Abstract
Acute respiratory distress syndrome (ARDS) in COVID-19 has been associated with catastrophic inflammation. We present measurements in humans and a new animal model implicating a role in danger-associated molecular patterns. Calprotectin (S100A8/A9) and high-mobility group box 1 (HMGB1) were measured in patients without/with ARDS, and admission calprotectin was associated with soluble urokinase plasminogen activator receptor (suPAR). An animal model was developed by intravenous injection of plasma from healthy or patients with COVID-19 ARDS into C57/BL6 mice once daily for 3 consecutive days. Mice were treated with one anti-S100A8/A9 antibody, the IL-1 receptor antagonist anakinra or vehicle, and Flo1-2a anti-murine anti-IL-1α monoclonal antibody or the specific antihuman IL-1α antibody XB2001 or isotype controls. Cytokines and myeloperoxidase (MPO) were measured in tissues. Calprotectin, but not HMGB1, was elevated in ARDS. Higher suPAR indicated higher calprotectin. Animal challenge with COVID-19 plasma led to inflammatory reactions in murine lung and intestines as evidenced by increased levels of TNFα, IL-6, IFNγ, and MPO. Lung inflammation was attenuated with anti-S100A8/A9 pre-treatment. Anakinra treatment restored these levels. Similar decrease was found in mice treated with Flo1-2a but not with XB2001. Circulating alarmins, specifically calprotectin, of critically ill COVID-19 patients induces tissue-specific inflammatory responses through an IL-1-mediated mechanism. This could be attenuated through inhibition of IL-1 receptor or of IL-1α.
Keywords: Calprotectin, Tumor necrosis factor, Interleukin-6, Interleukin-1, Soluble urokinase plasminogen activator receptor, Acute respiratory distress syndrome, Severe respiratory failure, Coronavirus diseases 2019
Introduction
As of April 2021, more than 2.3 million deaths have been attributed to the virus (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). Efforts are ongoing to elucidate what drives progression in COVID-19 cases from lower respiratory tract infections (LRTIs) to acute respiratory distress syndrome (ARDS). We have suggested that the deterioration from pneumonia to ARDS is the result of a complex immune dysregulation, which in about 25% of patients appears to be related to a macrophage activation like syndrome; in the remaining 75% of patients, the mechanism seems to involve an interleukin (IL)-6 receptor-dependent dysregulation, with an associated lymphopenia and a subset of cytokine overproducing monocytes that show decreased human leukocyte antigen-DR expression [1].
Recent data suggest that SARS-CoV-2 stimulates vicious pro-inflammatory responses inducing NLRP3 inflammasome activation in monocytes and macrophages [2]. In the open-label phase II study SAVE [3], patients with LRTI caused by SARS-CoV-2 and showing plasma concentrations of the biomarker soluble urokinase plasminogen activator receptor (suPAR) of 6 ng/mL or more were treated with the recombinant human IL-1 receptor antagonist anakinra. Anakinra is able to block both IL-1α and IL-1β bioactivity at the level of their common receptor [4] and prevent the development of ARDS.
The results of the SAVE trial led us to hypothesize that LRTI caused by SARS-CoV-2 can progress to ARDS. In this hypothesis, as a result of infection by SARS-CoV-2, patients overproduce IL-1α and other danger-associated molecular patterns (DAMPs) from their lung parenchyma. DAMPs trigger the release of IL-1β and in combination with IL-1α evoke strong innate immune pro-inflammatory responses which lead to ARDS [4]. In order to test our hypothesis, we used a three-step approach. In the first step, we measured and found higher concentrations of the DAMP calprotectin in the plasma of patients with ARDS than patients without ARDS. In the second step, we measured and found that DAMP concentrations in the plasma of patients with suPAR more than 6 ng/mL were significantly higher than that in patients with suPAR less than 6 ng/mL. In the third step, we studied a COVID-19-like animal model, where we prevented the development of strong pro-inflammatory responses through inhibition of IL-1 signaling via anakinra or anti-IL-1α antibodies.
Patients and Methods
Clinical Study
Plasma samples from a cohort of 60 patients with and without ARDS that was determined to be caused by SARS-CoV-2 and from 40 participants in the suPAR-guided Anakinra treatment for Validation of the risk and Early management of severe respiratory failure by COVID-19 (SAVE) trial were analyzed. Patients were enrolled after written informed consent was provided by themselves or by first-degree relatives in case of patients were unable to consent. All patients were adults of either sex with positive molecular testing of SARS-CoV-2 from respiratory secretions and with LRTI defined by the presence of diffuse infiltrates by chest X-ray or computed tomography of the chest. Exclusion criteria were (i) HIV-1 infection and (ii) neutropenia defined as less than 1,000 neutrophils/mm3. Blood was sampled on the day of hospital admission in the emergencies. For patients enrolled with ARDS, blood was collected within 24 h from mechanical ventilation. Control blood samples were collected from healthy volunteers (HVs) matched for age and gender.
The following clinical variables were recorded: (i) demographics; (ii) severity scores, namely Acute Physiology and Chronic Health Evaluation (APACHE) II score, Charlson's Comorbidity Index (CCI), and Sequential Organ Failure Assessment (SOFA) score; (iii) absolute blood cell counts, biochemistry, and blood gases; (iv) and progression into ARDS during the entire hospital stay. ARDS was defined as a ratio of partial oxygen pressure to the fraction of inspired oxygen below 150 mm Hg necessitating mechanical ventilation.
Five milliliters of whole blood were collected into a tube containing ethylenediaminetetraacetic acid and centrifuged. Calprotectin (S100A8/A9), high-mobility group box 1 (HMGB1), suPAR, and ferritin were measured by enzyme immunosorbent assays (InflaRx, Jena, Germany and Creative Diagnostics, Shirley, NY, USA; suPARnostic, ViroGates, Lyngby, Denmark, and ORGENTEC Diagnostika GmbH, Mainz, Germany, respectively); C-reactive protein was measured using a nephelometric assay (Behring, Berlin, Germany). The lowest limits of detection were 0.5 ng/mL for HMGB1, 1.1 ng/mL for suPAR, 0.2 mg/L for C-reactive protein, and 75 ng/mL for ferritin. RNA isolation of SARS-CoV-2 was performed with the QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany). Reverse transcription of the viral RNA into cDNA and enrichment of the sequences of interest via Real-time polymerase chain reaction was performed in the Rotor-Gene Q instrument (QIAGEN) using the SARS-CoV-2 reagent VIASURE (Certest Biotec SL, Zaragoza, Spain).
Animal Model of COVID-19 Pathogenic Inflammation
For the animal experiments, we studied 153 male and female C57Bl6 mice (7–8 weeks old). Mice were allowed to acclimate for 7 days before start of the experiments. They were housed in typical mouse cages, up to 5 mice per cage on a 12-h dark/light cycle and allowed free access to standard dry rodent diet and water. Analgesia was achieved with paracetamol suppositories to avoid interactions with the immune system.
