Summary
Centrosomal protein 120 (CEP120) is a 120 kDa centrosome protein that plays an important role in centrosome replication. Overexpression of CEP120 can lead to centrosome duplicate abnormality, which is closely associated with tumorigenesis and development. However, there are no reports on the relationship between CEP120 and tumors. In our study, overexpression of CEP120 promoted centrosome amplification in gastric cancer (GC), and the role of CEP120 in promoting GC progression was demonstrated in vitro and in vivo. We demonstrated that CEP120 promotes centrosome amplification and GC progression by promoting the expression and centrosome aggregation of the deubiquitinating enzyme USP54, maintaining the stability of PLK4 and reducing its ubiquitination degradation. In conclusion, the CEP120-USP54-PLK4 axis may play an important role in promoting centrosome amplification and GC progression, thus providing a potential therapeutic target for GC.
Subject areas: Molecular biology, Cell biology, Functional aspects of cell biology
Graphical abstract

Highlights
-
•
CEP120 is highly expressed in gastric cancer
-
•
CEP120 promotes centrosome amplification and progression of gastric cancer
-
•
CEP120 promotes gastric cancer progression by facilitating PLK4 deubiquitination
-
•
CEP120 facilitates PLK4 deubiquitination by regulating USP54
Molecular biology; Cell biology; Functional aspects of cell biology
Introduction
Gastric cancer (GC) is a serious threat to human health. Global cancer statistics from 2020 ranked GC fifth in incidence and fourth in mortality among malignant tumors.1 Most patients with GC are diagnosed at an advanced stage and have a poor prognosis. Therefore, exploring the mechanism of GC progression is of great importance for the treatment of GC.
A meaningful link between centrosome abnormalities and tumors has been proposed. As early as 1914, Boveri proposed the hypothesis that tumors often result from chromosomal segregation errors during cell division, and an abnormal centrosome number may be the source of such errors.2 Abnormal centrosome numbers are most commonly caused by centrosome amplification, which appears in almost all tumors and is closely related to tumorigenesis and progression.3 Centrosomal protein 120 (CEP120) is a key protein involved in centrosome replication; previous studies have shown that overexpression of CEP120 can lead to the production of additional centrioles.4 Our previous study showed that CEP120 is closely related to the proliferation and migration of cerebellar granulosa precursor cells.5 However, the relationship between CEP120 and tumors has not yet been reported.
PLK4 plays a key role in centrosome replication. Overexpression of PLK4 can lead to centrosome amplification and induce oncogene-like cell invasion in untransformed human breast epithelial cells,6 which is closely related to tumor occurrence and development.7 Some studies have proposed targeting PLK4 as a tumor therapy, and relevant PLK4 inhibitors are currently in clinical trials.8 Therefore, a more comprehensive understanding of the molecular mechanisms of PLK4 in tumor progression is essential for tumor therapy. In this study, we demonstrated that CEP120 promotes centrosome amplification and GC progression by promoting USP54 expression and centrosome aggregation to inhibit PLK4 ubiquitination and degradation. We observed that CEP120 is highly expressed in GC and plays a key role in its progression, which is of great significance for providing new research directions and therapeutic targets for GC.
Results
GC has higher expression of CEP120 and more common centrosome amplification
In our previous study, we successfully established a mouse model of CEP120 mutation, which demonstrated that CEP120 plays an important role in centrosome replication, proliferation, and migration of cerebellar granule neuron progenitors.5 To further explore its role in GC, we analyzed the data from The Cancer Genome Atlas database and found that CEP120 expression in patients with positive lymph node metastasis and distant metastasis was significantly higher than that in patients with negative lymph node metastasis or no distant metastasis, and patients with higher CEP120 expression had worse prognosis (Figure 1A). Kaplan-Meier curves of overall survival in GC patients from the Kaplan-Meier plotter (http://kmplot.com/analysis/) showed the same results (Figure 1B). We further detected the expression of CEP120 in GC and adjacent normal tissues by immunohistochemistry (IHC) and western blot (WB), and the results showed that CEP120 was significantly overexpressed in GC tissues (Figures 1C and 1D). What’s more, γ tubulin was upregulated in GC tissues together with CEP120, which suggested that the level of CEP120 increased in GC because of higher number of centrosomes (Figure 1D). WB results in GC cells and GES1 showed that CEP120 was highly expressed in most GC cell lines (Figure 1E). In addition, immunofluorescence (IF) with γ tubulin and CEP120 antibodies showed that CEP120 was localized in centrosomes, and centrosome amplification was observed in GC cells (Figure 1F). Quantitative analysis of centrosome amplification showed that centrosome amplification in SGC7901 and BGC823 cells was significantly higher than that in GES1 cells (Figure 1G). Together, these results suggested that CEP120 is highly expressed in GC and strongly associated with a poor prognosis.
Figure 1.
GC has higher expression of CEP120 and more common centrosome amplification
(A) Analyses of the association between CEP120 expression and N stage, M stage, and overall survival in GC patients derived from TCGA database. Data were expressed as mean ± SEM. Student’s t test was adopted to evaluate the association with N and M stage. Log rank test was adopted to evaluate the difference of prognosis. ∗p < 0.05.
(B) Survival curves of CEP120 expression in GC patients from Kaplan-Meier plotter.
(C) Representative IHC images of CEP120 expression levels in GC tissues and adjacent normal tissues. Scale bars, 100 μm.
(D) WB analysis of CEP120 and γ tubulin expression in GC specimens and adjacent normal tissues. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗∗∗p < 0.0001.
(E) WB and quantitative analysis of CEP120 expression in GC cells and GES1. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗∗p < 0.001, ns, not significant.
(F) Immunofluorescence analysis with CEP120 and γ tubulin antibodies in pHH3-negative GES1, SGC7901, and BGC823 cells. Scale bars, 5 μm.
