Abstract
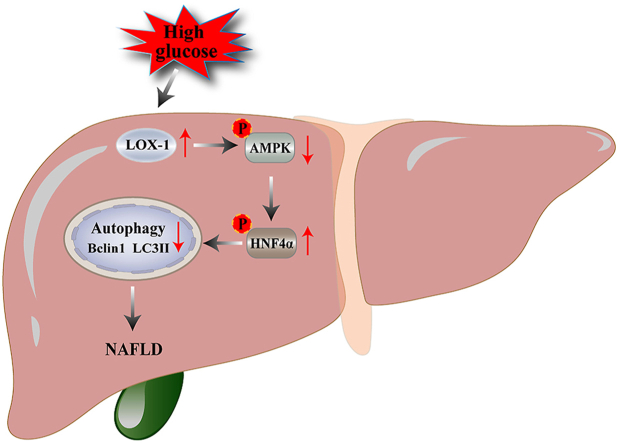
Type 2 diabetes mellitus (T2DM) combined with nonalcoholic fatty liver disease (NAFLD) is a common cause of death. Lectin-like oxidized low-density lipoprotein receptor-1 (LOX-1) is involved in the regulation of autophagy and associated with a variety of diseases, such as atherosclerosis, diabetes, and NAFLD. This study aimed to investigate the effect of LOX-1 on autophagy induced by high glucose levels in human liver sinusoidal endothelial cells (HLSECs) and whether it regulates autophagy through the adenosine monophosphate-activated protein kinase/hepatocyte nuclear factor 4α (AMPK/HNF4α) pathway. In this study, HLSECs cultured with high glucose medium showed increased expression of LOX-1, whereas autophagy was inhibited. High glucose levels decreased the AMPK phosphorylation, increased the HNF4α phosphorylation, and retained the HNF4α in the cytoplasm. By contrast, silencing of LOX-1 reversed the phenomenon induced by high glucose levels and restored the HNF4a localization. Taken together, our findings reveal a novel mechanism of high glucose-induced autophagy in HLSECs, namely, the LOX-1-mediated AMPK/HNF4α signaling pathway. Therefore, LOX-1 is an important target molecule for the regulation of autophagy in HLSECs.
Keywords: LOX-1, Autophagy, HLSECs, AMPK, HNF4α
Graphical abstract
LOX-1; Autophagy; HLSECs; AMPK; HNF4α.
1. Introduction
Type 2 diabetes mellitus (T2DM) is an important risk factor for nonalcoholic fatty liver disease (NAFLD) and accelerates the progression of steatosis in NAFLD. Liver sinusoidal endothelial cells (LSECs) are involved in lipid transport, and LSECs injury occurs during the prophase of NAFLD [1]. However, the association between LSECs injury and NAFLD progression remains unclear. Autophagy is a conserved intracellular lysosomal degradation mechanism that maintains the stability of the intracellular environment. Autophagy in a healthy liver ensures the quality control of organelles and regulates the levels of signaling proteins associated with liver metabolism [2]. Hepatic autophagy is dysregulated in patients with NAFLD and is associated with overall disease progression [3]. Patients with NAFLD, murine models of NAFLD, and human hepatocytes with excessive levels of lipids have impaired autophagy flux [4]. However, the mechanisms by which impaired autophagy facilitates the development of NAFLD remain unclear.
LOX-1 is a type II transmembrane glycoprotein. Under physiological conditions, the LOX-1 expression level is low, but is upregulated by oxidative stress, inflammation, low shear stress, and other environments. Hence, LOX-1 is associated with many pathological events and is involved in the development of autophagy in endothelial cells [5]. HNF4α is a major transcriptional activator of several genes in the liver. HNF4α deficiency disrupts hepatic lipid homeostasis by deregulating the genes involved in lipid metabolism [6]. AMPK is a key kinase that regulates autophagy and is an upstream kinase of HNF4α [7]. However, it remains uncertain whether LOX-1 regulates the AMPK/HNF4α signaling pathway and is involved in autophagy in HLSECs. Therefore, in the present study, we hypothesized that high glucose (HG) levels may impair HLSECs autophagy and that LOX-1 mediates this process through the AMPK/HNF4α signaling pathway.
2. Materials and methods
2.1. Reagents and antibodies
3-MA (HY-19312), BI-6015 (HY-108469), Compound C (HY-13418A) were purchased from Med Chem Express (USA). Cell counting kit-8 (CCK-8) was purchased from APExBio (USA). Antibodies against AMPK (bs-1115R), p-AMPK (bs-14318R), HNF4α (bs-3828R), p-HNF4α (bs-4001R), LC3 (bs-8878R), beclin-1 (bs-1353R), HRP-conjugated secondary antibodies were purchased from Bioss Biotechnology Co., Ltd. (Beijing, China). LOX-1 (DF6522) and β-actin (AF7018) were obtained from Affinity Biosciences (USA).
2.2. Cell culture
HLSECs were purchased from Wuhan Saios Biotechnology Co., LTD (Wuhan, China). The cells were cultured in low-glucose (5.5 mM) primary endothelial cell culture media (Wuhan, China) at 37 °C in a humidified atmosphere containing 5% CO2. The medium required for the cells was changed every 2 days. In order to depict HG, the cells were cultured in growth media containing 25 mM glucose. All experiments were conducted using five generations of cells.