In order to develop a model of COVID-19-like inflammatory illness, mice were treated intravenously (i.v.) with 100 μL of plasma from patients with ARDS due to SARS-CoV-2 infection for 3 consecutive days; control mice were treated with 100 μL of plasma from HVs for 3 consecutive days. On the fourth day, mice were sacrificed by subcutaneous (sc) injection of 300 mg/kg ketamine, followed by cervical dislocation. Under sterile conditions, a midline abdominal incision was performed. Then, segments of the liver, lower lobe of the right lung, right kidney, ileum (ca. 1 cm distally to the pyloric sphincter), and colon (ca. 1 cm proximally of the anal ring) were excised and collected into sterile tubes with 1 mL NaCl 0.9%. The intestinal samples were thoroughly flushed with NaCl 0.9% for removal of feces before storage. The samples were weighed and homogenized. Plasma coming from 10 different patients and six HVs was used for animal challenge. Samples were not pooled, and one sample from one individual was used to challenge one mouse.
In order to study the modulation of innate and adaptive immune responses, peritoneal macrophages and splenocytes were isolated. On animal sacrifice, the peritoneal cavity was washed with 10 mL of cold phosphate-buffered saline containing 2 mmol/L of ethylenediaminetetraacetic acid, and peritoneal cells were retrieved by peritoneal lavage. Segments of spleens were gently squeezed and passed through a sterile filter (250 mm, 12–13 cm, ALT-ERCHEM CO., Athens, Greece) for the collection of splenocytes. After three serial washings, spleen cells and peritoneal cells were counted on Neubauer plates with trypan blue for exclusion of dead cells. Peritoneal cells were incubated at a density of 5 × 106 cells/mL in sterile 24-well plates in RPMI-1640, supplemented with 2 mM glutamine, 100 U/mL of penicillin G, and 0.1 mg/mL of streptomycin, in the absence or presence of 10 ng/mL lipopolysaccharide of Escherichia coli. After 24 h of incubation at 37°C in 5% CO2, the plates were centrifuged, and the supernatants were collected. Splenocytes were incubated at a density of 5 × 106 cells/mL in sterile 24-well plates in RPMI-1640, supplemented with 2 mM glutamine, 10% fetal bovine serum, 100 U/mL of penicillin G, and 0.1 mg/mL of streptomycin in the absence or presence of 5 × 105 cfu/mL heat-killed Candida albicans or 1 μg/mL of recombinant SARS-CoV-2 S1 protein (Sigma-Aldrich, St. Louis, MO, USA). After 5 days of incubation at 37°C in 5% CO2, the plates were centrifuged, and the supernatants were collected. Concentrations of tumor necrosis factor alpha (TNFα), interferon gamma (IFNγ), IL-6, IL-17A, and IL-22 in supernatants from tissue samples and spleen cells were measured in duplicate by an enzyme immunosorbent assay (Thermo Fisher Scientific, Massachusetts, USA) according to the manufacturer's instructions. The lowest detections limits were as follows: for TNFα, 19 pg/mL; for IFNγ, 16 pg/mL; for IL-6, 10 pg/mL; for IL-17A, 8 pg/mL; and for IL-22, 16 pg/mL.
Lung segments were fixed with formalin and embedded in paraffin and tissue sections were stained with hematoxylin and eosin. Lung pathology was scored from 0 to 3 for the degree of infiltration of neutrophils in the alveoli and in the interstitial space and for the degree of thickening of the alveolar septa using the scoring suggested by Aeffner [5]. Scoring of each tissue sample represented the total score of ten different high microscopic power fields. Scoring was done by two expert pathologists, unaware of the treatment group of each animal.
Myeloperoxidase (MPO) activity in all collected tissues was determined. Tissue segments were homogenized with T-PER® (Thermo Fisher Scientific) and centrifuged at 10,000 rpm at 4°C. Then the homogenates were incubated in wells of a 96-well plate at 37°C with 4.2 mM tetramethylbenzidine (Serva, Heidelberg, Germany), 2.5 mM citrate, 5 mM NaH2PO4, and 1.18 mM H2O2 pH 5.0 at a final volume of 150 μL. After 5 min, the reaction was terminated by adding 50 mL 0.18 M H2SO4. Absorbance was read at 450 nm against blank wells. Results were adjusted for tissue sample protein content on Bradford assay (Sigma-Aldrich), and they were expressed as MPO units/mg protein/g.
Experiments were repeated in mice receiving in addition to i.v. plasma: (i) 100 μL sc of 0.9% NaCl and 100 μL intraperitoneally (i.p.) of 0.9% NaCl; (ii) 100 μL sc of 10 mg/kg anakinra (Swedish Orphan Biovitrum, Stockholm, Sweden) and 100 μL i.p. of 0.9% NaCl; (iii) 100 μL sc of 0.9% NaCl and 100 μL i.p. of 2.5 mg/kg dexamethasone [6] (Sigma-Aldrich); or (iv) 100 μL sc of 10 mg/kg anakinra and 100 μL i.p. of 2.5 mg/kg dexamethasone once daily for 3 consecutive days. In a different set of experiments, mice received in addition to i.v. plasma: (i) 100 μL i.p. of 0.9% NaCl; (ii) 100 μL i.p. of 1,000 μg/mL [4], anti-murine IL-1α antibody Flo1-2a or human IL-1α antibody XB2001, or murine isotype control (XBiotech, Austin, TX, USA). In separate experiments, mice were receiving 1 h before each day of i.v. treatment with plasma from patients with ARDS from COVID-19 i.p. either N/S 0.9% or 1.5 mg/kg of MRP8/14 (S100A8/A9) Chicken Polyclonal Antibody (Origene, Rockville, MD, USA). The dose of MRP8/14 (S100A8/A9) Chicken Polyclonal Antibody was selected by previous experiments [7].
To further investigate the mechanism of the hyperinflammation, transcriptomics analysis was done from lung and spleen sections. RNA-seq libraries were prepared with the NebNext Ultra II Directional RNA Library Prep Kit for Illumina. Library size distribution and quality control were performed using the Agilent bioanalyzer DNA1000 chip, and quantitation was done with the qubit HS spectrophotometric method. Between 13 and 63 million 100 bp single-end reads were generated per sample, using the Illumina NovaSeq 6000 sequencer. Bioinformatics analyses were performed using the Galaxy Suite [8]. The quality of sequencing reads was assessed using the FastQC algorithm (Galaxy Version 0.72 + galaxy1). To evaluate RNA integrity at the transcript level, transcript integrity number (Galaxy Version 2.6.4.1) from RseQC package was calculated [9]. Calculation of the distribution of sequencing reads over genomic features was performed using the Read Distribution option (Galaxy Version 2.6.4.1) from RseQC package. FASTQ files were aligned to the mouse genome version mm9 with the use of HISAT2 (Galaxy Version 2.1.0 + galaxy7) [10] using the Reverse Strand parameter. Sequencing reads falling into genes were counted with HTSeq count algorithm (Galaxy Version 0.9.1) [11] using the Union mode and Reverse Stranded options. Differentially expressed genes were identified with the DESeq2 package (Galaxy Version 2.11.40.6 + galaxy1) [12] using a fold change cutoff of 1.5 and adjusted p values less than 0.05.
In separate experiments, mice treated with plasma of HVs or COVID-19 plasma for 3 days were challenged on the fourth-day i.p. with 1 × 107 cfu/mL E. coli or Acinetobacter baumannii. Same experiments were performed with and without treatment with anakinra before bacterial challenge. Survival was recorded every 12 h for 7 days.
