Abstract
Capsular polysaccharide (CPS) is a major virulence factor in Streptococcus pneumoniae. CPS gene clusters of S. pneumoniae types 4, 6B, 8, and 18C were sequenced and compared with those of CPS types 1, 2, 14, 19F, 19A, 23F, and 33F. All have the same four genes at the 5′ end, encoding proteins thought to be involved in regulation and export. Sequences of these genes can be divided into two classes, and evidence of recombination between them was observed. Next is the gene encoding the transferase for the first step in the synthesis of CPS. The predicted amino acid sequences of these first sugar transferases have multiple transmembrane segments, a feature lacking in other transferases. Sugar pathway genes are located at the 3′ end of the gene cluster. Comparison of the four dTDP-l-rhamnose pathway genes (rml genes) of CPS types 1, 2, 6B, 18C, 19F, 19A, and 23F shows that they have the same gene order and are highly conserved. There is a gradient in the nature of the variation of rml genes, the average pairwise difference for those close to the central region being higher than that for those close to the end of the gene cluster and, again, recombination sites can be observed in these genes. This is similar to the situation we observed for rml genes of O-antigen gene clusters of Salmonella enterica. Our data indicate that the conserved first four genes at the 5′ ends and the relatively conserved rml genes at the 3′ ends of the CPS gene clusters were sites for recombination events involved in forming new forms of CPS. We have also identified wzx and wzy genes for all sequenced CPS gene clusters by use of motifs.
Streptococcus pneumoniae is an important human pathogen causing invasive diseases such as pneumonia, bacteremia, and meningitis. Control of pneumococcal diseases is being complicated by the increasing prevalence of antibiotic-resistant strains and the suboptimal clinical efficacy of existing vaccines. The capsular polysaccharide (CPS) of S. pneumoniae is essential for virulence because it protects S. pneumoniae from the nonspecific defence system of the host organism. All fresh isolates from patients with pneumococcal infection are encapsulated, and spontaneous nonencapsulated derivatives of such strains are almost completely avirulent (7).
Ninety different CPS types have been identified so far by immunological and chemical techniques (31). Each has a structurally distinct CPS, composed of repeating oligosaccharide units joined by glycosidic linkages. CPS production requires a complex pathway, including synthesis of the activated monosaccharide nucleotide precursors, sequential transfer of each sugar to a lipid carrier to form repeating CPS units, and subsequent polymerization, export, and attachment to the cell surface. The exception to this is type 3 CPS, which has a very simple CPS structure and appears to be synthesized by a processive transferase (6). Classic genetic studies carried out by Austrian et al. (8) demonstrated that the genes encoding CPS biosynthesis are closely linked on the S. pneumoniae chromosome. The first complete CPS gene cluster sequence was published in 1995 (4, 21) and, after a recent series of publications, sequences are available for CPS types 1, 2, 3, 14, 19F, 19A, 19B, 23F, 33F, and 37. In all cases the gene cluster is between the dexB and aliA genes (28), although types 3 and 37 are special cases (see Results and Discussion). As found in some other polysaccharide antigen gene clusters (11, 27, 33, 40, 41, 45, 66, 68, 79), the S. pneumoniae capsule locus has a cassette-like organization with type-specific CPS genes being flanked by genes that are common to all or many CPS types. The cassette-like arrangement of CPS genes may allow S. pneumoniae to change CPS types since the regions of homology can mediate the recombinational exchange (9, 13–16, 55). Further characterization of other S. pneumoniae CPS gene clusters can provide the detailed knowledge needed to expand our understanding of the origins of variation in capsular polysaccharides. Our previous work on O antigen, a polymorphic polysaccharide on the surface of most gram-negative bacteria, has suggested that interspecies gene transfer could be a general mechanism for generating polymorphism. Studies on the S. pneumoniae CPS, which is as polymorphic as O antigen, will facilitate our understanding of the origin of the extensive polymorphism of the bacterial cell polysaccharides.
Studies on the genetic determinants of CPSs would also contribute to unravelling the CPS biosynthetic pathways, which could, for example, open up the prospect for the design of inhibitors capable of obstructing the expression of this virulence factor.
