Abstract
Triple negative breast cancer (TNBC) is an aggressive subtype, associated with early metastatic recurrence and worse patient outcomes. TNBC patient tumors express molecular markers of the epithelial to mesenchymal transition (EMT) but its role during spontaneous TNBC metastasis in vivo remains incompletely understood. In this study, we demonstrate that spontaneous TNBC tumors from a genetically engineered mouse model (GEMM), multiple patient-derived xenografts (PDXs), and archival patient samples exhibit large populations of hybrid E/M cells in vivo that lead invasion ex vivo. We found that the mesenchymal marker vimentin promotes invasion and represses metastatic outgrowth. We next tested the requirement for five EMT transcription factors and observed distinct patterns of utilization during invasion and colony formation. These differences suggest a sequential activation of multiple EMT molecular programs during the metastatic cascade. Consistent with this model, our longitudinal single-cell RNA analysis detected 3 different EMT-related molecular patterns. We observed cancer cells progressing from epithelial to hybrid E/M and strongly mesenchymal patterns during invasion and from epithelial to a hybrid E/M pattern during colony formation. We next investigated the relative epithelial vs. mesenchymal state of cancer cells in both GEMM and patient metastases. In both contexts, we observed heterogeneity between and within metastases in the same individual. Strikingly, we observed a complex spectrum of epithelial, hybrid E/M, and mesenchymal cell states within metastases, suggesting there are multiple successful molecular strategies for distant organ colonization. Taken together, our results demonstrate an important and complex role for EMT programs during TNBC metastasis.
One Sentence Summary:
Triple negative breast cancer utilizes a vimentin-dependent hybrid EMT to generate metastases with diverse epithelial and mesenchymal characteristics.
Introduction:
Breast cancer is a common and heterogeneous disease with several subtypes associated with different clinical behaviors (1-4). Despite significant advances in diagnosis and treatment, metastasis remains the main driver of breast cancer mortality (1). Metastasis is a multistep process by which cancer cells invade into their surrounding stroma, enter and survive in the circulation, seed distant organs, and grow into new tumors (5). A deeper understanding of the cellular and molecular mechanism of metastasis formation is essential to develop new therapies against metastatic disease. We previously demonstrated that luminal breast cancer metastases can arise from the collective dissemination of keratin 14-expressing (K14+) tumor cell clusters (6, 7). In this study, we investigate the mechanisms of metastasis in triple negative breast cancer (TNBC). TNBC is characterized by the lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). TNBC is typically associated with a basal molecular phenotype (80% concordance) (8). However, molecular analyses have consistently revealed that TNBC is a highly heterogeneous disease, composed of multiple subtypes with pronounced transcriptomic heterogeneity (9-11). Deeper insight into the molecular mechanisms regulating TNBC metastasis is urgently needed as distant recurrence occurs frequently and rapidly in these subtypes and there are few options for targeted therapy (12, 13).
Multiple groups have reported mesenchymal gene expression in TNBC, suggesting an epithelial-mesenchymal transition (EMT) (14-20). The EMT model postulates that cancer cells downregulate epithelial gene expression and acquire mesenchymal features that allow them to invade, access the circulation, and survive in transit to seed distant sites (21, 22). Conversely, the reverse process of mesenchymal-epithelial transition (MET) is thought to be advantageous for metastatic outgrowth (23-27). However, despite the presence of mesenchymal gene expression, the requirement for and clinical relevance of EMT and MET during in vivo TNBC metastasis remains unclear (16, 17, 28). The main experimental platform for testing the role of EMT during spontaneous metastasis in vivo has been genetically engineered mouse models (GEMMs). In breast cancer, the most frequently studied GEMMs utilize the mouse mammary tumor virus (MMTV) LTR to drive expression of the polyoma middle T oncogene (MMTV-PyMT) or ERBB2 (MMTV-Neu); both model luminal breast cancer (29-34). To our knowledge, the in vivo role of EMT and MET in spontaneous TNBC models has not been reported.
In this study, we characterize GEMM, patient derived xenograft (PDX) TNBC models and both fresh and archival TNBC patient tumor tissues for markers of EMT. Using integrated ex vivo and in vivo analyses, we interrogate the functional role of the EMT effector vimentin in different stages of the metastatic cascade both in 3D culture and in vivo. We then use single cell RNA sequencing (scRNA-seq) to reveal dynamics in EMT molecular programs during different metastatic processes ex vivo and we isolate the distinct contributions of different EMT transcription factors during invasion and colony formation. Finally, we characterize the intra- and inter-tumoral heterogeneity of EMT states in both GEMM and patient TNBC metastases.
Results:
TNBC patient primary tumors contain hybrid E/M cancer cells and hybrid E/M gene expression correlates with poor patient outcome.
We started by investigating the presence of EMT markers in TNBC patient primary tumors by using a tissue microarray (TMA) enriched for TNBC. We first performed immunofluorescence against E-cadherin (Ecad) and vimentin (Vim), the most commonly used markers for epithelial and mesenchymal phenotypes, respectively (35). We observed that TNBC primary tumors present a higher percentage of hybrid E/M areas, expressing both Ecad and Vim, compared to the other breast cancer subtypes (Figure 1A-B). Details by breast cancer subgroups are presented in Figure S1A. We also observed a high percentage of hybrid E/M area in the ER−/PR+/HER2− subgroup. Despite this average increase, we observed substantial heterogeneity within the TNBC subgroup, with the Ecad+/Vim+ areas varying between 14% to 90%. We then asked whether the expression of Ecad, Vim, or both markers could have clinical relevance. Analysis of the relapse free survival (RFS) within untreated basal-like breast cancer patients revealed that Ecad alone is not sufficient to predict patient survival, whereas Vim alone does predict significantly worse RFS (Hazard Ratio: 1.62, p value 0.05). Notably, dual high expression of Ecad and Vim even more effectively stratifies patient survival (Hazard Ratio: 1.95, p value 0.0079) (Figure S1B). These results demonstrate that TNBC primary tumors frequently contain hybrid E/M cells, reveal that combined expression of Ecad and Vim is correlated with reduced patient relapse free survival, and suggest a functional role for hybrid EMT in TNBC metastasis.
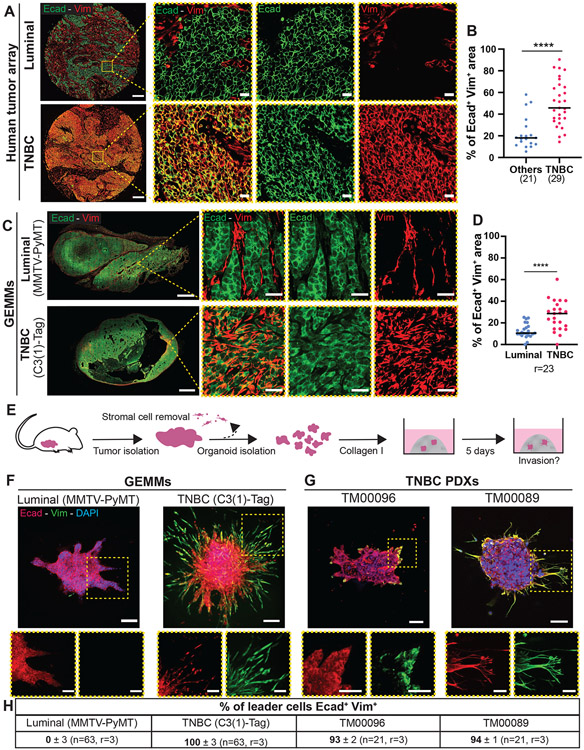
Figure 1: Primary TNBC tumors from patients and animal models contain hybrid E/M cells that lead invasion.
A) Representative epifluorescence images of human primary breast tumors stained with Ecad and Vim. Scale bar is 200 μm and 20 μm (insets).
B) Percentage of Ecad+ Vim+ double positive area in human breast tumors (n=21 non-TNBC, n= 29 TNBC tumors). Percentage calculated by pixel colocalization analysis. Each dot corresponds to a tumor, bars are medians. ****P<0.0001 (Mann-Whitney).
C) Representative epifluorescence (whole tumor, left) and confocal image (zoom, right) of MMTV-PyMT and C3(1)-Tag primary tumors stained with Ecad and Vim. Scale bars are 2000 μm and 25 μm for (insets).
D) Percentage of Ecad+ Vim+ area in whole MMTV-PyMT and C3(1)-Tag primary tumors. Percentage calculated by pixel colocalization analysis. Each dot corresponds to a tumor, bars show the medians. ****P<0.0001 (two side T-Test)
E) Schema of organoid isolation and invasion assay. The primary tumor is digested into tumor organoids and the stromal cells are removed by differential centrifugations. Each organoid is composed of 200 to 500 adherent cancer cells and embedded in 3D collagen I matrix. After 5 days, the organoids are fixed for immunostaining.
F, G) Representative confocal maximum intensity projections of whole organoids stained with anti-Ecad and anti-Vim antibodies and DAPI in GEMMs (F) and TNBC PDXs (G). Scale bars are 100 μm and 50 μm (insets).
H) Frequency of leader cells expressing Ecad and Vim with the median of the percentage and the standard error of mean (SEM).
C3(1)-Tag GEMM and TNBC PDX primary tumors present a high fraction of hybrid E/M cells.