Statistics
Categorical data were presented as frequencies and quantitative variables as mean ± SE. Comparisons between groups were done using the Fisher exact test for categorical data. Comparison for quantitative data was performed using the Mann-Whitney U test for two-group comparisons and the one-way ANOVA with the Bonferroni correction for multiple group comparison. Correlations were performed using Spearman's rank of order. Any p value below 0.05 was considered statistically significant.
Results
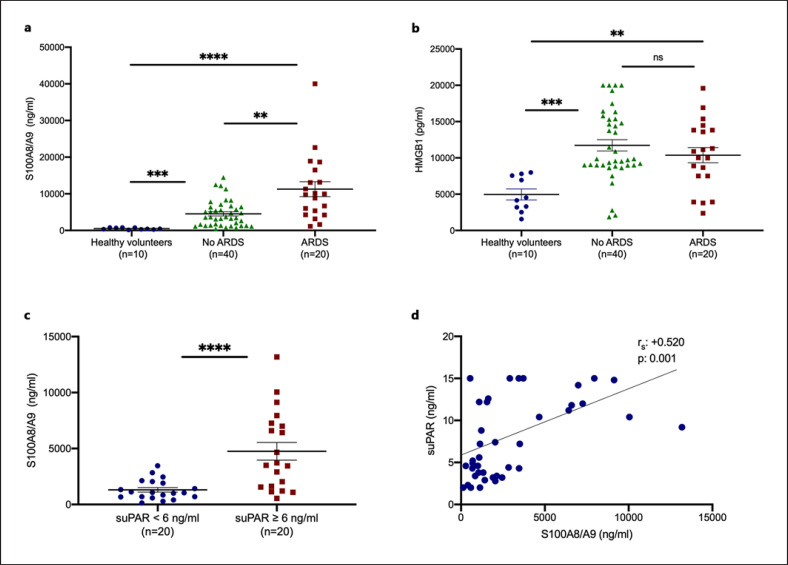
Forty consecutive patients with COVID-19 who did not develop ARDS during their hospital stay and 20 consecutive patients with ARDS were analyzed. Their demographics are shown in Table 1. Samples of 10 HVs were analyzed as healthy controls. Serum calprotectin levels were significantly higher among patients with COVID-19 and ARDS (Fig. 1a). HMGB1 concentrations did not differ between patients with COVID-19 with and without ARDS (Fig. 1b). Elevated calprotectin levels COVID-19-related ARDS raised the question whether calprotectin is released early among patients and may serve as a marker for patients who will progress to ARDS.
Table 1.
Baseline clinical and laboratory characteristics of patients according to the development of ARDS due to pneumonia by the SARS-CoV-2 coronavirus
| No ARDS (n = 40) | ARDS (n = 20) | p value | |
|---|---|---|---|
| Age, years, mean ± SD# | 57.3±14.85 | 66.60±11.45 | 0.020# |
| Male gender, n (%) | 33 (82.5) | 17 (85.0) | 0.559** |
| CCI (mean ± SD)# | 1.85±1.66 | 3.55±2.56 | 0.012# |
| APACHE II score (mean ± SD)# | 5.50±3.11 | 10.05±5.07 | 0.001# |
| SOFA (mean ± SD)# | 1.87±1.66 | 3.35±1.06 | 0.001# |
| Main comorbidities, n (%) | |||
| Type 2 diabetes mellitus | 6 (15.0) | 6 (30.0) | 0.189** |
| Congestive heart failure | 1 (2.5) | 2 (10.0) | 0.255** |
| Coronary heart disease | 1 (2.5) | 4 (20.0) | 0.038** |
| Chronic renal disease | 1 (2.5) | 0 (0) | 0.667** |
| COPD | 4 (10.0) | 3 (15.0) | 0.676** |
| Laboratory values (mean±SD) | |||
| White blood cells, mm3 | 6,098.46±2,102.44 | 8,758.00±3,992.08 | 0.003# |
| Absolute neutrophil counts, mm3 | 4,535.52±2,110.35 | 7,437.75±3,762.13 | 0.001# |
| Absolute lymphocyte counts, mm3 | 1,065.98±419.86 | 810.04±426.46 | 0.025# |
| Platelets ×103, mm3 | 191.2±85.6 | 178.3±54.9 | 0.389# |
| CRP, mg/L | 124.55±100.73 | 206.38±85.96 | 0.014# |
| Fibrinogen, mg/dL | 510.59±172.91 | 611.20±199.78 | 0.262# |
| Ferritin, ng/mL | 1,136.05±1,022.05 | 1,288.83±573.13 | 0.381# |
| Procalcitonin, ng/mL | 0.25±0.39 | 0.38±0.46 | 0.280# |
| D-dimers, ng/mL | 599.76±364.72 | 763.00±702.42 | 0.464# |
| AST, U/L | 41.32±26.62 | 64.80±108.08 | 0.502# |
| ALT, U/L | 39.33±34.24 | 52.80±74.30 | 0.822# |
| Creatinine, mg/dL | 0.89±0.18 | 1.29±1.16 | 0.006# |
| Total bilirubin, mg/dL | 0.59±0.54 | 5.25±18.34 | 0.211# |
SD, standard deviation; APACHE, Acute Physiology and Chronic Health Evaluation; CCI, Charlson's Comorbidity Index; SOFA, Sequential Organ Failure; COPD, chronic obstructive pulmonary disease; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ARDS, acute respiratory distress syndrome; CRP, C-reactive protein.
Comparison by the Fischer exact test.
Comparison by the Mann-Whitney U test.
Fig. 1.
Calprotectin is increased in patients with ARDS due to COVID-19. a, b Concentrations of calprotectin (S100A8/A9) and HMGB1 were measured in the plasma from HVs, from patients with COVID-19 who did not develop ARDS during follow-up on the day of hospital admission, and from patients with ARDS by COVID-19 on the first day of ARDS. Comparison by the Mann-Whitney U test; * p < 0.05; ** p < 0.01; *** p < 0.001; ****; p < 0.0001. c, d suPAR was measured in 40 patients screened for participation in the SAVE trial (3). None of the patients with suPAR less than 6 ng/mL developed ARDS, whereas 6 patients (30%) with suPAR 6 ng/mL or more developed ARDS. c Concentrations of calprotectin in 20 patients with suPAR less than 6 ng/mL and in 20 patients with suPAR 6 ng/mL or more. Comparison by the Mann-Whitney U test; * p < 0.05. d Correlation of levels of suPAR with plasma calprotectin on day of admission Spearman rank correlation coefficients (rs), interpolation lines, and p values are provided. ns, nonsignificant.
suPAR was previously identified as an early predictor of ARDS progression in patients with at least 6 ng/mL serum levels in the SAVE trial [3]. To study if the early increase of suPAR denotes increases of calcprotectin, calprotectin was measured in 40 patients screened for inclusion in the SAVE trial; 20 patients with suPAR less than 6 ng/mL and 20 patients with suPAR concentrations ≥6 ng/mL (Table 2). None of the patients with suPAR less than 6 ng/mL developed ARDS; 30% of those with suPAR ≥6 ng/mL progressed into ARDS. Remarkably, patients with suPAR concentrations ≥6 ng/mL had significantly higher concentrations of calprotectin (Fig. 1c). A significant positive correlation between calprotectin and suPAR was demonstrated (Fig. 1d). These findings indicate that both suPAR and calprotectin are reliable prognostic biomarkers in predicting the development of severe respiratory failure in COVID-19. These findings also suggest that calprotectin released at early stages of clinical infection is involved in mediating the progression of pneumonia into ARDS and that suPAR indicates the presence of increased calprotectin.