2.3. Cell viability assay
The CCK-8 assay was used to assess the viability of HLSECs. The cells were seeded at a density of 3 × 103/cm2 in 100 μL of primary endothelial cell culture medium in 96-well plates. After an overnight incubation at 37 °C, the HLSECs were treated with various concentrations of the autophagy inhibitor 3-MA (0, 2, 4, 6, 8, 10 mM), AMPK inhibitor Compound C (0, 1, 5, 10, 15 or 20 μM), and HNF4α inhibitor BI-6015 (0, 0.1, 0.2, 0.4, 0.6, 0.8, 1, 1.2 μM) for 6 h. After adding 10 μL of CCK-8 solution to each well, incubation lasted for 1 h. The optical density was measured at 490 nm using a microplate reader (Thermo Fisher Scientific, China).
2.4. Lentivirus transfection
The LOX-1 messenger ribonucleic acid (mRNA) sequence was obtained from the National Center for Biotechnology Information database (ID: ΜM_138648). The GV493 vector, encoding an enhanced green fluorescent protein, GV493 was double digested with the restriction enzymes NheI and AgeI and linked to a polymerase chain reaction (PCR) fragment encoding LOX-1 (top strand: 5′-CCGTTCTGGATTGGATTGCAT-3′; bottom strand:5′-CCGTTCTGGATTGGATTGCAT-3′) in the presence of T4 DNA ligase. To construct the lentivirus, the 293T cells were co-transfected with GV367, pHelper1.0, pHelper2.0, and control vectors, according to the manufacturer's instructions. After 48 h of transfection, the virus was obtained in the cell supernatant, and its titer was measured fluorometrically. The virus titer concentration was 3.0 × 109 TU/ml in the recombinant lentivirus LOX-1 (LV-LOX-1). Similarly, the control lentiviral cells (LV-CON), which only expressed eGFP, were produced as a negative control at a concentration of 1.5 × 109 TU/ml.
2.5. Transmission electron microscopy (TEM)
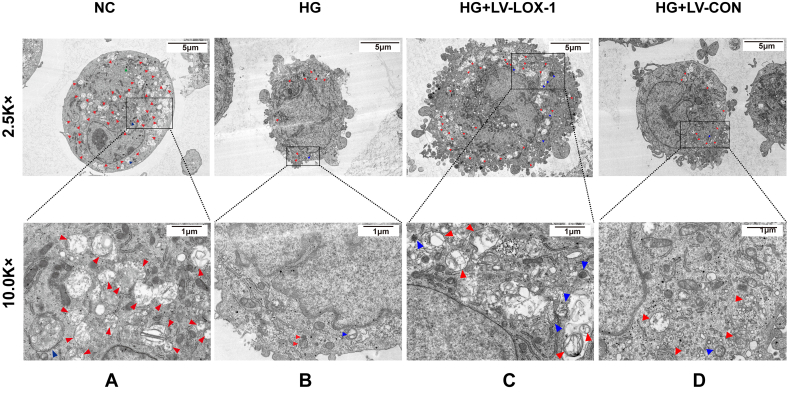
HLSECs were collected by centrifugation, washed twice with phosphate-buffered saline (PBS) and fixed with 2% glutaraldehyde and 2.5% glutaraldehyde for 6 h at 4 °C. The agarose blocks with samples were not exposed to light, post-fixed with 1% OsO4 (Ted Pella I) in 0.1 M PBS (pH 7.4) for 2 h at room temperature, dehydrated with a graded series of ethanol and absolute acetone and then with pure EMBed 812 for 5–8 h, and embedded in resin. The 70 nm ultrathin tissue sections were stained with 2% uranium acetate saturated alcohol solution to avoid light staining for 8 min, rinsed in 70% ethanol three times, and then rinsed in ultrapure water three times. The tissue sections were exposed to 2.6% lead citrate for 8 min to avoid CO2 staining. The cuprum grids were observed under TEM (Hitachi, HT7800), and images were taken.
2.6. Immunofluorescence
After 48 h of cell culture, the cells were fixed with 4% paraformaldehyde (Boster Bio, China) for 15 min at room temperature. The cells were washed three times with PBS for 5 min each and permeabilized with 0.3% Triton X-100 (Solarbio Life Science, China) for 20 min. The cells were washed again with PBS. The treated cells were incubated in 10% bovine serum albumin for 30 min at room temperature and then incubated with HNF4α antibody (1:200) overnight at 4 °C. The samples were incubated with fluorescent secondary antibody (1:500) for 2 h and kept in a light-proof environment, stained with 4′,6-diamidino-2-phenylindole (DAPI) (Solarbio Life Science, China) for 5 min, and washed three times with PBS. Images were captured and analyzed using a microscope (Olympus, Japan).
