Abstract
Background and aims:
Liver injury may persist in patients with HBV receiving antiviral therapy who have ongoing transcription and translation. We sought to assess ongoing HBV transcription by serum HBV RNA, translation by serum hepatitis B core related antigen (HBcrAg), and their associations with hepatic HBsAg and HBcAg staining in patients coinfected with HBV and HIV.
Methods:
This is a cross-sectional study of 110 adults coinfected with HBV and HIV who underwent clinical assessment and liver biopsy. Immunohistochemistry (IHC) was performed for HBsAg and HBcAg. Viral biomarkers included quantitative HBsAg, HBV RNA, and HBcrAg.
Results:
Participants’ median age was 49 years (male, 93%; Black, 51%; HBeAg+, 65%), with suppressed HBV DNA (79%) and undetectable HIV RNA (77%) on dually active antiretroviral therapy. Overall, HBV RNA and HBcrAg were quantifiable in 81% and 83%, respectively (96% and 100% in HBeAg+, respectively). HBcAg staining was detected in 60% and HBsAg in 79%. Higher HBV RNA was associated with higher HBcAg and HBsAg IHC grades (both p < 0.0001). The HBsAg membranous staining pattern was significantly associated with higher HBV-RNA and HBcrAg levels.
Conclusion:
HBcAg and HBsAg IHC staining persisted despite viral suppression, and IHC grades and staining patterns correlated with markers of transcription (HBV RNA) and translation (HBcrAg). These data indicate that apparent HBV suppression is associated with residual transcription and translation that could contribute to liver pathology. Additional antiviral strategies directed to HBV protein expression may be useful to ameliorate liver injury.
INTRODUCTION
Although the diagnosis of HBV infection relies on the use of serological and molecular virological tests, liver biopsy remains an indispensable tool to accurately assess disease grade (inflammation), stage (fibrosis), and steatosis. In addition, immunohistochemistry (IHC) and immunofluorescence for staining HBcAg and HBsAg, first used in patients with HBV infection in the 1970s,[1-3] may provide additional information on viral transcription and translation. HBcAg staining can be nuclear, cytoplasmic or both, whereas HBsAg is typically cytoplasmic or membranous.[4]
The presence of HBcAg and HBsAg staining in HBV-infected hepatocytes is linked to ongoing viral replication and transcription. Consequently, HBV-DNA suppression, HBeAg or HBsAg seroconversion, whether spontaneous or antiviral treatment–related, are associated with lower expression of HBV antigens by IHC.[5]
HBV RNA may exist as unspliced and spliced RNA species, but that detected in serum likely represents pregenomic RNA, the replicative template that is reverse-transcribed to minus strand DNA, which in turn is used to synthesize plus strand DNA to form a mature relaxed circular DNA. Studies have shown that HBV-RNA levels are only negligibly altered with the use of nucleos(t)ide analogues (NAs) and are highly correlated with HBV-DNA levels.[6,7]
Hepatitis B core-related antigen (HBcrAg), also detected in serum, shares a 149 amino-acid sequence with HBeAg, HBcAg, and a 22-kilodalton core-related protein encoded by the precore/core gene.[8] The synthesis of the core-related protein is independent of HBV-DNA formation. This biomarker has been shown to correlate, in untreated and treated patients, with serum HBV DNA, intrahepatic HBV DNA, and covalently closed circular DNA (cccDNA) levels.[9] There is limited information on the relationship between serum-based HBV RNA and HBcrAg, with HBcAg and HBsAg staining in liver tissue.
We performed a cross-sectional analysis of a geographically diverse North American sample of patients coinfected with HBV and HIV receiving dually active antiretroviral therapy (ART), to assess serum markers of HBV transcription—HBV RNA and HBcrAg and translation—HBcrAg, with hepatic HBsAg and HBcAg expression.
PATIENTS AND METHODS
Adult patients coinfected with HBV and HIV were recruited from eight Hepatitis B Research Network (HBRN) sites in the United States and Canada. Inclusion and exclusion criteria for this study have been previously published.[10] All included participants were at least 18 years old, and anti-HIV and HBsAg positive for at least 6 months. Although prior resolved infection with HCV or HDV were allowed, detectable HCV RNA less than 6 months from entry, decompensated cirrhosis, or HCC were exclusion criteria. The institutional review board at each center approved the protocol, and participants gave written informed consent. This study is registered at ClinicalTrials.gov (NCT01924455).
Standardized research assessments were conducted by study personnel based on a common research protocol. Data and central laboratory results were entered by study coordinators and transmitted to the HBRN Data Coordinating Center (University of Pittsburgh), which managed data collection and analysis of the study.
Demographic data
Age, sex, race/ethnicity, continent of birth, education level, risk factors for HIV and HBV, duration of infections, and medication use were self-reported at enrollment. Duration of HIV and/or HBV, as well as current and past ART, could not be verified satisfactorily in many subjects due to the fragmented care received from different health providers at various sites between diagnosis of HIV and HBV, and enrollment to this study. The self-reported Alcohol Use Disorder Identification Test (AUDIT) was used to categorize risk of alcohol use disorder: a score of 8–15 defined increased risk, and a score of ≥16 defined high risk.[11] Clinical assessment included height and weight, from which body mass index was calculated, and waist circumference.
Laboratory data
Complete blood count, clusters of differentiation 4 (CD4)/CD8, basic metabolic panel, aspartate aminotransferase, alanine aminotransferase (ALT), and alkaline phosphatase were determined by a local laboratory. Normal ALT was defined as ≤30 U/L in men and ≤19 U/L in women.[12] HIV stage (1–4) was defined by CD4 count at entry (≥500, 350–499, 200–349, and <200 cells/mm3, respectively).[13]
Histologic data
Liver biopsies were evaluated for inflammation and fibrosis using the histological activity index (HAI) and Ishak scoring systems by a committee of study pathologists blinded to the clinical data. The entire biopsy was examined under low power, and the areas of interest were further examined under high power. Liver biopsies were considered adequate if a minimum of three portal tracts were present. Most (93%) liver biopsies were >10 mm in length; 72% were >15 mm. Limited histologic data from this cohort were previously published.[14-16]
Immunostaining
Unstained slides were used for detection for HBsAg and HBcAg by IHC. The immunostaining was performed with the Roche Ventana BenchMark ULTRA System without antigen retrieval. Simultaneous positive and negative control samples were run in parallel. Inclusion-like, granularcytoplasmic, and membranous staining were categorized as HBsAg IHC patterns, while nuclear, cytoplasmic with nuclear predominance, and nuclear with cytoplasmic predominant staining were categorized as HBcAg IHC patterns. Immunostaining was graded according to the percentage of positive cells. Reported grades are as follows: A, no positive hepatocytes; B, <10% positive hepatocytes; and C, ≥10% positive hepatocytes. Grade C is a combination of three original categories: 10%–50%, >50%–90%, and >90% positives, which were collapsed due to very small frequencies.