Table 2.
Baseline clinical and laboratory characteristics of patients with pneumonia by SARS-CoV-2 coronavirus according to the measured levels of circulating suPAR
| suPAR <6 ng/mL (n = 20) |
suPAR ≥6 ng/mL (n = 20) |
p value | |
| Age, years, mean±SD# | 46.25±17.63 | 68.30±9.27 | 6×10–5# |
| Male gender, n (%) | 13 (65.0) | 13 (65.0) | 1.000* |
| CCI (mean±SD)# | 3.56±1.62 | 5.13±0.99 | 0.022# |
| APACHE II score (mean±SD)# | 7.28±3.43 | 6.69±5.25 | 0.953# |
| SOFA (mean±SD)# | 0.86±1.07 | 2.61±1.20 | 0.004# |
| Progression into ARDS, n (%) | 0 (0.0) | 6 (30) | 3×10–6* |
| Main comorbidities, n (%) | |||
| Type 2 diabetes mellitus | 1 (5.0) | 4 (20.0) | 0.342* |
| Congestive heart failure | 0 (0) | 1 (5.0) | 0.500* |
| Coronary heart disease | 1 (5.0) | 3 (15.0) | 0.658* |
| COPD | 0 (0) | 1 (5.0) | 0.506* |
| Laboratory values (mean±SD) | |||
| White blood cells, mm3 | 6,007.14±1,500.55 | 7,578.88±3,201.64 | 0.220# |
| Absolute neutrophil counts, mm3 | 3,573.30±1,723.14 | 5,833.80±2,988.62 | 0.055# |
| Absolute lymphocyte counts, mm3 | 2,233.86±1,872.39 | 1,203.26±614.70 | 0.198# |
| Platelets, ×103, mm3 | 261.8±133.8 | 212.1±68.2 | 0.574# |
| CRP, mg/L | 35.69±53.43 | 89.19±82.45 | 0.083# |
| Fibrinogen, mg/dL | 352.00±59.24 | 612.22±187.67 | 0.003# |
| Ferritin, ng/mL | 283.41±244.73 | 882.90±595.75 | 0.024# |
| Procalcitonin, ng/mL | 0.06±0.05 | 0.36±0.36 | 0.133# |
| D-dimers, ng/mL | 421.67±324.10 | 1,407.79±3,251.55 | 0.207# |
| AST, U/L | 22.57±6.68 | 46.44±29.49 | 0.041# |
| ALT, U/L | 27.14±14.37 | 38.67±27.78 | 0.458# |
| Creatinine, mg/dL | 0.92±0.27 | 0.97±0.34 | 0.883# |
| Total bilirubin, mg/dL | 0.259±0.11 | 1.54±2.06 | 0.001# |
SD, standard deviation; APACHE, Acute Physiology and Chronic Health Evaluation; CCI, Charlson's Comorbidity Index; SOFA, Sequential Organ Failure; ARDS, acute respiratory distress syndrome; COPD, chronic obstructive pulmonary disease; AST, aspartate aminotransferase; ALT, alanine aminotransferase; CRP, C-reactive protein.
Comparison by the Fischer exact test.
Comparison by the Mann-Whitney U test.
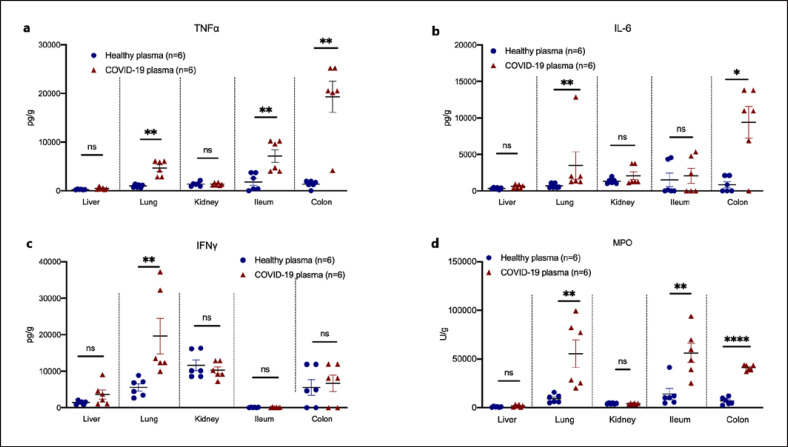
In order to investigate the mechanism by which calprotectin induces the hyperinflammation related to COVID-19-associated respiratory failure, we sought to design a COVID-19 murine model. Mice were challenged with plasma either from HVs or from patients with ARDS of COVID-19. This model has never been studied before and leads to substantial neutrophil infiltration in the lung alveoli and interstitial space (online suppl. Fig. S1; see www.karger.com/doi/10.1159/000524560 for all online suppl. material) similar to that developing after the challenge of mice hetero-expressing angiotensin-converting enzyme 2 (ACE2) by SARS-CoV-2 [13]. Plasma of COVID-19 patients led to organ-specific increase of cytokine concentrations. More precisely, TNFα and MPO activity were increased in the lung, ileum, and colon of mice challenged with COVID-19 plasma; IL-6 was upregulated in the lung and colon; and IFNγ was upregulated in the lung (Fig. 2a–d). These findings demonstrated a compartmentalized inflammatory response in mice after challenging with COVID-19 patient plasma.
Fig. 2.
A COVID-19-like animal model of compartmentalized hyperinflammation. C57Bl6 mice were injected i.v. with plasma of HVs or patients with ARDS due to COVID-19 for 3 consecutive days. Mice were sacrificed on day 4. Tissue concentrations of TNFα (a), IL-6 (b), IFNγ (c), and MPO activity (d) were determined. Comparison by the Mann-Whitney U test; * p < 0.05; ** p < 0.01; **** p < 0.0001. ns, nonsignificant.