2.7. Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Takara, Japan). In accordance with the manufacturer's protocol, quantitative reverse transcription PCR (qRT-PCR) was conducted using their respective RNA as the template with an Evo M-MLV RT Mix Kit (with gDNA Clean) and a Premix Pro Taq HS qPCR Kit (SYBR Green) (Accurate Biotechnology (Hunan) Co., Ltd). Each sample was amplified in triplicate using an sds7500 instrument (Thermo Fisher Scientific, New York, USA). The levels of target gene messenger ribonucleic acid (mRNA) were normalized against the levels of the endogenous control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and presented as 2−△△Ct. The primer sequences for qRT-PCR analysis were presented in Table 1.
Table 1.
Primer sequences for qRT-PCR.
| Gene | Forward (5′-3′) | Reserve (5′-3′) |
|---|---|---|
| LOX-1 (ΜM_001172632.2) | GAAGAGAGTAGCAAATTGTTCAGGA | ATAAGTGGGGCATCAAAGGAGA |
| LC3II (ΜM_022818.5) | TCCGAGAAGACCTTCAAACAGC | AAGAAGGCTTGGTTAGCATTGAG |
| Beclin-1 (ΜM_001313998.2) | AGGAGTTGCCGTTGTACTGTTCT | GTGTCTTCAATCTTGCCTTTCTCC |
| GAPDH (ΜM_002046) | GGAAGCTTGTCATCAATGGAAATC | TGATGACCCTTTTGGCTCCC |
2.8. Protein preparation and Western blot analysis
Total protein was extracted using radio immunoprecipitation assay lysis buffer (Millipore, China) containing protease and phosphatase inhibitors (MedChemExpress, USA). The protein concentrations were determined using bicinchoninic acid protein assay. Equal concentrations of proteins were separated using 10% or 12% sodium dodecyl-sulfate polyacrylamide electrophoresis gels (Boster Bio, China) and were transferred to polyvinylidene fluoride membranes. After blocking with 5% non-fat milk, the membranes were incubated overnight at 4 °C with antibodies. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature. The protein bands were incubated in an enhanced chemiluminescence kit (Boster Bio, China) and placed in a gel imaging system for exposure (Beyotime, Shanghai, China). Finally, the densitometric of each band was measured using ImageJ software and normalized to the levels of β-actin.
2.9. Statistical analyses
GraphPad Prism 9 and SPSS 24 software were used to perform the data analyses. Quantitative data were expressed as mean ± standard deviation (SD). Two groups were compared using Student's t-test, while multiple samples were compared using one-way analysis of variance. The experiments were performed at least in triplicate. The P values of < 0.05, < 0.01, and < 0.001 were considered significant.
3. Results
3.1. High glucose increased the expression of LOX-1 and attenuated autophagy in HLSECs
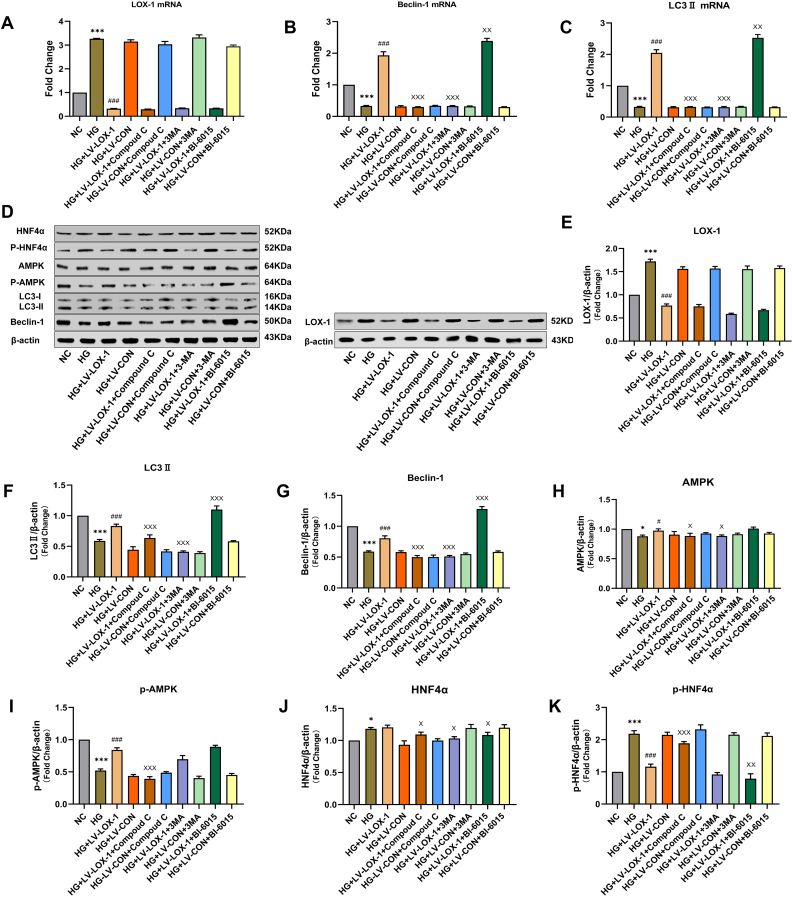
To determine whether and how high levels of glucose affect the autophagic activity in HLSECs, we cultured HLSECs in a medium with normal (5.5 mM) and high doses (25 mM) of glucose for 72 h. After inducing autophagy, microtubule-associated protein 1-light chain 3 (LC3) was processed into its cytoplasmic form, LC3I, and later modified into the membrane-bound form, LC3II. LC3II is localized to autophagosomes and is proportional to the number of autophagosomes [8]. Beclin-1 restored the autophagic activity in disrupted yeast, becoming one of the first genes identified to positively regulate autophagy [9]. We used these two markers to assess the autophagic activity in cultured HLSECs. Western blot and qRT-PCR analyses indicated that the HG group showed a decreased abundance of LC3II (Figure 1C, G) and Beclin-1 (Figure 1B, F) compared with that in the normal control (NC) group. TEM is the commonly used method for detecting autophagy. TEM results showed that after HG intervention, the number of autolysosomes and autophagosomes significantly decreased compared with that in the NC group (Figure 2B). Furthermore, LOX-1 expression levels increased after 72 h of incubation with high concentrations of glucose (Figure 1A, E). Therefore, these results suggest that HG increases the LOX-1 expression levels and impairs the autophagic activity in HLSECs.
