Abstract
With accumulating dysregulated circular RNAs (circRNAs) in pathological processes, the regulatory functions of circRNAs, especially circRNAs as microRNA (miRNA) sponges and their interactions with RNA-binding proteins (RBPs), have been widely validated. However, the collected information on experimentally validated circRNA–disease associations is only preliminary. Therefore, an updated CircR2Disease database providing a comprehensive resource and web tool to clarify the relationships between circRNAs and diseases in diverse species is necessary. Here, we present an updated CircR2Disease v2.0 with the increased number of circRNA–disease associations and novel characteristics. CircR2Disease v2.0 provides more than 5-fold experimentally validated circRNA–disease associations compared to its previous version. This version includes 4201 entries between 3077 circRNAs and 312 disease subtypes. Secondly, the information of circRNA–miRNA, circRNA–miRNA–target, and circRNA–RBP interactions has been manually collected for various diseases. Thirdly, the gene symbols of circRNAs and disease name IDs can be linked with various nomenclature databases. Detailed descriptions such as samples and journals have also been integrated into the updated version. Thus, CircR2Disease v2.0 can serve as a platform for users to systematically investigate the roles of dysregulated circRNAs in various diseases and further explore the posttranscriptional regulatory function in diseases. Finally, we propose a computational method named circDis based on the graph convolutional network (GCN) and gradient boosting decision tree (GBDT) to illustrate the applications of the CircR2Disease v2.0 database. CircR2Disease v2.0 is available at http://bioinfo.snnu.edu.cn/CircR2Disease_v2.0 and https://github.com/bioinforlab/CircR2Disease-v2.0.
Keywords: circRNA, circRNA–disease association, Graph convolutional network, Gradient boosting decision tree, Machine learning
Introduction
Circular RNAs (circRNAs) are a type of endogenous non-coding RNAs formed through back-splicing or lariat events in genes [1]. Owing to their circular characteristics, circRNAs are difficult to be degraded by exonucleases and are more stable than linear RNAs [2]. Accumulating evidence indicates that circRNAs play significant roles in physiological and pathological processes by acting as microRNA (miRNA) and RNA-binding protein (RBP) sponges, regulators of transcription and splicing [3], and translators to produce polypeptides. Currently, the changes in circRNA expression profiles in various diseases, such as cancer [4], Alzheimer’s disease [5], and cardiovascular disease [6], have attracted increasing attention. Thus, circRNAs may have significant application as diagnostic biomarkers for complex diseases.
Recently, several circRNA-related databases have been constructed, such as circBase [7], CircInteractome [8], and starBase [9]. These mainly provide basic circRNA annotation and functional information. The published circRNA-related databases are listed in Table 1. Although some disease-related circRNA databases have been developed, such as Circ2Traits [11] and CSCD [20], the circRNAs in these databases are mainly analyzed using next-generation sequencing technology and bioinformatics tools. To further understand the roles of circRNAs in diseases, it is necessary to determine the associations between circRNAs and diseases. Therefore, the first version of CircR2Disease database was constructed to collect 739 high-quality associations between 661 circRNAs and 100 diseases, prior to March 31, 2018 [33]. CircRNADisease [34], Circ2Disease [35], and Circad [36] databases were subsequently established, and these databases contained 354, 273, and 1388 circRNA–disease associations, respectively. Owing to the increasing reports on circRNA–disease associations, the collected circRNA–disease associations are not suffiently thorough. Furthermore, the regulatory functions of circRNAs, especially as miRNA sponges and RBPs, have been widely validated. Therefore, an updated CircR2Disease database for collecting and integrating more resources focusing on circRNAs as disease biomarkers is urgently required.
Table 1.
The published databases related to circRNAs
| Database | Link | Function | Ref. |
|---|---|---|---|
| circBase | http://www.circbase.org/ | The circRNA information predicted by large-scale studies, including the circRNA sequences, etc. | [7] |
| Circbank | http://www.circbank.cn/ | Provide a new nomenclature system and collect other five features of circRNAs, including miRNA binding sites, conservation across species, m6A modification, mutations in circRNAs, and predicted ORFs and IRESs | [10] |
| Circ2Traits | http://gyanxet-beta.com/circdb | The disease-related miRNA–circRNA–mRNA–lncRNA interaction networks | [11] |
| starBase v2.0 | http://starbase.sysu.edu.cn/ | Systematically identify the RNA–RNA and protein–RNA interaction networks | [9] |
| CircInteractome | https://circinteractome.nia.nih.gov | Predict the interactions between miRNAs and circRNAs with 109 RBPs, and the database also focus on IRESs and ORFs | [8] |
| circNet | http://syslab5.nchu.edu.tw/CircNet | Focus on the tissue-specific circRNA expression profiles and circRNA–miRNA–mRNA regulatory networks | [12] |
| deepBase2.0 | http://biocenter.sysu.edu.cn/deepBase/ | Comparison of non-coding RNAs including sRNA, lncRNA, and circRNA across 19 species | [13] |
| circRNADb | http://reprod.njmu.edu.cn/circrnadb | Report exon splicing information of exon circRNAs and predict IRESs | [14] |