(G) Quantitative analysis of centrosome amplification cell percent in pHH3-negative GES1, SGC7901, and BGC823 cells. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
CEP120 promotes centrosome amplification and progression of GC
To investigate the role of CEP120 in centrosome amplification and GC progression, we established GC cell lines with stable overexpression (SGC7901-CEP120) and knockdown (BGC823-shCEP120). Overexpression and knockdown efficiencies were verified by WB (Figure 2A). IF results showed that overexpression of CEP120 significantly promoted centrosome amplification, which was inhibited by knockdown of CEP120 (Figure 2B). The results of the cell counting kit-8 (CCK8) and colony formation assays showed that overexpression of CEP120 significantly promoted the proliferation of GC cells, while knockdown inhibited it (Figures 2C and 2D). Likewise, the results of the transwell and wound-healing assays showed that overexpression of CEP120 significantly promoted the migration and invasion of GC cells, whereas knockdown restrained it (Figures 2E and 2F). To confirm the effect of CEP120 on GC proliferation and metastasis in vivo, we performed mouse xenograft assay and mouse lung metastasis assay. As shown in Figures 2G and 2H, overexpression of CEP120 significantly increased the subcutaneous tumor growth and the number of lung metastatic foci, which were obviously inhibited by the knockdown of CEP120. In conclusion, these results demonstrated the important role of CEP120 in the proliferation and metastasis of GC in vitro and in vivo.
Figure 2.
CEP120 promotes centrosome amplification and progression of GC
(A) WB analysis of CEP120 expression in SGC7902 cells transfected with Lenti-CEP120 and BGC823 cells transfected with Lenti-shCEP120.
(B) Immunofluorescence and quantitative analysis of centrosome amplification percent in CEP120-overexpressed SGC7901 and CEP120-knockdown BGC823 of pHH3-negative cells. Scale bars, 5 μm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗p < 0.01.
(C) CCK8 assay and (D) Colony formation assay in CEP120-overexpressed SGC7901 and CEP120-knockdown BGC823 cells. Scale bars, 1 cm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant.
(E) Transwell assay and quantitative analysis of migratory and invasive abilities of CEP120-overexpressed SGC7901 and CEP120-knockdown BGC823 cells. Scale bars, 50 μm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
(F) Wound-healing assay and quantitative analysis of migration capacity of CEP120-overexpressed SGC7901 and CEP120-knockdown BGC823 cells. Scale bars, 100 μm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
(G) General image and growth curve of subcutaneous xenograft of SGC7901 cells after CEP120 overexpression and BGC823 cells after CEP120 knockdown. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(H) Fluorescence image and quantitative analysis of mouse lung metastasis model with CEP120-overexpressed SGC7901 and CEP120-knocked-down BGC823 cells. Scale bars, 1 cm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗p < 0.05.
CEP120 regulates centrosome amplification and GC progression by controlling PLK4 ubiquitination
PLK4 plays a key role in centrosome amplification.9,10 Therefore, we investigated whether the regulation of centrosome amplification by CEP120 is related to PLK4. We detected the protein and mRNA levels of PLK4 in GC cells and GC specimens and found that CEP120 and PLK4 showed a significant positive correlation at the protein level (Figures 3A–3C); however, there was no significant correlation at the RNA level (Figure 3D). Therefore, we speculated that CEP120 might regulate the protein degradation of PLK4. To investigate whether CEP120 regulates the stability of PLK4, we measured the abundance of PLK4 in cycloheximide (CHX)-treated GC cells. We found that overexpression of CEP120 significantly increased the stability and half-life of PLK4 in SGC7901 cells, whereas knockdown of CEP120 significantly disrupted the stability of PLK4 in BGC823 cells (Figure 3E). The autophagy-lysosome and ubiquitin-proteasome pathways are the primary protein degradation pathways. We treated GC cells with the ubiquitin-proteasome pathway inhibitor MG132 and autophagy-lysosome pathway inhibitor chloroquine diphosphate (CQ). And we found that MG132 could reverse the decreased protein level of PLK4 by CEP120 knockdown (Figure 3F), suggesting that CEP120 mainly regulated the ubiquitination pathway of PLK4. In addition, we found that overexpression of CEP120 significantly inhibited PLK4 ubiquitination, whereas knockdown of CEP120 promoted PLK4 ubiquitination (Figure 3G). These results suggested that CEP120 enhanced the stability of PLK4 by inhibiting its ubiquitination.
Figure 3.
CEP120 regulates centrosome amplification and GC progression by controlling PLK4 ubiquitination
(A) WB analysis of PLK4 expression after CEP120 overexpression and knockdown.
(B) WB results of PLK4 and CEP120 expression in GC tissues.
(C) The correlation of PLK4 and CEP120 protein level in GC tissues. p value derived from pearson correlation analysis.
(D) The correlation of PLK4 and CEP120 mRNA level in GC tissues. p value derived from Pearson correlation analysis.
(E) CHX chase analysis of PLK4 protein half-life after CEP120 overexpression and knockdown.
(F) WB analysis of PLK4 expression in BGC823 cells treated with MG132 or CQ after CEP120 knockdown.
(G) Results of the ubiquitination level of PLK4 after CEP120 overexpression and knockdown examined by ubiquitination assay.
We then used PLK4-specific siRNA or plasmids to further investigate whether CEP120 regulates centrosome amplification and GC progression in a PLK4-dependent manner. Silence of PLK4 was found to significantly reverse the enhanced centrosome amplification, and the proliferative, migratory, invasive capacities of CEP120-overexpressed cells, while overexpression of PLK4 rescued the reduced centrosome amplification proportion, and proliferative, migratory, invasive capacities of CEP120-knocked-down cells (Figures 4A–4D). In conclusion, CEP120 regulates centrosome amplification and GC progression by controlling PLK4 ubiquitination.
Figure 4.
CEP120 regulates centrosome amplification and GC progression by controlling PLK4 ubiquitination
SGC7901 cells with stable overexpression of CEP120 were transfected with siPLK4 or siNC, and BGC823 cells with stable knockdown of CEP120 were transfected with PLK4 plasmid or vector.
