Abstract
Staphylococcus aureus is an important pathogen causing various infections, including − as most frequently isolated bacterium − cutaneous infections. Keratinocytes as the first barrier cells of the skin respond to S. aureus by the release of defense molecules such as cytokines and antimicrobial peptides. Although several pattern recognition receptors expressed in keratinocytes such as Toll-like and NOD-like receptors have been reported to detect the presence of S. aureus, the mechanisms underlying the interplay between S. aureus and keratinocytes are still emerging. Here, we report that S. aureus induced gene expression of CYP1A1 and CYP1B1, responsive genes of the aryl hydrocarbon receptor (AhR). AhR activation by S. aureus was further confirmed by AhR gene reporter assays. AhR activation was mediated by factor(s) <2 kDa secreted by S. aureus. Whole transcriptome analyses and real-time PCR analyses identified IL-24, IL-6, and IL-1beta as cytokines induced in an AhR-dependent manner in S. aureus-treated keratinocytes. AhR inhibition in a 3D organotypic skin equivalent confirmed the crucial role of the AhR in mediating the induction of IL-24, IL-6, and IL-1beta upon stimulation with living S. aureus. Taken together, we further highlight the important role of the AhR in cutaneous innate defense and identified the AhR as a novel receptor mediating the sensing of the important skin pathogen S. aureus in keratinocytes.
Keywords: Host defense, Keratinocytes, Aryl hydrocarbon receptor, Staphylococcus aureus
Introduction
Staphylococcus aureus is a gram-positive, coagulase-positive bacterium that forms biofilms and causes opportunistic infections in various tissues, including skin [1]. S. aureus is temporarily found on human skin where its presence is associated with a higher risk for subsequent infections [2, 3]. Cutaneous colonization and infection with S. aureus is also a typical hallmark of the chronic inflammatory skin disease atopic dermatitis (AD) and AD skin is more frequently colonized by S. aureus than healthy skin [4].
Sensing of S. aureus by keratinocytes is the prerequisite to initiate a rapid defense response by the release of innate defense factors such as antimicrobial peptides and cytokines [5, 6]. Although several pattern recognition receptors such as Toll-like receptor TLR-2 and NOD-like receptor NOD2 have been implicated in the recognition of S. aureus by keratinocytes [7, 8], the detailed mechanisms underlying the sensing of S. aureus by keratinocytes are still emerging.
In a previous study, we found evidence that the skin commensal Staphylococcus epidermidis activates the aryl hydrocarbon receptor (AhR) in keratinocytes [9]. The AhR is a ligand-activated transcription factor involved in xenobiotic metabolism, epidermal barrier formation, immune signaling, and immune cell differentiation [10, 11, 12]. AhR is activated upon binding of various low-molecular-weight ligands; the receptor is expressed in various tissues, particularly high expression is found in the liver and in barrier organs such as gut and skin [13, 14]. There is increasing evidence that the AhR plays a major role in host defense [11, 13, 15]. The AhR can be activated by metabolites of bacteria such as Pseudomonas aeruginosa [10] or members of the skin microbiota, such as Malassezia yeasts [16]. Although the role of the AhR in cutaneous defense is still emerging there is growing evidence that it plays an important role in skin-microbe interaction [14]. Several reports have shown that the AhR is crucial for the maintenance of skin barrier function [17, 18]. AhR activation by coal tar or the AhR activator tapinarof has been reported to ameliorate AD symptoms by restoring the skin barrier [18, 19]. In addition, activation of the AhR by microbial tryptophan metabolites has been associated with attenuation of inflammation in AD patients [20]. On the other hand, it has been reported that AhR expression in AD skin correlated with the severity of AD symptoms [21].
In this study, we provide evidence that the AhR in keratinocytes is activated by S. aureus and that the expression of several inflammatory cytokines induced by S. aureus is mediated by the AhR. This strengthens the role of the AhR as an innate microbial sensor and a mediator of the innate immune defense of human skin.
Materials and Methods
Keratinocyte Cell Culture and Stimulation
Normal human primary keratinocytes (NHEKs), pooled from four donors (Promocell, Heidelberg, Germany) were cultured in Keratinocyte Growth Medium 2 (KGM2; Promocell), including supplements and CaCl2 at 37°C/5% CO2 in 24-well plates until post-confluency. S. aureus skin-derived clinical isolates (identity verified by MALDI-TOF mass spectrometry; MALDI Biotyper; Bruker, Billerica, MA, USA) and S. aureus ATCC 8325-4 were grown on blood agar plates for 24 h and then inoculated into tryptic soy broth and grown under agitation for 16–18 h at 37°C. 250 µL of the bacterial suspension was inoculated into 7 mL tryptic soy broth and further grown for 3–4 h. Bacteria were centrifuged for 5 min at 4,500 g, the pellet was washed with 7 mL phosphate-buffered saline (PBS) and then the OD600 was adjusted to 0.2 in KGM2 medium (without supplements, with CaCl2) corresponding to approx. 1.7 × 107 bacteria/mL. This suspension was diluted 1:2 with KGM2 and each well of NHEKs was stimulated with 300 μL. 3 h after the start of the stimulation, the medium was discarded, NHEKs were washed once with PBS and incubated with 300 μL KGM2 supplemented with 200 μg/mL gentamicin sulfate to kill any remaining extracellular bacteria. NHEKs were stimulated for another 14–16 h and then the medium was removed, centrifuged at 12,000 g for 5 min, and stored at −80°C for ELISA analyses. Keratinocytes were also stimulated with S. aureus culture supernatants and size filtrated supernatants (prepared as described below). After stimulation with living bacteria or bacterial culture supernatants, keratinocytes were washed with PBS and used for RNA isolation.