| TSCD | http://gb.whu.edu.cn/TSCD | Collect tissue-specific circRNAs in tissues of human and mouse, circRNA–miRNA interactions, predicted RBP binding sites, etc. | [15] |
| PlantcircBase | http://ibi.zju.edu.cn/plantcircbase | Plant circRNAs | [16] |
| PlantCircNet | http://bis.zju.edu.cn/plantcircnet | The expression profiles of plant circRNAs, circRNA–miRNA–gene regulatory networks, etc. | [17] |
| circlncRNAnet | http://120.126.1.61/circlnc/index.php | The co-expression network of lncRNA and circRNA, and the information of miRNA binding and RBP binding sites | [18] |
| exoRBase | http://www.exoRBase.org | The circRNAs, lncRNAs, and mRNAs in human blood exosomes | [19] |
| CSCD | http://gb.whu.edu.cn/CSCD | Provide cancer-specific circRNAs and circRNA–miRNA interactions; predict cellular location, RBP binding sites, and ORFs | [20] |
| CIRCpedia v2 | http://www.picb.ac.cn/rnomics/circpedia | Focus on circRNA annotation, expression comparison, and conservation between humans and mice | [21] |
| CircFunBase | http://bis.zju.edu.cn/CircFunBase | Collect experimentally validated and predicted functions of circRNAs, and circRNA-associated miRNAs and RBPs in plants and animals | [22] |
| CropCircDB | http://genome.sdau.edu.cn/crop/ | Investigate circRNAs under stress condition in maize and rice | [23] |
| GreenCircRNA | http://greencirc.cn/ | Plant circRNAs acting as miRNA decoys, and their potential networks involving circRNA–miRNA–mRNA in the corresponding species | [24] |
| VirusCircBase | http://www.computationalbiology.cn/ViruscircBase/home.html | Focus on the survey of circRNAs from viral species | [25] |
| CircAtlas | http://circatlas.biols.ac.cn/ | Describe multiple conservation, co-expression, and circRNA annotation and prioritization | [26] |
| MiOncoCirc | https://mioncocirc.github.io/ | The first database to be composed primarily of circRNAs directly detected in tumor tissues | [27] |
| circRic | https://hanlab.uth.edu/cRic/ | Characterize circRNA expression profiles; analyze the circRNA biogenesis regulators, the effect of circRNAs on drug response, the association of circRNAs with mRNAs, proteins, and mutations, etc. | [28] |
| LncRNADisease 2.0 | http://www.rnanut.net/lncrnadisease/ | Host disease-associated lncRNAs and circRNAs | [29] |
| riboCIRC | http://www.ribocirc.com | Provide computationally predicted ribosome-associated circRNAs and experimentally verified translated circRNAs | [30] |
| ClinVAR | http://soft.bioinfo-minzhao.org/circvar | Collect SNPs and small insertions and deletions in putative circRNA regions | [31] |
| TransCirc | https://www.biosino.org/transcirc/ | Provide comprehensive evidence supporting the translation potential of circRNAs | [32] |
Note: ORF, open reading frame; IRES, internal ribosome entry site; RBP, RNA-binding protein; lncRNA, long non-coding RNA; SNP, single nucleotide polymorphism.
In this study, we updated the CircR2Disease database to version 2.0 (named CircR2Disease v2.0), aiming to provide a comprehensive resource and web tool to clarify the relationships between circRNAs and diseases in diverse species. The updated CircR2Disease v2.0 includes 4201 entries of circRNA–disease associations retrieved from 2449 existing publications prior to May 15, 2021, exceeding the previous version by over 5-fold. In addition, the updated version of CircR2Disease database also includes high-quality information on genes producing circRNAs, circRNA–miRNA interactions, miRNA–miRNA target interactions, and circRNA-binding proteins. Furthermore, we proposed a computational method called circDis to predict the potential circRNA–disease associations as an example of the application of CircR2Disease v2.0. circDis was developed by combining graph convolutional network (GCN) and gradient boosting decision tree (GBDT). The results thus indicate that the updated CircR2Disease v2.0 can contribute to predicting circRNA–disease associations and exploring the roles of the circRNAs in disease diagnosis, treatment, and prognosis.
Data expansion and pre-processing
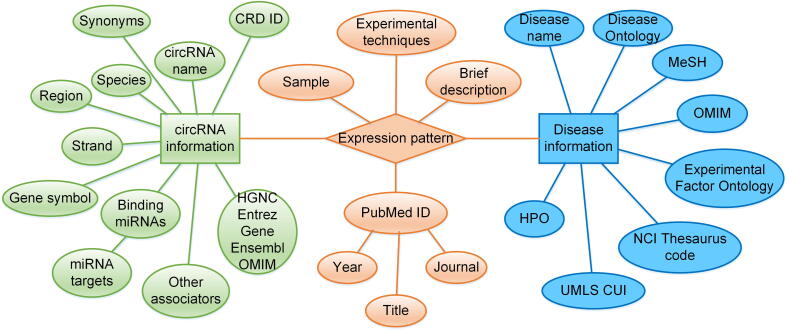
CircR2Disease v2.0 is an updated database for the increased information on circRNA–disease associations, and the details for each entry are depicted in Figure 1. First, the PubMed database was used to search relevant literature using keywords, such as “circular disease” and “circRNA cancer” (prior to May 15, 2021). Moreover, the literature was rescreened to retrieve more information on the CircR2Disease database. Secondly, entries between dysregulated circRNAs and diseases were collected, supported by strong experimental techniques (qRT-PCR, Western blot, luciferase reporter assay, and others). In addition, circRNAs with validated or predicted functions as miRNA sponges, circRNA–miRNA–target interactions, circRNA-related associators in the literature were also manually curated. Thirdly, the circRNA names and disease names were normalized to multiple nomenclature resources. As circRNAs from different studies may have more than one name, we normalized them using circBase database IDs. Furthermore, external links and corresponding IDs for the host genes of circRNAs were provided, including HUGO Gene Nomenclature Committee (HGNC), Entrez Gene, Ensembl, and Online Mendelian Inheritance in Man (OMIM) databases. As for disease names, we provided multiple disease nomenclature database links, including disease ontology (DO), medical subject headings (MeSH), OMIM, experimental factor ontology (EFO), NCI thesaurus code, Unified Medical Language System concept unique identifier (UMLS CUI), and human phenotype ontology (HPO).
