Abstract
The Network Makeup Artist (NORMA) is a web tool for interactive network annotation visualization and topological analysis, able to handle multiple networks and annotations simultaneously. Precalculated annotations (e.g., Gene Ontology, Pathway enrichment, community detection, or clustering results) can be uploaded and visualized in a network, either as colored pie-chart nodes or as color-filled areas in a 2D/3D Venn-diagram-like style. In the case where no annotation exists, algorithms for automated community detection are offered. Users can adjust the network views using standard layout algorithms or allow NORMA to slightly modify them for visually better group separation. Once a network view is set, users can interactively select and highlight any group of interest in order to generate publication-ready figures. Briefly, with NORMA, users can encode three types of information simultaneously. These are 1) the network, 2) the communities or annotations of interest, and 3) node categories or expression values. Finally, NORMA offers basic topological analysis and direct topological comparison across any of the selected networks. NORMA service is available at http://norma.pavlopouloslab.info, whereas the code is available at https://github.com/PavlopoulosLab/NORMA.
Keywords: Network annotation, Visualization, Topological analysis, Community detection, Functional enrichment visualization
Introduction
In the biomedical field, networks are widely used to capture the interactions and associations between bioentities (e.g., proteins, genes, small molecules, metabolites, ligands, diseases, and/or drugs) in order to unravel how complex systems operate [1]. Despite the great variety of visualization tools to efficiently represent such networks [2], [3], visualization still remains a bottleneck as views often become incomprehensive due to a large number of nodes and edges. Therefore, tools for the visualization of communities or areas of interest in a network emerge. Popular state-of-the-art tools include, Cytoscape [4], Cytoscape.js web tool [5], Gephi [6], Pajek [7], Ondex [8], Proviz [9], VisANT [10], Medusa [11], Osprey [12], Tulip [13], Arena3D [14], Arena3Dweb [15], and Graphia (Kajeka) and BioLayout Express [16]. Nevertheless, most of these tools mostly focus on interactivity, layout, and network exploration and lack efficiency in network annotation visualization.
ClueGO [17], for example, a Cytoscape plugin [18], partially solves this problem by using pie-chart nodes to visualize the non-redundant biological terms for large clusters of genes in either a functionally grouped network [19] or a KEGG/BioCarta network [20]. Moreover, Enrichment Map [21] uses similar approaches to visualize gene-set enrichment results. In addition, STRING [22] uses pie-chart nodes to overlay Gene Ontology (GO), KEGG [20], and Reactome [23] pathways on a protein–protein interaction (PPI) network [24]. Osprey mainly builds data-rich graphical representations from GO interaction data and is bound to the BioGRID repository [25]. NAViGaTOR [26], offers among others, a GO annotated matrix view as well as a matrix layout to represent groupings. DyCoNet [27], a Gephi plugin, allows community detection and visualization. CellNetVis [28] is a web tool for the visualization of biological networks placing nodes in the cellular components they belong to. ClusterProfiler [29] is an R package that automates the process of biological-term classification and the enrichment analysis of gene clusters.
Many of these tools try to partially address the annotation visualization problem by using node colors or pie-chart nodes. However, they mainly focus on integrating networks with knowledge from existing repositories. In addition, they require familiarity, are bound to certain databases or are offered in a command-line version.
In this study, we present The Network Makeup Artist (NORMA), a highly interactive web tool that is specially designed to visualize communities and network annotations using alternative representations. NORMA is designed for non-experts and is a tool of general-purpose.
Method
NORMA is a handy web tool for interactive network annotation, visualization, and topological analysis and is able to handle multiple networks and annotations simultaneously. In its current version, the main interface is split into four tabs, namely, upload, network, annotation, and topology.
The upload tab
Users can upload three different file types as input. These are the network file, the annotation file, and the node coloring file.
The network file is an obligatory, 2-column or 3-column (unweighted and weighted, respectively), tab-delimited file containing all connections of an undirected network. For simplicity, self-loops and multiple-edges are eliminated automatically.
The annotation file is an obligatory, 2-column, tab-delimited file, which contains information about the defined groups. The first column contains the group names, whereas the second column contains the node names in that group. This file may contain results from a functional enrichment analysis (e.g., GO or KEGG/Reactome pathways) or a clustering analysis (e.g., MCL output [30], [31]).
The node coloring file is an optional, 2-column, tab-delimited file, in which users may directly assign a color to a node in order to encode different types of information. An example could be to highlight gene expression values (e.g., red for a down-regulated gene or green for an up-regulated gene) or a node category (e.g., yellow for category-1 and blue for category-2). In this file, the first column contains the node names, whereas the second column contains the colors as names or hex codes (e.g., red, green, yellow, blue, orange, #00ff00, #ff0000, #ffff00).
In NORMA, users can upload multiple network and annotation files simultaneously. Εach time a network or an annotation file is being uploaded, a name can be given first. Once the network and annotation files have been named and uploaded, users can visualize any network and overlay any of the selected annotations. Users can remove indifferent annotations or networks at any time. Network and annotation file contents are always shown as interactive tables. In these tables, one can search by suffix from the search field.
