | GSM6112671 |
| Intact Female 5 | Ventral Spinal Cord - IF5 | 244 | 43.6 | 97.88 | GSM6112672 |
| Intact Female 6 | Ventral Spinal Cord - IF6 | 244 | 38.7 | 97.77 | GSM6112673 |
| OVX Female 1 | Ventral Spinal Cord - XF1 | 162 | 50.5 | 97.47 | GSM6112674 |
| OVX Female 2 | Ventral Spinal Cord - XF2 | 162 | 46.6 | 97.65 | GSM6112675 |
| OVX Female 3 | Ventral Spinal Cord - XF3 | 161 | 41.1 | 97.75 | GSM6112676 |
| OVX Female 4 | Ventral Spinal Cord - XF4 | 162 | 50.7 | 97.56 | GSM6112677 |
| OVX Female 5 | Ventral Spinal Cord - XF5 | 162 | 39.6 | 97.51 | GSM6112678 |
| OVX Female 6 | Ventral Spinal Cord - XF6 | 162 | 37.6 | 97.59 | GSM6112679 |
| Intact Male 1 | Ventral Spinal Cord - IM1 | 243 | 43.7 | 97.25 | GSM6112658 |
| Intact Male 2 | Ventral Spinal Cord - IM2 | 162 | 39.8 | 97.72 | GSM6112659 |
| Intact Male 3 | Ventral Spinal Cord - IM3 | 244 | 39.3 | 97.87 | GSM6112660 |
| Intact Male 4 | Ventral Spinal Cord - IM5 | 162 | 47.7 | 97.82 | GSM6112661 |
| Intact Male 5 | Ventral Spinal Cord - IM6 | 243 | 44.3 | 97.53 | GSM6112662 |
| GDX Male 1 | Ventral Spinal Cord - XM1 | 162 | 44.4 | 97.65 | GSM6112663 |
| GDX Male 2 | Ventral Spinal Cord - XM2 | 162 | 50.9 | 97.43 | GSM6112664 |
| GDX Male 3 | Ventral Spinal Cord - XM4 | 161 | 44.0 | 97.71 | GSM6112665 |
| GDX Male 4 | Ventral Spinal Cord - XM5 | 162 | 42.7 | 97.93 | GSM6112666 |
| GDX Male 5 | Ventral Spinal Cord - XM6 | 162 | 46.1 | 97.75 | GSM6112667 |
Fig. 2.
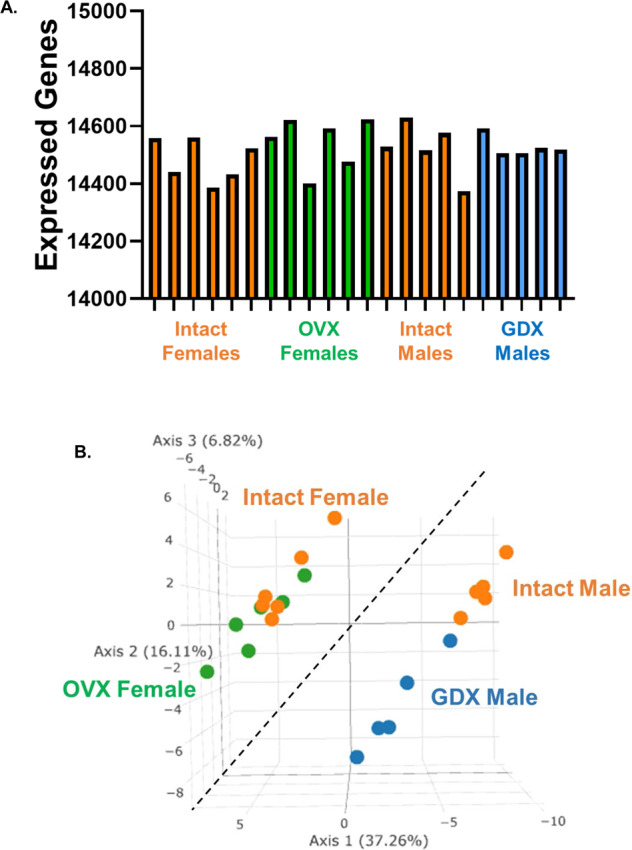
RNA-Seq data analysis. After quality check, reads were mapped to the rat genome (Rnor_6.0.98) using Hisat2 (v2.1.0). The number of genes expressed per sample is shown in (A). For this plot a gene was considered expressed if it had expression equal to or higher than 1 read per million. Principle component analysis (PCA; B) illustrates the clustering of female and male samples. The first three principal components are shown, and the percent of total variation explained by each component is shown in the axis titles. Samples with similar characteristics appear close to each other, and samples with dissimilar characteristics are farther apart.