Figure 1.
The effect of high glucose on the expression levels of LOX-1, Beclin-1, and LC3II in HLSECs transfected by recombinant LV-LOX-1. HLSECs were treated with high glucose (HG), HG + recombinant lentivirus LOX-1 (LV-LOX-1), and HG + control lentiviral cells (LV-CON) for 72 h. (A–C) The mRNA expression levels of LOX-1, Beclin-1, and LC3II in HLSECs within 72 h after treatment with HG (25 mM), LV-LOX-1, and LV-CON. (D) The protein expression levels of each group were detected using western blotting (see original blots in Supplemental Figure 1D). (E–G) Western blotting analysis of the protein expression levels of LOX-1, Beclin-1, and LC3II in HLSECs within 72 h after treatment with HG and/or LV-LOX-1 and LV-CON. Western blot and qRT-PCR analyses were used to normalize the levels of GAPDH and β-actin (n = 3 independent experiments). All results are expressed as mean ± SD. ∗∗∗P < 0.001 versus the NC group; ###P < 0.001 versus HG + LV-CON group.
Figure 2.
Lentiviral transfection effects on autophagy in HLSECs with high glucose. TEM evaluation of the number of lysosomes, autophagosomes, and autolysosomes in HLSECs treated with HG, HG + LV-LOX-1 and HG + LV-CON for 72 h. In the figure, red arrows represent autolysosomes, blue arrows represent autophagosomes, and green arrows represent lysosomes (n = 6 independent experiments). The scale bars in 2.5 K× represent 5 μm, 10 K× is the magnified TEM images of the portions of 2.5 K× (solid lines), where the scale bar is 1 μm.
3.2. Lentiviral LOX-1-RNAi vector successfully decreasing the expression levels of LOX-1 in high glucose-induced HLSECs
We evaluated the effect of HG on the LOX-1 and autophagy-related protein expression levels and found that LOX-1 is closely associated with autophagy. For this purpose, we constructed a LOX-1-RNAi lentiviral vector to investigate the potential mechanisms linking LOX-1 with the reduction of HG-induced autophagy. HLSECs were infected with LV-LOX-1 and LV-CON in an HG environment; after 72 h, the fluorescence images showed that the green fluorescence intensity was significantly enhanced, and the infection efficiency of HLSECs transfected with LV-LOX-1 was 90% (Figure 3). Additionally, we quantified the LOX-1 mRNA and protein levels in HG-induced LV-LOX-1 (HG + LV-LOX-1) and HG-induced control (HG + LV-CON)-transfected HLSECs using qRT-PCR (Figure 1A) and western blotting (Figure 1E). Results revealed that HLSECs transfected with LOX-1-RNAi expressed significantly less LOX-1 compared with the control cells.
Figure 3.
Lentiviral transfection of HLSECs. HLSECs were transfected in high glucose conditions for 72 h; the infection rate of LV-LOX-1 and LV-CON cells at a multiplicity of infection of 50 was observed under a fluorescence microscope (n = 3 independent experiments). The scale bar in each image represents 100 μm.
3.3. LOX-1 silencing increased the level of autophagy in high glucose-induced HLSECs
We investigated the effect of LOX-1 silencing on LC3II, Beclin-1, and autophagosomes in transfected cells (HG + LV-LOX-1 or HG + LV-CON) within 72 h. Results revealed that LOX-1 silencing significantly increased the mRNA and protein expression levels of LC3II and Beclin-1 compared with those of the HG + LV-CON group (Figure 1B, C, F, and G). In addition, autolysosomes and autophagosomes increased after LOX-1 silencing compared with that of the HG + LV-CON group (Figure 2C). These results suggest that high levels impair the autophagy in HLSECs, at least in part, by increasing the LOX-1 expression levels.