GEMMs are the main experimental platform to investigate spontaneous metastasis formation in vivo. To our knowledge, the role of EMT and MET has not been investigated in TNBC GEMMs. We chose to focus on the C3(1)-Tag mouse model of basal-TNBC to assay for the presence of hybrid E/M cells (33, 36). C3(1)-Tag mice express a transgene containing the 5' flanking region of the C3(1) component of the rat prostate steroid binding protein (PSBP) fused to the simian virus 40 large T-antigen (ag) (36). Essentially all mice develop invasive carcinomas, with sporadic metastasis to the lungs (37). Previous gene expression profiling studies have demonstrated that C3(1)-Tag tumors cluster with the basal-like subgroup of breast cancer, which largely although not exclusively overlaps with the TNBC subtype (33, 36). As a luminal comparison, we used the MMTV-PyMT GEMM, which is the most studied breast cancer GEMM for EMT (29-32, 34). MMTV-PyMT mice develop invasive mammary tumors that metastasize efficiently to the lungs (38). These tumors resemble the luminal B subtype in patients (33, 36). Immunofluorescence of Ecad and Vim within whole sections from 23 MMTV-PyMT and C3(1)-Tag tumors revealed that C3(1)-Tag primary tumors present a higher percentage of Ecad+/Vim+ areas than MMTV-PyMT (Figure 1C-D). It is important to note that this area-based measurement will overestimate the representation of double positive cells as an area can be positive due to the presence of, for example, adjacent Ecad−/Vim+ stromal fibroblasts and Ecad+/Vim− epithelial phenotype cancer cells. To define quantitatively the percentage of Ecad+/Vim+ cells within these models, we used a fluorescence-activated cell sorting (FACS) strategy. Briefly, we isolated fresh organoids from our GEMMs, used differential centrifugations to remove the stromal cell population, as previously described (6, 7, 39, 40), dissociated them into single cells, and fixed and double stained them with Ecad and Vim (Figure S1C). We observed that 85% of MMTV-PyMT cancer cells are Ecad+/Vim− and 10% are Ecad+/Vim+, consistent with previous studies (30, 31, 41). We detected a higher proportion of hybrid E/M cells within C3(1)-Tag cancer cells, with 32% Ecad+/Vim+ cells. The remaining 60% of cells were Ecad+/Vim−. For both MMTV-PyMT and C3(1)-Tag, we detected fewer than 5% Ecad− cells, indicating a low proportion of mesenchymal cells (Figure S1D-F).
The C3(1)-Tag TNBC GEMM model presented striking inter-tumor heterogeneity with varying proportions of hybrid E/M area (Figure 1C-D), as well as intra-tumor heterogeneity with both Ecad+/Vim− and Ecad+/Vim+ cells observed in the same tumor (Figure S2A). We conclude that these hybrid E/M cells are cancer cells because they stain positive for the SV40 antigen by immunofluorescence (Figure S2A and S2B). We noticed that Ecad localization differs between Ecad+/Vim− areas and Ecad+/Vim+ areas, in which Ecad localized to the cell-cell junctions or intracellularly, respectively (Figure 1C and S2A). Vim and Ecad immunostaining were validated using C3(1)-Tag Vim knock-out and C3(1)-Tag Ecadfl/fl mice (Figure S2C and S2D). To further confirm the hybrid E/M states of cancer cells within C3(1)-Tag primary tumors, we stained with other EMT markers (Figure S2E). We observed multiple hybrid E/M cell states with cancer cells expressing the epithelial markers Ecad and EpCAM with the mesenchymal marker N-cadherin (Ncad), with or without keratin 14 (K14) and Vim. This result indicates that within the same tumors, cancer cells occupy multiple hybrid E/M states indicating large heterogeneity at individual cell level.
We next evaluated hybrid E/M markers in two primary tumor TNBC patient-derived xenografts (PDXs) (Jackson Laboratory TM00096 and TM00089). These TNBC PDX models exhibited a high, though variable, percentage of double positive Ecad+/Vim+ area (Figure S3A-B). We noted that these primary tumors present large necrotic area. We also detected the expression of EpCAM, alpha smooth muscle actin (αSMA), and Ncad within these tumors (Figure S3C). These markers were sufficient to reveal a diversity of hybrid E/M states, such as EpCAM+/Vim+/Ncad+ cells with or without αSMA expression. Since αSMA is considered a late EMT marker, its coexpression with EpCAM and E-cadherin reveals that TNBC cancer cells can combine early and late EMT markers in unexpected ways. Taken together, our data reveal that C3(1)-Tag GEMM, and TNBC PDX primary tumors contain a heterogeneous mix of cancer cells in diverse hybrid E/M states, suggesting that these are clinically relevant models to evaluate the role of EMT in TNBC metastasis.
Ecad+/Vim+ cells lead invasion in TNBC models and patients.
We next sought to test whether hybrid E/M cells participate in invasion using 3D culture assays. Briefly, we freshly isolated organoids from primary tumors and embedded them in collagen I to induce invasion (Figure 1E). After 5 days, organoids were stained with Ecad and Vim. MMTV-PyMT organoids had few Ecad+/Vim+ cells and the leader cells were typically Vim−. In contrast, C3(1)-Tag organoids contained a large fraction of Ecad+/Vim+ cells that led the invasive strands (Figure 1F-H). We then validated these results in TNBC PDXs and TNBC patient primary tumors. Similar to C3(1)-Tag organoids, 93% and 94% of leader cells in organoids from two TNBC PDX models were Ecad+/Vim+ (TM00096 and TM00089, respectively) (Figure 1G-H). Next, we analyzed the expression of Ecad and Vim in freshly isolated TNBC patient organoids (Figure S3D) and observed that hybrid E/M cells led invasion in 3D culture assays established from three different TNBC patient tumors. The presence of hybrid E/M invasive leader cells across TNBC GEMM, PDX, and patient primary tumors provides strong correlative support for the hypothesis that hybrid E/M cells drive the invasive phenotype in TNBC.
Vimentin is required for invasion and metastasis formation in TNBC models.
We previously showed that K14+ cancer cells frequently lead invasion in luminal breast tumors and that its expression is required for invasion both in 3D culture and in vivo (6). Since we consistently observed Vim+ cancer cells leading invasion from TNBC breast tumors, we hypothesized that Vim is required for TNBC invasion. We first evaluated five different short hairpin RNAs (shRNAs) targeting Vim and selected the two hairpins with the highest knockdown (KD) efficiency (Figures S4A-E). To test the requirement for Vim in invasion, we cultured non-targeting-shRNA (NT-shRNA), shVim #1, and shVim #2 transduced tumor organoids in 3D collagen I (Figure 2A). Vim knockdown blocked invasion of TNBC GEMM C3(1)-Tag organoids and TNBC PDX organoids without affecting invasion of luminal MMTV-PyMT organoids (Figure 2B-C, Movie 1). Knock-down efficiency across experiments and representative western blots are presented in Figure S4F-I. We noted variability in the invasive response in Vim knockdown organoids, with some organoids still able to invade vigorously into collagen I in the Vim KD condition. To distinguish a Vim-independent invasion mechanism from invasion led by persisting Vim+ cells, we immunostained these organoids for Vim. We observed a striking correlation between the presence of Vim+ cells and organoid invasive behavior; even very few Vim+ cells were sufficient to lead invasion (Figure S4J). These data support our hypothesis that Vim is essential for TNBC invasion in 3D culture in both GEMM and PDX models.
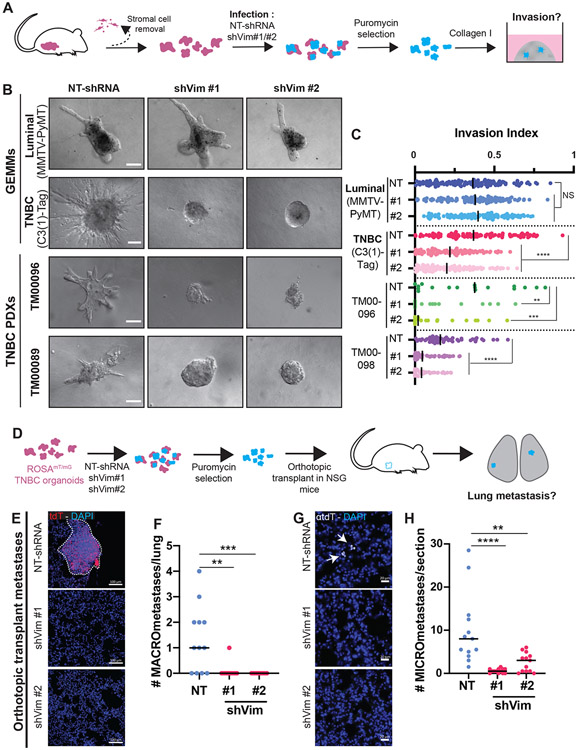
Figure 2: Vimentin is required for TNBC invasion and orthotopic metastasis formation.
A) Schema of collagen I invasion assay with shRNA knockdown. After isolation, organoids are infected with either non-targeting shRNA (NT-shRNA) or either of two shRNA sequences against vimentin (shVim #1 and shVim #2). After puromycin selection to eliminate non-infected cells, organoids are plated into collagen I.
B) Representative DIC images of organoids infected with NT-shRNA, shVim #1, or shVim #2 after 5 days in collagen I matrix. Scale bars are 100 μm.
C) Quantification of organoid invasion represented in B. Each dot corresponds to one organoid, bars show the medians. NS: not significant, ****P<0.0001, **P=0.0065 and ***P=0.0002 (Kruskal-Wallis). n=926 organoids, r=3 biological replicates.
D) Schema of transplant assay to study metastasis. Fluorescent organoids expressing membrane tdTomato are isolated and infected with lentivirus expression NT-shRNA, shVim #1, or shVim#2. After puromycin selection organoids are orthotopically transplanted into the cleared mammary fat pad of NSG mice. After 6-8 weeks, when the primary tumors are 20 mm, lungs are collected.
E) Representative confocal images of lung sections stained with DAPI. Metastasis are detected by membrane tomato (tdT) signal. Scale bars are 100 μm.
F) Quantification of the number of MACROmetastases (>100 cells). Each dot corresponds to the number of macrometastases in whole lungs. Bars show the medians. **P=0.0019, ***P=0.0006 (Kruskal-Wallis).
G) Representative epifluorescence of MICROmetastases in the lungs. Micrometastases were detected by anti-tdT immunostaining and DAPI. Scale bars are 20 μm. Arrows indicate single cell micrometastases. N=35 mice, r=3 biological replicates.
H) Number of MICROmetastases per lung section (20 μm thickness). Each dot corresponds to the number of micrometastases in a lung section. Bars show medians. **P=0.0082, ****P<0.0001 (Kruskal-Wallis). N=39 sections, r=3 biological replicates.