We report here the sequences of CPS gene clusters for types 4, 6B, 8, and 18C. These CPSs are all included in the 23-valent vaccine (65), and the pneumococci of CPS types 6, 4, 8, and 18 are ranked second, eighth, ninth, and tenth, respectively, among sterile-site isolates (69). We also present data on the diversity of CPS genes based on comparisons of our sequence data and also published data on other CPS types.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains WCH35 (type 4), WCH18 (type 6B), WCH56 (type 8) and WCH94 (type 18C) were obtained from James C. Paton, Molecular Microbiology Unit, Women's and Children's Hospital, North Adelaide, Australia. All are clinical isolates. S. pneumoniae strains were routinely grown in Todd-Hewitt broth with 0.5% yeast extract or on blood agar. Plasmids were maintained in Escherichia coli K-12 strain DH10B (10).
Construction of random DNase I bank.
Chromosomal DNA used as template for PCR was prepared using the Wizard DNA preparation kits from Promega. Oligonucleotides 1430 (5′-TGTCCAATGAAGAGCAAGACTTGACAGTAG) and 1402 (5′-CAATAATGTCACGCCCGCAAGGGCAAGT) were based on dexB and aliA of type 19F S. pneumoniae and were used to PCR amplify CPS gene clusters by long-PCR using the Expand Long Template PCR System from Boehringer. PCR cycles were as follows: denaturation at 94°C for 10 s, annealing at 55 to 58°C for 30 s, and extension at 68°C for 15 min. The long-PCR products were subjected to DNase I digestion and cloned into pGEM-T to make a bank by the method described by Wang and Reeves (78).
Sequencing and analysis.
DNA template for sequencing was prepared using the 96-well-format plasmid DNA miniprep kit from Advanced Genetic Technologies Corp. and the procedure developed by The Institute for Genome Research (TIGR) (73). Sequencing was carried out by Sydney University and The Prince Alfred Macromolecular Analysis Centre using an Applied Biosystem model 377A automated DNA sequencing system and The ABI Dye Terminator Cycle Sequencing Kit. Sequence data were assembled and analyzed using the Australian National Genomic Information Service (ANGIS), which incorporates several sets of programs (63). Sequence data were assembled using programs PHRAP, PHRED, and CONSED (29). Sequence databases were searched using BLAST (1). The programs BESTFIT (20) and MULTICOMP (61) were used for pairwise sequence comparison. The programs BLOCKMAKER (30) and PSI-BLAST (2) were used to detect motifs among sets of related proteins. We used the algorithm described by Eisenberg et al. (23) to identify potential transmembrane segments from the amino acid sequence of hydrophobic proteins. Phylogenetic trees were constructed by the neighbor-joining method (67) using PHYLIP (version 3.4, written by J. Felsenstein, Department of Genetics, University of Washington, Seattle). Intragenic recombination was detected by the Stephens' test (71).
Nucleotide sequence accession numbers.
The DNA sequences of S. pneumoniae CPS type 4, 6B, 8, and 18C CPS gene clusters have been deposited in GenBank under accession numbers AF316639, AF316640, AF316641, and AF316642, respectively.
RESULTS AND DISCUSSION
DNA sequence of gene clusters for S. pneumoniae CPS types 4, 6B, 8, and 18C.