Figure 1.
Overview of the entry information curated in CircR2Disease v2.0
The information of CircR2Disease v2.0 consists of three parts, circRNA information, disease information, and other information. circRNA information includes CRD ID, circRNA name, synonyms, species, region, strand, gene symbol, interacted miRNAs, miRNA targets, other associators, external links for gene symbol including HGNC, Entrez gene, Ensembl, and OMIM. Disease information includes disease name, external links for disease names including disease ontology, MeSH, OMIM, EFO, NCI Thesaurus code, UMLS CUI, and HPO. Other information includes expression pattern, sample, experimental techniques, brief description, Pubmed ID, year, title, and journal. CRD ID, the ID of CircR2Disease v2.0; HGNC, HUGO Gene Nomenclature Committee; OMIM, Online Mendelian Inheritance in Man; MeSH, medical subject headings; EFO, experimental factor ontology; NCI, National Cancer Institute; UMLS CUI, Unified Medical Language System concept unique identifier; HPO, human phenotype ontology.
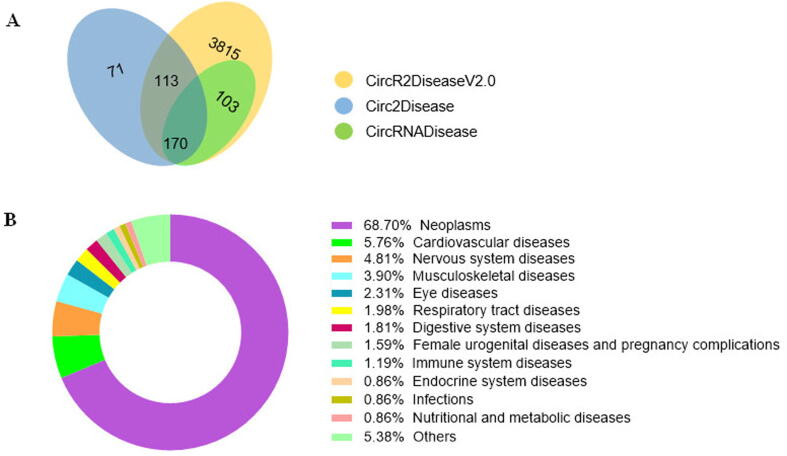
Although four databases have been constructed for collecting experimentally supported circRNA–disease associations, the entries are kept in the updated CircR2Disease v2.0 database. Comparison of CircR2Disease v2.0 with CircR2Disease and three other databases indicated that the CircR2Disease v2.0 database provides the largest number of associations and features (Table 2). In CircR2Disease v2.0, because the Circad database is not available, we collected strong experimentally validated associations recorded in CircRNADisease and Circ2Disease. In addition, we plotted the distribution of circRNA–disease entries in CircRNADisease, Circ2Disease, and CircR2Disease v2.0. We then found that CircRNADisease has 71 circRNA–disease associations only detected by microarray or RNA-seq techniques (Figure 2). Furthermore, according to the disease hierarchy based on MeSH [37], we conducted a survey on the circRNA–disease entries in CircR2Disease v2.0. The results revealed that neoplasms are the most thoroughly investigated disease subtypes (68.7%), followed by cardiovascular diseases (5.76%) and nervous system diseases (4.81%) (Figure 2).
Table 2.
The features of current circRNA–disease association databases
| Feature | CircR2Disease v2.0 | CircR2Disease | CircRNADisease | Circ2Disease | Circad |
|---|---|---|---|---|---|
| circRNA–disease associations | 4201 | 725 | 354 | 273 | 1388 |
| circRNAs | 3077 | 661 | 330 | 237 | 1292 |
| Gene symbols | 1611 | 445 | 236 | 211 | 706 |
| Disease subtypes | 312 | 100 | 48 | 54 | 129 |
| circRNAs regulated by miRNAs | 1496 | – | 148 | 44 | – |
| circRNAs as miRNA sponges | 1108 | – | 35 | 40 | – |
| circRNA-binding proteins | 283 | – | 22 | 20 | – |
| Disease name IDs | DO, MeSH, OMIM, EFO, HPO, etc. | – | – | – | – |
| Gene symbol IDs | HGNC, Entrez Gene, Ensembl, OMIM | – | – | – | – |
Note: DO, disease ontology; MeSH, medical subject headings; OMIM, Online Mendelian Inheritance in Man; EFO, experimental factor ontology; HPO, human phenotype ontology; HGNC, HUGO Gene Nomenclature Committee. –, not available.
Figure 2.
Information on CircR2Disease v2.0
A. Distribution of circRNA–disease associations in three databases. B. Distribution of disease entries in CircR2Disease v2.0.
Improved user interface
In this study, we used the SQL server to store and manage the data in CircR2Disease v2.0. The website was built using the “.Net”, a C# web framework (version 4.5). CircR2Disease v2.0 is available at http://bioinfo.snnu.edu.cn/CircR2Disease_v2.0 and https://github.com/bioinforlab/CircR2Disease-v2.0, and the previous version CircR2Disease database is still in service at http://bioinfo.snnu.edu.cn/CircR2Disease/.