The network tab
This tab consists of two sub-tabs dedicated to network analysis and visualization. These are the interactive network and the automated community detection.
The interactive network sub-tab offers a dynamic network visualization in its simplest form with the use of the vizNetwork library. Nodes are connected with undirected edges, whereas their coordinates are calculated using vizNetwork’s default force-directed layout. The network is fully interactive as zooming, dragging, and panning is allowed either by using the mouse or the navigation buttons. Nodes can be selected and dragged anywhere on the plane, whereas the first neighbors of any node can be highlighted upon selection. The network view is automatically updated when a different network from the ones that have been uploaded is selected.
The automated community detection sub-tab offers methods for automatic community detection. This is particularly important in cases where users do not have any external pre-calculated results to provide. The algorithms that are currently offered are: Fast-Greedy [32], which tries to find densely connected communities via directly optimizing a modularity score; Louvain [33], which implements a multi-level modularity optimization algorithm for finding community structures, and is based on the modularity measure and a hierarchical approach; Label-Propagation [34], a fast, nearly linear time algorithm for detecting community structures in a network, which is done by labeling the vertices with unique labels and then updating them by majority voting in the neighborhood of the vertex; Walktrap [35], which tries to find densely connected subgraphs in a graph via random walks, with the idea that short random walks tend to restrict themselves in the same community; and Betweenness [36] where clustering is made by ‘breaking’ the bridges which connect the densely connected regions of a network.
Once a community detection method has been selected, users can see the results as interactive and searchable tables or as static plots for an at-a-glance view. In order for users to take advantage of NORMA’s advanced interactive visualization capabilities, the automatically generated annotations must be first exported and then imported as annotation input files.
The annotation tab
This tab is the NORMA’s strongest feature and is used to visualize annotated networks in an easy and user-friendly way. Annotated are considered the networks which are accompanied by predefined clusters, communities, subgraphs, regions, or neighborhoods. Here, users can select between any of the uploaded networks or annotation files and visualize them in combination (dropdown selection lists). This tab consists of four sub-tabs. These are convex hulls, 3D convex hulls, pie-chart nodes, and Venn diagrams.
Convex hulls
In this sub-tab, the selected network is initially visualized after applying any of the offered layout algorithms. Color-filled convex hulls are then used to highlight communities in a Venn-diagram-like view. Notably, a node might belong to more than one group. Groups are highlighted with visually distinct colors, while transparency is used to efficiently highlight the overlapping regions.
3D convex hulls
Like in 2D convex hulls, in this sub-tab, the selected network is initially visualized in 3D space with the help of plotly library, whereas 3D shaded volumes are used to highlight communities in a 3D Venn-diagram-like view.
Pie-chart nodes
Here, the selected network is initially visualized after applying any of the offered layout algorithms, and nodes are then visualized as pie-charts. A pie chart is divided into slices to illustrate the groups a node belongs to. If a node, for example, belongs to four groups, then the pie chart will consist of four equal slices colored with distinct colors. Nodes that do not belong to any group are marked gray.
Node coloring
Often, one might want to assign certain colors to nodes in order to encode certain types of information. In a gene expression network, for example, one might want to highlight the up- and down-regulated genes. Once an expression file has been uploaded, nodes in the “convex hulls” tab will be filled with the color of interest, whereas nodes in the “pie-chart nodes” tab will appear with a colored border. As node coloring is an optional feature, one can enable or disable this functionality at any time (selection box).
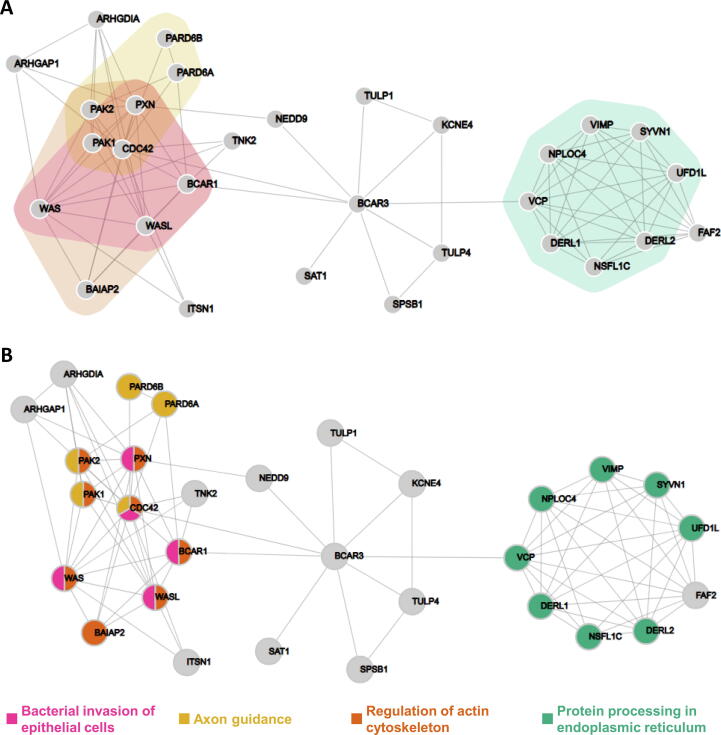
To briefly demonstrate this functionality, we queried the STRING database for the human BCAR3 gene and retrieved its interactions. We used all STRING channels (Text-mining, Experiments, Databases. Co‑expression, Neighborhood, Gene Fusion, and Co‑occurrence) using the medium confidence threshold (0.4) and allowed no more than 10 interactors (first neighbors). After exporting STRING’s KEGG analysis results and importing them into the NORMA application, we highlighted four pathways. These are the protein processing in endoplasmic reticulum (genes involved: DERL1, DERL2, NPLOC4, NSFL1C, SYVN1, UFD1L, VCP, and VIMP), the regulation of actin cytoskeleton (genes involved: BAIAP2, BCAR1, CDC42, PAK1, PAK2, PXN, WAS, and WASL), the axon guidance (genes involved: CDC42, PAK1, PAK2, PARD6A, and PARD6B), and the bacterial invasion of epithelial cells (genes involved: BCAR1, CDC42, PXN, WAS, and WASL). The network, along with the four selected pathways, is shown in Figure 1.