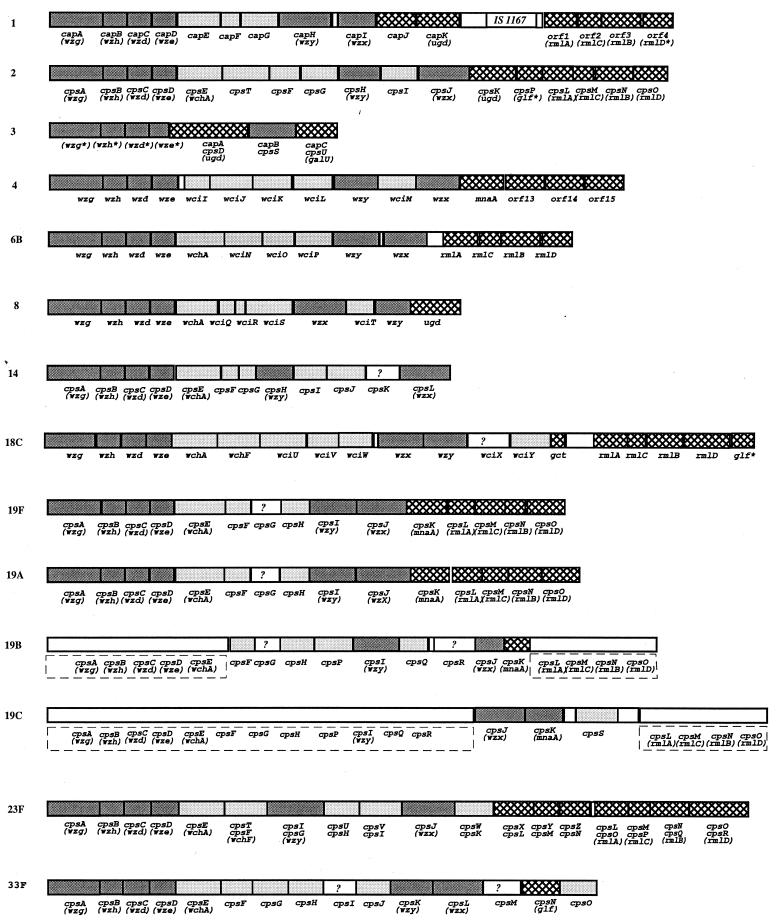
S. pneumoniae CPS gene clusters responsible for capsule biosynthesis are generally located between dexB and aliA, two genes that do not participate in capsule formation. The CPS gene clusters from S. pneumoniae strains of CPS types 4, 6B, 8, and 18C were PCR amplified and sequenced. For S. pneumoniae CPS types 4, 6B, 8, and 18C, sequences of 20,558 bp (15 genes), 17,160 bp (14 genes), 13,746 bp (12 genes), and 21,648 bp (18 genes), respectively, were obtained (Fig. 1). For each gene cluster all open reading frames (ORFs) have the same transcriptional direction from dexB to aliA.
FIG. 1.
Comparison of the CPS gene clusters of S. pneumoniae types 1, 2, 3, 4, 6B, 8, 14, 18C, 19F, 19A, 19B, 19C, 23F, and 33F. The structure for type 4, 6B, 8, and 18C gene clusters are based on our sequence data. We have named CPS genes sequenced according to the BPGN system. For previously sequenced gene, the original names are used, and in cases where they have the same functions as one of our newly sequenced genes, the new names are given in brackets. The capsule type number usually included within the gene names has been omitted as redundant here. Patterns:  ,
,  ,
,  , □?, □, processing genes, transferase genes, nucleotide sugar biosynthesis genes, unallotted genes, and noncoding regions, respectively. Gene names without gene boundaries in types 19B and 19C indicate regions characterized only by Southern hybridization.
, □?, □, processing genes, transferase genes, nucleotide sugar biosynthesis genes, unallotted genes, and noncoding regions, respectively. Gene names without gene boundaries in types 19B and 19C indicate regions characterized only by Southern hybridization.
During the course of this study, the type 8 CPS gene cluster has also been sequenced by another group (52) and that of type 4 can be found in the currently unannotated TIGR genome database (http://www.tigr.org.) as part of type 4 S. pneumoniae genome. The two sequences are very similar to ours. For the type 4 CPS gene cluster, the major difference is that the first 348 bases, part of dexB in isolate WCH35 match no sequence in the corresponding region of contig spn_9 but match a sequence in contig spn_131 (5,394 to 5,047 bp), implying a sequence inserted between two parts in the TIGR isolate. The remaining 20,210 bp are almost identical in the two sequences with only 4 bp differences. There is at least threefold high-quality sequence coverage for each query region in our sequence, and the differences probably arise from sequence variation among isolates of type 4 S. pneumoniae. For type 8, 18 base differences were found in the 13,746-bp DNA, with 6 in the 5′ intergenic region and 12 within CPS genes. Again, there is at least threefold high-quality sequence coverage for each query region in our sequence, and the observed differences again probably represent variation among different isolates of type 8 S. pneumoniae.
Gene nomenclature.