Viral data
Qualitative HBsAg, qualitative HBeAg, HBV DNA, HIV RNA, and HBV genotypes from local laboratories were recorded. HIV RNA had a limit of detection (LOD) of 20 copies/ml. Additionally, the HBRN Central Laboratory (University of Washington) tested qualitative and quantitative HBsAg (qHBsAg), HBeAg and quantitative HBeAg (qHBeAg), and HBV DNA. qHBsAg and qHBeAg were tested using the Roche Diagnostics Elecsys platform with a LOD of 0.05 IU/ml and 0.3 IU/ml, respectively. HBV DNA was tested with the Roche COBAS TaqMan assay with a LOD of 10 IU/ml and a lower limit of quantification (LLOQ) of 20 IU/ml. HBV DNA was categorized as unquantifiable (<20 IU/ml), suppressed but quantifiable (≥20 to <1000 IU/ml), or not suppressed (≥1000 IU/ml). HBV-HIV suppression status was categorized as suppressed (HBV DNA < 1000 IU/ml, HIV RNA < 400 copies/ml), incomplete suppression (HBV DNA ≥ 1000 IU/ml, HIV RNA < 400 copies/ml), and not suppressed (HBV DNA ≥ 1000 IU/ml, HIV RNA ≥ 400 copies/ml). Serum for central testing was stored at the National Institute of Diabetes and Digestive and Kidney Diseases bio-repository at −70°C.
Viral markers
HBcrAg serum concentrations were measured using a chemiluminescence enzyme immunoassay (Lumipulse G HBcrAg assay; Fujirebio Europe). The assay has a linear measurement with a LLOQ of 3.0 and upper limit of quantification (ULOQ) of 6.7 log10 U/ml, as no international standard is currently available. As recommended by the manufacturer, dilution was not performed for samples with a concentration ≥ 6.8 log10 U/ml. Due to the high percentage of unquantifiable HBcrAg values, the data were evaluated as an ordinal variable: <3.0, 3.0 to <5.0, 5.0 to <6.8, and ≥6.8 log10 U/ml.
Pregenomic HBV-RNA assay: HBV RNA was isolated from plasma and amplified as described by Butler et al. [17] using the m2000 system (Abbott Molecular; Department of Infectious Diseases, Abbott Diagnostics). The LLOQ in this assay is 1.65 log10 U/ml. Levels below quantification (<1.65 log10 U/ml) were randomly imputed with a number between 0.01 and 1.64 log10 U/ml. Nondetected HBV-RNA levels were set to 0.
Statistical analysis
Descriptive statistics were used to summarize demographic, clinical, and virological characteristics. Frequencies and percentages are reported for categorical data. Medians and interquartile range (IQR) are reported for continuous data. As appropriate, associations between variable pairs are presented graphically using paneled boxplots (continuous-categorical), scatter plots (both continuous), and stacked bar plots (both categorical). Spearman rank correlation coefficient was used to assess the relationship between two continuous variables (e.g., HBV-DNA and HBV-RNA levels). Jonckheere-Terpstra trend test was used for comparing a categorical or continuous variable across an ordinal variable (e.g., HBV-DNA level by staining grade or HBcrAg categories). Kruskal-Wallis test (nonparametric analysis of variance) was used for comparing a continuous variable across categories. Chi-square test or its exact version was used to assess the significance of the association between two categorical variables.
To examine the association among demographic, clinical, and virological data with IHC grades, simple and multivariable logistic regression models with generalized logit link were used with IHC grade as the outcome (separately for HBcAg and HBsAg stains). Results are presented as ORs with 95% CIs and p values. qHBeAg and HBV-DNA levels (log10 IU/ml) were limited to participants with quantifiable HBeAg and DNA, respectively, and excluded from multivariable models. The multivariable models considered age, sex, race/ethnicity, ALT, platelets, estimated HIV duration, HIV stage, detectable (vs. undetectable) HIV RNA, estimated HBV duration, HBeAg positivity, qHBsAg level (log10 IU/ml), HBV-DNA categories, and HBV-HIV suppression status. Due to low representation, Asian non-Hispanic race/ethnicity was collapsed with the “other” category. Variables that had p > 0.10 were removed using a stepwise variable selection method. Next, associations of the serum biomarkers, HBV RNA and HBcrAg, with IHC grades, and their addition to the multivariable models, were evaluated.
Analyses were conducted using SAS version 9.4 (SAS Institute) and R version 3.5.3143. All reported p-values were two-sided; p-values less than 0.05 were statistically significant.
RESULTS
Cohort characteristics, demographics, and laboratory tests
At the end of enrollment, 139 participants attended at least one study assessment. Later, 4 were discovered to be HBsAg-negative via central laboratory testing, leaving 135 HBsAg-positive participants, of whom 110 had a liver biopsy within 12 months of enrollment with a central reading by study pathologists, required for this report (Figure S1). Among the 110, liver biopsies were performed a median (IQR) of 21 (1–47) days after the date of the enrollment research evaluation.
The characteristics of the analysis cohort are summarized in Table 1. The median (IQR) age of the participants was 49 (44–55) years; 92.7% were male; 32.1% were non-Hispanic White; and 51.4% were non-Hispanic Black. The median (IQR) duration of HIV infection was 20 (11–25) years, whereas that of HBV co-infection was 15 (8–22) years. Most participants had stage 1 (72.0%) or 2 (13.4%) HIV disease, and 8.5% had stage 4 disease. Most (97.2%) participants were currently on HBV therapy as part of ART; 94.5% were on ART including an anti-HBV nucleotide and NAs (i.e., 84.5%, tenofovir alone or in combination with other ART; 10.0%, entecavir with or without lamivudine), and 2.7% on lamivudine alone. Of the 2.7% not on HBV therapy, one had a history of tenofovir use but was not on any antiviral therapy at the time of the baseline assessment; one was on ritonavir, darunavir, emtricitabine, and dolutegravir; and one was on ritonavir alone. The median (IQR) ALT was 27 (19–39) U/L with 54.5% having a normal ALT. The median (IQR) platelet count was 200 (175–238) k/mm.[3] HBeAg was positive in 65.0% with a median (IQR) qHBeAg level of 15 (2–249) IU/ml. Median (IQR) qHBsAg level was 1578 (379–7765) IU/ml and median (IQR) HBV DNA was 986 (57–26,344) IU/ml among those with quantifiable levels. HBV DNA was unquantifiable in 56.3%, suppressed but quantifiable in 22.3%, and not suppressed in 21.4%. HIV RNA was undetectable (<20 copies/ml) in 77.0%. HBV HIV were suppressed (HBV DNA < 1000 IU/ml, HIV RNA < 400 copies/ml) in 77.4%; HBV HIV were not suppressed in 8.6%; and an additional 14.0% had incomplete suppression of HBV (HBV DNA ≥ 1000 IU/ml) but suppressed HIV (HIV RNA < 400 copies/ml). Further details on patient characteristics as well as histological features of this cohort have been reported previously.[14-16]
TABLE 1.