Figure 1.
Visualization of BCAR3 interactors from STRING with four highlighted KEGG pathways
A. Color-filled convex hulls. B. Pie-chart nodes. The networks were generated based on the gene expression data.
Layouts
Users can choose between several layout algorithms to visualize a network. In its current version, NORMA offers some of the layout algorithms offered by the igraph library [37], including Fruchterman-Reingold [38], which places nodes on a plane using the force-directed layout algorithm developed by Fruchterman and Reingold; Random, which places the vertices of the graph on a plane uniformly using random coordinates; Circle, which places vertices on a circle, ordered by their vertex IDs; Kamada-Kawai [39], which places the vertices on a plane by simulating a physical model of springs; Reingold-Tilford [40], a tree-like layout suitable for trees, hierarchies, or graphs without many cycles; LGL, a force-directed layout suitable for larger graphs; Graphopt, a force-directed layout algorithm which scales relatively well to large graphs; Gem [41], which places vertices on a plane using the GEM force-directed layout algorithm; Star, which places vertices of a graph on a plane, according to the simulated annealing Davidson and Harel algorithm; and Grid, which places vertices on a rectangular grid.
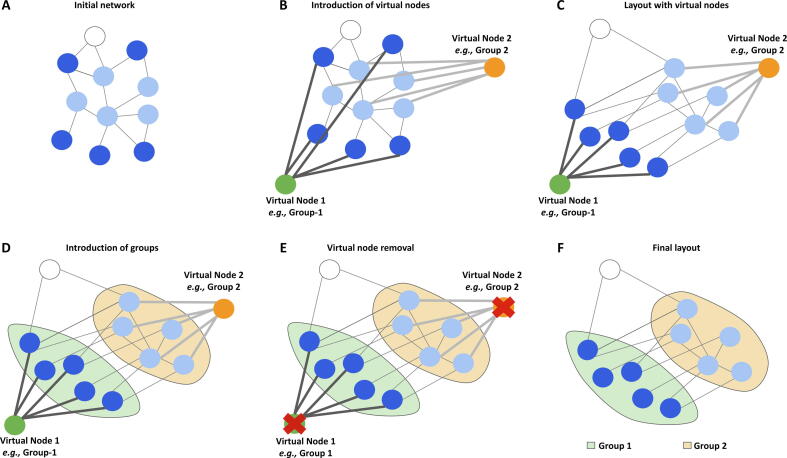
For better clarity, NORMA gives the option to slightly modify the selected layout in order to make groups as distinct as possible and avoid unnecessary overlaps which may occur from the original layout. To do this, NORMA starts by introducing one virtual node per group. Then, it connects these virtual nodes with any node which belongs to this group by assigning a heavyweight. Through this way, the virtual nodes will attract the nodes they relate to regardless of the selected layout. After the completion, all of the virtual nodes are then removed. The whole process is schematically shown in Figure 2.
Figure 2.
NORMA’s layout process
A. NΟRMA starts with a network using random coordinates. B. Virtual nodes are introduced (one per group) and are connected with heavyweights to the nodes they relate to. C. A selected igraph layout is applied, including the virtual nodes. Due to the heavyweight, virtual nodes are expected to attract the nodes they relate to. D. Once the selected layout algorithm has converged, the color-filled convex hulls are introduced. E. Virtual nodes are removed. F. The final network view.
Interactivity and visualization
NORMA gives a variety of options for the creation of optimal custom views. Network zoom in/out and panning functionalities are offered while users can interactively drag any node and place it anywhere on the plane. In addition to the visualized networks, groups are shown in an interactive table whose rows are colored accordingly. By selecting one or more groups, one can show the convex hulls as well as the pie-chart nodes of interest. Colored groups (rows) in the table correspond to colored groups in the offered views and vice versa. In addition, users have the option to show and hide the labels or only keep the labels of the selected groups. Labels below a certain zoom level are hidden for clarity. Finally, sliders to adjust node and label size as well as the network size are offered.