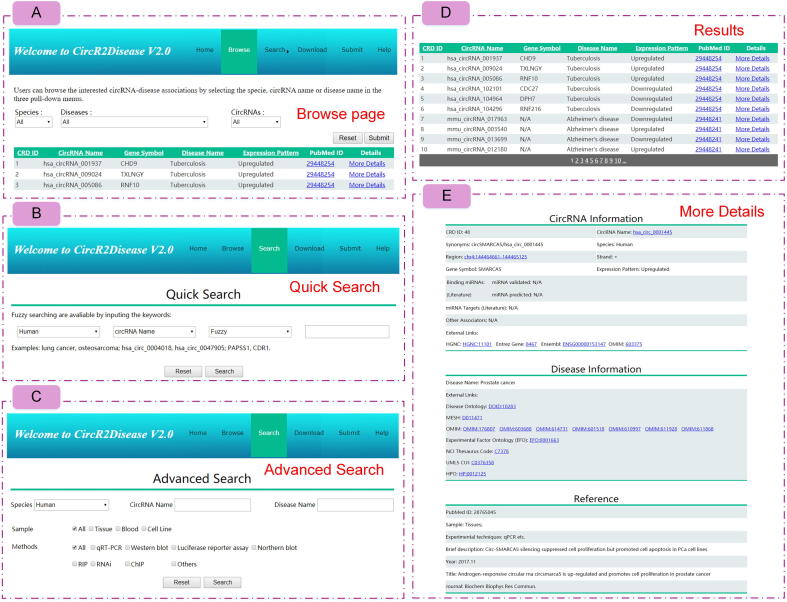
CircR2Disease v2.0 provides a novel web service for users to browse, search, download, and submit circRNA–disease associations (Figure 3). From the “Browse” page, users can view the entries of interest regarding circRNA–disease associations by selecting the species, circRNAs, or diseases in three pull-down menus (Figure 3A). The “Search” page is divided into “Quick Search” and “Advanced Search”. On the “Quick Search” page, users can search by the circRNA name, disease name, or gene symbol (Figure 3B). On the “Advanced Search” page, the sample or experiment information can be restricted to obtain a systemic search (Figure 3C). As a result, CircR2Disease v2.0 returns a brief table with seven items, including CRD ID (the ID of CircR2Disease v2.0), circRNA name, gene symbol, disease name, expression pattern, and Pubmed ID (Figure 3D). When clicking the hyperlink of ‘More Details’ in the result table on the ‘‘Browse” page, the entry details are displayed with three main parts, circRNA information (CRD ID, circRNA name, synonyms, species, region, strand, gene symbol, expression pattern, binding miRNAs, miRNA targets, other associators, and external links for gene symbol including HGNC, Entrez gene, Ensembl, and OMIM), disease information (disease name, and external links for disease names including disease ontology, MeSH, OMIM, EFO, NCI thesaurus code, UMLS CUI, and HPO), and reference (Pubmed ID, sample, experimental techniques, brief description, year, title, and journal) (Figure 3E). In addition, users can submit novel experimentally validated circRNA–disease associations on the “Submit” page. The “Download” page allows users to obtain the data in CircR2Disease v2.0. On the “Help” page, a detailed tutorial on how to use the database is available.
Figure 3.
Schematic workflow for CircR2Disease v2.0
A. Users could browse circRNA–disease associations by selecting the species, disease names, or individual circRNAs with circBase ID, circRNA name, and host gene. B. Interface of the “Quick Search” model in a fuzzy or exact manner. C. Interface of the “Advanced Search” model. D. Results of the “Browse” or “Search” page. E. Detailed information of the circRNA–disease entries.
Interactions of biomolecules in CircR2Disease v2.0
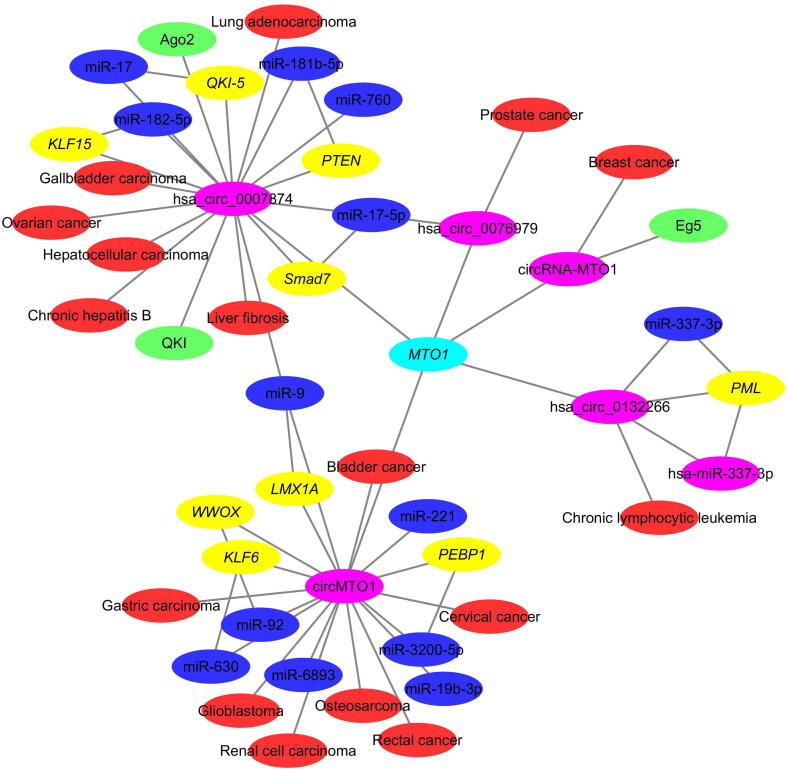
The CircR2Disease v2.0 database includes 4201 circRNA–disease entries, 3077 circRNAs (1496 circRNAs are regulated by miRNAs, and 1108 circRNAs are validated as miRNA sponges), 1611 genes, 312 diseases, and 283 circRNA-binding proteins (Table 2). Besides the experimentally verified circRNA–disease associations, the database also includes high-quality information on genes producing circRNAs, circRNA–miRNA interactions, miRNA–miRNA target interactions, and circRNA-binding proteins. Therefore, these relationships could be used to study the pathogenesis of complex diseases. In this section, circRNA–gene, circRNA–miRNA, circRNA–miRNA–target, and circRNA–RBP interactions are integrated into the circRNA–disease interaction network. For example, the hsa_circ_0007874, circRNA-MTO1, circMTO1, hsa_circ_0132266, and hsa_circ_0076979 are derived from the MTO1 gene locus (Figure 4). By facilitating the analysis of more relationships among the biomolecules involved in diseases, this database is useful for predicting circRNA–disease associations, as well as for exploring the roles of other related biomolecules in complex diseases.