(A) Immunofluorescence and quantitative analysis of the effect of PLK4 on CEP120-induced centrosome amplification percent in pHH3-negative SGC7901 and BGC823 cells. Scale bars, 5 μm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) CCK8 and (C) Colony formation assay of the effect of PLK4 on CEP120-induced cell proliferation ability. Scale bars, 1 cm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗∗p < 0.001.
(D) Transwell assay of the role of PLK4 on CEP120-induced migratory and invasive capacities. Scale bars, 50 μm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
USP54 combines with PLK4 and mediates its deubiquitination
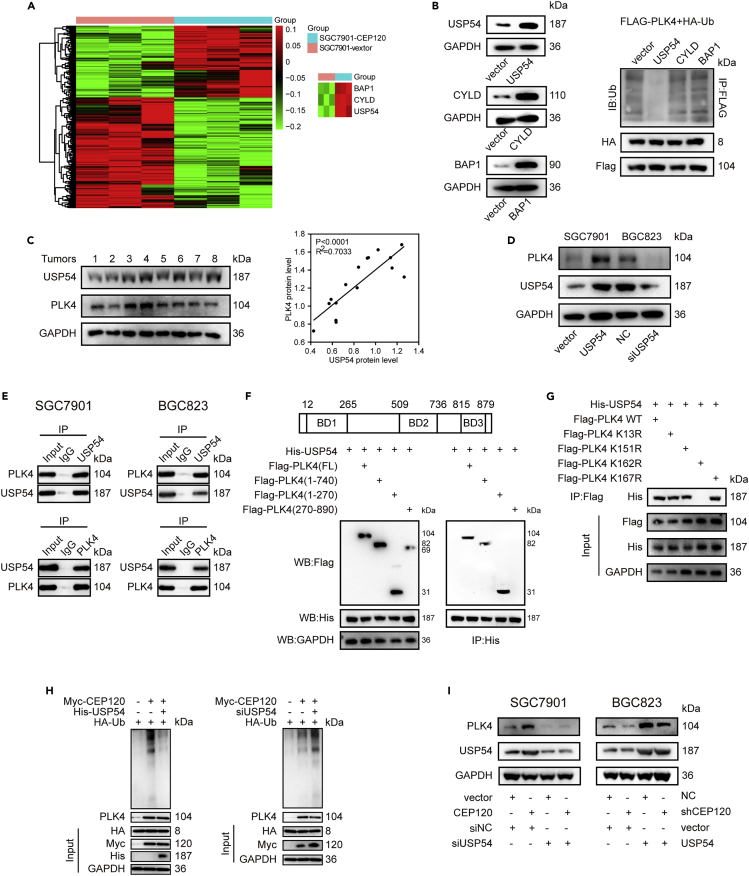
Since CEP120 is unlikely to directly mediate PLK4 deubiquitination, we hypothesized that some deubiquitinating enzymes (DUBs) may be involved in CEP120-induced PLK4 deubiquitination. Therefore, RNAs extracted from the control group and CEP120-overexpressed SGC7901 cells were sequenced and analyzed, and three DUBs were screened out from the differentially expressed genes (Figure 5A). Subsequently, we explored the effects of these three DUBs on PLK4 ubiquitination, and the results showed that only USP54 significantly reduced PLK4 ubiquitination (Figure 5B). PLK4 protein level in GC specimens was significantly positively correlated with USP54 protein level (Figure 5C). These results suggested that USP54 is a potential deubiquitinating enzyme of PLK4.
Figure 5.
USP54 combines with PLK4 and mediates its deubiquitination
(A) Microarray analysis of mRNAs extracted from SGC7901-vector and SGC7901-CEP120 cells.
(B) Results of the ubiquitination level of PLK4 in HEK293T cells, which were co-transfected with Flag-PLK4, HA-Ub, and one of the three DUBs.
(C) WB results of USP54 and PLK4 expression in GC tissues. p value derived from Pearson correlation analysis.
(D) WB results of PLK4 and USP54 after USP54 overexpression or knockdown.
(E) CoIP analysis of the interaction between PLK4 and USP54 in SGC7901 and BGC823 cells.
(F) HEK 293T cells were co-transfected with His-USP54 and Flag-PLK4 full length or Flag-PLK4 (1-740), Flag-PLK4 (1-270), and Flag-PLK4 (270-890) constructs and harvested for WB and IP analysis.
(G) HEK 293T cells were co-transfected with His-USP54 and Flag-PLK4 wildtype or Flag-PLK4 lysine residue mutants and harvested for IP analysis.
(H) Ubiquitination level of PLK4 after USP54 overexpression or knockdown examined by ubiquitination assay.
(I) WB results of PLK4 in SGC7901 cells transfected with Lenti-CEP120 and siUSP54 and BGC823 cells transfected with Lenti-shCEP120 and USP54 plasmid.
We then used USP54-specific small interfering RNA (siRNA) and overexpression plasmid to detect the effect of USP54 on PLK4 protein level. As shown in Figure 5D, USP54 overexpression significantly increased PLK4 protein level, whereas USP54 knockdown significantly decreased PLK4 protein level. Coimmunoprecipitation (CoIP) experiments confirmed the interaction between USP54 and PLK4 (Figure 5E). In order to find out the detailed interacting region of PLK4 and USP54, we generated three truncation mutants of PLK4 according to the location of binding domain (BD), namely Flag-PLK4 (1-740), Flag-PLK4 (1-270), and Flag-PLK4 (270-890). The immunoprecipitation (IP) results showed that PLK4 bound with USP54 via BD1 (Figure 5F). It is reported that the deubiquitinating site is usually at the lysine residue.11 In order to further explore the specific deubiquitinating site of PLK4, we found four potential deubiquitinating sites of lysine residue at the position of 13/151/162/167 in PLK4 BD1 through the protein lysine modification database. Subsequently, we substituted lysine residue at the position of 13/151/162/167 by arginine residue to generate four mutants and transfected them with USP54 plasmid. The results showed that the mutation of K162 hindered the combination of USP54 and PLK4, indicating that K162 was the potential deubiquitinating site of PLK4 (Figure 5G). Moreover, we found that the ubiquitination level of PLK4 decreased significantly after USP54 overexpression. Conversely, USP54 knockdown significantly increased PLK4 ubiquitination (Figure 5H). We further conducted rescue experiments to explore whether the regulation of PLK4 ubiquitination by CEP120 is related to USP54. We found that knockdown of USP54 could reverse the increased PLK4 protein level by CEP120 overexpression, while overexpression of USP54 could rescue the reduced PLK4 protein level by CEP120 knockdown (Figure 5I). These results indicated that CEP120 regulates PLK4 ubiquitination via USP54.
