Abstract
BY-kinases (for bacterial tyrosine kinases) constitute a family of protein tyrosine kinases that are highly conserved in the bacterial kingdom and occur most commonly as essential components of multicomponent assemblies responsible for the biosynthesis, polymerization, and export of complex polysaccharides involved in biofilm or capsule formation. BY-kinase function has been attributed to a cyclic process involving formation of an oligomeric species, its disassembly into constituent monomers, and subsequent reassembly, depending on the overall phosphorylation level of a C-terminal cluster of tyrosine residues. However, the relationship of this process to the active/inactive states of the enzyme and the mechanism of its integration into the polysaccharide production machinery remain unclear. Here, we synthesize the substantial body of biochemical, cell-biological, structural, and computational data, acquired over the nearly 3 decades since the discovery of BY-kinases, to suggest means by which they fulfill their physiological function. We propose a mechanism involving temporal coordination of the assembly/disassembly process with the autokinase activity of the enzyme and its ability to be dephosphorylated by its counteracting phosphatase. We speculate that this temporal control enables BY-kinases to function as molecular timers that coordinate the diverse processes involved in the synthesis, polymerization, and export of complex sugar derivatives. We suggest that BY-kinases, which deploy distinctive catalytic domains resembling P-loop nucleoside triphosphatases, have uniquely adapted this ancient fold to drive functional processes through exquisite spatiotemporal control over protein–protein interactions and conformational changes. It is our hope that the hypotheses proposed here will facilitate future experiments targeting these unique protein kinases.
Keywords: protein kinase, P-loop enzyme, bacterial tyrosine kinase, EPS, CPS
Abbreviations: BY-kinase, bacterial tyrosine kinase; CD, catalytic domain; CPS, capsular polysaccharide; CS, closed state; EPS, exopolysaccharide; LMW-PTP, low molecular weight protein tyrosine phosphatase; O-motif, oligomerization motif; OS, open state; PD, periplasmic domain; TAD, transactivation domain
Tyrosine phosphorylation, long recognized as a ubiquitous mode of signal transduction in higher eukaryotes (1, 2), is now known to also play a significant regulatory role in bacterial cells (3, 4). In the bacterial kingdom, this post-translational modification is largely driven by members of the BY-kinase (for bacterial tyrosine kinase) family (5, 6, 7) that are widely conserved in both Gram-positive and Gram-negative species (8) but have no known eukaryotic or archeal orthologs. While BY-kinases have been shown to contribute to diverse cellular processes (9, 10, 11), their most significant, and best studied, role is in the assembly and export of complex polysaccharides that are key components of biofilms (12, 13) or virulence-determining capsules (9, 14, 15) in many bacterial species. In a manner reminiscent of higher eukaryotes (16), BY-kinases appear capable of integrating into complex signaling networks, which also include serine/threonine kinases (17), to drive critical physiological processes.
In the 25 years that have elapsed since the assignment of Acinetobacter johnsonii Ptk (18, 19) as a bona fide protein tyrosine kinase, and a “founding” member of the BY-kinase family, a wealth of information on these enzymes has emerged from structural, biochemical, and cell-biological studies, reviewed in several excellent treatises (5, 6, 7, 20, 21). Despite the considerable progress, many critical aspects of the functional regulation of BY-kinases remain poorly understood. Recently, in silico approaches (22, 23) have begun to provide insight into hitherto unknown regulatory features of these unique enzymes. Here, we will draw on these recent results, from both theory and experiment, to interpret the available biochemical, cell-biological, and functional data on BY-kinases to suggest a mechanism for their activation and activity in the context of bacterial physiology, specifically in the synthesis and export of complex polysaccharides. We will also utilize this review as a vehicle to propose hypotheses that are amenable to future experimental validation/testing.
Escherichia coli Wzc, the archetypal (see later) BY-kinase, has been extensively studied using a variety of biophysical and biochemical approaches in the context of the K-12 and K30 strains by the Grangeasse and Whitfield groups among others. Wzc proteins from these two E. coli strains are very similar (sequence identity ∼52%) but are integrated into slightly different macromolecular assemblies that bridge the inner and outer membranes. The polysaccharide products are also slightly different in the two cases with the exopolysaccharide (EPS) colanic acid being produced in K-12 (24), and capsular polysaccharides (CPSs) utilized in forming group 1 capsules being generated in K30 (25). However, the overarching mechanisms underlying Wzc regulation appear to be identical in these two cases (we use Wzc interchangeably later unless explicitly stated). Furthermore, broad mechanistic insights obtained from studies on E. coli Wzc are expected to be relevant across the bacterial kingdom. Therefore, in our discussion later, we will rely heavily on data for E. coli Wzc, invoking available results from other systems as and when necessary.
An ancient fold for a new function
In a manner characteristic of most Gram-negative BY-kinases (Table 1), E. coli Wzc comprises an N-terminal “transactivation” domain (TAD) (26) and C-terminal catalytic domain (CD). The TAD consists of a two-pass transmembrane domain responsible for inner membrane localization and a periplasmic domain (PD) that is proposed to interact with the outer membrane transporter, Wza (27). The enzymatic function is encoded within the CD, which is fully cytosolic, and terminates in a flexible tail containing a cluster of tyrosine residues (the Y-cluster) (28, 29) (Fig. 1A) that are sites for intermolecular autophosphorylation. Both the number and sequential positions of the Y-cluster tyrosine residues are variable across the BY-kinase family. Contrasting Wzc, in many Gram-positive bacteria (Table 1), most notably, the firmicutes, the TAD and CD occur as two separate proteins, encoded by flanking genes, which must associate to form a functional membrane-anchored complex (30). This latter group has been termed firmicute- or F-type, contrasting the proteobacterial- or P-type BY-kinases (20) like Wzc. In addition, some bacterial genomes (8) contain so-called “orphan” CDs with no obvious TAD partners (Table 1). It has been speculated that these orphan CDs are activated by the TADs of another F-type BY-kinase also present in the cell (31). Interestingly, some of the orphan CDs are predicted to contain transmembrane segments (Table 1) suggesting nonstandard arrangements that likely represent species-specific variations on a common theme.
Table 1.
| P-type | F-type | Orphan CD with transmembrane domainsc | Orphan CD | Total | |
|---|---|---|---|---|---|
| Gram-positive | |||||
| Actinomycetota (Actinobacteria) | 1588 | 2 | 265 | 46 | 1901 |
| Bacillota (Firmicutes) | 281 | 441 | 133 | 5025 | 5880 |
| Mycoplasmatota (Tenericutes) | — | — | 1 | 39 | 40 |
| Deinococcota (Deinococcus-thermus) | 2 | — | 3 | — | 5 |
| Gram-negative | |||||
| Abditibacteriota | 2 | — | 1 | — | 3 |
| Acidobacteriota (Acidobacteria) | 424 | — | 74 | 238 | 736 |
| Aquificota (Aquificae) | 2 | — | 2 | — | 4 |
| Armatimonadota (Armatimonadetes) | 17 | — | 9 | 2 | 28 |
| Atribacterota | 1 | — | — | 2 | 3 |
| Bacteroidota (Bacteroidetes) | 792 | 3 | 237 | 20 | 1052 |
| Balneolota (Balneolaeota) | 13 | — | 6 | 19 | |
| Calditrichota (Calditrichaeota) | 30 | — | 10 | 7 | 47 |
| Chlorobiota (Chlorobi) | 34 | — | 4 | — | 38 |
| Chloroflexota (Chloroflexi) | 126 | 2 | 29 | 69 | 226 |
| Chrysiogenota (Chrysiogenetes) | — | — | 1 | 1 | |
| Cyanobacteria | 537 | — | 75 | 4 | 616 |
| Elusimicrobiota (Elusimicrobia) | 19 | — | 4 | 2 | 25 |
| Euryarchaeota (Euryarchaeota) | 2 | — | — | 4 | 6 |
| Fibrobacterota (Fibrobacteres) | 82 | — | 9 | 1 | 92 |
| Fusobacteriota (Fusobacteria) | — | 3 | — | 14 | 17 |
| Gemmatimonadota (Gemmatimonadetes) | 115 | — | 60 | 14 | 189 |
| Ignavibacteriota (Ignavibacteriae) | 46 | — | 20 | — | 66 |
| Kiritimatiellota (Kiritimatiellaeota) | 12 | — | 3 | — | 15 |
| Lentisphaerota (Lentisphaerae) | 19 | — | 12 | 1 | 32 |
| Nitrospirota (Nitrospirae) | 34 | — | 9 | 24 | 67 |
| Planctomycetota (Planctomycetes) | 256 | — | 215 | 21 | 492 |
| Pseudomonadota (Proteobacteria) | 10,741 | 28 | 422 | 592 | 11,783 |
| Rhodothermota (Rhodothermaeota) | 7 | — | 3 | — | 10 |
| Spirochaetota (Spirochaetes) | 22 | — | — | 2 | 24 |
| Thermodesulfobacteriota (Thermodesulfobacteria) | — | — | — | 3 | 3 |
| Thermotogota (Thermotogae) | 41 | — | 1 | — | 42 |
| Verrucomicrobiota (Verrucomicrobia) | 308 | 236 | 11 | 555 | |
| Unclassified/unknown | 286 | 1 | 79 | 173 | 539 |
| Total | 15,839 | 480 | 1922 | 6315 | 24,556 |
Based on sequence dataset in BYKdb release 67.0 (8).