Figure 4.
Relationships among the MTO1 gene, circRNAs derived from the MTO1 gene locus, miRNAs, miRNA targets, and circRNA-binding proteins
Pink nodes represent circRNAs, red nodes represent diseases, blue nodes represent miRNAs, yellow nodes represent miRNA targets, green nodes represent circRNA-binding proteins, and edges represent relationships among circRNAs, diseases, and other biomolecules.
Applications of CircR2Disease v2.0
The collected data resources could be widely used to predict the circRNA–disease associations, depict the high-quality miRNA–circRNA associations, predict the circRNA-binding proteins, etc. In addition, because circRNAs are a class of non-coding RNAs, the information of CircR2Disease v2.0 could also be utilized to predict the miRNA–disease and long non-coding RNA (lncRNA)–disease associations [38]. For example, experimentally validated circRNA–disease associations are widely applied to design computational methods for predicting relationships between circRNAs and diseases, such as PWCDA [39], KATZHCDA [40], iCircDA-MF [41], DWNN-RLS [42]. In this section, we propose a computational method named circDis based on GCN and GBDT to mine potential circRNA–disease associations.
Prediction of circRNA–disease associations with GCN and GBDT
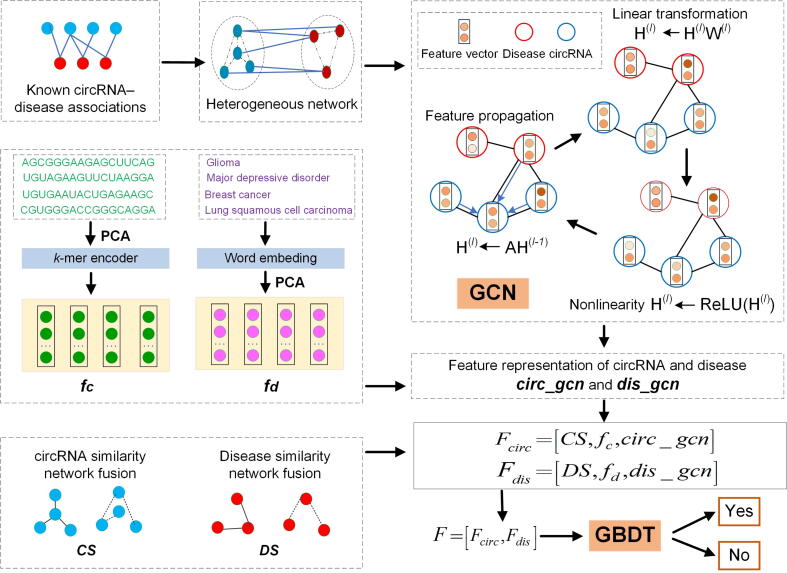
In this section, we present the GCN- and GBDT-based framework named circDis, developed to predict potential circRNA–disease associations. The flow chart of the circDis method is shown in Figure 5. First, GCN is used to extract circRNA features and disease features from the bipartite graph of known circRNA–disease associations. Meanwhile, we calculate circRNA functional similarity, disease semantic similarity, and Gaussian interaction profile (GIP) kernel similarity for circRNAs and diseases. Second, the circRNA sequence information and disease names are converted to the node features of a network using k-mer encoding and word embedding techniques, respectively. Finally, the feature vectors of circRNA–disease pairs can be obtained based on circRNA features and disease features; we then apply a GBDT classifier to predict the potential circRNA–disease associations.
Figure 5.
Schematic of the circDis model for predicting circRNA–disease associations
Firstly, GCN is used to extract circRNA features and disease features from the bipartite graph of known circRNA–disease associations. Meanwhile, circRNA functional similarity, disease semantic similarity, and GIP kernel similarity for circRNAs and diseases are calculated. Secondly, the circRNA sequence information and disease names are converted to the node features of a network using k-mer encoding and word embedding techniques, respectively. Then, the feature vectors of circRNA–disease pairs can be obtained based on circRNA features and disease features. Finally, we apply a GBDT classifier to predict the potential circRNA–disease associations. GCN, graph convolutional network; PCA, principal component analysis; GBDT, gradient boosting decision tree; ReLU, Rectified Linear Unit.
Similarities in circRNA and disease associations
The gold standard circRNA–disease associations were derived from CircR2Disease v2.0, and the human associations between circRNAs and diseases were retained when the circRNAs matched the circRNA IDs in the circBase database [7]. Here, we extracted 2099 experimentally validated circRNA–disease associations from CircR2Disease v2.0, which involved 1577 circRNAs and 202 diseases. We defined the adjacency matrix I to represent the circRNA–disease associations. If there was an association between circRNA i and disease j, Ii,j was considered equal to 1; otherwise, it was considered equal to 0.