In some experiments, the AhR was inhibited by using the AhR inhibitor CH-223191 (Cayman Chemicals). To this end, NHEKs were preincubated with 10 μM CH-223191 for 1–1.5 h before the start of the stimulation and then stimulated in the presence of 10 μM CH-223191. 0.1% DMSO served as vehicle control.
Production of Bacterial Culture Supernatants
S. aureus was adjusted to an OD 600 nm of 0.2 in KGM2 medium as described above. Eight milliliters of this suspension was filled into sterile petri dishes and incubated for 24 h at 37°C. Subsequently, the bacteria suspension was harvested and centrifuged for 5 min at 8,500 g. The supernatant was sterile filtered (0.2 μm pore size) and stored at −20°C until use in stimulation experiments. For size filtration, the supernatant was applied to 2 kDa centrifugal concentrators (Vivaspin 15 R Hydrosart filter device; Sartorius, Göttingen, Germany) and centrifuged for 1 h at 3,000 g according to the suppliers' protocol. The >2 kDa concentrate was washed three times with KGM2. Filtrate and concentrate were used for stimulation of NHEKs diluted 1:2 in KGM2.
AhR Gene Reporter Luciferase Assay
To test nuclear translocation and binding of the AhR to AhR-responsive elements, the firefly luciferase reporter plasmid pGUDLUC6.1 (generously gifted by M. Denison, U.C. Davis) was used. This plasmid contains 4 AhR-responsive elements and no other known regulatory elements [22]. 300 ng of this plasmid together with 30 ng of a renilla luciferase control plasmid (pGL4.74[hRluc/TK]; Promega, Madison, WI, USA) were transfected in keratinocytes (24 wells, cultured with 400 μL KGM2) using the transfection reagent Fugene HD (Promega). 24 h after transfection, cells were stimulated with S. aureus as described above. After stimulation, cells were lysed with passive lysis buffer (Promega), and firefly and renilla luciferase activities were determined using the Dual Luciferase assay system (Promega). Specific AhR-luciferase activity was determined by normalizing the firefly luciferase activity to renilla luciferase activity.
AhR siRNA Experiments
NHEKs were transfected at 50%–70% confluency with 1 μL HiPerfect transfection reagent (Qiagen) and 5 nM of either AhR-specific “SilencerSelect” siRNA (s1199) or nonsilencing control siRNA (4390844) purchased from Life Technologies (Carlsbad, CA, USA). After 24 h of incubation with the siRNA, medium was changed and cells were grown for 3 additional days until stimulation.
3D Organotypic Skin Equivalent
The organotypic 3D skin equivalent was constructed as previously described [23]. The skin equivalent was preincubated with 10 μM CH-223191 or the corresponding volume of DMSO as a solvent control for 1–1.5 h. Stimulation with S. aureus SA 129 was done by application of approximately 1.2 × 108 CFU/mL in 20 μL of KGM2 without supplements onto the skin equivalent. Stimulation was done for approximately 24 h at 37°C/5% CO2.
Real-Time PCR Analysis
Total RNA of the keratinocytes was isolated using the reagent Crystal RNAmagic according to the manufacturer's protocol (Biolabproducts, Hamburg, Germany). 0.5 μg of the isolated RNA was reverse transcribed to cDNA using an oligo dT primer and 12.5 units of reverse transcriptase mix (PrimeScript RT Reagent Kit; TaKaRa Bio, Saint-Germain-en-Laye, France). cDNA corresponding to 10 ng total RNA served as the template in a real-time PCR. Real-time PCR was performed with the QuantStudio 3 System (BD Biosciences) using SYBR Premix Ex Taq II mix (TaKaRa Bio) as described [8]. The following intron-spanning primers were used: IL-1β: 5′-AAG CCC TTG CTG TAG TGG TG-3′ (forward primer) and 5′-GAA GCT GAT GGC CCT AAA CA-3′ (reverse primer); CYP1A1: 5′-CAC CAT CCC CCA CAG CAC-3′ (forward primer) and 5′-ACA AAG ACA CAA CGC CCC TT-3′ (reverse primer); CYP1B1: 5′-TAT CAC TGA CAT CTT CGG CG-3′ (forward primer) and 5′-CTG CAC TCG AGT CTG CAC AT-3′ (reverse primer); IL-24: 5′-GTT CCC CAG AAA CTG TGG GA-3 (forward primer) and 5′-CGAGACGTTCTGCAGAACC-3′ (reverse primer); IL-6: 5′-GGT ACA TCC TCG ACG GCA TCT-3′ (forward primer) and 5′-GTG CCT CTT TGC TGC TTT CAC-3′ (reverse primer). Standard curves were produced for each primer set with serial dilutions of cDNA. All quantifications were normalized to the housekeeping gene RPL38 (ribosomal protein L38) using the primer pair: 5′-TCA AGG ACT TCC TGC TCA CA-3′ (forward primer) and 5′-AAA GGT ATC TGC TGC ATC GAA-3′ (reverse primer).
Whole Transcriptome Sequencing
In this pilot study, human primary keratinocytes were stimulated with S. aureus clinical isolate SA 179 for 20 h in the presence or absence of the AhR inhibitor CH-223191 (n = 5 of each treatment). Total RNA was isolated with the NucleoSpin RNA Kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's protocol. RNA libraries were prepared and sequenced on a HiSeq4000 (Illumina, San Diego, CA, USA) and analyzed as described recently [24].
Immunostaining
The organotypic 3D skin equivalent was fixed by formalin and embedded in paraffin. For immunostaining goat anti-IL-24 antibody (R&D Systems, Minneapolis, MN, USA) 1:100 diluted in TBS/1% BSA was used as primary antibody. Biotinylated rabbit anti-goat (Jackson ImmunoResearch, UK) 1:500 diluted in TBS/1% BSA served as second antibody. Blocking was done with TBS/12% BSA. For development, the Vectastain Elite ABC Komplex (PK-6100) followed by the Vector Nova Red-Substrate (SK-4800; both Vector Laboratories, USA) was used. Slides were counterstained with hematoxylin and mounted with Eukitt (Poly(butyl methacrylate-co-methyl methacrylate); O. Kindler, Freiburg, Germany).
