Abstract
INTRODUCTION:
Variants in the tau gene (MAPT) region are associated with breast cancer in women and Alzheimer disease (AD) among persons lacking APOE ε4 (ε4-).
METHODS:
To identify novel genes associated with tau-related pathology, we conducted two genome-wide association studies (GWAS) for AD, one among 10,340 ε4- women in the Alzheimer’s Disease Genetics Consortium (ADGC) and another in 31 members (22 women) of a consanguineous Hutterite kindred.
RESULTS:
We identified novel associations of AD with MGMT variants in ADGC (rs12775171, odds ratio [OR]=1.4, P=4.9×10−8) and Hutterites (rs12256016 and rs2803456, OR=2.0, P=1.9×10−14). Multi-omics analyses showed that the most significant and largest number of associations among the SNPs, DNA-methylated CpGs, MGMT expression, and AD-related neuropathological traits were observed among women. Futhermore, pcHi-C analyses revealed long-range interactions of the MGMT promoter with MGMT SNPs and CpG sites.
DISCUSSION:
These findings suggest that epigenetically-regulated MGMT expression is involved in AD pathogenesis, especially in women.
Keywords: genome-wide association study, MGMT, tau, gene expression, methylation
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder and the most common cause of dementia, affecting over 5.8 million individuals in the U.S [1]. Although the APOE ε4 allele is the most established and strongest genetic risk factor for AD occurring after age 65, only ∼40% of AD cases carry this allele compared to ∼13.7% of the population, implying that other genes contribute to the genetic architecture of this disease [2]. A recent meta-analysis of genome-wide association studies (GWAS) of AD identified genome-wide significant associations with 42 novel and 33 previously identified loci [3], but a large proportion of genetic risk remains unexplained. This may be accounted for by the contribution of rare variants (which are difficult to identify from GWAS even in large samples), epigenetic mechanisms and/or interactions between genes and other factors (e.g., sex) [4].
To identify additional novel AD loci, we performed in parallel two complementary GWAS. One approach focused on dementia in the Hutterites, a population isolate of central European ancestry. The second approach was predicated on epidemiological and pathological evidence suggesting a link between AD and breast cancer (BC). The locus including MAPT which encodes the tau protein has been associated with AD risk among persons lacking the APOE ε4 allele [5], as well as with the BC risk in women [6] and the age at onset of breast/ovarian cancer among women carrying BRCA1/2 mutations [7]. Another study found that persons with mild cognitive impairment (MCI) or AD had low expression of BRCA1 in the brain, and that the abundance of Aβ42 oligomers was negatively associated with low Brca1 expression in neuronal cells in hAPP mice [8]. Low Brca1 expression in these mice was also associated with increased DNA damage, neuronal shrinkage, and synaptic plasticity impairments, which caused learning and memory deficits [8]. To test the hypothesis that other genes involved in tau-related pathology may be associated with AD risk among women, and especially those who lack the APOE ε4 allele, we also performed a GWAS for AD in ε4 non-carrier women included in datasets from the Alzheimer Disease Genetics Consortium (ADGC). Top results were further evaluated by analysis of genetic, transcriptome, and methylome data, as well as information obtained from promoter capture (pc) Hi-C in induced pluripotent stem cell (iPSC)-derived neurons and measures of AD-related proteins obtained from autopsied brain tissue.
2. Methods
2.1. Alzheimer’s Disease Genetics Consortium Cohorts
2.1.1. Subjects and Data.
This study utilized information for 10,304 women lacking the APOE ε4 allele (ε4-), which included 3,399 AD cases and 6,905 control subjects from 30 GWAS datasets assembled by ADGC (Table S1). These subjects were selected from a larger ADGC sample including women having the ε4 allele (ε4+, 5,592 AD cases, 2,537 controls), ε4+ men (4,050 AD cases, 1,670 controls) and ε4- men (2,697 AD cases, 4,851 controls). Details of the datasets, phenotyping, quality control (QC) procedures applied to the genotype data, imputation and analysis of population substructure are provided in the Supplementary Information and have also been previously described [9]. Studies of the individual cohorts were approved by the appropriate Institutional Review Boards, and written informed consent for all subjects was provided on behalf of themselves or for substantially cognitively impaired subjects, by a caregiver, legal guardian or other proxy.
2.1.2. Genome-wide association and pathway analyses.
Association of AD with the imputed dosage for each autosomal SNP was tested in each dataset using logistic regression models that included age and the first four principle components of ancestry (PCs) as covariates. Models for the analysis of the ADGC8 dataset, which includes eight aggregated datasets each containing < 50 subjects (see Supplementary Information), also included dummy variables to account for the multiple genotyping arrays across the constituent datasets. Results from the individual datasets were combined by meta-analysis using inverse variance weighting as implemented in the METAL software [10]. Heterogeneity of pooled estimates was quantified by computing Cochrane’s Q-test [11], which is implemented in METAL. Genes located near top-ranked SNPs (P<1.0×10−4) were included in analyses described in the Supplementary Information to discern potential AD-related biological pathways.
2.2. Hutterite Cohort
2.2.1. Subject Ascertainment.
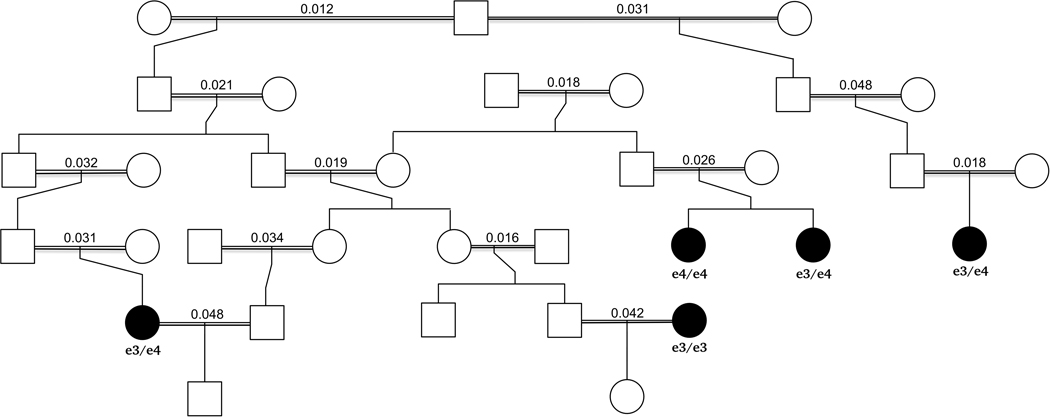
DNA and genotypes were available for five (all women) of 29 AD cases identified in a large Hutterite kindred. We then selected controls from among the 41 Hutterites who were over age 85 years at the time of our study with no reported dementia and with genotype data available. This yielded 26 controls (9 males, 17 females). There was one sibling pair among the five cases (Figure 1), four sibling pairs among the 26 controls, and no sibling pairs between the case and control groups. Additional details of the entire kindred, as well as diagnostic procedures and familial relationships among the subjects included in this study, are presented in the Supplementary Information. These studies were approved by the University of Chicago IRB.
Figure 1.
Minimum pedigree linking the five cases (shown as filled black symbols) in the Hutterites. APOE genotypes are shown below each case and the kinship coefficient of the parents are shown over the double horizontal bars connecting the parents of each case (equivalent to the inbreeding coefficient of each case). The double horizontal bars indicate that the marriage was consanguineous. The minimum pedigree includes only those relatives needed to connect the five cases in the minimum number of steps.