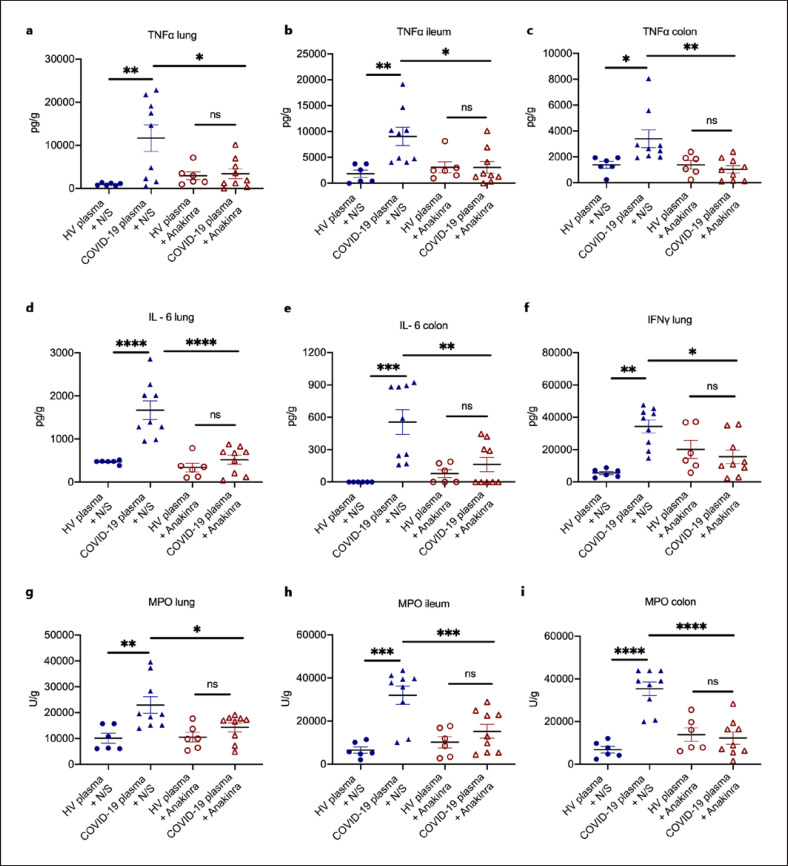
DAMPs, like calprotectin, act on monocytes and macrophages and tend to stimulate intense IL-1 responses. To provide evidence on the contribution of IL-1 stimulation, mice challenged with plasma from HVs or COVID-19 patients were treated with N/S or with anakinra; the latter blocks both human and murine IL-1α and ΙL-1β. Treatment with anakinra significantly reduced tissue concentrations of TNFα, IL-6, IFN-γ, and MPO activity (Fig. 3a–i). This effect of anakinra was partially attenuated when dexamethasone was co-administered (online suppl. Fig S2a–d).
Fig. 3.
IL-1 inhibition attenuates the compartmentalized hyperinflammation in a COVID-19-like murine model. In a COVID-19-like infection model, C57Bl6 mice were challenged i.v. with plasma of HVs or patients with ARDS due to COVID-19 for 3 consecutive days. In separate experiments, on each day of plasma challenge, mice were treated with anakinra, which inhibits human and murine IL-1, or vehicle. Mice were sacrificed on day 4. a–c TNFα, IL-6 (d, e), IFNγ (f), and MPO activity (g–i) was determined in tissues. Comparison by the Mann-Whitney U test; * p < 0.05; ** p < 0.01. ns nonsignificant.
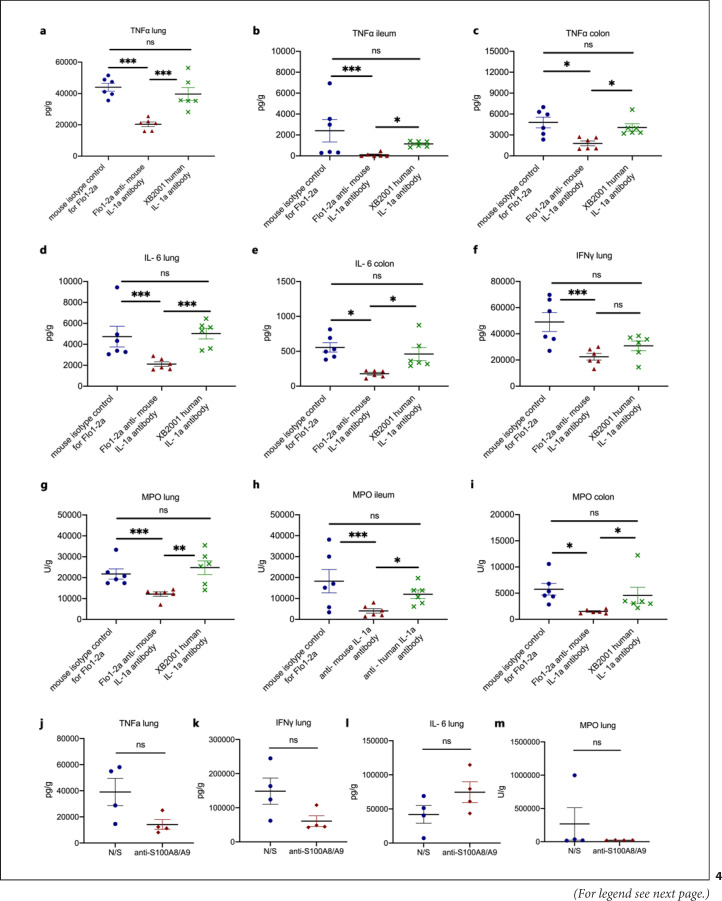
However, the observed inflammation may have been mediated through human IL-1α present in the plasma of COVID-19 patients, or it may be attributed to DAMPs other than S100A8/A9 (calprotectin). To further clarify this, we run two sets of experiments. In the first set of experiments, mice challenged with COVID-19 plasma were treated with the anti-murine-specific anti-IL-1α antibody Flo1-2a and with the human-specific antihuman IL-1α antibody XB2001, respectively. Treatment with Flo1-2a significantly reduced tissue concentrations of TNFα, IL-6, IFNγ, and MPO activity in contrast to treatment with XB2001 (Fig. 4a–i). In the second set of experiments, we showed that pre-treatment with anti-S10A8/A9 attenuates lung inflammation (Fig. 4j). These findings suggest that (i) the host-derived mouse IL-1α and not any injected human IL-1α and (ii) calprotectin are main inducers of the compartmentalized inflammation.
Fig. 4.
Murine IL-1α and S100A8/A9 drive the hyperinflammation caused by SARS-CoV-2. In a COVID-19-like infection model, C57Bl6 mice were challenged i.v. with plasma of patients with ARDS due to COVID-19 for 3 consecutive days. In separate experiments, on each day of plasma challenge, mice were treated with Flo1-2a anti-murine IL-1α antibody or XB2001 human IL-1α antibody or murine isotype control for Flo1-2a or pre-treated with anti-S100A8/A9. Mice were sacrificed on day 4. a–c TNFα in the lung, ileum, and colon and modulation by blockade of human or murine IL-1α. d, e IL-6 in the lung, ileum, and colon and modulation by blockade of human or murine IL-1α. f IFNγ in the lung, ileum, and colon and modulation by blockade of human or murine IL-1α. g–i MPO activity in the lung, ileum, and colon and modulation by blockade of human or murine IL-1α. j TNFα in the lung and modulation by blockade of S100A8/A9. k IFNγ in the lung and modulation by blockade of S100A8/A9. l IL-6 in the lung and modulation by blockade of S100A8/A9. m MPO activity in the lung and modulation by blockade of S100A8/A9. Comparison by the Mann-Whitney U test; * p < 0.05; ** p < 0.01; *** p < 0.001. ns, nonsignificant.
