Abstract
Goose astrovirus (GoAstV), goose parvovirus (GPV), and goose circovirus (GoCV) infections have similar symptoms, such as severe diarrhea, and cause serious economic losses to the goose industry globally. Therefore, it is necessary to develop a rapid and accurate method for the differential diagnosis of the 3 viruses. In this study, a TaqMan probe-based multiplex reverse transcription-qualitative polymerase chain reaction (RT-qPCR) method was established and optimized for simultaneous detection of the three viruses. Three pairs of specific primers and probes were designed considering the conserved sequences of ORF2, VP3, and Rep of GoAstV, GPV, and GoCV, respectively. Singleplex real-time RT-qPCR detected a minimum of 10 copies of these genes, while multiplex real-time RT-qPCR detected a minimum of 100 copies. The correlation coefficients exceeded 0.99, and the amplification efficiency was 80 to 100%. The assay had high sensitivity, specificity, and repeatability. In 85 tissue samples, GoAstV and GPV were the main pathogens and demonstrated co-infection. This assay provides a rapid, efficient, specific, and sensitive tool for the detection of GoAstV, GPV, and GoCV. This can facilitate disease management and epidemiological surveillance.
Key words: goose astrovirus, goose circovirus, goose parvovirus, multiplex real-time RT-qPCR, TaqMan probe
Introduction
Since November 2016, goose gout has been reported in goose farms in several provinces of China, such as Jiangsu, Hunan, and Fujian, and this has caused serious damage to the goose-rearing industry (Chen et al., 2020). The disease is characterized by high morbidity, mortality, a low feed conversion rate, and growth retardation (Yu et al., 2018). It mainly affects goslings aged 1 to 15 d, leading to severe diarrhea and white excrement. The kidney and liver show urate deposition (An et al., 2020). The disease is caused by goose astrovirus (GoAstV), which is a nonenveloped, single-stranded RNA virus (Arias and DuBois, 2017). The genome length of GoAstV varies from 6100 to 7900 nt and consists of three open reading frames (ORF1a, ORF1b, and ORF2), a short 5ʹ-untranslated region, a 3ʹ-untranslated region, and a polymeric A-tail. ORF1a and ORF1b encode nonstructural proteins with conserved sequences in their overlapping regions (An et al., 2020). ORF2 encodes nucleocapsid proteins associated with virus formation; these proteins are involved in cell surface receptor recognition and immune response (Yin et al., 2021).
Another goose disease, gosling plague, is an exudative inflammatory bowel disease caused by goose parvovirus (GPV). This is an acute, highly infectious, and fatal disease, also known as Dermorelli's disease (Gough et al., 2005; Yang et al., 2010). The main signs are a short beak, protruding tongue, slow growth, short or thick tibias, and the main symptom is watery diarrhea (Yu et al., 2016; Luo et al., 2018). Goslings at 20 d of age are susceptible to the disease, which spreads rapidly and has a high mortality rate (reaching >95% within 5 d) (Yu et al., 2018). The GPV is a small, nonenveloped, single-stranded DNA virus, with a genome length of approximately 5.1 Kb. It consists of 2 open reading frames, ORF1 and ORF2; ORF1 encodes the Ns protein, and ORF2 encodes three structural proteins: VP1, VP2, and VP3 (Deng et al., 2014; Shao et al., 2014).
Goose circovirus (GoCV), which causes growth retardation and high mortality in geese, was first discovered in Germany in 1999 (Yang et al., 2020). In 2004, GoCV was detected in Zhejiang Province and first reported in mainland China (Yu et al., 2007; Stenzel et al., 2018). Subsequently, the existence of GoCV has been reported in other regions of China, where it caused serious economic losses. Owing to suppressed immunity, mild air sac turbidity or serositis due to mixed infections may occur in infected geese (Ting et al., 2021). The GoCV is a small nonenveloped DNA virus. The genome length is 1821 Kb and is composed of 4 reading frames: ORFv1, ORFc1, ORFc2, and ORFv2. ORFv1 encodes the replication-related protein Rep, while ORFc1 encodes the capsid protein (Scott et al., 2006; Rosario et al., 2017).
In recent years, TaqMan-probe-based real-time quantitative polymerase chain reaction (qPCR) assays have been successfully established for the simultaneous detection of GoAstV and GPV (Wan et al., 2019). Similarly, a TaqMan-probe-based real-time qPCR assay has been established for GoCV detection (Yang et al., 2020). However, TaqMan-probe-based simultaneous detection of GoAstV, GPV, and GoCV has not yet been reported. Studies have shown that cross-infection with GoAstV, GPV, and GoCV can occur. Goose parvovirus was detected in the gut of geese with severe gout, and it was shown that the co-infection of GoAstV and GPV increases the severity of gout (Liu et al., 2020a). Additionally, it was found that GPV and GoCV caused co-infections in ducks (Liu et al., 2020b). All 3 viruses cause severe diarrhea in geese. Therefore, it is necessary to develop a rapid, specific, and accurate method for the simultaneous detection of GoAstV, GPV, and GoCV, as this will be a useful tool for epidemiological research and disease control.