2.2.2. Genome-wide association study.
Details about genotyping and imputation are included in the Supplementary Information. Based on the fact that there are few (if any) examples of autosomal recessive causes of late onset AD and because, other than one pair of sisters, all other AD cases were relatively unrelated, we hypothesized that the genetic risk for late onset AD in the Hutterites was autosomal dominant and carried on a haplotype that was identical by descent in the affected individuals. A GWAS for AD was performed with 4,807,330 variants that had no missing genotypes in the five AD cases using a mixed effects logistic model as implemented in GEMMA [12] following procedures outlined in the Supplementary Information.
2.3. Multi-omics Data Analyses
Methylation array and post QC normalized gene expression data derived from the dorsolateral prefrontal cortex (DLPFC) tissue obtained from neuropathologically examined brains (399 AD cases and 376 controls) donated by Religious Orders Study and the Rush Memory and Aging Project (ROSMAP) participants were obtained from the CommonMind Consortium Portal ((https://www.synapse.org/#!Synapse:syn3388564) to further examine associations with the top-ranked GWAS findings. Data for AD-related neuropathologic traits were obtained from the same source and included amyloid-beta (Abeta) and phospho-tau (ptau) measured by immunohistochemistry and NFT measured by microscopic examination as described elsewhere (www.radc.rush.edu) [13]. We also evaluated gene expression data and semi-quantitative measures of tau pathology (Braak stage) and neuritic amyloid plaques derived from from dorsolateral prefrontal cortex tissue obtained from 177 participants (58 autopsy-confirmed AD cases and 119 controls) of the Framingham Heart Study (FHS) [14, 15]. Sample sizes for analyses of each type of omics data are presented in Tables S2 and S3 for the ROSMAP and FHS datasets, respectively. Statistical methods used to analyze these data are presented in the Supplementary Information.
2.4. Cell type-specific expression analysis in human brain
We investigated cell type-specific expression of the top-ranked candidate AD gene identified in the GWAS using a single cell RNA-sequencing (scRNA-Seq) dataset derived from the temporal lobe of 8 healthy adults [16] and in two single nuclei RNA-Seq (snRNA-Seq) datasets derived from the dorsolateral prefrontal cortex (DLPFC) region. One of the PFC region datasets included 48 individuals with varying degrees of AD pathology [17] and the other included 12 AD cases and 9 controls [18]. Additional details about the sc/snRNA-Seq datasets and analysis are described in the Supplementary Information.
2.5. Promoter-capture Hi-C
Because regulatory elements such as enhancers can be located at large genomic distances from their target genes, we performed pcHi-C using data obtained from hypothalamic neurons differentiated from iPSCs to link the promoters of genes at the top-ranked AD locus identified by GWAS to potential enhancers. Details of our protocols the tissue culture of iPSCs, differentiation of human hypothalamic neurons, and in situ pcHi-C library preparation and and data processing are presented in the Supplementary Information.
3. Results
3.1. Genome-wide analyses in two cohorts identify the same novel AD locus
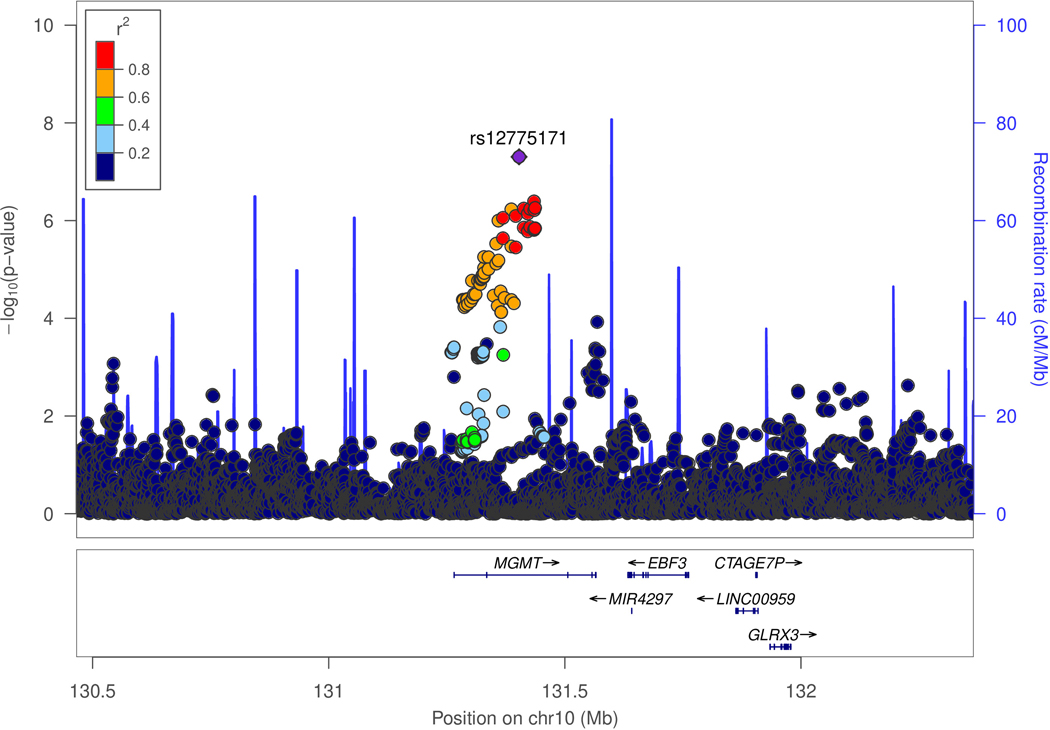
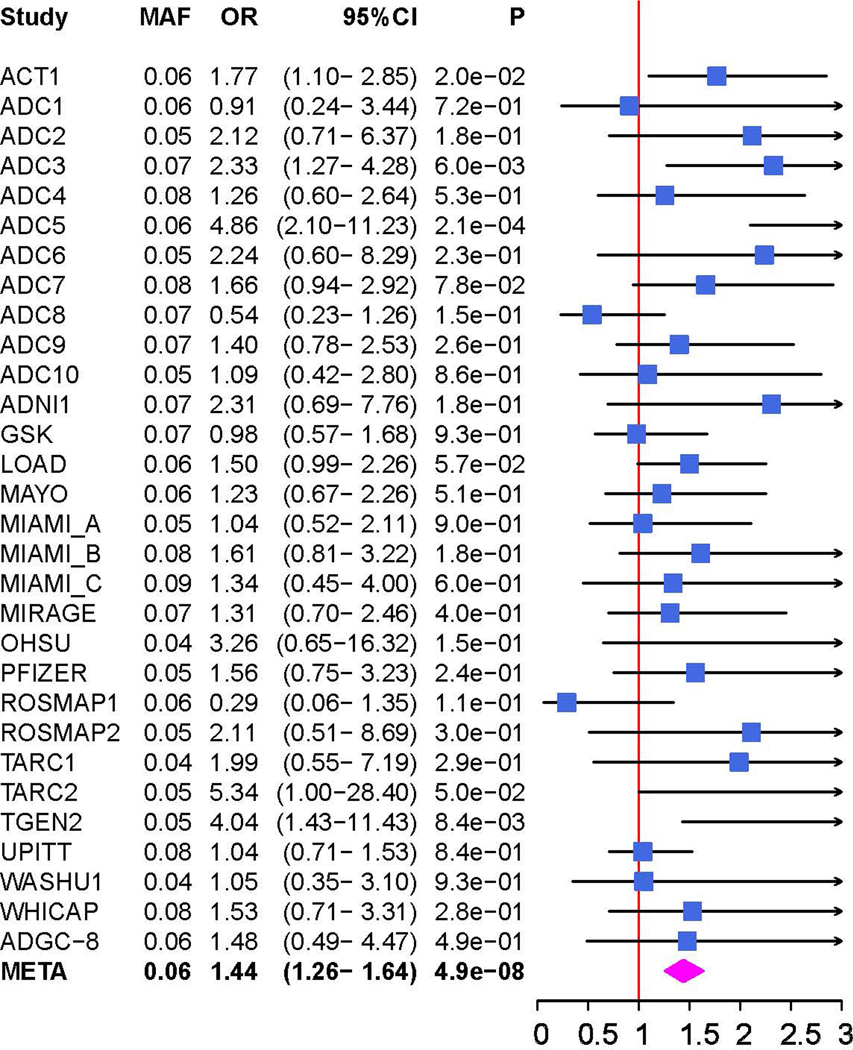
There was little genomic inflation among ε4- women in the ADGC dataset (λ=1.082, Figure S1). Genome-wide significant (GWS; P<5.0×10−8) associations were identified for two previously established AD loci (BIN1 and APOE) and one novel locus, MGMT (rs12775171, MAF=6%, odds ratio [OR]=1.4, P=4.9×10−8; Table 1 and Figure 2A) and the previously known AD loci BIN1 (best SNP: rs11680911, OR=1.2, P=2.2×10−8) and APOE (best SNP: rs390082, OR=0.6, P=7.0×10−15). Because rs390082 is in linkage disequilibrium (LD) with rs7412 (r2=0.68) that determines the APOE ε2 allele but not with rs429358 (r2<0.01) that determines APOE ε4, the direction and magnitude of effect for rs7412 is comparable to that for ε2, and subjects in this subgroup do not have ε4, our observed association in the APOE locus likely reflects the protective influence of ε2 [19]. The effect direction of rs12775171 was consistent in 26 of 30 ADGC datasets (Figure 2B) and there was not significant heterogeneity in the magnitude of effect on AD risk (Cochran Q-Test χ2=37.31, P=0.13). The association with MGMT was not observed in ε4+ women or men with or without the ε4 allele (Figure S2).