CEP120 regulates USP54 expression and its centrosome aggregation
We analyzed the mRNA correlation between USP54 and CEP120 using the Gene Expression Profiling Interactive Analysis database and found that the mRNA level of USP54 was significantly positively correlated with CEP120 (Figure 6A). The mRNA levels of USP54 and CEP120 in GC specimens were further detected by quantitative real-time PCR (RT-qPCR), and the results confirmed an obvious correlation between USP54 and CEP120 (Figure 6B). Similarly, overexpression of CEP120 increased USP54 protein level, whereas knockdown of CEP120 decreased USP54 protein level (Figure 6C). In addition, we found a high overlap between the localization of CEP120 and USP54 in GC cells through IF assay. Overexpression of CEP120 promoted the relative fluorescence intensity of USP54 in centrosomes, whereas knockdown of CEP120 weakened it (Figure 6D).
Figure 6.
CEP120 regulates USP54 expression and its centrosome aggregation
(A) Analysis of correlation between USP54 and CEP120 mRNA in GC from GEPIA database. p value derived from Pearson correlation analysis.
(B) Analysis of correlation between USP54 and CEP120 mRNA in GC tissues. p value derived from Pearson correlation analysis.
(C) WB results of USP54 after CEP120 overexpression and knockdown.
(D) Immunofluorescence analysis of the localization and fluorescence intensities of USP54 and CEP120 in pHH3-negative cells. Scale bars, 5 μm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗p < 0.05, ∗∗∗p < 0.001.
(E) CCK8 and (F) Colony formation assay were used to verify the effect of USP54 on CEP120-induced cell proliferation ability. Scale bars, 1 cm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(G) Transwell assay was used to verify the role of USP54 on CEP120-induced migratory and invasive capacities. Scale bars, 50 μm. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
To further explore whether CEP120 regulates GC proliferation, invasion, and metastasis in a USP54-dependent manner, we conducted rescue experiments using siUSP54 or USP54 plasmids. The results showed that silence of USP54 significantly reversed the enhanced proliferative, migratory, and invasive abilities of CEP120-overexpressed cells, whereas USP54 overexpression restored the reduced proliferative, migratory, and invasive abilities of CEP120-knocked-down cells (Figures 6E–6G).
Finally, to verify the important role of CEP120 in the progression of GC in clinical settings, we collected clinicopathological and prognostic data of 56 patients and detected the expression of CEP120 in GC specimens by RT-qPCR. We found that the expression of CEP120 in patients with stage III–IV disease was significantly higher than that in patients with stage I–II disease. Moreover, patients with higher CEP120 expression had a worse prognosis (Figures 7A–7C). The results above suggested that CEP120 promotes USP54 expression and centrosome aggregation, which inhibits the ubiquitination degradation of PLK4 and increases PLK4 protein level, thus promoting centrosome amplification and GC progression (Figure 7D).
Figure 7.
Higher expression of CEP120 was associated with clinical GC progression and worse prognosis
(A) RT-qPCR analysis of CEP120 mRNA expression level in GC tissues of different stage patients. Data were expressed as mean ± SEM. p value derived from Student’s t test. ∗∗p < 0.01.
(B) Analysis of the association between CEP120 expression and clinicopathological character. p value derived from χ2 test.
(C) Kaplan-Meier curves of the correlation between CEP120 expression and overall survival rates in GC patients. p value derived from log rank test.
(D) The schematic diagram of molecular mechanism by which CEP120 promotes centrosome amplification and GC progression.
Discussion
The centrosome, as the microtubule-organizing center of the cell, is involved in the assembly of the mitotic spindle.12 Centrosome amplification has long been proposed to contribute to cell division disorder, aneuploidy, and genomic instability, which appears in almost all tumors and may be a universal marker of tumors.3 Centrosome amplification even appears in the early stages of tumors13,14 and is closely related to tumor genesis and progression.15 Therefore, exploration of centrosome amplification is of great significance for early diagnosis and the investigation of new therapeutic targets for tumors. Centrosome protein disorders are closely related to centrosome amplification and tumor progression. Reportedly, overexpression of CEP131 can promote centrosome amplification and the progression of colon cancer.16 Overexpression of CEP72 can promote the aggressiveness of bladder urothelial tumors.17 Our study found that CEP120 was significantly overexpressed in GC. In vitro and in vivo experiments confirmed that abnormally high expression of CEP120 promoted centrosome amplification and GC progression. In terms of mechanism, we found that CEP120 promotes centrosome amplification and GC progression by maintaining the protein stability of PLK4 through USP54-mediated PLK4 deubiquitination.
CEP120 was firstly reported to regulate interkinetic nuclear migration and neurogenesis.18 Subsequently, CEP120 was found to play a key role in centriole assembly and is one of the proteins required for centrosome replication and overduplication.19 Further studies have found that CEP120 is closely related to centrosomal protein 4.1-associated protein (CPAP) and jointly regulates the process of centriole elongation. Overexpression of CEP120 or CPAP can lead to the production of additional centrioles.4,20 Our previous study showed that CEP120 plays an important role in centrosome replication and is necessary for the proliferation and migration of cerebellar granule neuron progenitors.5 A recent study using exome sequencing found that the CEP120 mutation increased the risk of pregnancy-derived aneuploidy, and ectopic expression of CEP120: P. arg947His led to reduced spindle microtubule nucleation efficiency and increased incidence of aneuploidy,21 which is one of tumor characteristics, suggesting that CEP120 may be closely related to the development of tumors. However, the correlation between CEP120 and tumors has not been reported. In our study, we found that overexpression of CEP120 promoted centrosome amplification and progression of GC, which is of great significance for discovering the potential function of CEP120 and exploring new therapeutic targets for GC.