3.4. Effects of 3-MA, BI-6015, and Compound C on the viability of HLSECs
We investigated the relationship between LOX-1 and the AMPK/HNF4α signaling pathway. The cells (HG + LV-LOX-1 or HG + LV-CON) were treated with 0, 1, 5, 10, 15 or 20 μM of Compound C; 0, 1, 5, 10, 15, or 20 mM of 3-MA; and 0, 0.1, 0.2, 0.4, 0.6, 0.8, 1, and 1.2 μM of BI-6015 for 6 h, 12 h, 24 h, 48 h, respectively, to determine the effective treatment concentrations of 3-MA, BI-6015, and Compound C. Subsequently, the CCK-8 assay was used for assessment. In our study, the cell viability decreased with increasing doses and durations (Figure 4A–C). The concentrations of 3-MA (>6 mM), Compound C (>10 μM), and BI-6015 (>0.6 μM) were significantly inhibitory and cytotoxic. After treatment with 10 mM of 3-MA, 20 μM of Compound C, and 1.2 μM of BI-6015 for 48 h, the viability rates of HLSECs were significantly reduced to 73.4%, 73.9%, and 82.2% in the control group, respectively. Hence, 6 mM of 3-MA (48.9%), 10 μM of Compound C (48.7%), and 0.6 μM of BI-6015 (49.3%) were used to incubate the cells for 24 h in the subsequent intervention experiments.
Figure 4.
Effect of 3-MA, Compound C, and BI-6015 treated on the viability in HLSECs. Cell viability was determined by CCK-8 assay (n = 3 independent experiments). (A) The effect of Compound C (0, 1, 5, 10, 15, 20 μM) treatment on HLSECs viability (control is untreated group). (B) The effect of BI-6015 (0, 0.1, 0.2, 0.4, 0.6, 0.8, 1, 1.2 μM) treatment on HLSECs viability (control is untreated group). (C) The effect of 3-MA (0, 2, 4, 6, 8, 10 mM) treatment on HLSECs viability (control is untreated group).
3.5. Effect of LOX-1 silencing on the expression of AMPK and HNF4α
AMPK is a key factor that affects autophagy. Hence, we examined whether HG intervention affected the AMPK activity in HLSECs. The increased phosphorylation at threonine Ser356 on the AMPK subunit induced in HLSECs was significantly decreased after HG treatment. Compared with the HG + LV-CON group, LOX-1 silencing increased the protein level of p-AMPK (Figure 6I), but the protein level of AMPK was not significantly changed in the HG + LV-LOX-1 group (Figure 6H). An AMPK downstream factor, HNF4α, is involved in regulating lipid metabolism in the liver. HNF4α is also required to maintain the structure of LSECs [10]. We examined the total protein expression levels of HNF4α in HLSECs and found that HG intervention cells were comparable to normal cells (Figure 6J), however, further examination revealed that HNF4α was retained in the cytoplasm (Figure 5), and western blotting showed increased levels of p-HNF4α (Ser313) expression (Figure 6K). LOX-1 silencing decreased the p-HNF4α expression levels and restored the subcellular localization of p-HNF4α compared with the HG + LV-CON group (Figures 5 and 6K).
Figure 6.
LOX-1 silencing increased autophagy of HLSECs through AMPK/HNF4α signaling pathway. All groups were treated with 25 mM of glucose for 72 h, except for the normal control group. The cells in the transfection group HG + LV-LOX-1 or HG + LV-CON cells were blocked with AMPK (10 μM), HNF4α (0.6 μM), or autophagy (6 mM) inhibitors. The untreated intact samples were run as a control in each experiment. (A–C) HLSECs were cultured under the above conditions for 72 h, and the mRNA expression levels of LOX-1, Beclin-1, and LC3II in each group were determined by qRT-PCR. (D) The protein expression levels of each group were determined using western blotting, and β-actin was used as a loading control (see original blots in Supplemental Figure 6D). (E–K) Western blotting analyzed the relative protein expression of LOX-1, Beclin-1, LC3II, AMPK, p-AMPK, HNF4α, and p-HNF4α in each group (n = 3 independent experiments). All data are expressed as mean ± SD. from three independent experiments. ∗P < 0.05, ∗∗∗P < 0.01 versus the NC group; #P < 0.05, ###P < 0.001 versus HG + LV-CON group; xP < 0.05, xxP < 0.05, xxxP < 0.01 versus HG + LV-LOX-1 group.
Figure 5.
Effect of high glucose and LOX-1 silencing on HNF4α localization in HLSECs. HLSECs were treated with HG, HG + LV-LOX-1 and HG + LV-CON for 72h. The fixed cells were stained with HNF4α primary antibody and overlaid with goat anti-rabbit secondary antibody (green) and then with DAPI (blue) (n = 3 independent experiments). The scale bar in each image represents 50 μm.
3.6. LOX-1 silencing increased the autophagy through the HNF4α signaling pathway in high glucose-induced HLSECs
We examined the levels of HNF4α signaling to determine the molecular mechanism by which LOX-1 mediates impaired autophagy induced by HG. BI-6015 (0.6 μM), an inhibitor of HNF4α, further increased the protein and mRNA expression levels of LC3II and Beclin-1 in the HG + LV-LOX-1 + BI-6015 group (Figure 6B, C, F, and G), but had no significant effect on the expression levels of LOX-1, AMPK, and p-AMPK (Figure 6A, E, H, and I). This finding implies that HNF4α is a downstream pathway for LOX-1 and AMPK in the regulation of autophagic signaling. Taken together, LOX-1 silencing increased the autophagic activity by inhibiting HNF4α signaling in HG-induced HLSECs.