Metastasis is a systemic phenotype and therefore the requirement for Vim in metastasis must be tested in vivo. Accordingly, we next investigated the consequences of Vim knock-down in orthotopic transplant-based metastasis assays. Briefly, C3(1)-Tag tumor organoids were isolated from fluorescently labeled advanced carcinomas (ROSAmT/mG C3(1)-Tag), transduced with either NT-shRNA, shVim #1, or shVim #2 lentivirus, and then orthotopically transplanted into the cleared mammary fat pads of immunodeficient nod scid gamma (NSG) mice (Figure 2D). Analysis of primary tumor sections revealed that the proportion of Ecad+/Vim+ areas was reduced from a median of 72% in control tumors to 20% and 30% for shVim #1 and shVim #2, respectively (Figure S5A-B). Primary tumor growth tended to increase in Vim KD conditions, with some variation across mice, possibly due to varying efficiency of Vim KD in vivo (Figure S5C-D). To test the requirement for Vim in metastasis, we analyzed the lungs of transplant recipient mice. We observed a significant reduction in macrometastasis (>100 cells) number (Figure 2E-F, S5E), as well as micrometastasis (<100 cells) number in the Vim KD groups (Figure 2G-H). These results reveal that the acquisition of a key marker of the hybrid E/M state, vimentin, is required for spontaneous metastasis formation in vivo in this TNBC GEMM.
Vimentin represses TNBC metastatic outgrowth.
Several studies suggest that cancer cells must lose or downregulate mesenchymal gene expression and revert to epithelial gene expression to form macrometastases (23-25, 27). However, the role of MET during in vivo TNBC metastasis remains unclear. We therefore decided to investigate the role of Vim during colony formation ex vivo and during metastatic outgrowth in vivo. We previously demonstrated that our ex vivo colony formation assay models key aspects of the survival and proliferative processes involved in metastatic seeding and outgrowth (7, 40). We used this assay to assess the consequences of Vim KD in cancer cells freshly isolated from luminal MMTV-PyMT GEMM, TNBC C3(1)-Tag GEMM, and TNBC PDX. Briefly, we cultured NT-shRNA, shVim #1, and shVim #2 infected cancer cell clusters in 3D Matrigel environment and assessed their capacity to form colonies (Figure 3A). Vim KD did not affect colony formation of luminal MMTV-PyMT clusters. In contrast, Vim KD increased colony formation of C3(1)-Tag and TNBC PDX clusters (Figure 3B-C), indicating that Vim expression suppresses ex vivo TNBC colony formation.
Figure 3: Vimentin represses colony formation ex vivo and tail vein experimental metastasis in vivo.
A) Schema of colony formation assay using shRNA. After isolation, organoids are infected with either NT-shRNA or either of two shRNA sequences against vimentin (shVim #1 and shVim #2). After puromycin selection to eliminate non-infected cells, organoids are dissociated into clusters and embedded in Matrigel.
B) Representative DIC images of colonies infected with NT-shRNA, shVim #1, or shVim #2 after 5 days (GEMM) or 10 days (PDX) in Matrigel. Scale bars are 100 μm.
C) Number of colonies per well. Each dot represents the number of colonies in one well, bars show the medians. NS = not significant, **P=0.008, ***P<0.001 ****P<0.0001 (one-way ANOVA). n=89, r=3.
D) Tail vein assay schema to study the late steps of the metastatic cascade. Fluorescent organoids are infected with NT-shRNA, shVim #1, or shVim #2. After selection, organoids are dissociated into clusters and injected into the tail vein of NSG mice. After 4 weeks, the number of metastases in the lungs is assessed.
E) Representative epifluorescence images of whole lungs. Macrometastases indicated by arrows are detected by tdT signal.
F) Number of macrometastases per lung. Each dot corresponds to a lung. Bars show medians. **P=0.0036, ***P=0.0004 (Kruskal-Wallis). n=24 mice, r=2 biological replicates.
Our ex vivo experiments demonstrate a requirement for Vim in invasion and a complete block of invasion would be expected to block metastasis by preventing cancer cells from reaching distant organs. Hence, our orthotopic metastasis experiments do not necessarily evaluate the role of Vim or MET later in the metastatic cascade. We therefore next utilized a tail vein-based experimental metastasis assay to specifically interrogate the later stages of cell seeding and metastatic outgrowth. Briefly, fluorescently labeled organoids (ROSAmT/mG C3(1)-Tag) were isolated and infected with lentivirus expressing either NT-shRNA, shVim #1, or shVim #2. After selection, organoids were dissociated into clusters and injected into the tail vein of NSG mice (Figure 3D). Consistent with our ex vivo colony formation assay results, we observed a significant increase in the number of macrometastases in Vim knock-down groups (Figure 3E-F). These results demonstrate that Vim suppresses metastatic outgrowth in vivo, suggesting it is advantageous for TNBC cancer cells to downregulate Vim expression during distant metastasis formation.
scRNA-seq reveals that C3(1)-Tag cancer cells progress from epithelial to hybrid E/M states during both invasion and colony formation.
Our results indicate that the acquisition of the mesenchymal marker Vim is required for TNBC invasion and that it also represses metastatic outgrowth. These contrasting effects of Vim expression on metastatic efficiency at different stages suggests contextual dynamics in EMT molecular programs. However, the stage-specific molecular regulation of EMT and Vim expression in TNBC remains incompletely understood. To better identify the EMT programs and molecular alterations associated with invasion and colony formation, we next performed scRNA-seq on organoids over time in our ex vivo assays. The main advantage of this approach is that we are able to analyze transcriptional dynamics during specific metastatic processes in a constant cell population. Briefly, C3(1)-Tag organoids were isolated and directly plated into collagen I matrix or further dissociated into clusters and embedded into Matrigel. Single cell profiling was performed at day 0 (12 hours in matrix), day 3, and day 5 in collagen and Matrigel. A challenge for time course experiments with scRNA-seq is that the variation in library preparation between days can introduce a technical bias in the data resulting in a batch effect, therefore we also profiled a cell line control (4T1) with a consistent transcriptional profile over time (Figure 4A). Uniform Manifold Approximation and Projection (UMAP) of the uncorrected data showed that the organoid samples in collagen and Matrigel both cluster together and separately from the 4T1 cells (Figure S6A). 4T1 cells cluster together for day 0 and day 5 but not for day 3, and there is a similar separation by day observed in the organoid data, consistent with this anticipated batch effect. We corrected for batch effects and observed that we retain transcriptional heterogeneity in the organoid culture while successfully removing the technical sources of variation that cause the 4T1 cells to cluster separately between time points (Figure S6A).
Figure 4: Cancer cells transition to hybrid E/M states and require different EMT TFs during invasion and colony formation.
A) Schema of the longitudinal single-cell RNA sequencing experiment. After isolation, C3(1)-Tag organoids are embedded in collagen I or dissociated into clusters and embedded in Matrigel. After 12h, 3 days and 5 days, organoids are extracted from the matrix and dissociated into single cells. Single tumor cells and 4T1 cells, used as a reference for batch correction, are barcoded per condition before flow sorting for live cells using propidium iodide. Live cells are next used for 10X Genomics barcoding and library preparation. All conditions are then sequenced together.
B) UMAP representation of the sequenced cells from invasion and colony formation assay. Each dot corresponds to single cell and the colors indicate the day the cells were extracted from the matrix.
C) Distribution of the proportion of Ecad+ and/or Vim+ cells by matrix and day.
D) Schema for shRNA-based gene knock-down in the invasion and colony formation assays. After isolation, organoids are infected with either NT-shRNA or a pool of 3 shRNA sequences against a specific gene. After puromycin selection to eliminate non-infected cells, organoids are either directly lysed to extract mRNA for qPCR analysis, embedded into collagen I, or dissociated into clusters and embedded in Matrigel. After 5 days, the invasion or the number of colonies is measured.
E) mRNA fold change, assayed by qRT-PCR, of Ecad (Cdh1) and Vim gene expression in C3(1)-Tag organoids knocked-down for Grhl2, Zeb2, Foxc2, Zeb1 and Ovol1. Histogram of mean with SEM. r ≥3. * P<0.05, ***P<0.005 (paired T-test, two sided on the deltaCT value between NT-shRNA and shRNA against specific gene).
F) Representative confocal images of maximum intensity projection of whole C3(1)-Tag organoids stained with Vim. GFP indicates infected cells. Scale bars are 100 μm.
G) Quantification of organoid invasion represented in F. Each dot corresponds to one organoid, bars show the medians. **P=0.0016, ***P=0.0003, ****P<0.0001 (Kruskal-Wallis), n=163 organoids, r=3 biological replicates.
H) Representative epifluorescence images of maximum intensity projection of C3(1)-Tag colonies. GFP indicates infected cells. Scale bars are 500 μm.
I) Number of colonies per well. Each dot represents the number of colonies in one well, bars show the medians. *P=0.0267, **P=0.034 (Kruskal-Wallis), n=67 wells, r=3 biological replicates.
Representation of cancer cells from the organoid model on the two-dimensional, batch-corrected UMAP embedding identifies shared transcriptional signatures between the collagen and Matrigel conditions but still separates days, indicating a transition of cancer cell states over time during invasion and colony formation (Figure 4B). We then looked at Ecad and Vim expression in these cells. During invasion, we observed a decrease in Ecad expression and an increase in Vim expression (Figure S6B). We detected a decrease in Ecad+/Vim− cells over time and an increase in Ecad−/Vim+ and Ecad+/Vim+ cells, with the majority of cancer cells at Day 5 being Ecad+/Vim+ (Figure 4C). Unexpectedly, we also observed a decrease in Ecad expression during colony formation (Figure S6B). Both the Ecad−/Vim+ and Ecad+/Vim− populations decreased over time, while the Ecad+/Vim+ cells increased over time (Figure 4C). These results demonstrate that the majority of TNBC cancer cells acquire a hybrid E/M state during both invasion and colony formation.
Different EMT transcription factors are implicated in invasion vs colony formation.