Phyla are named according to the International Code of Nomenclature for Prokaryotes; historical synonyms are shown in parantheses.
Transmembrane segments predicted by DeepTMHMM (biolib.com/DTU/DeepTMHMM/).
Figure 1.
Structural organization of a functional BY-kinase.A, structure of full-length Escherichia coli K30 Wzc (K540M mutant; Protein Data Bank code: 7NHR; a P-type BY-kinase) showing the periplasmic domain (PD), the two-pass transmembrane domain (TMD) that is embedded in the bacterial inner membrane, and the cytoplasmic catalytic domain (CD). The TMD and PD together form the transactivation domain (TAD). The TAD and CD are separate proteins in F-type BY-kinases. The middle inset shows a representative monomer from the octameric assembly indicating the N-terminal transmembrane helix (TM1) that leads to the PD followed by a second transmembrane helix (TM2) that terminates in the CD. The top right inset shows a view of the CD octamer looking up from the cytoplasm into the periplasmic space; the TAD is hidden for ease in visualization. The C-terminal Y-cluster is colored red, and the constituent tyrosine residues are shown in stick representation. Shown on the bottom right panel is a representative trimer extracted from the octameric assembly that illustrates the enzyme–substrate dual role of each CD. A tyrosine from the Y-cluster of monomer a is inserted into the active site of monomer b, which in turn inserts a tyrosine from its own Y-cluster into the active site of monomer c. Thus, b can simultaneously act both as an enzyme (with respect to a) and as a substrate (with respect to c). B, interactions at the intermonomer interface of the CD of E. coli K-12 Wzc (K540M mutant; Protein Data Bank code: 3LA6) involving conserved O-motif residues E508, R511, and R514 on α2 (yellow) at the first interface (I1) with residues on α7 (light blue) and α9 (dark blue) that form the second interface (I2). The two interacting monomers (designated a and b) are colored pink and purple. Key interface residues are shown as spheres. C, the interaction of the E. coli K-12 Wzc CD with its cognate phosphatase, the LMW-PTP Wzb (teal); this interaction also involves α2 on the former and the O-motif therein. Selected interactions at the interface are illustrated with the residues involved (Wzb residues are indicated using three-letter codes) shown as spheres. D, the characteristic P-loop fold of Wzc CD is shown indicating the conserved motifs (key side chains are shown in stick representation; the motifs they belong to are indicated by the corresponding subscripts): Walker-A (533GVSPSIGKT541, teal; conserved residues are indicated in bold font), Walker-A’ (558VLLIDCDMR566, orange), and Walker-B (639VLIDTPP645, purple). Also indicated are α2, α7, and α9, colored as in (B). The Y-cluster tyrosine residues (Y705, Y708, Y710, Y711, Y713, and Y715) are also indicated (in red as sticks). Y705 (indicated by the “∗”) is neither autophosphorylated nor can it optimally access the Wzb active site. A substrate tyrosine residue (Y715S) from the Y-cluster of a flanking CD docked at the active site is also shown. BY-kinase, bacterial tyrosine kinase; LMW-PTP, low molecular weight protein tyrosine phosphatase; O-motif, oligomerization motif.
The BY-kinase CD possesses an inherent ability to self-associate, assembling into a ring-shaped octamer both in isolated form (30, 32) and in the context of the full-length protein (33) when the Y-cluster is dephosphorylated or sparsely phosphorylated. Within this octameric assembly, individual CD monomers associate in a front-to-back arrangement through a conserved EX2RX2R sequence (oligomerization motif [O-motif]; where X is any residue) on a helical segment (α2), which defines an interaction interface (I1) on one monomer, and engages helices α7 and α9, which comprise the complementary interface (I2) on a flanking monomer (Fig. 1B). This arrangement, with C8 rotational symmetry, enables each monomer to act as an enzyme, hosting a Y-cluster tyrosine from one flanking monomer at its active site, while simultaneously acting as a substrate by inserting a tyrosine from its own Y-cluster into the active site of the other flanking monomer (Fig. 1A, right panel). In Wzc, the α2 helix and the O-motif therein is also utilized to dock its counteracting low molecular weight protein tyrosine phosphatase (LMW-PTP) Wzb (Fig. 1C) (34, 35).
The BY-kinase CD, illustrated by several structures, for example, of E. coli Wzc (32, 33) and Etk (36), Staphylococcus aureus CapB (30), Vibrio cholerae VpsO (13), and others, bears no structural resemblance to the dual-lobed fold characteristic of eukaryotic protein kinases (37), deploying instead a variation on the P-loop scaffold (38) seen in NTPases. The P-loop fold, a primordial structural module that predates the emergence of the last universal common ancestor (39), is also utilized by many small-molecule kinases (40) but is exceedingly rare for a protein kinase; the dual-acting serine kinase/phosphorylase, HprK/P, found in some Gram-positive bacteria, being the only other known example (41). The BY-kinase active site (Fig. 1D) is lined by variants of the nucleoside-binding Walker-A (GX5GKS/T) and Walker-B (φ4DX2P; where φ is a hydrophobic residue) motifs (42). Also present is a so-called Walker-A′ motif (φ4DXDXR) reminiscent of the DXD sequences seen in some P-loop kinases, for example, shikimate and gluconate kinases (40). The second aspartate within the Walker-A′ motif has been proposed to play a critical role in recognizing the tyrosine substrate and driving chemistry (32). Mutations of conserved residues within the Walker motifs alter both the intrinsic ATPase activity and the autokinase activity in Wzc (23, 32, 43) and other BY-kinases (44, 45) (Table 2). This suggests that BY-kinases utilize an NTPase-like mechanism to transfer the γ-phosphate of ATP to a substrate −OH, which in this case is housed on a tyrosine rather than on water. It is therefore apparent that BY-kinases have evolved elaborate mechanisms establishing the primacy of their autokinase function over their ATPase activity as explored in greater detail later.
Table 2.
Influence of key mutations on the kinase and ATPase activities of select BY-kinases
| BY-kinase | Mutations | ATPase activity | Kinase activity | Reference |
|---|---|---|---|---|
| Bacillus subtilis YwqD | D81A, D83A (Walker-A′) | Greatly reduced | Abolished | Mijakovic (45) |
| Y225F/Y227F/Y228F (Y-cluster) | Enhanced | N/A | Mijakovic (45) | |
| Sinorhizobium meliloti ExoP | G588V, G588E, K589I (Walker-A) | Abolished | Abolished | Niemeyer (44) |
| E. coli K-12 Wzc | D562A, D564A (Walker-A′) | Reduced | Reduced | Soulat (43) |
| Y569N, Y569F | Reduced | Reduced | Soulat (43) | |
| K540M (Walker-A) | Abolished (Hajredini) | Reduced (Bechet) |
Assembly and disassembly, hallmarks of a functional cycle
Wzc function (32, 33), and that of BY-kinases (30) in general, has been ascribed to a cyclic assembly/disassembly process coupled to the phosphorylation state of the Y-cluster. As noted previously, formation of an octameric ring provides an efficient means of facilitating intramolecular autophosphorylation of the Y-cluster by allowing each monomer to simultaneously serve as both enzyme and substrate. The octameric ring disassembles into its constituent monomers after several rounds of autophosphorylation on the Y-cluster. Disassembly depends on the overall level of Y-cluster phosphorylation rather than the phosphorylation state of any specific tyrosine or set of tyrosine residues therein (28, 46). Reassembly occurs once the Y-cluster is sufficiently dephosphorylated by its counteracting phosphatase, which in the case of Wzc is the LMW-PTP Wzb (47), and the cycle reinitiates. This oscillatory assembly/disassembly process integrates into an elaborate multiprotein assembly (see, e.g., Fig. 4 of Whitfield (25) for an illustration) responsible for polysaccharide synthesis, polymerization, and export. The mechanism of this integration and the precise role of Wzc therein remain poorly defined.