Table 1.
Genome-wide significant associations with AD risk in the ADGC datasets among women lacking the APOE ε4 allele
| SNP | CHR | Gene | EA | EAF | OR | (95% CI) | P-value | HetChiSq | HetPVal |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| rs11680911 | 2 | BIN1 | C | 0.35 | 1.21 | (1.13–1.29) | 2.22×10−8 | 32.11 | 0.31 |
| rs12775171 | 10 | MGMT | G | 0.06 | 1.44 | (1.26–1.64) | 4.95×10−8 | 37.31 | 0.14 |
| rs111278892 | 19 | ABCA7 | G | 0.15 | 1.30 | (1.18–1.44) | 7.24×10−8 | 21.00 | 0.64 |
| rs390082 | 19 | APOE | G | 0.08 | 0.58 | (0.51–0.67) | 6.99×10−15 | 25.25 | 0.56 |
EA = effect allele; NEA = non-effect allele; EAF = effect allele frequency; OR = odds ratio; CI = confidence interval; HetChiSq = chi-square test statistic for heterogeneity; HetPVal = heterogeneity test P-value
Figure 2.
Genetic association of Alzheimer disease (AD) with MGMT in the ADGC datasets (A and B) and in the Hutterites (C). A. Locus-zoom plot showing the association of AD with SNPs in the MGMT region in the ADGC datasets. The SNP with the lowest P-value (rs12775171) is indicated with a purple diamond. Computed estimates of linkage disequilibrium (r2) of SNPs in this region with rs1277517 are color-coded according to the key. B. Forest plot of the association of AD with rs12775171 among the APOE ε4- women within each ADGC dataset and in the toal sample (META). The ADGC-8 dataset includes eight individual GWAS datasets (ACT2, BIOCARD, CHAP2, EAS, NBB, RMAYO, UKS, and WASHU2) each containing < 50 subjects. The odds ratio (shaded symbol) and 95% confidence interval (represented by horizontal line) are plotted on the x-axis. C. Locus-zoom plot showing the association of AD with SNPs in the MGMT region in the Hutterites. Significance level is shown as the -log10(p-value). Computed estimates of linkage disequilibrium (r2) of SNPs in this region with the two most significant SNPs, rs12256016 (left panel) and rs2803456 (right panel), are color-coded according to the key. Recombination rates among adjacent SNPs are indicated with vertical blue lines.
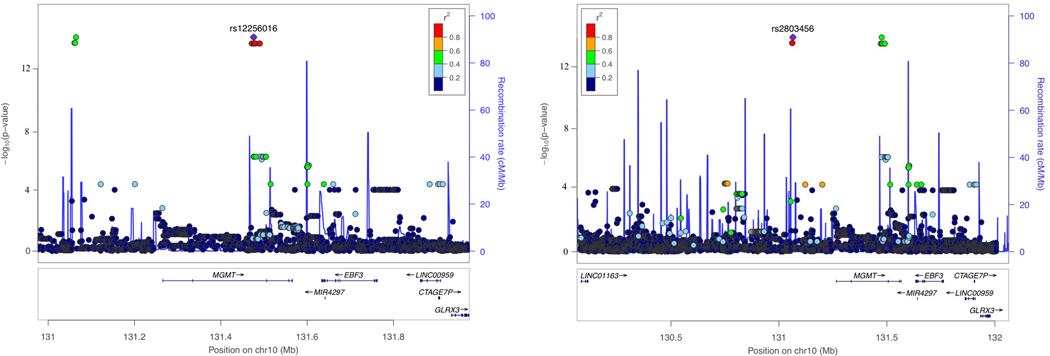
The Hutterite GWAS revealed 418 variants at 22 loci reaching GWS, after adjusting for genomic inflation (λ=1.16) (Figure S3A-B; Table S4), but the P-value at only one locus (MGMT) as small or smaller than the 200 permuted GWASs (lead SNPs = rs2803456 and rs12256016, OR=2.02, P=1.89×10−14) (Figure S3C; Table S4). This indicates that the likelihood of observing this significant an association by chance alone is <0.005. There were 14 GWS SNPs at this locus, including three variants approximately 159kb upstream of the MGMT transcription start site and 11 variants in an intron of MGMT (Figure 2C; Table 2). The minor alleles for all 14 variants were present in all AD cases and absent in all 26 controls. One case was homozygous for the risk alleles and the other four were heterozygotes. In a larger sample of Hutterites (n=1,653) who were genotyped, there was strong LD among the 11 intronic and among the three upstream variants (average pairwise r2=0.97 and 0.96, respectively), but only modest LD between the intronic and upstream variants (average pairwise r2=0.54), similar to LD patterns observed in HapMap samples of European ancestry. The 14 risk alleles occurred on a single haplotype that was present at a frequency of 0.06 in the larger Hutterite population. These combined observations suggest the possibility that these variants have independent effects on AD risk. Minor allele frequencies of these 14 SNPs, except for rs12256016, in the larger Hutterite sample were not noticeably different from those observed in other populations of European ancestry (Table 2).
Table 2.