Our functional analysis identified distinct roles for Vim during different stages of metastasis and our scRNA-seq data reveal distinct changes in EMT marker expression during invasion and colony formation. Taken together, these results led us to hypothesize that EMT transcription factors (TFs) would regulate distinct biological processes during metastasis. To test this hypothesis, we selected EMT TFs for functional validation based on the correlation of epithelial and mesenchymal genes with Vim expression during invasion and colony formation at the single cell level (Figure S6C). We identified Zeb1, Zeb2, and Foxc2, all TFs known to promote EMT, as strongly positively correlated with Vim expression. Conversely, Grhl2 and Ovol1, two TFs known to positively regulate epithelial gene expression, were strongly anti-correlated with Vim expression. We then tested the functional role of these EMT TFs in our invasion and colony assays using a shRNA knock-down strategy (Figure 4D). The knockdown efficiency for each gene as well as the impact of the KD on Ecad and Vim mRNA expression is indicated in Figure 4E. We found that Grhl2 knockdown results in increased invasion (Figure 4F-G), decreased expression of epithelial genes (such as Cdh1 (Ecad), Krt14, and Dsp), and increased expression of mesenchymal genes (such as Zeb1 and Vim) (Figure S7A). Grhl2 is therefore an invasion suppressor that contributes to the hybrid E/M state by modulating both epithelial and mesenchymal gene expression. Conversely, knockdown of either Zeb2 or Foxc2 reduces invasion (Figure 4F-G) and mesenchymal gene expression without impacting epithelial gene expression (Figure S7B-C). Intriguingly, knockdown of either Zeb1 or Ovol1 did not affect invasion (Figure 4F-G) despite associated decreases in mesenchymal gene expression (Figure S7D-E). We conclude that Zeb2 and Foxc2 are invasion promoters that selectively regulate mesenchymal components of the hybrid E/M state.
We next evaluated the requirement for these TFs in colony formation. Surprisingly, knockdown of Grhl2, Foxc2, or Zeb2 did not impact colony formation. However, Zeb1 or Ovol1 knockdown increased colony formation (Figure 4H-I) without increasing epithelial gene expression (Figure S7D-E). To summarize, our results demonstrate that Grhl2, Foxc2, and Zeb2 regulate invasion without impacting colony formation, whereas Zeb1 and Ovol1 represses colony formation without impacting invasion. These data establish the concept of sequential utilization of different EMT TFs to regulate distinct modules of mesenchymal gene expression during different metastatic processes ex vivo. They suggest the activation of multiple EMT programs during these two key steps of the metastatic cascade.
Single cell transcriptome analysis reveals multiple EMT programs during invasion and colony formation.
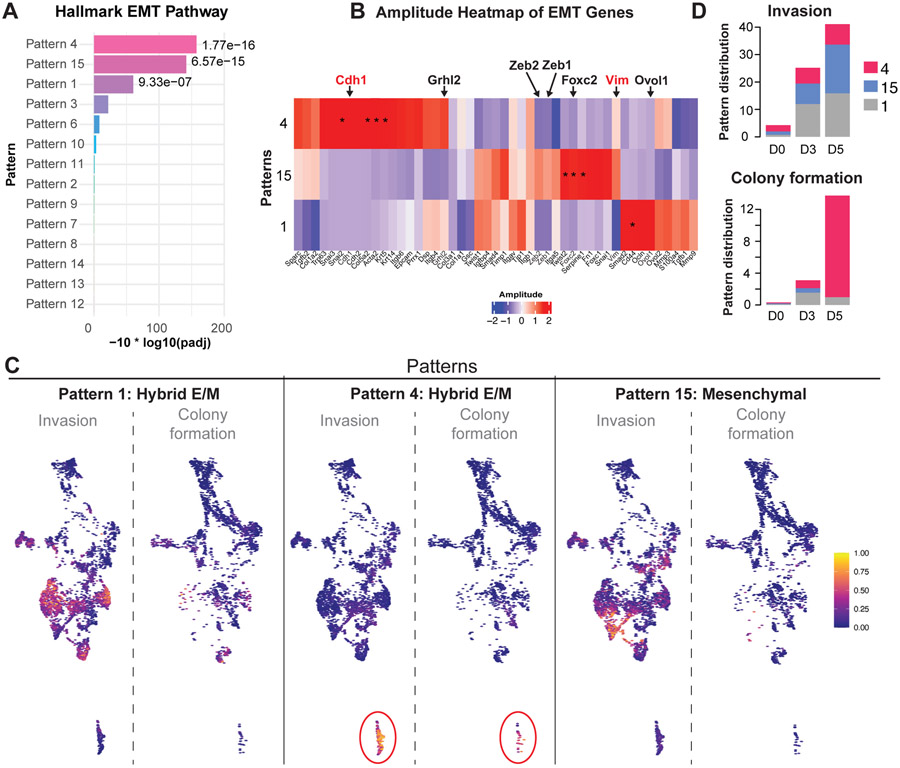
The contrasting requirements for these EMT TFs during invasion and colony formation and the diverse sets of mesenchymal genes that they regulate suggests that they activate different EMT molecular programs. To characterize these EMT programs at single cell level, we next performed a whole transcriptome characterization of our scRNA-seq data. To define transcriptional signatures associated with these programs, we apply the Bayesian non-negative matrix factorization method CoGAPS (42). In contrast to clustering, CoGAPS models co-expression of genes in multiple biological processes by estimating two associated matrices. The first matrix, the amplitude matrix, contains columns of gene weights defining transcriptional signatures learned from the data while the second matrix, the pattern matrix, defines cell weights corresponding to the usage of each of these signatures in each cell. We applied this approach to estimate 14 patterns from the scRNA-seq data. We observed one pattern that is enriched for cells in the control 4T1 cell line (pattern 5), while the remaining patterns are enriched either in the organoid data alone or are enriched in cells from both models (Figure S8A). We then identified marker genes associated with these patterns. using the ranking of unique gene association with patterns from the CoGAPS patternMarker statistic (43), excluding the 4T1 cell line patterns 5 from analysis. Gene set analysis of these marker genes with the Hallmark gene sets in MSigDB (44) finds three patterns strongly enriched for EMT (patterns 1, 4, and 15; Figure 5A). In addition, two patterns (patterns 8 and 9) are significantly enriched for the G2M cell cycle pathway and are distinct from the EMT patterns (Figure S8B). These results demonstrate that CoGAPS is effectively isolating the gene expression changes related to cell proliferation from the transcriptional signature during invasion and colony formation. Evaluation of the gene weights and pattern marker statistics in a subset of EMT genes identified that pattern 15 presents mostly mesenchymal genes including Vim, Foxc2, Zeb1 and Zeb2. Conversely, both patterns 1 and 4 are associated with a hybrid E/M state of epithelial and mesenchymal markers consistent with the predominance of double positive cells observed in the culture (Figure 5B-C). Interestingly, we observed an increase over time in cells associated with the hybrid E/M pattern 4 in both invasion and colony formation (Figure 5D), validating at the single cell transcriptome level that cancer cells transition to hybrid E/M states during both processes. However, we also detected an increase over time in cells associated with the hybrid E/M pattern 1 and mesenchymal pattern 15 during invasion. Taken together, our results reveal that TNBC cancer cells transition to more mesenchymal states during both invasion and colony formation and that these transitions involved more diverse EMT patterns during invasion than colony formation.
Figure 5: scRNA-seq reveals different EMT programs during invasion and colony formation.
A) Barplot representing the p-value of the enrichment of hallmark EMT pathway for the genes associated with patterns learned from the single-cell data with CoGAPS Bayesian non-negative matrix factorization.
B) Heatmap of weights of genes associated with patterns learned from the single-cell data with CoGAPS Amplitude matrix for EMT genes. *indicates unique association of genes with each pattern with the CoGAPS patternMarker statistic.
C) Cell weights for each of the four CoGAPS patterns associated with the EMT pathway plotted on the UMAP embedding of the scRNA-seq data.
D) Distribution of the proportion of cells associated with each of the four CoGAPS patterns associated with the EMT pathway by matrix and day.
C3(1)-Tag metastases are heterogeneous and express mesenchymal markers.
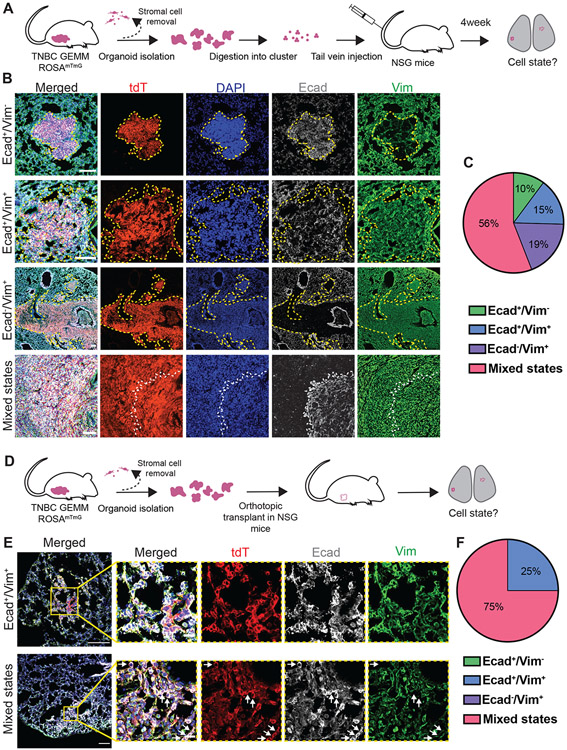
We had anticipated that hybrid E/M TNBC cancer cells would transition to a more purely epithelial state to form metastases. Since our scRNA-seq analysis instead revealed that TNBC cancer cells transition to a hybrid E/M state during colony formation ex vivo, we next hypothesized that TNBC metastases in vivo would present hybrid E/M characteristics. To test this hypothesis, we characterized lung metastases from the C3(1)-Tag GEMM for their expression of epithelial and mesenchymal markers. As spontaneous metastases are rare in this model (<15%) (37), we dissociated freshly isolated tdTomato+ organoids into clusters and single-cells and injected them into the tail vein of NSG mice (Figure 6A). Consistent with the diversity of single cell states in the scRNA-seq data from our colony formation assays, we observed heterogenous EMT phenotypes within lung metastases (Figure 6B-C). We detected few epithelial phenotype metastases (10% Ecad+/Vim−) or purely hybrid E/M metastases (15% Ecad+/Vim+). Surprisingly, we observed metastases that lost Ecad expression and maintained Vim expression, suggesting a strongly mesenchymal character (19% Ecad−/Vim+). However, most metastases (56%) contained a mixture of cell states, with both Ecad+/Vim+ and Ecad−/Vim+ cancer cells. We next investigated the correlation of Ecad with other markers in these metastases. To do so, we immunostained consecutive sections of lung metastases with different epithelial and mesenchymal markers (Figure S9A). We observed that Ecad− cancer cells in metastases were frequently EpCAM−, Pan-cytokeratin+, and K14+, indicating retention of some epithelial features. These cells were also frequently Fsp1+, αSMA+, and Ncad+, indicating expression of multiple epithelial and mesenchymal markers. We also detected Ecad+ cells expressing the late EMT marker αSMA, providing further support for both the concept of hybrid E/M states combining early and late EMT markers and for the concept that cancer cells with strongly mesenchymal gene expression can form macrometastases in distant organs.