Statistics
Statistical analyses were performed with GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). D'Agostino & Pearson test was used to analyze the distribution of the data. Normally distributed data were analyzed by t test (comparison of two groups) or ANOVA with Sidak's multiple comparisons test. Otherwise, a nonparametric Mann-Whitney test (comparison of two groups) or Kruskal-Wallis test with Dunn's multiple comparisons test was used. A p value <0.05 was considered statistically significant.
Results
S. aureus Bacteria Induce AhR-Luciferase Reporter Activity
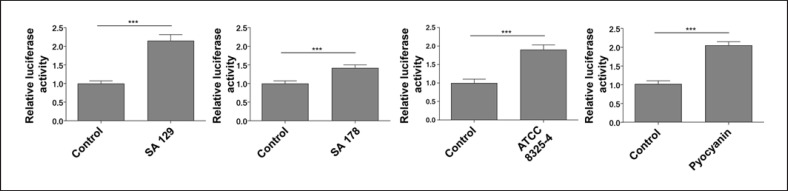
To analyze if S. aureus can activate the AhR, we transfected NHEKs with an AhR-luciferase reporter plasmid and stimulated the cells with different S. aureus strains: the clinical isolate SA 129 from the skin of a healthy person, the clinical isolate SA 178 from lesional skin of an AD patient and the ATCC reference strain 8325-4. All strains increased AhR reporter luciferase activity in comparison to unstimulated NHEKs (shown in Fig. 1). For strain, SA 129 and ATCC 8325-4 this increase was similar to reporter luciferase activity in NHEKs stimulated with the AhR activator pyocyanin [10] which was used as a positive control in this experiment.
Fig. 1.
S. aureus induces AhR-luciferase reporter activity. NHEKs were transfected with an AhR firefly luciferase reporter plasmid (pGudLuc6.1) and a renilla luciferase control plasmid (hRLuc/TK). Two days later the cells were stimulated with living S. aureus (clinical isolates SA 129 and SA 178, ATCC strain 8325-4 and 6.25 μM pyocyanin as positive control). AhR activation was determined by measuring luciferase activity, which was calculated as the ratio of firefly and renilla luciferase activities. Shown are means + SEM (n = 12–18 stimulations, ***p < 0.001, Mann-Whitney U test).
S. aureus Induces AhR Target Gene Expression in Primary Keratinocytes
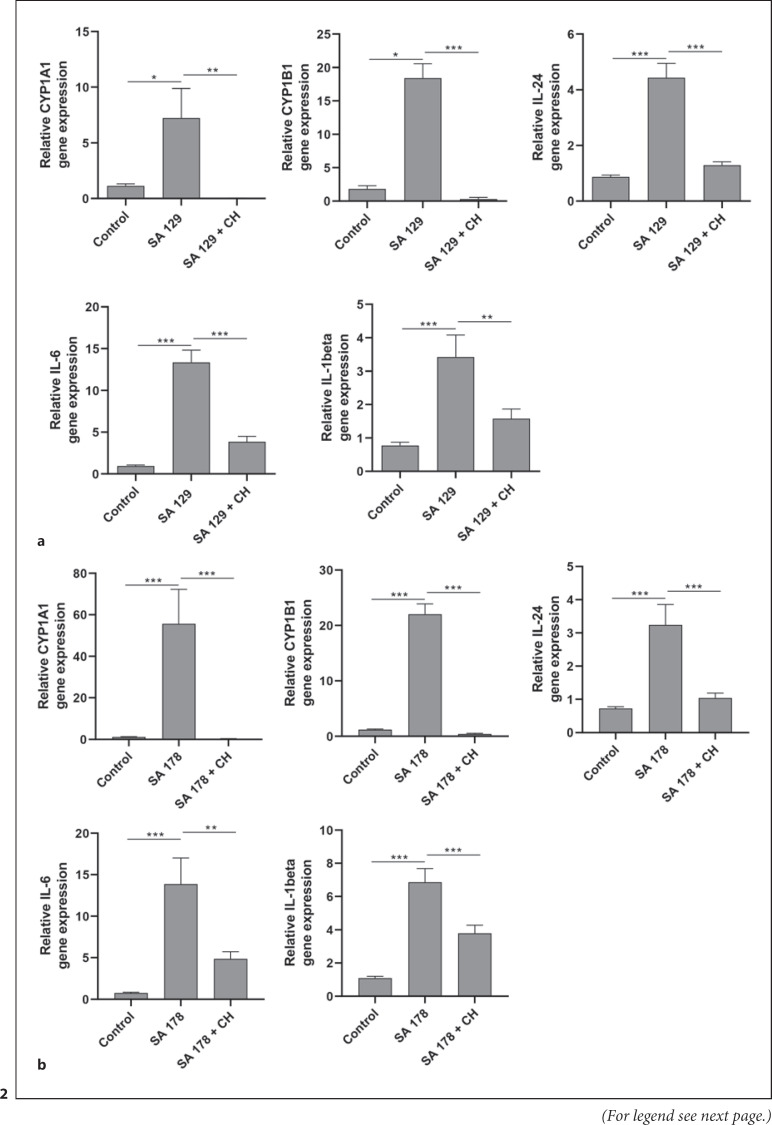
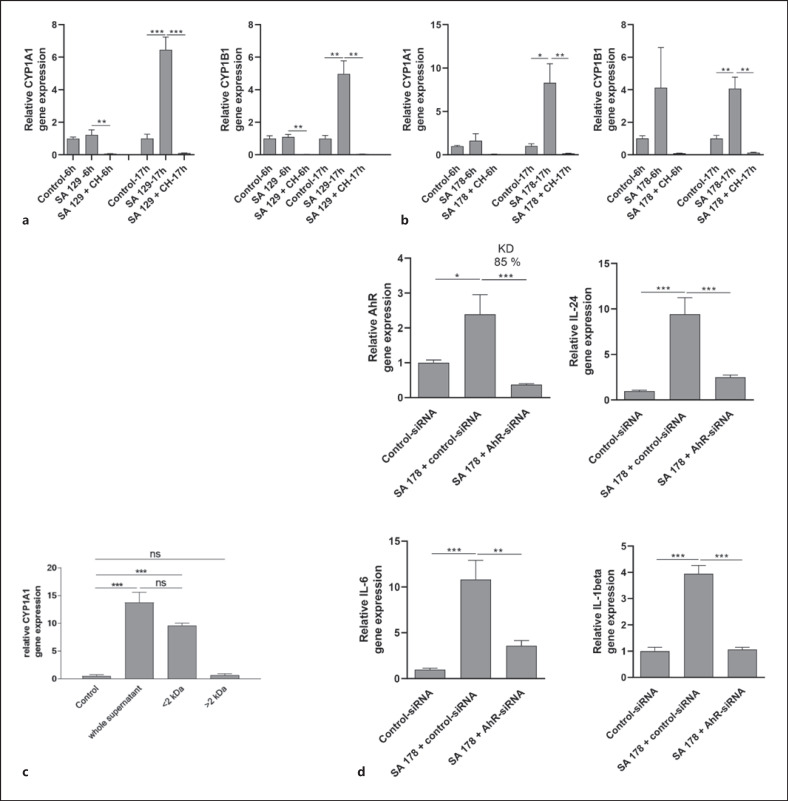
Stimulation of NHEKs with the clinical isolates SA 129 and SA 178 induced the AhR-responsive genes CYP1A1 and CYP1B1. This induction was completely abrogated in the presence of the AhR inhibitor CH-223191 (shown in Fig. 2). To gain further insight into the potential influence of the AhR in S. aureus-induced genes we performed a whole transcriptome analysis of NHEKs stimulated with S. aureus clinical isolate SA 178 in the presence or absence of the AhR inhibitor CH-223191. This approach