Centrosome replicates once per cell cycle, which is controlled by serine/threonine protein kinase PLK4.9,10 Centrosomes cannot be formed with the knockout of PLK4,22 and overexpression of PLK4 can lead to centrosome amplification and tumorigenesis.23 PLK4 is highly expressed in various tumors and is involved in tumor formation, invasion, and metastasis.7,24,25 Several studies on the inhibition of PLK4 as tumor treatment are in the clinical trial stage.8,26 The stability of PLK4 is regulated by different molecules in different biological processes.27,28,29 Here, we found that CEP120 regulates PLK4 protein level through the deubiquitinating enzyme USP54 in GC cells. Overexpression of CEP120 promotes the expression and centrosome aggregation of USP54, which combines with PLK4 and reduces its ubiquitination degradation, thus promoting centrosome amplification and GC progression. This discovery may provide new drug targets for inhibiting PLK4 in tumor therapy.
Ubiquitination is a highly dynamic process that can be reversed by DUBs, which cleave isopeptide bonds linking ubiquitin and target proteins.30 Among DUBs, ubiquitin-specific proteases (USPs) are the most abundant group and play important roles in various biological processes, including DNA repair, cell growth, and apoptosis, and are predicted to be associated with cancers.31 However, the role of USPs in tumorigenesis remains largely unclear. Fraile et al. found that USP54 is highly expressed in colorectal cancer and is related to the proliferation, invasion, and migration of colorectal cancer. In addition, high expression of USP54 is significantly related to the low survival rate of patients with colorectal cancer.32 Previous studies have reported that some DUBs reduce PLK4 ubiquitination via different biological processes.27,29 Our study identified USP54 as a novel deubiquitinating enzyme of PLK4 in GC. Overexpression of USP54 could rescue the reduced PLK4 protein level and GC progression, which were triggered by CEP120 knockdown. These results suggested that USP54 plays a key role in the CEP120-mediated stabilization of PLK4 and is expected to become a novel target for the treatment of GC.
In conclusion, we revealed the key role of the CEP120-USP54-PLK4 axis in centrosome amplification and GC progression. Therefore, impeding this axis may be an effective strategy for GC treatment.
Limitations of the study
How CEP120 regulates USP54 expression and its aggregation in centrosome has not been figured out in this study. Our previous study showed that the role of CEP120 in the proliferation and migration of cerebellar granule neuron progenitors is closely related to the hedgehog signaling pathway.5 Therefore, we speculate that CEP120 may be associated with the regulation of USP54 through the hedgehog signaling pathway, which is the focus of our next research.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CEP120 | Abcam | Cat#ab230239; RRID: NA |
| CEP120 | Thermo Fisher | Cat#PA5-55985; RRID: AB_2639665 |
| GAPDH | Proteintech | Cat#60004-1-Ig; RRID: AB_2107436 |
| γ tubulin | Proteintech | Cat#66320-1-Ig; RRID: AB_2857350 |
| PLK4 | Abclonal | Cat#A9863; RRID: AB_2771714 |
| USP54 | Affinity | Cat#DF9595; RRID: AB_2842791 |
| CYLD | Proteintech | Cat#11110-1-AP; RRID: AB_10915966 |
| BAP1 | Proteintech | Cat#10398-1-AP; RRID: AB_2180460 |
| Myc | Abcam | Cat#ab32; RRID: AB_303599 |
| HA | Abcam | Cat#ab1424; RRID: AB_301017 |
| Ub | Abcam | Cat#ab134953; RRID: AB_2801561 |
| Flag | Abcam | Cat#ab205606; RRID: AB_2916341 |
| His | Abcam | Cat#ab18184; RRID: AB_444306 |
| pHH3 | Proteintech | Cat#66863-1-Ig; RRID: AB_2882201 |
| Biological samples | ||
| Gastric cancer tissue of patient | Department of Gastrointestinal Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology | NA |
| Adjacent normal tissue of patient | Department of Gastrointestinal Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology | NA |
| Chemicals, peptides, and recombinant proteins | ||
| Cycloheximide | Selleck | Cat#S7418 |
| MG132 | Selleck | Cat#S2619 |
| Chloroquine diphosphate | Selleck | Cat#S4157 |
| TRIzol | Invitrogen | Cat#15596018 |
| Lipofectamine 3000 | Invitrogen | Cat#L3000075 |
| Critical commercial assays | ||
| Ubiquitylation Assay Kit | Abcam | Cat#ab139471 |
| RT-PCR kit | Takara | Cat#RR014A |
| Cell counting kit-8 | Dojondo | Cat#CK04 |
| Q5 Site-Directed Mutagenesis Kit | New England BioLabs | Cat#E0554S |
| Deposited data | ||
| Raw RNAseq data | This paper | GEO: GSE217567 |
| Experimental models: Cell lines | ||
| SGC7901 | Cobioer Biosciences | Cat#CBP60500; RRID: CVCL_0520 |
| BGC823 | Cobioer Biosciences | Cat#CBP60477; RRID: CVCL_3360 |
| AGS | Cobioer Biosciences | Cat#CBP60476; RRID: CVCL_0139 |
| MKN45 | Cobioer Biosciences | Cat#CBP60488; RRID: CVCL_0434 |
| SNU-719 | Cobioer Biosciences | Cat#CBP60511; RRID: CVCL_5086 |
| GES1 | Cobioer Biosciences | Cat#CBP60512; RRID: CVCL_EQ22 |
| Experimental models: Organisms/strains | ||
| Mouse: BALB/c male nude mice (four weeks old) | Beijing Huafukang | NA |
| Oligonucleotides | ||
| Primer sequence of CEP120: (forward: 5-AGTTGGCTACTGATCCTGTGG-3; reverse: 5-GGAGTTTGATAGGAGTACGCTGT-3) | This paper | NA |
| Primer sequence of PLK4: (forward: 5- AAGCTCGACACTTCATGCACC-3; reverse: 5- GCATTTTCAGTTGAGTTGCCAG-3) | This paper | NA |
| Primer sequence of USP54: (forward: 5- GATGTTTGCACCTCGAAGCTC-3; reverse: 5- TCCCATGCACTTGTGAGTTGT-3) | This paper | NA |
| CEP120 shRNA: 5-gaTTATTTGACTCGCCTGATA-3 | This paper | NA |
| siPLK4: 5-GGACCUUAUUCACCAGUUACU-3 | This paper | NA |
| siUSP54: 5-GGAAGGAUGUGGUGACCAAAU-3 | This paper | NA |
| Recombinant DNA | ||
| GV703 | Genechem | https://www.genechem.com.cn/index/supports/zaiti_info.html?id=485 |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com |
| SPSS 20.0 | IBM | https://www.ibm.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chuanqing Wu (wucq2014@hust.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell culture and cell lines
The GC cell lines SGC7901, BGC823, AGS, MKN45, and SNU-719, and the gastric mucosal epithelial cell line GES1 were purchased from the Cobioer Biosciences Co., Ltd (Nanjing, China), which have been tested and authenticated by DNA short tandem repeat test. All cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA) at 37 °C in an incubator of 5% CO2 and 95% atmosphere.