Figure 6: TNBC GEMM metastases are heterogeneous and typically maintain mesenchymal gene expression.
A) Schema of tail vein experimental metastasis assay to characterize EMT states during colony formation in vivo. C3(1)-Tag fluorescent organoids expressing membrane tdTomato are isolated, digested into single cells and clusters and injected into the tail vein of NSG mice. Lungs are collected at 4 weeks and immunostained for epithelial and mesenchymal markers.
B) Representative epifluorescence images of the different metastasis states detected in lung sections stained for Ecad and Vim. Scale bars are 100 μm.
C) Proportion of macrometastases from tail vein experimental metastasis assays composed of Ecad+/Vim− cells, Ecad+/Vim+ cells, Ecad−/Vim+ cells, and mixed states (composed of cells in two or three of the previous categories). N=21 mice, r=5 biological replicates.
D) Scheme of orthotopic transplant assay to study metastases in vivo. C3(1)-Tag fluorescent organoids expressing membrane tdTomato are isolated and transplanted into the cleared mammary fat pad of NSG mice. Lungs are collected after 6-8 weeks, when primary tumors reach 20 mm.
E) Representative confocal images of the different metastasis states detected in lung sections stained for Ecad and Vim. Scale bars are 100 μm and 20 μm (insets). White arrows indicate epithelial cells (Ecad+/Vim−).
F) Proportion of orthotopic macrometastases composed of Ecad+/Vim− cells, Ecad+/Vim+ cells, Ecad−/Vim+ cells, and mixed states (composed of cells in two or three of the previous categories). n=6 mice, r=2 biological replicates.
It is possible that the EMT cell states observed in experimental metastases derived from tail vein injection of cancer cells could differ systematically from those that would predominate under spontaneous conditions. Accordingly, we next characterized the epithelial and mesenchymal characteristics of metastases generated in an orthotopic transplant assay (Figure 6D). We detected hybrid E/M metastases expressing both Ecad and Vim and metastases containing a mixture of epithelial and E/M cell states (Figure 6E-F). We did not detect strongly epithelial (Ecad+/Vim−) or strongly mesenchymal (Ecad−/Vim+) metastases. However, this difference may be attributable to the limited number of metastases that could be analyzed in this assay due to the low spontaneous metastatic burden of this model. Taken together, our results reveal that both experimental and spontaneous metastases in the C3(1)-Tag TNBC GEMM model frequently retain hybrid E/M or strongly mesenchymal character and that there is substantial cell state heterogeneity among metastases in the same animal and between mice.
TNBC patient metastases are heterogenous and express the mesenchymal marker vimentin.
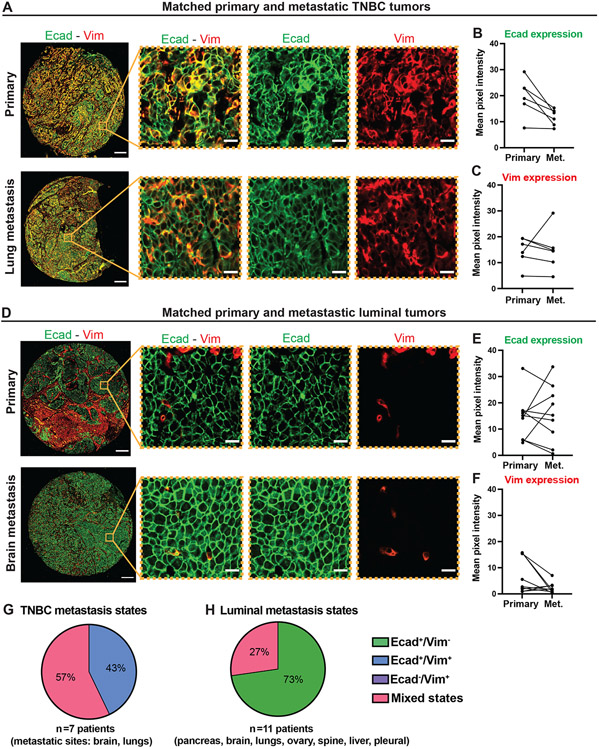
Since our observation of the retention of mesenchymal character in TNBC GEMM metastases was surprising, we next assayed for mesenchymal gene expression in TNBC patient metastases. To do so, we characterized a TMA containing paired primary and metastatic tumors from individual patients with TNBC or luminal breast cancer (45, 46). For each patient, we quantified the pixel intensity of Ecad and Vim staining in the primary and metastatic tumors. In TNBC patient samples, Ecad expression decreased and Vim expression remained consistent between primary tumors and metastases (Figure 7A-C). In luminal breast cancer patient samples, Ecad expression varied between the primary tumors and metastases and Vim was either absent in the cancer cells or decreased in the metastases relative to the primary tumors (Figure 7D-F). We next used these samples to evaluate the relationship between Ecad subcellular localization and epithelial vs. mesenchymal cell state in this TMA (47). We classified Ecad localization as membrane-localized, intracellular, or mixed (both membrane and intracellular observed) (Figure S10A). In TNBC tumors, Ecad localization was predominantly intracellular or mixed (Figure S10B). In luminal breast cancer tumors, Ecad localization was predominantly at the membrane or mixed (Figure S10B). Ecad localization was consistent between the primary and metastatic tumors for most patients. These results in clinical samples provide validation of the conclusions of our TNBC GEMM analyses and support the concept of predominantly epithelial character of luminal breast cancer metastases and retention of mesenchymal character in TNBC metastases.
Figure 7: EMT states in paired patient primary and metastatic TNBC or luminal tumors.
A) Representative epifluorescence images of paired primary and metastatic TNBC tumors. Scale bars are 200 μm and 20 μm (insets).
B, C) Quantification of the mean pixel intensity of Ecad (B) and Vim (C) staining in cancer cells from primary and metastatic TNBC tumors from individual patients. Each dot corresponds to the mean pixel intensity of cancer cells from a tumor. n=5 primary tumors and 6 metastases.
D) Representative epifluorescence images of paired primary and metastatic luminal tumors. Scale bars are 200 μm and 20 μm (insets).
E, F) Quantification of the mean pixel intensity of Ecad (E) and Vim (F) staining in cancer cells from primary and metastatic luminal tumors from individual patients. Each dot corresponds to the mean pixel intensity of cancer cells from a tumor. n=8 primary tumors and 9 metastases.
G, H) Proportion of patient TNBC (G) and luminal (H) metastases composed of Ecad+/Vim− cells, Ecad+/Vim+ cells, Ecad−/Vim+ cells, and mixed states (composed of cells in two or three of the previous categories).
We then classified the cancer cell states in patient TNBC and luminal metastases. Among the 7 TNBC metastases analyzed, we observed that 3 (43%) metastases were composed of Ecad+/Vim+ cells only. The remaining 4 metastases contained mixed cell states with either Ecad+/Vim− and Ecad+/Vim+ cells, or Ecad+/Vim−, Ecad+/Vim+, and Ecad−/Vim+ cells (Figure 7G). We detected these Ecad+/Vim+ and mixed states in both lung and brain sites. Our results indicate that, similar to the C3(1)-Tag metastases, TNBC patient metastases are heterogenous and maintain mesenchymal gene expression, suggesting that a complete MET is not always observed during TNBC metastasis formation. In contrast, most luminal metastases (73%) were Ecad+/Vim− (Figure 7H). The remaining metastases (27%) from brain and spine presented a mix of Ecad+/Vim− cells with Ecad+/Vim+ cells.
Taken together, our results reveal that luminal primary and metastatic tumors are typically epithelial in character, while TNBC tumors display heterogeneous hybrid E/M cells states in both primary and distant sites, rather than reverting to purely epithelial gene expression. The mesenchymal character and the greater cell state diversity observed in TNBC may be a factor contributing to the treatment resistance commonly observed in metastatic TNBC.
Discussion:
TNBC is an aggressive breast cancer subtype with frequent early metastasis and an urgent need for new therapies. TNBC is known to express EMT molecular markers (14-20) and our study sought to determine the functional relationship between EMT and spontaneous TNBC metastasis. We based our conclusions on analysis of both TNBC GEMM and PDX models and both fresh and archival patient TNBC tumors. We consistently observed that primary TNBC tumors exhibit large populations of hybrid E/M cells in vivo and that hybrid E/M TNBC cells led invasion into collagen I ex vivo. We detected a much lower abundance of hybrid E/M cells in either luminal GEMM or patient luminal breast cancer samples and their invasion was typically led by Vim− epithelial phenotype cancer cells. We demonstrated that Vim is required for invasion of TNBC GEMM and PDX models ex vivo and for orthotopic metastasis in the C3(1)-Tag model. In contrast, Vim knockdown did not affect invasion in the luminal MMTV-PyMT GEMM, suggesting a specific requirement for Vim in TNBC metastasis.
At the molecular level, longitudinal single-cell RNA sequencing revealed that primary TNBC cancer cells progress towards diverse hybrid and mesenchymal states during both invasion and colony formation. Analysis of the epithelial and mesenchymal genes whose expression correlated with Vim in single cells revealed multiple EMT TFs. These TFs were then functionally evaluated using lentiviral shRNA in both invasion and colony formation assays. Knock-down of either Zeb2 or Foxc2 inhibited invasion without affecting colony formation whereas knock-down of Zeb1 or Ovol1 did not affect invasion but increased colony formation. At the transcriptional level, knockdown of each of these TFs decreased distinct sets of mesenchymal genes without effect on epithelial gene expression. This was surprising for Ovol1 as it has been previously associated with MET and with positive regulation of epithelial gene expression (48), suggesting a different function for Ovol1 in this TNBC model. Knockdown of Grhl2 increased invasion without effect on colony formation, decreased epithelial gene expression, and regulated distinct sets of mesenchymal gene either positively or negatively, as previously described (49, 50). The key concepts that emerge are that Grhl2 is responsible for maintaining epithelial character and that distinct TFs are selectively regulating different modules of mesenchymal gene expression to generate varied EMT molecular programs to accomplish different cellular processes of metastasis. We next used an innovative machine learning approach, CoGAPS, to identify three distinct EMT-related molecular patterns associated with these TFs during invasion and colony formation. It will be important for subsequent studies to extend this analysis to additional EMT TFs, such as Prrx1 and Twist1, which has been described to play a critical role in the generation of hybrid E/M cells and MET during metastasis formation (23, 25). Follow up studies will be required to determine which of these EMT patterns are implicated in the different steps of the metastatic cascade in vivo.