identified several S. aureus-induced genes whose induction was inhibited by blocking the AhR through CH-223191 (shown in online suppl. Table 1; see www.karger.com/doi/10.1159/000524033 for all online suppl. material). Based on this analysis, we have chosen the cytokines IL-24 and IL-6 for further verification by real-time PCR because the transcriptome sequencing revealed a high S. aureus-induced expression of IL-24 and IL-6, which was inhibited in the presence of the inhibitor CH-223191. In addition, we analyzed the expression of IL-1beta because our previous study showed an AhR-dependent induction of IL-1beta in keratinocytes stimulated with S. epidermidis [9]. Real-time PCR analyses revealed induction of IL-24, IL-6, and IL-1beta in primary keratinocytes treated with S. aureus isolates SA 129 and SA 178. This induction was inhibited in the presence of the specific AhR inhibitor CH-223191 (shown in Fig. 2).
Fig. 2.
S. aureus clinical isolates induce AhR target gene expression. NHEKs were stimulated with two living clinical S. aureus isolates SA 129 (a) and SA 178 (b) with or without the AhR inhibitor CH-223191. Relative gene expression of the AhR-responsive genes CYP1A1 and CYP1B1 as well as the cytokines IL-24, IL-6, and IL-1beta was analyzed by real-time PCR. Shown are cumulative data (means + SEM; n = 9 (a) and n = 15 (b); *p < 0.05, **p < 0.01, ***p < 0.001).
To evaluate if activation of the AhR pathway is a general feature of S. aureus we screened various S. aureus isolates for their capacity to induce CYP1A1 gene induction in primary keratinocytes. This revealed that most strains induced CYP1A1 gene expression (shown in online suppl. Fig. S1).
S. aureus Induces AhR Target Gene Expression in 3D Skin Equivalents
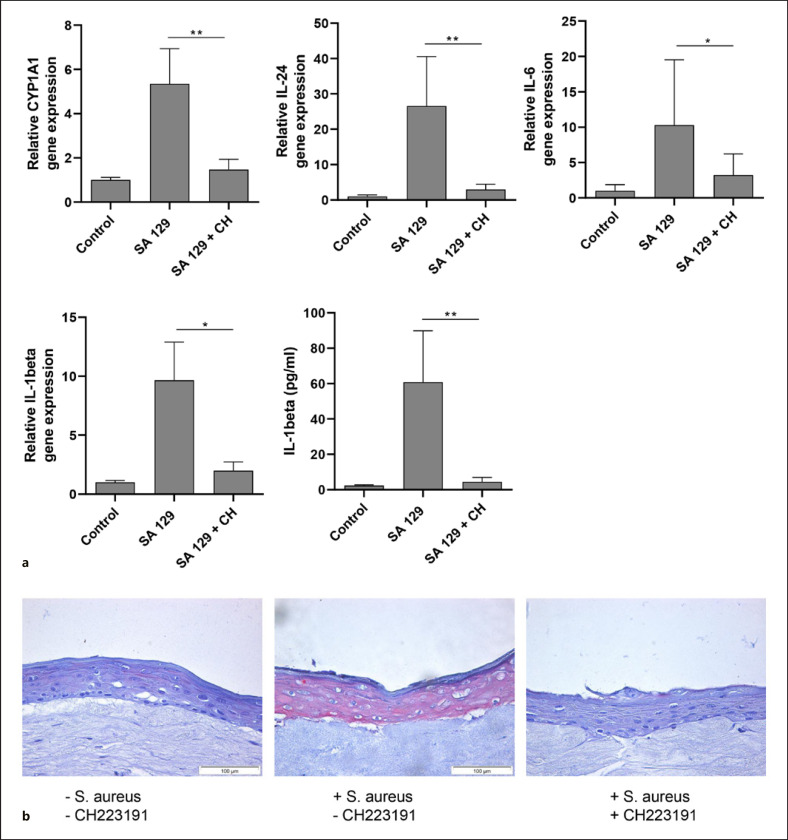
We next stimulated 3D skin equivalents with living S. aureus SA 129 in the presence or absence of the AhR inhibitor CH-223191 and analyzed gene expression by real-time PCR. In line with the results obtained in the 2D culture, S. aureus induced gene expression of the AhR-responsive genes CYP1A1 and CYP1B1 as well as the cytokines IL-24, IL-6, and IL-1beta. This induction was inhibited by the AhR inhibitor CH-223191. IL-1beta protein secretion was also induced by S. aureus and inhibited by CH-223191 (shown in Fig. 3a). Moreover, IL-24 immunostaining of the 3D skin equivalent exposed to S. aureus revealed an intensified IL-24 immunoreactivity, which was reduced by co-treatment with CH-223191 (Fig. 3b).