In the present study, a TaqMan-probe-based multiplex reverse transcription-qPCR (RT-qPCR) method was established and optimized for simultaneous detection of these 3 viruses.
MATERIALS AND METHODS
Primers and Probes
GoAstV strain AH/2018 (accession number: MN099162), GPV strain WX3 (accession number: MK333463), and GoCV strain AH/2019 (accession number: MN756799) were isolated from the liver and kidney tissues of diseased geese from Anhui Province. Primers and TaqMan probes used for real-time qPCR detection were designed using Primer Premier 5 (Premier Biosoft Intl., Canada) and Beacon Designer 7 software (America). The primers and probes used are listed in Table 1.
Table 1.
Primers and probes designed for real-time reverse transcription quantitative polymerase chain reaction.
| Virus | Primer/Probe | Sequence(5’-3’) | Target gene | Size(bp) |
|---|---|---|---|---|
| GoAstVVV | Forward | ACAGCAGGATTTATCAGA | ||
| Reverse | CCTTGTCCAGTTGTATTC | ORF2 | 109 | |
| Probe | FAM-CACCGAACACGCCACTACA-BHQ1 | |||
| GPV | Forward | TGAAGAAGTTCCTTTCCATA | ||
| Reverse | CTGCTGTCTACCTCATTG | VP3 | 108 | |
| Probe | CY5-CTCATTCACAGGACTTAGACAGGC-BHQ2 | |||
| GoCV | Forward | TACCTTCGAGTCGGTGAA | ||
| Reverse | TGACGCAATCGCTCTAAG | Rep | 155 | |
| Probe | HEX-AGGTCGCTCCAGTGATCTCT-BHQ1 |
Duck plague virus (DPV, AV122 vaccine strain), duck circovirus (DCV, accession number: MY646346), novel goose parvovirus (NGPV, accession number: MT646163), duck Tembusu virus (DTMUV, accession number: KM102539.1), duck hepatitis A virus (DHAV-1, accession number: KU923754.1), duck hepatitis B virus (DHBV, accession number: MZ048741), and avian adenovirus 4 (FADV-4, accession number: MN781665) were preserved in our laboratory.
A total of 85 waterfowl tissue samples with clinical features such as uric acid deposition and enteritis were collected and analyzed from goose farms in different areas of Anhui Province, China. All samples were stored at –80°C until used.
DNA/RNA Nucleic Acid Extraction and Reverse Transcription
Tissue samples preserved at −80°C were added to phosphate buffered saline (PBS) at a ratio of 1:5 and were homogenized in a mortar. After repeated freeze thaw cycles (3 times) and centrifugation at 10,000 × g for 10 min, nucleic acid was extracted from the supernatant using the TIANamp Virus DNA/RNA Kit (Tiangen, Beijing, China). According to the manufacturer's instructions, the GoAstV RNA genome was reverse-transcribed using the FastKing cDNA First-Strand Synthesis Kit (Tiangen). After identification on PCR testing and gene sequencing, the viral genome cDNA was stored at –80°C before use.
Construction, Culture, and Purification of Standard Plasmids
The primers and probes used in this study were synthesized by China General Biology Co, Ltd. (Anhui, China). After amplification using a conventional PCR, reaction conditions were as follows: GoAstV: predenaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 58.7°C for 30 s, extension at 72°C for 10 min, 40 cycles; GPV: predenaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 59.3°C for 30 s, extension at 72°C for 10 min, 40 cycles; GoCV: predenaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 59.0°C for 30 s, extension at 72°C for 10 min, 40 cycles. The ORF2, VP3, and Rep genes of GoAstV, GPV, and GoCV were inserted into the pMD19-T cloning vector (Takara Biotechnology Company, Dalian, China). The reaction mixture included 4.5 μL of the target gene, 0.5 μL of the pMD19-T clone vector, and 5 μL of Solution I. Ligation proceeded at 16°C for 2 to 3 h.
The constructed recombinant plasmid was then transformed into TrerisDH5α chemoreceptor cells (Qingdao, China). Of the DH5α cells, 30 μL was added to 5 μL of the recombinant plasmid solution. Samples were placed in an ice bath for 30 min and then heat-shocked for 90 s at 42°C. Thereafter, 600 μL liquid LB medium was added and the samples were incubated at 37°C for 150 s. Then, after shaking at 220 r/min for 50 min at 37°C, samples were centrifuged at 6,000 r/min for 3 min; 500 μL supernatant was discarded, and the pellet was resuspended in the remaining 100 μL. The culture was sterile in a solid medium containing ampicillin (Amp) and incubated overnight at 37°C. Single colonies were selected and inoculated in LB liquid medium containing Amp for undergoing shaking culture at 180 rpm at 37°C for 4 h. Then, a small scale plasmid extraction kit (Beyotime Biotechnology, Hiamen, China) was used for plasmid extraction according to the manufacturer's instructions.