Both in ex vivo colony forming assays and within in vivo metastases, TNBC GEMM and TNBC patient cancer cells exhibited a diverse range of epithelial, hybrid E/M and mesenchymal markers. This diversity suggests several important areas for future study. First, the observation of hybrid E/M character in TNBC patient metastases suggests that a complete MET is not required for metastasis formation. While challenging to test with single gene perturbations, it will therefore be important to evaluate the requirement for MET in both luminal and basal-like cancer models. Second, the detection of epithelial, hybrid, and mesenchymal metastases in the same mouse suggests that there may be multiple metastatic molecular programs operating in parallel. Third, the coexpression of epithelial markers (ie, Ecad and EpCAM) in the same cancer cell as mesenchymal markers thought to be specific to late stages of EMT (ie, αSMA and vimentin) suggests that the diversity of EMT states within tumors is wider than previously appreciated from some cell line based studies.
Our results align with previous studies that detected EMT phenotypes in various spontaneous tumors, such as squamous cell carcinoma (51), prostate cancer (52), HER2-enriched breast cancer (53), luminal breast cancer (30, 32), or head and neck cancer (54). However, relatively fewer studies have evaluated the requirement for EMT-related genes in metastasis formation in spontaneous cancer models. In breast cancer GEMMs, the functional role of EMT has been evaluated in luminal models (30, 32, 34, 55). These studies demonstrated important roles of the mesenchymal markers tenascin C and Ncad and the EMT TF Snail1 for metastasis formation. Likewise, while multiple in vitro studies have used cancer cell lines to demonstrate that Vim regulates cell shape and migration (56-59), its role in metastasis formation in vivo was uncertain. For instance, lineage tracing strategies suggest that Vim is not expressed during luminal breast cancer metastasis (30, 32). It will be important for future studies to evaluate the functional modulation and behavior characteristics of metastasis with epithelial-predominant status vs hybrid E/M status, both within breast cancer subtypes and across primary human cancer types. ER−/PR+/HER2− primary tumors also presented high percentage of hybrid E/M cells and so it is a logical next subtype to evaluate.
Given their consistent dual expression of Ecad and Vim in their invasive cancer cells, we note that we previously demonstrated that E-cad suppresses invasion and promotes colony formation in the C3(1)-Tag GEMM and in one of the TNBC PDX used in the present study (TM00089) (40). Therefore, both epithelial and mesenchymal gene expression play essential roles in invasion and colony formation in TNBC GEMM and PDX models, a strict demonstration of the requirement for hybrid EMT in TNBC metastasis. Follow-up studies will be required to understand at the biochemical level how cell adhesion and the intermediate filament cytoskeleton interact to confer metastatic properties on cancer cells.
While our data suggests that MET is not always required for TNBC metastasis, we acknowledge that multiple studies have revealed an essential role MET in outgrowth in distant organs of other cancer types, including models of squamous cell carcinoma (23-27). Consistent with this concept, knockdown of vimentin in a tail vein experimental metastasis resulted in an increased number of metastases, suggesting that downregulation of Vim is advantageous to TNBC cells in distant organs. Surprisingly, both TNBC GEMM and TNBC patient metastases typically contained cancer cells positive for vimentin and other mesenchymal genes. This apparent contradiction might be explained by additional selective pressures in the distant organ that outweigh the advantage conferred by reducing vimentin levels. Alternately, it could indicate the existence of MET-independent metastatic strategies, as proposed by Somarelli et al in prostate cancer cell lines (60). Furthermore, it will be interesting to determine whether the intra- and inter-metastasis cell state heterogeneity we observe in TNBC GEMM and TNBC patient metastases result from cooperation between cancer cells in different cell states, as previously demonstrated in melanoma (61). Our data also suggest that TNBC cancer cells maintain significant phenotypic plasticity in distant organs, potentially explaining the treatment resistance that frequently develops in TNBC patients.
In conclusion, we demonstrated that TNBC cancer cells metastasize using a hybrid EMT and that TNBC primary tumors and metastases exhibit significant intra- and inter-tumoral heterogeneity in EMT programs. Further studies will be required to determine the mechanism regulating transitions between different EMT programs in both the primary and distant sites. The presence of different EMT cell states in the same tumor suggests that different cancer cells would have different responses to chemotherapy (62), potentially explaining the chemoresistance and frequent early relapse that is observed in TNBC patients (63-65). Our results further suggest that it will be important to evaluate anti-EMT therapeutic strategies in TNBC. Finally, the GEMM and PDX TNBC models used are commercially available and thus could provide a powerful resource for identifying the targetable effectors of EMT driven metastasis.
Supplementary Material
movie S1: Vimentin suppresses C3(1)-Tag cancer cell invasion ex vivo
Figure S1: E-cadherin and vimentin expression in patients primary breast cancers and in C3(1)-Tag GEMM.
Figure S2: TNBC GEMM hybrid E/M characteristics.
Figure S3: E-cadherin and vimentin expression in primary TNBC PDXs tumors and TNBC patient-derived organoids.
Figure S4: Vimentin small hairpin RNA validation.
Figure S5: Vimentin is required for C3(1)-Tag metastasis formation.
Figure S6: scRNA-seq batch correction and gene expression.
Figure S7: Impact of EMT TFs knock-down on EMT-related gene expression.
Figure S8: CoGAPS patterns distribution by condition and hallmark of cell cycle.
Figure S9: Epithelial and mesenchymal markers in TNBC GEMM metastases.
Figure S10: E-cadherin localization in patient primary tumors and metastases.
Acknowledgments:
We thank the TEMTIA community for critical feedback on our research. We thank all members of the Ewald Laboratory for critical discussions and specifically A. Fraser for his help with the single-cell RNA sequencing experiment, M. Haynes for technical assistance with mouse colony maintenance, and V. Padmanaban for providing the C3(1)-Tag Ecadfl/fl primary tumors. We thank H. Hao and L.D. Orzolek from the Microarray and Deep Sequencing Core Facility for technical assistance and J. Bader for statistical advice. We thank C.S. McGinnis and Z.J. Gartner for the MULTI-seq single-cell RNA sequencing protocol and their help with the analysis. We thank J. Zhu for his advice on flow sorting. We thank N. Aceto, M. Saini and I. Krol for their help with preliminary experiments related to this study. We thank G. Stein-O’Brien for her critical comments on the manuscript.
Funding:
A.J.E. received support for this project through grants from: the Breast Cancer Research Foundation (BCRF-20-048), Twisted Pink Foundation, Hope Scarves, the Jayne Koskinas Ted Giovanis Foundation, the Cindy Rosencrans Fund for Triple Negative Breast Cancer Research, and the National Institutes of Health / National Cancer Institute (U01CA217846, U54CA268083, 3P30CA006973).
Footnotes
Competing interests:
AJE has unlicensed patents related to the use of K14 as a biomarker in breast cancer and to the use of antibodies as cancer treatments. AJE's spouse is an employee of Immunocore. EJF is a consultant for Champions Oncology and is on the Scientific Advisory Board of Viosera Therapeutics. The other authors do not have competing interests.