Genome-wide significant associations with AD risk in the MGMT region among the Hutterites
| SNP | CHR | EA | EAF in Hapmap | EAF in Hutterites | OR (95% CI) | P-value |
|---|---|---|---|---|---|---|
|
| ||||||
| rs2803458 | 10 | T | 0.11 | 0.1 | 2.00 (1.78, 2.25) | 4.76×10−14 |
| rs11596752 | 10 | G | 0.11 | 0.1 | 2.00 (1.78, 2.25) | 4.76×10−14 |
| rs2803456 | 10 | G | 0.11 | 0.097 | 2.02 (1.80, 2.26) | 1.89×10−14 |
| rs11016864 | 10 | G | 0.15 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
| rs12256016 | 10 | G | 0.29 | 0.097 | 2.02 (1.80, 2.26) | 1.89×10−14 |
| rs56880178 | 10 | G | 0.15 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
| rs56800605 | 10 | C | 0.15 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
| rs12255679 | 10 | G | 0.15 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
| rs60541995 | 10 | G | 0.18 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
| rs68109323 | 10 | A | 0.15 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
| rs67372222 | 10 | A | 0.15 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
| rs12245575 | 10 | C | 0.15 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
| rs11016878 | 10 | G | 0.15 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
| rs35617552 | 10 | C | 0.16 | 0.1 | 2.01 (1.79, 2.26) | 3.83×10−14 |
EA = effect allele; EAF = effect allele frequency; OR = odds ratio; CI = confidence interval
Next, we compared the top-ranked MGMT SNPs identified in the ADGC (rs12775171) and the Hutterite (rs12256016 and rs2803456) cohorts. The three SNPs are not in LD with each other (r2<0.1 for all pairwise comparisons) in the ADGC samples as well as in the European ancestry subset of the 1000 Genomes reference. The top-ranked SNPs in the Hutterites were not associated with AD risk in the ADGC datasets (P>0.26; Table S5). Although rs12775171 is common (MAF=0.06) in European ancestry populations, it was not imputed in the Hutterite sample because it had an MAF of 0.0085 which is below the SNP filter threshold of 0.05. The AD risk-associated rs12775171 allele in the ADGC dataset was not present in any of the five cases or 26 controls.
3.2. eQTL and Pathway Analyses
Analysis of HaploReg eQTL data [20] for MGMT and its adjacent genes, EBF3 and GLRX3, revealed that proxy SNPs in high LD (r2>0.8) with AD risk alleles of rs12775171, rs11016864, and rs2803456 were significantly associated with expression of MGMT only in whole blood (best eQTL: rs12248703 proxy for rs12256016; P=3.56×10−7) (Table S6). The top-ranked SNPs were nominally associated with expression levels of MGMT and EBF3 in multiple tissues including several brain regions (best eQTL: rs2803456 for MGMT, P=0.01 in the caudate) in the GTEx database [21]. Pathway analysis that was seeded with genes containing the top-ranked SNPs from the ADGC GWAS among ε4- women found that the most significant pathway was lipopolysaccharide (LPS)/IL-1 mediated inhibition of retinoid-X-receptors (RXR) function (P=7.7×10−4), which includes MGMT, as well as APOE and APOC1 (Table S7).
3.3. Multi-Omics Analysis Links MGMT Expression and Methylation to AD Risk and AD-related Neuropathology
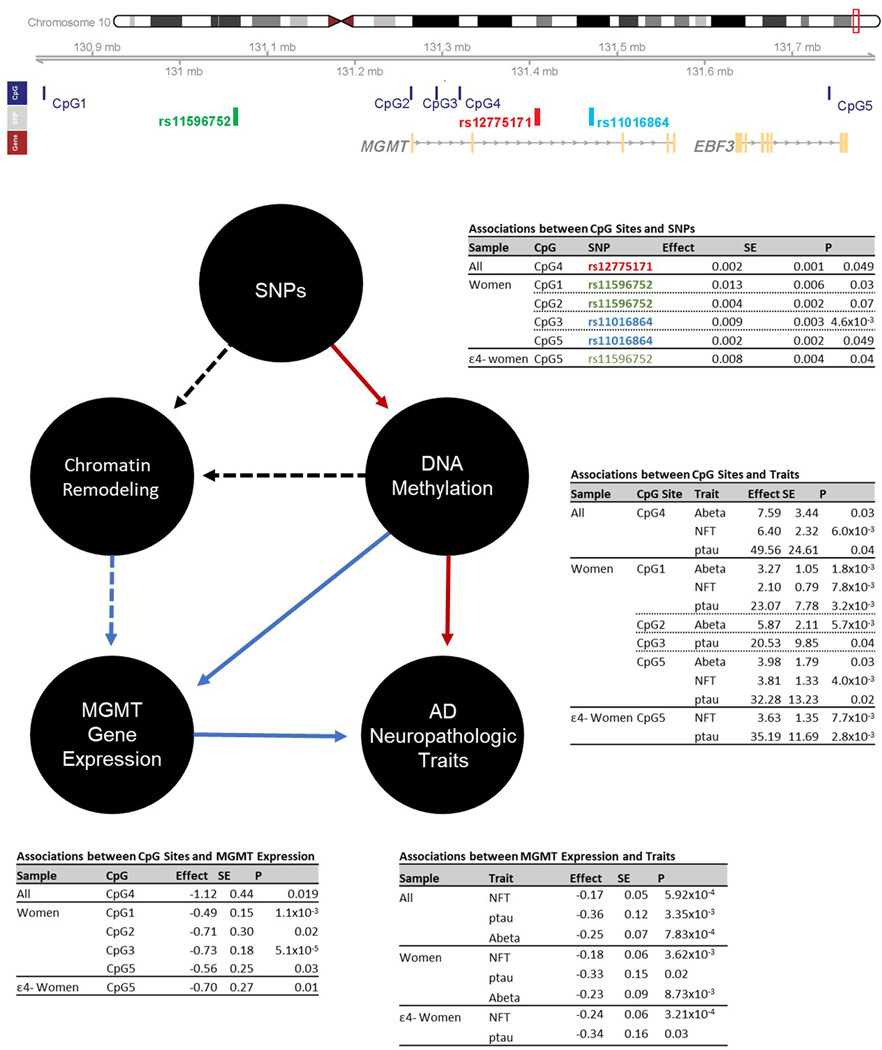
Because the strongest eQTL associations for the top-ranked SNPs were observed with MGMT, we hypothesized that MGMT is the gene at this locus functionally related to AD risk. A previous epigenetic study of glioblastoma reported that DNA hypermethylation in the MGMT promoter region leads to lower expression and subsequently defective function of MGMT [22], suggesting that this epigenetic mechanism may explain at least in part the observed association of MGMT SNPs with AD risk. To test this hypothesis, we evaluated associations between AD-associated MGMT SNPs (rs12775171 and rs11596752 and rs11016864 as proxies for rs2803456 and rs12256016) and methylation levels of CpG sites at the MGMT locus together with MGMT gene expression and measures of Abeta, ptau, and NFT in 399 AD and 376 control autopsied brains from ROSMAP.
The AD risk associated alleles for the three MGMT SNPs were associated with hypermethylation of 105 CpGs in the MGMT region in at least one APOE ε4 status and gender subgroup (Table S8), but methylation levels of only five of these CpGs – cg07646467 (CpG1), cg09450835 (CpG2), cg02634492 (CpG3), cg05596517 (CpG4), and cg01419164 (CpG5) – were both negatively associated with MGMT expression (Table S9) and positively associated with the neuropathological traits (Table S10) in the entire sample, among women, or specifically among ε4- women (Figure 3, Tables S11-S13. We therefore focused on these five CpG sites.