Construction of Standard Curves
A NanoVue spectrophotometer (GE Healthcare, Chicago, IL) was used to measure the concentration of the recombinant plasmid, and the copy number of the recombinant plasmid was calculated using the following formula:
To establish a standard curve for individual goose viruses, each plasmid was diluted 10-fold from 107 to 101 copies/μL.
Singleplex and Multiplex Real-Time Fluorescence qPCR
All fluorescence real-time RT-qPCR assays were performed in 20 μL reaction volumes. For amplification of GoAstV, GPV, and GoCV with singleplex fluorescence real-time RT-qPCR, the reaction mixtures contained 10 μL of 2 × TaqMan Fast qPCR premix (Sangon Biotech, Shanghai, China), 0.4 μL of the upstream and downstream primers, 0.2 μL probe, 1 μL plasmid template, and sterilized ddH2O to a final volume of 20 μL. All reactions were performed on a LightCycler 96 RT-qPCR instrument (Roche; Basel, Switzerland). The reaction conditions were as follows: predenaturation at 94°C for 3 min, denaturation at 94°C for 5 s, and annealing and extension at 60°C for 30 s, for 40 cycles.
For multiplex RT-qPCR, 2 × TaqMan Fast qPCR premix (10 μL) was combined with all primers, probes, templates, and sterile ddH2O, as for singleplex reactions, to a final volume of 20 μL. The concentrations of each primer and probe for GoAstV, GPV, and GoCV were optimized to achieve a better yield. The number of amplification cycles for multiplex RT-qPCR was the same as that for singleplex RT-qPCR.
Sensitivity, Specificity, and Repeatability of Multiplex RT-qPCR
To analyze the sensitivity of the established multiplex RT-qPCR assay, the linearized standard plasmids prepared above were serially diluted 10 times in sterile ddH2O. The final concentrations ranged between 107 and 101 copies/μL. These diluted standard plasmids were used as templates for RT-qPCR amplification.
To evaluate the specificity of the established multiplex RT-qPCR, the cDNA of seven other waterfowl viruses, DPV, DCV, NGPV, DTMUV, DHAV-1, DHBV, and FADV-4, were used as amplification templates. Water without nuclease was used as the negative control template.
To evaluate repeatability, a 10-fold serial dilution of a standard template was used to detect the coefficient of variation (R2) of RT-qPCR. All tests were repeated three times to assess repeatability within the test. To evaluate repeatability between tests, the tests were repeated three times at different locations.
Sample Testing
A total of 85 visceral samples from different areas of Anhui Province, China, were collected from geese experiencing diarrhea, enteritis, and uric acid deposition. All samples were diluted with sterile PBS and homogenized in a mortar. After three freeze–thaw cycles, samples were centrifuged at 10,000 × g for 10 min. Nucleic acid was extracted from the supernatant using the TIANamp Virus DNA/RNA Kit. According to the manufacturer's instructions, the RNA genome in the tissue of the diseased material was reverse-transcribed using the FastKing cDNA First-Strand Synthesis Kit (Tiangen, Beijing, China), and the extracted total DNA was used as the template. The cDNAs of all clinical samples were detected using the multiplex RT-qPCR assay developed in this study.
Results
Single Real-Time Quantitative RT-qPCR Detection of a Single Virus
To develop a multiplex real-time RT-qPCR assay, singleplex real-time RT-qPCR assays were first established with different fluorescent-labeled target probes (GoAstV gene: FAM, GPV gene: Cy5, and GoCV gene: HEX). A NanoVue spectrophotometer was used to measure the concentration of the recombinant plasmid, and the copy number of each viral plasmid was calculated as 2.6 × 1010 for GoAstV, 2.8 × 1010 for GPV, and 2.6 × 1010 for GoCV. The standard curve for each virus was established using a 10-fold serial dilution of the linearized plasmid, representing 108 to 101 copies of the target gene. The singleplex real-time RT-qPCR assay was successfully established for each virus, with a detection limit of approximately 10 copies. All the standard curves showed good correlation coefficients and amplification effects, with a good linear relationship between the cyclic threshold (Ct) and the logarithm of plasmid copy number. The coefficient of variation (R2) value and amplification efficiency were 0.9961 and 96% (GoAstV), 0.9965 and 81% (GPV), and 0.996 and 83% (GoCV), respectively. The standard curve of GoAstV was indicated by y = -3.4135x + 35.027 (Figure 1), that of GPV by y =-3.8593x + 38.377 (Figure 2), and that of GoCV by y = -3.8058x + 39.945 (Figure 3). This indicates that the single real-time RT-qPCR assay for each virus was effective and reliable.
Figure 1.