Venn diagrams
Users are allowed to choose between any pair of nodes and visualize the common groups they belong to, both as a Venn diagram and as a table. As it is not in the scope of NORMA to provide more complex Venn diagrams (e.g., more than three groups), users are encouraged to visit other online applications [42], [43], [44], [45].
The topology tab
Inspired by NAP’s [46] and Cytoscape’s topological network analysis features, in this tab, NORMA offers automated topological analysis and direct comparison of topological features between two or more selected networks. The topological measures that are offered by the igraph library are as follows: Number of Edges (the number of connections in the network), Number of Nodes (the number of nodes in the network), Density (the ratio of the number of edges to the number of possible edges in a graph), Average Path Length (the average number of steps needed to go from one node to another), Clustering Coefficient (a metric which shows if the network has the tendency to form clusters), Modularity (a function which calculates how modular a given division of a graph into subgraphs is), Average Eccentricity (the distance from a particular vertex to all other vertices in the graph is first calculated; among those distances, eccentricity is the highest one), Average Number of Neighbors (the total number of neighbors per node divided by the number of nodes), Centralization Betweenness (it is an indicator of the average centrality in a network, and is equal to the number of shortest paths from all vertices to all others that pass through that node; Betweenness centrality quantifies the number of times a node acts as a bridge along the shortest path between two other nodes), and Centralization Degree (the average value of the number of links incident upon a node across all nodes of a network).
The topology tab is divided into two sub-tabs: the summaries and the comparative plots. The “summaries” sub-tab shows the aforementioned topological measures in a numerical form as a table. Users can select one or more topological measures of interest and expand the table accordingly. Notably, this can be done for one network at a time upon selection (dropdown selection list). In the “comparative plots” sub-tab, users can directly compare the topological features of two or more networks simultaneously. In contrast to the previous tab, users are allowed to select one topological feature at a time (radio buttons) but as many networks as they like (checkboxes). Once two or more networks and one topological feature have been selected, direct comparisons can be made by the generated bar charts. A slider to adjust the chart height is offered.
Implementation
NORMA’s backend is implemented in R, and its frontend in Shiny. Network analysis features are based on the igraph library. Network interactivity and visualization are implemented in d3.js. The 3D network and convex hull visualization are offered by plotly.
A general overview
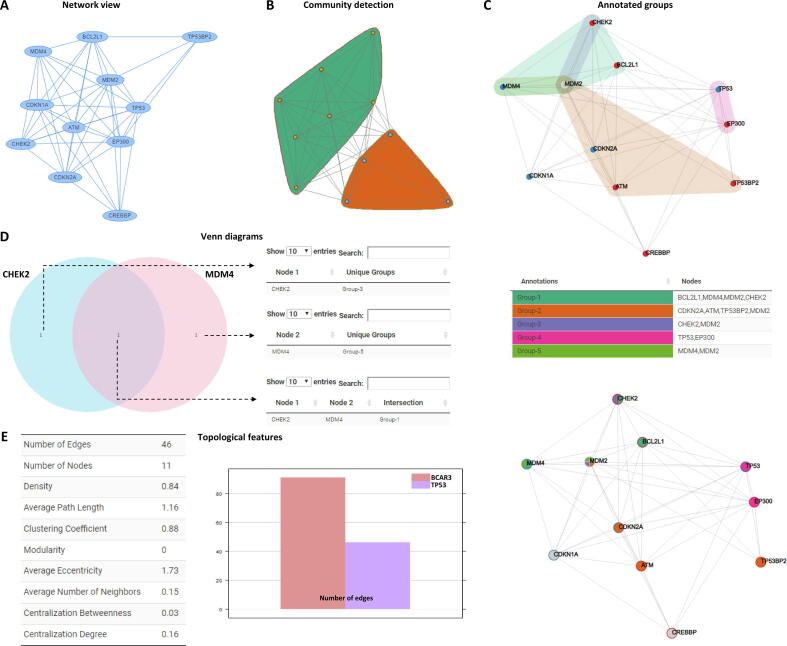
In order to summarize NORMA’s functionality, we briefly show its features in Figure 3. The network has been generated by querying STRING for human TP53 gene (default settings). Figure 3A shows the unweighted network in its simplest form. Figure 3B shows a static version of the same network clustered with NORMA’s Louvain clustering algorithm. Figure 3C shows the network and its groups (manually and randomly assigned) both as color-filled convex hulls and as pie-chart nodes. Groups are also shown as a table (legend), and colors between the network views and the table are consistent. In both views, node coloring encodes additional information (full color in the case of convex hulls or border color in the case of pie-chart nodes). Figure 3D shows a Venn diagram to highlight the common groups between two selected nodes. Figure 3E shows the topological network analysis of the TP53 network and a direct comparison (e.g., the number of edges) with the BCARC3 network described in Figure 1.
Figure 3.