Figure 3.
Association summary statistics among Omics data including MGMT SNPs (or their proxies) significantly associated with AD risk in the ADGC (colored in red) and Hutterite (colored in green or blue) datasets, methylation of adjacent CpG sites, MGMT expression, and AD-related neuropathological traits in the total ROSMAP sample, as well as in subgroups stratified sex and APOE ε4 carrier status (ε4+ or ε4-). Only significant results are shown. Rs11596752 and rs11016864 are proxy SNPs for rs2803456 and rs12256016 (LD r2 > 0.8). CpG IDs are serially numbered 1–5 based on their base pair positions: cg07646467 (CpG1), cg09450835 (CpG2), cg02634492 (CpG3), cg05596517 (CpG4), and cg01419164 (CpG5). Red and blue arrows indicate positive and negative effect directions, respectively. Dashed black arrows signify inferred relationships based on the pcHi-C results assuming DNA containing AD-associated alleles render the chromatin to be less accessible leading to reduced MGMT expression.
The largest number and most significant associations of CpG methylation and MGMT SNPs were observed among women, but these findings were specific to particular CpG-SNP combinations among ε4+ or ε4- women (Figure 3, Table S11). Although MGMT expression level was associated with DNAm levels at all five CpG sites, the most significant associations of DNAm at a particular CpG site with a MGMT SNP and expression was observed at CpG3 among women (rs11016864: P=4.1×10−4, expression: P=5.1×10−5), with most of the evidence derived from the ε4- women (rs11016864: P=1.3×10−4, expression: P=8.9×10−5, Tables S11-S12). Several MGMT CpG sites were also significantly associated with severity of AD-related neuropathology. In particular, in the total sample, CpG1 was significantly associated with both Abeta (P=4.35×10−5) and ptau (P=3.83×10−4), with comparable effect sizes in women and men (Figure 3, Table S13). Other CpG sites were associated with Abeta in the total sample (CpG5, P=0.0048) or in women (CpG2, P=0.0057), with NFT in the total sample (CpG4, P=0.0060) and among ε4- women (CpG5, P=0.0076), and with ptau in ε4- women (CpG5, P=0.0028), although none of these results surpassed the multiple test correction threshold of P=0.0021 (0.05 / (3 SNPs * 2 independent AD-related neuropathological traits * 4 independent subsets defined by sex and APOE ε4 status)).
MGMT expression was inversely correlated with Abeta (P=7.83×10−4) and with NFT in the total sample (P=5.92×10−4) and among ε4- women in particular (P=3.21×10−4) (Figure 3 and Table 3). However, we did not observe significant association of MGMT expression with clinically-defined AD status in any group (Table 3). The findings with tau pathology were confirmed in several strata of the FHS sample (total [P=0.047], women [P=0.008], ε4- women [P=0.003]), but not in other APOE/sex FHS subgroups (P≥0.43) (Table S14).
Table 3.
Association of MGMT gene expression with clinically-defined AD and AD-related neuropathologic traits in ROSMAP.
| Subjects | Phenotype | N | Beta | SE | P-value |
|---|---|---|---|---|---|
|
| |||||
| All | AD status | 261 | −0.08 | 0.07 | 0.24 |
| NFT | 601 | −0.17 | 0.05 | 0.00059 | |
| ptau | 595 | −0.36 | 0.12 | 0.0034 | |
| Abeta | 595 | −0.25 | 0.07 | 0.00078 | |
|
| |||||
| Women | AD status | 179 | −0.12 | 0.09 | 0.15 |
| NFT | 385 | −0.18 | 0.06 | 0.0036 | |
| ptau | 381 | −0.33 | 0.15 | 0.02 | |
| Abeta | 381 | −0.23 | 0.09 | 0.0087 | |
|
| |||||
| ε4- Women | AD status | 131 | −0.14 | 0.09 | 0.15 |
| NFT | 289 | −0.24 | 0.06 | 0.00032 | |
| ptau | 288 | −0.34 | 0.16 | 0.03 | |
| Abeta | 288 | −0.20 | 0.10 | 0.05 | |
|
| |||||
| ε4+ Women | AD status | 48 | 0.01 | 0.18 | 0.94 |
| NFT | 96 | −0.01 | 0.15 | 0.93 | |
| ptau | 93 | −0.29 | 0.32 | 0.36 | |
| Abeta | 93 | −0.30 | 0.15 | 0.04 | |
|
| |||||
| Men | AD status | 82 | 0.01 | 0.13 | 0.95 |
| NFT | 216 | −0.15 | 0.08 | 0.07 | |
| ptau | 214 | −0.38 | 0.22 | 0.09 | |
| Abeta | 214 | −0.31 | 0.13 | 0.02 | |
|
| |||||
| ε4- Men | AD status | 59 | −0.04 | 0.15 | 0.77 |
| NFT | 159 | −0.14 | 0.07 | 0.06 | |
| ptau | 158 | −0.47 | 0.23 | 0.04 | |
| Abeta | 158 | −0.41 | 0.16 | 0.01 | |
|
| |||||
| ε4+ Men | AD status | 23 | −0.08 | 0.21 | 0.69 |
| NFT | 57 | −0.13 | 0.21 | 0.55 | |
| ptau | 56 | 0.01 | 0.52 | 0.98 | |
| Abeta | 56 | 0.11 | 0.20 | 0.60 | |
Effect = effect estimate; SE = standard error; significant results (Padjusted < 0.0042) are in bold.
3.4. Cell-type Expression of MGMT in brain
Analysis of data from the temporal lobe of 8 healthy brains revealed that MGMT was expressed primarily in neurons, astrocytes, and endothelial cells (Figure S4A). However, we found in two independent snRNA-seq datasets derived from DLPFC from AD cases and controls that MGMT expression was greater in microglia and endothelial cells than other cell types (Figure S4B and S4C). Notably, expression in endothelial cells was higher in controls than AD cases in both datasets, whereas the pattern of association of expression in microglia differed between the datasets.
3.5. Interaction of the MGMT Promoter Region with AD-associated SNPs and Methylation Sites
PcHi-C contact maps from development-specific stages revealed a complex regulatory landscape that included long-range interactions between the promoter of MGMT and SNPs in strong LD (r2>0.8) with the lead SNPs in both the ADGC and Hutterite GWAS, as well as with CpG1, CpG4, and CpG5 (Figure 4A). All of these interactions were dependent on developmental context and restricted to specific temporal windows. In hypothalamic precursor cells, the MGMT promoter interacted with the regions containing SNPs in high LD with the top-ranked MGMT SNPs, as well as with CpG1 and CpG5. In early development cells, the MGMT promoter interacted with the Hutterite intronic SNPs and another ADGC SNP, and in late development cells, the MGMT promoter interacted with yet another ADGC SNP and CpG4. In addition, the EBF3 promoter specifically interacted with SNPs near the leading SNPs in the ADGC GWAS in precursor cells and with CpG1 in early and late development cells. Both MGMT and EBF3 expression was detected in all development stages, with MGMT most highly expressed in precursor cells and EBF3 most highly expressed in early development cells (Table S15). Overall, the pcHi-C contact map suggests physical interactions across this approximately 1 Mb region that bring the MGMT and EBF3 genes into close proximity, which may enable genetic variation (SNPs) and epigenetic variation (CpGs) to have development stage-specific effects on the regulation of these genes.