Fig. 3.
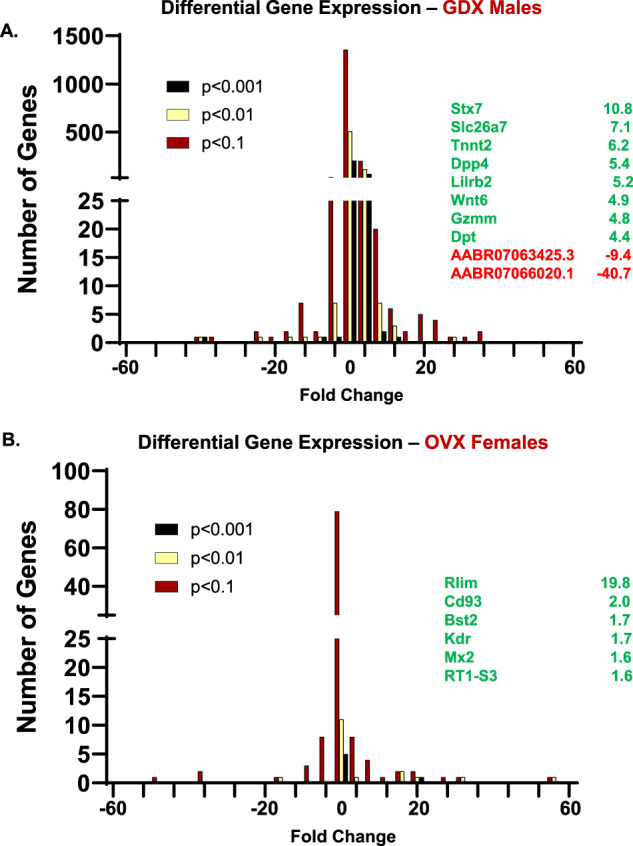
Differential Gene Expression following GDX. Removal of the primary sex organs induced transcriptomic changes in the ventral cervical spinal cord. The frequency distribution of DEGs relative to their fold-change is plotted at three levels of statistical stringency (p < 0.1; p < 0.01; p < 0.0001) in male rats following GDX (A) and female rats following OVX (B). Included are the top 10 most differentially expressed genes at p < 0.001 for males, and the 6 genes upregulated in females at p < 0.001.
Data Records
Tables 1, 2 provide specific information related to each sample used in this study. Complete RNA-seq data were deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GSE202381)20.
Technical Validation
As our samples represent heterogeneous populations of cells in the spinal cord, we eliminated the dorsal spinal tissue in each sample to reduce variability and focus our analysis on the area of primary interest, the ventral cervical spinal cord. The quality of RNA was examined using an Agilent BioAnalyzer 2100 and all samples showed high RNA integrity (Avg. RIN 8.2) appropriate for deep sequencing. RNA-seq data quality was determined using FastQC. Average input fragment size per sample was ~188 bp with a mean read depth of 44.36 million (Table 1). Subsequently, a high percentage of reads were mapped to the reference rat genome (97.69% alignment; Table 1). PCA plots for the 4 groups are provided in Fig. 2.
Supplementary information
Acknowledgements
The authors would like to acknowledge the contribution of the UMN Genomics Center, in particular, John Garbe, Veronica Tonnell, and Elyse Froehling. This work was supported by funding from the NIH National Heart, Lung, and Blood Institute (R01HL146477-01A1), The Craig H. Neilsen Foundation, and the University of Minnesota Division of Physical Therapy. Figure one was created using Biorender.com.
Author contributions
S.M. collected and processed the tissue and prepared figures. J.E.A. processed data and assisted with sequencing analysis. A.R. assisted with data analysis. B.J.D. supervised all aspects of the study and wrote the manuscript. All authors participated in careful editing of manuscript.
Code availability
No custom code was generated or used for analysis of the data presented.
Competing interests
The Author’s declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-022-01917-y.
References
- 1.Delchev, S. & Georgieva, K. in Sex Hormones in Neurodegenerative Processes and Diseases (ed. G. Drevenšek) 10.5772/intechopen.71140 (IntechOpen, 2017).