Data and materials availability:
All data associated with this study are in the paper or supplementary materials. The scRNA-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE149299 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149299)
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer Statistics, 2021. CA Cancer J Clin 71, 7–33 (2021); published online EpubJan ( 10.3322/caac.21654). [DOI] [PubMed] [Google Scholar]
- 2.Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H, Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol 28, 1684–1691 (2010); published online EpubApr 1 (JCO.2009.24.9284 [pii] 10.1200/JCO.2009.24.9284). [DOI] [PubMed] [Google Scholar]
- 3.Dang TT, Prechtl AM, Pearson GW, Breast cancer subtype-specific interactions with the microenvironment dictate mechanisms of invasion. Cancer Res 71, 6857–6866 (2011); published online EpubNov 1 (0008-5472.CAN-11-1818 [pii] 10.1158/0008-5472.CAN-11-1818). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kast K, Link T, Friedrich K, Petzold A, Niedostatek A, Schoffer O, Werner C, Klug SJ, Werner A, Gatzweiler A, Richter B, Baretton G, Wimberger P, Impact of breast cancer subtypes and patterns of metastasis on outcome. Breast Cancer Res Treat, (2015); published online EpubMar 18 ( 10.1007/s10549-015-3341-3). [DOI] [PubMed] [Google Scholar]
- 5.Gupta GP, Massagué J, Cancer Metastasis: Building a Framework. Cell 127, 679–695 (2006); published online Epub2006/11/17/ ( 10.1016/j.cell.2006.11.001). [DOI] [PubMed] [Google Scholar]
- 6.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ, Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 155, 1639–1651 (2013); published online EpubDec 19 (S0092-8674(13)01480-3 [pii] 10.1016/j.cell.2013.11.029). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung KJ, Padmanaban V, Silvestri V, Schipper K, Cohen JD, Fairchild AN, Gorin MA, Verdone JE, Pienta KJ, Bader JS, Ewald AJ, Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci U S A 113, E854–863 (2016); published online EpubFeb 16 ( 10.1073/pnas.1508541113). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D, How basal are triple-negative breast cancers? Int J Cancer 123, 236–240 (2008); published online EpubJul 1 ( 10.1002/ijc.23518). [DOI] [PubMed] [Google Scholar]
- 9.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SAW, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, Mills GB, Lau CC, Brown PH, Comprehensive Genomic Analysis Identifies Novel Subtypes and Targets of Triple-Negative Breast Cancer. Clinical Cancer Research 21, 1688–1698 (2015) 10.1158/1078-0432.Ccr-14-0432). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y-Z, Ma D, Suo C, Shi J, Xue M, Hu X, Xiao Y, Yu K-D, Liu Y-R, Yu Y, Zheng Y, Li X, Zhang C, Hu P, Zhang J, Hua Q, Zhang J, Hou W, Ren L, Bao D, Li B, Yang J, Yao L, Zuo W-J, Zhao S, Gong Y, Ren Y-X, Zhao Y-X, Yang Y-S, Niu Z, Cao Z-G, Stover DG, Verschraegen C, Kaklamani V, Daemen A, Benson JR, Takabe K, Bai F, Li D-Q, Wang P, Shi L, Huang W, Shao Z-M, Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 35, 428–440.e425 (2019); published online Epub2019/03/18/ ( 10.1016/j.ccell.2019.02.001). [DOI] [PubMed] [Google Scholar]
- 11.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, Moses HL, Sanders ME, Pietenpol JA, Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One 11, e0157368 (2016) 10.1371/journal.pone.0157368). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta GK, Collier AL, Lee D, Hoefer RA, Zheleva V, Siewertsz van Reesema LL, Tang-Tan AM, Guye ML, Chang DZ, Winston JS, Samli B, Jansen RJ, Petricoin EF, Goetz MP, Bear HD, Tang AH, Perspectives on Triple-Negative Breast Cancer: Current Treatment Strategies, Unmet Needs, and Potential Targets for Future Therapies. Cancers (Basel) 12, 2392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Zhan Z, Yin X, Fu S, Deng X, Targeted Therapeutic Strategies for Triple-Negative Breast Cancer. Frontiers in Oncology 11, (2021); published online Epub2021-October-28 ( 10.3389/fonc.2021.731535). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foulkes WD, Smith IE, Reis-Filho JS, Triple-Negative Breast Cancer. New England Journal of Medicine 363, 1938–1948 (2010) 10.1056/NEJMra1001389). [DOI] [PubMed] [Google Scholar]
- 15.Garrido-Castro AC, Lin NU, Polyak K, Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discovery 9, 176–198 (2019) 10.1158/2159-8290.Cd-18-1177). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang MH, Kim HJ, Kim EJ, Chung YR, Park SY, Expression of epithelial-mesenchymal transition–related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Human Pathology 46, 1267–1274 (2015); published online Epub2015/09/01/ ( 10.1016/j.humpath.2015.05.010). [DOI] [PubMed] [Google Scholar]
- 17.Cheung SY, Boey YJY, Koh VCY, Thike AA, Lim JCT, Iqbal J, Tan PH, Role of epithelial–mesenchymal transition markers in triple-negative breast cancer. Breast Cancer Research and Treatment 152, 489–498 (2015); published online Epub2015/08/01 ( 10.1007/s10549-015-3485-1). [DOI] [PubMed] [Google Scholar]
- 18.Sarrió D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J, Epithelial-Mesenchymal Transition in Breast Cancer Relates to the Basal-like Phenotype. Cancer Research 68, 989 (2008) 10.1158/0008-5472.CAN-07-2017). [DOI] [PubMed] [Google Scholar]
- 19.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA, Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121, 2750–2767 (2011); published online Epub07/01/ ( 10.1172/JCI45014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge Ø, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale A-L, Brown PO, Botstein D, Molecular portraits of human breast tumours. Nature 406, 747–752 (2000); published online Epub2000/08/01 ( 10.1038/35021093). [DOI] [PubMed] [Google Scholar]
- 21.Greenburg G, Hay ED, Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J Cell Biol 95, 333–339 (1982); published online EpubOct ( 10.1083/jcb.95.1.333). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay ED, An Overview of Epithelio-Mesenchymal Transformation. Cells Tissues Organs 154, 8–20 (1995) 10.1159/000147748). [DOI] [Google Scholar]
- 23.Ocaña OH, Córcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA, Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724 (2012); published online EpubDec 11 ( 10.1016/j.ccr.2012.10.012). [DOI] [PubMed] [Google Scholar]
- 24.Esposito M, Mondal N, Greco TM, Wei Y, Spadazzi C, Lin S-C, Zheng H, Cheung C, Magnani JL, Lin S-H, Cristea IM, Sackstein R, Kang Y, Bone vascular niche E-selectin induces mesenchymal–epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nature Cell Biology 21, 627–639 (2019); published online Epub2019/05/01 ( 10.1038/s41556-019-0309-2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J, Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012); published online EpubDec 11 ( 10.1016/j.ccr.2012.09.022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E, Localized and reversible TGFβ signalling switches breast cancer cells from cohesive to single cell motility. Nature Cell Biology 11, 1287–1296 (2009); published online Epub2009/11/01 ( 10.1038/ncb1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Pozo Martin Y, Park D, Ramachandran A, Ombrato L, Calvo F, Chakravarty P, Spencer-Dene B, Derzsi S, Hill CS, Sahai E, Malanchi I, Mesenchymal Cancer Cell-Stroma Crosstalk Promotes Niche Activation, Epithelial Reversion, and Metastatic Colonization. Cell Rep 13, 2456–2469 (2015) 10.1016/j.celrep.2015.11.025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou S, Sun X, Yu L, Zhou R, Li A, Li M, Yang W, Differential expression and clinical significance of epithelial-mesenchymal transition markers among different histological types of triple-negative breast cancer. J Cancer 9, 604–613 (2018) 10.7150/jca.19190). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, Schäfer R, van Diest P, Voest E, van Oudenaarden A, Vrisekoop N, van Rheenen J, Plasticity between Epithelial and Mesenchymal States Unlinks EMT from Metastasis-Enhancing Stem Cell Capacity. Cell Rep 14, 2281–2288 (2016) 10.1016/j.celrep.2016.02.034). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, Schwabe RF, Vahdat LT, Altorki NK, Mittal V, Gao D, Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015); published online EpubNov 26 ( 10.1038/nature15748). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, Weinberg RA, Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525, 256–260 (2015); published online EpubSep 10 ( 10.1038/nature14897). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Lv Z, Zhang S, Wang Z, He L, Tang M, Pu W, Zhao H, Zhang Z, Shi Q, Cai D, Wu M, Hu G, Lui KO, Feng J, Nieto MA, Zhou B, Genetic Fate Mapping of Transient Cell Fate Reveals N-Cadherin Activity and Function in Tumor Metastasis. Developmental Cell 54, 593–607.e595 (2020) 10.1016/j.devcel.2020.06.021). [DOI] [PubMed] [Google Scholar]
- 33.Pfefferle AD, Herschkowitz JI, Usary J, Harrell JC, Spike BT, Adams JR, Torres-Arzayus MI, Brown M, Egan SE, Wahl GM, Rosen JM, Perou CM, Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol 14, R125 (2013); published online EpubNov 12 ( 10.1186/gb-2013-14-11-r125). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lüönd F, Sugiyama N, Bill R, Bornes L, Hager C, Tang F, Santacroce N, Beisel C, Ivanek R, Bürglin T, Tiede S, van Rheenen J, Christofori G, Distinct contributions of partial and full EMT to breast cancer malignancy. Developmental Cell 56, 3203–3221.e3211 (2021); published online Epub2021/12/06/ ( 10.1016/j.devcel.2021.11.006). [DOI] [PubMed] [Google Scholar]
- 35.Lu W, Kang Y, Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev Cell 49, 361–374 (2019); published online EpubMay 6 ( 10.1016/j.devcel.2019.04.010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, Backlund MG, Yin Y, Khramtsov AI, Bastein R, Quackenbush J, Glazer RI, Brown PH, Green JE, Kopelovich L, Furth PA, Palazzo JP, Olopade OI, Bernard PS, Churchill GA, Van Dyke T, Perou CM, Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8, R76 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green JE, Shibata MA, Yoshidome K, Liu ML, Jorcyk C, Anver MR, Wigginton J, Wiltrout R, Shibata E, Kaczmarczyk S, Wang W, Liu ZY, Calvo A, Couldrey C, The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene 19, 1020–1027 (2000); published online EpubFeb 21 ( 10.1038/sj.onc.1203280). [DOI] [PubMed] [Google Scholar]