NORMA’s overview
A. The TP53 network. B. Community detection using the Louvain algorithm. C. Upper: convex hulls with colored nodes. Lower: pie-chart nodes with border colors. Middle: groups visualized in an interactive table (group selection/isolation is allowed). D. Venn diagram showing common groups between any pair of selected nodes. E. Left: basic topological analysis of the TP53 network. Right: bar chart to directly compare one topological feature between two networks. The networks were generated based on the gene expression data.
Results
In order to demonstrate NORMA’s capabilities in a real case study, we used the experimental data described in [47]. Null tau mutants in Drosophila were used to demonstrate broad changes in their brain proteome with mass-spectrometry. To briefly describe the protocol used in this study, tauKO mutant flies [48] and Cantonised-w1118 control flies were cultured on standard wheat-flour-sugar food supplemented with soy flour and CaCl2, at 25 °C in 50%–70% relative humidity in a 12 h light/dark cycle. Three to four biological and two technical replicas from each genotype were used with each biological replica counting 10 brains. The samples were digested following the filter-aided sample preparation (FASP) method using spin filter devices with a 10 kDa cutoff (Sartorius, VN01H02) [49]. The peptide products were detected by an LTQ Orbitrap XL mass spectrometer (ThermoFisher Scientific, Waltham, MA) as described elsewhere [50]. The mass spectral files (raw files) were processed using MaxQuant software (version 1.5.3.30) [51] against the complete UniProt proteome of Drosophila melanogaster (downloaded April 1, 2016; 42,456 entries) and a common contaminant database by the Andromeda search engine. For the calculation of the protein abundances, label-free quantification (LFQ) was performed with both “second peptides” and “match between run” options enabled. Statistical analysis was performed using Perseus (version 1.5.3.2). The label-free quantified proteins were subjected to statistical analysis with the Student's t-test (permutation-based P value with 0.05 cutoff).
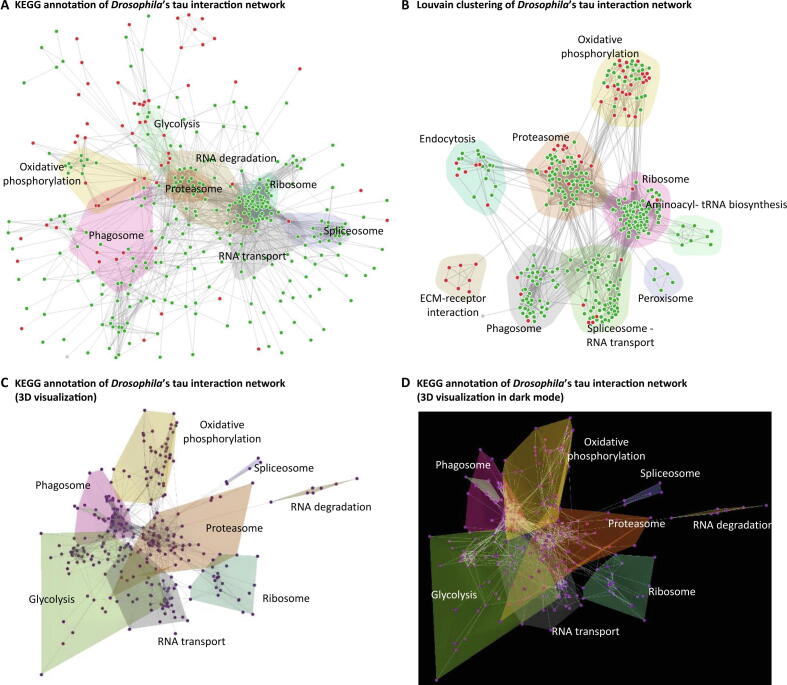
Starting from the identified 503 differentially expressed proteins, we used STRING to extract the connections among 358 of them and performed STRING’s KEGG analysis to highlight several important pathways (Figure 4). Up- and down-regulated proteins are marked in green and red accordingly. The most affected pathways upon Drosophila tau (dTau) loss in the central brain are shown as color-filled convex hulls and involve molecular processes reported altered in Tauopathies. Tau, for example, is known to bind to ribosomes in the brain and thus impair their function by reducing protein synthesis [52], an effect enhanced in Tauopathy brains [53]. In agreement, loss of dTau, resulted in elevated translation and protein synthesis as reflected by the increased expression of translation machinery components, including initiation factors, proteins that control gene expression or silencing, and ribosomal proteins. Spliceosome disruption and altered pre-mRNA processing are also emerging as potential contributors to Tau-mediated neurodegeneration [54]. On the other hand, transgenic Tau elevation by expression of human Tau in the fly central nervous system, triggered the reduction of multiple spliceosomal proteins [55]. In accordance with these results, several RNA-binding proteins and spliceosome components are more abundant in the mutant. Congruently with translation, proteins mediating proteasomal catabolism are equally elevated. Interestingly, proteasome activation can be viewed as an adaptive response to support elevated protein synthesis by increasing the size of the intracellular amino acid pool [56]. Finally, proteins implicated in energy-metabolism (oxidoreductases) and responsiveness to oxidative stress, appear as differentially regulated in the mutants, with about half of them attenuated by its loss. This is consistent with the reported accumulation of oxidative damage markers in Tauopathy patients [57] and the effect of oxidative stress in mediating tau-induced neurotoxicity in flies [58] (Figure 4A).