Mouse models
The mouse experiments were approved by the Institutional Animal Care and Use Committee of the Huazhong University of Science and Technology. BALB/c male nude mice (four weeks old) were purchased from Beijing Huafukang (Beijing, China) and fed by professional animal keepers in sterile animal rooms following the recommended procedures of National Institutes of Health guide for the care and use of laboratory animals.
Method details
GC specimens
A total of 56 specimens were obtained from patients with GC who did not undergo radiotherapy or chemotherapy before surgery. The diagnosis of GC was confirmed based on the pathological results of the surgical specimens. All procedures were conducted in accordance with the principles of the Declaration of Helsinki and were approved by the Ethics Committee of Union Hospital, Huazhong University of Science and Technology. An informed consent was signed by all patients before participating in the study.
Cell culture and reagents
The GC cell lines SGC7901, BGC823, AGS, MKN45, and SNU-719, and the gastric mucosal epithelial cell line GES1 were purchased from the Cobioer Biosciences Co., Ltd (Nanjing, China). All cells were cultured in RPMI-1640 medium supplemented with 10% FBS (Gibco, Grand Island, NY, USA). Cycloheximide, proteasome inhibitor MG132, and chloroquine diphosphate were purchased from Selleck Chemicals (Houston, TX, USA).
Western blot and immunoprecipitation analysis
RIPA lysis buffer was used for isolation of the total protein from tissues and cells. The separation of protein samples was conducted by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, which were then transferred to PVDF membranes. The membranes were blocked with 5% milk in TBST buffer for 1h before incubated with primary antibodies in 4 °C overnight. Detailed antibody information is provided in Table S1. Then the membranes were washed for 3 times in TBST before incubated with secondary antibody for the next day, which were finally visualized with enhanced chemiluminescence system (Millipore). Each experiment was repeated three times.
In vitro ubiquitination assay
HEK293T and GC cells were transiently transfected with specific plasmids or siRNAs and HA-Ub, and were then treated with 20 μM MG132 for 6h. RIPA lysis buffer was used to extract cell lysates of the transfected cells, and the ubiquitination assay was performed with Ubiquitylation Assay Kit (Abcam) according to the manufacture’s protocol.
Quantitative real-time polymerase chain reaction
Total RNA from cells or specimens was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. RT-qPCR was performed using a RT-PCR kit (TaKaRa, Kyoto, Japan) according to the manufacturer’s instructions. The primer sequences are listed as follows: CEP120, (forward: 5-AGTTGGCTACTGATCCTGTGG-3; reverse: 5-GGAGTTTGATAGGAGTACGCTGT-3), PLK4, (forward: 5-AAGCTCGACACTTCATGCACC-3; reverse: 5-GCATTTTCAGTTGAGTTGCCAG-3), USP54, (forward: 5-GATGTTTGCACCTCGAAGCTC-3; reverse: 5-TCCCATGCACTTGTGAGTTGT-3).
Cell proliferation assay
The CCK8 reagent (Dojondo Laboratories, Kumamoto, Japan) was used to perform cell proliferation assays according to the manufacturer’s instructions; 96-well plates were planted with 2 × 103 cells per well, and the cells were incubated with CCK8 for 2h at 24, 48, 72, and 96h.
Colony formation assay
In total, 1000 cells were planted in 6-well plates and cultured in the corresponding medium for two weeks. We used 0.1% crystal violet to stain the cell colonies.
Cell migration and invasion assays
Wound-healing assay and transwell assay were used to assess the migratory and invasive capacities of the GC cells. For wound-healing assay, 1.5 × 105 cells were planted in 6-well plate and incubated for 24h to form a confluent monolayer. Then a sterile plastic micropipette tip was used to scratch for a straight-edged “wound” in each well. After washed with phosphate buffered saline (PBS) for 3 times, the well was added with FBS-free medium. The images were acquired with a light microscope at 100x at the time the “wound” was created and 48h of incubation. For migration assay, 5 × 104 cells were planted on the upper chamber with FBS-free medium and the lower chamber was filled with 25% FBS medium. After incubated for 24h, the cells on the upper side of the membrane were softly removed with a cotton swab and the cells on the bottom side of the membrane were fixed with paraformaldehyde and stained with 0.1% crystal violet, the images of which were finally acquired with a light microscope at 200x. For invasion assay, the bottom of the upper chamber was coated with Matrigel (BD Bioscience, USA) and the procedures were the same as migration assay.
