Abstract
The molecular processes underlying human health and disease are highly complex. Often, genetic and environmental factors contribute to a given disease or phenotype in a non-additive manner, yielding a gene–environment (G × E) interaction. In this work, we broadly review current knowledge on the impact of gene–environment interactions on human health. We first explain the independent impact of genetic variation and the environment. We next detail well-established G × E interactions that impact human health involving environmental toxicants, pollution, viruses, and sex chromosome composition. We conclude with possibilities and challenges for studying G × E interactions.
Subject terms: Population genetics, Immunology
Introduction
For centuries, clinicians and scientists have sought to understand the etiology of disease. While some diseases can be traced back to a single factor, the etiology of complex diseases is more difficult to discern, in part due to the combinatorial nature of various contributing factors. One such factor contributing to disease risk is an individual’s genetics, with some individuals inheriting specific genetic variants that either (1) directly trigger disease pathogenesis or (2) work in concert with other factors and/or other genetic variants to increase disease risk. In many cases, Environmental exposures, defined here as pathogens, chemicals, and additional external factors, have also been shown to contribute to disease. While epidemiological studies can identify associative relationships between exposure to environmental factors and disease pathogenesis, not all individuals who are exposed to a specific environmental factor develop disease. Likewise, not all individuals who inherit particular genetic variants develop disease. For the vast majority of diseases, it is apparent that combinations of synergistic or antagonistic factors are important to disease risk. Such “Gene by Environment” (G × E) interactions are the focus of this review.
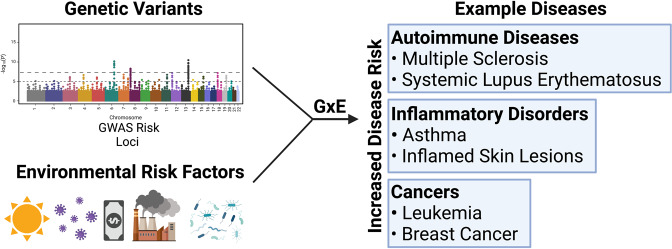
Prior reviews have described G × E interactions in nonhuman organisms such as yeasts [1], or in specific human disease contexts, such as inflammatory diseases or particular psychological conditions [2–7]. Others have focused on models of GxE interactions, such as Ottman [8] and Kauffman and Demenais [9] who collectively proposed four models for G × E interactions: (1) the risk genotype exacerbates the effect of the environmental risk factor; (2) exposure to the environmental risk factor exacerbates the effect of the risk genotype; (3) both the environmental risk factor and risk genotype are required to increase disease risk, and (4) the environmental risk factor and the genotype each have some effect on disease risk, and risk is higher when they occur together than when they occur alone. This review focuses on the latter model, where a synergistic relationship between environmental risk factors and genetic factors increases disease risk (Fig. 1). Herein, we broadly review G × E in the context of human health with a focus on how particular genetic and environmental factors synergistically increase disease risk. We first describe instances where genetic and environmental risk factors can independently potentiate disease. We then examine how these two factors can work together to increase disease risk.
Fig. 1. Gene × environment (G × E) interactions involve synergy between environmental risk factors and genetic variants.
Some G × E interactions can increase the risk of disease. A model of G × E interaction originally defined in Ottman [8] and further refined in Kauffmann and Demenais [9] is depicted where the genetic risk variants and one or more environmental risk factor synergistically affect disease risk.
Genetic etiology of disease
In many cases, genetic variants impact phenotypes that contribute to disease pathology. Historically, family studies have been used to measure the contribution of genetics to a particular trait through studies comparing disease risk in monozygotic twins (100% shared genetic identity), dizygotic twins (50% shared genetic identity), and siblings (also 50% shared genetic identity) [10]. Significant differences between monozygotic and dizygotic disease concordance establish strong genetic heritability. For example, three independent twin studies have identified a monozygotic twin concordance for Crohn’s disease of ~50% compared to ~3–4% disease concordance in dizygotic twins [11–13]. The difference in concordance of monozygotic and dizygotic twins also highlights the possible role of epistatic risk factors that require interactions between multiple genetic variants to increase disease risk [14]. The over ten-fold increase in Crohn’s disease concordance in siblings with identical DNA sequence compared to siblings sharing 50% genetic identity indicates a large genetic component to disease risk.
Diseases with a genetic component can be monogenic (i.e., caused by a single rare mutation), complex (i.e., caused by the cumulative effect of multiple genetic events and/or the environment), or both. For example, cystic fibrosis is a monogenic condition caused by a mutation in the CFTR gene [15, 16], with additional genetic variants impacting disease presentation and severity [17]. However, many “causal” disease mutations are incompletely penetrant, with some individuals carrying the mutation expressing various degrees of symptoms and some individuals not expressing the disease at all [18]. For example, only 10–30% of people with damaging mutations in the gene encoding complement component 2 (C2) develop systemic lupus erythematosus (lupus) [19]. Instead, most disease manifestations are the result of complex etiologies, with many relatively common genetic polymorphisms (i.e., allele frequencies greater than 1%) contributing to disease risk in an additive manner [20]. Indeed, most patients with lupus do not have monogenetic disease [21], with the most common form of monogenic lupus (mutations in the TREX1 gene) contributing to only 0.5 to 2% of adult lupus cases [22].
Identifying genetic risk loci
Genome-wide association studies (GWAS) have emerged as the predominant tool for the systematic, genome-wide identification of disease-associated genetic risk variants. Such studies genotype thousands of cases and controls to identify statistically significant genetic associations between particular variants and a given disease phenotype [23]. The most recently published GWAS “catalog” contains over 5000 independent GWAS datasets that describe more than 70,000 variant-trait associations [24]. While such studies are helpful for identifying disease risk loci for further functional analysis, they have shortcomings. First, study participant environments are not standardized, and thus, potential environmental effects are not well controlled. Second, only a small fraction of the variants identified in a given GWAS are causal, due to linkage disequilibrium; specifically, the particular variant(s) directly studied (“tagged”) in the GWAS may simply be in strong linkage disequilibrium with the variant that is functionally influencing the phenotype [25]. Finally, GWAS to date have focused disproportionately (>78%) on individuals of European descent [26]. Additional studies focused on nonwhite populations are thus needed to obtain a clearer picture of the disease spectrum across ancestries [27, 28]. Despite these limitations, the increasing availability of GWAS data has enabled researchers to pursue novel hypotheses and design targeted studies to further investigate functional roles between a particular variant and its associated phenotype.
Functional interpretation of genetic risk loci
While GWAS have identified many genetic risk loci, additional studies are required to elucidate the causative molecular mechanisms underlying disease. Genetic variants identified in GWAS can increase disease risk through multiple means, including changing the amino acid of a protein, altering gene regulatory mechanisms, impacting RNA splicing, and affecting translation rates. For example, the identification of loss-of-function nonsynonymous coding genetic variants within the filaggrin (FLG) gene in ~10% of atopic dermatitis patients [29] led to further studies revealing the role of FLG in the development of a healthy epidermis [30, 31] and atopic dermatitis pathogenesis [32]. In particular, studies in cells from patients with FLG mutations and in mice lacking a functional FLG helped shape the current hypothesis that defects in the skin barrier lead to allergic sensitization and the progression of atopic dermatis in infancy toward asthma and allergic disease in later childhood, commonly known as the atopic march [33]. More recent studies have focused on the role of FLG mutation-mediated skin barrier defects in skin dysbiosis and Staphylococcus colonization [34–36], illustrating how genetic studies can lead to previously unexplored avenues and, ultimately, targeted studies to ascribe functional contributions of variants to disease processes.
Mutations at the FLG locus demonstrate how a change in the genetic code can contribute to disease processes by altering the amino acid sequence of a protein, which may affect its structure and/or function as a consequence. However, it should be noted that variants do not need to occur in coding regions to elicit a phenotype. In addition to coding mutations, variants in noncoding regions also make substantial contributions to disease risk by altering gene regulatory mechanisms. Indeed, GWAS variants for many diseases are highly enriched within noncoding genomic regions [37]. Such variants likely alter gene regulatory mechanisms, leading to genotype-dependent variability in gene expression levels that contribute to disease risk [37, 38], often by altering transcriptional elements (e.g., promoters or distal regulatory elements such as enhancers) or posttranscriptional elements (e.g., regions controlling mRNA splicing or mRNA localization) [39]. One important way in which noncoding variants affect regulatory element function is through the alteration of transcription factor (TF) DNA binding interactions. Both amino acid-altering variants located within the TF proteins themselves and noncoding variants located within TF genomic binding sites can alter TF-DNA binding specificity or affinity [40–42]. For example, an obesity-associated intronic genetic variant in the FTO locus leads to genotype-dependent binding of the ARID5B TF, resulting in a doubling of the expression of the IRX3 and IRX5 genes, a genotype-dependent increase in energy-storing adipocytes, and a decrease in energy-dissipating adipocytes [43]. In addition to genetic variants that alter TF-based transcriptional regulatory mechanisms, genetic variants can also alter posttranscriptional regulatory mechanisms by affecting the binding of RNA binding proteins [44], microRNAs [45–47], or long noncoding RNAs [48].
Modern computational and statistical methods have enabled the robust assessment of genotype-dependent biology. Recently, quantitative trait locus (QTL) and allelic imbalance analyses have emerged as powerful tools for identifying genetic variants with genotype-dependent biological effects [49]. QTL studies identify genotype-dependent biology by quantitatively comparing a particular phenotype (e.g., the expression level of a gene) across many individuals as a function of the genotype of each individual [50]. Such associations can be used to describe the effects of genetic variants on cell biology, meaning that QTLs and GWAS data can collectively inform numerous aspects of disease mechanisms, including the nomination of likely causal disease variants [49]. For example, histone QTLs are highly enriched within autoimmune disease risk haplotypes in cell types relevant for disease etiology, implicating differential epigenetic mechanisms at multiple genomic loci [51].