Figure 4.
Drosophila’s tau interaction network
A. Eight major KEGG pathways highlighted. B. Louvain clustering and functional annotation with DAVID application. C. The previous result was visualized by NORMA in 3D space. D. The latter in dark mode. In (A) and (B), up- and down-regulated proteins are marked in green and red, respectively.
To partially regenerate the annotation in Figure 4A, we used the NORMA’s Louvain community detection algorithm to cluster the network based on its topology only and the DAVID [59] application to perform pathway and GO enrichment analyses. In Figure 4B, results revealed nine distinct communities, which are highlighted. Within these communities, 4/22 proteins are involved in endocytosis, 21/75 in the proteasome, 3/5 in the peroxisome, 43/85 in the ribosome, 15/58 in the spliceosome, 15/49 in oxidative phosphorylation, 4/12 in the extracellular matrix (ECM) receptor, 8/47 in the phagosome, and 7/8 in aminoacyl-tRNA biosynthesis. Figure 4C and 4D show the same information as in Figure 4B in 3D space in light and dark modes, respectively.
Discussion
NORMA is a web tool for network annotation visualization. It is able to handle multiple annotations and networks simultaneously and is made to aid researchers in producing high-quality publication-ready figures as well as to explore novel hypotheses. In its current version, NORMA is not bound to a certain database or repository, and in the future, it will come with alternative visualization options (e.g., circos-like diagrams with edge bundling support). At the moment, NORMA’s online version supports networks of up to 10,000 nodes, a limitation that can be removed in its offline version. We believe that due to its interactivity and ease of use, NORMA is a handy tool for selective annotation visualization designed for non-experts without prior familiarity with command-line tools or scripting. To highlight NORMA’s advantages, we compared it with other applications in Table S1 and used it to show how it can complement alternative software like the ClueGO (Figure S1). The dataset used was a Gallus gallus BioGrid [25] PPI analyzed by DAVID functional annotation tool.
Code availability
The code is available at https://github.com/PavlopoulosLab/NORMA.
Data availabiltiy
NORMA service is available at http://norma.pavlopouloslab.info.
CRediT author statement
Mikaela Koutrouli: Methodology, Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Evangelos Karatzas: Software, Validation, Writing - review & editing. Katerina Papanikolopoulou: Formal analysis, Validation, Writing - review & editing. Georgios A. Pavlopoulos: Conceptualization, Supervision, Methodology, Writing - original draft, Writing - review & editing. All authors have read and approved the final manuscript.
Competing interests
Authors declare that there is no conflict of interest.
Acknowledgments
Georgios A. Pavlopoulos and Mikaela Koutrouli were supported by the Operational Program Competitiveness, Entrepreneurship, and Innovation, NSRF 2014-2020 (Grant No. MIS 5002562, co-financed by Greece and the European Union (European Regional Development Fund). Georgios A. Pavlopoulos was also supported by the Hellenic Foundation for Research and Innovation (HFRI) under the “First Call for HFRI Research Projects to support Faculty members and Researchers and the procurement of high-cost research equipment grant” (Grant No. 1855-BOLOGNA). Evangelos Karatzas was supported by the Action Strengthening Human Resources, Education and Lifelong Learning, 2014–2020, co-funded by the European Social Fund (ESF) and the Greek State (Grant No. MIS 5000432). Katerina Papanikolopoulou was supported by a grant from the Stavros Niarchos Foundation to the Biomedical Sciences Research Center “Alexander Fleming”, as part of the initiative of the Foundation to support the Greek research center ecosystem. We thank Dr. Yorgos Sofianatos for his valuable feedback and Dr. Alexandros Dimopoulos for the server setup.
Handled by Edwin Wang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2021.02.005.
Supplementary material
The following are the Supplementary data to this article:
Analysis of a Gallus Gallus PPI IntAct network A. A ClueGO analysis showing the GO Cellular Components as a network. B. The GO Cellular Component Categories from DAVID functional enrichment web tool. C. NORMA’s pie chart view highlighting groups as pie-chart nodes. D. NORMA’s view is highlighting some groups as color-filled areas. E. NORMA’s view, highlighting some groups as color-filled volumes in 3D view. F. The same as before in dark mode.
References
- 1.Koutrouli M., Karatzas E., Paez-Espino D., Pavlopoulos G.A. A guide to conquer the biological network era using graph theory. Front Bioeng Biotechnol. 2020;8:34. doi: 10.3389/fbioe.2020.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavlopoulos G.A., Wegener A.L., Schneider R. A survey of visualization tools for biological network analysis. BioData Min. 2008;1:12. doi: 10.1186/1756-0381-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavlopoulos G.A., Malliarakis D., Papanikolaou N., Theodosiou T., Enright A.J., Iliopoulos I. Visualizing genome and systems biology: technologies, tools, implementation techniques and trends, past, present and future. Gigascience. 2015;4:38. doi: 10.1186/s13742-015-0077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–311. doi: 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. Proceedings of the International AAAI Conference on Web and Social Media 2009;3:361–2.