We further constructed the circRNA functional similarity matrix CF and disease semantic similarity matrix DSe. The circRNA functional similarity matrix was computed according to the previous method proposed by Wang et al. [43], whereas the disease sematic similarity matrix was calculated based on the DOSE tool [44]. Considering that some circRNAs or diseases have no similarity scores in CF and DSe, we calculated the GIP kernel similarity for circRNAs (CG) and diseases (DG), respectively. We used the matrix CG and matrix DG to replace the zero values in the similarity matrices CF and DSe, respectively. The circRNA fusion similarity (CS) and disease fusion similarity (DS) are defined as follows:
| (1) |
| (2) |
By integrating the circRNA similarity network, disease similarity network, and circRNA–disease association network, we constructed a circRNA–disease heterogeneous network.
Extracted features of circRNAs and diseases using GCN
In previously proposed GCN [45], the number of graph edges follows a linear scale, and hidden layer representations that encode both local graph structure and features of nodes are learned. In this study, GCN was employed to learn the latent representation of circRNAs and diseases by combining the circRNA–disease association matrix I. The circRNA–disease heterogeneous network A is defined as follows:
| (3) |
where matrix A is a symmetric matrix, and each node in the heterogeneous network can connect with itself. Then, the normalized symmetric adjacent matrix of matrix A is calculated as:
| (4) |
where D is the degree matrix, and its diagonal elements are:
| (5) |
The forward propagation of this network followed the method described by Kipf [45], which is defined as follows:
| (6) |
where Hl−1 indicates the features of the (l−1) layer. Specially, H0 is an identity matrix, with the diagonal elements 1. The latent embedding vectors of circRNAs and diseases are defined as circ_gcn and dis_gcn, respectively.
In this study, two-layer GCN was used to obtain the circRNA and disease features. The dimensions of the two layers are 32 and 16, respectively. To prevent overfitting, the dropout rate was introduced, which can select some neurons randomly, hide or discard them temporarily, and then train and optimize them. We set the dropout rate as 0.05 and the learning rate as 0.001. Finally, we obtained the latent embedding vector of circRNA, circ_gcn, and the latent embedding vector of disease, dis_gcn, both with dimensions of 8.
Construction of feature vectors for circRNA–disease pairs
To better enrich the features of circRNAs and diseases, circRNA sequence features and disease name features were also introduced. The circRNA sequences were extracted from the circBase database [7], and the circRNA sequence features were converted to their feature vectors with 3-mer encoding. Disease name features were transformed into word vectors using bidirectional encoder representations from transformers (BERT) [46]. We then utilized principal component analysis (PCA) to obtain low-dimensional circRNA fc and disease fd feature vectors. Specifically, the dimensions of both fc and fd are 8.
Finally, three types of circRNA features and three types of disease features were obtained. The three types of circRNA (disease) features include circRNA (disease) fusion feature, low-dimensional circRNA (disease) feature, and embedding vector of circRNA (disease) extracted from the circRNA–disease bipartite network with GCN. The feature matrices of circRNAs and diseases can be defined as and , respectively. The feature vector of circRNA ci and disease dj can be defined as follows:
| (7) |
where Fi,j is the feature vector of the association between circRNA ci and disease dj. The matrix F is used to indicate the feature of all circRNA–disease pairs. [·] indicates the connection of two vectors.
Training the circDis model
GBDT is the combination of decision trees and gradient boosting. GBDT inherits many advantages of decision trees, but also improves on their disadvantages. As the trees used by GBDT are all low complexity trees, the variance is very small. Integration of multiple decision trees through gradient promotion can finally solve the problem of overfitting [47]. GBDT is an additive model composed of multiple cart decision regression trees, and can be described as follows:
| (8) |
where M indicates the total training times of the model, and a weak classifier is generated each time. The model of step m can be expressed as:
| (9) |
where represents the current model, and GBDT determines the next weak classifier by minimizing the parameters.
| (10) |
where N is the number of samples, L (·) represents the loss function.
In the iteration of GBDT, a weak learner is found based on the strong learner obtained in the previous iteration to minimize the loss function in this round. In other words, the core of GBDT is that each tree learns the results and residuals of all previous trees. This residual is an accumulated amount that can be obtained after adding the predicted value.
GBDT is a mature algorithm that has been integrated into the machine library in Python. In this study, we used GBDT to classify circRNA–disease pairs by feeding the final feature into the GBDT classifier. The main parameters of GBDT, learning_rate, max_depth, and min_samples_leaf, are set to 0.1, 3, and 30, respectively.