Characteristics of participants coinfected with HBV and HIV
| Demographics and clinical characteristics |
N = 110c |
|---|---|
| Age (IQR in years) | 49 (44:55) |
| Male | 102 (92.7%) |
| Race/ethnicity | N = 109 |
| Non-Hispanic White | 35 (32.1%) |
| Non-Hispanic Black | 56 (51.4%) |
| Other | 18 (16.5%) |
| Estimated duration of HIV infection (years) | N = 99 |
| 20 (11:25) | |
| HIV stage | N = 82 |
| 1 (CD4 ≥ 500 cells/mm3) | 59 (72.0%) |
| 2 (CD4 350–499 cells/mm3) | 11 (13.4%) |
| 3 (CD4 200–349 cells/mm3) | 5 (6.1%) |
| 4 (CD4 < 200 cells/mm3) | 7 (8.5%) |
| Estimated duration of HBV infection (years) | N = 86 |
| 14.5 (8:22) | |
| HBV treatment | |
| Nonea | 3 (2.7%) |
| Lamivudine alone | 3 (2.7%) |
| Tenofovir alone or in combination | 93 (84.5%) |
| Lamivudine and entecavir | 8 (7.3%) |
| Entecavir alone | 3 (2.7%) |
| Laboratory characteristics | |
| ALT (U/L) | 27 (19: 39) |
| Normal ALT (≤30 U/L for male, ≤19 U/L for female) | 60 (54.5%) |
| Platelets (×1000/mm3) | N = 108 |
| 200 (175:238) | |
| HIV RNA undetectable (<20 copies/ml) | N = 100 |
| 77 (77.0%) | |
| HBeAg+ | N = 103 |
| qHBeAg (IU/ml) in those quantifiable | 67 (65.0%) |
| N = 60 | |
| 15.0 (1.9:248.9) | |
| qHBsAg (IU/ml) | N = 108 |
| 1578.0 (379.2:7764.9) | |
| HBV-DNA categories | N = 103 |
| Unquantifiable (<20 IU/ml) | 58 (56.3%) |
| Suppressed but quantifiable (20 to <1000 IU/ml) | 23 (22.3%) |
| Not suppressed (≥1000 IU/ml) | 22 (21.4%) |
| HBV-DNA level (IU/ml) in those quantifiable | N = 45 |
| 986 (57:26,344) | |
| HBV-DNA and HIV-RNA suppression status, n (%) | N = 93 |
| Suppressed (HBV DNA < 1000 IU/ml, HIV RNA < 400 copies/ml) | 72 (77.4%) |
| Incomplete suppression (HBV DNA ≥ 1000 IU/ml, HIV RNA < 400 copies/ml) | 13 (14.0%) |
| Not suppressed (HBV DNA ≥ 1000 IU/ml,b HIV RNA ≥ 400 copies/ml) | 8 (8.6%) |
| Assays | N = 108 |
| HBV RNA quantifiable (≥1.65 log10 IU/ml) | 87 (80.6%) |
| HBV RNA (log10 U/ml) | 3.63 (1.85: 5.62) |
| HBcrAg (log10 U/ml) categories | |
| <3.0 (LLOQ) | 16 (14.8%) |
| 3.0 to <5.0 | 32 (29.6%) |
| 5.0–6.8 | 33 (30.6%) |
| ≥6.8 (ULOQ) | 27 (25.0%) |
Abbreviations: ALT, alanine aminotransferase; CD4, clusters of differentiation 4; IQR, interquartile range; LLOQ, lower limit of quantification; qHBeAg, quantitative HBeAg; qHBsAg, quantitative HBsAg; ULOQ, upper limit of quantification.
One participant with a history of tenofovir use was not on any antiviral therapy at the time of the baseline assessment. One was on ritonavir, darunavir, emtricitabine, and dolutegravir. Another was on ritonavir alone.
Participants with HBV DNA < 1000 IU/ml, HIV RNA ≥ 400 copies/ml (n = 2) we evaluated; 1 was categorized as not suppressed, although HBV-DNA level was below the threshold (869 IU/ml), while 1 with very higher HIV RNA but low HBV DNA was set to missing.
Data reported in 110 patients unless otherwise indicated. Numbers represent median (IQR) for continuous variables and n (%) for categorical variables, unless otherwise stated.
HBcAg staining
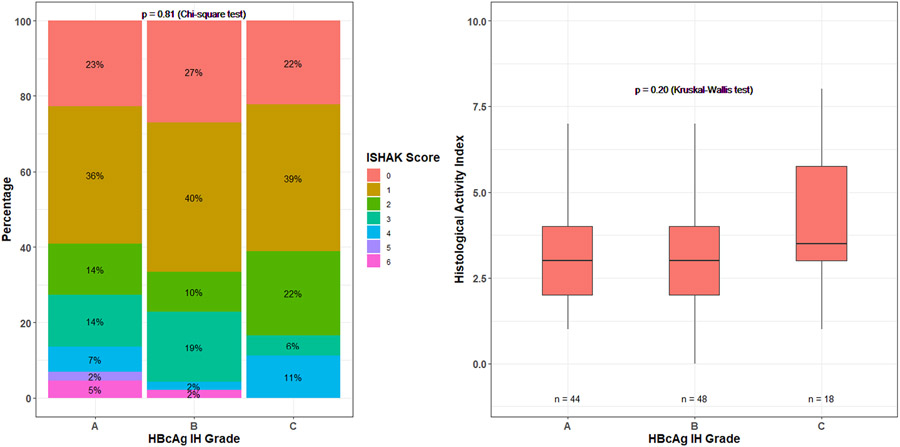
Among the 110 biopsies, HBcAg IHC grades were as follows: Grade A, 44 (40%); Grade B, 48 (44%); and Grade C, 18 (16%). Of the 66 biopsies with detectable HBcAg staining (Grades B and C), 13 (20%) had only a nuclear pattern, 33 (50%) had predominantly nuclear and cytoplasmic patterns, and the remaining 20 (30%) had predominantly cytoplasmic and nuclear patterns. Males were more likely to have Grade A or B staining compared with females (Table S1A). Higher grade of HBcAg IHC was associated with HBcAg positivity, higher qHBeAg levels, with qHBsAg HBV-DNA categories and HBV RNA (all p ≤ 0.001) (Table S1B). Higher grade of HBcAg staining was associated with a predominantly cytoplasmic and nuclear pattern, whereas lower grade of HBcAg was associated with a nuclear only pattern of staining (p = 0.001) (Table S1C). However, there was no association between grades of HBcAg staining and HAI and fibrosis scores (Figure 1). There were no significant demographic, clinical, and serological characteristics across nuclear and cytoplasmic staining patterns (data not shown).