- 38.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW, Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 163, 2113–2126 (2003); published online EpubNov ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Padmanaban V, Grasset EM, Neumann NM, Fraser AK, Henriet E, Matsui W, Tran PT, Cheung KJ, Georgess D, Ewald AJ, Organotypic culture assays for murine and human primary and metastatic-site tumors. Nature Protocols, (2020); published online Epub2020/07/20 ( 10.1038/s41596-020-0335-3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padmanaban V, Krol I, Suhail Y, Szczerba BM, Aceto N, Bader JS, Ewald AJ, E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573, 439–444 (2019); published online EpubSep ( 10.1038/s41586-019-1526-3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer KR, Altorki NK, Mittal V, Gao D, Fischer et al. reply. Nature 547, E5–E6 (2017) 10.1038/nature22817). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein-O’Brien GL, Clark BS, Sherman T, Zibetti C, Hu Q, Sealfon R, Liu S, Qian J, Colantuoni C, Blackshaw S, Goff LA, Fertig EJ, Decomposing Cell Identity for Transfer Learning across Cellular Measurements, Platforms, Tissues, and Species. Cell Systems 8, 395–411.e398 (2019); published online Epub2019/05/22/ ( 10.1016/j.cels.2019.04.004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein-O'Brien GL, Carey JL, Lee WS, Considine M, Favorov AV, Flam E, Guo T, Li S, Marchionni L, Sherman T, Sivy S, Gaykalova DA, McKay RD, Ochs MF, Colantuoni C, Fertig EJ, PatternMarkers & GWCoGAPS for novel data-driven biomarkers via whole transcriptome NMF. Bioinformatics (Oxford, England) 33, 1892–1894 (2017) 10.1093/bioinformatics/btx058). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov Jill P., Tamayo P, The Molecular Signatures Database Hallmark Gene Set Collection. Cell Systems 1, 417–425 (2015) 10.1016/j.cels.2015.12.004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cimino-Mathews A, Hicks JL, Illei PB, Halushka MK, Fetting JH, De Marzo AM, Park BH, Argani P, Androgen receptor expression is usually maintained in initial surgically resected breast cancer metastases but is often lost in end-stage metastases found at autopsy. Human pathology 43, 1003–1011 (2012) 10.1016/j.humpath.2011.08.007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cimino-Mathews A, Ye X, Meeker A, Argani P, Emens LA, Metastatic triple-negative breast cancers at first relapse have fewer tumor-infiltrating lymphocytes than their matched primary breast tumors: a pilot study. Human pathology 44, 2055–2063 (2013) 10.1016/j.humpath.2013.03.010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aiello NM, Maddipati R, Norgard RJ, Balli D, Li J, Yuan S, Yamazoe T, Black T, Sahmoud A, Furth EE, Bar-Sagi D, Stanger BZ, EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev Cell 45, 681–695.e684 (2018); published online EpubJun 18 ( 10.1016/j.devcel.2018.05.027). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roca H, Hernandez J, Weidner S, McEachin RC, Fuller D, Sud S, Schumann T, Wilkinson JE, Zaslavsky A, Li H, Maher CA, Daignault-Newton S, Healy PN, Pienta KJ, Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS One 8, e76773–e76773 (2013) 10.1371/journal.pone.0076773). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cieply B, Farris J, Denvir J, Ford HL, Frisch SM, Epithelial–Mesenchymal Transition and Tumor Suppression Are Controlled by a Reciprocal Feedback Loop between ZEB1 and Grainyhead-like-2. Cancer Research 73, 6299 (2013) 10.1158/0008-5472.CAN-12-4082). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cieply B, Riley P, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM, Suppression of the Epithelial–Mesenchymal Transition by Grainyhead-like-2. Cancer Research 72, 2440 (2012) 10.1158/0008-5472.CAN-11-4038). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D, Moers V, Lemaire S, De Clercq S, Minguijon E, Balsat C, Sokolow Y, Dubois C, De Cock F, Scozzaro S, Sopena F, Lanas A, D'Haene N, Salmon I, Marine JC, Voet T, Sotiropoulou PA, Blanpain C, Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018); published online EpubApr ( 10.1038/s41586-018-0040-3). [DOI] [PubMed] [Google Scholar]
- 52.Ruscetti M, Quach B, Dadashian EL, Mulholland DJ, Wu H, Tracking and Functional Characterization of Epithelial–Mesenchymal Transition and Mesenchymal Tumor Cells during Prostate Cancer Metastasis. Cancer Research 75, 2749 (2015) 10.1158/0008-5472.CAN-14-3476). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, Farias EF, Condeelis J, Klein CA, Aguirre-Ghiso JA, Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 540, 588–592 (2016); published online Epub2016/12/01 ( 10.1038/nature20609). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, Deschler DG, Varvares MA, Mylvaganam R, Rozenblatt-Rosen O, Rocco JW, Faquin WC, Lin DT, Regev A, Bernstein BE, Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 171, 1611–1624.e1624 (2017) 10.1016/j.cell.2017.10.044). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran HD, Luitel K, Kim M, Zhang K, Longmore GD, Tran DD, Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res 74, 6330–6340 (2014); published online EpubNov 1 ( 10.1158/0008-5472.Can-14-0923). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilles C, Polette M, Zahm JM, Tournier JM, Volders L, Foidart JM, Birembaut P, Vimentin contributes to human mammary epithelial cell migration. J Cell Sci 112 (Pt 24), 4615–4625 (1999); published online EpubDec ( [DOI] [PubMed] [Google Scholar]
- 57.Mendez MG, Kojima S-I, Goldman RD, Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J 24, 1838–1851 (2010) 10.1096/fj.09-151639). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C-Y, Lin H-H, Tang M-J, Wang Y-K, Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget 6, 15966–15983 (2015) 10.18632/oncotarget.3862). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vuoriluoto K, Haugen H, Kiviluoto S, Mpindi JP, Nevo J, Gjerdrum C, Tiron C, Lorens JB, Ivaska J, Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 30, 1436–1448 (2011); published online Epub2011/03/01 ( 10.1038/onc.2010.509). [DOI] [PubMed] [Google Scholar]
- 60.Somarelli JA, Schaeffer D, Marengo MS, Bepler T, Rouse D, Ware KE, Hish AJ, Zhao Y, Buckley AF, Epstein JI, Armstrong AJ, Virshup DM, Garcia-Blanco MA, Distinct routes to metastasis: plasticity-dependent and plasticity-independent pathways. Oncogene 35, 4302–4311 (2016); published online Epub2016/08/01 ( 10.1038/onc.2015.497). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campbell NR, Rao A, Hunter MV, Sznurkowska MK, Briker L, Zhang M, Baron M, Heilmann S, Deforet M, Kenny C, Ferretti LP, Huang T-H, Perlee S, Garg M, Nsengimana J, Saini M, Montal E, Tagore M, Newton-Bishop J, Middleton MR, Corrie P, Adams DJ, Rabbie R, Aceto N, Levesque MP, Cornell RA, Yanai I, Xavier JB, White RM, Cooperation between melanoma cell states promotes metastasis through heterotypic cluster formation. Developmental Cell 56, 2808–2825.e2810 (2021); published online Epub2021/10/25/ ( 10.1016/j.devcel.2021.08.018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Las Rivas J, Brozovic A, Izraely S, Casas-Pais A, Witz IP, Figueroa A, Cancer drug resistance induced by EMT: novel therapeutic strategies. Archives of Toxicology 95, 2279–2297 (2021); published online Epub2021/07/01 ( 10.1007/s00204-021-03063-7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kast K, Link T, Friedrich K, Petzold A, Niedostatek A, Schoffer O, Werner C, Klug SJ, Werner A, Gatzweiler A, Richter B, Baretton G, Wimberger P, Impact of breast cancer subtypes and patterns of metastasis on outcome. Breast Cancer Res Treat 150, 621–629 (2015); published online EpubApr ( 10.1007/s10549-015-3341-3). [DOI] [PubMed] [Google Scholar]
- 64.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K, Metastatic behavior of breast cancer subtypes. J Clin Oncol 28, 3271–3277 (2010); published online EpubJul 10 (JCO.2009.25.9820 [pii] 10.1200/JCO.2009.25.9820). [DOI] [PubMed] [Google Scholar]
- 65.Reis-Filho JS, Tutt AN, Triple negative tumours: a critical review. Histopathology 52, 108–118 (2008); published online EpubJan (HIS2889 [pii] 10.1111/j.1365-2559.2007.02889.x). [DOI] [PubMed] [Google Scholar]
- 66.Guy CT, Cardiff RD, Muller WJ, Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12, 954–961 (1992); published online EpubMar ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maroulakou IG, Anver M, Garrett L, Green JE, Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci U S A 91, 11236–11240 (1994); published online EpubNov 8 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L, A global double-fluorescent Cre reporter mouse. genesis 45, 593–605 (2007); published online Epub2007/09/01 ( 10.1002/dvg.20335). [DOI] [PubMed] [Google Scholar]
- 69.Grasset EM, Bertero T, Bozec A, Friard J, Bourget I, Pisano S, Lecacheur M, Maiel M, Bailleux C, Emelyanov A, Ilie M, Hofman P, Meneguzzi G, Duranton C, Bulavin DV, Gaggioli C, Matrix Stiffening and EGFR Cooperate to Promote the Collective Invasion of Cancer Cells. Cancer Research 78, 5229 (2018) 10.1158/0008-5472.CAN-18-0601). [DOI] [PubMed] [Google Scholar]
- 70.McGinnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, Hu JL, Murrow LM, Weissman JS, Werb Z, Chow ED, Gartner ZJ, MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nature Methods 16, 619–626 (2019); published online Epub2019/07/01 ( 10.1038/s41592-019-0433-8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Melsted P, Booeshaghi AS, Liu L, Gao F, Lu L, Min KH, da Veiga Beltrame E, Hjörleifsson KE, Gehring J, Pachter L, Modular, efficient and constant-memory single-cell RNA-seq preprocessing. Nature Biotechnology 39, 813–818 (2021); published online Epub2021/07/01 ( 10.1038/s41587-021-00870-2). [DOI] [PubMed] [Google Scholar]
- 72.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome Biology 19, 15 (2018); published online Epub2018/02/06 ( 10.1186/s13059-017-1382-0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J, The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019); published online EpubFeb ( 10.1038/s41586-019-0969-x). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dorrity MW, Saunders LM, Queitsch C, Fields S, Trapnell C, Dimensionality reduction by UMAP to visualize physical and genetic interactions. Nat Commun 11, 1537 (2020); published online EpubMar 24 ( 10.1038/s41467-020-15351-4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haghverdi L, Lun ATL, Morgan MD, Marioni JC, Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nature Biotechnology 36, 421–427 (2018); published online Epub2018/05/01 ( 10.1038/nbt.4091). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson WE, Li C, Rabinovic A, Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007); published online EpubJan ( 10.1093/biostatistics/kxj037). [DOI] [PubMed] [Google Scholar]
- 77.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD, The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012); published online EpubMar 15 ( 10.1093/bioinformatics/bts034). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sherman TD, Gao T, Fertig EJ, CoGAPS 3: Bayesian non-negative matrix factorization for single-cell analysis with asynchronous updates and sparse data structures. BMC Bioinformatics 21, 453 (2020); published online Epub2020/10/14 ( 10.1186/s12859-020-03796-9). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
movie S1: Vimentin suppresses C3(1)-Tag cancer cell invasion ex vivo
Figure S1: E-cadherin and vimentin expression in patients primary breast cancers and in C3(1)-Tag GEMM.
Figure S2: TNBC GEMM hybrid E/M characteristics.
Figure S3: E-cadherin and vimentin expression in primary TNBC PDXs tumors and TNBC patient-derived organoids.
Figure S4: Vimentin small hairpin RNA validation.
Figure S5: Vimentin is required for C3(1)-Tag metastasis formation.
Figure S6: scRNA-seq batch correction and gene expression.
Figure S7: Impact of EMT TFs knock-down on EMT-related gene expression.
Figure S8: CoGAPS patterns distribution by condition and hallmark of cell cycle.
Figure S9: Epithelial and mesenchymal markers in TNBC GEMM metastases.
Figure S10: E-cadherin localization in patient primary tumors and metastases.