Fig. 3.
S. aureus induces AhR target gene expression in 3D skin equivalents. a 3D skin equivalents were stimulated for 20–24 h with living S. aureus clinical isolate SA 129 in the presence or absence of the AhR inhibitor CH-223191 (CH). Gene expression of the AhR-responsive genes CYP1A1 and CYP1B1 as well as the cytokines IL-24, IL-6, and IL-1beta was analyzed by real-time PCR. IL-1beta protein secretion was measured by ELISA. Shown are cumulative data of 5 skin equivalents (means + SEM; *p < 0.05, **p < 0.01). b Protein expression of IL-24 was analyzed by immunohistochemistry.
S. aureus Culture Supernatants Induce AhR Target Gene Expression in Primary Keratinocytes
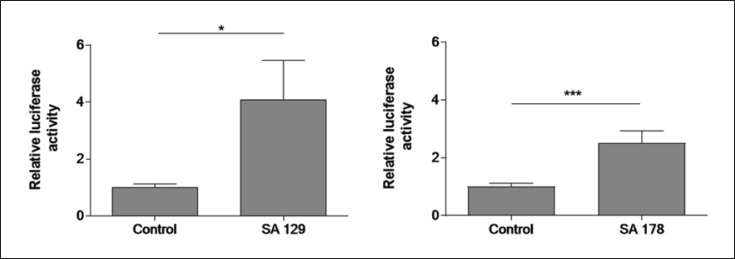
We next sought to determine whether the observed AhR-dependent S. aureus-mediated induction of AhR target genes was mediated by factor(s) released by S. aureus. To this end, we transfected NHEKs with an AhR-luciferase reporter plasmid and stimulated the cells with culture supernatants of S. aureus isolates SA 129 and SA178. This revealed an enhanced luciferase activity indicating activation of the AhR (shown in Fig. 4).
Fig. 4.
S. aureus culture supernatants induce AhR-luciferase reporter activity. NHEKs were transfected with an AhR firefly luciferase reporter plasmid (pGudLuc6.1) and a renilla luciferase control plasmid (hRLuc/TK). 48 h later the cells were stimulated with culture supernatants (1:5 dilution) of S. aureus clinical isolates SA 129 and SA 178 for 16–18 h. AhR activation was determined by measuring luciferase activity, which was calculated as the ratio of firefly and renilla luciferase activities. Shown are means + SEM (n = 13 [SA 129] and n = 16 [SA 178]; *p < 0.05, ***p < 0.001, Mann-Whitney U test).
Subsequently, we stimulated NHEKs with culture supernatants of S. aureus SA 129 and SA 178 for 6 h and 17 h and analyzed gene expression of the AhR-responsive genes CYP1A1 and CYP1B1 by real-time PCR. Induction was seen only after 17 h (shown in Fig. 5a, b). Stimulation of the NHEKs with <2 and >2 kDa ultrafiltrates of S. aureus culture supernatants revealed induction of CYP1A1 only with the <2 kDa ultrafiltrate. This induction was blocked by CH-223191 (shown in Fig. 5c). These data indicate that the AhR-inducing activity is present in the <2 kDa ultrafiltrate.
Fig. 5.
S. aureus culture supernatants induce AhR target gene expression. NHEKs were stimulated with culture supernatants of S. aureus isolates SA 129 (a) and SA 178 (b) for 6 h and 17 h with or without the AhR inhibitor CH-223191 (CH). c NHEKs were stimulated with culture supernatants of S. aureus isolates SA 178, either whole non-filtered supernatant or supernatant with a molecular weight < or >2 kDa. d NHEKs were transfected with a control siRNA and an AhR-specific siRNA and stimulated with culture supernatants of SA 178. Relative gene expression was analyzed by real-time PCR. Bars are means + SEM of three (a–c) or six (d) stimulations (*p < 0.05, **p < 0.01, ***p < 0.001).
We next inhibited the expression of the AhR in NHEKs by transfection of the cells with an AhR-specific siRNA. This revealed a knockdown of AhR expression of 85% (shown in Fig. 5d). Stimulation of the AhR-siRNA-treated NHEKs with culture supernatant of S. aureus SA 178 revealed decreased induction of IL-24, IL-6, and L-1beta (shown in Fig. 5d). These data show that S. aureus secretes factor(s) that induce the cytokines IL-24, IL-6, and L-1beta in an AhR-dependent manner.