FIGURE 1.
Ishak and histological activity index scores by HBcAg immunohistochemistry (IHC) grade.
In multivariable analysis with a stepwise variable selection, only higher qHBsAg level and higher HBV-DNA category were independently associated with the odds of higher HBcAg staining grades, such that compared with Grade A, each log increase in qHBsAg level was associated with 1.1 (95% CI: 0.7–1.7) times higher odds of having grade B HBcAg staining and 5.4 (95% CI: 1.6–17.8) times higher odds of having grade C HBcAg staining (p = 0.02). The odds of having Grade B and C versus Grade A staining, respectively, were 6.8 (95% CI: 1.7–26.4) and 22.7 (95% CI: 1.7–297.5) times higher for those with suppressed but quantifiable HBV DNA versus unquantifiable levels. Estimates were similar for the comparison of not suppressed versus unquantifiable HBV DNA (p = 0.046).
HBsAg staining
Among the 110 biopsies, HBsAg IHC staining were as follows: Grade A, 23 (21%); Grade B, 58 (53%); and Grade C, 29 (26%). Of the 87 biopsies positive for HBsAg hepatocytes, 95% and 41%, respectively, had granular cytoplasmic staining of scattered and contiguous type, whereas 66% had inclusion-like and 31% membranous staining patterns. Participants with Grade A HBsAg staining were significantly older (median = 54 years) compared to participants with Grades B (median = 50 years) and C (median = 46 years) (p = 0.004). No statistically significant differences were observed in other demographic characteristics (Table S2A). Like HBcAg IHC, higher levels of qHBeAg, qHBsAg, and HBV DNA were associated with higher grades of HBsAg staining (all p ≤ 0.01) (Table S2B). Higher grade of HBsAg staining was associated with a membranous staining pattern (p < 0.0001) and inclusion-like pattern (p = 0.003), but not a granular cytoplasmic staining pattern (p = 0.20) (Table S2C). No statistically significant differences were detected between the HBsAg staining patterns and the demographic characteristics or biochemical variables for inclusion-like and contiguous staining patterns (data not shown). However, the HBsAg membranous staining was associated with younger age, non-Hispanic Black race/ethnicity, and higher levels qHBeAg, qHBsAg, and HBV DNA (all p ≤ 0.03) (Table S3A,B).
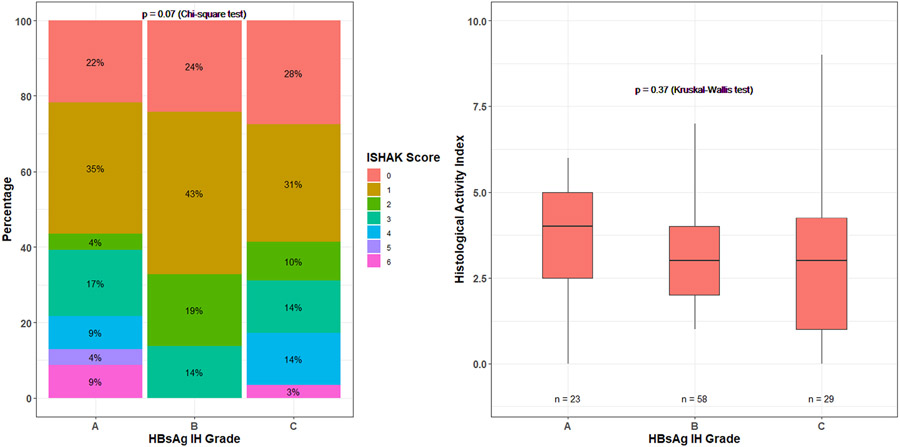
In multivariable analysis with a stepwise variable selection, only qHBsAg level was independently associated with the odds of higher HBsAg staining grades, such that each log increase in qHBsAg level was associated with 3.7 (95% CI: 1.83–7.30) times higher odds of having Grade B HBsAg IHC and 19.8 (95% CI: 7.1–55.0) times higher odds of having Grade C HBsAg IHC compared with Grade A (p < 0.001). Because qHBsAg is expected to correlate with HBsAg staining, we conducted a sensitivity analysis leaving out qHBsAg level as a candidate independent variable in multivariable analysis. In this model, only higher HBV-DNA category was significantly associated with higher HBsAg staining grades (Table 3). There was no association between grades of HBsAg staining and HAI and fibrosis scores (Figure 2).
TABLE 3.
Association between participant features and the HBsAg staining positivity grade*from multinomial logistic regression model
| Participant feature | OR (95% CI)a |
p Value | |
|---|---|---|---|
| HBsAg staining Grade B vs. A |
HBsAg staining Grade C vs. A |
||
| Univariable analysis | |||
| Age (per 10 year) | 0.71 (0.40, 1.27) | 0.42 (0.21, 0.83) | 0.03 |
| Male (ref = female) | 2.08 (0.23, 18.80) | 1.63 (0.14, 19.18) | 0.80 |
| Race/ethnicity (ref = non-Hispanic White) | |||
| Non-Hispanic Black | 0.60 (0.19, 1.86) | 1.19 (0.33, 4.28) | 0.23 |
| Other | 0.69 (0.17, 2.73) | 0.15 (0.01, 1.64) | |
| ALT (per ULN) | 1.05 (0.54, 2.03) | 1.74 (0.90, 3.34) | 0.07 |
| Platelet (per 1000/mm3) | 1.01 (0.999, 1.02) | 1.004 (0.99, 1.01) | 0.21 |
| Estimated duration of HIV (per year) | 0.996 (0.94, 1.05) | 0.99 (0.93, 1.06) | 0.97 |
| HIV stage ≥ 2 (ref = 1) | 0.48 (0.15, 1.55) | 0.63 (0.17, 2.40) | 0.47 |
| Detectable HIV RNA (ref = <20 copies/ml) | 0.53 (0.15, 1.88) | 1.77 (0.49, 6.34) | 0.10 |
| Estimated duration of HBV (per year) | 0.999 (0.94, 1.06) | 0.99 (0.93, 1.06) | 0.96 |
| HBeAg positive (ref = negative) | 4.88 (1.69, 14.10) | 4.38 (1.33, 14.45) | 0.01 |
| qHBeAg (log10 IU/ml) in those quantifiable | 1.08 (0.45, 2.57) | 2.42 (0.95, 6.15) | 0.01 |
| qHBsAg (log10 IU/ml) | 3.65 (1.83, 7.30) | 19.81 (7.14, 54.96)b | <0.0001 |
| HBV DNA (ref = unquantifiable) | |||
| Suppressed but quantifiable | 1.59 (0.45, 5.71) | 3.31 (0.76, 14.38) | 0.02 |
| Not suppressed | 2.13 (0.41, 11.12) | 11.33 (2.07, 62.10) | |
| HBV DNA (per log10 IU/ml) in those quantifiable | 0.92 (0.57, 1.48) | 1.18 (1.051, 2.41) | 0.29 |
| HBV DNA and HIV RNA (vs. suppressed) | 0.044 | ||
| Incomplete suppression | 2.25 (0.25, 20.67) | 9.00 (0.99, 81.93) | |
| Not suppressed | c | c | |
| Multivariable analysis | |||
| The only variable that was retained in the multivariable logistic model is the continuous qHBsAg (log10 IU/ml). Therefore, the estimates of OR and CI remain the same as that reported in the previous univariable analysis. | |||
Because of very small or null sample sizes in some groups, the OR estimates either do not exist or are unreliable.