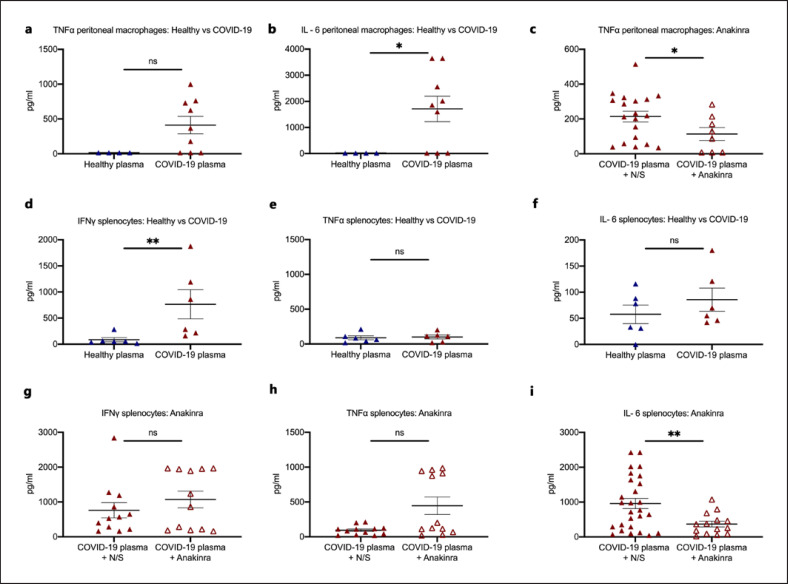
Treatment with COVID-19 plasma led to an increase of ex vivo production capacity of peritoneal macrophages for TNFα and IL-6 and of splenocytes for IFNγ. The production capacity of peritoneal macrophages for TNFα and of splenocytes for IL-6 was decreased in mice treated with anakinra (Fig. 5; online suppl. Fig S3). Ex vivo stimulation of splenocytes with the SARS-CoV-2 spike protein failed to evoke an efficient cytokine production (online suppl. Fig S4a–c).
Fig. 5.
Activation of peritoneal macrophages and splenocytes and modulation through IL-1 inhibition. a, b In a COVID-19-like infection model, C57Bl6 mice were treated i.v. with plasma of HVs or patients with ARDS due to COVID-19 for 3 consecutive days. Mice were sacrificed on day 4. Peritoneal macrophages were isolated and incubated with bacterial LPS for the production of TNFα and IL-6. c In a COVID-19-like infection model, C57Bl6 mice were treated i.v. with plasma of patients with ARDS due to COVID-19 with or without treatment with the IL-1 receptor inhibitor anakinra for 3 consecutive days. Mice were sacrificed on day 4. Peritoneal macrophages were isolated and incubated with bacterial LPS for the production of TNFα. d–f In a COVID-19-like infection model, C57Bl6 mice were treated i.v. with plasma of HVs or patients with ARDS due to COVID-19 for 3 consecutive days. Mice were sacrificed on day 4. Splenocytes were isolated and incubated with heat-killed C. albicans for the production of IFNγ, TNFα, and IL-6. e–i In a COVID-19-like infection model, C57Bl6 mice were treated i.v. with plasma of patients with ARDS due to COVID-19 with or without treatment with the IL-1 receptor inhibitor anakinra for 3 consecutive days. Mice were sacrificed on day 4. Splenocytes were isolated and incubated with heat-killed C. albicans for the production of IFNγ, TNFα, and IL-6. Comparison by the Mann-Whitney U test; * p < 0.05; ** p < 0.01. LPS, lipopolysaccharide; ns nonsignificant.
Bioanalysis revealed four genes to be upregulated in the lung, namely Cyp2a5, Nfe2, Pcx, and Tcap, and one gene downregulated in the spleen, namely Ctgf. Cyp2a5 is involved in retinol metabolism and Pcx in the citrate cycle. Ctgf is mediating Apelin and Hippo which are implicated in cardiac fibrosis and cellular apoptosis (online suppl. Fig. S5).
The number of transcripts of core SARS-CoV-2 genes ORF1ab and N were indirectly provided by the cycles (Ct) of polymerase chain reaction positivity in plasma samples that were used for the induction of the COVID-19-like hyperinflammation in the murine model. No correlation was found between Cts and the levels of TNFα, IL-6, IFNγ, and MPO in the lung of COVID-19 patients with ARDS due to COVID-19 (online suppl. Fig. S6). This suggests that the hyperinflammation is independent of the viral load.
In order to mimic hospital-acquired secondary bacterial infections among COVID-19 patients in mice, mice treated for 3 consecutive days with plasma from HV or COVID-19 plasma were further challenged with A. baumannii or E. coli. Survival was significantly lower after challenging with A. baumannii compared to E. coli but only in mice challenged with COVID-19 plasma (Fig. 6a). Observations coming from our prospective cohort of 134 ARDS patients revealed higher incidence of secondary infections by A. baumannii when compared to other Gram-negative bacteria (online suppl. Table S1; online suppl. Fig. S7). This further supports the immunological similarities of the developed animal model to the human infection. Treatment with anakinra did not significantly alter mortality after challenge with either A. baumannii or E. coli (Fig. 6b, c).
Fig. 6.
COVID-19-like hyperinflammation is associated with susceptibility to A. baumannii infections. a C57Bl6 mice were injected i.v. with plasma of HVs or patients with ARDS due to COVID-19 for 3 consecutive days. On day 4, mice were challenged i.p. with E. coli or A. baumannii. Survival comparison by the log-rank test and the respective p value is provided. b, c C57Bl6 mice were injected i.v. with plasma of patients with ARDS due to COVID-19 for 3 consecutive days. On each day of plasma challenge, half of the mice were treated with anakinra. On day 4, mice were challenged i.p. with E. coli (b) or A. baumannii (c). Survival comparison by the log-rank test and the respective p value is provided.
Discussion
This study presented evidence that severe respiratory failure in COVID-19 patients is possibly triggered by DAMPs, most likely calprotectin, which are released early after the interaction of SARS-CoV-2 with host cells. It may well be the case that calprotectin is this specific DAMP since circulating levels of HMGB1, another DAMP, do not follow those of calprotectin. DAMPs lead to a compartmentalized inflammation through enhancement of the production of pro-inflammatory cytokines IL-1α and IL-1β.
Calprotectin, a heterodimer of S100A8 and S100A9, is an intracellular protein secreted by leukocytes. The upregulation of calprotectin expression has been associated with various inflammatory processes such as inflammatory bowel disease, rheumatoid arthritis, spondyloarthritis, and cardiovascular disease in type 2 diabetes mellitus [14, 15, 16, 17]. Additionally, calprotectin is crucial for neutrophil accumulation and inflammation in the lung parenchyma during bacterial community-acquired pneumonia [18]. Recently, high levels of calprotectin in samples of COVID-19 patients were correlated with disease severity and were associated with increased levels of inflammatory cytokines and chemokines and myelopoiesis of abnormal myeloid cells [19].