GWAS identify groups of genetic variants that are inherited together as a genetic haplotype. Genetic association is often insufficient to narrow these variants down to the small group that actually change biological processes in a genotype-dependent manner. Additional computational methods can nominate causal disease variants by identifying risk alleles that alter the binding of particular regulatory proteins. An example of a current method is Measurement of Allelic Ratios Informatics Operator (MARIO), which identifies allele-dependent binding of regulatory proteins at heterozygous variants [52] by examining allelic imbalance in the reads of a functional genomics experiment. A substantial benefit of computational methods such as MARIO is that they can largely bypass the need for large sample sizes used in QTL analyses. However, such methods are also limited by their dependence on contextually-relevant datasets obtained through methods such as chromatin immunoprecipitation sequencing (ChIP-seq). While this information may be available for diseases that have been well-studied, sequencing datasets for regulatory proteins in less-common diseases may not be as abundant or available. Therefore, continued development of new analytical methodologies should be pursued in order to deliver the full benefits of large-scale data analysis to a broader range of topics in human health and disease. Collectively, QTLs, allelic imbalance analyses, and other advances in functional genomics methodologies have enabled researchers to go beyond risk variant identification to discover potential molecular mechanisms of disease underlying genetic associations.
Environmental etiology of diseases
As a part of the environment, organisms are continuously exposed to a myriad of external factors that shape health and disease. Many environmental factors that individuals are routinely exposed to have been associated with disease risk, including the use and consumption of various substances, such as tobacco and alcohol, as well as exposure to ultraviolet light [53–56]. Environmental data can be collected prospectively through cohort studies or retrospectively through medical records, surveys, or government records. For example, the addresses reported by a child across numerous trips to the emergency department for asthma treatment can be converted into geocodes that allow quantification of exposure to air pollution near interstate highways and provide information on the median home price and salary [57–59].
The environment itself is constantly changing. Industrialization and urbanization adversely affect human health [60, 61], while climate change in turn threatens to alter the way humans live and interact with the environment [62]. The coronavirus disease-19 (COVID-19) pandemic has demonstrated how quickly the environment can change, with mitigation measures in place to combat the pandemic drastically changing the prevalence of influenza and other infectious diseases [63, 64]. In this section, we focus on major components of the environment and how they impact human health: the microbiome, pollutants and environmental toxicants, viral infection, climate change, and psychosocial and economic factors. While recent reports have comprehensively reviewed hallmarks of environmental insults (e.g., [65]), this section will highlight diseases that impact the immune system.
The microbiome
The microbiome includes all of the microbes that reside on and inside the human body. Contributing up to 100 trillion cells in adults, the microbiome plays critical roles in development, nutrition, digestion, and immunity [66–69]. In this section, we introduce the microbiome in the context of responding to external exposures, with an emphasis on disease risk.
The microbiome plays a critical role in immunity by providing protection against allergic disease. Lifestyle changes in recent decades, especially in high-income and industrialized countries, have led to a decrease in the incidence of infections while increasing the incidence of autoimmune and allergic diseases [70]. According to the hygiene hypothesis, this increase in allergic and autoimmune disease prevalence in recent decades is attributable to a decrease in infectious disease incidence, particularly in developed nations. While this concept is strongly supported by epidemiological data, the mechanisms driving this relationship are not as well characterized [71, 72]. Some gut microbiota confer protection against food allergen sensitization through the activation of genes involved in innate immunity [73, 74]. For example, germ-free mice transplanted with microbiota from a healthy infant were protected from an allergic response when challenged with cow’s milk allergen β-lactoglobulin, while the transfer of microbiota from infants allergic to cow’s milk resulted in a response [74]. Transcriptional analyses of intestinal epithelial cells from germ-free mice, healthy mice, and mice colonized with cow’s milk allergen identified unique transcriptomic signatures among the three groups. Notably, genes involved in epithelial repair and metabolism were expressed as a function of the type of colonization. In a separate study, Stefka et al. found an increased proportion of regulatory T cells in the colonic lamina propria and elevated concentrations of fecal immunoglobulin A of mice colonized with Clostridia-containing microbiota, mechanistically demonstrating how colonization with these microbiota provides protection against allergies [73]. Furthermore, intestinal epithelial cells from colonized mice also showed an increase in the expression of genes with functions in the innate immune system compared to germ-free mice. The identities of these genes further elucidate how the microbiota confer protection for disease. For example, regenerating islet-derived 3 beta (REG3B), which encodes an antimicrobial peptide that regulates mucosal microbiota composition, is upregulated in colonized mice. Moreover, clostridia colonization induces the immunological cytokine IL-22, resulting in intestinal epithelial cell-mediated production of antimicrobial peptides and protection of the intestinal epithelial barrier by increasing the number of mucus-producing goblet cells. These findings collectively suggest that microbiota may induce specific, immunologically relevant gene expression signatures that help protect against allergic disease.
While the underlying molecular and microbial mechanisms remain to be fully characterized, these exemplary studies highlight the critical role of commensal microbiota in shaping the immune system and, subsequently, their contributions to the modulation of susceptibility for multiple immune-related diseases. Notably, allergic diseases are just one of many phenotypes impacted by the microbiome, and the microbiome itself is affected by many other environmental factors including diet, birth mode, exposure to antibiotics, and age [75, 76].
Pollutants and environmental toxicants
Environmental exposures are complex and rely on several factors that include, but are not limited to, time, geographic region, and route of exposure. In the United States, residential proximity to sites containing environmental hazards has been associated with potential reduction in life expectancy [77] and multiple adverse health outcomes that have been extensively reviewed [78]. More broadly, the Lancet Commission on pollution and health reported that total pollution exposure was a leading risk factor for global estimated deaths in their analysis of the 2019 Global Burden of Diseases, Injuries, and Risk Factors Study data [79], attributing pollution to ~9 million deaths [79]. Given the magnitude of these disease burden estimates, understanding how environmental toxicants elicit adverse effects serves as a critical step for identifying populations at risk, reducing offending sources of emissions, and improving public health. In this section, we briefly highlight connections between environmental exposures and adverse health outcomes with an emphasis on ambient air pollution and water contamination.
The World Health Organization (WHO) has estimated that exposure to ozone and ambient air pollution, defined in their global burden estimates as particulate matter with a diameter less than or equal to 2.5 µm (PM2.5), can be attributed to 4 to 9 million global deaths annually [80]. The composition of PM2.5 is diverse and may depend on its source of introduction, with combustion-related activities of energy production, energy use, and industrial processes being notable sources of anthropogenic contribution [81]. Studies investigating the associations between PM2.5 and adverse health outcomes have implicated the particulate matter (and its composition [82]) in the initiation and progression of multiple diseases including cardiovascular disease [83–86], asthma [87–89], and lung cancer [90, 91], with the International Agency for Research on Cancer classifying both outdoor pollution and its relevant constituents as carcinogenic to humans [92]. This is particularly concerning given that the trend toward increased urbanization and anthropogenic activity has been positively correlated with changes in PM2.5 concentrations [93], reinforcing the need for air quality monitoring and pollution reduction initiatives.
Ingestion of contaminated food and water are additional routes of exposure, with UN-Water estimating that two billion people lacked safely managed drinking water services in 2020 [94]. Although many different types of contamination contribute to overall risk, here we introduce a subset of metals that have been consistently identified as significant environmental contaminants. In particular, arsenic is currently considered to be a global issue that may expose between 94 million and 220 million people to high concentrations of the metal through groundwater sources [95]. Chronic arsenic poisoning, also known as arsenicosis, frequently manifests in the form of skin lesions such as melanosis and keratosis [96], and has been causally associated with multiple cancers [97]. While arsenic contamination is often the result of natural processes, industrial wastewater and improper disposal methods of other metals have been attributed to the development of adverse health outcomes, including Minimata disease (methylmercury) [98] and Itai-itai disease (cadmium) [99].
Together, these examples provide a brief introduction to the intrinsic link that exists between environmental contaminants and human health. However, while some of the cases presented here implicate a single compound in elevated concentrations as the source of toxicity, this is not necessarily representative of the challenges that are faced when studying environmental toxicants in the context of health and disease. For instance, individuals are more likely to be subject to complex mixtures that occur in lower concentrations, resulting in chronic exposure at levels that may not elicit immediate effects. In addition, increases in chemical manufacturing have led to the use of thousands of chemicals lacking adequate toxicity assessments. Thus, a paradigm exists in which the general population is potentially exposed to more compounds through widespread, albeit low, exposures across all stages of development that may alter their disease risk and potentiate adverse health outcomes later in life [100].
Viral infection
Viruses, defined as infectious particles comprised of genetic material (DNA or RNA) surrounded by either a protein coat or membrane [101], represent an important component of the environment that is present virtually worldwide. In addition to directly causing diseases such as HIV and shingles, viral infections can increase the risk for a variety of noninfectious diseases, including cancers, allergic diseases, and autoimmune diseases [102–104].
Approximately two million cancer cases annually result from infectious agents, including viruses [105]. Human papillomavirus (HPV), Epstein-Barr virus (EBV), hepatitis B, and hepatitis C (HCV) can cause metastatic transformation of specific cell types originating in a variety of organs. The relationship between infection of high-risk HPV types and anogenital cancers, particularly cervical cancer, is well characterized, with recent studies demonstrating a causal role for HPV in head and neck cancers as well as cancers of the vulva, vagina, penis, and anus [106]. Similarly, HCV infection is associated with hepatocellular carcinoma and subtypes of non-Hodgkin lymphoma, with recent studies suggesting that HCV could also increase the risk of bile duct cancers and diffuse large B-cell lymphoma [107, 108]. EBV infection is associated with several B-cell lymphoproliferative disorders, such as Burkitt lymphoma, Hodgkin disease, systemic non-Hodgkin lymphoma, primary central nervous system lymphoma, and nasopharyngeal carcinoma [109–113]. Some cancers, such as cervical cancer, are related to viral infections acquired during infancy or childhood (for example HPV) that can impact cancer onset later in life. Preventing infections via vaccination can significantly reduce the risk of many of these lethal cancers. A nationwide study in Sweden with over 1.5 million participants showed that quadrivalent HPV vaccine use substantially reduces the risk of invasive cervical cancer [114, 115].
Viral infection can also contribute to the development of multiple allergic diseases [116]. Respiratory viruses have been found to account for 85% of asthma exacerbations in both adults and school-aged children [117]. In particular, respiratory syncytial virus is a risk factor for the development of bronchiolitis and asthma [117–119]. While the exact molecular mechanisms mediating the epidemiological associations of viruses and asthma are not fully understood, they likely involve virus-induced damage of the airways, changes to immune cell activity, modifications to the bacterial microbiome, and additional virulence factors [116]. Causality in association studies is challenged by disease-associated physiological changes in viral defense systems. For example, asthma is associated with rhinovirus infection, and it is challenging to decipher from these statistical associations if patients at risk for asthma have disease-specific physiology that make them more susceptible to rhinoviruses or if rhinovirus infection leads to asthma-specific physiology [120–122].