Standard curve analysis of a standard plasmid containing a gene of goose circovirus (A) Concentrations ranging from 2.60 × 108 to 2.60 × 101 copies/μL; (B) y = -3.4135x + 35.027; the coefficient of variation (R2) = 0.9961; Eff = 96%.
Figure 2.
Standard curve analysis of a standard plasmid containing a gene of goose parvovirus (A) Concentrations ranging from 2.80 × 108 to 2.80 × 101 copies/μL; (B) y = −3.8593x + 38.377; the coefficient of variation (R2) = 0.9965; Eff = 81%.
Figure 3.
Standard curve analysis of a standard plasmid of goose circovirus (A) Concentrations ranging from 2.60 × 108 to 2.60 × 101 copies/μL; (B) y = −3.8058x + 39.945; the coefficient of variation (R2) = 0.996; Eff = 83%.
Establishment of a Multiplex Real-Time Fluorescence RT-qPCR Detection Method
To establish a multiplex real-time RT-qPCR method, all primers, probes, and serially diluted plasmid standards for GoAstV, GPV, and GoCV were mixed with 2 × TaqMan Fast qPCR premix and sterile ddH2O in a reaction system of 20 μL. Finally, the concentrations of each primer and probe were optimized. The optimal concentration of primers and probes were as follows: GoAstV, 1.4 μL primer and 0.6 μL probe; GPV, 0.4 μL primer and 0.2 μL probe; and GoCV, 0.2 μL primer and 0.1 μL probe. The results showed that multiplex RT-qPCR could efficiently detect the target genes of all three viruses with high correlation values. All standard curves showed good correlation coefficients and amplification effects, as follows: GoAstV, R2 = 0.999, Eff % = 178.0; GPV, R2 = 0.997, Eff% = 200.0, and GoCV, R2 = 0.994, Eff% = 161.0. This multiplex RT-qPCR assay had a detection limit of approximately 100 copies per virus per reaction.
Specificity of Multiplex Real-Time Fluorescence qRT-qPCR Detection
To evaluate the specificity of this multiplex RT-qPCR assay, we used DNA/cDNA templates from seven other major waterfowl viruses as templates for amplification. The cDNA of GoAstV, GPV, and GoCV was used as a positive control, and nuclease-free water was used as a negative control. The GoAstV, GPV, and GoCV cDNAs were successfully detected. However, no positive signals were detected for DPV, DCV, NGPV, DTMUV, DHAV-1, DHBV, and FADV-4, and the negative control, indicating the high specificity of the multiplex RT-qPCR system based on TaqMan probes, as shown in Figure 4.
Figure 4.
Specificity of real-time reverse transcription-quantitative polymerase chain reaction using multiple TaqMan probes Blue, red, and black indicate positive templates of GoAstV, GPV, and GoCV, respectively. There was no specific curve for duck plague virus, duck circovirus, novel goose parvovirus, duck Tembusu virus, duck hepatitis A virus, duck hepatitis B virus, avian adenovirus 4, and double-distilled water.
Sensitivity of Multiplex Real-Time Fluorescence RT-qPCR Detection
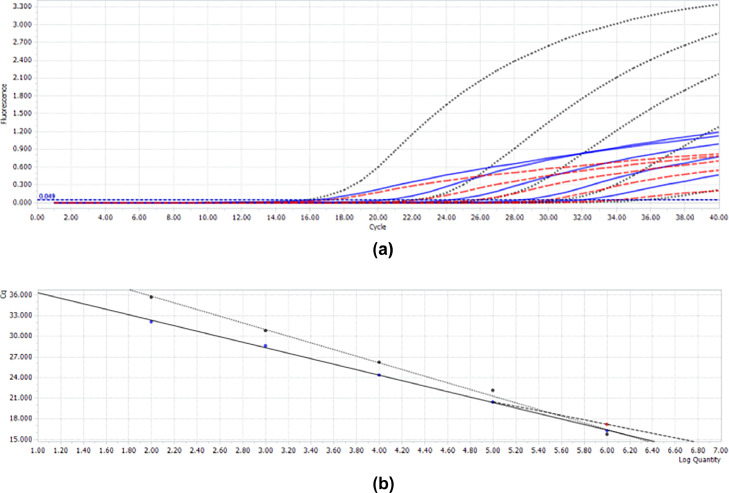
To determine the sensitivity of the multiplex real-time RT-qPCR, 3 linearized plasmids were 10-fold serially diluted and used in the multiplex assay. The results showed that 102 copies of GoAstV, GPV, and GoCV could be detected simultaneously, with Ct values of 32.08, 34.04, and 35.66, respectively. However, dilutions containing 101 copies of each target were not detected using the same amplification system, as shown in Figure 5 and Table 2.
Figure 5.