In this study, we suggest calprotectin as a potential first element in the pathway leading to ARDS in COVID-19 patients. Indeed, we found greater levels in patients with ARDS compared to patients without ARDS, whereas the use of one calprotectin blocking antibody in the mice experiments attenuated lung pro-inflammatory responses. On the contrary, HMGB1, an additional alarmin, which has been associated with sepsis pathogenesis and outcome [20], did not significantly differ among patients with and without ARDS due to COVID-19. The attenuation of pro-inflammatory animal responses with anakinra and with the specific mouse anti-IL-1α antibody supports that the studied animal model is driven through excess IL-1 responses, most probably where calprotectin plays a major role. SARS-CoV-2 stimulates the NLRP3 inflammasome, leading to activation of caspase-1 and of pyroptosis. This primes the release of calprotectin from the intracellular space [21]. Calprotectin stimulates the production of pro-inflammatory cytokines through the activation of Toll-like receptor-4 and MyD88 [22] or the receptor for advanced glycation end-products and may prime for intracellular pro-IL-1β [23] which is subsequently cleaved by SARS-CoV-2-activated NLRP3 inflammasome [2] into IL-1β. Anakinra prevents the inflammatory action of both IL-1α and IL-1β by blocking their cellular receptor. IL-1α released during cell death is characterized by an autocrine function enabling an amplifying loop of inflammation [24]. The excess stimulation of IL-1 cascade is compatible with the observed downregulation of Ctgf in splenocytes. The dependence of Ctgf expression by calprotectin, IL-1α, and IL-1β has been shown in airway smooth muscle cells [25], in bronchial epithelial cells [26] and in fibroblasts [27]. Moreover, suPAR which is an early predictor of respiratory deterioration in COVID-19 [3, 28] was indicative of higher calprotectin levels, suggesting that increased suPAR can be the biomarker of DAMP-induced pathways in COVID-19.
SARS-CoV-2 uses the cellular surface protein ACE2 to bind and enter cells. However, the mouse ACE2 does not effectively bind the viral spike protein [29]. Therefore wild-type mice cannot be directly used as a model to gain insight in COVID-19 pathophysiology. Mice over-expressing ACE2 have been used as a strategy to tackle this problem [30]. While it is accepted that the interaction of the spike protein of SARS-CoV-2 with human ACE2 is the first step for viral invasion in host cells of the upper and lower respiratory tract [31], the ability of the virus to directly infect other human tissues, where ACE2 is also expressed, such as the small intestine, kidneys, and the heart, is a matter of dispute [32]. In addition, we also need to understand the pure immunological mechanisms of hyperinflammation in COVID-19. This is introducing the need of COVID-19 models like the one developed after animal challenge with human plasma. The model simulates the inflammatory reaction in human SARS-CoV-2 infection since it provides compartmentalized hyperinflammation of the lung.
Surprisingly, we found that the attenuation of tissue pro-inflammatory responses in our model was more pronounced by anakinra than by the anakinra and dexamethasone combination. This may be due to the lack of target specificity in the mode of action on dexamethasone. In our experiments, the stimulation of isolated splenocytes by purified protein S of SARS-CoV-2 did not yield any significant cytokine production. This may have one or more explanations: (i) mouse splenocytes are lacking ACE2 which acts as the receptor; (ii) splenocytes are downregulated for cytokine production through the S protein at that stage of compartmentalized hyperinflammation; and (iii) when compartmentalized hyperinflammation is mediated through calprotectin immune activation the S protein ceases to be an important immune trigger.
In the study, we measured only concentrations of S100A8/A9 (calprotectin) and HMGB1 and not of other DAMPs like mitochondrial DNA, urate, and adenosine triphosphate. However, the significant differences in the concentrations of S100A8/A9 in patients with ARDS than in patients without ARDS and the attenuation of lung pro-inflammatory responses in mice treated with anti-S100A8/A9 and mouse-specific anti-IL-1α fully support a role of calprotectin and endogenous IL-1 as the drivers of the compartmentalized inflammation.
Our findings provide evidence that progression from LRTI by SARS-CoV-2 to ARDS is a process which is mediated by DAMPs, like IL-1α and calprotectin, released following cell destruction. The early detection of the presence of circulating DAMPs and blockade through IL-1a inhibition may represent a viable therapeutic strategy. This is the hallmark of the efficacy of anakinra − guided by the biomarker suPAR − which led to a significant decrease in the incidence of ARDS in the SAVE trial [3]. Further experimentation, using the proposed animal model, is necessary in order to associate the circulating DAMPs to known COVID-19-related pathological changes, such as lung leak and epithelial damage.
Statement of Ethics
The clinical study received approval 30/20 by the National Ethics Committee of Greece; approval IS 021-20 by the National Organization for Medicines of Greece. Samples from patients enrolled in the suPAR-guided Anakinra treatment for Validation of the risk and Early management of severe respiratory failure by COVID-19 (SAVE) trial (EudraCT number 2020-001466-11; National Ethics Committee approval 38/20; National Organization for Medicines approval ISO 28/20; ClinicalTrials.gov registration NCT04357366) were also included in the analysis.
Patients were enrolled after written informed consent was provided by themselves or by first-degree relatives in the case of patients who were unable to consent. Animal experiments were conducted in the Unit of Animals for Medical and Scientific purposes of the University General Hospital “Attikon” (Athens, Greece). All experiments were licensed from the Greek veterinary directorate under the protocol number 471955/06-07-2020.
Conflict of Interest Statement
J. Eugen-Olsen is a cofounder, shareholder, and CSO of ViroGates A7S, Denmark, and is named inventor on patents on suPAR owned by Copenhagen University Hospital Hvidovre, Denmark. He is granted by the Horizon 2020 European Grant RISCinCOVID. J. Simard is the CEO and founder of XBiotech. M.G. Netea is a scientific founder of TTxD and received research grants from GSK and ViiV Healthcare. E.J. Giamarellos-Bourboulis has received honoraria from AbbVie USA, Abbott CH, InflaRx GmbH, MSD Greece, XBiotech Inc., and Angelini Italy; independent educational grants from AbbVie, Abbott, Astellas Pharma Europe, Axis Shield, bioMérieux Inc, InflaRx GmbH, Sobi, and XBiotech Inc.; and funding from the FrameWork 7 program HemoSpec (granted to the National and Kapodistrian University of Athens), the Horizon 2020 Marie-Curie Project European Sepsis Academy (granted to the National and Kapodistrian University of Athens), and the Horizon 2020 European Grant ImmunoSep (granted to the Hellenic Institute for the Study of Sepsis).
Funding Sources
The study was funded in part by the Hellenic Institute for the Study of Sepsis and in part by the Horizon 2020 grant RISCinCOVID. The funders had no role in the design, collection, analysis, and interpretation of data, writing, and decision to publish.
Author Contributions
George Renieris, Dionyssia-Eirini Droggiti, Theano Anriopoulou, Victoria-Marina Spanou, and Dionyssios Kafousopoulos performed animal experiments. George Renieris and Eleni Karakike collected and analyzed the patient data. Theologia Ghavogianni performed serum analysis, MPO, and cytokine measurements. Emmanouil Stylianakis performed bioanalysis. George Renieris, Eleni Karakike, Mihai G. Netea, Jesper Eugen-Olsen, and Evangelos J. Giamarellos-Bourboulis analyzed the data. John Simard provided the used antibodies. George Renieris drafted the manuscript under the supervision of Evangelos J. Giamarellos-Bourboulis with all authors contributing to writing and providing feedback. Mihai G. Netea, Jesper Eugen-Olsen, John Simard, and Evangelos J. Giamarellos-Bourboulis conceived ideas and oversaw the research program.