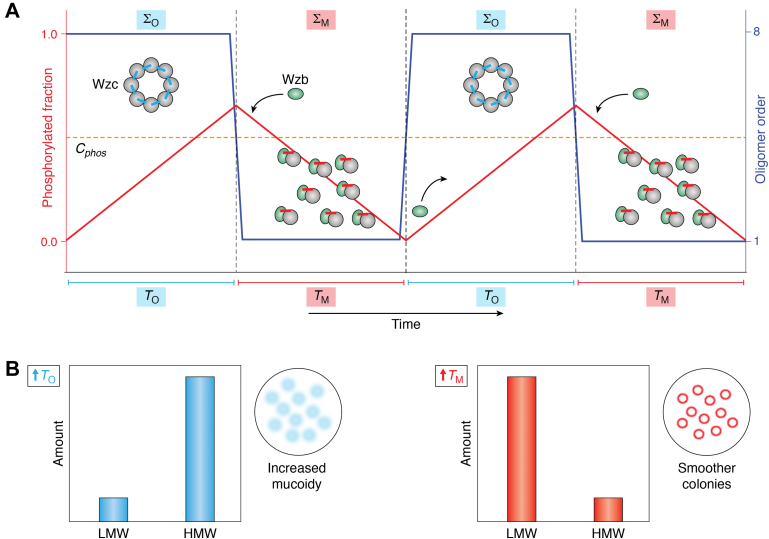
In its simplest conceptualization, the cyclic process may be conceived as involving transitions between two discrete functional species/states (Fig. 2A), designated ΣΟ and ΣΜ. For ΣΟ, CDs with dephosphorylated Y-clusters assemble into an octameric ring; ΣΜ comprises monomeric CDs with highly phosphorylated Y-clusters. Individual monomers that comprise ΣΟ adopt conformations that allow the efficient engagement of ATP–Mg2+ and are thus chemistry capable, that is, “active” (discussed in detail later). This assembly allows the Y-cluster tyrosine residues to be successively autophosphorylated at the expense of ATP (generating ADP), thereby enhancing the overall level of phosphorylation. This is accompanied by increasing intermonomer repulsion because of the accumulation of negative charges within the constituent Y-clusters. Once the level of phosphorylation exceeds a certain critical threshold (Cphos), estimated to be an average of about four phosphorylated tyrosine residues per monomer in Wzc (33), the electrostatic repulsion leads to dissociation of the octameric ring into its constituent monomers generating ΣΜ. As noted previously, Wzc oligomerization and its interaction with Wzb both involve I1 (Fig. 1, B and C) and the O-motif therein. Therefore, these two sets of interactions cannot occur simultaneously. Since Wzc is housed in the bacterial inner membrane, the resultant enhancement in local concentration favors intermonomer interactions over the diffusion-controlled association with the wholly cytosolic Wzb. Additional mechanisms that could potentially serve to exclude Wzb from interacting with CDs assembled within the ΣΟ species may also be conceived. For example, π–π stacking and hydrogen bonding between the tyrosine residues have been shown to result in fibril formation (48). Given that the Wzc active site can accommodate only a single tyrosine at any given time, formation of a viscous gel-like phase-separated state (49) resulting from interactions between exposed and still unphosphorylated Y-cluster tyrosines to further spatially exclude Wzb, in a manner somewhat reminiscent of that seen in T-cell receptor signaling (50), is not beyond the realm of possibility. It is worth testing this scenario through future experiments. Irrespective of the detailed mechanisms operative, it may be reasonably argued that the engagement of Wzc by Wzb can occur with any level of efficiency only after the Cphos threshold has been exceeded and the octamer has disassembled, that is, in the context of ΣΜ. It is also notable that Wzc CD monomers (ΣΜ) adopt conformations that are not chemistry compatible, that is, “inactive” (discussed later). This provides an additional level of control facilitating Y-cluster dephosphorylation catalyzed by Wzb while preventing the competing (and unproductive) rephosphorylation of the Y-cluster through transient interactions between monomers (or perhaps through intramolecular phosphoryl transfer). Furthermore, since the Wzc I1 is protected by the associated Wzb through an interaction that is further enhanced by the engagement of a phosphorylated Y-cluster tyrosine at the Wzb active site and the resultant avidity (∼10 μM affinity estimated for interactions outside the Wzb active site and an additional ∼250 μM for the interaction of a phosphotyrosine with the Wzb catalytic elements) (35), it is possible that dephosphorylation continues to levels below the Cphos threshold (hysteresis), perhaps to completion in some cases. Sufficient reduction in Y-cluster phosphorylation levels ultimately regenerates the octameric species (ΣΟ) and reinitiates the cyclic process. This proposed bistable behavior and accompanying hysteresis is not unusual in systems regulated through multisite phosphorylation/dephosphorylation cycles (51). It is to be mentioned here, that in some Gram-positive bacteria, the phosphatase partner of the BY-kinase is not an LMW-PTP but rather belongs to the polymerase and histidinol phosphatase family, for example, Streptococcus pneumoniae CpsB or S. aureus CapC (52). Precisely, how these phosphatases engage their cognate BY-kinase CDs is not known. Additional studies, perhaps like those described for the Wzc/Wzb pair (34, 35), are necessary to establish the universality of the mechanism described previously on the precise mechanistic role of phosphatases in the context of the assembly/disassembly process.
Figure 2.
A simple model for BY-kinase function.A, schematic representation of the effects of autophosphorylation and dephosphorylation (red line) on the temporal distribution of the octameric (ΣO) and monomeric (ΣM) species (dark blue line). The left ordinate indicates the fraction of phosphorylated Y-cluster tyrosine residues; the right ordinate indicates the oligomer order. Autophosphorylation occurs at a given rate within the octameric state until the Y-cluster phosphorylation levels exceed a critical threshold (Cphos, dashed orange line), following which the octameric ring dissociates into its constituent monomers. The phosphatase (light green) can then engage the kinase and perform multiple rounds of Y-cluster dephosphorylation, following which the oligomer reassembles and the cycle repeats (two cycles are shown). Assuming fixed rates of autophosphorylation and dephosphorylation (in reality, these rates are likely to be nonlinear functions of the level of Y-cluster phosphorylation), the system cycles between ΣO and ΣM; the corresponding lifetimes of TO and TM, respectively, define the periodicity of the process. B, increasing the lifetime of ΣO (relative to the lifetime of ΣM), that is, enhancement of TO (relative to TM) shifts the distribution toward high molecular weight (HMW) polysaccharides and increased mucoidy (blurred circles in the inset); a larger TM (relative to TO) results in increased production of low molecular weight (LMW) polysaccharides and smoother colonies (sharp circles in the inset). BY-kinase, bacterial tyrosine kinase.
Altered cycle, modified product
As mentioned earlier, Wzc is part of a complex multiprotein machine responsible for the synthesis, polymerization, and export of polysaccharides (24, 25) utilized in capsule or biofilm formation. It has been shown that mutations that perturb the phosphorylation/dephosphorylation cycle and thereby the associated disassembly/assembly process in Wzc, and other BY-kinases (see later), strongly influence the properties of the polysaccharide product. A mechanism for potentially achieving this effect is through alteration of the relative lifetimes of the ΣΟ and ΣΜ species (thereby altering the periodicity of the assembly/disassembly process) to perturb the coordination between the synthesis of the sugar subunits and their subsequent polymerization. While the details of how Wzc integrates into this scheme remain obscure, mutations that putatively stabilize it (53), or other BY-kinases (13, 15, 44, 54, 55, 56, 57) (Table 3), in states that mimic ΣΟ or ΣΜ, provide important clues. Y-cluster Y-to-F mutants or catalytically dead (or compromised) variants that prevent (or impair) Y-cluster phosphorylation are expected to stabilize ΣΟ (increase TO in Fig. 2A). Mutations that mimic the enhanced negative charge on the Y-cluster because of phosphorylation (Y-cluster Y-to-E mutants), or variants with modified O-motifs impaired in the ability to oligomerize, would lead to a stabilized ΣΜ (increased TM). Analysis of the available functional data (Table 3) suggests that, in general, putative ΣΟ-stabilizers shift the product distribution toward high molecular weight polysaccharides or yield bacterial colonies that display enhanced mucoidy (indicative of increased levels of high molecular weight polymers). In contrast, mutations that are expected to stabilize ΣΜ result in a higher proportion of low molecular weight polysaccharides and smoother bacterial colonies. These observations seem to suggest that increasing TΟ results in aberrant extension of the polymer chains, whereas increasing TΜ results in reduced polymerization (Fig. 2B). These correlations are, expectedly, not perfect, given the simplicity of the model shown in Figure 2A, the diversity of experimental conditions, and system/strain-dependent idiosyncrasies (see Table 3). For example, E. coli K-12, cells expressing wildtype Wzc, produce EPS with a broad size distribution in 600 to 2000 kDa range. In contrast, ΣΟ-stabilizers (Y-cluster Y-to-F or Y569F mutants; Y569 plays a significant functional role described later) result in a more uniform distribution centered around ∼1350 kDa, whereas putative ΣΜ-stabilizers (Y-cluster Y-to-E mutants) are unable to produce EPS, a similar phenotype as the cells expressing a construct missing the catalytic Walker-A lysine (K540M) that would be expected to stabilize ΣΟ (53). Despite these nuances, it would not be too bold to speculate that the cyclic processes attributed to BY-kinase function serve to coordinate the polysaccharide synthesis, oligomerization, and perhaps, the export processes.
Table 3.