Genes were named (Fig. 1) according to the bacterial polysaccharide gene nomenclature (BPGN) system (62) (www.microbio.usyd.edu.au/BPGD/default.htm), a system applicable to all species that distinguishes different classes of genes and provides a single name for all genes of a given function. The names also comply with the Demerec scheme (19) generally applied in bacteria, including S. pneumoniae. The BPGN system is widely used but not previously in S. pneumoniae, and to facilitate comparisons we have included BPGN names in parentheses in Fig. 1, where the genes are also present in one of the four new gene clusters, and in the text for some general comments.
General organization.
The general organization of the four new clusters is similar to that of other S. pneumoniae CPS clusters except for those of types 3 and 37 (Fig. 1). They are located on the chromosome between dexB and aliA, with a cassette-like structure: type-specific genes are flanked by highly homologous genes common to all or many gene clusters. The 5′ portion contains four conserved genes thought to be involved in the regulation and export of CPS. This is followed by a central type-specific region, which encodes glycosyltransferases, a CPS polymerase and a CPS repeat unit flippase. The 3′ region encodes enzymes for the biosynthesis of activated monosaccharide precursors, some of which are common to several CPS types, e.g., those encoding dTDP–l-rhamnose and UDP-glucuronic acid synthesis. Although genes for pathway enzymes generally follow the structure and assembly genes, there are cases where this does not apply absolutely, e.g., a biosynthesis pathway gene (cps19cK) preceding a transferase gene (cps19cS) in the type 19C CPS gene cluster (Fig. 1). The type 37 CPS gene cluster is not shown in Fig. 1 since its homopolymer CPS biosynthesis is driven by a single gene (tts) located far apart from the usual location (46): its redundant cryptic “classical” CPS gene cluster is virtually identical to that of type 33F with some genes inactivated by mutations.
wzg, wzh, wzd, and wze genes are conserved in all CPS types.
The first four genes of the four CPS gene clusters are very similar to those of other S. pneumoniae CPS gene clusters. The functions of their deduced protein products are not yet established (28). The homologues of Orf3 and Orf4 have been reviewed by Paulsen et al. (57), the closest being in Streptococcus suis, Streptococcus salivarius, Streptococcus thermophilus, Streptococcus agalactiae, and Staphylococcus aureus capsule gene clusters, where the genes are adjacent and in the same order as in S. pneumoniae. The products of each pair of genes are homologues to the N- and C-terminal parts of proteins encoded by genes found in polysaccharide gene clusters of gram-negative bacteria. These larger proteins and the pairs of smaller proteins, which are seen as equivalent, are in the MPA1 class of the Paulsen et al. (57) classification and are thought to be involved in polysaccharide export. The genes for MPA1 proteins of gram-negative bacteria have been named wzc (22), and for the orf3 and orf4 we propose wzd and wze to comply with the Demerec system and the principle of consistent nomenclature for bacterial polysaccharide genes. Wzd proteins are very similar to the N-terminal part of Wzc proteins, possessing two transmembrane segments and a cytoplasmic loop, whereas Wze proteins are similar to the C-terminal part of Wzc proteins, bearing an ATP-binding motif.
All S. pneumoniae CPS gene clusters have similar genes for orf1 and likewise for orf2. The same pair of genes is also present at the 5′ end of several other Streptococcus spp. capsule gene clusters (AF118389, Y17218, Y27241, Y17221, AB028896, Z98171, AF053346, and X94980), and it seems appropriate to give them a common name suitable for more general use; we propose wzg and wzh. S. pneumoniae Wzg proteins show approximately 26% identity to Bacillus subtilis LytR, which is involved in the regulation of the autolysin (lytABC) operon (42). Hence, Wzg proteins have been predicted to play a role in the regulation of CPS synthesis. The function of Wzh is unknown.
Nucleotide sugar biosynthesis genes.
Nucleotide sugar biosynthesis genes are generally located in the 3′ portion of CPS gene clusters. However, genes encoding enzymes necessary for the synthesis of sugar with housekeeping functions are generally not found in CPS gene clusters but the functions are provided from other pathways. Of the sugars present in the capsules studied, Glc is generally transferred from UDP-Glc, synthesized by a housekeeping gene of which we found a copy in the partial nucleotide sequence of the genome of type 4 S. pneumoniae, being an orf (from 8,550 to 9,449 bp of contig sp_113) that encodes a protein 44% identical to GalU of E. coli, the glucosyl-1-phosphate uridylyltransferase for the synthesis of UDP-Glc. We also found an orf (from 3,900 to 5,000 bp of contig sp_66) that encodes a protein 53% identical to GalE of E. coli, the UDP–glucose-4-epimerase for the conversion of UDP-Glc to UDP-Gal, and an orf (from 902,051 to 903,055 bp of contig sp_3475) encoding a protein 56% identical to Gne (previously also named GalE) of Yersinia enterocolitica O8, now shown to be a 4-epimerase for conversion of UDP-GlcNAc to UDP-GalNAc (M. Skurnik, personal communication). It is therefore as expected that none of these genes were found in the gene clusters for CPSs contain these residues.