- 7.Mrvar A., Batagelj V. Analysis and visualization of large networks with program package Pajek. Complex Adapt Syst Model. 2016;4:6. [Google Scholar]
- 8.Köhler J., Baumbach J., Taubert J., Specht M., Skusa A., Rüegg A., et al. Graph-based analysis and visualization of experimental results with ONDEX. Bioinformatics. 2006;22:1383–1390. doi: 10.1093/bioinformatics/btl081. [DOI] [PubMed] [Google Scholar]
- 9.Iragne F., Nikolski M., Mathieu B., Auber D., Sherman D. ProViz: protein interaction visualization and exploration. Bioinformatics. 2005;21:272–274. doi: 10.1093/bioinformatics/bth494. [DOI] [PubMed] [Google Scholar]
- 10.Hu Z., Hung J.H., Wang Y., Chang Y.C., Huang C.L., Huyck M., et al. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 2009;37:W115–W121. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavlopoulos G.A., Hooper S.D., Sifrim A., Schneider R., Aerts J. Medusa: a tool for exploring and clustering biological networks. BMC Res Notes. 2011;4:384. doi: 10.1186/1756-0500-4-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitkreutz B.J., Stark C., Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auber D., Archambault D., Bourqui R., Delest M., Dubois J., Lambert A., et al. In: Encycl Soc Netw Anal Min. Alhajj R., Rokne J., editors. Springer; New York: 2017. Tulip 5; pp. 1–28. [Google Scholar]
- 14.Pavlopoulos G.A., O'Donoghue S.I., Satagopam V.P., Soldatos T.G., Pafilis E., Schneider R. Arena3D: visualization of biological networks in 3D. BMC Syst Biol. 2008;2:104. doi: 10.1186/1752-0509-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karatzas E., Baltoumas F.A., Panayiotou N.A., Schneider R., Pavlopoulos G.A. Arena3Dweb : interactive 3D visualization of multilayered networks. Nucleic Acids Res. 2021;49:W36–W45. doi: 10.1093/nar/gkab278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theocharidis A., van Dongen S., Enright A.J., Freeman T.C. Network visualization and analysis of gene expression data using BioLayout Express(3D) Nat Protoc. 2009;4:1535–1550. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- 17.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., et al. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito R., Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Lotia S., et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gene Ontology Consortium The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuda S., Yamada T., Hamajima M., Itoh M., Katayama T., Bork P., et al. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isserlin R., Merico D., Voisin V., Bader G.D. Enrichment Map – a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res. 2014;3:141. doi: 10.12688/f1000research.4536.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koutrouli M., Hatzis P., Pavlopoulos G.A. In: Systems Medicine. Wolkenhauer O., editor. Academic Press; Oxford: 2021. Exploring networks in the STRING and Reactome database; pp. 507–520. [Google Scholar]
- 25.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown K.R., Otasek D., Ali M., McGuffin M.J., Xie W., Devani B., et al. NAViGaTOR: network analysis, visualization and graphing toronto. Bioinformatics. 2009;25:3327–3329. doi: 10.1093/bioinformatics/btp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kauffman J., Kittas A., Bennett L., Tsoka S., Ben-Jacob E. DyCoNet: a Gephi plugin for community detection in dynamic complex networks. PLoS One. 2014;9:e101357. doi: 10.1371/journal.pone.0101357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heberle H., Carazzolle M.F., Telles G.P., Meirelles G.V., Minghim R. Cell NetVis: a web tool for visualization of biological networks using force-directed layout constrained by cellular components. BMC Bioinformatics. 2017;18:395. doi: 10.1186/s12859-017-1787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azad A., Pavlopoulos G.A., Ouzounis C.A., Kyrpides N.C., Buluç A. HipMCL: a high-performance parallel implementation of the Markov clustering algorithm for large-scale networks. Nucleic Acids Res. 2018;46:e33. doi: 10.1093/nar/gkx1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clauset A., Newman M.E.J., Moore C. Finding community structure in very large networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 33.Blondel V.D., Guillaume J.L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008;2008:P10008. [Google Scholar]
- 34.Raghavan U.N., Albert R., Kumara S. Near linear time algorithm to detect community structures in large-scale networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:036106. doi: 10.1103/PhysRevE.76.036106. [DOI] [PubMed] [Google Scholar]
- 35.Walkowiak K. A unified approach to survivability of connection-oriented networks. In: Yolum pInar, Güngör T, Gürgen F, Özturan C, editors. Computer and Information Sciences - ISCIS 2005. Heidelberg: Springer Berlin Heidelberg; 2005, p.3–12.