Effects of BY-kinase mutations on observed phenotype
| BY-kinase | Mutation | Kinase activity | Phenotype | Proposed to stabilize | Reference |
|---|---|---|---|---|---|
| V. cholerae VpsO | K551A (Walker-A) | Abolished | Rougher colonies, higher EPS levels | ΣΟ | Schwechheimer (13) |
| VpsU C12S (inactive cognate LMW-PTP) | — | Smooth colonies, lower EPS levels | ΣΜ | ||
| E519A/R522A/R525A (O-motif) | Reduced | Smooth colonies, lower EPS levels | ΣΜ | ||
| Y720F/Y721F/Y726F/Y727F (Y-cluster) | — | Smooth colonies, lower EPS levels | ΣO | ||
| E. coli K-12 Wzc | Y708E/Y710E/Y11E/Y713E/Y715E (Y-cluster) | — | No EPS | ΣΜ | Obadia (53) |
| ΔY-cluster | — | No EPS | ΣΜ | ||
| K540M (Walker-A) | Abolished | No EPS | ΣΟ | ||
| Y708F/Y710F/Y11F/Y713F/Y715F (Y-cluster) | — | Uniform EPS size distribution ∼1350 kDa (600–2000 kDa in WT) | ΣΟ | ||
| Y569F | Reduced | Uniform EPS size distribution ∼1350 kDa (600–2000 kDa in WT) | ΣΟ | ||
| S. pneumoniae CpsD | Y215F/Y218F/Y221F (Y-cluster) | — | Decreased levels of LMW EPS, no effect on total abundance | ΣΟ | Nakamoto (15) |
| Y215E/Y218E/Y221E (Y-cluster) | — | Increased levels of LMW EPS, no effect on total abundance | ΣΜ | ||
| Acinetobacter baumannii Wzc | Y712F/Y714F/Y716F/Y718F/Y720F/Y722F (Y-cluster) | — | Increased levels of high molecular weight (HMW) CPS | ΣΟ | Geisinger (54) |
| D569N (Walker-A′) | Reduced | Increased levels of HMW CPS | ΣΟ | ||
| K547Q (Walker-A) | Not affected | Increased levels of HMW CPS | ΣΟ | ||
| D649N (Walker-B) | Not affected | Increased levels of HMW CPS | ΣΟ | ||
| Burkholderia cepacia BceF | ΔBceD (deletion of cognate LMW-PTP) | — | Reduced viscosity suggestive of LMW EPS | ΣΜ | Ferreira (55) |
| Acinetobacter lwoffii Wzc | Y712F/Y714F/Y716F/Y718F/Y720F (Y-cluster) | — | Increased mucoidy, enhanced levels of higher MW EPS | ΣΟ | Nakar (56) |
| B. subtilis EpsB | Y225F/Y227F (Y-cluster) | — | Hyperwrinkled colonies | ΣΟ | Esholz (57) |
| Y225E/Y227E (Y-cluster) | — | Flat colonies | ΣΜ |
Open and closed, inactive and active
As was mentioned earlier, Wzc CD monomers are compatible with chemistry when part of ΣΟ, and inactive as ΣΜ. This provides additional temporal separation between the autophosphorylation and dephosphorylation events. Enhanced sampling molecular dynamics simulations suggest that monomeric Wzc CD accesses distinct conformations in its various liganded states (22). These conformations are distinguished by global changes involving a displacement of helix α4 and rotations of helices α2 and α3 with respect to the protein core (Fig. 3A). These changes are best represented in a two-dimensional coordinate system comprising an angle θ and a rise |h| (Fig. 3B) (22). Projection of the structural ensembles onto θ−|h| space reveals two dominant conformations, an open state (OS) with high values of both θ and |h| and a closed state (CS), with low values of both variables. While apo Wzc CD exists almost exclusively in the OS, the presence of ADP–Mg2+or ATP–Mg2+, but not ADP alone, results in a substantial population shift to the CS, suggesting that this transition is induced by Mg2+, rather than by nucleotide (Fig. 3C) (22). The OS is not compatible with formation of the closed octameric ring. This property is clearly illustrated by the catalytically inactive K540M mutant that populates an OS that is significantly more extended than for the wildtype enzyme when free of ligands and transitions almost completely into a CS when bound to ATP–Mg2+ (Fig. 3D, top panels). In line with these predictions, this mutant exists in a monomeric state at low Mg2+ levels despite containing a dephosphorylated Y-cluster; the presence of excess Mg2+ (also ATP–Mg2+or ADP–Mg2+) results in appearance of the oligomeric species (Fig. 3D, bottom panel) (23).
Figure 3.
Coupling between global and local conformations in the Wzc CD.A, global conformational changes that drive the transition from the closed state (CS, cyan) to the open state (OS, lime) are represented by the red arrows. B, definition of the 2-dimensional frame of reference that parses the OS and CS. P1: center of mass (COM, indicated by colored spheres in all cases) of the Cα atoms of the first two turns of the N terminus of α3 (541–547; Escherichia coli K-12 Wzc numbering, brown); P2: COM of Cα atoms of the penultimate turn (548–552, red) from the C terminus of α3; P3: COM of Cα atoms of the 569 to 573 segment (pink) on α4; and P4: COM of the Cα atoms of the first turn of the N terminus of α2 (505–509, green). The structures are rotated and translated to place P1 and P2 along the Cartesian z-axis and then rotated about the z-axis to place P3 on the x-axis in the positive direction. The |h| coordinate is then defined as the absolute value of the z-projection of P1; θ is defined as the angle between the x-axis and the xy-projection of a vector directed from P1 to P4. C, probability density plots of the conformational ensembles obtained through enhanced sampling MD simulations of apo Wzc CD (top left), and the corresponding ATP–Mg2+ (top right), ADP–Mg2+ (bottom left), and ADP (bottom right) complexes in θ−|h| space. The green circle represents the position occupied by the crystallographic monomers. Regions corresponding to the OS and CS are indicated. An additional “hyperclosed state” (HS), which contains structural features characteristic of a fully active state, sampled only in the ATP–Mg2+ complex, is also shown. D, probability density plots of the structural ensembles of the K540M mutant of Wzc CD in its unliganded state (top left), and its ATP–Mg2+ (top right) complex, are represented in θ−|h| space; the plots for the corresponding WT cases are shown as green contours with the positions of key states indicated. The lower plot shows gel filtration traces of the untreated mutant CD and that incubated with excess Mg2+. E, OS/CS transitions are correlated with local rearrangements at the active site. In the CS, the Walker-B D642 forms a hydrogen bond (lime dashed lines) with the Walker-A T541 enabling coordination of the catalytically essential Mg2+ ion (not shown for visual ease). This interaction is broken in the OS with the Walker-A K540 establishing interactions with the D642 side chain and the T643 backbone in a manner that neither allows optimal coordination of Mg2+ nor the availability of K540 for chemistry. Parts of this figure were adapted from Hajredini et al. (22, 23); © the authors, some rights reserved; exclusive licensee AAAS. Distributed under a Creative Commons Attribution noncommercial license 4.0 (CC BY-NC). CD, catalytic domain; MD, molecular dynamics.
The global conformational changes that distinguish the CS and OS are also correlated with local rearrangements of conserved elements at the active site (Fig. 3E). Within the CS, the conserved Walker-B aspartate (D642 in E. coli K-12 Wzc) interacts with the Walker-A signature threonine (T541) to yield a configuration necessary for the optimal coordination of the catalytically essential Mg2+ cation (23), suggesting a state that encodes features compatible with chemistry. This active-site geometry appears to be a universal characteristic of the CSs of P-loop enzymes (58). Perhaps the geometric constraints on such a state are defined by the ability to yield a Michaelis complex appropriate for NTP hydrolysis. In contrast, in the OS, the Walker-B aspartate (D642) traps the catalytic Walker-A lysine (K540) in a salt bridge, with the latter also forming a hydrogen bond with the backbone of a residue (T643) that immediately follows the Walker-B aspartate. This alignment is incompatible with Mg2+, and therefore ATP–Mg2+ (ATP is mostly Mg2+ bound, see discussion later), coordination suggesting that the OS represents an inactive state. It is notable that while all P-loop enzymes populate states that are more open than the reactive CSs described previously, the former are stabilized by diverse sets of interactions that, unlike the universal conservation of the defining features of the latter, are dependent on the family/class of P-loop enzyme. This suggests that diversification of the open and inactive states has enabled adaptation of the basic P-loop fold to different functional contexts by the acquisition of family specific structural detail (58). This feature, that is, similar active-state geometries with divergent inactive-state geometries, has been proposed for eukaryotic protein kinases (59), and we suspect that this may also hold in other large enzyme families.
As noted previously, the presence of ATP–Mg2+ leads to a shift of population into the CS for Wzc CD monomers. However, in such cases, the CS (while in ΣM) ensemble comprises structures that show significant distortions at the interfaces I1 and I2 that are involved in oligomer formation (transition to ΣO, Fig. 4A). These include a significant bending of α2 (I1) or the partial unfolding of α7 and/or α9 (I2) (22), suggesting that formation of the CS upon ATP–Mg2+ binding within a monomeric (ΣΜ) species results in a loss of several hydrogen bonds at the two interaction interfaces. This instability may be resolved by ejection of ATP–Mg2+ and transition back to the OS where α2, α7, and α9 are intact. One may, therefore, attribute the tendency of monomeric Wzc to form the OS to the inherent helical propensity of the two interaction interfaces. Oligomer formation (ΣΟ) stabilizes these interfaces through interactions between flanking monomers, resulting in a stable CS that can then efficiently engage ATP-Mg2+. One can therefore conceptualize the CS formed in the presence of ATP–Mg2+ in the CD monomers (ΣΜ) as “mechanically strained.” This strain is “released” in the oligomer (ΣΟ) through intermonomer interactions and resultant stabilization of the interfaces, I1 and I2 (Fig. 4B). For the Wzc CD, the CS may also be stabilized in the context of the monomer (ΣΜ) by engineering “strain-releasing” mutations. Such a variant is illustrated by the “G4-construct” generated by the in silico replacement of the 523 to 526 segment at the C-terminus of α2 by glycine residues, thereby introducing disorder but away from the region that participates directly in oligomer formation (Fig. 4C) (22).
Figure 4.