- 2.Dougherty BJ, Kopp ES, Watters JJ. Nongenomic Actions of 17-beta Estradiol Restore Respiratory Neuroplasticity in Young Ovariectomized Female Rats. J Neurosci. 2017;37:6648–6660. doi: 10.1523/JNEUROSCI.0433-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srivastava DP, et al. Rapid estrogen signaling in the brain: implications for the fine-tuning of neuronal circuitry. J Neurosci. 2011;31:16056–16063. doi: 10.1523/JNEUROSCI.4097-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Xiao X, Zhang XM, Zhao ZQ, Zhang YQ. Estrogen facilitates spinal cord synaptic transmission via membrane-bound estrogen receptors: implications for pain hypersensitivity. J Biol Chem. 2012;287:33268–33281. doi: 10.1074/jbc.M112.368142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielecki B, et al. Unexpected central role of the androgen receptor in the spontaneous regeneration of myelin. Proc Natl Acad Sci USA. 2016;113:14829–14834. doi: 10.1073/pnas.1614826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussain R, et al. The neural androgen receptor: a therapeutic target for myelin repair in chronic demyelination. Brain. 2013;136:132–146. doi: 10.1093/brain/aws284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, et al. Endocrine Therapy for the Functional Recovery of Spinal Cord Injury. Front Neurosci. 2020;14:590570. doi: 10.3389/fnins.2020.590570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byers JS, et al. Neuroprotective effects of testosterone on motoneuron and muscle morphology following spinal cord injury. J Comp Neurol. 2012;520:2683–2696. doi: 10.1002/cne.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sengelaub DR, et al. Protective Effects of Estradiol and Dihydrotestosterone following Spinal Cord Injury. J Neurotrauma. 2018;35:825–841. doi: 10.1089/neu.2017.5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sengelaub DR, Xu XM. Protective effects of gonadal hormones on spinal motoneurons following spinal cord injury. Neural Regen Res. 2018;13:971–976. doi: 10.4103/1673-5374.233434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acosta MC, Copley PA, Harrell JR, Wilhelm JC. Estrogen signaling is necessary for exercise-mediated enhancement of motoneuron participation in axon regeneration after peripheral nerve injury in mice. Dev Neurobiol. 2017;77:1133–1143. doi: 10.1002/dneu.22501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto, A. Androgen stimulates neuronal plasticity in the perineal motoneurons of aged male rats. J Comp Neurol430, 389–395 10.1002/1096-9861(20010212)430:3<389::AID-CNE1038>3.0.CO;2-3 (2001). [DOI] [PubMed]
- 13.Liu NJ, et al. Plasticity of Signaling by Spinal Estrogen Receptor α, κ-Opioid Receptor, and Metabotropic Glutamate Receptors over the Rat Reproductive Cycle Regulates Spinal Endomorphin 2 Antinociception: Relevance of Endogenous-Biased Agonism. J Neurosci. 2017;37:11181–11191. doi: 10.1523/JNEUROSCI.1927-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zabka AG, Behan M, Mitchell GS. Long term facilitation of respiratory motor output decreases with age in male rats. J Physiol. 2001;531:509–514. doi: 10.1111/j.1469-7793.2001.0509i.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zabka AG, Mitchell GS, Behan M. Ageing and gonadectomy have similar effects on hypoglossal long-term facilitation in male Fischer rats. J Physiol. 2005;563:557–568. doi: 10.1113/jphysiol.2004.077511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zabka AG, Mitchell GS, Behan M. Conversion from testosterone to oestradiol is required to modulate respiratory long-term facilitation in male rats. J Physiol. 2006;576:903–912. doi: 10.1113/jphysiol.2006.114850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barok R, Grittner JML, Dougherty BJ. The long-term impact of ovariectomy on ventilation and expression of phrenic long-term facilitation in female rats. Exp Physiol. 2021;106:2002–2012. doi: 10.1113/EP089546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIntosh D, Dougherty BJ. Development of ventilatory long-term facilitation is dependent on estrous cycle stage in adult female rats. Respir Physiol Neurobiol. 2019;264:1–7. doi: 10.1016/j.resp.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Dubal DB, Broestl L, Worden K. Sex and gonadal hormones in mouse models of Alzheimer’s disease: what is relevant to the human condition? Biol Sex Differ. 2012;3:24. doi: 10.1186/2042-6410-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller S, Abrahante J, Roopra A, Dougherty B. 2022. GEO. //identifiers.org/geo/GSE202381 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Miller S, Abrahante J, Roopra A, Dougherty B. 2022. GEO. //identifiers.org/geo/GSE202381 [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
No custom code was generated or used for analysis of the data presented.