Immunofluorescence assay
The IF assay was performed with specific antibodies against CEP120 (1:100) (Thermo Scientific, PA5-55985, USA), γ tubulin (1:100) (Proteintech, 66320-1-lg, Wuhan, China) and USP54 (1:100) (Affinity Biosciences, DF9595, Jiangsu, China). Phospho-Histone H3 (pHH3) (1:100) (Proteintech, 66863-1-Ig, Wuhan, China) was incubated simultaneously to distinguish the G1 phase cells. Briefly, the cells were fixed with −20 °C methanol for 10min after washed with PBS, which were then subjected to permeation with 0.5% triton-X100-PBS for 5min. After that, the cells were blocked with 10% goat serum for 0.5h and incubated with the primary antibodies at 4 °C overnight. The second day, the cells were incubated with second antibodies for 1h and DAPI for 5min. Fluorescence images were acquired using a Zeiss confocal laser scanning microscope (LSM 780 with Airyscan; Carl Zeiss). The proportion of centrosome amplification was counted in more than five microscopic fields of pHH3 negative cells and averaged.
Immunohistochemistry
IHC analysis was performed with CEP120 antibody (1:100) (Abcam, ab230239, England). Gastric cancer and adjacent normal tissues were embedded with hard wax, sliced, dewaxed and rehydrated, which were then counterstained with hematoxylin after incubated with primary and second antibodies. Images were acquired under a light microscope at 400x.
Transfection
Human Lenti-shCEP120-GFP (CEP120 shRNA 5-gaTTATTTGACTCGCCTGATA-3, Genechem, Shanghai, China), Lenti-CEP120-GFP (Genechem), and their controls were used in our study. Stable cell lines were obtained by selection with 4 μg/mL puromycin after lentiviral infection. WB and RT-qPCR analyses were conducted to assess transfection efficiency. The siRNAs used for PLK4 and USP54 were as follows: siPLK4, 5-GGACCUUAUUCACCAGUUACU-3; siUSP54, 5-GGAAGGAUGUGGUGACCAAAU-3. Q5 Site-Directed Mutagenesis Kit (New England BioLabs) was used to generate PLK4 mutants according to the manufacturer protocol. Plasmids or siRNA transfections were conducted using Lipofectamine 3000 (Invitrogen), and the transfection efficiency was confirmed by WB.
Xenograft assay
The mouse experiments were approved by the Institutional Animal Care and Use Committee of the Huazhong University of Science and Technology. The care and use of mice followed the recommended procedures of National Institutes of Health guide. BALB/c male nude mice (four weeks old) were purchased from Beijing Huafukang (Beijing, China) and randomly separated into four groups. To develop subcutaneous xenograft models, 5×106 cells (lentiviral infectious SGC7901-vector, SGC7901-CEP120, BGC823-NC, BGC823-shCEP120) in 100 μL of PBS were subcutaneously injected into the groin of the mice. Tumor length and width were recorded every four days after inoculation, and the tumor volume was calculated. All mice were sacrificed four weeks after inoculation. For the metastasis assay, 4×106 cells (lentiviral infectious SGC7901-vector, SGC7901-CEP120, BGC823-NC, BGC823-shCEP120) in 150 μL of PBS were injected into the tail veins of mice, and all mice were sacrificed six weeks after the injection. The lungs of the mice were dissected to obtain metastatic images.
Quantification and statistical analysis
All in vitro experiments were performed at least three times. The results are presented as the mean ± SEM. Statistical analyses were performed using GraphPad Prism 8 or SPSS 20.0. Comparisons of quantitative variables between groups were conducted using Student’s t-test. The association between target gene expression and clinicopathological features was evaluated using the χ2 test. Log-rank test was adopted to assess the difference of prognostic curve. Pearson correlation analysis was used to evaluate the correlation of expression level. The differences were considered significant at ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001, and ∗∗∗∗p<0.0001, ns, not significant.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 82073324, 81874184, 82000512) and scientific research project of Hubei Health Commission (No. WJ2019Q035). The authors thank Editage for editing grammar, spelling, and other common errors. Thank Fangqi Chen for the help with figure layout.
Author contributions
Conceptualization: Z.C.G., M. X. X., and W. C.Q.; Formal Analysis: Z.C.G. and M. X. X.; Investigation: Z.C.G., M. X. X., W.G.X., Z.X.X., H.P., and C.X.; Writing – Original Draft: Z.C.G.; Writing – Review & Editing: Z.C.G., M. X. X., W.D.S., L.Y., R.T., Z.W.K., T.K.X., and W.C.Q.; Funding Acquisition: T.K.X. and W.C.Q.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Ethics approval statement
This study was approved by the Ethics Committee of Union Hospital, Huazhong University of Science and Technology, and all procedures were conducted in accordance with the principles of the Declaration of Helsinki. The mouse experiments were approved by the Institutional Animal Care and Use Committee of the Huazhong University of Science and Technology. The care and use of mice followed the recommended procedures of National Institutes of Health guide.
Patient consent statement
An informed consent was signed by all patients before participating in the study.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105745.