Structural destabilization in the CS of monomeric Wzc CD.A, major conformations selected from within the CS ensemble obtained through enhanced sampling MD simulations of monomeric Wzc CD bound to ATP–Mg2+. These structures are characterized by distortions of the two oligomerization interfaces including a significant bending (red arrows) of α2 (yellow; I1; left panel) or a partial unfolding (red circles) of α7 (light blue; I2) and/or α9 (dark blue; I2; right panel). ATP–Mg2+ is not shown for ease in visualization. B, classical MD simulations of the ATP–Mg2+ complex of Wzc CD (red) as part of a trimeric “pseudoring” (used in lieu of the octamer for computational convenience) where positional restraints are applied to the flanking monomers (cyan) to mimic the octameric ring (top left) or in its monomeric state (top right). The corresponding projections of structures from representative 100 ns classical MD simulations onto θ−|h| space are shown on the lower panels; the points are color coded according to the simulation time stamps. The black circle indicates the reference value in the crystal structure of the octamer. The CS is stabilized in the pseudoring preventing transition to the OS. C, the CS for the Wzc CD in the monomeric state may be stabilized through the introduction of a 4-glycine sequence (G4) at the C terminus of the α2 away from the O-motif (G4-construct). The structural ensemble of the ATP–Mg2+ complex (top) of the G4-construct obtained through enhanced sampling MD simulations populates a stable CS in which the constituent structures display intact oligomerization interfaces free from distortions in α2, α7, or α9. The disordered G4 region at the C terminus of α2 is indicated by the orange circle; the positions of the conserved O-motif residues are also indicated in green for reference. The bottom panel shows the probability density plot of the G4-construct–ATP–Mg2+ complex projected onto in θ-|h| space. The structure seen in crystallo (representative of the CS) is shown as a green circle. Parts of this figure were adapted with permission from Hajredini et al. (22, 23); © the authors, some rights reserved; exclusive licensee AAAS. Distributed under a Creative Commons Attribution noncommercial license 4.0 (CC BY-NC). CD, catalytic domain; CS, closed state; MD, molecular dynamics; O-motif, oligomerization motif; OS, open state.
An intricate dance in space and time
Based on the observations described previously, it is possible to propose a comprehensive model for the regulation of Wzc (as a representative BY-kinase) through Y-cluster phosphorylation (shown schematically in Fig. 5A). Oligomerization (ΣΟ) results in a stable CS and formation of the associated critical hydrogen bond between the Walker-B aspartate (D642 in E. coli K-12 Wzc) and the Walker-A serine/threonine (T541) allowing optimal engagement of ATP–Mg2+ and the exchange of the bound ADP through mass action. It is worth noting that in the cellular context, ATP, rather than ADP, is significantly more likely to be bound to Mg2+, given the ∼20-fold higher Mg2+ affinity of the former compared with the latter (60). Formation of the CS also generates additional structural features that are conducive to chemistry (Fig. 5B). ATP (ATP–Mg2+) engages the CS with its triphosphate moiety trending toward a reactive high-energy all-eclipsed conformation (23) known to promote hydrolysis (61, 62). Furthermore, formation of the CS (and of the so-called hyper-CS, which represents a greater degree of closure, only observed in the ATP–Mg2+ complex, and may be considered an extreme case of the CS; Fig. 3C) enables the interaction of a conserved tyrosine (Y569) with the α-phosphate of ATP, and ordering of a basic element, the so-called RK-cluster (32), to allow the insertion of a positively charged residue (R490) therein into the active site. These events serve to promote the desolvation of ATP (23), a feature that makes a favorable free-energy contribution to promote hydrolysis (63, 64). The basic RK-cluster residue (R490) also appears to play a role equivalent to the so-called “arginine finger” that has been shown to “activate” the γ-phosphate of NTP in P-loop NTPases (62, 65). Furthermore, oligomerization and engagement of a substrate Y-cluster tyrosine at the active site prior to the stable binding of ATP–Mg2+ and the consequent displacement of water prevents futile ATP hydrolysis through the intrinsic ATPase activity (43). This sequence of events ensures phosphotransfer to tyrosine rather than to water, thereby establishing the dominance of the kinase activity over the ATPase function of Wzc. Following multiple rounds of phosphorylation at the expense of ATP and increasing negative charge on the Y-clusters, the oligomer becomes progressively unstable, finally dissociating into its constituent monomers (ΣΜ) once the Cphos threshold (Fig. 2A) is exceeded. While the exchange of the ADP with ATP (ATP–Mg2+) can occur in optimal fashion within ΣΟ, it can no longer take place within ΣΜ that now adopts an OS. Thus, Wzc CD monomers remain stably bound to the ADP product from the last round of phosphorylation prior to disassembly. Mg2+ can no longer be optimally coordinated because of the loss of the D642–T541 hydrogen bond in the OS. Furthermore, the presence of an unobstructed I1 interface allows Wzb to dock onto Wzc and catalyze Y-cluster dephosphorylation. Once sufficiently dephosphorylated, the monomers can transit back into ΣΟ through formation of the octameric ring to reinitiate the cycle. Thus, activity is driven by coordinated conformational transitions on multiple length scales ranging from supramolecular events (assembly and disassembly), at the level of the tertiary fold (CS and OS), and locally at the active site (side-chain reorientation, realignment of secondary elements, and exclusion of solvent).
Figure 5.
Global and local features in the regulation of Wzc activity.A, in clockwise fashion starting from the point indicated by the red “∗.” In the oligomer (ΣΟ), individual Wzc CD monomers (indicated by the green outlines) with dephosphorylated Y-clusters (cyan sticks) exist in the CS. As shown in the inset, the two oligomerization interfaces, I1 (α2 in yellow) and I2 (α7 and α9 in light and dark blue, respectively), are stabilized through intermonomer interactions though some disorder persists (red arcs) at the C terminus of α2 away from the O-motif. The CS allows formation of the characteristic D642B–T541A hydrogen bond (Fig. 3E) and thereby the stable engagement of ATP–Mg2+ (orange) that displaces the bound ADP (purple) from the active site. Binding of ATP–Mg2+, multiple rounds of Y-cluster phosphorylation (indicated by progressively darker shades of orange), and nucleotide exchange within ΣΟ ultimately results in the Cphos threshold being exceeded and disassembly of the oligomer into its constituent monomers (ΣΜ). These monomers exist in the OS (monomers shown with dark blue outlines) bound to ADP but unable to engage Mg2+ (and therefore ATP–Mg2+). While a CS (CSM, shown in the inset) may be forced within ΣΜ by the presence of ATP–Mg2+, such a state shows instability in I1 (through bending of α2; green arrows) or I2 (through partial unfolding of the α7 and/or α9). The RK-cluster (in brown) is disordered in ΣΜ; its ordering for a stabilized CS facilitates the formation of a reactive state. In ΣΜ, stable engagement with Wzb (shown in green) through I1 initiates Y-cluster dephosphorylation. ΣΟ formation can occur after several rounds of Wzb-mediated dephosphorylation, and the functional cycle can be reinitiated. It is likely that the assembly/disassembly process also involves partially assembled/disassembled states (dashed boxes) representing a deviation from the simple two-state scheme presented in Figure 2. Some individual monomers in such cases also likely contain features that deviate (pink outlines) from those that are characteristic of the OS and CS, perhaps containing properties of both in addition to unique ones. B, engagement within the CS facilitates the formation of a reactive conformation of ATP. ATP is shown as a Newman projection with its α-, β-, and γ-phosphate moieties indicated by blue, yellow, and red lines, respectively. The Walker-A lysine (K540A, scarlet sphere) and the catalytic Mg2+ ion (green circle), lock the β- and γ-phosphates within the same plane. Rotation of the α-phosphate is enabled through its engagement with Y569, thereby facilitating formation of a high-energy all-eclipsed conformation. The ordering of the RK-cluster and insertion of R490 (cyan circle) therein, together with Y569, and a substrate tyrosine (from the Y-cluster of a flanking monomer; red), into the active site facilitate desolvation (dark blue dashes) providing favorable free energy contributions to overcome the activation barrier for chemistry. The precise sequence of the conformational events mentioned that lead to the reactive state remains unclear. Parts of this figure were adapted with permission from Hajredini et al. (22); © the authors, some rights reserved; exclusive licensee AAAS. Distributed under a Creative Commons Attribution noncommercial license 4.0 (CC BY-NC). CD, catalytic domain; CS, closed state; O-motif, oligomerization motif; OS, open state.