Viral infections have also been linked to several inflammatory and autoimmune conditions. Some viruses, for example, can induce inflammation that causes tissue damage, as is the case in coxsackievirus B3-induced autoimmune myocarditis [123]. Infection with SARS-CoV-2, the virus that causes coronavirus disease 2019 (COVID-19), has recently been implicated in the development of autoimmune diseases, including Kawasaki disease, pediatric inflammatory multisystemic syndrome, coagulopathy, antiphospholipid syndrome, and Guillain–Barre syndrome [124, 125]. Recent studies also suggest that some severe cases of COVID-19 may be exacerbated by the presence of autoantibodies against type I interferons, meaning that autoimmunity due to autoantibodies made by the adaptive immune system may impair innate antiviral immunity [126]. EBV infection in particular has been associated with a host of autoimmune diseases, including systemic lupus erythematosus (SLE), multiple sclerosis (MS), and rheumatoid arthritis [127–129]. Patients with SLE and MS have a statistically elevated viral load and decreased EBV-driven cell-mediated immunity compared to healthy controls, suggesting that these patients have poorer control over EBV replication [130, 131]. Thus, viral infections have been shown to be powerful drivers of disease, with two recent studies providing highly compelling evidence that EBV infection is causative for MS [132, 133]. Taken together, these examples highlight the need to better understand how viral infections act in concert with disease risk variants to increase risk for diseases with complex etiologies.
Climate change
Long-term shifts in temperature and weather patterns due to human activity have both directly and indirectly increased the prevalence of disease [134]. For example, changing temperature and weather patterns are directly accelerating the allergy epidemic by altering concentrations of pollens that exacerbate allergy symptoms [135]. Further, climate change impacts the distribution of vector-borne pathogens, altering the length of transmission seasons and the duration that immunologically naïve populations are exposed to infectious diseases [136]. For example, current models predict that climate change across the world will lead to a climate more suitable for dengue and arbovirus transmission [137]. Indeed, climate change has led to recent outbreaks of dengue, West Nile fever, and chikungunya in Europe [138]. Some countries in sub-Saharan Africa are accustomed to high levels of malaria transmission and thus have developed effective tools to control transmission [137]—such interventions might need to be applied more widely. However, interventions are currently not available for blocking transmission of viruses such as arboviruses and dengue. This leaves populations impacted by climate change-induced virus exposure vulnerable to epidemic-level spread and morbidity.
Climate change can also indirectly increase disease susceptibility by altering socioeconomic factors that leave individuals vulnerable to disease. A recent study showed that warming temperatures and increasing rainfall variability due to climate change adversely affect food security and diet diversity. Such effects are particularly strong in low-income regions, leading to increased malnutrition and impaired childhood development [139, 140]. Thus, the broad impacts of climate change are expected to contribute to disease prevalence both directly and indirectly, highlighting the need to take into consideration how a rapidly changing environment may affect the public health of global communities.
Racism, stress, and economic factors
Healthcare disparities are defined as preventable differences in health outcomes that negatively impact groups of people with shared socioeconomic or demographic features. Such differences in disease risk are often driven by environmental exposures [141–145]. Occupational and general environmental exposure to toxicants (e.g., lead in drinking water and paint), rates of nicotine use, and access to high-quality primary care are examples of environmental exposures that impact racial and socioeconomic groups disproportionally [146, 147]. Stress and trauma are well-established environmental risk factors for diseases ranging from heart disease to anxiety that disproportionately impact people who are Black [146, 147]. In a recent study, Resztak et al. developed an approach to derive transcriptional signatures from peripheral blood RNA-seq samples of asthmatic children in the metropolitan Detroit area that were correlated with various psychosocial factors. Among other findings, the authors reported that psychosocial factors altered the expression of 169 genes that have been causally linked to asthma or allergic disease and concluded that the modulation of the immune system may serve as an important mediator between these factors and asthma risk [148]. The deeper implications of this study suggest that molecular-based approaches, when coupled with statistical modeling techniques, could be used to better understand how extrinsic environmental factors may play a disproportionate role in the health and wellbeing of an individual. Taken together, these studies demonstrate that the health of an individual is intricately shaped by their surroundings. Institutional discrimination, which can include socioeconomic status and systemic racism, may dramatically alter the extrinsic factors of an individual’s environment, which may subsequently increase the risk of particular adverse health outcomes. It is therefore imperative for the research community to identify cohorts that are representative of our diverse communities and use current methodologies to identify additional causes of adverse health outcomes. As molecular-based approaches continue to improve, novel techniques may serve as an avenue for identifying previously unknown risk factors, which could then pave the way for developing solutions that would improve public health and close the gap in healthcare disparities.
Gene × environment interactions
While genetic and environmental factors can independently increase the risk of disease, the interactions between these risk factors (G × E) also have a profound influence on human health. An expanding number of studies have found that disease risk variants impact environmental risk factors, with the implication being that environmental exposures can elicit an altered response in the context of genetic risk variants [149–151]. Identification of G × E interactions and their contributions to disease etiology provides a more comprehensive understanding of the mechanisms driving risk for many human diseases. In this section, we discuss major categories of environmental factors currently implicated in G × E mechanisms.
Environmental toxicants
There are many forms of environmental toxicants, and they can influence many diseases. Toxicants represent a special class of G × E interactions because the relationship between the two components is bidirectional—the environment can directly alter the genotype of an individual (i.e., through a somatic mutation), and toxicant metabolism can be affected by inherited genetic variants (i.e., through germline inherited polymorphisms) [152]. Toxicants that cause cancer are called carcinogens. An increase in the amount of carcinogens in the environment has contributed to a global increase in cancer incidence [100]. When carcinogens directly contribute to tumor development, often through a combination of somatic mutations and epigenetic modifications, these changes can directly result in genotype-dependent alterations impacting DNA repair mechanisms or gene regulatory mechanisms [153].
There are numerous ways that environmental toxicants can interact with germline polymorphisms affecting the uptake, metabolism, and transport of toxic compounds. For example, genetic variants associated with arsenic metabolism at the 10q24.32 locus near AS3MT are associated with inefficient arsenic metabolism and subsequent toxic arsenic exposure [154]. Similarly, a missense variant in the FTCD gene has been proposed to affect the efficiency of arsenic metabolism, potentially by reducing the availability of methyl groups involved in its detoxification [155]. Arsenic contamination in drinking water sources is considered to be a widespread problem and it has been estimated that over 100 million people worldwide are exposed to concentrations exceeding WHO-recommended limits [156]. As a consequence, arsenic is expected to be a significant contributor to disease burden. Paul et al. provide an in-depth review supporting the role of genetic variation in arsenic-induced toxicity, suggesting that the effects of arsenic on the health of an individual have a genotype-dependent component that may account for differences in disease outcome [157]. Like other toxicants, arsenic-related toxicities depend on a multitude of factors including the concentration of the metal, the length of exposure, and the efficiency of its detoxification pathways within the body.
The metabolism of other heavy metals provides additional support for the hypothesis that common polymorphisms contribute to diseases through G × E mechanisms [158]. For example, metallothioneins are metal-binding proteins that regulate metal distribution and help protect cells against heavy metal toxicity. A genetic variant in the core promoter of metallothionein 2A (MT2A) affects the expression level of MT2Am, which is inversely correlated with the accumulation of cadmium and copper in sinonasal inverted papilloma tissues [159]. Genetic polymorphisms in genes involved in heavy metal metabolism are of significant public health importance because most individuals experience chronic exposure to some level of heavy metals [160].
Pollution
The worldwide pollution crisis continues to negatively impact human health. In the example of asthma, both outdoor and indoor air pollution can interact with genetic variants to increase disease risk [161]. A G × E study in mice demonstrated that the magnitude of airway hyperreactivity in response to diesel exhaust particles is dependent upon genotypes at the Dapp1 locus [162]. In humans, GWAS identified a G × E interaction between diesel exhaust-elicited airway hyperreactivity and a locus on chromosome 3 encoding DAPP1 [162]. Asthma prevalence is higher among low-income African-American children, who are more likely to reside near highways and industrial areas. The health disparity of asthma can thus be partially attributed to the fact that pollution exposure disproportionately affects low-income populations. Because the currently known genetic and environmental risk factors cannot fully explain the risk of asthma, there is a tremendous need to further delineate additional G × E interactions.
Viruses
Viruses interact with their hosts on many levels. When human cells encounter viruses, the pattern recognition and adaptive immune receptors lead to immunological responses aimed at clearing viral infection. Some viruses infect cells without killing them, transitioning to a latent infection. In latency, the virus continues to produce low levels of certain genes, including those encoding transcriptional regulators that interact with the virus and host genomes. The most well-studied GxE viral mechanisms involve viral transcriptional regulatory proteins that interact with the human genome at disease risk variants and alter human gene expression. For example, Epstein-Barr nuclear antigen 2 (EBNA2) regulates human gene expression levels by mimicking activated Notch [163]. Similar to Notch, EBNA2 can influence gene expression by impacting chromatin looping, chromatin accessibility, and human TF binding [164–166]. The genetic locations of these epigenetic effects are highly enriched for autoimmune genetic risk variants. For example, EBNA2 binding events intersect nearly half of known lupus and MS risk loci [52, 164]. Similarly, EBNA2-dependent altered chromatin accessibility and looping events are highly enriched for autoimmune genetic risk variants [164]. Complementary analyses in the same study demonstrate that EBNA2-dependent binding, chromatin accessibility, and chromatin looping at genetic autoimmune disease risk variants are often genotype-dependent [164]. EBNA2 and other EBV proteins are amongst the most highly studied viral transcriptional regulators [167], and it is likely that many other virally encoded transcriptional regulators interact with the human genome at disease risk variants to mediate GxE effects on transcription, cell biology, and disease risk.