We have found as discussed below the genes needed for the biosynthesis of activated precursors of all other residues in the representative gene clusters for types 4, 6B, 8, and 18C CPSs.
dTDP–l-Rha biosynthesis genes.
We expect genes for dTDP–l-Rha in types 6B and 18C CPS gene clusters. Four genes from type 6B (orf11–14) and type 18C (orf15–18) are identified as rmlA (glucose-1-phosphate thymidylyltransferase), rmlC (dTDP–4-keto-6-deoxyglucose-3,5 epimerase), rmlB (dTDP–glucose-4,6-dehydratase), and rmlD (dTDP-l- rhamnose synthase), respectively, by the high level of identity to other rml genes, and particularly those of S. pneumoniae 19F (identity from 90 to 99%) (51).
UDP–d-Man2NAc biosynthesis gene.
We expect a gene for the biosynthesis of UDP–d-Man2NAc in type 4 CPS synthesis. orf12 shows 65.9% identity to cps19fK of type 19F S. pneumoniae CPS gene cluster, which can complement an E. coli mnaA (wecB) mutation (51). MnaA is a UDP–N-acetylglucosamine 2-epimerase for the conversion of UDP-GlcNAc to UDP-Man2NAc. Also, Orf12 has 57.2% identity to Cap5P and Cap8P of S. aureus, which complement the same E. coli mnaA mutation (68). All of these polysaccharides contain d-Man2NAc; we therefore conclude that orf12 encodes a UDP–N-acetylglucosamine 2-epimerase and have named it mnaA.
UDP–l-Fuc2NAc biosynthesis genes.
We expect genes for UDP–l-Fuc2NAc in type 4 CPS synthesis. orf13, orf14, and orf15 of type 4 encode proteins that, along their entire lengths, are highly similar to products encoded by three clustered genes from five other polysaccharide biosynthesis loci. Orf13, Orf14, and Orf15 show, respectively, 69.0, 77.0, and 73.3% identity to CapE, CapF, and CapG of S. aureus; 62.0, 42.0, and 51.5% identity to WbjB, WbjC, and WbjD of Pseudomonas aeruginosa; 64.0, 58.3, and 78.8% identity to WcgJ, WcgK, and WcgL of B. fragilis; and 67.8, 40.5, and 49.5% identity to the corresponding gene products from E. coli. These proteins are respectively encoded by genes in the loci for the S. aureus types 5 and 8 capsules (68), P. aeruginosa O11 O antigen (18), Bacteroides fragilis polysaccharide B (17), and E. coli O26 O-antigen locus (M. D'Souza, L. Wang, and P. R. Reeves, unpublished data), all three genes being consecutive and in the same order in the five species. Five of the six polysaccharides have known structures which include l-Fuc2NAc (26, 34, 37, 48, 49) and that of B. fragilis polysaccharide B includes l-Fuc2NAc (L. Comstock, personal communication). We propose that the three genes encode enzymes for the biosynthesis of the l-Fuc2NAc precursor, thought to be UDP–l-Fuc2NAc (70).