3.7. LOX-1 silencing increased the autophagic activity via the AMPK/HNF4α pathway in high glucose-induced HLSECs
To further clarify the potential mechanism of LOX-1-mediated autophagy, we investigated the regulation of AMPK signaling (upstream kinases of HNF4α). The cells (HG + LV-LOX-1 or HG + LV-CON) were treated with an AMPK inhibitor (Compound C, 10 μM) for 24 h. Compound C significantly decreased the expression levels of LC3II and Beclin-1 in the HG + LV-LOX-1 + Compound C group (Figure 6B, C, F, and G). Furthermore, Compound C significantly increased the phosphorylation of HNF4α at Ser313 (Figure 6K), but it did not affect the expression levels of LOX-1 (Figure 6A, E). These results indicate that AMPK is an HNF4α upstream factor in the regulation of autophagy and that AMPK/HNF4α is involved in autophagy in LV-LOX-1-induced HLSECs. Western blotting showed a significant increase in the levels of p-AMPK and p-HNF4α compared with that of the total AMPK and HNF4α, implying that the silencing of LOX-1 affected the AMPK/HNF4α pathway by modifying the phosphorylation in HLSECs. Furthermore, 3-MA (6 mM) inhibited the expression of LC3II and Beclin-1 in the HG + LV-LOX-1+3-MA group (Figure B, C, F and G). However, 3-MA did not affect the expression of LOX-1, HNF4α, and AMPK (Figure 6E and H–K), suggesting that the LOX-1-mediated autophagic pathway AMPK/HNF4α is located upstream of autophagy. In summary, HG decreases the autophagy by inhibiting AMPK/HNF4α signaling, while silencing of LOX-1 activates the AMPK/HNF4α signaling pathway to increase the HG-induced autophagy.
4. Discussion
T2DM and NAFLD have common pathological causes, including glucose and lipid metabolism abnormalities [11]. The mechanisms leading to the onset of NALFD are complex; in recent years, autophagy is involved in regulating the development of NAFLD. At the cellular and animal levels, the dysregulation of hepatic autophagy is associated with steatosis in patients with NAFLD [12]. The autophagic activity and lysosomal function are impaired in the livers of patients with NAFLD [13]. LSECs are the most abundant non-parenchymal cells and gatekeepers of lipid entry into the liver [1]. Recently, the absence of autophagy in LSECs induced liver injury and inflammation in a mouse model of NAFLD [14]. In patients with severe NAFLD, the number and size of autophagic vesicles are fewer and smaller, respectively, in LSECs compared with those in patients with simple steatosis [14]. However, the molecular mechanism underlying autophagy in LSECs has not been clearly elucidated. Therefore, we explored how LOX-1 regulates autophagy in HLSECs in vitro.
LOX-1, originally identified in bovine aortic endothelial cells, belongs to the class E scavenger receptor subfamily [15]. It is involved in atherosclerosis formation. LOX-1 is associated with cancer, obesity, and NAFLD [16]. The serum levels of LOX-1 in patients with NAFLD are significantly higher than those in healthy individuals [17]. The LOX-1 expression at the gene and protein levels increased at HG concentrations (5.5–30 mM) [18]. In this study, HG (25 mM) was used to treat HLSECs. Similarly, LOX-1 protein and mRNA expression levels increased in treated HLSECs.
Autophagy plays a crucial role in maintaining cellular homeostasis, mainly by removing damaged and redundant organelles, invading pathogens, and proteins that cannot be degraded by proteasomes. Autophagy regulates the expression of resident liver cells such as hepatocytes, stellate cells, and LSECs [19]. In HLSECs, treatment with HG significantly decreased the levels of Beclin-1, LC3II, and autophagosomes, suggesting that HG impaired autophagic activity. Lipophagy is a type of autophagy that delays lipid degeneration by blocking lipid droplet accumulation [20]. Thus, the activation of hepatic autophagy may be a therapeutic strategy for NAFLD. To further investigate the effect of LOX-1 on the autophagic activity in HLSECs, we developed an LV-LOX-1-RNAi vector; LV-LOX-1-RNAi transfection increased the expression levels of Beclin-1 and LC3II, and the number of autophagosomes in HG-interfered HLSECs. These data implied that autophagy and LOX-1 are opposing processes. Therefore, LOX-1 mediates autophagy in HLSECs.
The HNF4 family is involved in regulating the transcription of lipid genes, among which HNF4α regulates multiple metabolic pathways in the liver [21]. HNF4α is expressed in the kidneys, pancreas, colon, and small intestine since it initially existed in the liver. We found that HNF4α was also expressed in HLSECs and localized in the nucleus. Targeted knockout HNF4α mice exhibit hepatic lipid accumulation [22]. However, HG treatment did not affect the protein expression levels of HNF4α [23]. A recent study found that HNF4α expression was significantly upregulated in the endothelial cells of rodents and patients with T2DM [24]. We assume that HG leads to the dysfunction of HNF4α, which in turn disrupts the autophagic activity in HLSECs. We observed an increased protein expression of p-HNF4α and an aberrant cytoplasmic retention of HNF4α in HLSECs cultured with HG, which was reversed by LOX-1 silencing. Therefore, the dysregulation of HNF4α cellular localization by HG levels is mediated by LOX-1-mediated phosphorylation of HNF4α. Furthermore, BI-6015 further enhanced autophagy in HG + LV-LOX-1 transfected cells. Our results suggest that LOX-1 silencing prevents the nuclear translocation of HNF4α and increases the function of HLSECs autophagy by activating the HNF4α signaling pathway.