Genetic modifiers of infectious diseases
In addition to increasing disease risk through interaction with human regulatory elements at disease risk variants, mechanisms initiated by viruses and other infectious agents can also be impacted by rare mutations in key regulators of the immune response. For example, the broad spectrum of disease severity in response to infection with pathogens such as SARS-CoV-2 and influenza is due in part to host genetic variation at loci encoding regulators of antiviral cytokines and innate pattern recognition receptors. Rare mutations in the interferon regulatory factor 7 (IRF7) gene, a key regulator of antiviral type I Interferons (IFN-I), have been shown to underlie cases of severe influenza COVID-19 pneumonia. Similarly, whole-exome sequencing on an otherwise healthy child with influenza-induced life-threatening acute respiratory distress syndrome (ARDS) revealed two compound heterozygous mutations in IRF7, resulting in very little IFN-I production in response to influenza infection [168]. IRF7 is activated primarily by stimulation of endosomal Toll-Like Receptors (TLRs), resulting in phosphorylation and nuclear localization where IRF7 regulates IFN-I gene expression [169]. Mechanistically, the two mutant loss-of-function alleles result in IRF7 protein that (1) localizes to the nucleus without phosphorylation and (2) does not localize to the nucleus following phosphorylation, respectively. Surprisingly, this patient’s adaptive responses (as measured through B and T cell responses to infection) were normal, suggesting that the life-threatening disease was caused by a blunting of the innate response due to the mutations in the two copies of IRF7. In the case of COVID-19, 3.5% of patients with life-threatening COVID-19 pneumonia in one study had genetic defects in TLR-3 and IRF7-dependent signaling pathways of IFN-I [170]. Moreover, deficiencies in the IFN-I pathway are estimated to contribute to nearly 10% of pediatric COVID-19 hospitalizations, despite this age group being classified as low risk for severe disease [171]. These examples illustrate how inherited deficiencies in regulators of the immune response to infection translate to severe outcomes for relatively common diseases.
While some rare loss-of-function mutations can result in life-threatening infection, such effects are likely virus-specific and host-cell intrinsic. A recent study in otherwise healthy humans with inherited IRF7 deficiency showed that while affected individuals were highly susceptible to infections of the respiratory tract, these patients mounted strong immune responses to other pathogens and even retained strong adaptive immune responses to respiratory viruses [172]. Overall, numerous studies have identified specific mutations that can confer risk for severe disease from specific infectious agents, emphasizing the need for future studies that comprehensively identify genetic variants that are impacted by pathogens that can be used to identify patients who are potentially vulnerable.
Immunological syndromes and somatic genetic mutations in genes associated with pathogen sensing
The etiology of inflammatory syndromes and diseases that arise in adulthood can be challenging to identify. Somatic genetic mutations that occur after zygote formation have been found to drive some of these complex inflammatory disorders [173]. For example, somatic mutations in NLRP3, which encodes an important intracellular sensor of infection, have numerous links to autoinflammatory syndromes [174–178]. Schnitzler’s syndrome is a rare adult-onset autoinflammatory disease that invovles both hematological and rheumatological features, and 90% of patients with Schnitzler’s syndrome who also develop macroglobulinemia carry a somatic mutation in the Toll-like receptor adapter MYD88 [179]. Patients with Schnitzler’s syndrome who develop a non-malignant expansion of hematopoietic stem cells have somatic mutations in TET2 and U2AF1 that are involved in transcriptional and splicing regulation and can impact the production of reactive oxygen species that can trigger the NLRP3-driven inflammasome [180].
With heterogenous symptoms and clinical presentations, recruiting a sufficient number of patients to assess genetic causes of disease is a challenge. Beck et al. addressed this challenge by sequencing the exomes of patients with late-onset inflammatory syndromes that involved peripheral blood abnormalities and were not responsive to treatment [181]. Despite the clinical heterogeneity, somatic deleterious mutations in UBA1 were identified in a subset of male patients. UBA1 encodes the E1 enzyme that initiates ubiquitylation, with systemic inflammation resulting from deletion of this gene in zebrafish [181]. Subsequent experiments demonstrated that the myeloid cells (neutrophils and monocytes) and myeloid progenitor stem cells but not the lymphocytes (B and T cells) carried the somatic mutation. In many cases, it is only when the genetic etiology of these inflammatory diseases are appropriately identified that effective treatments are provided. Each of the examples above involve somatic mutations that disrupt sensors and adapters of pathogen detection, and the impact of infection in these patients is yet to be fully elucidated.
With inflammatory disease at the junction between the gut microbiome and human gastrointestinal track, somatic mutations have also been studied in the context of Crohn’s Disease and Ulcerative Colitis. Indeed, somatic mutations in the colonic crypts of patients with these inflammatory bowel diseases are found at a rate 2.4-fold higher than in controls [182]. In particular, an accumulation of somatic mutations in genes known to be important in the pathogenesis of IBD was observed, including those in the IL17 signaling pathway [182].
Hormones and sex chromosome composition
An individual’s sex chromosome composition can play a significant role in immune responses and disease severity. However, most pharmaceutical interventions, including most drugs and vaccines, are given without regard to an individual’s sex chromosomes. Because the words male and female can refer to both sex and gender, we focus on chromosomes and hormones in this section of the review. Individuals with two X chromosomes and those with one X chromosome differ in immunological responses to foreign and self antigens. These differences subsequently contribute to variations in susceptibility to infectious diseases, incidence of autoimmune diseases, and responses to vaccines [183]. Indeed, individuals with an XY karyotype are more likely to die from COVID-19 than XX individuals. While such a bias in mortality is consistent with other infections, the specific underlying mechanisms are not fully understood [184]. Moreover, XX individuals typically develop more robust antibody responses and adverse reactions to vaccines [185]. Reasons likely include differences in sex steroid hormones (e.g., estrogen and testosterone) and differences in adaptive immune responses, with XX individuals exhibiting greater antibody responses and elevated humoral and cell-mediated immunity compared to XY individuals. In addition, several immune-related genes encoding proteins such as the IL-2 receptor and multiple Toll-like receptors (TLRs) are encoded on the X chromosome. Epidemiological studies in genetically diverse mice and cohorts of patients with XXX, XXY, or X0 to study the role of sex chromosomes independent of sex hormones have been of great utility for understanding sex chromosome dependent diseases [186–191]. It is critical to consider disease risk and potential G × E mechanisms in the context of sex differences that influence immune responses.
Possibilities and challenges for studying G × E interactions
The identification and characterization of G × E interactions in humans is crucial to combating human disease [192]. Individuals are born with the genetic variants that they inherit from their parents, and these variants are not easily manipulated. However, many environmental exposures are modifiable or preventable through public policy initiatives, vaccines, and/or lifestyle choices. Likewise, targeting of G × E interactions, for example through genome-editing, holds the promise to enable the development of preventative strategies and therapies. Recent advances in machine learning-based approaches to G × E studies offer one promising solution for learning new G × E mechanisms [148, 193]. However, such methods are still relatively in their infancy, and numerous challenges remain for discovering how the environment works in the context of DNA variation to increase disease risk.
Multiple testing burden is a major challenge in G × E research because of the large sample size required to obtain statistically meaningful associations. A simple solution is to use biologically guided hypotheses to limit the search space, e.g., by limiting analysis to a particular pathway. Another solution is to limit the amount of genetic variation while assessing multiple environmental exposures. For example, a recent study exposed induced pluripotent stem cells derived from six individuals to a variety of treatments to study the environmental effects on allelic gene expression [194]. Because allelic expression was used as a measurement of G × E interactions, a smaller sample size could be used to interrogate environmental exposures. Continued development of additional solutions to the multiple testing problem remains critical.
The environment is difficult to measure and quantify consistently. Many G × E studies address this challenge by developing scores to rank and prioritize environmental exposures. In a recent study, variance quantitative trait locus (vQTL) analysis was performed by associating particular genetic variants associated with phenotypic variability for over 5 million genetic variants in 300,000 individuals. These efforts identified 75 vQTLs highly enriched for G × E effects [195]. This study demonstrates that G × E interactions can be identified without direct measurement of environmental exposures in a large set of samples. The development of additional methodologies will be necessary to quantify specific environmental exposures, identified from large public resources, that increase disease risk.
Conclusions
The etiology of human disease is complex, with genetic, environmental, and G × E contributors. Studies aimed at genetic or environmental contributors individually can miss important G × E interactions that contribute to disease. A growing body of work has produced compelling evidence linking G × E interactions to a wide range of human diseases. The availability of new datasets with genetic and environmental measurements, in conjunction with the development of novel analytical approaches, will enable the discovery of additional G × E interactions. These discoveries will ultimately lead to impactful interventions that improve human health.
Acknowledgements
We thank Katelyn Dunn and Carmy Forney for helpful feedback.
Author contributions
Conceptualization: SJV, MTW, and LCK; Writing—original draft preparation: SJV, MTW, and LCK; Writing—review and editing: SJV, AVH, KCMFV, MTW, and LCK; Supervision: MTW and LCK; Project administration: MTW and LCK; Funding acquisition: MTW and LCK.