Discussion
The role of the AhR in cutaneous defense is still emerging. Recent studies reporting that bacteria such as P. aeruginosa [10] and S. epidermidis as well as Malassezia yeasts [16] activate the AhR strengthen the hypothesis that the AhR may serve as an ancient pattern recognition receptor. Moreover, a recent mouse study has shown that murine skin lacking AhR signaling displayed enhanced epidermal barrier defects. Interestingly, topical colonization with a mix of defined bacterial skin commensals (S. epidermidis, S. hemolyticus, S. warneri, Micrococcus luteus, Corynebacterium aurimucosum) restored epidermal barrier function. This study highlights an important role of the AhR in the epidermal barrier-microbiota interplay and provides further evidence of a crucial role of the AhR in bacterial sensing [25].
In the present study, we show for the first time that the AhR is involved in the recognition of the important skin pathogen S. aureus by keratinocytes. Various S. aureus strains were able to induce expression of the AhR-responsive gene CYP1A1 in keratinocytes indicating that S. aureus in general has the capacity to activate the AhR. Thus, the AhR may play a major role in the interplay of keratinocytes and S. aureus and may act as a pattern recognition receptor to sense the presence of S. aureus. Our data show that the AhR-activating factor(s) released by S. aureus has/have a molecular weight <2 kDa which is in line with the characteristics of small aromatic hydrocarbons as AhR ligands. It is known that tryptophan metabolites act as ligands of the AhR [26] and such tryptophan-derived AhR ligands may be produced by S. aureus, a hypothesis that remains to be proven. A recent study showed that peptidoglycan, a bacterial ligand of toll-like receptor-2 (TLR-2), led to increased CYP1A1 gene expression in keratinocytes indicating activation of AhR signaling. Given the size of peptidoglycan, it is rather unlikely that it serves as a direct AhR ligand. Accordingly, the authors of that study assume that peptidoglycan may indirectly activate AhR signaling through stimulated production of endogenous AhR ligands [27].
There is increasing evidence that therapeutically targeting the AhR may ameliorate skin-associated inflammatory scenarios as seen in the chronic inflammatory skin diseases psoriasis and AD [28]. On the other hand, AhR expression is induced in psoriasis and AD [29], and mice constitutively overexpressing AhR in keratinocytes reveal a disturbed epidermal barrier and increased inflammation that resembled typical AD [30]. It has been hypothesized that under specific pro-inflammatory conditions AhR signaling might be compromised and thus restoration of AhR signaling by AhR agonist may offer a beneficial treatment strategy. In other conditions where an environmental over-activation of the AhR takes place, it would be preferable to dampen AhR signaling. This may also play a role in the prevention of skin cancer and skin aging [31].
We have shown that S. aureus induces IL-24 in keratinocytes, a process that required activation of the AhR. This implies that activation of the AhR in general may lead to increased IL-24 levels. In line with these data, AhR agonists increased IL-24 in an AhR-dependent manner in lung cells and thus IL-24 may contribute to the inflammatory effects of environmental AhR agonists [32]. Moreover, the AhR agonist tapinarof induced the secretion of IL-24 in keratinocytes and IL-24 negatively regulated expression of the skin barrier proteins filaggrin and loricrin [33]. Despite these inhibitory effects of tapinarof-induced IL-24 on filaggrin and loricrin, both proteins were surprisingly induced in keratinocytes treated with tapinarof [33]. IL-24 activated also the JAK1-STAT3 and MAPK pathways in keratinocytes and induced the secretion of pro-inflammatory mediators IL-8, PGE2, and MMP-1 [34]. In transgenic mice that overexpressed IL-24 in the skin, abnormal epidermal differentiation and proliferation were observed accompanied by increased chemokine production and macrophage infiltration [35]. Accordingly, it has been suggested that topical tapinarof application may promote IL-24 expression by keratinocytes thus promoting skin inflammation [33]. Another study suggested that cytokines targeting the IL-20 receptors type I and II, including IL-24 promote cutaneous S. aureus infection in a mouse model by downregulating IL-1beta and IL-17A dependent pathways. As mentioned in the introduction, increased susceptibility for cutaneous S. aureus colonization is associated with AD [4]. Interestingly, elevated IL-24 levels are present in the lesional skin of AD patients [36]. Moreover, a recent transcriptome study using skin biopsies revealed that AhR gene expression positively correlated with AD disease severity scores [21]. Together, these data suggest that activation of the AhR by AhR agonists may trigger inflammatory processes by increased production of IL-24. Our results imply that activation of the AhR by S. aureus may promote S. aureus-mediated inflammatory processes by increased AhR-dependent production of IL-24, a process that may be relevant in AD and other skin infections. Similarly, we also found an increased AhR-dependent induction of IL-6 and IL-1beta in S. aureus-treated keratinocytes. Both cytokines have been also implicated in the pathogenesis of AD. Thus, an AhR-mediated inflammatory response triggered by S. aureus may contribute to skin inflammation in AD. On the other hand, IL-1beta induces human beta-defensin-2 in keratinocytes and human beta-defensin-2 protected against skin damage mediated by an S. aureus protease [37]. Therefore, the AhR-dependent IL-1beta induction by S. aureus may also have beneficial effects to control S. aureus-related harmful effects. Further studies are required to decipher the exact role of the AhR in AD and other inflammatory skin diseases. Since immune cells such as T cells, macrophages, and dendritic cells also functionally express the AhR it is also interesting to evaluate a potential AhR response of such cells against S. aureus [28].
In summary, our study highlights an important role of the AhR in sensing the important skin pathogen S. aureus by keratinocytes. This provides further evidence for the crucial role of the AhR in innate defense. Future studies have to show whether interference with cutaneous AhR signaling may offer therapeutic options to treat or prevent infectious skin diseases.