Grade A, no positive hepatocytes; Grade B, <10% positive hepatocytes; and Grade C, >10% positive hepatocytes.
Because of none or few in categories, the OR becomes infinite.
FIGURE 2.
Ishak and histological activity index scores by HBsAg IHC grade.
Viral markers of HBV: HBV RNA and HBcrAg levels
HBV RNA was quantifiable (≥1.65 log10 U/ml) in 87 of 108 (81%) participants. The median (IQR) HBV-RNA level was 3.63 (1.85, 5.62) log10 U/ml (Table 1). HBcrAg was below LLOQ (<3.00 log10 U/ml) in 16 of 108 (15%) participants and above ULOQ (>6.8 log10 U/ml) in 27 of 108 (25%) (Table 1). Among those with quantifiable levels, the median (IQR) HBcrAg levels were 5.35 (3.70, 6.75) log10 U/ml.
Associations between HBV-RNA and HBcrAg levels with HBcAg/HBsAg IHC
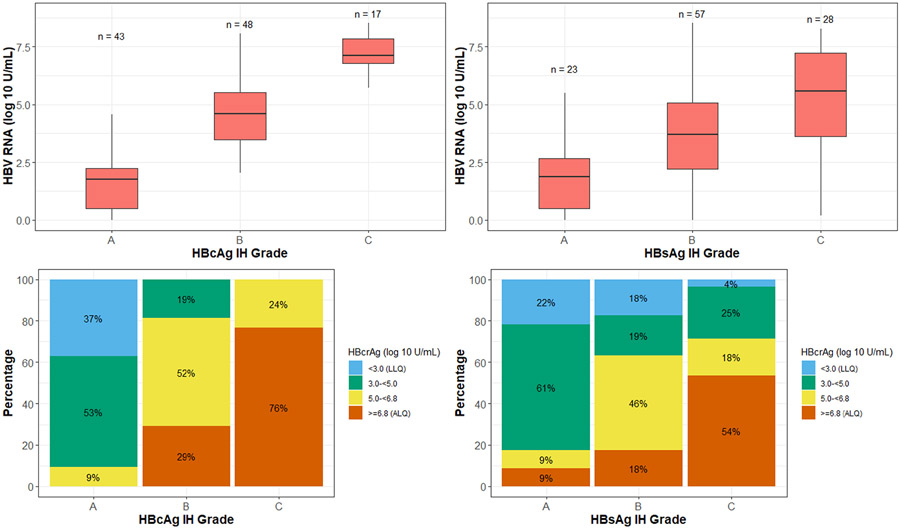
Higher levels of HBV RNA were significantly associated with higher HBcAg and HBsAg staining grades (both p < 0.001; Figure 3 top row). For hepatic HBcAg staining grades A, B and C, corresponding median HBV-RNA levels were 1.8, 4.6, and 7.1 log10 U/ml, respectively. For hepatic HBsAg staining grades A, B and C, corresponding median HBV-RNA levels were 1.9, 3.7, and 5.6 log10 U/ml, respectively (Figure 3, top row). HBcrAg levels followed a similar pattern (Figure 3, bottom row). Higher levels of HBcrAg were associated with more hepatocytes staining positive for HBcAg and HBsAg (Figure 3, bottom row). For hepatic HBcAg staining grades A, B and C, percentage of participants with HBcrAg levels above the limit of quantification (≥6.8 log10 U/ml) were 0%, 29% and 76%, respectively; for hepatic HBsAg staining grades A, B and C, these percentages were 9%, 18% and 54%, respectively.
FIGURE 3.
HBV RNA and hepatitis B core antigen (HBcrAg) by HBcAg IHC and HBsAg IHC grades. Top left: HBV RNA by HBcAg IHC grade. Top right: HBV RNA by HBsAg IHC grade. Bottom left: HBcrAg by HBcAg IHC grade. Bottom right: HBcrAg by HBsAg IHC grade.
There were 24 hepatic HBcAg Grade B and 1 Grade C participants with HBV DNA below detectable levels. Of them, 7 had HBV RNA in the 1.65–3.29 log10 range, 14 in the 3.30–4.94 log10 range, and 3 in the 4.95–6.59 log10 range. The distribution of these 24 patients for HBcrAg was as follows: 5 in the 3.00–4.99 log10 range, 16 in the 5.00–6.79 log10 range, and 3 in the ≥6.8 log10 range.
The HBcAg staining patterns (nuclear and cytoplasmic) were not significantly associated with HBV RNA and HBcrAg levels (data not shown). From the HBsAg staining patterns, only the membranous staining pattern was significantly associated with higher HBV RNA (p = 0.001), with median HBV RNA 6.1 log10 U/ml for membranous staining compared with 3.7 log10 U/ml for others (Figure S2, left panel). Similarly, higher HBcrAg level was significantly associated with HBsAg membranous staining (Figure S2, right panel).
To assess whether markers HBV RNA and HBcrAg improve the prediction of HBcAg and HBsAg staining grading, we added these markers to the multivariable models presented in Table 2 (HBcAg IHC) and Table 3 (HBsAg IHC). Consistent with the strong relationship between HBV-RNA levels and HBcAg staining grading observed in Figure S2, when HBV-RNA level was added to the HBcAg IHC grading model, no other variables remained statistically significant. For every log10 U/ml increase in HBV RNA, the odds of having HBcAg staining grade B versus A were 3.2 (95% CI: 2.0–5.0) times higher, and of having HBcAg IHC grade C versus Awere 12.2 (95% CI: 5.3–28.4) times higher (p < 0.001). However, for the HBsAg staining model, markers HBV RNA and HBcrAg were not statistically significant in the presence of qHBsAg levels.
TABLE 2.