Establishment of multiple real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (A) Under optimal amplification conditions, the optimized goose astrovirus (GoAstV), goose parvovirus (GPV), and goose circovirus (GoCV) amplification curves were generated using real-time RT-qPCR with TaqMan probes. Blue indicates the standard curve for GoAstV; red for, GPV; and black for, GoCV. (B) Concentrations ranging from 2.80 × 106 to 2.80 × 102 copies/μL; y = −3.998x + 40.28; the coefficient of variation (R2) = 0.9991; Eff = 178.0%; concentrations ranging from 2.80 × 106 to 2.80 × 102 copies/μL; y = −3.322x + 37.08; R2 = 0.9973; Eff = 200.0%, and concentrations ranging from 2.80 × 106 to 2.80 × 102 copies/μL; y = −4.853x + 45.50; R2 = 0.9937; Eff = 161.0%, respectively.
Table 2.
Sensitivity of multiple reverse transcription quantitative polymerase chain reaction.
| Templates (copies/gene) | Cq Value |
||
|---|---|---|---|
| GAstV | GPV | GoCV | |
| 106 | 16.23 | 17.15 | 15.71 |
| 105 | 20.41 | 22.24 | 22.23 |
| 104 | 24.35 | 25.88 | 26.18 |
| 103 | 28.59 | 29.98 | 30.76 |
| 102 | 32.08 | 34.04 | 35.66 |
| 101 | ND | ND | ND |
| NTC | ND | ND | ND |
Repeatability of Multiplex Real-Time RT-qPCR Detection
To estimate both intra- and inter-assay reproducibility of the multiplex RT-qPCR assay, a 10-fold serial dilution of pooled linearized plasmids was analyzed in triplicate. For intra-assay reproducibility tests, standard plasmids were simultaneously amplified in replicate. For inter-assay reproducibility tests, standard curves were obtained at three different times, using a different batch of the standard templates. As shown in Tables 3, 4, and 5, GoAstV intra-assay test of R2 was 0.07 to 0.85% and 0.10 to 0.84% for the inter-assay test. GPV amplification exhibited coefficients of variation of 0.23 to 1.69% and 0.27 to 1.62% for the inter-assay and intra-assay tests, respectively. GoCV amplification exhibited coefficients of variation of 0.52 to 1.40% and 0.42 to 2.03% for the inter- and intra-assay tests, respectively. These results indicated that the TaqMan-probe-based multiplex real-time RT-qPCR assay established in this study is both repeatable and reliable.
Table 3.
Intra- and inter-assay coefficients of variation of goose astrovirus.
| Standard copies/ul | Intra-assay variability |
Inter-assay variability |
||||
|---|---|---|---|---|---|---|
| Mean | SD | CV(%) | Mean | SD | CV(%) | |
| 2.6 × 106 | 16.26 | 0.026 | 0.16 | 16.22 | 0.021 | 0.10 |
| 2.6 × 105 | 20.52 | 0.015 | 0.07 | 20.32 | 0.171 | 0.84 |
| 2.6 × 104 | 24.30 | 0.210 | 0.85 | 24.20 | 0.031 | 0.15 |
| 2.6 × 103 | 28.34 | 0.031 | 0.11 | 28.30 | 0.056 | 0.20 |
| 2.6 × 102 | 32.64 | 0.025 | 0.08 | 32.25 | 0.215 | 0.67 |
| 2.6 × 101 | ND | ND | ND | ND | ND | ND |
| NTC | ND | ND | ND | ND | ND | ND |
Table 4.
Intra- and inter-assay coefficients of variation of goose parvovirus.
| Standard copies/uL | Intra-assay variability |
Inter-assay variability |
||||
|---|---|---|---|---|---|---|
| Mean | SD | CV(%) | Mean | SD | CV(%) | |
| 2.8 × 106 | 17.19 | 0.040 | 0.23 | 17.2 | 0.046 | 0.27 |
| 2.8 × 105 | 22.22 | 0.071 | 0.32 | 22.13 | 0.188 | 0.85 |
| 2.8 × 104 | 25.77 | 0.100 | 0.32 | 25.58 | 0.359 | 1.40 |
| 2.8 × 103 | 29.40 | 0.499 | 1.69 | 29.69 | 0.482 | 1.62 |
| 2.8 × 102 | 33.95 | 0.096 | 0.28 | 34.12 | 0.202 | 0.59 |
| 2.8 × 101 | ND | ND | ND | ND | ND | ND |
| NTC | ND | ND | ND | ND | ND | ND |
Table 5.