Statement of Ethics
The isolation of bacteria derived from human skin was approved by the local Ethical Committee of the Medical Faculty at Kiel University (AZ: A 104/06). All study participants gave their written consent. Primary cells were purchased from Promocell (Heidelberg, Germany). The company followed the strictest European and international ethical standards (https://promocell.com/about-us/compliance), including the Declaration of Helsinki. All donors signed an informed consent form.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This study was supported by grants from the German Research Foundation given to J. Harder (HA 3386/5-1/-2) and in parts by funding of the medical faculty of the University of Kiel.
Author Contributions
Eva-Lena Stange, Franziska Rademacher, Regine Gläser, and Jürgen Harder conceived and designed the experiments. Eva-Lena Stange, Franziska Rademacher, Katharina Antonia Drerup, Nina Heinemann, and Lena Möbus performed the experiments and acquired the data. Eva-Lena Stange, Franziska Rademacher, Lena Möbus, Regine Gläser, and Jürgen Harder analyzed the data and prepared the figures. Eva-Lena Stange, Franziska Rademacher, Regine Gläser, and Jürgen Harder wrote the paper. All authors discussed the results and commented on the manuscript.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgments
The authors would like to thank Heilwig Hinrichs and Cornelia Wilgus for their excellent technical assistance. We thank Dr. M.S. Denison (University of California, Davis, CA, USA) for his generous gift of the pGUDLUC6.1 vector. We thank Dr. S. Schubert (Institute for Infection Medicine, Kiel, Germany) for her help to verify the identity of the bacteria by MS-analyses.
Eva-Lena Stange and Franziska Rademacher contributed equally to this work.
Funding Statement
This study was supported by grants from the German Research Foundation given to J. Harder (HA 3386/5-1/-2) and in parts by funding of the medical faculty of the University of Kiel.
References
- 1.Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021 Dec;12((1)):547–69. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, Vos MC, et al. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol. 2009 Jan;9((1)):32–47. doi: 10.1016/j.meegid.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Mistry RD. Skin and soft tissue infections. Pediatr Clin North Am. 2013 Oct;60((5)):1063–82. doi: 10.1016/j.pcl.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Totte JE, van der Feltz WT, Hennekam M, van Belkum A, van Zuuren EJ, Pasmans SG. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol. 2016 Oct;175((4)):687–95. doi: 10.1111/bjd.14566. [DOI] [PubMed] [Google Scholar]
- 5.Kopfnagel V, Harder J, Werfel T. Expression of antimicrobial peptides in atopic dermatitis and possible immunoregulatory functions. Curr Opin Allergy Clin Immunol. 2013 Oct;13((5)):531–6. doi: 10.1097/ACI.0b013e328364ddfd. [DOI] [PubMed] [Google Scholar]
- 6.Bitschar K, Wolz C, Krismer B, Peschel A, Schittek B. Keratinocytes as sensors and central players in the immune defense against Staphylococcus aureus in the skin. J Dermatol Sci. 2017 Sep;87((3)):215–20. doi: 10.1016/j.jdermsci.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Menzies BE, Kenoyer A. Signal transduction and nuclear responses in Staphylococcus aureus-induced expression of human beta-defensin 3 in skin keratinocytes. Infect Immun. 2006 Dec;74((12)):6847–54. doi: 10.1128/IAI.00389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth SA, Simanski M, Rademacher F, Schröder L, Harder J. The pattern recognition receptor NOD2 mediates Staphylococcus aureus-induced IL-17C expression in keratinocytes. J Invest Dermatol. 2014 Feb;134((2)):374–80. doi: 10.1038/jid.2013.313. [DOI] [PubMed] [Google Scholar]
- 9.Rademacher F, Simanski M, Hesse B, Dombrowsky G, Vent N, Glaser R, et al. Staphylococcus epidermidis activates aryl hydrocarbon receptor signaling in human keratinocytes: implications for cutaneous defense. J Innate Immu. 2019;11((2)):125–35. doi: 10.1159/000492162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moura-Alves P, Fae K, Houthuys E, Dorhoi A, Kreuchwig A, Furkert J, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014 Aug 28;512((7515)):387–92. doi: 10.1038/nature13684. [DOI] [PubMed] [Google Scholar]
- 11.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–32. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 12.Rothhammer V, Quintana FJ. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol. 2019 Mar;19((3)):184–97. doi: 10.1038/s41577-019-0125-8. [DOI] [PubMed] [Google Scholar]
- 13.Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev. 2015;67((2)):259–79. doi: 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- 14.van den Bogaard EH, Esser C, Perdew GH. The aryl hydrocarbon receptor at the forefront of host-microbe interactions in the skin: a perspective on current knowledge gaps and directions for future research and therapeutic applications. Exp Dermatol. 2021 Oct;30((10)):1477–83. doi: 10.1111/exd.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cella M, Colonna M. Aryl hydrocarbon receptor: linking environment to immunity. Semin Immunol. 2015 Sep;27((5)):310–4. doi: 10.1016/j.smim.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magiatis P, Pappas P, Gaitanis G, Mexia N, Melliou E, Galanou M, et al. Malassezia yeasts produce a collection of exceptionally potent activators of the Ah (dioxin) receptor detected in diseased human skin. J Invest Dermatol. 2013 Aug;133((8)):2023–30. doi: 10.1038/jid.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas K, Weighardt H, Deenen R, Kohrer K, Clausen B, Zahner S, et al. Aryl hydrocarbon receptor in keratinocytes is essential for murine skin barrier integrity. J Invest Dermatol. 2016 Nov;136((11)):2260–9. doi: 10.1016/j.jid.2016.06.627. [DOI] [PubMed] [Google Scholar]