Figure 4.
Regulatory landscape of MGMT locus. A. Chromatin looping interactions emanating from the MGMT and EBF3 promoters in human iPSCs-derived neurons at three stages of differentiation. PCHi-C interactions are displayed as gray arcs. Interactions between MGMT and EBF3 promoters with SNPs are represented by arcs highlighted in blue (Hutterites) and orange (ADGC). Green arcs highlight interactions between MGMT and EBF3 promoters with CpGs. The yellow and green strip highlight the interaction with SNPs and CpGs, respectively. B. Magnified view of the MGMT locus with chromatin state annotations from the Roadmap Epigenomics Project (REP). Colored bars indicate chromatin state annotations from tissues profiled by the REP, including neuronal cells: neuronal progenitors (epigenome identifiers E007), brain anterior caudate (E068), and brain germinal matrix (E070); and non-neuronal cells: IMR90 fetal lung (E017), small intestine (E109), primary T-cells (E043 and E045), fetal heart (E083), and pancreatic islets (E087).
To extend the pcHi-C findings derived from hypothalamic neurons to brain regions more impacted by AD, we investigated interactions between the MGMT promoter region and AD-associated MGMT SNPs using publicly available Hi-C data serived from the hippocampal and DLPFC regions (http://3dgenome.fsm.northwestern.edu) [23]. These analyses showed that the MGMT promoter interacts with a region that includes one of the AD-associated SNPs in the Hutterites, rs12256016, in both brain regions, but not with the other SNPs (Figure S5).
To further predict the cell types in which the MGMT variants may act as regulatory elements, we examined chromatin state maps [24] of the MGMT locus in different cell types (brain, heart, intestine, pancreas, lung, and blood cells). Those data revealed that the six of the nine variants that physically interacted with MGMT or EBF3 promoters map within accessible chromatin regions in human neuronal tissues, as profiled by the Roadmap Epigenomics Consortium. Interestingly, five of these variants (rs11596752, rs35890176, rs61859925, rs56800605, and rs12255679) seem to modulate enhancer activity exclusively in brain (Figure 4B).
4. Discussion
4.1. A new locus for AD implicated by two independent uniquely designed GWAS
Motivated by previously reported AD genetic associations with SNPs in the MAPT region among persons lacking the APOE ε4 allele [5], as well as by epidemiological and pathological links between AD and BC [6–8], we conducted a GWAS among ε4- women in the ADGC and identified one novel GWS association at MGMT. This novel locus finding was bolstered by a parallel GWAS in five AD cases (who happened to be all women) and 26 controls from a consanguineous Hutterite kindred that also found GWS associations with SNPs on an extended haplotype that includes MGMT. The ability to detect a genome-wide significant association in a small dataset reflects the increased power afforded by founder populations with reduced genetic variation and reduced environmental heterogeneity, as evidenced by our prior studies in a consanguineous group of Israeli-Arabs leading to the first robust associations of AD with ACE and SORL1 [25, 26]. Because rs12775171, the most significantly associated SNP in the ADGC, is not in LD with either Hutterite SNP (r2≤0.001) in the ADGC dataset, it is plausible that multiple MGMT variants are functionally related to AD. The AD/MGMT association finding was supported by results from comparisons of MGMT SNPs, MGMT gene expression and methylation data obtained from brain, and quantified brain measures of AD-related proteins. In support of our hypothesis that other genes involved in tau-related pathology may be associated with AD risk among women, and especially those who lack APOE ε4, we found that MGMT expression is associated with neurofibrillary tangles specifically among ε4(−) women (Table 3). Moreover, the pcHi-C data indicate that all three associated variants themselves or variants in high LD reside within chromatin accessible regions that physically interact with the promoter of MGMT in neuronal tissues. Taken together, our results indicate AD risk in women can be attributed in part to genetic variants that regulate the expression of MGMT in developing neurons.
4.2. MGMT is functionally linked to AD, especially among women
MGMT encodes O6-methylguanine-DNA methyltransferase, a DNA repair protein that protects cells from apoptotic effects of alkylating agents by removing alkyl groups from the 6-O-methylguanine [27]. MGMT is epigenetically inactivated via hypermethylation of CpG islands that span roughly 1,000 bps around the MGMT transcription start site [28–30] in glioblastomas [31], as well as in breast cancer tissue [32, 33]. A link between MGMT and AD was suggested by a previous study that reported decreased MGMT protein levels in CSF of AD patients compared to matched healthy controls [34]. Coppede et al [35] compared methylation of CpG sites in the promoter region of eight DNA repair genes including MGMT in a sample of 56 AD cases and 55 matched controls, but did not observe any significant differences in the MGMT promoter region, likely due to the small sample size and focus on blood rather than disease-relevant regions in brain.
The female-specific association of AD with MGMT identified in both the ADGC and Hutterites is consistent with the most significant associations with omics data and AD-related pathology, which were also observed predominantly in women. Intriguingly, sexual dimorphic genetic and epigenetic associations with MGMT have been previously reported. Methylation levels in the MGMT promoter region are different between men and women in lung tumor cells [36], a pattern that manifests clinically: glioblastoma survival rates vary by sex and degree of methylation in the MGMT promoter region [37, 38]. In this study, we found that the methylation levels at the MGMT locus were significantly associated with the AD-associated MGMT SNPs and MGMT expression, as well as with AD-related neuropathologic traits. In particular, the most significant associations of methylation levels with both MGMT expression and AD-associated SNPs involved CpG3, which is located near the MGMT transcription start site. Interestingly, hypermethylation of CpG2 and CpG3 have been linked to occurrence and progression of glioblastoma [28–30]. These findings suggest that inactivation of MGMT via epigenetic regulation in a sex-specific manner may promote AD pathogenetic mechanisms and underlie the associations discovered in our GWAS. This idea is supported by results from pcHi-C in iPSC-derived neurons showing long-range interactions between the MGMT promoter and SNPs in strong LD with the top-ranked SNPs in both datasets as well as with CpG sites near MGMT. Moreover, chromatin state maps from the Roadmap Epigenomics Consortium indicate that the SNPs in the MGMT locus may modulate regulatory activity specifically in neuronal cell types and tissues. Together, our omics findings suggest that these SNPs may affect the function of distal enhancers thereby influencing MGMT expression via chromatin loops [39]. Our results suggest that EBF3 is not the causal gene at this locus (see Supplementary Information).
4.3. Links Between APOE and MGMT
Insight into the role of APOE in the AD/MGMT association may be gleaned from a recent glioblastoma study showing that increased methylation level at the MGMT promoter, which is a well-known predictor for immunotheraphy response and survival rate in glioblastoma patients [40], is significantly associated with increased APOE expression, which has been observed in AD brains [41] . Furthermore, our pathway analysis suggests that MGMT and APOE are involved in LPS/IL-1 mediated inhibition of RXR function. RXR signaling, which is associated with synaptic plasticity in the hippocampal region and remyelination in the central nervous system, has been considered as a therapeutic target for neurodegenerative diseases including AD and Parkinson disease [42–44].