Intra- and inter-assay coefficients of variation of goose circovirus.
| Standard copies/uL | Intra-assay variability |
Inter-assay variability |
||||
|---|---|---|---|---|---|---|
| Mean | SD | CV(%) | Mean | SD | CV(%) | |
| 2.6 × 106 | 15.90 | 0.168 | 1.05 | 16.03 | 0.325 | 2.03 |
| 2.6 × 105 | 22.11 | 0.131 | 1.40 | 21.95 | 0.401 | 1.83 |
| 2.6 × 104 | 26.18 | 0.135 | 0.52 | 26.19 | 0.111 | 0.42 |
| 2.6 × 103 | 30.45 | 0.311 | 1.02 | 30.57 | 0.163 | 0.53 |
| 2.6 × 102 | 33.71 | 0.295 | 0.88 | 33.65 | 0.335 | 0.99 |
| 2.6 × 101 | ND | ND | ND | ND | ND | ND |
| NTC | ND | ND | ND | ND | ND | ND |
Sample Testing
A total of 85 tissue samples from waterfowl showing uric acid deposition and enteritis were collected from goose farms in different regions of Anhui Province, China, and analyzed using our multiplex RT-qPCR assay as well as conventional PCR. As shown in Table 6, the positivity rate for GoAstV was 42.35% (36/85) on real-time fluorescence RT-qPCR testing and 15.29% (13/85) on conventional PCR testing. The positivity rate for GPV was 56.47% (48/85) on real-time fluorescence RT-qPCR and 41.18% (35/85) on conventional PCR testing. The positivity rate for GoCV was 61.18% (52/85) on real-time fluorescence RT-qPCR and 44.71% (38/85) on conventional PCR testing. The combined infection rates of GoAstV and GPV were 28.24% (24/85) and 17.65% (15/85), respectively. The combined infection rates of GPV and GoCV were 23.53% (20/85) and 11.76% (10/85), respectively, and the combined infection rates of GoAstV and GoCV were 21.18% (18/85) and 5.88% (5/85), respectively. The joint infection rates of 3 viruses were 10.59% (9/85) and 4.71% (4/85).
Table 6.
Positive rate of goose astrovirus (GoAstV), goose parvovirus (GPV) and goose circovirus (GoCV) in clinical samples by quantitative real-time polymerase chain reaction (qPCR) and conventional PCR (cPCR).
| Positive rate (༅) |
|||
|---|---|---|---|
| Virus | Total clinical samples | qPCR | cPCR |
| GoAstV | 85 | 42.35% (36/85) | 15.29% (13/85) |
| GPV | 85 | 56.47% (48/85) | 41.18% (35/85) |
| GoCV | 85 | 61.18% (52/85) | 44.71% (38/85) |
| GoAstV and GPV co-infection | 85 | 28.24% (24/85) | 17.65% (15/85) |
| GPV and GoCV co-infection | 85 | 23.53% (20/85) | 11.76% (10/85) |
| GoAstV and GoCV co-infection | 85 | 21.18% (18/85) | 5.88% (5/85) |
| GoAstV, GPV and GoCV co-infection | 85 | 10.59% (9/85) | 4.71% (4/85) |
DISCUSSION
We developed a TaqMan probe-based multiplex RT-qPCR assay that allows simultaneous detection of three viruses that adversely affect the health of geese (GoAstV, GPV, and GoCV). This assay could detect a minimum of 100 copies of each virus. The amplification efficiency as 80 to 100%, and the assay showed high sensitivity, specificity, and repeatability.
Astroviruses were first reported in feces of human infants in 1975. In November 2016, the virus was found in dead poultry in Shandong, Jiangsu, and Anhui provinces in China. Astroviruses mainly cause acute gastroenteritis in humans and animals alike and, occasionally, cause encephalitis. Astrovirus infection in poultry is associated with many diseases. The rate of infection was usually high (up to 80%), and the mortality rates reached a peak of 50% (Niu et al., 2018). Additionally, severe urate deposition was found in the viscera of dead goslings infected with Astrovirus (Yin et al., 2021). Goose parvovirus, first identified in Hungary in 1967, causes an acute, highly contagious, and fatal disease commonly known as gosling plague (Derzsy, 1967; Gough et al., 2005). Goose circovirus can cause lymphocyte loss and immunosuppression and is also linked to the outbreak of many diseases (Soike et al., 1999).
Studies by Liu and Hong et al. reported cases of natural co-infection of GPV and GoAstV in goslings in the middle east region of China (Liu et al., 2019; Liu et al., 2020a). Chiu et al. reported co-infection of GoCV and GPV in southern Taiwan, China (Ting et al., 2021). Various methods for the detection of GoAstV, GPV, and GoCV such as loop-mediated isothermal amplification (Yang et al., 2010; Wozniakowski et al., 2012; He et al., 2020), enzyme-linked immunosorbent assay (Fan et al., 2013), colloidal gold immunochromatography (Yu et al., 2018), and real-time quantitative PCR (Wan et al., 2019; Yang et al., 2020; Yi et al., 2022) have been established. Among these detection methods, real-time PCR has high specificity and sensitivity. To date, no studies have reported simultaneous co-infection of GoAstV, GPV, and GoCV. This study aimed to develop a method for the simultaneous detection of GoAstV, GPV, and GoCV for epidemiological investigations and disease control.