On this basis we propose a putative four-step (three-enzyme) biosynthetic pathway for UDP–l-Fuc2NAc from UDP-GlcNAc. The proposed pathway begins with 2-epimerization of UDP-GlcNAc to UDP–d-Man2NAc by the protein products of orf13. Orf13 and Orf14, together with their homologues encoded by the other five gene clusters, have consensus NAD-binding domains (GxxGxxG) near their N termini, which is thought to be important for the activity of epimerases and dehydratases. Orf14 shares 42% similarity over the whole length with Gmd of E. coli K-12 colanic acid biosynthesis gene cluster (72). Gmd is a dehydratase catalyzing the formation of GDP–4-keto-6-deoxy-d-mannose from GDP–d-Man. Thus, we propose that Orf13 carries out the epimerization step (first step) and Orf14 carries out the dehydration step (second step) to form UDP–4-keto-6-deoxy-d-Man2NAc. The third step is to form UDP–l-Fuc2NAc from UDP–4-keto-6-deoxy-d-Man2NAc by two reactions: epimerization and oxidoreduction both by Orf15. Orf15 shares 45% similarity over the whole length with Fcl (3), an enzyme with both epimerization and oxidoreduction activities for the final two steps in formation of GDP–l-Fuc from GDP–4-keto-6-deoxy-d-mannose.
This proposal is different from the one put forward by Lee and Lee (43). In support of our proposal, we note that in both proposals the pathway starts with UDP–d-Man2NAc, synthesized by the well-documented mnaA gene. However, mnaA is not present in P. aeruginosa O11 or E. coli O26, which lack UDP-Man2NAc in the polysaccharide. Since orf13, orf14, and orf15 are the only genes common to all known gene clusters for repeats with l-Fuc2NAc, we prefer a three-enzyme pathway from UDP-GlcNAc. However, a drawback in our proposed pathway is that we assign the same function, 2-epimerization of UDP-GlcNAc to UDP–d-Man2NAc, to orf13 and mnaA in CPS4 of S. pneumoniae and the two homologous genes in CAP5 and CAP8 of S. aureus. Therefore, until there is direct confirmation of a pathway, we are not allocating final gene names but simply using ORF names for the three genes thought by both groups to be involved in synthesis of UDP–l-Fuc2NAc.
UDP-GlcA biosynthesis genes.
We expect a gene for UDP-glucuronic acid synthesis in type 8 S. pneumoniae. Orf12 shares high similarity with many published UDP–glucose-6-dehydrogenases (Ugd), which convert UDP-glucose to UDP-glucuronic acid. We have named this gene ugd. The homologues in the CPS1, CPS2, and CPS3 of S. pneumoniae gene clusters have a high level of similarity (76.3, 90.6, and 61.6% identity, respectively) to the CPS8 gene, and those of CPS1 and CPS3 have been shown by experiment to encode UDP–glucose-6-dehydrogenases (5, 53). Type 1, 2, 3, and 8 CPSs all have glucuronic acid in their structures, and all have ugd in the gene clusters.
CDP-glycerol biosynthesis gene.
We expect a gene for the synthesis of CDP–glycerol-1-phosphate in S. pneumoniae type 18C. orf14 shows 61.9% similarity at the amino acid level with TagD, which was described as a glycerol-3-phosphate cytidyl transferase in the biosynthesis of the major cell wall teichoic acid of B. subtilis (56). However, it should be noted that glycerol-1-phosphate and glycerol-3-phosphate are different names for the one structure, and we are using the designation glycerol-1-phosphate and the corresponding designation for the linkages in the polysaccharides. Glycerol-1-phosphate is the side-branch residue linked to galactose of 18C CPS. We propose that orf14 is responsible for the synthesis of CDP–glycerol-1-phosphate; we have named it gct, and we suggest that the same name could be used in place of tagD.
A nonfunctional UDP-galactopyranose mutase gene (glf).
Two segments from positions 20539 to 20877 and from positions 20850 to 21353 at the end of the type 18C CPS gene cluster if translated show 63.6 and 51.8% identities with the N and C termini, respectively, of Glf, a UDP-galactopyranose mutase from E. coli, an enzyme for the conversion of UDP-galactopyranose (UDP-Galp) to UDP-galactofuranose (UDP-Galf) (54). It is likely that they are nonfunctional relics of a glf gene, which may once have been part of the CPS gene cluster of the ancestor of the type 18C strain(s).
Genes encoding the first transferases.
The orf5 genes of CPS types 6B, 8, and 18C encode proteins with high sequence and hydrophobicity profile similarities. They are also very similar (71 to 95% identity) to the corresponding gene of CPS type 14, shown to encode a transferase transferring glucose-1-phosphate to undecaprenol phosphate. We suggest that these genes, which we have named wchA, have the same function as in CPS14. All are similar to WbaP, a galactosyl-1-phosphate transferase of the Salmonella enterica O-antigen gene cluster catalyzing the first step of O-antigen synthesis (33, 77). They and WbaP differ from most other transferases by having a unique hydrophobic profile, with four and one predicted transmembrane segments, respectively, in their N- and C-terminal domains.