- 18.Furue M, Hashimoto-Hachiya A, Tsuji G. Aryl hydrocarbon receptor in atopic dermatitis and psoriasis. Int J Mol Sci. 2019 Oct 31;20((21)):5424. doi: 10.3390/ijms20215424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Bogaard EH, Bergboer JG, Vonk-Bergers M, van Vlijmen-Willems IM, Hato SV, van der Valk PG, et al. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. J Clin Invest. 2013 Feb;123((2)):917–27. doi: 10.1172/JCI65642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J, Luo Y, Zhu Z, Zhou Y, Sun L, Gao J, et al. A tryptophan metabolite of the skin microbiota attenuates inflammation in patients with atopic dermatitis through the aryl hydrocarbon receptor. J Allergy Clin Immunol. 2019 Jun;143((6)):2108–19 e12. doi: 10.1016/j.jaci.2018.11.036. [DOI] [PubMed] [Google Scholar]
- 21.Mobus L, Rodriguez E, Harder I, Stolzl D, Boraczynski N, Gerdes S, et al. Atopic dermatitis displays stable and dynamic skin transcriptome signatures. J Allergy Clin Immunol. 2021 Jan;147((1)):213–23. doi: 10.1016/j.jaci.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Long WP, Pray-Grant M, Tsai JC, Perdew GH. Protein kinase C activity is required for aryl hydrocarbon receptor pathway-mediated signal transduction. Mol Pharmacol. 1998 Apr;53((4)):691–700. doi: 10.1124/mol.53.4.691. [DOI] [PubMed] [Google Scholar]
- 23.Rademacher F, Simanski M, Schröder L, Mildner M, Harder J. The role of RNase 7 in innate cutaneous defense against Pseudomonas aeruginosa. Exp Dermatol. 2017 Mar;26((3)):227–33. doi: 10.1111/exd.13166. [DOI] [PubMed] [Google Scholar]
- 24.Bayer A, Wijaya B, Rademacher F, Mobus L, Preuss M, Singh M, et al. Platelet-released growth factors induce genes involved in extracellular matrix formation in human fibroblasts. Int J Mol Sci. 2021 Sep 29;22((19)) doi: 10.3390/ijms221910536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uberoi A, Bartow-McKenney C, Zheng Q, Flowers L, Campbell A, Knight SAB, et al. Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host Microbe. 2021 Aug 11;29((8)):1235–48 e8. doi: 10.1016/j.chom.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szelest M, Walczak K, Plech T. A new insight into the potential role of tryptophan-derived AhR ligands in skin physiological and pathological processes. Int J Mol Sci. 2021 Jan 22;22((3)):1104. doi: 10.3390/ijms22031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Cheng B, Ju Q, Sun BK. AhR regulates peptidoglycan-induced inflammatory gene expression in human keratinocytes. J Innate Immun. 2021 Aug 5;:1–11. doi: 10.1159/000517627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Gallego N, Sanchez-Madrid F, Cibrian D. Role of AHR ligands in skin homeostasis and cutaneous inflammation. Cells. 2021 Nov 15;10((11)):3176. doi: 10.3390/cells10113176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HO, Kim JH, Chung BY, Choi MG, Park CW. Increased expression of the aryl hydrocarbon receptor in patients with chronic inflammatory skin diseases. Exp Dermatol. 2014 Apr;23((4)):278–81. doi: 10.1111/exd.12350. [DOI] [PubMed] [Google Scholar]
- 30.Tauchi M, Hida A, Negishi T, Katsuoka F, Noda S, Mimura J, et al. Constitutive expression of aryl hydrocarbon receptor in keratinocytes causes inflammatory skin lesions. Mol Cell Biol. 2005 Nov;25((21)):9360–8. doi: 10.1128/MCB.25.21.9360-9368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haarmann-Stemmann T, Esser C, Krutmann J. The janus-faced role of aryl hydrocarbon receptor signaling in the skin: consequences for prevention and treatment of skin disorders. J Invest Dermatol. 2015 Nov;135((11)):2572–6. doi: 10.1038/jid.2015.285. [DOI] [PubMed] [Google Scholar]
- 32.Luo YH, Kuo YC, Tsai MH, Ho CC, Tsai HT, Hsu CY, et al. Interleukin-24 as a target cytokine of environmental aryl hydrocarbon receptor agonist exposure in the lung. Toxicol Appl Pharmacol. 2017 Jun 1;324:1–11. doi: 10.1016/j.taap.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Vu YH, Hashimoto-Hachiya A, Takemura M, Yumine A, Metamora Y, Nakahara T, et al. IL-24 negatively regulates keratinocyte differentiation induced by tapinarof, an aryl hydrocarbon receptor modulator: implication in the treatment of atopic dermatitis. Int J Mol Sci. 2020 Dec 10;21((24)):9412. doi: 10.3390/ijms21249412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin SHE, Choi D, Chun NJ, Noh M. Keratinocyte-derived IL-24 plays a role in the positive feedback regulation of epidermal inflammation in response to environmental and endogenous toxic stressors. Toxicol Appl Pharmacol. 2014 Oct 15;280((2)):199–206. doi: 10.1016/j.taap.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 35.He M, Liang P. IL-24 transgenic mice: in vivo evidence of overlapping functions for IL-20, IL-22, and IL-24 in the epidermis. J Immunol. 2010 Feb 15;184((4)):1793–8. doi: 10.4049/jimmunol.0901829. [DOI] [PubMed] [Google Scholar]
- 36.Metamora Y, Nunomura S, Nanri Y, Ogawa M, Yoshihara T, Masuoka M, et al. The IL-13/periostin/IL-24 pathway causes epidermal barrier dysfunction in allergic skin inflammation. Allergy. 2018 Sep;73((9)):1881–91. doi: 10.1111/all.13437. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, McHugh BJ, Qureshi A, Campopiano DJ, Clarke DJ, Fitzgerald JR, et al. IL-1beta-induced protection of keratinocytes against Staphylococcus aureus-secreted proteases is mediated by human beta-defensin 2. J Invest Dermatol. 2017 Jan;137((1)):95–105. doi: 10.1016/j.jid.2016.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.