AMPK signaling is a key link between the regulation of cellular energy metabolism and autophagy [7]. Previous studies have demonstrated impaired autophagy and reduced AMPK activity in NAFLD patients [25]. Similarly, we demonstrated that the level of p-AMPK expression was downregulated in HG-treated HLSECs, whereas LOX-1 silencing reversed the downregulation of p-AMPK. Furthermore, the HG + LV-LOX-1 transfected cells showed decreased protein levels and mRNA expression of LC3II and Beclin-1 after treatment with Compound C. According to our results, LOX-1 silencing restores autophagy by activating the AMPK signaling pathway in HLSECs. AMPK directly activates p-HNF4α and inhibits its transcriptional activity [26]. In HG + LV-LOX-1-treated HLSECs, treatment with Compound C increased the expression level of p-HNF4α; however, BI-6015 was not significantly associated with the changes in the expression of AMPK and p-AMPK. These results suggested that AMPK is an HNF4α upstream factor in the regulation of autophagy. In addition, treatment with BI-6015 and Compound C had no effect on LOX-1, which proves that LOX-1 is located upstream of the AMPK/HNF4α signaling pathway. Based on these results, our use of autophagy inhibitors (3-MA) to unsurprisingly reduce autophagy in HLSECs, but largely unaffected the expression levels of LOX-1, AMPK, and HNF4α in HLSECs, further suggesting that LOX-1-mediated AMPK/HNF4α signaling is located upstream of autophagy. In short, LOX-1 silencing successfully increased the autophagy in HLSECs by activating the AMPK and subsequently inhibiting the HNF4α signaling. Potential limitations of the present study, first, we lacked detection of the nuclear/cytoplasmic protein distribution of HNF4α. Second, the outcome of impaired autophagy in HLSECs was not further observed. Finally, we used LOX-1 interference techniques that failed to completely knock out LOX-1.
5. Conclusions
In summary, LOX-1 triggers autophagy in HLSECs through the AMPK/HNF4α signaling pathway in vitro. We also determined the localization of HNF4α in HLSECs and established a link with LOX-1. Furthermore, our findings shed light on LOX-1-mediated autophagy in HG-induced HLSECs, which lays the foundation for elucidating the pathogenesis of autophagy in diabetic fatty liver. Subsequently, we will continue to validate our findings at the animal and clinical levels to deepen our research.
Declarations
Author contribution statement
Qidang Duan: Conceived and designed the experiments; Performed the experiments; Wrote the paper.
Huiling Si; Na Zhang: Performed the experiments.
Limin Tian; Jumei Qiu; Jing Yu: Analyzed and interpreted the data.
Jing Liu: Contributed reagents, materials, analysis tools or data.
Qi Zhang: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Jing Liu was supported by National Natural Science Foundation of China [81760151].
Qi Zhang was supported by National Natural Science Foundation of China [81960173], National Research Incubation Project of Gansu Provincial People's Hospital [19SYPYB-4], Lanzhou Health and Wellness Commission [2021005].
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare no competing interests.
Additional information
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2022.e12385.
Contributor Information
Jing Liu, Email: liujing551108@126.com.
Qi Zhang, Email: lzzq78@126.com.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
10-Beclin1.tif.
Supplemental Figure 6D-Beclin1
10-LC3.tif.
Supplemental Figure 6D-LC3
10-LOX-1.tif.
Supplemental Figure 6D-LOX-1
AMPKa1.tif.
Supplemental Figure 6D-AMPK
Beclin1.tif.

Supplemental Figure 1D-Beclin1
HNF4a.tif.
Supplemental Figure 6D-HNF4a
LC3.tif.

Supplemental Figure 1D-LC3
LOX1.tif.

Supplemental Figure 1D-LOX-1
P-AMPKa1.tif.
Supplemental Figure 6D-P-AMPK
P-HNF4a.tif.
Supplemental Figure 6D-P-HNF4a
renamed_81f02.tif.

Supplemental Figure 6D-β-actin
renamed_87eba.tif.
Supplemental Figure 6D-β-actin
renamed_381c4.tif.

Supplemental Figure 1D-β-actin
renamed_fb81f.tif.