4.4. Study Limitations
Our study has several limitations. First, despite consistent results regarding the association of MGMT with AD risk in two very distinct cohorts, these findings should be replicated in other GWAS datasets especially because the Hutterite sample is small and the variants associated with AD in each dataset differ. Arguably, the association patterns between the Hutterites who are descended from a central European population should be similar to those observed in the ADGC dataset comprised of persons of European ancestry. However, the association of AD with distinct SNPs across the datasets may be explained by allelic heterogeneity such that the functional variant accounting for the association signal in each dataset is unique. This explanation is consistent with the observation that the top-ranked SNP in the ADGC dataset is not in LD with the AD-associated SNPs in the Hutterites. Moreover, because of the Hutterite cultural norms, neuropsychological and imaging studies were not performed to substantiate a diagnosis of AD. As a result, we can not formally differentiate between AD and other forms of dementia in the Hutterites. However, the high degree of relatedness among the Hutterites due to a founder effect, and their cultural, geographic and environmental homogeneity likely afforded improved statistical power compared to genetic studies of large outbred populations. Second, our findings from analyses of omics data also require replication in additional larger samples, noting, however, that the association of MGMT expression with tau pathology among women, and ε4- women in particular, was observed in two independent sets of neuropathologically examined brains (ROSMAP and FHS). Third, at present, there is no apparent explanation for the specificity of the AD/MGMT association in the ADGC GWAS datasets to women lacking APOE ε4, a finding that is not supported by the association pattern in the Hutterites, in which four of the five AD cases, although all women, were ε4 carriers. However, this could be due to the very small number of AD cases in the Hutterite sample, genetic and environmental background differences between Hutterites and the ADGC cohorts, or the possibility that there is more than one mechanism mediating risk at this locus. Fourth, MGMT expression was not associated with clinically defined AD status (Table 3). However, this may be explained by the much smaller sample size of AD cases and controls (a binary outcome) compared to the sample available for analysis of quantitative measures of AD-related proteins which also included subjects with MCI. Alternatively, this may reflect the reduced accuracy of the clinical diagnosis of AD compared to measures of AD-related proteins which are strongly correlated with the neuropathological diagnosis. Such an idea is supported by a recent observation that effect sizes of APOE ε2 and ε4 alleles for AD risk are significantly higher in neuropathologically characterized cases and controls compared to clinically-defined groups [18]. Lastly, our primary pcHi-C data that support a role for MGMT expression in the etiology of AD were based on studies in iPSC-derived hypothalamic neurons, which may not be the primary neuronal cells involved in AD. However, previous studies have shown that gene expression patterns are associated with chromatin structures that are highly conserved across cell types [45–48], especially among closely related cell types [49] such as different types of neurons. The shared nature of chromatin organization identified by our pcHi-C study is corroborated by orthogonal data from the Roadmap Epigenomics Consortium showing that five of the AD-associated variants map within open chromatin accessible regions only in brain and not in other cells types or tissues (heart, intestine, pancreas, lung, and blood cells [50]. Moreover, our analysis of pcHi-C data derived from brain regions most impacted by AD (hippocampus and DFPLC) confirmed an interaction of one of the top-ranked AD-associated MGMT SNPs with the MGMT promoter region.
4.5. Conclusions
We identified a novel association of AD with genetic variants in MGMT among women lacking the APOE ε4 allele in the ADGC cohorts and with genetic variants within and upstream of MGMT in Hutterite women regardless of APOE ε4 carrier status. Multi-omics analysis suggested that epigenetically regulated expression of MGMT, which is involved in DNA damage repair function, is significantly associated with the development of the hallmark AD proteins, amyloid-β and tau, especially in women.
Supplementary Material
Acknowledgments
We thank Nicholas O’Neill for his advice regarding analysis of cell type-specific expression data and the Hutterites for their continued support of our studies. This study was supported by the National Institutes of Health grants R01 HD21244, R01 HL085197, RF1 AG057519, R01 AG069453, R01 AG048927, U19 AG068753, U01 AG062602, U01 AG058654, and R01 AG062634. The Alzheimer’s Disease Genetics Consortium supported the collection of samples used in this study through National Institute on Aging (NIA) grants U01 AG032984 and RC2 AG036528. GWAS data used in this study were prepared, archived, and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (U24 AG041689) and the National Alzheimer Coordinating Center (NACC) at the University of Washington (U01 AG016976). Phenotype data for the subjects included in the GWAS were collected with support from the following grants: NIA LOAD (Columbia University), U24 AG026395, U24 AG026390, R01AG041797; Banner Sun Health Research Institute P30 AG019610; Boston University, P30 AG013846, U01 AG10483, R01 CA129769, R01 MH080295, R01 AG017173, R01 AG025259, R01 AG048927, R01AG33193, R01 AG009029; Columbia University, P50 AG008702, R37 AG015473, R01 AG037212, R01 AG028786; Duke University, P30 AG028377, AG05128; Emory University, AG025688; Group Health Research Institute, UO1 AG006781, UO1 HG004610, UO1 HG006375, U01 HG008657; Indiana University, P30 AG10133, R01 AG009956, RC2 AG036650; Johns Hopkins University, P50 AG005146, R01 AG020688; Massachusetts General Hospital, P50 AG005134; Mayo Clinic, P50 AG016574, R01 AG032990, KL2 RR024151; Mount Sinai School of Medicine, P50 AG005138, P01 AG002219; New York University, P30 AG08051, UL1 RR029893, 5R01AG012101, 5R01AG022374, 5R01AG013616, 1RC2AG036502, 1R01AG035137; North Carolina A&T University, P20 MD000546, R01 AG28786–01A1; Northwestern University, P30 AG013854; Oregon Health & Science University, P30 AG008017, R01 AG026916; Rush University, P30 AG010161, R01 AG019085, R01 AG15819, R01 AG17917, R01 AG030146, R01 AG01101, RC2 AG036650, R01 AG22018; TGen, R01 NS059873; University of Alabama at Birmingham, P50 AG016582; University of Arizona, R01 AG031581; University of California, Davis, P30 AG010129; University of California, Irvine, P50 AG016573; University of California, Los Angeles, P50 AG016570; University of California, San Diego, P50 AG005131; University of California, San Francisco, P50 AG023501, P01 AG019724; University of Kentucky, P30 AG028383, AG05144; University of Michigan, P50 AG008671; University of Pennsylvania, P30 AG010124; University of Pittsburgh, P50 AG005133, AG030653, AG041718, AG07562, AG02365; University of Southern California, P50 AG005142; University of Texas Southwestern, P30 AG012300; University of Miami, R01 AG027944, AG010491, AG027944, AG021547, AG019757; University of Washington, P50 AG005136, R01 AG042437; University of Wisconsin, P50 AG033514; Vanderbilt University, R01 AG019085; and Washington University, P50 AG005681, P01 AG03991, P01 AG026276. ROSMAP data were generated with support from NIA grants P30‐AG10161, R01‐AG17917, R01‐AG36042, and U01‐AG46152.
Abbreviations
- Abeta
beta-amyloid
- AD
Alzheimer disease
- ADGC
Alzheimer’s Disease Genetics Consortium
- APOE
apolipoprotein E
- BC
breat cancer
- FHS
Framingham Heart Study
- GWAS
genome-wide association study
- GWS
genome-wide significant
- iPSC
induced pluripotent stem cell
- MAF
minor allele frequency
- NFT
neurofibrillary tangles
- OR
odds ratio
- pcHi-C
promoter capture Hi-C
- PMI
post mortem interval
- ptau
phospho-tau
- RIN
RNA integrity number
- ROSMAP
Religious Orders Study and Memory and Aging Project
- SNP
single nucleotide polymorphism
Footnotes
Conflicts of Interest: The authors declare they have no conflicts of interest
Data Availability
ADGC GWAS data and summarized results are available from the National Institutue on Aging Genetics of Alzheimer Disease Storage site (NIAGADS; https://www.niagads.org). Summary statistics for the Hutterites GWAS are deposited in dbGaP (accession: phs000185).