Funding
This research was funded by National Institutes of Health (NIH) R01 DK107502, R01 AI148276, U01 HG011172, and P30 AR070549 to LCK; R01 HG010730, R01 NS099068, R01 GM055479, U01 AI130830, U01 AI150748, R01 AI141569, and P01 AI150585 to MTW; R01 AR073228 and R01 AI024717 to MTW and LCK. Funding was also provided by CCHMC ARC Award 53632 to MTW and LCK.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Matthew T. Weirauch, Email: Matthew.Weirauch@cchmc.org
Leah C. Kottyan, Email: Leah.Kottyan@cchmc.org
References
- 1.Yadav A, Sinha H. Gene-gene and gene-environment interactions in complex traits in yeast. Yeast. 2018;35:403–16. doi: 10.1002/yea.3304. [DOI] [PubMed] [Google Scholar]
- 2.Renz H, von Mutius E, Brandtzaeg P, Cookson WO, Autenrieth IB, Haller D. Gene-environment interactions in chronic inflammatory disease. Nat Immunol. 2011;12:273–7. doi: 10.1038/ni0411-273. [DOI] [PubMed] [Google Scholar]
- 3.Manuck SB, McCaffery JM. Gene-environment interaction. Annu Rev Psychol. 2014;65:41–70. doi: 10.1146/annurev-psych-010213-115100. [DOI] [PubMed] [Google Scholar]
- 4.Simonds NI, Ghazarian AA, Pimentel CB, Schully SD, Ellison GL, Gillanders EM, et al. Review of the gene-environment interaction literature in cancer: what do we know? Genet Epidemiol. 2016;40:356–65. doi: 10.1002/gepi.21967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6:287–98. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 6.Halldorsdottir T, Binder EB. Gene x environment interactions: from molecular mechanisms to behavior. Annu Rev Psychol. 2017;68:215–41. doi: 10.1146/annurev-psych-010416-044053. [DOI] [PubMed] [Google Scholar]
- 7.McAllister K, Mechanic LE, Amos C, Aschard H, Blair IA, Chatterjee N, et al. Current challenges and new opportunities for gene-environment interaction studies of complex diseases. Am J Epidemiol. 2017;186:753–61. doi: 10.1093/aje/kwx227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ottman R. An epidemiologic approach to gene-environment interaction. Genet Epidemiol. 1990;7:177–85. doi: 10.1002/gepi.1370070302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kauffmann F, Demenais F. Gene-environment interactions in asthma and allergic diseases: challenges and perspectives. J Allergy Clin Immunol. 2012;130:1229–40. doi: 10.1016/j.jaci.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 10.Sahu M, Prasuna JG. Twin studies: a unique epidemiological tool. Indian J Community Med. 2016;41:177–82. doi: 10.4103/0970-0218.183593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halfvarson J, Bodin L, Tysk C, Lindberg E, Jarnerot G. Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124:1767–73. doi: 10.1016/S0016-5085(03)00385-8. [DOI] [PubMed] [Google Scholar]
- 12.Orholm M, Binder V, Sorensen TI, Rasmussen LP, Kyvik KO. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol. 2000;35:1075–81. doi: 10.1080/003655200451207. [DOI] [PubMed] [Google Scholar]
- 13.Thompson NP, Driscoll R, Pounder RE, Wakefield AJ. Genetics versus environment in inflammatory bowel disease: results of a British twin study. BMJ. 1996;312:95–6. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G, Yang E, Brinkmeyer-Langford CL, Cai JJ. Additive, epistatic, and environmental effects through the lens of expression variability QTL in a twin cohort. Genetics. 2014;196:413–25. doi: 10.1534/genetics.113.157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drumm ML, Wilkinson DJ, Smit LS, Worrell RT, Strong TV, Frizzell RA, et al. Chloride conductance expressed by delta F508 and other mutant CFTRs in Xenopus oocytes. Science. 1991;254:1797–9. doi: 10.1126/science.1722350. [DOI] [PubMed] [Google Scholar]
- 16.Dalemans W, Barbry P, Champigny G, Jallat S, Dott K, Dreyer D, et al. Altered chloride ion channel kinetics associated with the delta F508 cystic fibrosis mutation. Nature. 1991;354:526–8. doi: 10.1038/354526a0. [DOI] [PubMed] [Google Scholar]
- 17.Sepahzad A, Morris-Rosendahl DJ, Davies JC. Cystic fibrosis lung disease modifiers and their relevance in the new era of precision medicine. Genes. 2021;12:562. [DOI] [PMC free article] [PubMed]
- 18.Antonarakis SE. The search for allelic variants that cause monogenic disorders or predispose to common, complex polygenic phenotypes. Dialogues Clin Neurosci. 2001;3:7–15. doi: 10.31887/DCNS.2001.3.1/seantonarakis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macedo AC, Isaac L. Systemic lupus erythematosus and deficiencies of early components of the complement classical pathway. Front Immunol. 2016;7:55. doi: 10.3389/fimmu.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–8. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langefeld CD, Ainsworth HC, Cunninghame Graham DS, Kelly JA, Comeau ME, Marion MC, et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat Commun. 2017;8:16021. doi: 10.1038/ncomms16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almlof JC, Nystedt S, Leonard D, Eloranta ML, Grosso G, Sjowall C, et al. Whole-genome sequencing identifies complex contributions to genetic risk by variants in genes causing monogenic systemic lupus erythematosus. Hum Genet. 2019;138:141–50. doi: 10.1007/s00439-018-01966-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam V, Patel N, Turcotte M, Bosse Y, Pare G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–84. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 24.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–12. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bush WS, Moore JH. Chapter 11: genome-wide association studies. PLoS Comput Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson RE, Kuchenbaecker K, Walters RK, Chen CY, Popejoy AB, Periyasamy S, et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell. 2019;179:589–603. doi: 10.1016/j.cell.2019.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kottyan L, Spergel JM, Cianferoni A. Immunology of the ancestral differences in eosinophilic esophagitis. Ann Allergy Asthma Immunol. 2019;122:443–4. doi: 10.1016/j.anai.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentley AR, Callier SL, Rotimi CN. Evaluating the promise of inclusion of African ancestry populations in genomics. NPJ Genom Med. 2020;5:5. doi: 10.1038/s41525-019-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 30.Luger T, Amagai M, Dreno B, Dagnelie MA, Liao W, Kabashima K, et al. Atopic dermatitis: role of the skin barrier, environment, microbiome, and therapeutic agents. J Dermatol Sci. 2021;102:142–57. doi: 10.1016/j.jdermsci.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Brown SJ, McLean WH. One remarkable molecule: filaggrin. J Invest Dermatol. 2012;132(3 Pt 2):751–62. doi: 10.1038/jid.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drislane C, Irvine AD. The role of filaggrin in atopic dermatitis and allergic disease. Ann Allergy Asthma Immunol. 2020;124:36–43. doi: 10.1016/j.anai.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Bantz SK, Zhu Z, Zheng T. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. 2014;5: 202. [DOI] [PMC free article] [PubMed]
- 34.Clausen ML, Edslev SM, Norreslet LB, Sorensen JA, Andersen PS, Agner T. Temporal variation of Staphylococcus aureus clonal complexes in atopic dermatitis: a follow-up study. Br J Dermatol. 2019;180:181–6. doi: 10.1111/bjd.17033. [DOI] [PubMed] [Google Scholar]
- 35.Simpson EL, Villarreal M, Jepson B, Rafaels N, David G, Hanifin J, et al. Patients with atopic dermatitis colonized with Staphylococcus aureus have a distinct phenotype and endotype. J Invest Dermatol. 2018;138:2224–33. doi: 10.1016/j.jid.2018.03.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baurecht H, Ruhlemann MC, Rodriguez E, Thielking F, Harder I, Erkens AS, et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J Allergy Clin Immunol. 2018;141:1668–76.e16. doi: 10.1016/j.jaci.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 37.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–51. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.French JD, Edwards SL. The role of noncoding variants in heritable disease. Trends Genet. 2020;36:880–91. doi: 10.1016/j.tig.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Deplancke B, Alpern D, Gardeux V. The genetics of transcription factor DNA binding variation. Cell. 2016;166:538–54. doi: 10.1016/j.cell.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Fuxman Bass JI, Sahni N, Shrestha S, Garcia-Gonzalez A, Mori A, Bhat N, et al. Human gene-centered transcription factor networks for enhancers and disease variants. Cell. 2015;161:661–73. doi: 10.1016/j.cell.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrera LA, Vedenko A, Kurland JV, Rogers JM, Gisselbrecht SS, Rossin EJ, et al. Survey of variation in human transcription factors reveals prevalent DNA binding changes. Science. 2016;351:1450–4. doi: 10.1126/science.aad2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee DSM, Ghanem LR, Barash Y. Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat Commun. 2020;11:527. doi: 10.1038/s41467-020-14404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–6. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 46.Gholami M, Larijani B, Sharifi F, Hasani-Ranjbar S, Taslimi R, Bastami M, et al. MicroRNA-binding site polymorphisms and risk of colorectal cancer: A systematic review and meta-analysis. Cancer Med. 2019;8:7477–99. doi: 10.1002/cam4.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatlen A, Helmy M, Marco A. PopTargs: a database for studying population evolutionary genetics of human microRNA target sites. Database. 2019;2019:baz102. [DOI] [PMC free article] [PubMed]
- 48.Kulkarni S, Lied A, Kulkarni V, Rucevic M, Martin MP, Walker-Sperling V, et al. CCR5AS lncRNA variation differentially regulates CCR5, influencing HIV disease outcome. Nat Immunol. 2019;20:824–34. doi: 10.1038/s41590-019-0406-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Consortium GT. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye Y, Zhang Z, Liu Y, Diao L, Han L. A multi-omics perspective of quantitative trait loci in precision medicine. Trends Genet. 2020;36:318–36. doi: 10.1016/j.tig.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Pelikan RC, Kelly JA, Fu Y, Lareau CA, Tessneer KL, Wiley GB, et al. Enhancer histone-QTLs are enriched on autoimmune risk haplotypes and influence gene expression within chromatin networks. Nat Commun. 2018;9:2905. doi: 10.1038/s41467-018-05328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harley JB, Chen X, Pujato M, Miller D, Maddox A, Forney C, et al. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat Genet. 2018;50:699–707. doi: 10.1038/s41588-018-0102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costenbader KH, Karlson EW. Cigarette smoking and autoimmune disease: what can we learn from epidemiology? Lupus. 2006;15:737–45. doi: 10.1177/0961203306069344. [DOI] [PubMed] [Google Scholar]
- 54.Rehm J. The risks associated with alcohol use and alcoholism. Alcohol Res Health. 2011;34:135–43. [PMC free article] [PubMed] [Google Scholar]