Supplemental Figure 1D-β-actin
References
- 1.Pandey E., Nour A.S., Harris E.N. Prominent receptors of liver sinusoidal endothelial cells in liver homeostasis and disease. Front. Physiol. 2020;11:873. doi: 10.3389/fphys.2020.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrnes K., Blessinger S., Bailey N.T., et al. Therapeutic regulation of autophagy in hepatic metabolism. Acta Pharm. Sin. B. 2022;12:33–49. doi: 10.1016/j.apsb.2021.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian H., Chao X., Williams J., et al. Autophagy in liver diseases: a review. Mol. Aspect. Med. 2021 doi: 10.1016/j.mam.2021.100973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.González-Rodríguez A., Mayoral R., Agra N., et al. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179. doi: 10.1038/cddis.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding Z., Liu S., Deng X., et al. Hemodynamic shear stress modulates endothelial cell autophagy: role of LOX-1. Int. J. Cardiol. 2015;184:86–95. doi: 10.1016/j.ijcard.2015.01.065. [DOI] [PubMed] [Google Scholar]
- 6.Tachmatzidi E.C., Galanopoulou O., Talianidis I. Transcription control of liver development. Cells. 2021;10 doi: 10.3390/cells10082026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou G., Myers R., Li Y., et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanida I., Ueno T., Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funderburk S.F., Wang Q.J., Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parviz F., Matullo C., Garrison W.D., et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 11.Targher G., Corey K.E., Byrne C.D., et al. The complex link between NAFLD and type 2 diabetes mellitus - mechanisms and treatments. Nat. Rev. Gastroenterol. Hepatol. 2021;18:599–612. doi: 10.1038/s41575-021-00448-y. [DOI] [PubMed] [Google Scholar]
- 12.Scorletti E., Carr R.M. A new perspective on NAFLD: focusing on lipid droplets. J. Hepatol. 2022;76:934–945. doi: 10.1016/j.jhep.2021.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Carotti S., Aquilano K., Zalfa F., et al. Lipophagy impairment is associated with disease progression in NAFLD. Front. Physiol. 2020;11:850. doi: 10.3389/fphys.2020.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammoutene A., Biquard L., Lasselin J., et al. A defect in endothelial autophagy occurs in patients with non-alcoholic steatohepatitis and promotes inflammation and fibrosis. J. Hepatol. 2020;72:528–538. doi: 10.1016/j.jhep.2019.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Plato A., Willment J.A., Brown G.D. C-type lectin-like receptors of the dectin-1 cluster: ligands and signaling pathways. Int. Rev. Immunol. 2013;32:134–156. doi: 10.3109/08830185.2013.777065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balzan S., Lubrano V. LOX-1 receptor: a potential link in atherosclerosis and cancer. Life Sci. 2018;198:79–86. doi: 10.1016/j.lfs.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 17.Ozturk O., Colak Y., Senates E., et al. Increased serum soluble lectin-like oxidized low-density lipoprotein receptor-1 levels in patients with biopsy-proven nonalcoholic fatty liver disease. World J. Gastroenterol. 2015;21:8096–8102. doi: 10.3748/wjg.v21.i26.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L., Sawamura T., Renier G. Glucose enhances endothelial LOX-1 expression: role for LOX-1 in glucose-induced human monocyte adhesion to endothelium. Diabetes. 2003;52:1843–1850. doi: 10.2337/diabetes.52.7.1843. [DOI] [PubMed] [Google Scholar]
- 19.Sun X., Harris E.N. New aspects of hepatic endothelial cells in physiology and nonalcoholic fatty liver disease. Am. J. Physiol. Cell Physiol. 2020;318:C1200–C1213. doi: 10.1152/ajpcell.00062.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Lopez N., Singh R. Autophagy and lipid droplets in the liver. Annu. Rev. Nutr. 2015;35:215–237. doi: 10.1146/annurev-nutr-071813-105336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatzis P., Talianidis I. Regulatory mechanisms controlling human hepatocyte nuclear factor 4alpha gene expression. Mol. Cell Biol. 2001;21:7320–7330. doi: 10.1128/MCB.21.21.7320-7330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thymiakou E., Othman A., Hornemann T., et al. Defects in high density lipoprotein metabolism and hepatic steatosis in mice with liver-specific ablation of hepatocyte nuclear factor 4A. Metab. Clin. Exp. 2020;110 doi: 10.1016/j.metabol.2020.154307. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Chaudhari S., Ren Y., et al. Impairment of hepatic nuclear factor-4α binding to the Stim1 promoter contributes to high glucose-induced upregulation of STIM1 expression in glomerular mesangial cells. Am. J. Physiol. Ren. Physiol. 2015;308:F1135–F1145. doi: 10.1152/ajprenal.00563.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chai X., Yan J., Gao Y., et al. Endothelial HNF4α potentiates angiogenic dysfunction via enhancement of vascular endothelial growth factor resistance in T2DM. J. Cell. Biochem. 2019;120:12989–13000. doi: 10.1002/jcb.28570. [DOI] [PubMed] [Google Scholar]
- 25.Smith B.K., Marcinko K., Desjardins E.M., et al. Treatment of nonalcoholic fatty liver disease: role of AMPK. Am. J. Physiol. Endocrinol. Metab. 2016;311:E730–E740. doi: 10.1152/ajpendo.00225.2016. [DOI] [PubMed] [Google Scholar]
- 26.Hong Y.H., Varanasi U.S., Yang W., et al. AMP-activated protein kinase regulates HNF4alpha transcriptional activity by inhibiting dimer formation and decreasing protein stability. J. Biol. Chem. 2003;278:27495–27501. doi: 10.1074/jbc.M304112200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.