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary material. Further inquiries can be directed to the corresponding author. The sequence libraries generated in this study are publicly available through the National Center for Biotechnology Information (NCBI) gene expression omnibus (GEO) under the accession number GSE199001.
Supplementary Material
Supplementary data
Acknowledgments
We thank Zhongli Xu, Katja Schubert, and Marlen Hermann (InflaRx GmbH, Jena, Germany) for the measurements of calprotectin in patient samples as well as for their critical review of the manuscript.
Funding Statement
The study was funded in part by the Hellenic Institute for the Study of Sepsis and in part by the Horizon 2020 grant RISCinCOVID. The funders had no role in the design, collection, analysis, and interpretation of data, writing, and decision to publish.
References
- 1.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodrigues TS, de Sá KSG, Ishimoto AY, Becerra A, Oliveira S, Almeida L, et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med. 2021;218:e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyriazopoulou E, Panagopoulos P, Metallidis S, Dalekos GN, Poulakou G, Gatselis N, et al. An open label trial of anakinra to prevent respiratory failure in COVID-19. eLife. 2021;10:e66125. doi: 10.7554/eLife.66125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Luca A, Smeekens SP, Casagrande A, Iannitti R, Conway KL, Gresnigt MS, et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci U S A. 2014;111:3526–31. doi: 10.1073/pnas.1322831111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aeffner F, Bolon B, Davis IC. Mouse models of acute respiratory distress syndrome: a review of analytical approaches, pathologic features, and common measurements. Toxicol Pathol. 2015;43:1074–92. doi: 10.1177/0192623315598399. [DOI] [PubMed] [Google Scholar]
- 6.Xu T, Qiao J, Zhao L, He G, Li K, Wang J, et al. Effect of dexamethasone on acute respiratory distress syndrome induced by the H5N1 virus in mice. Eur Respir J. 2009;33:852–60. doi: 10.1183/09031936.00130507. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Wei L, Wang J, Qin Z, Wang J, Lu Y, et al. Suppression colitis and colitis-associated colon cancer by anti-S100a9 antibody in mice. Front Immunol. 2017;8:1774. doi: 10.3389/fimmu.2017.01774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–5. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 10.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders S, Pyl PT, Huber W. HTSeq: a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong W, Yang J, Bi Z, He C, Lei H, Yu W, et al. A mouse model for SARS-CoV-2-induced acute respiratory distress syndrome. Signal Transduct Target Ther. 2021;6((1)):1. doi: 10.1038/s41392-020-00451-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukunaga S, Kuwaki K, Mitsuyama K, Takedatsu H, Yoshioka S, Yamasaki H, et al. Detection of calprotectin in inflammatory bowel disease: fecal and serum levels and immunohistochemical localization. Int J Mol Med. 2018;41:107–18. doi: 10.3892/ijmm.2017.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azramezani Kopi T, Shahrokh S, Mirzaei S, Asadzadeh Aghdaei H, Amini Kadijani A. The role of serum calprotectin as a novel biomarker in inflammatory bowel diseases: a review study. Gastroenterol Hepatol Bed Bench. 2019;12:183–9. [PMC free article] [PubMed] [Google Scholar]
- 16.Jarlborg M, Courvoisier DS, Lamacchia C, Martinez Prat L, Mahler M, Bentow C, et al. Serum calprotectin: a promising biomarker in rheumatoid arthritis and axial spondyloarthritis. Arthritis Res Ther. 2020;22:105. doi: 10.1186/s13075-020-02190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oosterwijk MM, Bakker SJL, Nilsen T, Navis G, Laverman GD. Determinants of increased serum calprotectin in patients with type 2 diabetes mellitus. Int J Mol Sci. 2020;21:8075. doi: 10.3390/ijms21218075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotsiou OS, Papagiannis D, Papadopoulou R, Gourgoulianis KI. Calprotectin in lung diseases. Int J Mol Sci. 2021;22((4)):1706. doi: 10.3390/ijms22041706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–8.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karakike E, Adami ME, Lada M, Gkavogianni T, Koutelidakis IM, Bauer M, et al. Late peaks of HMGB1 and sepsis outcome: evidence for synergy with chronic inflammatory disorders. Shock. 2019;52:334–9. doi: 10.1097/SHK.0000000000001265. [DOI] [PubMed] [Google Scholar]
- 21.Zheng M, Williams EP, Malireddi RKS, Karki R, Banoth B, Burton A, et al. Impaired NLRP3 inflammasome activation/pyroptosis leads to robust inflammatory cell death via caspase-8/RIPK3 during coronavirus infection. J Biol Chem. 2020;295:14040–52. doi: 10.1074/jbc.RA120.015036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai SY, Segovia JA, Chang TH, Morris IR, Berton MT, Tessier PA, et al. DAMP molecule S100A9 acts as a molecular pattern to enhance inflammation during influenza A virus infection: role of DDX21-TRIF-TLR4-MyD88 pathway. PLoS Pathog. 2014;10:e1003848. doi: 10.1371/journal.ppat.1003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogl T, Stratis A, Wixler V, Völler T, Thurainayagam S, Jorch SK, et al. Autoinhibitory regulation of S100A8/S100A9 alarmin activity locally restricts sterile inflammation. J Clin Invest. 2018;128:1852–66. doi: 10.1172/JCI89867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantovani A, Dinarello CA, Molgora M, Garlanda C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity. 2019;50:778–95. doi: 10.1016/j.immuni.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie S, Sukkar MB, Issa R, Oltmanns U, Nicholson AG, Chung KF. Regulation of TGF-beta 1-induced connective tissue growth factor expression in airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2005;288:L68–76. doi: 10.1152/ajplung.00156.2004. [DOI] [PubMed] [Google Scholar]
- 26.Zhou T, Yu Q, Lin H, Wang Z, Fu G, Lei L, et al. The role of CTGF in inflammatory responses induced by silica particles in human bronchial epithelial cells. Lung. 2019;197:783–91. doi: 10.1007/s00408-019-00272-x. [DOI] [PubMed] [Google Scholar]
- 27.Rohde D, Schön C, Boerries M, Didrihsone I, Ritterhoff J, Kubatzky KF, et al. S100A1 is released from ischemic cardiomyocytes and signals myocardial damage via toll-like receptor 4. EMBO Mol Med. 2014;6:778–94. doi: 10.15252/emmm.201303498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rovina N, Akinosoglou K, Eugen-Olsen J, Hayek S, Reiser J, Giamarellos-Bourboulis EJ. Soluble urokinase plasminogen activator receptor (suPAR) as an early predictor of severe respiratory failure in patients with COVID-19 pneumonia. Crit Care. 2020;24:187. doi: 10.1186/s13054-020-02897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muñoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, et al. Animal models for COVID-19. Nature. 2020;586:509–15. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–9. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bost P, Giladi A, Liu Y, Bendjelal Y, Xu G, David E, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–88.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary material. Further inquiries can be directed to the corresponding author. The sequence libraries generated in this study are publicly available through the National Center for Biotechnology Information (NCBI) gene expression omnibus (GEO) under the accession number GSE199001.