- 36.Newman M.E.J., Girvan M. Finding and evaluating community structure in networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 37.Csardi G., Nepusz T. The igraph software package for complex network research. InterJournal Complex Systems. 2006;1695:1–9. [Google Scholar]
- 38.Fruchterman T.M.J., Reingold E.M. Graph drawing by force-directed placement. Softw Pract Exp. 1991;21:1129–1164. [Google Scholar]
- 39.Kamada T., Kawai S. An algorithm for drawing general undirected graphs. Inf Process Lett. 1989;31:7–15. [Google Scholar]
- 40.Reingold E.M., Tilford J.S. Tidier drawings of trees. IEEE Trans Softw Eng. 1981;SE-7:223–228. [Google Scholar]
- 41.Frick A., Ludwig A., Mehldau H. In: Graph Draw. Tamassia R., Tollis I.G., editors. Springer; Heidelberg: 1995. A fast adaptive layout algorithm for undirected graphs (extended abstract and system demonstration) pp. 388–403. [Google Scholar]
- 42.Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hulsen T., de Vlieg J., Alkema W. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kestler H.A., Muller A., Gress T.M., Buchholz M. Generalized Venn diagrams: a new method of visualizing complex genetic set relations. Bioinformatics. 2005;21:1592–1595. doi: 10.1093/bioinformatics/bti169. [DOI] [PubMed] [Google Scholar]
- 45.Pirooznia M., Nagarajan V., Deng Y. GeneVenn – a web application for comparing gene lists using Venn diagrams. Bioinformation. 2007;1:420–422. doi: 10.6026/97320630001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theodosiou T., Efstathiou G., Papanikolaou N., Kyrpides N.C., Bagos P.G., Iliopoulos I., et al. NAP: the network analysis profiler, a web tool for easier topological analysis and comparison of medium-scale biological networks. BMC Res Notes. 2017;10:278. doi: 10.1186/s13104-017-2607-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papanikolopoulou K., Roussou I.G., Gouzi J.Y., Samiotaki M., Panayotou G., Turin L., et al. Drosophila tau negatively regulates translation and olfactory long-term memory, but facilitates footshock habituation and cytoskeletal homeostasis. J Neurosci. 2019;39:8315–8329. doi: 10.1523/JNEUROSCI.0391-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burnouf S., Grönke S., Augustin H., Dols J., Gorsky M.K., Werner J., et al. Deletion of endogenous tau proteins is not detrimental in Drosophila. Sci Rep. 2016;6:23102. doi: 10.1038/srep23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 50.Terzenidou M.E., Segklia A., Kano T., Papastefanaki F., Karakostas A., Charalambous M., et al. Novel insights into SLC25A46-related pathologies in a genetic mouse model. PLoS Genet. 2017;13:e1006656. doi: 10.1371/journal.pgen.1006656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 52.Meier S., Bell M., Lyons D.N., Rodriguez-Rivera J., Ingram A., Fontaine S.N., et al. Pathological tau promotes neuronal damage by impairing ribosomal function and decreasing protein synthesis. J Neurosci. 2016;36:1001–1007. doi: 10.1523/JNEUROSCI.3029-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piao Y.S., Hayashi S., Wakabayashi K., Kakita A., Aida I., Yamada M., et al. Cerebellar cortical tau pathology in progressive supranuclear palsy and corticobasal degeneration. Acta Neuropathol (Berl) 2002;103:469–474. doi: 10.1007/s00401-001-0488-2. [DOI] [PubMed] [Google Scholar]
- 54.Apicco D.J., Zhang C., Maziuk B., Jiang L., Ballance H.I., Boudeau S., et al. Dysregulation of RNA splicing in tauopathies. Cell Rep. 2019;29:4377–4388.e4. doi: 10.1016/j.celrep.2019.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsieh Y.C., Guo C., Yalamanchili H.K., Abreha M., Al-Ouran R., Li Y., et al. Tau-mediated disruption of the spliceosome triggers cryptic RNA splicing and neurodegeneration in Alzheimer’s disease. Cell Rep. 2019;29:301–316.e10. doi: 10.1016/j.celrep.2019.08.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y., Nicholatos J., Dreier J.R., Ricoult S.J.H., Widenmaier S.B., Hotamisligil G.S., et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Markesbery W.R., Lovell M.A. Four-hydroxynonenal, a product of lipid peroxidation, is increased in the brain in Alzheimer’s disease. Neurobiol Aging. 1998;19:33–36. doi: 10.1016/s0197-4580(98)00009-8. [DOI] [PubMed] [Google Scholar]
- 58.Dias-Santagata D., Fulga T.A., Duttaroy A., Feany M.B. Oxidative stress mediates tau-induced neurodegeneration in Drosophila. J Clin Invest. 2007;117:236–245. doi: 10.1172/JCI28769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of a Gallus Gallus PPI IntAct network A. A ClueGO analysis showing the GO Cellular Components as a network. B. The GO Cellular Component Categories from DAVID functional enrichment web tool. C. NORMA’s pie chart view highlighting groups as pie-chart nodes. D. NORMA’s view is highlighting some groups as color-filled areas. E. NORMA’s view, highlighting some groups as color-filled volumes in 3D view. F. The same as before in dark mode.