References
- [1].2021 Alzheimer’s disease facts and figures. Alzheimers Dement. 2021;17:327–406. [DOI] [PubMed] [Google Scholar]
- [2].Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bellenguez C, Küçükali F, Jansen I, Andrade V, Moreno-Grau S, Amin N, et al. New insights on the genetic etiology of Alzheimer’s and related dementia. medRxiv. 2020:2020.10.01..20200659 [Google Scholar]
- [4].Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21:108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Suberbielle E, Djukic B, Evans M, Kim DH, Taneja P, Wang X, et al. DNA repair factor BRCA1 depletion occurs in Alzheimer brains and impairs cognitive function in mice. Nat Commun. 2015;6:8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. [DOI] [PubMed] [Google Scholar]
- [12].Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious Orders Study and Rush Memory and Aging Project. J Alzheimers Dis. 2018;64:S161–S89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Panitch R, Hu J, Chung J, Zhu C, Meng G, Xia W, et al. Integrative brain transcriptome analysis links complement component 4 and HSPA2 to the APOE epsilon2 protective effect in Alzheimer disease. Mol Psychiatry. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jun GR, You Y, Zhu C, Meng G, Chung J, Panitch R, et al. Protein phosphatase 2A, complement component 4, and APOE genotype linked to Alzheimer’s disease using a systems biology approach. medRxiv. 2020:2020.11.20.20235051. [Google Scholar]
- [16].Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112:7285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lau SF, Cao H, Fu AKY, Ip NY. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2020;117:25800–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Reiman EM, Arboleda-Velasquez JF, Quiroz YT, Huentelman MJ, Beach TG, Caselli RJ, et al. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat Commun. 2020;11:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Christmann M, Kaina B. Epigenetic regulation of DNA repair genes and implications for tumor therapy. Mutat Res Rev Mutat Res. 2019;780:15–28. [DOI] [PubMed] [Google Scholar]
- [23].Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Meng Y, Baldwin CT, Bowirrat A, Waraska K, Inzelberg R, Friedland RP, et al. Association of polymorphisms in the Angiotensin-converting enzyme gene with Alzheimer disease in an Israeli Arab community. Am J Hum Genet. 2006;78:871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Warren CL, Kratochvil NCS, Hauschild KE, Foister S, Brezinski ML, Dervan PB, et al. Defining the sequence-recognition profile of DNA-binding molecules. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:867–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bienkowski M, Berghoff AS, Marosi C, Wöhrer A, Heinzl H, Hainfellner JA, et al. Clinical Neuropathology practice guide 5–2015: MGMT methylation pyrosequencing in glioblastoma: unresolved issues and open questions. Clin Neuropathol. 2015;34:250–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Everhard S, Tost J, El Abdalaoui H, Crinière E, Busato F, Marie Y, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11:348–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Malley DS, Hamoudi RA, Kocialkowski S, Pearson DM, Collins VP, Ichimura K. A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol. 2011;121:651–61. [DOI] [PubMed] [Google Scholar]
- [31].Alvarez RH, Valero V, Hortobagyi GN. Emerging targeted therapies for breast cancer. J Clin Oncol. 2010;28:3366–79. [DOI] [PubMed] [Google Scholar]
- [32].An N, Shi Y, Ye P, Pan Z, Long X. Association between MGMT promoter methylation and breast cancer: a meta-analysis. Cell Physiol Biochem. 2017;42:2430–40. [DOI] [PubMed] [Google Scholar]
- [33].Asiaf A, Ahmad ST, Malik AA, Aziz SA, Rasool Z, Masood A, et al. Protein expression and methylation of MGMT, a DNA repair gene and their correlation with clinicopathological parameters in invasive ductal carcinoma of the breast. Tumour Biol. 2015;36:6485–96. [DOI] [PubMed] [Google Scholar]
- [34].Oláh Z, Kálmán J, Tóth ME, Zvara Á, Sántha M, Ivitz E, et al. Proteomic analysis of cerebrospinal fluid in Alzheimer’s disease: wanted dead or alive. J Alzheimers Dis. 2015;44:1303–12. [DOI] [PubMed] [Google Scholar]
- [35].Coppedè F, Tannorella P, Stoccoro A, Chico L, Siciliano G, Bonuccelli U, et al. Methylation analysis of DNA repair genes in Alzheimer’s disease. Mech Ageing Dev. 2017;161:105–11. [DOI] [PubMed] [Google Scholar]
- [36].Lai J-C, Wu J-Y, Cheng Y-W, Yeh K-T, Wu T-C, Chen C-Y, et al. O6-methylguanine-DNA methyltransferase hypermethylation modulated by 17β-estradiol in lung cancer cells. Anticancer Research. 2009;29:2535–40. [PubMed] [Google Scholar]
- [37].Franceschi E, Tosoni A, Minichillo S, Depenni R, Paccapelo A, Bartolini S, et al. The prognostic roles of gender and O6-methylguanine-DNA methyltransferase methylation status in glioblastoma patients: The female power. World Neurosurg. 2018;112:e342–e7. [DOI] [PubMed] [Google Scholar]
- [38].Smits A, Lysiak M, Magnusson A, Rosell J, Söderkvist P, Malmström A. Sex disparities in MGMT promoter methylation and survival in glioblastoma: Further evidence from clinical cohorts. J Clin Med. 2021;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sobreira DR, Joslin AC, Zhang Q, Williamson I, Hansen GT, Farris KM, et al. Extensive pleiotropism and allelic heterogeneity mediate metabolic effects of IRX3 and IRX5. Science. 2021;372:1085–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang Z, Guo X, Gao L, Wang Y, Ma W, Xing B. Glioblastoma cell differentiation trajectory predicts the immunotherapy response and overall survival of patients. Aging (Albany NY). 2020;12:18297–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Linnertz C, Anderson L, Gottschalk W, Crenshaw D, Lutz MW, Allen J, et al. The cis-regulatory effect of an Alzheimer’s disease-associated poly-T locus on expression of TOMM40 and apolipoprotein E genes. Alzheimers Dement. 2014;10:541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Huang JK, Jarjour AA, Nait Oumesmar B, Kerninon C, Williams A, Krezel W, et al. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat Neurosci. 2011;14:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Skerrett R, Malm T, Landreth G. Nuclear receptors in neurodegenerative diseases. Neurobiol Dis. 2014;72 Pt A:104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nam KN, Mounier A, Fitz NF, Wolfe C, Schug J, Lefterov I, et al. RXR controlled regulatory networks identified in mouse brain counteract deleterious effects of Aβ oligomers. Scientific Reports. 2016;6:24048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. [DOI] [PubMed] [Google Scholar]
- [48].Montefiori LE, Sobreira DR, Sakabe NJ, Aneas I, Joslin AC, Hansen GT, et al. A promoter interaction map for cardiovascular disease genetics. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ADGC GWAS data and summarized results are available from the National Institutue on Aging Genetics of Alzheimer Disease Storage site (NIAGADS; https://www.niagads.org). Summary statistics for the Hutterites GWAS are deposited in dbGaP (accession: phs000185).