- 55.Sample A, He YY. Mechanisms and prevention of UV-induced melanoma. Photodermatol Photoimmunol Photomed. 2018;34:13–24. doi: 10.1111/phpp.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330:460–1. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beck AF, Huang B, Wheeler K, Lawson NR, Kahn RS, Riley CL. The child opportunity index and disparities in pediatric asthma hospitalizations across one Ohio metropolitan area, 2011-2013. J Pediatr. 2017;190:200–6.e1. doi: 10.1016/j.jpeds.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson LH, Beck AF, Kahn RS, Huang B, Ryan PH, Olano KK, et al. Characteristics of pediatric emergency revisits after an asthma-related hospitalization. Ann Emerg Med. 2017;70:277–87. doi: 10.1016/j.annemergmed.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Auger KA, Kahn RS, Simmons JM, Huang B, Shah AN, Timmons K, et al. Using address information to identify hardships reported by families of children hospitalized with asthma. Acad Pediatr. 2017;17:79–87. doi: 10.1016/j.acap.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Popkin BM. Nutrition, agriculture and the global food system in low and middle income countries. Food Policy. 2014;47:91–6. doi: 10.1016/j.foodpol.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuddus MA, Tynan E, McBryde E. Urbanization: a problem for the rich and the poor? Public Health Rev. 2020;41:1. doi: 10.1186/s40985-019-0116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rocque RJ, Beaudoin C, Ndjaboue R, Cameron L, Poirier-Bergeron L, Poulin-Rheault RA, et al. Health effects of climate change: an overview of systematic reviews. BMJ Open. 2021;11:e046333. doi: 10.1136/bmjopen-2020-046333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olsen SJ, Azziz-Baumgartner E, Budd AP, Brammer L, Sullivan S, Pineda RF, et al. Decreased influenza activity during the COVID-19 pandemic—United States, Australia, Chile, and South Africa, 2020. Morb Mortal Wkly Rep. 2020;69:1305–9. doi: 10.15585/mmwr.mm6937a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiymet E, Boncuoglu E, Sahinkaya S, Cem E, Celebi MY, Duzgol M, et al. Distribution of spreading viruses during COVID-19 pandemic: effect of mitigation strategies. Am J Infect Control. 2021;49:1142–5. doi: 10.1016/j.ajic.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peters A, Nawrot TS, Baccarelli AA. Hallmarks of environmental insults. Cell. 2021;184:1455–68. doi: 10.1016/j.cell.2021.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iweala OI, Nagler CR. The microbiome and food allergy. Annu Rev Immunol. 2019;37:377–403. doi: 10.1146/annurev-immunol-042718-041621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tasnim N, Abulizi N, Pither J, Hart MM, Gibson DL. Linking the gut microbial ecosystem with the environment: does gut health depend on where we live? Front Microbiol. 2017;8:1935. doi: 10.3389/fmicb.2017.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karl JP, Hatch AM, Arcidiacono SM, Pearce SC, Pantoja-Feliciano IG, Doherty LA, et al. Effects of psychological, environmental and physical stressors on the gut microbiota. Front Microbiol. 2018;9:2013. doi: 10.3389/fmicb.2018.02013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin Exp Immunol. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lambrecht BN, Hammad H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat Immunol. 2017;18:1076–83. doi: 10.1038/ni.3829. [DOI] [PubMed] [Google Scholar]
- 72.Bach JF. The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat Rev Immunol. 2018;18:105–20. doi: 10.1038/nri.2017.111. [DOI] [PubMed] [Google Scholar]
- 73.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci USA. 2014;111:13145–50. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. 2019;25:448–53. doi: 10.1038/s41591-018-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin J, Pellinen JC, Galetta SL. Subacute progressive ptosis, ophthalmoplegia, gait instability, and cognitive changes. JAMA Neurol. 2018;75:1284–5. doi: 10.1001/jamaneurol.2018.1588. [DOI] [PubMed] [Google Scholar]
- 76.Wu SE, Hashimoto-Hill S, Woo V, Eshleman EM, Whitt J, Engleman L, et al. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature. 2020;586:108–12. doi: 10.1038/s41586-020-2604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kiaghadi A, Rifai HS, Dawson CN. The presence of Superfund sites as a determinant of life expectancy in the United States. Nat Commun. 2021;12:1947. doi: 10.1038/s41467-021-22249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brender JD, Maantay JA, Chakraborty J. Residential proximity to environmental hazards and adverse health outcomes. Am J Public Health. 2011;101:S37–52. doi: 10.2105/AJPH.2011.300183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fuller R, Landrigan PJ, Balakrishnan K, Bathan G, Bose-O’Reilly S, Brauer M, et al. Pollution and health: a progress update. Lancet Planet Health. 2022;6:e535–47. doi: 10.1016/S2542-5196(22)00090-0. [DOI] [PubMed] [Google Scholar]
- 80.World Health Organization. (2021). WHO global air quality guidelines: particulate matter (PM2.5 and PM10), ozone, nitrogen dioxide, sulfur dioxide and carbon monoxide: executive summary. World Health Organization. https://apps.who.int/iris/handle/10665/345334. License: CC BY-NC-SA 3.0 IGO. [PubMed]
- 81.McDuffie EE, Martin RV, Spadaro JV, Burnett R, Smith SJ, O’Rourke P, et al. Source sector and fuel contributions to ambient PM2.5 and attributable mortality across multiple spatial scales. Nat Commun. 2021;12:3594. doi: 10.1038/s41467-021-23853-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franklin M, Koutrakis P, Schwartz P. The role of particle composition on the association between PM2.5 and mortality. Epidemiology. 2008;19:680–9. doi: 10.1097/EDE.0b013e3181812bb7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hayes RB, Lim C, Zhang Y, Cromar K, Shao Y, Reynolds HR, et al. PM2.5 air pollution and cause-specific cardiovascular disease mortality. Int J Epidemiol. 2020;49:25–35. doi: 10.1093/ije/dyz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alexeeff SE, Liao NS, Liu X, Van Den Eeden SK, Sidney S. Long-term PM2.5 exposure and risks of ischemic heart disease and stroke events: review and meta-analysis. J Am Heart Assoc. 2021;10:e016890. doi: 10.1161/JAHA.120.016890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Malik AO, Jones PG, Chan PS, Peri-Okonny PA, Hejjaji V, Spertus JA. Association of long-term exposure to particulate matter and ozone with health status and mortality in patients after myocardial infarction. Circ Cardiovasc Qual Outcomes. 2019;12:e005598. doi: 10.1161/CIRCOUTCOMES.119.005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Madrigano J, Kloog I, Goldberg R, Coull BA, Mittleman MA, Schwartz J. Long-term exposure to PM2.5 and incidence of acute myocardial infarction. Environ Health Perspect. 2013;121:192–6. doi: 10.1289/ehp.1205284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.James C, Bernstein DI, Cox J, Ryan P, Wolfe C, Jandarov R, et al. HEPA filtration improves asthma control in children exposed to traffic-related airborne particles. Indoor Air. 2020;30:235–43. doi: 10.1111/ina.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mirabelli MC, Vaidyanathan A, Flanders WD, Qin X, Garbe P. Outdoor PM2.5, ambient air temperature, and asthma symptoms in the past 14 days among adults with active asthma. Environ Health Perspect. 2016;124:1882–90. doi: 10.1289/EHP92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rosenquist NA, Metcalf WJ, Ryu SY, Rutledge A, Coppes MJ, Grzymski JJ, et al. Acute associations between PM2.5 and ozone concentrations and asthma exacerbations among patients with and without allergic comorbidities. J Expo Sci Environ Epidemiol. 2020;30:795–804. doi: 10.1038/s41370-020-0213-7. [DOI] [PubMed] [Google Scholar]
- 90.Lepeule J, Laden F, Dockery D, Schwartz J. Chronic exposure to fine particles and mortality: an extended follow-up of the Harvard Six Cities study from 1974 to 2009. Environ Health Perspect. 2012;120:965–70. doi: 10.1289/ehp.1104660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Turner MC, Krewski D, Pope CA, 3rd, Chen Y, Gapstur SM, Thun MJ. Long-term ambient fine particulate matter air pollution and lung cancer in a large cohort of never-smokers. Am J Respir Crit Care Med. 2011;184:1374–81. doi: 10.1164/rccm.201106-1011OC. [DOI] [PubMed] [Google Scholar]
- 92.Humans IWGotEoCRt. Outdoor air pollution. IARC Monogr Eval Carcinog Risks Hum. 2016;109:9–444. [PMC free article] [PubMed] [Google Scholar]
- 93.Han L, Zhou W, Li W. Fine particulate (PM2.5) dynamics during rapid urbanization in Beijing, 1973-2013. Sci Rep. 2016;6:23604. doi: 10.1038/srep23604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.United Nations. Summary Progress Update 2021: SDG 6—water and sanitation for all. 2021.
- 95.Podgorski J, Berg M. Global threat of arsenic in groundwater. Science. 2020;368:845–50. doi: 10.1126/science.aba1510. [DOI] [PubMed] [Google Scholar]
- 96.Saha KC. Diagnosis of arsenicosis. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2003;38:255–72. doi: 10.1081/ESE-120016893. [DOI] [PubMed] [Google Scholar]
- 97.Humans IWGotEoCRt. Arsenic, metals, fibres, and dusts. IARC Monogr Eval Carcinog Risks Hum. 2012;100(Pt C):11–465. [PMC free article] [PubMed] [Google Scholar]
- 98.Ekino S, Susa M, Ninomiya T, Imamura K, Kitamura T. Minamata disease revisited: an update on the acute and chronic manifestations of methyl mercury poisoning. J Neurol Sci. 2007;262:131–44. doi: 10.1016/j.jns.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 99.Aoshima K. [Itai-itai disease: cadmium-induced renal tubular osteomalacia]. Nihon Eiseigaku Zasshi. 2012;67:455–63. [DOI] [PubMed]
- 100.Soffritti M, Belpoggi F, Esposti DD, Falcioni L, Bua L. Consequences of exposure to carcinogens beginning during developmental life. Basic Clin Pharm Toxicol. 2008;102:118–24. doi: 10.1111/j.1742-7843.2007.00200.x. [DOI] [PubMed] [Google Scholar]
- 101.Herrero-Uribe L. Viruses, definitions and reality. Rev Biol Trop. 2011;59:993–8. [PubMed] [Google Scholar]
- 102.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smatti MK, Cyprian FS, Nasrallah GK, Al Thani AA, Almishal RO, Yassine HM. Viruses and autoimmunity: a review on the potential interaction and molecular mechanisms. Viruses. 2019;11:762. [DOI] [PMC free article] [PubMed]
- 104.Martorano LM, Grayson MH. Respiratory viral infections and atopic development: from possible mechanisms to advances in treatment. Eur J Immunol. 2018;48:407–14. doi: 10.1002/eji.201747052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–15. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 106.Serrano B, Brotons M, Bosch FX, Bruni L. Epidemiology and burden of HPV-related disease. Best Pr Res Clin Obstet Gynaecol. 2018;47:14–26. doi: 10.1016/j.bpobgyn.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 107.Mahale P, Torres HA, Kramer JR, Hwang LY, Li R, Brown EL, et al. Hepatitis C virus infection and the risk of cancer among elderly US adults: a registry-based case-control study. Cancer. 2017;123:1202–11. doi: 10.1002/cncr.30559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Di Bisceglie AM. Hepatitis B and hepatocellular carcinoma. Hepatology. 2009;49(5 Suppl):S56–60. doi: 10.1002/hep.22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hochberg D, Middeldorp JM, Catalina M, Sullivan JL, Luzuriaga K, Thorley-Lawson DA. Demonstration of the Burkitt’s lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo. Proc Natl Acad Sci USA. 2004;101:239–44. doi: 10.1073/pnas.2237267100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kleinschmidt-DeMasters BK, Damek DM, Lillehei KO, Dogan A, Giannini C. Epstein Barr virus-associated primary CNS lymphomas in elderly patients on immunosuppressive medications. J Neuropathol Exp Neurol. 2008;67:1103–11. doi: 10.1097/NEN.0b013e31818beaea. [DOI] [PubMed] [Google Scholar]