Real-time fluorescence RT-qPCR using TaqMan probes is a rapid, highly specific, highly sensitive, and reproducible tool for virus identification and detection. To establish a highly specific multiplex real-time RT-qPCR for the detection and differential diagnosis of gosling enteroviruses, we designed primers and TaqMan probes targeting highly conserved regions of GoAstV, GPV, and GoCV genes, based on bioinformatics analysis of each virus. We demonstrated that each primer set and probe could only detect the target gene and not any other target, indicating high specificity. In singleplex real-time RT-qPCR, our TaqMan probe sets had a lower limit of detection of 10 copies of GoAstV, GPV, and GoCV gene templates. However, the detection limit for each target gene in our multiplex real-time RT-qPCR assay was approximately 100 copies, indicating that the sensitivity of multiple real-time RT-qPCR was lower than that of the corresponding singleplex real-time RT-qPCR, which may be owing to competition among primers, probes, templates, and reagents. Multiplex RT-qPCR analyses of clinical diarrhea samples revealed that GoAstV and GPV were the main co-infections in geese in the sampled region.
In conclusion, we developed a multiplex real-time RT-qPCR assay using TaqMan probes and demonstrated high specificity and sensitivity for simultaneous detection and differential diagnosis of GoAstV, GPV, and GoCV infections. This real-time RT-qPCR assay is of great significance for the prevention and treatment of gosling enterovirus diseases and epidemiological investigations.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
This work was supported financially by the Hefei Rural Revitalization Science and Technology Expert Assistance Team.
Compliance With Ethical Standards: All experiments were compliant with the ethical standards of Anhui Agricultural University.
Author Contributions: Ju Yu and Junwei Zou conceived of the study, carried out the experiment and drafted the manuscript. Xuan Liu, Ying Pan, Yuanyuan Mu and Shuyan Li participated in the data collection and analysis. Juhua Wang and Fazhi Xu participated in statistical analysis. Yong Wang conceived of the study, revising the manuscript critically. All authors have read and approved the final manuscript.
Disclosures
The authors declare that they have no conflict of interest.
References
- An D., Zhang J., Yang J., Tang Y., Diao Y. Novel goose-origin astrovirus infection in geese: the effect of age at infection. Poult. Sci. 2020;99:4323–4333. doi: 10.1016/j.psj.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C.F., DuBois R.M. The astrovirus capsid: a review. Viruses. 2017;9:15. doi: 10.3390/v9010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang B., Yan M., Diao Y., Tang Y. First report of a novel goose astrovirus outbreak in Cherry Valley ducklings in China. Transbound. Emerg. Dis. 2020;67:1019–1024. doi: 10.1111/tbed.13418. [DOI] [PubMed] [Google Scholar]
- Deng S.X., Cai M.S., Cui W., Huang J.L., Li M.L. Evaluation of the immune response in Shitou geese (Anser anser domesticus) following immunization with GPV-VP1 DNA-based and live attenuated vaccines. Vet Q. 2014;34:180–184. doi: 10.1080/01652176.2014.966173. [DOI] [PubMed] [Google Scholar]
- Derzsy D. A viral disease of goslings. I. Epidemiological, clinical, pathological and aetiological studies. Acta Vet. Acad. Sci. Hung. 1967;17:443–448. [PubMed] [Google Scholar]
- Fan J.H., Zuo Y.Z., Yang Z., Pei L.H. The development of an indirect ELISA for the detection of antibodies to goose parvovirus in blood serum. Lett. Appl. Microbiol. 2013;57:26–32. doi: 10.1111/lam.12070. [DOI] [PubMed] [Google Scholar]
- Gough D., Ceeraz V., Cox B., Palya V., Mato T. Isolation and identification of goose parvovirus in the UK. Vet. Rec. 2005;156:424. doi: 10.1136/vr.156.13.424. [DOI] [PubMed] [Google Scholar]
- He D., Yang J., Jiang X., Lin Y., Chen H., Tang Y., Diao Y. A quantitative loop-mediated isothermal amplification assay for detecting a novel goose astrovirus. Poult. Sci. 2020;99:6586–6592. doi: 10.1016/j.psj.2020.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Hu D., Zhu Y., Xiong H., Lv X., Wei C., Liu M., Yin D., He C., Qi K., Wang G. Coinfection of parvovirus and astrovirus in gout-affected goslings. Transboundary Emerg. Dis. 2020;67:2830–2838. doi: 10.1111/tbed.13652. [DOI] [PubMed] [Google Scholar]
- Liu J., Yang X., Hao X., Feng Y., Zhang Y., Cheng Z. Effect of goose parvovirus and duck circovirus coinfection in ducks. J. Vet. Res. 2020;64:355–361. doi: 10.2478/jvetres-2020-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zhao Y., Hu D., Huang X., Xiong H., Qi K., Liu H. Clinical and histologic characterization of co-infection with astrovirus and goose parvovirus in goslings. Avian Dis. 2019;63:731–736. doi: 10.1637/aviandiseases-D-19-00110. [DOI] [PubMed] [Google Scholar]