In CPS type 4, orf5 encodes a 211-amino-acid protein, which show 35.8 and 38.4% sequence identities, respectively, to the C-terminal domains of WchA and WbaP, with a very similar hydrophobic profile containing one potential transmembrane segment. It has been shown that WbaP is a bifunctional protein, with the C-terminal half having the transferase function (77). We propose that orf5 of CPS type 4, which we have named wciI, also encodes the first glycosyltransferase. The substrate is not clear, but we believe it is not UDP-glucose since the type 4 CPS contains no glucose. Immediately upstream of orf5 is a “orf”, which if translated gives a 44-residue polypeptide with 20% identity to the N-terminal half of WbaP. Thus, it is likely that in CPS type 4 the wciI gene was derived by deletion from a larger gene which included a functional N-terminal domain.
Genes encoding other transferases.
In CPS types 4, 6B, 8, and 18C we detected four, three, three, and five additional putative transferase genes, respectively, in addition to the first transferase gene (Table 1). In CPS type 8, a pair of genes, wciQ and wciR, probably encode two proteins for one glycosyltransferase activity, with one being an enhancer, as proposed for the S. pneumoniae CPS14 and Lactococcus lactis exopolysaccharide loci (39, 75, 76). The total number of transferases genes is thus consistent with our expectation based on the number of linkages (Fig. 2), and the potential assignments are discussed below.
TABLE 1.
Properties of putative transferase genes in S. pneumoniae type 4, 6B, 8, and 18C CPS gene clusters
| CPS type | Gene | Gene size (bp) | No. of amino acids | Predicted protein mass (kDa) | % G+C | Highest-scoring homologue(s) | Organism(s) | % Amino acid identity/similarity | Proposed function |
|---|---|---|---|---|---|---|---|---|---|
| 4 | wciI | 636 | 211 | 24.0 | 33.3 | Cps14E | S. pneumoniae | 35.8/62.1 | Glycosyl-1-phosphate-transferase |
| WbaP | S. enterica | 38.4/62.1 | |||||||
| 4 | wciJ | 1,230 | 409 | 46.9 | 38 | Cap5L | S. aureus | 25.1/54.1 | α-1,3-l-Fuc2NAc transferase |
| Cap8L | S. aureus | 24.3/52.2 | |||||||
| 4 | wciK | 1,077 | 358 | 42.6 | 27.8 | Cap8H | S. aureus | 32.6/56.8 | β-1,3-d-Man2NAc transferase |
| 4 | wciL | 1,119 | 372 | 42.7 | 31.4 | RfbF | C. hyoilei | 25.4/51.1 | Glycosyltransferase |
| 4 | wciM | 1,068 | 355 | 40.8 | 32.2 | WcaK | E. coli K-12 | 20.3/45.2 | Pyruvate transferase |
| 6B | wchA | 1,368 | 455 | 52.0 | 38.1 | Cps14E | S. pneumoniae | 70.7/86.5 | Glucosyl-1-phosphate transferase |
| 6B | wciN | 945 | 314 | 36.6 | 27.3 | WaaR | E. coli | 24.6/50.2 | Glycosyltransferase |
| 6B | wciO | 720 | 239 | 27.0 | 31.1 | SpsI | A. aerolicus | 27.7/57.8 | Ribitol-1-phosphate transferase |
| 6B | wciP | 987 | 328 | 38.7 | 31.2 | RgpBc | S. mutans | 31.4/57.8 | Glycosyltransferase |
| 8 | wchA | 1,368 | 455 | 51.8 | 38.4 | Cps14E | S. pneumoniae | 70.3/86.8 | Glycosyl-1-phosphate transferase |