Associations between participant features and the HBcAg staining positivity from multinomial logistic regression model
| Participant feature | OR (95% CI)b |
p Value | |
|---|---|---|---|
| HBcAg staining Grade B vs. Aa |
HBcAg staining Grade C vs. Aa |
||
| Univariable analysis | |||
| Age (per year) | 0.97 (0.61, 1.56) | 0.54 (0.28, 1.03) | 0.14 |
| Male (ref = Female) | 2.87 (0.287, 28.63) | 12.29 (1.27, 119.25) | 0.052 |
| Race/ethnicity (ref = non-Hispanic White) | |||
| Non-Hispanic Black | 0.79 (0.32, 1.97) | 1.67 (0.45, 6.29) | 0.77 |
| Other | 1.06 (0.31, 3.57) | 1.001 (0.15, 6.85) | |
| ALT (per ULN) | 1.02 (0.59, 1.77) | 1.96 (1.12, 3.43) | 0.02 |
| Platelet (per 1000/mm3) | 1.01 (0.997, 1.01) | 0.999 (0.99, 1.01) | 0.31 |
| Estimated duration of HIV (per year) | 0.97 (0.93, 1.02) | 0.97 (0.92, 1.03) | 0.42 |
| HIV stage ≥ 2 (ref = 1) | 0.73 (0.26, 2.06) | 0.32 (0.06, 1.68) | 0.40 |
| Detectable HIV RNA (ref = <20 copies/ml) | 2.65 (0.76, 9.28) | 8.75 (2.19, 35.02) | 0.01 |
| Estimated duration of HBV (per year) | 1.02 (0.97, 1.07) | 0.998 (0.94, 1.07) | 0.77 |
| HBeAg-positive (ref = negative) | 48.00 (12.93, 178.21) | c | <0.0001 |
| qHBeAg (per log10 IU/ml) in those quantifiable | 1.54 (0.45, 5.29) | 13.24 (2.67, 65.59) | 0.0003 |
| qHBsAg (per log10 IU/ml) | 1.37 (0.93, 2.01) | 10.44 (3.93, 27.78) | <0.0001 |
| HBV DNA (per log10 IU/ml) in those quantifiable | 0.91 (0.55, 1.49) | 1.36 (1.57, 4.36) | 0.08 |
| HBV DNA (ref = unquantifiable) | |||
| Suppressed but quantifiable | 5.54 (1.63, 18.84) | 34.00 (3.01, 383.83) | <0.0001 |
| Not suppressed | 3.45 (0.81, 14.74) | 134.00 (12.88, >999) | |
| HBV-DNA and HIV-RNA suppression status (ref = suppressed) | |||
| Incomplete suppression | 4.12 (0.44, 38.83) | 54.40 (5.56, 532.42) | 0.0003 |
| Not suppressed | 2.06 (0.18, 23.8) | 27.20 (2.51, 295.05) | |
| Multivariable analysis | |||
| qHBsAg (per log10 IU/ml) | 1.13 (0.73, 1.74) | 5.41 (1.64, 17.81) | 0.02 |
| HBV DNA (ref = unquantifiable) | |||
| Suppressed but quantifiable | 6.75 (1.73, 26.40) | 22.68 (1.73, 297.48) | 0.03 |
| Not suppressed | 2.82 (0.59,13.53) | 12.93 (0.85,196.79) | |
Abbreviation: ULN, upper limit of normal.
Grade A, no positive hepatocytes; Grade B, <10% positive hepatocytes; and Grade C, >10% positive hepatocytes.
Because of very small or null sample sizes in some groups, the OR estimates are unreliable.
Among participants who are negative for HBeAg, only 4 had Grade B and none had grade C, so the OR becomes infinite for the second category.
DISCUSSION
We performed a cross-sectional analysis to assess the degree and pattern of HBcAg and HBsAg staining on liver biopsy and their associations with virological markers, including two markers, serum HBV RNA and HBcrAg, among a racially diverse cohort of patients coinfected with HBV and HIV, most of whom were dually virally suppressed on ART. HBcAg was detected in 60% and HBsAg in 79% of liver biopsies despite the finding that 80% of the cohort were HBV virally suppressed (HBV DNA < 1000 IU/ml), with no specific staining pattern observed related to virological characteristics. On multivariate analysis, greater HBcAg staining was strongly associated with higher HBV-RNA levels, whereas greater HBsAg IHC grades were associated with higher qHBsAg levels.
Previous IHC studies conducted over four decades ago used less-sensitive/specific immunostaining and virological methods. These studies, performed among untreated, HBV mono-infected HBeAg-positive and HBeAg-negative patients reported that the degree of staining and distribution was related to phase of chronic hepatitis B (CHB) (and therefore level of viremia) but there was overlap among phases.[18-21] Thus, a nuclear pattern of HBcAg staining was predominantly seen among HBeAg-positive patients in the immune-tolerant phase and associated with high viral replication. Cytoplasmic staining was primarily seen on biopsies obtained from patients with HBeAg-positive and HBeAg-negative immune-active CHB, also associated with high viral replication and inflammatory activity, whereas HBcAg staining was typically absent among patients with inactive CHB with low viremia. It is unclear whether there is any clinical significance between nuclear and cytoplasmic staining for HBcAg. It has been speculated that a shift of staining from nuclear to cytoplasmic pattern may be due to hepatocyte damage and regeneration, and cytoplasmic/membrane HBcAg expression might be a target of immune response.
In contrast, HBsAg staining was unrelated to CHB phase and usually detected to varying degrees among all phases of CHB. Whether the different patterns of HBsAg staining have any clinical significance has not been established. It has been suggested that membranous staining pattern for HBsAg may reflect active viral replication and correlates with high serum HBV-DNA levels.[18] Excess HBsAg accumulation in the hepatocytes could lead to inclusion-like staining pattern, recognized in hematoxylin and eosin–stained sections as “ground-glass” hepatocytes. The current study is unique in that it was conducted in a cohort coinfected with HBV and HIV on ART, which included an anti-HBV agent. The major HBcAg pattern on liver biopsy was nuclear predominant and cytoplasmic, probably because two-thirds of the cohort were HBeAg-positive. Among the 40% who were negative for hepatic HBcAg, we are uncertain whether this was due to receipt of ART, the duration of ART, or whether they were inactive carriers before ART was initiated, as we did not have a pretreatment liver biopsy or knowledge of phase of CHB before treatment, or finally due to sampling error. Nevertheless, a relatively high proportion of participants continued to have hepatic HBcAg expression despite long-term viral suppression. Whether this is a feature unique to patients coinfected with HBV HIV is unknown, as we did not have access to a HBV mono-infected cohort on long-term ART for comparison. HBsAg staining was more prevalent compared with HBcAg staining being detected in 79% of biopsies. Among biopsies positive for HBsAg, a cytoplasmic pattern of staining was present in almost all, together to some degree, with the other staining patterns.