Supplemental information
Data and code availability
-
•
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession number is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci. 2008;121:1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 3.Chan J.Y. A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci. 2011;7:1122–1144. doi: 10.7150/ijbs.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Y.N., Wu C.T., Lin Y.C., Hsu W.B., Tang C.J.C., Chang C.W., Tang T.K. CEP120 interacts with CPAP and positively regulates centriole elongation. J. Cell Biol. 2013;202:211–219. doi: 10.1083/jcb.201212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C., Yang M., Li J., Wang C., Cao T., Tao K., Wang B. Talpid3-binding centrosomal protein Cep120 is required for centriole duplication and proliferation of cerebellar granule neuron progenitors. PLoS One. 2014;9:e107943. doi: 10.1371/journal.pone.0107943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godinho S.A., Picone R., Burute M., Dagher R., Su Y., Leung C.T., Polyak K., Brugge J.S., Théry M., Pellman D. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 2014;510:167–171. doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maniswami R.R., Prashanth S., Karanth A.V., Koushik S., Govindaraj H., Mullangi R., Rajagopal S., Jegatheesan S.K. PLK4: a link between centriole biogenesis and cancer. Expert Opin. Ther. Targets. 2018;22:59–73. doi: 10.1080/14728222.2018.1410140. [DOI] [PubMed] [Google Scholar]
- 8.Veitch Z.W., Cescon D.W., Denny T., Yonemoto L.M., Fletcher G., Brokx R., Sampson P., Li S.W., Pugh T.J., Bruce J., et al. Safety and tolerability of CFI-400945, a first-in-class, selective PLK4 inhibitor in advanced solid tumours: a phase 1 dose-escalation trial. Br. J. Cancer. 2019;121:318–324. doi: 10.1038/s41416-019-0517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nigg E.A., Holland A.J. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 2018;19:297–312. doi: 10.1038/nrm.2017.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zitouni S., Nabais C., Jana S.C., Guerrero A., Bettencourt-Dias M. Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 2014;15:433–452. doi: 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
- 11.Clague M.J., Urbé S., Komander D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019;20:338–352. doi: 10.1038/s41580-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 12.Nigg E.A., Raff J.W. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 13.Goepfert T.M., Adigun Y.E., Zhong L., Gay J., Medina D., Brinkley W.R. Centrosome amplification and overexpression of aurora A are early events in rat mammary carcinogenesis. Cancer Res. 2002;62:4115–4122. [PubMed] [Google Scholar]
- 14.Pihan G.A., Wallace J., Zhou Y., Doxsey S.J. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- 15.Mittal K., Ogden A., Reid M.D., Rida P.C.G., Varambally S., Aneja R. Amplified centrosomes may underlie aggressive disease course in pancreatic ductal adenocarcinoma. Cell Cycle. 2015;14:2798–2809. doi: 10.1080/15384101.2015.1068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D.H., Ahn J.S., Han H.J., Kim H.M., Hwang J., Lee K.H., Cha-Molstad H., Ryoo I.J., Jang J.H., Ko S.K., et al. Cep131 overexpression promotes centrosome amplification and colon cancer progression by regulating Plk4 stability. Cell Death Dis. 2019;10:570. doi: 10.1038/s41419-019-1778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., Dong P., Wei W., Jiang L., Guo S., Huang C., Liu Z., Chen J., Zhou F., Xie D., Liu Z. Overexpression of CEP72 promotes bladder urothelial carcinoma cell aggressiveness via epigenetic CREB-mediated induction of SERPINE1. Am. J. Pathol. 2019;189:1284–1297. doi: 10.1016/j.ajpath.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Xie Z., Moy L.Y., Sanada K., Zhou Y., Buchman J.J., Tsai L.H. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahjoub M.R., Xie Z., Stearns T. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J. Cell Biol. 2010;191:331–346. doi: 10.1083/jcb.201003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comartin D., Gupta G.D., Fussner E., Coyaud É., Hasegan M., Archinti M., Cheung S.W.T., Pinchev D., Lawo S., Raught B., et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 2013;23:1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Tyc K.M., El Yakoubi W., Bag A., Landis J., Zhan Y., Treff N.R., Scott R.T., Tao X., Schindler K., Xing J. Exome sequencing links CEP120 mutation to maternally derived aneuploid conception risk. Hum. Reprod. 2020;35:2134–2148. doi: 10.1093/humrep/deaa148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348:1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serçin Ö., Larsimont J.C., Karambelas A.E., Marthiens V., Moers V., Boeckx B., Le Mercier M., Lambrechts D., Basto R., Blanpain C. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat. Cell Biol. 2016;18:100–110. doi: 10.1038/ncb3270. [DOI] [PubMed] [Google Scholar]
- 24.Liao Z., Zhang H., Fan P., Huang Q., Dong K., Qi Y., Song J., Chen L., Liang H., Chen X., et al. High PLK4 expression promotes tumor progression and induces epithelial-mesenchymal transition by regulating the Wnt/β-catenin signaling pathway in colorectal cancer. Int. J. Oncol. 2019;54:479–490. doi: 10.3892/ijo.2018.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosario C.O., Kazazian K., Zih F.S.W., Brashavitskaya O., Haffani Y., Xu R.S.Z., George A., Dennis J.W., Swallow C.J. A novel role for Plk4 in regulating cell spreading and motility. Oncogene. 2015;34:3441–3451. doi: 10.1038/onc.2014.275. [DOI] [PubMed] [Google Scholar]
- 26.Parsyan A., Cruickshank J., Hodgson K., Wakeham D., Pellizzari S., Bhat V., Cescon D.W. Anticancer effects of radiation therapy combined with Polo-Like Kinase 4 (PLK4) inhibitor CFI-400945 in triple negative breast cancer. Breast. 2021;58:6–9. doi: 10.1016/j.breast.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Čajánek L., Glatter T., Nigg E.A. The E3 ubiquitin ligase Mib1 regulates Plk4 and centriole biogenesis. J. Cell Sci. 2015;128:1674–1682. doi: 10.1242/jcs.166496. [DOI] [PubMed] [Google Scholar]
- 28.Cunha-Ferreira I., Bento I., Pimenta-Marques A., Jana S.C., Lince-Faria M., Duarte P., Borrego-Pinto J., Gilberto S., Amado T., Brito D., et al. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 2013;23:2245–2254. doi: 10.1016/j.cub.2013.09.037. [DOI] [PubMed] [Google Scholar]
- 29.Yang X.D., Li W., Zhang S., Wu D., Jiang X., Tan R., Niu X., Wang Q., Wu X., Liu Z., et al. PLK4 deubiquitination by Spata2-CYLD suppresses NEK7-mediated NLRP3 inflammasome activation at the centrosome. EMBO J. 2020;39:e102201. doi: 10.15252/embj.2019102201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komander D., Clague M.J., Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 31.Mason S.D., Joyce J.A. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraile J.M., Campos-Iglesias D., Rodríguez F., Español Y., Freije J.M.P. The deubiquitinase USP54 is overexpressed in colorectal cancer stem cells and promotes intestinal tumorigenesis. Oncotarget. 2016;7:74427–74434. doi: 10.18632/oncotarget.12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession number is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.