Dynamic regions as hot spots for adaptation
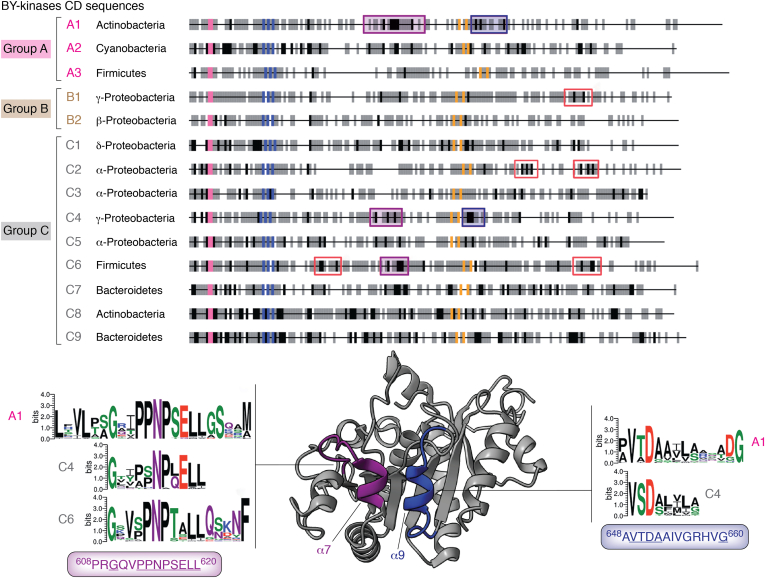
BY-kinases show mutation rates that are higher than those of other bacterial genes (31). Overall, there is sparse conservation outside the Walker motifs and for specific residues with known functional roles (e.g., Y569, discussed previously). Nevertheless, BY-kinase CD sequences clustered using protein similarity network analysis (66) parse into distinct groups each with multiple subgroups containing signature motifs beyond the broadly conserved “catalytic” residues (Fig. 6; the readers are referred to the study by Shi et al. (31) for further detail). A subset of defining features of these motifs is retained within some groups, but not others, suggesting that these regions have differentiated within the subgroups and across groups of BY-kinase CDs. Interestingly, some of these conserved elements overlap with those that display significant conformational variability between functional states, for example, the CS and OS, discussed previously. A prime example of such a region includes helices α7 and α9 that form I2 (Fig. 6) and comprise regions of instability within the CS (CSM in Fig. 5A) of the monomeric (ΣΜ) species.
Figure 6.
Dynamic regions as hot spots for adaptation in BY-kinases. Clustering of 796 BY-kinase CD sequences results in three large groups—A (labeled in magenta), B (brown), and C (gray). Group A comprises three subgroups—A1 (mostly Actinobacteria, dominant constituent phyla indicated in all cases using historical phyla names), A2 (Cyanobacteria), and A3 (Firmicutes). Group B comprises two subgroups—B1 (Gammaproteobacteria) and B2 (Betaproteobacteria). Group C comprises nine subgroups—C1 (Deltaproteobacteria), C2 (Alphaproteobacteria), C3 (Alphaproteobacteria), C4 (Gammaproteobacteria), C5 (Alphaproteobacteria), C6 (Firmicutes), C7 (Bacteroidetes), C8 (Actinobacteria), and C9 (Bacteroidetes). Each of these groupings display specific signature motifs (strongly conserved positions are indicated by black and gray bars; conserved motifs are indicated by the red, dark blue, and purple boxes) beyond the universally conserved Walker motifs (indicated by the colored bars; Walker-A: pink, Walker-A’: orange, and Walker-B: blue). Key features of some of these motifs are conserved across some groups but not others. Some of these motifs overlap with dynamic regions, for example, α7 (purple; similar in A1, C4, and C6) and α9 (dark blue; similar in A1 and C4). The corresponding sequence logos are indicated for each case, and that from the Escherichia coli (K-12) Wzc sequence is shown below; residues with high level of conservation are underlined. Parts of this figure have been modified with permission from Shi et al. (31). BY-kinase, bacterial tyrosine kinase; CD, catalytic domain.
A possible role of mutations localized within key conformationally labile regions could be to impart fitness in pathogenic species by altering the stabilities of the OS and CS, thereby affecting the relative lifetimes of ΣΜ and ΣΟ and allowing rapid adaption to environmental cues. It has been shown that Wzc mutations enable drug-resistant strains of Klebsiella pneumoniae to adapt to the urinary tract by regulating mucoidy (67). G569, which lies on α4 (rigid body motions of α4 distinguish the CS and OS; Fig. 3A), was found to be extensively mutated (G569C, G569V, and G569D). These modifications were causative for enhanced mucoidy, a characteristic that we attribute to a stabilized ΣΟ. Indeed, these mutations also resulted in decreased kinase activity, a reduction that would yield an increased TO in our simple model (Fig. 2). Furthermore, an insertion at the C-terminal end of α2 (at position 526) was also found to enhance mucoidy without reducing kinase activity (67). It is notable that this insertion coincides with the region modified to generate the G4-construct that stabilizes the CS in E. coli Wzc (22), as discussed previously (Fig. 4C). It is conceivable that this insertion, by increasing disorder in this region, produces a similar effect in K. pneumoniae Wzc of stabilizing the CS and thereby ΣΟ.
A molecular timer for polysaccharide production
As noted previously, perturbations in phosphorylation/dephosphorylation-coupled oscillations between the octameric (ΣΟ) and monomeric (ΣΜ) species modify the physiological output. Specifically, it appears that modulation of BY-kinase activity alters the size distribution of the polysaccharide product suggesting a change in the balance between the synthesis of constituent sugar units and their polymerization within the multicomponent machine (24, 25). One may therefore speculate that the proposed switch-like behavior driven by Y-cluster phosphorylation could potentially provide a timing mechanism to maintain coherence between these events. We propose that BY-kinase activity serves the role of a “molecular timer” that coordinates the various processes involved in the generation of the polysaccharide product. We suggest that this coordination is achieved by modulating the lifetimes of the octameric (ΣΟ) and monomeric (ΣΜ) forms of the enzyme to enable adaptation to specific conditions in a particular bacterium under altered environmental pressures (short evolutionary timescales) or in different bacterial species (long evolutionary timescales). These lifetimes are related to the degree of instability introduced into the oligomeric form by each successive Y-cluster phosphorylation event and on the corresponding autophosphorylation and dephosphorylation rates. One could conceive of several ways by which this could potentially be achieved. For example, changes in the rates of phosphorylation and/or dephosphorylation would affect TO and/or TM (Fig. 2B). Alternatively, modifications in the relative stabilities of the CS or the OS and therefore the ability to generate ΣΟ or ΣΜ species with increased/decreased Cphos thresholds could also provide similar outcomes. For example, a CS stabilized through the introduction of disorder at the C terminus of α2, as in the G4-construct of Wzc mentioned previously (22), likely generates a ΣΟ that would require a higher Cphos threshold for dissociation compared with the wildtype enzyme.
An intriguing example that suggests functional regulation by modulation of the relative lifetimes of the ΣΟ and ΣΜ species is provided by the Xanthomonas campestris inner-membrane protein, GumC. However, in this case, modulation is achieved through a unique phosphorylation-independent mechanism (68). GumC contains a cytosolic domain that closely resembles a BY-kinase CD but lacks the Y-cluster. Overexpression of GumC results in xanthan molecules with increased chain lengths suggestive of a stabilized ΣΟ and an enhanced TO. While the detailed biochemical characteristics of this system are not yet known, it is conceivable that activity in GumC is regulated through clustering in a manner reminiscent of the scaffolding function of eukaryotic pseudokinases (indeed it would not be too much of a stretch to classify GumC as a “pseudo-BY-kinase”) (69). Thus, enhanced local concentration from increased expression of this membrane-anchored protein potentially stabilizes the oligomeric ΣΟ and yields the characteristic phenotype. The ΣΟ species can be “dissolved” through reduced expression levels and decreased local concentration.
Looking back, looking forward
Here, we have attempted to synthesize the large body of cell-biological, biochemical, structural, and computational data into a coherent framework to interpret the mechanisms underlying the activation and activity of BY-kinases. We suggest that BY-kinases integrate into the polysaccharide synthesis machinery by functioning as molecular timers that serve to synchronize the synthesis and polymerization of complex carbohydrates in a manner unique to a given bacterial species. Such “timing control” has been suggested for other multisite phosphorylation-driven systems (70). Several observations raise the tantalizing possibility of such a role for BY-kinase function. In S. aureus, activity of the functional complex comprising the catalytic CapB and its membrane-anchored TAD partner, CapA, appears to synchronize capsule assembly and cell wall biosynthesis (71). In S. pneumoniae, the phosphorylation/dephosphorylation cycle of the BY-kinase CpsD is required for the appropriate coupling of cell division and capsule synthesis (14). However, this idea of a molecular clock remains in the realm of speculation pending verification through precisely designed future experiments. In this regard, a defining set of measurements could involve obtaining quantitative estimates of the cellular lifetimes of the ΣO and ΣM species (and perhaps additional ones in more comprehensive models), the influence of alterations (through mutations like those described previously) thereon, and resultant changes in the nature of the polysaccharide product.
In the aforementioned discussions, we have largely focused on interactions within the cytosol, that is, those involving the BY-kinase CD and the catalytic activity encoded therein. There is mounting evidence that extracytoplasmic events also affect the physiological outcomes. For example, despite close similarities, E. coli K-12 Wzc is incapable of substituting for the native enzyme in E. coli K30 cells apparently decoupling CPS synthesis from their display on the cell surface suggesting altered export properties (72). It has been proposed that the outer membrane transporter Wza is gated through interactions with the Wzc PD in the periplasm (33). Deletion of the so-called motif 3 (33), a helical segment in the Wzc PD that is a likely interaction partner of Wza, results in a phenotype that is similar to that of a Wza deletion strain without affecting the ability to form the octameric species or perturbing kinase activity (33, 73). Whether and how these intracytoplasmic and extracytoplasmic events communicate with each other remains unknown. It is possible that the extracytoplasmic unit, necessarily part of a functional assembly involving a BY-kinase (as a single protein in case of a P-type or as a separate TAD protein for an F-type BY-kinase), could serve as a sensor to elicit activating/deactivating conformational changes in the CD by means of the transmembrane domain, perhaps also influenced by the lipid membrane. Such a mode of communication has been suggested for the eukaryotic receptor tyrosine kinase, epidermal growth factor receptor (74, 75). Indeed, observation of EPS-mediated regulation of the activity of the BY-kinase EpsB through its membrane-bound TAD EpsA in the Gram-positive B. subtilis (57) hints at such possibilities. One can, however, expect a larger diversity in the roles of the extracellular/PDs and their interactions compared with those within the cytosol across the bacterial kingdom. A comparison of the AlphaFold-predicted structures (76, 77) of the TADs of representative P-type and F-type BY-kinases shows more variability compared with the corresponding CDs. These differences could, in some cases, be related to the presence/absence of a protein partner (e.g., Wza for Wzc). It has also been suggested that in addition to stimulating kinase function, the TAD may itself encode specific enzymatic activity, for example, for one of the two CapA variants in S. aureus, CapA1 (71).