A key finding is that serum HBV RNA was the best virological correlate of hepatic HBcAg staining on multivariate analysis. Indeed, there was a stepwise increase in HBV-RNA levels with the HBcAg IHC grades on biopsy. This is not surprising, as it is believed that the source of HBV RNA detected in serum is the pregenomic RNA, which encodes the core protein, as well as serving as the template for HBV replication.[22] In all cases, HBV RNA and HBcrAg were detectable in the serum of participants with HBcAg staining, independent of HBV-DNA level, suggesting that although viral replication was absent, the cccDNA was transcriptionally active, likely explaining the presence of HBcAg. The clinical implication of this finding remains unclear; however, some studies suggest that these patients (i.e., virally suppressed but transcriptionally active) may be at higher risk for hepatitis flares following withdrawal of NAs therapy.[6,23]
Although there were strong associations between degree of HBsAg IHC grades and HBV-RNA and HBcrAg levels, on multivariate analysis, higher serum HBsAg level was the only virological correlate associated with greater HBsAg IHC grades. This is likely because the HBsAg is encoded by the HBsAg gene and not the pregenomic RNA, and both viral genes are differentially regulated. Multiple patterns of HBsAg staining were observed with the cytoplasmic scattered or ground-glass pattern—the most commonly observed. Several studies suggest that this pattern results from the retention of HBsAg within the Golgi-intermediate compartment of the endoplasmic reticulum.[24-26] None of the observed patterns of HBsAg staining were correlated with virological features, with the exception of the membranous pattern, which was associated with higher HBV-DNA and HBV-RNA levels. The association of higher viral replication with a membranous pattern of HBsAg staining has been previously reported in studies of HBV mono-infection.[18] It was postulated that membranous staining pattern reflects the presence of Dane particles being secreted from the hepatocyte membrane.[27] It is unclear why this would persist in the presence of low/undetectable HBV-DNA levels due to antiviral therapy, unless it also represents the non-HBV DNA containing spherical and filamentous HBsAg particles.
Of interest, 40% and 21% of biopsies were negative for HBcAg and HBsAg staining, yet low levels of HBV RNA and HBcrAg were detectable in serum, suggesting the presence of ongoing viral transcription. Whether this represents sampling error on the biopsy or the presence of viral antigen too low to detect with current IHC techniques, or that only a small proportion of transcriptionally active hepatocytes can efficiently translate viral proteins, is unknown. There is concern that ongoing viral transcription in the absence of viral replication may result in continued liver damage. However, we found no association between HBV-RNA and HBcrAg levels with either degree of hepatic inflammation or fibrosis. Similarly, there was no association between the amount of HbcAg and HBsAg staining with either degree of hepatic inflammation or fibrosis. Due to the cross-sectional design, we could not comment on disease progression. These findings differ from those reported in a mono-infected cohort treated with entecavir, in whom HBV-RNA levels were shown to correlate with histological scores for grading and staging, but again this study could not look at disease progression.[28] It is unlikely that the presence of HIV, which was adequately controlled in most cases, would explain the difference in results between our co-infected sample and this prior study of mono-infected patients. We speculate that duration of ART and suppression of viremia may account for the difference, as the coinfected cohort had been receiving ART before enrollment into the study. Previous studies have reported that HBV DNA is the strongest predictor of disease progression.[29]
There are several limitations to the analysis. We were unable to describe the IHC grades and pattern of HBcAg and HBsAg staining before treatment among the cohort, as biopsies were performed while participants were already on ART. Similarly, we did not have access to liver biopsies from patients monoinfected with HBV with viral suppression on NAs for comparison to assess whether the extent of intrahepatic HBV protein staining is more prominent in HIV coinfection versus HBV monoinfection. In addition, we could not comment on whether distribution of HBcAg and HBsAg staining was influenced by HBV genotype due to low/undetectable HBV-DNA levels in most participants. However, studies conducted among monoinfected patients suggest that the pattern of HBcAg and HBsAg is unrelated to HBV genotype. Similarly, we do not know whether distribution of HBcAg and HBsAg staining was influenced by the duration of ART and inhibition of replication, as data on medication history and HBV DNA before enrollment in the study was limited. Finally, the cross-sectional nature of the study precluded us from determining whether the observed staining intensity and pattern were due to the underlying phase of CHB before treatment or reflected suppression of replication.
Hepatic HBV viral antigen expression remains detectable in most patients with HBV-HIV coinfection despite treatment-related suppression of HBV replication. Circulating markers of HBV transcription, HBV RNA and HBcrAg, and translation of HBsAg were the best correlates of HBcAg and HBsAg staining, respectively, in adults with treated coinfection with HBV and HIV. Neither the degree of hepatic HBV viral antigen expression nor the levels of viral transcription correlated with severity of underlying liver disease. Whether persistent viral transcription in the absence of viral replication leads to disease progression, influences HBsAg clearance, HCC risk and ability to safely withdraw ART, at least in the population monoinfected with HBV, requires further clarification. Nevertheless, the finding of ongoing transcription in long-term virally suppressed patients would suggest a low likelihood of achieving functional cure and the need for more effective therapy.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Jeffrey Gersch for his assistance in testing the samples for HBV RNA.
FUNDING INFORMATION
Supported by the National Institute of Diabetes and Digestive and Kidney Diseases (R01-DK94818) as an ancillary study of the Hepatitis B Research Network; National Institutes of Health (K24AA022523, P30 DK043351, and R01 AI155140). This study is registered at ClinicalTrials.gov (NCT01924455).
Abbreviations:
- ALT
alanine aminotransferase
- ART
active antiretroviral therapy
- CHB
chronic hepatitis B
- HAI
histologic activity index
- HBcrAg
hepatitis B core related antigen
- HBRN
Hepatitis B Research Network
- IQR
interquartile range
- LLOQ
lower limit of quantification
- LOD
limit of detection
- NA
nucleos(t) ide analogue
- ULOQ
upper limit of quantification
Footnotes
CONFLICT OF INTEREST
Gavin Cloherty owns stock and is employed by Abbott. Mamta K. Jain received grants from Gilead, Merck, and Janssen. Mauricio Lisker-Melman is on the speakers’ bureau for AbbVie and Gilead. Mandana Khalili consults for and received grants from Gilead. She received grants from Intercept. Mark Sulkowski consults for Gilead and Antios. He consults for and received grants from Assembly Biosciences. Raymond T. Chung owns stock in Gilead, BMS, Janssen, AbbVie, Roche, and GSK. Richard K. Sterling received grants from Abbott, Roche, AbbVie, and Gilead. He is on the DSMB for Pfizer and AskBio. Abdus S. Wahed consults for Merck. Wendy C. King received grants from AbbVie.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
Data set used in this research paper is available upon request to richard.sterling@vcuhealth.org (coauthor of this study).