- 111.Levine PH, Ablashi DV, Berard CW, Carbone PP, Waggoner DE, Malan L. Elevated antibody titers to Epstein-Barr virus in Hodgkin’s disease. Cancer. 1971;27:416–21. doi: 10.1002/1097-0142(197102)27:2<416::AID-CNCR2820270227>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 112.Mitarnun W, Pradutkanchana J, Takao S, Saechan V, Suwiwat S, Ishida T. Epstein-barr virus-associated non-Hodgkin’s lymphoma of B-cell origin, Hodgkin’s disease, acute leukemia, and systemic lupus erythematosus: a serologic and molecular analysis. J Med Assoc Thai. 2002;85:552–9. [PubMed] [Google Scholar]
- 113.Wolf H, zur Hausen H, Becker V. EB viral genomes in epithelial nasopharyngeal carcinoma cells. Nat N Biol. 1973;244:245–7. doi: 10.1038/newbio244245a0. [DOI] [PubMed] [Google Scholar]
- 114.Drolet M, Benard E, Boily MC, Ali H, Baandrup L, Bauer H, et al. Population-level impact and herd effects following human papillomavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2015;15:565–80. doi: 10.1016/S1473-3099(14)71073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lei J, Ploner A, Elfstrom KM, Wang J, Roth A, Fang F, et al. HPV vaccination and the risk of invasive cervical cancer. N Engl J Med. 2020;383:1340–8. doi: 10.1056/NEJMoa1917338. [DOI] [PubMed] [Google Scholar]
- 116.Jartti T, Gern JE. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol. 2017;140:895–906. doi: 10.1016/j.jaci.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Edwards MR, Strong K, Cameron A, Walton RP, Jackson DJ, Johnston SL. Viral infections in allergy and immunology: how allergic inflammation influences viral infections and illness. J Allergy Clin Immunol. 2017;140:909–20. doi: 10.1016/j.jaci.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu P, Hartert TV. Evidence for a causal relationship between respiratory syncytial virus infection and asthma. Expert Rev Anti Infect Ther. 2011;9:731–45. doi: 10.1586/eri.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mejias A, Wu B, Tandon N, Chow W, Varma R, Franco E, et al. Risk of childhood wheeze and asthma after respiratory syncytial virus infection in full-term infants. Pediatr Allergy Immunol. 2020;31:47–56. doi: 10.1111/pai.13131. [DOI] [PubMed] [Google Scholar]
- 120.Djukanovic R, Harrison T, Johnston SL, Gabbay F, Wark P, Thomson NC, et al. The effect of inhaled IFN-beta on worsening of asthma symptoms caused by viral infections. A randomized trial. Am J Respir Crit Care Med. 2014;190:145–54. doi: 10.1164/rccm.201312-2235OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. 2005;201:937–47. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cakebread JA, Xu Y, Grainge C, Kehagia V, Howarth PH, Holgate ST, et al. Exogenous IFN-beta has antiviral and anti-inflammatory properties in primary bronchial epithelial cells from asthmatic subjects exposed to rhinovirus. J Allergy Clin Immunol. 2011;127:1148–54.e9. doi: 10.1016/j.jaci.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 123.Rose NR. Viral myocarditis. Curr Opin Rheumatol. 2016;28:383–9. doi: 10.1097/BOR.0000000000000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Galeotti C, Bayry J. Autoimmune and inflammatory diseases following COVID-19. Nat Rev Rheumatol. 2020;16:413–4. doi: 10.1038/s41584-020-0448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Saad MA, Alfishawy M, Nassar M, Mohamed M, Esene IN, Elbendary A. COVID-19 and autoimmune diseases: a systematic review of reported cases. Curr Rheumatol Rev. 2021;17:193–204. doi: 10.2174/1573397116666201029155856. [DOI] [PubMed] [Google Scholar]
- 126.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585.. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Balandraud N, Roudier J, Roudier C. Epstein-Barr virus and rheumatoid arthritis. Autoimmun Rev. 2004;3:362–7. doi: 10.1016/j.autrev.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 128.Almohmeed YH, Avenell A, Aucott L, Vickers MA. Systematic review and meta-analysis of the sero-epidemiological association between Epstein Barr virus and multiple sclerosis. PLoS ONE. 2013;8:e61110. doi: 10.1371/journal.pone.0061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hanlon P, Avenell A, Aucott L, Vickers MA. Systematic review and meta-analysis of the sero-epidemiological association between Epstein-Barr virus and systemic lupus erythematosus. Arthritis Res Ther. 2014;16:R3. doi: 10.1186/ar4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Draborg AH, Duus K, Houen G. Epstein-Barr virus in systemic autoimmune diseases. Clin Dev Immunol. 2013;2013:535738. doi: 10.1155/2013/535738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bar-Or A, Pender MP, Khanna R, Steinman L, Hartung HP, Maniar T, et al. Epstein-Barr virus in multiple sclerosis: theory and emerging immunotherapies. Trends Mol Med. 2020;26:296–310. doi: 10.1016/j.molmed.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bjornevik K, Cortese M, Healy BC, Kuhle J, Mina MJ, Leng Y, et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science. 2022;375:296–301. doi: 10.1126/science.abj8222. [DOI] [PubMed] [Google Scholar]
- 133.Lanz TV, Brewer RC, Ho PP, Moon JS, Jude KM, Fernandez D, et al. Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM. Nature. 2022;603:321–7. doi: 10.1038/s41586-022-04432-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rossati A. Global warming and its health impact. Int J Occup Environ Med. 2017;8:7–20. doi: 10.15171/ijoem.2017.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rothenberg ME. The climate change hypothesis for the allergy epidemic. J Allergy Clin Immunol. 2022;149:1522–4. doi: 10.1016/j.jaci.2022.02.006. [DOI] [PubMed] [Google Scholar]
- 136.Patz JA, Reisen WK. Immunology, climate change and vector-borne diseases. Trends Immunol. 2001;22:171–2. doi: 10.1016/S1471-4906(01)01867-1. [DOI] [PubMed] [Google Scholar]
- 137.Mordecai EA, Ryan SJ, Caldwell JM, Shah MM, LaBeaud AD. Climate change could shift disease burden from malaria to arboviruses in Africa. Lancet Planet Health. 2020;4:e416–e423. doi: 10.1016/S2542-5196(20)30178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Semenza JC, Paz S. Climate change and infectious disease in Europe: impact, projection and adaptation. Lancet Reg Health Eur. 2021;9:100230. doi: 10.1016/j.lanepe.2021.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arthur SS, Nyide B, Soura AB, Kahn K, Weston M, Sankoh O. Tackling malnutrition: a systematic review of 15-year research evidence from INDEPTH health and demographic surveillance systems. Glob Health Action. 2015;8:28298. doi: 10.3402/gha.v8.28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Phalkey RK, Aranda-Jan C, Marx S, Hofle B, Sauerborn R. Systematic review of current efforts to quantify the impacts of climate change on undernutrition. Proc Natl Acad Sci USA. 2015;112:E4522–9. doi: 10.1073/pnas.1409769112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Walker RJ, Strom Williams J, Egede LE. Influence of race, ethnicity and social determinants of health on diabetes outcomes. Am J Med Sci. 2016;351:366–73. doi: 10.1016/j.amjms.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shonkoff JP, Garner AS, Committee on Psychosocial Aspects of C, Family H, Committeeon Early Childhood A, Dependent C, et al. The lifelong effects of early childhood adversityand toxic stress. Pediatrics. 2012;129:e232–46. [DOI] [PubMed]
- 143.Levy JI, Quiros-Alcala L, Fabian MP, Basra K, Hansel NN. Established and emerging environmental contributors to disparities in asthma and chronic obstructive pulmonary disease. Curr Epidemiol Rep. 2018;5:114–24. doi: 10.1007/s40471-018-0149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Groover O, Morton ML, Janocko NJ, Teagarden DL, Villarreal HK, Drane DL, et al. Mind the gap: health disparities in families living with epilepsy are significant and linked to socioeconomic status. Epileptic Disord. 2020;22:782–9. doi: 10.1684/epd.2020.1229. [DOI] [PubMed] [Google Scholar]
- 145.Dupre ME, Nelson A, Lynch SM, Granger BB, Xu H, Churchill E, et al. Socioeconomic, psychosocial and behavioral characteristics of patients hospitalized with cardiovascular disease. Am J Med Sci. 2017;354:565–72. doi: 10.1016/j.amjms.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moncrief T, Beck AF, Simmons JM, Huang B, Kahn RS. Single parent households and increased child asthma morbidity. J Asthma. 2014;51:260–6. doi: 10.3109/02770903.2013.873806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kahn RS, Wise PH, Kennedy BP, Kawachi I. State income inequality, household income, and maternal mental and physical health: cross sectional national survey. BMJ. 2000;321:1311–5. doi: 10.1136/bmj.321.7272.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Resztak JA, Farrell AK, Mair-Meijers H, Alazizi A, Wen X, Wildman DE, et al. Psychosocial experiences modulate asthma-associated genes through gene-environment interactions. Elife. 2021;10:e63852. doi: 10.7554/eLife.63852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fave MJ, Lamaze FC, Soave D, Hodgkinson A, Gauvin H, Bruat V, et al. Gene-by-environment interactions in urban populations modulate risk phenotypes. Nat Commun. 2018;9:827. doi: 10.1038/s41467-018-03202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gage SH, Davey Smith G, Ware JJ, Flint J, Munafo MR. G = E: what GWAS can tell us about the environment. PLoS Genet. 2016;12:e1005765. doi: 10.1371/journal.pgen.1005765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dunn AR, O’Connell KMS, Kaczorowski CC. Gene-by-environment interactions in Alzheimer’s disease and Parkinson’s disease. Neurosci Biobehav Rev. 2019;103:73–80. doi: 10.1016/j.neubiorev.2019.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Carbone M, Amelio I, Affar EB, Brugarolas J, Cannon-Albright LA, Cantley LC, et al. Consensus report of the 8 and 9th Weinman Symposia on Gene x Environment Interaction in carcinogenesis: novel opportunities for precision medicine. Cell Death Differ. 2018;25:1885–904. doi: 10.1038/s41418-018-0213-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tabrez S, Priyadarshini M, Priyamvada S, Khan MS, Na A, Zaidi SK. Gene-environment interactions in heavy metal and pesticide carcinogenesis. Mutat Res Genet Toxicol Environ Mutagen. 2014;760:1–9. doi: 10.1016/j.mrgentox.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 154.Pierce BL, Kibriya MG, Tong L, Jasmine F, Argos M, Roy S, et al. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet. 2012;8:e1002522. doi: 10.1371/journal.pgen.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pierce BL, Tong L, Dean S, Argos M, Jasmine F, Rakibuz-Zaman M, et al. A missense variant in FTCD is associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet. 2019;15:e1007984. doi: 10.1371/journal.pgen.1007984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ravenscroft P, Brammer H, Richards KS. Arsenic pollution: a global synthesis. Chichester, U.K., Malden, MA: Wiley-Blackwell; 2009.