- Luo Q., Chen B., Xu J., Ma W., Lao C., Li Y., Tan J., Tang Y., Huang C., Liu W., Chen Z. Development of a SYBR green II real-time polymerase chain reaction for the clinical detection of the duck-origin goose parvovirus in China. Intervirology. 2018;61:230–236. doi: 10.1159/000495181. [DOI] [PubMed] [Google Scholar]
- Niu X., Tian J., Yang J., Jiang X., Wang H., Chen H., Yi T., Diao Y. Novel goose astrovirus associated gout in Gosling, China. Vet. Microbiol. 2018;220:53–56. doi: 10.1016/j.vetmic.2018.05.006. [DOI] [PubMed] [Google Scholar]
- Rosario K., Breitbart M., Harrach B., Segalés J., Delwart E., Biagini P., Varsani A. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 2017;162:1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- Scott A.N., Beckett A., Smyth J.A., Ball N.W., Palya V., Todd D. Serological diagnosis of goose circovirus infections. Avian Pathol. 2006;35:495–499. doi: 10.1080/03079450601087841. [DOI] [PubMed] [Google Scholar]
- Shao H., Lv Y., Ye J., Qian K., Jin W., Qin A. Isolation of a goose parvovirus from swan and its molecular characteristics. Acta Virol. 2014;58:194–198. doi: 10.4149/av_2014_02_194. [DOI] [PubMed] [Google Scholar]
- Soike D., Kohler B., Albrecht K. A circovirus-like infection in geese related to a runting syndrome. Avian Pathol. 1999;28:199–202. doi: 10.1080/03079459994939. [DOI] [PubMed] [Google Scholar]
- Stenzel T., Dziewulska D., Muhire B.M., Hartnady P., Kraberger S., Martin D.P., Varsani A. Recombinant goose circoviruses circulating in domesticated and wild geese in Poland. Viruses. 2018;10:107. doi: 10.3390/v10030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C.H., Lin C.Y., Huang Y.C., Liu S.S., Peng S.Y., Wang C.W., Wu H.Y. Correlation between goose circovirus and goose parvovirus with gosling feather loss disease and goose broke feather disease in southern Taiwan. J. Vet. Sci. 2021;22:e1. doi: 10.4142/jvs.2021.22.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C., Chen C., Cheng L., Liu R., Shi S., Fu G., Chen H., Fu Q., Huang Y. Specific detection and differentiation of classic goose parvovirus and novel goose parvovirus by TaqMan real-time PCR assay, coupled with host specificity. BMC Vet Res. 2019;15:389. doi: 10.1186/s12917-019-2090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniakowski G., Kozdrun W., Samorek-Salamonowicz E. Loop-mediated isothermal amplification for the detection of goose circovirus. Virol. J. 2012;9:110. doi: 10.1186/1743-422X-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yang R., Cheng A., Wang M., Fu L., Yang S., Zhang S., Yang L., Xu Z. A simple and rapid method for detection of Goose Parvovirus in the field by loop-mediated isothermal amplification. Virol. J. 2010;7:14. doi: 10.1186/1743-422X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.K., Yin D.D., Xu L., Liang Y.Q., Tu J., Song X.J., Shao Y., Liu H.M., Qi K.Z. A TaqMan-based quantitative real-time PCR assay for identification of the goose circovirus. Mol. Cell Probes. 2020;52 doi: 10.1016/j.mcp.2020.101564. [DOI] [PubMed] [Google Scholar]
- Yi Z., Ding R., Cao R., Sun W., Sun M., Dong Y., Rehana B., Bao E., Lv Y. Development of a duplex TaqMan real-time RT-PCR assay for simultaneous detection of goose astrovirus genotypes 1 and 2. J. Virol. Methods. 2022;306 doi: 10.1016/j.jviromet.2022.114542. [DOI] [PubMed] [Google Scholar]
- Yin D., Tian J., Yang J., Tang Y., Diao Y. Pathogenicity of novel goose-origin astrovirus causing gout in goslings. BMC Vet Res. 2021;17:40. doi: 10.1186/s12917-020-02739-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Ma X., Sheng Z., Qi L., Liu C., Wang D., Huang B., Li F., Song M. Identification of goose-origin parvovirus as a cause of newly emerging beak atrophy and dwarfism syndrome in ducklings. J. Clin. Microbiol. 2016;54:1999–2007. doi: 10.1128/JCM.03244-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wei L., Chen H., Niu X., Dou Y., Yang J., Wang Z., Tang Y., Diao Y. Development of colloidal gold-based immunochromatographic assay for rapid detection of goose parvovirus. Front. Microbiol. 2018;9:953. doi: 10.3389/fmicb.2018.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zhu C., Zheng X., He S., Liu X. Genome analysis and epidemiological investigation of goose circovirus detected in eastern China. Virus Genes. 2007;35:605–609. doi: 10.1007/s11262-007-0112-1. [DOI] [PubMed] [Google Scholar]