REFERENCES
- 1.Deodhar KP, Tapp E, Scheuer PJ. Orcein staining of hepatitis B antigen in paraffin sections of liver biopsies. J Clin Pathol. 1975;28:66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burns J. Immunoperoxidase localisation of hepatitis B antigen (HB) in formalin-paraffin processed liver tissue. Histochemistry. 1975;44:133–5. [DOI] [PubMed] [Google Scholar]
- 3.Akeyama T, Kamada T, Koyama M, Abe H. Distribution patterns of hepatitis B antigen in the liver. Immunofluorescence studies. Arch Pathol. 1974;98:252–6. [PubMed] [Google Scholar]
- 4.Tapp E, Jones DM. HBsAg and HBcAg in the livers of asymptomatic hepatitis B antigen carriers. J Clin Pathol. 1977;30:671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrillo RP, Brunt EM. Hepatic histologic and immunohistochemical changes in chronic hepatitis B after prolonged clearance of hepatitis B e antigen and hepatitis B surface antigen. Ann Intern Med. 1991;115:113–5. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol. 2016;65:700–10. [DOI] [PubMed] [Google Scholar]
- 7.van Bommel F, Bartens A, Mysickova A, Hofmann J, Krüger DH, Berg T, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61:66–76. [DOI] [PubMed] [Google Scholar]
- 8.Kimura T, Rokuhara A, Sakamoto Y, Yagi S, Tanaka E, Kiyosawa K, et al. Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J Clin Microbiol. 2002;40:439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Testoni B, Lebosse F, Scholtes C, Berby F, Miaglia C, Subic M, et al. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J Hepatol. 2019;70:615–25. [DOI] [PubMed] [Google Scholar]
- 10.Sterling RK, Wahed AS, King WC, Kleiner DE, Khalili M, Sulkowski M, et al. Spectrum of liver disease in hepatitis B virus (HBV) patients co-infected with human immunodeficiency virus (HIV): results of the HBV-HIV cohort study. Am J Gastroenterol. 2019;114:746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohn MJ, Babor TF, Kranzler HR. The Alcohol Use Disorders Identification Test (AUDIT): validation of a screening instrument for use in medical settings. J Stud Alcohol. 1995;56:423–32. [DOI] [PubMed] [Google Scholar]
- 12.Prati D, Taioli E, Zanella A, Torre ED, Butelli S, del Vecchio E, et al. Updated definitions of healthy ranges for serum alanine aminotransferase levels. Ann Intern Med. 2002;137:1–10. [DOI] [PubMed] [Google Scholar]
- 13.Manosuthi W, Ongwandee S, Bhakeecheep S, Leechawengwongs M, Ruxrungtham K, Phanuphak P, et al. Guidelines for antiretroviral therapy in HIV-1 infected adults and adolescents 2014, Thailand. AIDS Res Ther. 2015;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterling RK, King WC, Wahed AS, Kleiner DE, Khalili M, Sulkowski M, et al. Evaluating noninvasive markers to identify advanced fibrosis by liver biopsy in HBV/HIV co-infected adults. Hepatology. 2020;71:411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalili M, King WC, Kleiner DE, Jain MK, Chung RT, Sulkowski M, et al. Fatty liver disease in a prospective north american cohort of adults with HIV and hepatitis B Coinfection. Clin Infect Dis. 2021;73:e3275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sterling RK, King WC, Khalili M, Kleiner DE, Hinerman AS, Sulkowski M, et al. Performance of serum-based scores for identification of mild hepatic steatosis in HBV mono-infected and HBV-HIV co-infected adults. Dig Dis Sci. 2022;67:676–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler EK, Gersch J, McNamara A, Luk KC, Holzmayer V, de Medina M, et al. Hepatitis B virus serum DNA andRNA levels in nucleos(t)ide analog-treated or untreated patients during chronic and acute infection. Hepatology. 2018;68:2106–17. [DOI] [PubMed] [Google Scholar]
- 18.Hsu HC, Lai MY, Su IJ, Chen DS, Chang MH, Yang PM, et al. Correlation of hepatocyte HBsAg expression with virus replication and liver pathology. Hepatology. 1988;8:749–54. [DOI] [PubMed] [Google Scholar]
- 19.Chu CM, Liaw YF. Intrahepatic distribution of hepatitis B surface and core antigens in chronic hepatitis B virus infection. Hepatocyte with cytoplasmic/membranous hepatitis B core antigen as a possible target for immune hepatocytolysis. Gastroenterology. 1987;92:220–5. [DOI] [PubMed] [Google Scholar]
- 20.Naoumov NV, Portmann BC, Tedder RS, Ferns B, Eddleston ALWF, Alexander GJM, et al. Detection of hepatitis B virus antigens in liver tissue. A relation to viral replication and histology in chronic hepatitis B infection. Gastroenterology. 1990;99:1248–53. [DOI] [PubMed] [Google Scholar]
- 21.Chu CM, Liaw YF. Immunohistological study of intrahepatic expression of hepatitis B core and E antigens in chronic type B hepatitis. J Clin Pathol. 1992;45:791–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuh CH, Chang YL, Ting LP. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol. 1992;66:4073–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carey I, Gersch J, Wang B, Moigboi C, Kuhns M, Cloherty G, et al. Pregenomic HBV RNA and hepatitis B core-related antigen predict outcomes in hepatitis B e antigen-negative chronic hepatitis B patients suppressed on nucleos(t)ide analogue therapy. Hepatology. 2020;72:42–57. [DOI] [PubMed] [Google Scholar]
- 24.Winckler K, Junge U, Creutzfeldt W. Ground-glass hepatocytes in unselected liver biopsies. Ultrastructure and relationship to hepatitis B surface antigen. Scand J Gastroenterol. 1976;11:167–70. [PubMed] [Google Scholar]
- 25.Cohen C “Ground-glass” hepatocytes. S Afr Med J. 1975;49:1401–3. [PubMed] [Google Scholar]
- 26.Gerber MA, Hadziyannis S, Vissoulis C, Schaffner F, Paronetto F, Popper H. Electron microscopy and immunoelectronmicroscopy of cytoplasmic hepatitis B antigen in hepatocytes. Am J Pathol. 1974;75:489–502. [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada G, Sakamoto Y, Mizuno M, Nishihara T, Kobayashi T, Takahashi T, et al. Electron and immunoelectron microscopic study of Dane particle formation in chronic hepatitis B virus infection. Gastroenterology. 1982;83:348–56. [PubMed] [Google Scholar]
- 28.Wang J, Yu Y, Li G, Shen C, Meng Z, Zheng J, et al. Relationship between serum HBV-RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. J Hepatol. 2017. Sep 21. 10.1016/j.jhep.2017.08.021. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data set used in this research paper is available upon request to richard.sterling@vcuhealth.org (coauthor of this study).