- 157.Paul S, Majumdar S, Giri AK. Genetic susceptibility to arsenic-induced skin lesions and health effects: a review. Genes Environ. 2015;37:23. doi: 10.1186/s41021-015-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ng E, Lind PM, Lindgren C, Ingelsson E, Mahajan A, Morris A, et al. Genome-wide association study of toxic metals and trace elements reveals novel associations. Hum Mol Genet. 2015;24:4739–45. doi: 10.1093/hmg/ddv190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Starska K, Brys M, Forma E, Olszewski J, Pietkiewicz P, Lewy-Trenda I, et al. The effect of metallothionein 2A core promoter region single-nucleotide polymorphism on accumulation of toxic metals in sinonasal inverted papilloma tissues. Toxicol Appl Pharm. 2015;285:187–97. doi: 10.1016/j.taap.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 160.Joneidi Z, Mortazavi Y, Memari F, Roointan A, Chahardouli B, Rostami S. The impact of genetic variation on metabolism of heavy metals: genetic predisposition? Biomed Pharmacother. 2019;113:108642. doi: 10.1016/j.biopha.2019.108642. [DOI] [PubMed] [Google Scholar]
- 161.Johansson H, Mersha TB, Brandt EB, Khurana Hershey GK. Interactions between environmental pollutants and genetic susceptibility in asthma risk. Curr Opin Immunol. 2019;60:156–62. doi: 10.1016/j.coi.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Maazi H, Hartiala JA, Suzuki Y, Crow AL, Shafiei Jahani P, Lam J, et al. A GWAS approach identifies Dapp1 as a determinant of air pollution-induced airway hyperreactivity. PLoS Genet. 2019;15:e1008528. doi: 10.1371/journal.pgen.1008528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chiaramonte R, Calzavara E, Balordi F, Sabbadini M, Capello D, Gaidano G, et al. Differential regulation of Notch signal transduction in leukaemia and lymphoma cells in culture. J Cell Biochem. 2003;88:569–77. doi: 10.1002/jcb.10383. [DOI] [PubMed] [Google Scholar]
- 164.Hong T, Parameswaran S, Donmez OA, Miller D, Forney C, Lape M, et al. Epstein-Barr virus nuclear antigen 2 extensively rewires the human chromatin landscape at autoimmune risk loci. Genome Res. 2021;19;31:2185–98. [DOI] [PMC free article] [PubMed]
- 165.Wang L, Laing J, Yan B, Zhou H, Ke L, Wang C, et al. Epstein-Barr virus episome physically interacts with active regions of the host genome in lymphoblastoid cells. J Virol. 2020;94:e01390–20. [DOI] [PMC free article] [PubMed]
- 166.Keane JT, Afrasiabi A, Schibeci SD, Swaminathan S, Parnell GP, Booth DR. The interaction of Epstein-Barr virus encoded transcription factor EBNA2 with multiple sclerosis risk loci is dependent on the risk genotype. EBioMedicine. 2021;71:103572. doi: 10.1016/j.ebiom.2021.103572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu X, Hong T, Parameswaran S, Ernst K, Marazzi I, Weirauch MT, et al. Human virus transcriptional regulators. Cell. 2020;182:24–37. doi: 10.1016/j.cell.2020.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ciancanelli MJ, Huang SX, Luthra P, Garner H, Itan Y, Volpi S, et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–53. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lin R, Mamane Y, Hiscott J. Multiple regulatory domains control IRF-7 activity in response to virus infection. J Biol Chem. 2000;275:34320–7. doi: 10.1074/jbc.M002814200. [DOI] [PubMed] [Google Scholar]
- 170.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. [DOI] [PMC free article] [PubMed]
- 171.Zhang Q, Matuozzo D, Le Pen J, Lee D, Moens L, Asano T, et al. Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia. J Exp Med. 2022;219:e20220131. [DOI] [PMC free article] [PubMed]
- 172.Campbell TM, Liu Z, Zhang Q, Moncada-Velez M, Covill LE, Zhang P, et al. Respiratory viral infections in otherwise healthy humans with inherited IRF7 deficiency. J Exp Med. 2022;219:e20220202. [DOI] [PMC free article] [PubMed]
- 173.Solis-Moruno M, Batlle-Maso L, Bonet N, Arostegui JI, Casals F. Somatic genetic variation in healthy tissue and non-cancer diseases. Eur J Hum Genet. 2022 10.1038/s41431-022-01213-8. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 174.Tanaka N, Izawa K, Saito MK, Sakuma M, Oshima K, Ohara O, et al. High incidence of NLRP3 somatic mosaicism in patients with chronic infantile neurologic, cutaneous, articular syndrome: results of an International Multicenter Collaborative Study. Arthritis Rheum. 2011;63:3625–32. doi: 10.1002/art.30512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Poulter JA, Savic S. Genetics of somatic auto-inflammatory disorders. Semin Hematol. 2021;58:212–7. doi: 10.1053/j.seminhematol.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 176.Zhou Q, Aksentijevich I, Wood GM, Walts AD, Hoffmann P, Remmers EF, et al. Brief Report: Ccryopyrin-associated periodic syndrome caused by a myeloid-restricted somatic NLRP3 mutation. Arthritis Rheumatol. 2015;67:2482–6. doi: 10.1002/art.39190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mensa-Vilaro A, Teresa Bosque M, Magri G, Honda Y, Martinez-Banaclocha H, Casorran-Berges M, et al. Brief Report: Late-onset cryopyrin-associated periodic syndrome due to myeloid-restricted somatic NLRP3 mosaicism. Arthritis Rheumatol. 2016;68:3035–41. doi: 10.1002/art.39770. [DOI] [PubMed] [Google Scholar]
- 178.Rowczenio DM, Gomes SM, Arostegui JI, Mensa-Vilaro A, Omoyinmi E, Trojer H, et al. Late-onset cryopyrin-associated periodic syndromes caused by somatic NLRP3 Mosaicism-UK Single Center Experience. Front Immunol. 2017;8:1410. doi: 10.3389/fimmu.2017.01410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pathak S, Rowczenio DM, Owen RG, Doody GM, Newton DJ, Taylor C, et al. Exploratory study of MYD88 L265P, rare NLRP3 variants, and clonal hematopoiesis prevalence in patients with Schnitzler syndrome. Arthritis Rheumatol. 2019;71:2121–5. doi: 10.1002/art.41030. [DOI] [PubMed] [Google Scholar]
- 180.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–8. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Beck DB, Ferrada MA, Sikora KA, Ombrello AK, Collins JC, Pei W, et al. Somatic mutations in UBA1 and severe adult-onset autoinflammatory disease. N Engl J Med. 2020;383:2628–38. doi: 10.1056/NEJMoa2026834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nanki K, Fujii M, Shimokawa M, Matano M, Nishikori S, Date S, et al. Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature. 2020;577:254–9. doi: 10.1038/s41586-019-1844-5. [DOI] [PubMed] [Google Scholar]
- 183.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16:626–38. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 184.Scully EP, Haverfield J, Ursin RL, Tannenbaum C, Klein SL. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat Rev Immunol. 2020;20:442–7. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Klein SL, Marriott I, Fish EN. Sex-based differences in immune function and responses to vaccination. Trans R Soc Trop Med Hyg. 2015;109:9–15. doi: 10.1093/trstmh/tru167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sasidhar MV, Itoh N, Gold SM, Lawson GW, Voskuhl RR. The XX sex chromosome complement in mice is associated with increased spontaneous lupus compared with XY. Ann Rheum Dis. 2012;71:1418–22. doi: 10.1136/annrheumdis-2011-201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liu K, Kurien BT, Zimmerman SL, Kaufman KM, Taft DH, Kottyan LC, et al. X chromosome dose and sex bias in autoimmune diseases: increased prevalence of 47,XXX in systemic lupus erythematosus and Sjogren’s syndrome. Arthritis Rheumatol. 2016;68:1290–300. doi: 10.1002/art.39560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Harris VM, Sharma R, Cavett J, Kurien BT, Liu K, Koelsch KA, et al. Klinefelter’s syndrome (47,XXY) is in excess among men with Sjogren’s syndrome. Clin Immunol. 2016;168:25–29. doi: 10.1016/j.clim.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Itoh Y, Golden LC, Itoh N, Matsukawa MA, Ren E, Tse V, et al. The X-linked histone demethylase Kdm6a in CD4+ T lymphocytes modulates autoimmunity. J Clin Invest. 2019;129:3852–63. doi: 10.1172/JCI126250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Golden LC, Itoh Y, Itoh N, Iyengar S, Coit P, Salama Y, et al. Parent-of-origin differences in DNA methylation of X chromosome genes in T lymphocytes. Proc Natl Acad Sci USA. 2019;116:26779–87. [DOI] [PMC free article] [PubMed]
- 191.Palaszynski KM, Smith DL, Kamrava S, Burgoyne PS, Arnold AP, Voskuhl RR. A yin-yang effect between sex chromosome complement and sex hormones on the immune response. Endocrinology. 2005;146:3280–5. doi: 10.1210/en.2005-0284. [DOI] [PubMed] [Google Scholar]
- 192.Bookman EB, McAllister K, Gillanders E, Wanke K, Balshaw D, Rutter J, et al. Gene-environment interplay in common complex diseases: forging an integrative model-recommendations from an NIH workshop. Genet Epidemiol. 2011;35:217–25. doi: 10.1002/gepi.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gillberg J, Marttinen P, Mamitsuka H, Kaski S. Modelling GxE with historical weather information improves genomic prediction in new environments. Bioinformatics. 2019;35:4045–52. doi: 10.1093/bioinformatics/btz197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Findley AS, Monziani A, Richards AL, Rhodes K, Ward MC, Kalita CA, et al. Functional dynamic genetic effects on gene regulation are specific to particular cell types and environmental conditions. Elife. 2021;10:e67077. [DOI] [PMC free article] [PubMed]
- 195.Wang H, Zhang F, Zeng J, Wu Y, Kemper KE, Xue A, et al. Genotype-by-environment interactions inferred from genetic effects on phenotypic variability in the UK Biobank. Sci Adv. 2019;5:eaaw3538. doi: 10.1126/sciadv.aaw3538. [DOI] [PMC free article] [PubMed] [Google Scholar]