It is evident, that a lot is still unknown about the unique enzymes that are the BY-kinases, in terms of structure, mechanism, and the diversity therein. Answering these open questions, testing the many predictions made here, and refining the proposed hypotheses will involve extensive future experimentation through the application of techniques ranging from structural/biochemical studies of intact functional assemblies to cell-biological studies and single-molecule analyses combined with computational approaches. The design of these approaches can perhaps draw inspiration from those that have been crucial in providing critical insights into the regulation of the eukaryotic functional cousins of the BY-kinases, the eukaryotic receptor tyrosine kinases (74, 78). We expect that these studies will provide fertile grounds for BY-kinase research during this, the third decade since their annotation, and beyond, given the importance of these systems both in terms of fundamental biology and the opportunity for the design of novel antibacterial therapies (79).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by National Science Foundation grant MCB1937937. Drs Shi and Mijakovic (Chalmers University of Technology) are thanked for providing a high-resolution version of Figure 5 from Ref. (60), parts of which have been incorporated into Figure 6 of the present article. We thank Dr Luciana Giono for input in refining the figures.
Author contributions
F. H., S. A., and R. G. conceptualization; F. H., S. A., and R. G. writing–review & editing; F. H., S. A., and R. G. visualization; R. G. supervision; R. G. funding acquisition.
Edited by Wolfgang Peti
References
- 1.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter T. The genesis of tyrosine phosphorylation. CSH Persp. Biol. 2014;6:a020644. doi: 10.1101/cshperspect.a020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cozzone A.J., Grangeasse C., Doublet P., Duclos B. Protein phosphorylation on tyrosine in bacteria. Arch. Microbiol. 2004;181:171–181. doi: 10.1007/s00203-003-0640-6. [DOI] [PubMed] [Google Scholar]
- 4.Grangeasse C., Cozzone A.J., Deutscher J., Mijakovic I. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem. Sci. 2007;32:86–94. doi: 10.1016/j.tibs.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Bechet E., Guiral S., Torres S., Mijakovic I., Cozzone A.J., Grangeasse C. Tyrosine-kinases in bacteria: from a matter of controversy to the status of key regulatory enzymes. Amino Acids. 2009;37:499–507. doi: 10.1007/s00726-009-0237-8. [DOI] [PubMed] [Google Scholar]
- 6.Grangeasse C., Terreux R., Nessler S. Bacterial tyrosine-kinases: Structure-function analysis and therapeutic potential. Biochim. Biophys. Acta. 2010;1804:628–634. doi: 10.1016/j.bbapap.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 7.Grangeasse C., Nessler S., Mijakovic I. Bacterial tyrosine kinases: Evolution, biological function and structural insights. Philos. Trans. R. Soc. Lond. B. 2012;367:2640–2655. doi: 10.1098/rstb.2011.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jadeau F., Grangeasse C., Shi L., Mijakovic I., Deleage G., Combet C. BYKdb: the Bacterial protein tYrosine Kinase database. Nucl. Acids Res. 2012;40:D321–324. doi: 10.1093/nar/gkr915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst C.M., Braxton J.R., Rodriguez-Osorio C.A., Zagieboylo A.P., Li L., Pironti A., et al. Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nat. Med. 2020;26:705–711. doi: 10.1038/s41591-020-0825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolot M., Gorovits R., Silberstein N., Fichtman B., Yagil E. Phosphorylation of the integrase protein of coliphage HK022. Virology. 2008;375:383–390. doi: 10.1016/j.virol.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Mijakovic I., Petranovic D., Macek B., Cepo T., Mann M., Davies J., et al. Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucl. Acids Res. 2006;34:1588–1596. doi: 10.1093/nar/gkj514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danese P.N., Pratt L.A., Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 2000;182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwechheimer C., Hebert K., Tripathi S., Singh P.K., Floyd K.A., Brown E.R., et al. A tyrosine phosphoregulatory system controls exopolysaccharide biosynthesis and biofilm formation in Vibrio cholerae. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nourikyan J., Kjos M., Mercy C., Cluzel C., Morlot C., Noirot-Gros M.F., et al. Autophosphorylation of the bacterial tyrosine-kinase CpsD connects capsule synthesis with the cell cycle in Streptococcus pneumoniae. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamoto R., Kwan J.M.C., Chin J.F.L., Ong H.T., Flores-Kim J., Midonet C., et al. The bacterial tyrosine kinase system CpsBCD governs the length of capsule polymers. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2103377118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brognard J., Hunter T. Protein kinase signaling networks in cancer. Curr. Opin. Genet. Dev. 2011;21:4–11. doi: 10.1016/j.gde.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grangeasse C. Rewiring the pneumococcal cell cycle with serine/threonine- and tyrosine-kinases. Trends Microbiol. 2016;24:713–724. doi: 10.1016/j.tim.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Duclos B., Grangeasse C., Vaganay E., Riberty M., Cozzone A.J. Autophosphorylation of a bacterial protein at tyrosine. J. Mol. Biol. 1996;259:891–895. doi: 10.1006/jmbi.1996.0366. [DOI] [PubMed] [Google Scholar]
- 19.Grangeasse C., Doublet P., Vaganay E., Vincent C., Deleage G., Duclos B., et al. Characterization of a bacterial gene encoding an autophosphorylating protein tyrosine kinase. Gene. 1997;204:259–265. doi: 10.1016/s0378-1119(97)00554-4. [DOI] [PubMed] [Google Scholar]
- 20.Chao J.D., Wong D., Av-Gay Y. Microbial protein-tyrosine kinases. J. Biol. Chem. 2014;289:9463–9472. doi: 10.1074/jbc.R113.520015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mijakovic I., Grangeasse C., Turgay K. Exploring the diversity of protein modifications: special bacterial phosphorylation systems. FEMS Microbiol. Rev. 2016;40:398–417. doi: 10.1093/femsre/fuw003. [DOI] [PubMed] [Google Scholar]
- 22.Hajredini F., Piserchio A., Ghose R. Long-range dynamic correlations regulate the catalytic activity of the bacterial tyrosine kinase Wzc. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajredini F., Ghose R. An ATPase with a twist: a unique mechanism underlies the activity of the bacterial tyrosine kinase, Wzc. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abj5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid J., Sieber V., Rehm B. Bacterial exopolysaccharides: Biosynthesis pathways and engineering strategies. Front. Microbiol. 2015;6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 26.Jadeau F., Bechet E., Cozzone A.J., Deleage G., Grangeasse C., Combet C. Identification of the idiosyncratic bacterial protein tyrosine kinase (BY-kinase) family signature. Bioinformatics. 2008;24:2427–2430. doi: 10.1093/bioinformatics/btn462. [DOI] [PubMed] [Google Scholar]
- 27.Collins R.F., Beis K., Dong C., Botting C.H., McDonnell C., Ford R.C., et al. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2390–2395. doi: 10.1073/pnas.0607763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grangeasse C., Doublet P., Cozzone A.J. Tyrosine phosphorylation of protein kinase Wzc from Escherichia coli K12 occurs through a two-step process. J. Biol. Chem. 2002;277:7127–7135. doi: 10.1074/jbc.M110880200. [DOI] [PubMed] [Google Scholar]
- 29.Wugeditsch T., Paiment A., Hocking J., Drummelsmith J., Forrester C., Whitfield C. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J. Biol. Chem. 2001;276:2361–2371. doi: 10.1074/jbc.M009092200. [DOI] [PubMed] [Google Scholar]
- 30.Olivares-Illana V., Meyer P., Bechet E., Gueguen-Chaignon V., Soulat D., Lazereg-Riquier S., et al. Structural basis for the regulation mechanism of the tyrosine kinase CapB from Staphylococcus aureus. PLoS Biol. 2008;6:e143. doi: 10.1371/journal.pbio.0060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi L., Ji B., Kolar-Znika L., Boskovic A., Jadeau F., Combet C., et al. Evolution of bacterial protein-tyrosine kinases and their relaxed specificity toward substrates. Genome Biol. Evol. 2014;6:800–817. doi: 10.1093/gbe/evu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bechet E., Gruszczyk J., Terreux R., Gueguen-Chaignon V., Vigouroux A., Obadia B., et al. Identification of structural and molecular determinants of the tyrosine-kinase Wzc and implications in capsular polysaccharide export. Mol. Microbiol. 2010;77:1315–1325. doi: 10.1111/j.1365-2958.2010.07291.x. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y., Liu J., Clarke B.R., Seidel L., Bolla J.R., Ward P.N., et al. The molecular basis of regulation of bacterial capsule assembly by Wzc. Nat. Commun. 2021;12:4349. doi: 10.1038/s41467-021-24652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alphonse S., Djemil I., Piserchio A., Ghose R. Structural basis for the recognition of the bacterial tyrosine kinase Wzc by its cognate tyrosine phosphatase Wzb. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2201800119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Temel D.B., Dutta K., Alphonse S., Nourikyan J., Grangeasse C., Ghose R. Regulatory interactions between a bacterial tyrosine kinase and its cognate phosphatase. J. Biol. Chem. 2013;288:15212–15228. doi: 10.1074/jbc.M113.457804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee D.C., Zheng J., She Y.M., Jia Z. Structure of Escherichia coli tyrosine kinase Etk reveals a novel activation mechanism. EMBO J. 2008;27:1758–1766. doi: 10.1038/emboj.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor S.S., Radzio-Andzelm E. Three protein kinase structures define a common motif. Structure. 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 38.Saraste M., Sibbald P.R., Wittinghofer A. The P-loop - a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 39.Longo L.M., Jablonska J., Vyas P., Kanade M., Kolodny R., Ben-Tal N., et al. On the emergence of P-Loop NTPase and Rossmann enzymes from a beta-alpha-beta ancestral fragment. eLife. 2020;9 doi: 10.7554/eLife.64415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leipe D.D., Koonin E.V., Aravind L. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 41.Fieulaine S., Morera S., Poncet S., Mijakovic I., Galinier A., Janin J., et al. X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13437–13441. doi: 10.1073/pnas.192368699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequence in the alpha and beta subunits of ATP synthase, myosin, kinases and other ATP requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soulat D., Jault J.M., Geourjon C., Gouet P., Cozzone A.J., Grangeasse C. Tyrosine-kinase Wzc from Escherichia coli possesses an ATPase activity regulated by autophosphorylation. FEMS Microbiol. Lett. 2007;274:252–259. doi: 10.1111/j.1574-6968.2007.00841.x. [DOI] [PubMed] [Google Scholar]
- 44.Niemeyer D., Becker A. The molecular weight distribution of succinoglycan produced by Sinorhizobium meliloti is influenced by specific tyrosine phosphorylation and ATPase activity of the cytoplasmic domain of the ExoP protein. J. Bacteriol. 2001;183:5163–5170. doi: 10.1128/JB.183.17.5163-5170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mijakovic I., Poncet S., Boel G., Maze A., Gillet S., Jamet E., et al. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 2003;22:4709–4718. doi: 10.1093/emboj/cdg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paiment A., Hocking J., Whitfield C. Impact of phosphorylation of specific residues in the tyrosine autokinase, Wzc, on its activity in assembly of group 1 capsules in Escherichia coli. J. Bacteriol. 2002;184:6437–6447. doi: 10.1128/JB.184.23.6437-6447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vincent C., Doublet P., Grangeasse C., Vaganay E., Cozzone A.J., Duclos B. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J. Bacteriol. 1999;181:3472–3477. doi: 10.1128/jb.181.11.3472-3477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anand B.G., Prajapati K.P., Shekhawat D.S., Kar K. Tyrosine-Generated nanostructures initiate amyloid cross-seeding in proteins leading to a lethal aggregation trap. Biochemistry. 2018;57:5202–5209. doi: 10.1021/acs.biochem.8b00472. [DOI] [PubMed] [Google Scholar]
- 49.Mittag T., Pappu R.V. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell. 2022;82:2201–2214. doi: 10.1016/j.molcel.2022.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Markevich N.I., Hoek J.B., Kholodenko B.N. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J. Cell Biol. 2004;164:353–359. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Standish A.J., Morona R. The role of bacterial protein tyrosine phosphatases in the regulation of the biosynthesis of secreted polysaccharides. Antioxid. Redox Signal. 2014;20:2274–2289. doi: 10.1089/ars.2013.5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obadia B., Lacour S., Doublet P., Baubichon-Cortay H., Cozzone A.J., Grangeasse C. Influence of tyrosine-kinase Wzc activity on colanic acid production in Escherichia coli K12 cells. J. Mol. Biol. 2007;367:42–53. doi: 10.1016/j.jmb.2006.12.048. [DOI] [PubMed] [Google Scholar]
- 54.Geisinger E., Isberg R.R. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferreira A.S., Leitao J.H., Sousa S.A., Cosme A.M., Sa-Correia I., Moreira L.M. Functional analysis of Burkholderia cepacia genes bceD and bceF, encoding a phosphotyrosine phosphatase and a tyrosine autokinase, respectively: Role in exopolysaccharide biosynthesis and biofilm formation. Appl. Environ. Microbiol. 2007;73:524–534. doi: 10.1128/AEM.01450-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakar D., Gutnick D.L. Involvement of a protein tyrosine kinase in production of the polymeric bioemulsifier emulsan from the oil-degrading strain Acinetobacter lwoffii RAG-1. J. Bacteriol. 2003;185:1001–1009. doi: 10.1128/JB.185.3.1001-1009.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elsholz A.K., Wacker S.A., Losick R. Self-regulation of exopolysaccharide production in Bacillus subtilis by a tyrosine kinase. Genes Dev. 2014;28:1710–1720. doi: 10.1101/gad.246397.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hajredini F., Ghose R. Conserved link between catalytic site interactions and global conformation in P-loop enzymes. bioRxiv. 2022 doi: 10.1101/2022.07.13.499785. [preprint] [DOI] [Google Scholar]
- 59.Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 60.Gout E., Rebeille F., Douce R., Bligny R. Interplay of Mg2+, ADP, and ATP in the cytosol and mitochondria: unravelling the role of Mg2+ in cell respiration. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4560–E4567. doi: 10.1073/pnas.1406251111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matte A., Tari L.W., Delbaere L.T. How do kinases transfer phosphoryl groups? Structure. 1998;6:413–419. doi: 10.1016/s0969-2126(98)00043-4. [DOI] [PubMed] [Google Scholar]
- 62.Mann D., Teuber C., Tennigkeit S.A., Schroter G., Gerwert K., Kotting C. Mechanism of the intrinsic arginine finger in heterotrimeric G proteins. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E8041–E8050. doi: 10.1073/pnas.1612394113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takahashi H., Umino S., Miki Y., Ishizuka R., Maeda S., Morita A., et al. Drastic Compensation of electronic and solvation effects on atp hydrolysis revealed through large-scale QM/MM simulations combined with a theory of solutions. J. Phys. Chem. B. 2017;121:2279–2287. doi: 10.1021/acs.jpcb.7b00637. [DOI] [PubMed] [Google Scholar]
- 64.Kotting C., Kallenbach A., Suveyzdis Y., Wittinghofer A., Gerwert K. The GAP arginine finger movement into the catalytic site of Ras increases the activation entropy. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6260–6265. doi: 10.1073/pnas.0712095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komoriya Y., Ariga T., Iino R., Imamura H., Okuno D., Noji H. Principal role of the arginine finger in rotary catalysis of F1-ATPase. J. Biol. Chem. 2012;287:15134–15142. doi: 10.1074/jbc.M111.328153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y., Zagnitko O., Rodionova I., Osterman A., Godzik A. The FGGY carbohydrate kinase family: insights into the evolution of functional specificities. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khadka S., Ring B., Krzeminski L.R., Hathaway M., Walker R.S., Mobley H.L.T., et al. Regulation of Klebsiella pneumoniae mucoidy by the bacterial tyrosine kinase Wzc. bioRxiv. 2022 doi: 10.1101/2022.06.05.494587. [preprint] [DOI] [Google Scholar]
- 68.Galvan E.M., Ielmini M.V., Patel Y.N., Bianco M.I., Franceschini E.A., Schneider J.C., et al. Xanthan chain length is modulated by increasing the availability of the polysaccharide copolymerase protein GumC and the outer membrane polysaccharide export protein GumB. Glycobiol. 2013;23:259–272. doi: 10.1093/glycob/cws146. [DOI] [PubMed] [Google Scholar]
- 69.Mace P.D., Murphy J.M. There's more to death than life: noncatalytic functions in kinase and pseudokinase signaling. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salazar C., Brummer A., Alberghina L., Hofer T. Timing control in regulatory networks by multisite protein modifications. Trends Cell Biol. 2010;20:634–641. doi: 10.1016/j.tcb.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 71.Rausch M., Deisinger J.P., Ulm H., Muller A., Li W., Hardt P., et al. Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus. Nat. Commun. 2019;10:1404. doi: 10.1038/s41467-019-09356-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reid A.N., Whitfield C. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 2005;187:5470–5481. doi: 10.1128/JB.187.15.5470-5481.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drummelsmith J., Whitfield C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 2000;19:57–66. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Endres N.F., Das R., Smith A.W., Arkhipov A., Kovacs E., Huang Y., et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Srinivasan S., Regmi R., Lin X., Dreyer C.A., Chen X., Quinn S.D., et al. Ligand-induced transmembrane conformational coupling in monomeric EGFR. Nat. Commun. 2022;13:3709. doi: 10.1038/s41467-022-31299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucl. Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arkhipov A., Shan Y., Das R., Endres N.F., Eastwood M.P., Wemmer D.E., et al. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cozzone A.J. Bacterial tyrosine kinases: novel targets for antibacterial therapy? Trends Microbiol. 2009;17:536–543. doi: 10.1016/j.tim.2009.09.005. [DOI] [PubMed] [Google Scholar]