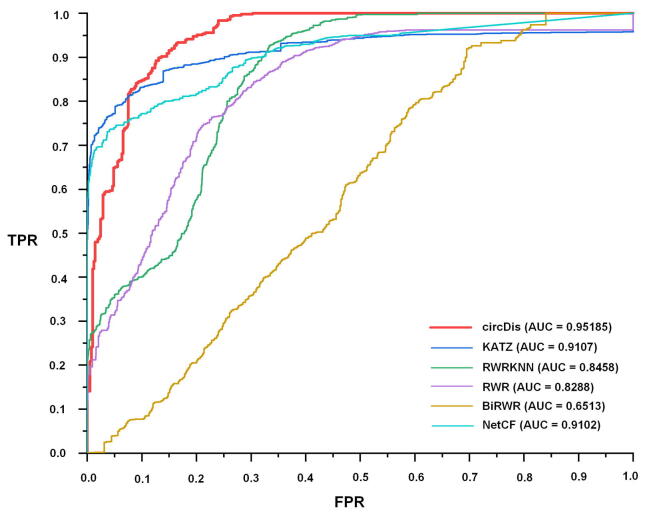
During the training process of the circDis model, we minimized the cross-entropy loss and utilized the 5-fold cross-validation to evaluate the performance of the proposed method. All samples are divided into five equal parts; each part is regarded as testing samples in turn, whereas the other parts are selected as training samples. The area under the receiver operating characteristic curve (AUC) is selected as the evaluation metric of model classification ability. As shown in Figure 6, the AUC value of circDis is 0.95185, which is better than five other methods, including KATZ, NetCF, RWR, BiRWR, and RWRKNN. Therefore, CircR2Disease v2.0 could be effectively utilized to predict potential circRNA–disease associations.
Figure 6.
ROC curves and AUC values of the circDis method
The ROC curves and AUC values of the circDis and other five methods. ROC, receiver operating characteristic; AUC, area under the receiver operating characteristic curve.
Conclusion
Information regarding associations between circRNAs and diseases is valuable information for clinical diagnosis, prognosis, and treatment. In this study, the CircR2Disease database was updated to CircR2Disease v2.0, including novel services and high-quality information on the latest circRNA–disease associations. Information of the experimentally validated circRNA–miRNA, circRNA–miRNA–target, and circRNA–RPB interactions from literature were also integrated. This circRNA-related information could provide effective materials to design computational models for mining potential circRNA–disease associations. In this study, we also proposed a computational model to predict potential associations between circRNAs and diseases, based on machine learning and multiple similarity features. Furthermore, the circRNA–disease associations and experimentally validated circRNA–miRNA interactions could be integrated to mine miRNA–disease associations. Therefore, we believe that CircR2Disease v2.0 can serve as a significant resource to study dysregulated circRNAs as disease mechanisms in the future.
Data availability
The CircR2Disease v2.0 database is available at http://bioinfo.snnu.edu.cn/CircR2Disease_v2.0, and also available at https://github.com/bioinforlab/CircR2Disease-v2.0.
CRediT author statement
Chunyan Fan: Data curation, Visualization, Writing - original draft. Xiujuan Lei: Conceptualization, Supervision, Writing - review & editing. Jiaojiao Tie: Methodology. Yuchen Zhang: Methodology, Visualization. Fang-Xiang Wu: Writing - review & editing. Yi Pan: Writing - review & editing. All authors have read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work is supported by the National Natural Science Foundation of China (Grant Nos. 61972451, 61672334, and 61902230) and the Fundamental Research Funds for the Central Universities (Grant No. GK201901010).
Handled by Fangqing Zhao
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Contributor Information
Xiujuan Lei, Email: xjlei@snnu.edu.cn.
Fang-Xiang Wu, Email: faw341@mail.usask.ca.
Yi Pan, Email: yipan@gsu.edu.
References
- 1.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 2.Su M., Xiao Y., Ma J., Tang Y., Tian B., Zhang Y., et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90. doi: 10.1186/s12943-019-1002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conn V.M., Hugouvieux V., Nayak A., Conos S.A., Capovilla G., Cildir G., et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 4.Bach D.H., Lee S.K., Sood A.K. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Y., Tan L., Wang X. Circular HDAC9/microRNA-138/Sirtuin-1 pathway mediates synaptic and amyloid precursor protein processing deficits in Alzheimer's disease. Neurosci Bull. 2019;35:877–888. doi: 10.1007/s12264-019-00361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B., Li G., Peng J., Ren L., Lei L., Ye H., et al. CircMACF1 attenuates acute myocardial infarction through miR-500b-5p-EMP1 axis. J Cardiovasc Transl Res. 2021;14:161–172. doi: 10.1007/s12265-020-09976-5. [DOI] [PubMed] [Google Scholar]
- 7.Glažar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA–ceRNA, miRNA–ncRNA and protein–RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu M., Wang Q., Shen J., Yang B.B., Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16:899–905. doi: 10.1080/15476286.2019.1600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosal S., Das S., Sen R., Basak P., Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y.C., Li J.R., Sun C.H., Andrews E., Chao R.F., Lin F.M., et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng L.L., Li J.H., Wu J., Sun W.J., Liu S., Wang Z.L., et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, lncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44:D196–D202. doi: 10.1093/nar/gkv1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Han P., Zhou T., Guo X., Song X., Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia S., Feng J., Lei L., Hu J., Xia L., Wang J., et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 16.Chu Q., Zhang X., Zhu X., Liu C., Mao L., Ye C., et al. PlantcircBase: a database for plant circular RNAs. Mol Plant. 2017;10:1126–1128. doi: 10.1016/j.molp.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Zhang P., Meng X., Chen H., Liu Y., Xue J., Zhou Y., et al. PlantCircNet: a database for plant circRNA-miRNA-mRNA regulatory networks. Database. 2017;2017:bax089. doi: 10.1093/database/bax089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S.M., Liu H., Huang P.J., Chang I.Y., Lee C.C., Yang C.Y., et al. circlncRNAnet: an integrated web-based resource for mapping functional networks of long or circular forms of noncoding RNAs. Gigascience. 2018;7:1–10. doi: 10.1093/gigascience/gix118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S., Li Y., Chen B., Zhao J., Yu S., Tang Y., et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106–D112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia S., Feng J., Chen K., Ma Y., Gong J., Cai F., et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46:D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong R., Ma X.K., Li G.W., Yang L. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinformatics. 2018;16:226–233. doi: 10.1016/j.gpb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng X., Hu D., Zhang P., Chen Q., Chen M. CircFunBase: a database for functional circular RNAs. Database. 2019;2019:baz003. doi: 10.1093/database/baz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Wang C., Guo B., Song K., Shi C., Jiang X., et al. CropCircDB: a comprehensive circular RNA resource for crops in response to abiotic stress. Database. 2019;2019:baz053. doi: 10.1093/database/baz053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Hao Z., Yin S., Li G. GreenCircRNA: a database for plant circRNAs that act as miRNA decoys. Database. 2020;2020:aaa039. doi: 10.1093/database/baaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai Z., Fan Y., Zhang Z., Lu C., Zhu Z., Jiang T., et al. VirusCircBase: a database of virus circular RNAs. Brief Bioinform. 2020;2020:bbaa052. doi: 10.1093/bib/bbaa052. [DOI] [PubMed] [Google Scholar]
- 26.Wu W., Ji P., Zhao F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020;21:101. doi: 10.1186/s13059-020-02018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan H., Xiang Y.u., Ko J., Li S., Jing Y., Zhu X., et al. Comprehensive characterization of circular RNAs in ∼ 1000 human cancer cell lines. Genome Med. 2019;11:55. doi: 10.1186/s13073-019-0663-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao Z., Yang Z., Huang Z., Zhou Y., Cui Q., Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019;47:D1034–D1037. doi: 10.1093/nar/gky905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Xie M., Wang Y., Yang L., Xie Z., Wang H. riboCIRC: a comprehensive database of translatable circRNAs. Genome Biol. 2021;22:79. doi: 10.1186/s13059-021-02300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao M., Qu H. circVAR database: genome-wide archive of genetic variants for human circular RNAs. BMC Genomics. 2020;21:750. doi: 10.1186/s12864-020-07172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang W., Ling Y., Zhang S., Xia Q., Cao R., Fan X., et al. TransCirc: an interactive database for translatable circular RNAs based on multi-omics evidence. Nucleic Acids Res. 2021;49:D236–D242. doi: 10.1093/nar/gkaa823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan C., Lei X., Fang Z., Jiang Q., Wu F.X. CircR2Disease: a manually curated database for experimentally supported circular RNAs associated with various diseases. Database. 2018;2018:bay044. doi: 10.1093/database/bay044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Z., Wang K., Wu F., Wang W., Zhang K., Hu H., et al. circRNA disease: a manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis. 2018;9:475. doi: 10.1038/s41419-018-0503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao D., Zhang L., Zheng M., Sun X., Lu Y., Liu P. Circ2Disease: a manually curated database of experimentally validated circRNAs in human disease. Sci Rep. 2018;8:11018. doi: 10.1038/s41598-018-29360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rophina M., Sharma D., Poojary M., Scaria V. Circad: a comprehensive manually curated resource of circular RNA associated with diseases. Database. 2020;2020:baaa019. doi: 10.1093/database/baaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipscomb C.E. Medical subject headings (MeSH) Bull Med Libr Assoc. 2000;88:265–266. [PMC free article] [PubMed] [Google Scholar]
- 38.Lei X., Mudiyanselage T.B., Zhang Y., Bian C., Lan W., Yu N., et al. A comprehensive survey on computational methods of non-coding RNA and disease association prediction. Brief Bioinform. 2020;2020:bbaa350. doi: 10.1093/bib/bbaa350. [DOI] [PubMed] [Google Scholar]
- 39.Lei X., Fang Z., Chen L., Wu F.X. PWCDA: path weighted method for predicting circRNA-disease associations. Int J Mol Sci. 2018;19:3410. doi: 10.3390/ijms19113410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan C., Lei X., Wu F.X. Prediction of circRNA-disease associations using KATZ model based on heterogeneous networks. Int J Biol Sci. 2018;14:1950–1959. doi: 10.7150/ijbs.28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei H., Liu B. iCircDA-MF: identification of circRNA-disease associations based on matrix factorization. Brief Bioinform. 2020;21:1356–1367. doi: 10.1093/bib/bbz057. [DOI] [PubMed] [Google Scholar]
- 42.Yan C., Wang J., Wu F.X. DWNN-RLS: regularized least squares method for predicting circRNA-disease associations. BMC Bioinformatics. 2018;19:520. doi: 10.1186/s12859-018-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D., Wang J.A., Lu M., Song F., Cui Q.H. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics. 2010;26:1644–1650. doi: 10.1093/bioinformatics/btq241. [DOI] [PubMed] [Google Scholar]
- 44.Yu G., Wang L.G., Yan G.R., He Q.Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics. 2015;31:608–609. doi: 10.1093/bioinformatics/btu684. [DOI] [PubMed] [Google Scholar]
- 45.Kipf T.N., Welling M. 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, April 24–26. 2017. Semi-supervised classification with graph convolutional networks. [Google Scholar]
- 46.Devlin J., Chang M.W., Lee K., Toutanova K. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers) Association for Computational Linguistics; Minneapolis: 2019. BERT: pre-training of deep bidirectional transformers for language understanding; pp. 4171–4186. [Google Scholar]
- 47.Zhang C., Zhang Y., Shi X., Almpanidis G., Fan G., Shen X. On incremental learning for gradient boosting decision trees. Neural Process Lett. 2019;50:957–987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The CircR2Disease v2.0 database is available at http://bioinfo.snnu.edu.cn/CircR2Disease_v2.0, and also available at https://github.com/bioinforlab/CircR2Disease-v2.0.