| 8 | wciQ | 498 | 165 | 19.5 | 33.8 | Cps14E | S. pneumoniae | 37.4/62.6 | Putative glycosyltransferase enhancer |
| EpsE | L. lactis | 36.8/59.4 | |||||||
| CpsIaE | S. agalactiae | 34.2/60.4 | |||||||
| CpsF | S. agalactiae | 34.9/60.4 | |||||||
| Orf5 | E. coli | 32.2/54.6 | |||||||
| 8 | wciR | 480 | 159 | 18.2 | 33.0 | Cps14G | S. pneumoniae | 32.9/58.2 | Glycosyltransferase |
| EpsF | L. lactis | 36.5/61.6 | |||||||
| CpsIaF | S. agalactiae | 19.3/44.2 | |||||||
| CpsG | S. agalactiae | 36.3/59.2 | |||||||
| Orf6 | E. coli | 36.2/60.4 | |||||||
| 8 | wciS | 1,065 | 354 | 40.0 | 31.0 | Cap33fG | S. pneumoniae | 25.9/51.8 | Glycosyltransferase |
| 8 | wciT | 729 | 242 | 28.4 | 30.2 | Cps14K | S. pneumoniae | 24.2/50.2 | Glycosyltransferase |
| Cap33fI | S. pneumoniae | 25.9/55.6 | |||||||
| 18C | wchA | 1,368 | 455 | 52.7 | 32.8 | Cps14E | S. pneumoniae | 94.9/97.5 | Glucosyl-1-phosphate-transferase |
| 18C | wchF | 1,173 | 390 | 44.7 | 36.6 | Cps23fF | S. pneumoniae | 90.7/93.2 | Rhamnosyl transferase |
| Cps23fT | S. pneumoniae | 90.7/93.2 | |||||||
| Cps2T | S. pneumoniae | 82.8/88.7 | |||||||
| 18C | wciU | 1,272 | 423 | 49.1 | 30.9 | WbdM | E. coli | 23.1/45.6 | Glycosyltransferase |
| 18C | wciV | 1,065 | 354 | 41.1 | 33.4 | Cap33fJ | S. pneumoniae | 35.8/57.2 | Glycosyltransferase |
| 18C | wciW | 912 | 303 | 35.8 | 32.4 | Cps14K | S. pneumoniae | 39.5/60.1 | Glycosyltransferase |
| Cap33fI | S. pneumoniae | 21.5/46.0 | |||||||
| 18C | wciY | 1,242 | 413 | 49.2 | 30.6 | TagF | B. subtilis | 25.7/51.7 | Glycerol-1-phosphate transferase |
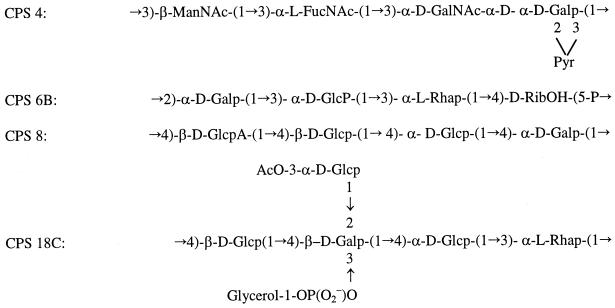
FIG. 2.
Structures of the repeat units of the CPSs of S. pneumoniae types 4 (34), 6B (74), 8 (35), and 18C (47, 58). Glc, glucose; Gal, galactose; Rha, rhamnose; GlcA, glucuronic acid; GalNAc, 2-acetamido-2-deoxy-d-galactose; ManNAc, 2-acetamido-2-deoxymanose; FucNAc, 2-acetamido-2,6-deoxygalactose; RibOH, ribitol.
WchF of CPS type 18C shares high-level identity with Orf6 (cps23fF/cps23fI) of type 23F (90.7% identity) and Orf6 (cps2T) of type 2 (82.8% identity), which are both putative rhamnosyl transferases for the addition of rhamnose to glucose. Thus, it is most likely that WchF is the expected rhamnosyl transferase. The high level of similarity of the three putative transferases suggests that all of the CPSs have the same rhamnose-glucose linkage. However, while rhamnose of types 2 and 23F CPSs was reported to be beta (32, 64), that of type 18C was reported to be alpha (47).
The assignments of the anomeric configuration of the rhamnose moieties of types 2 and 23F were strengthened by the fact that their Orf6 proteins group with other beta transferases in a retaining group of the Campbell et al. classification (12), in which glycosyltransferases can be classified according to the stereochemistry of the reaction substrates as either retaining or inverting enzymes. Therefore, on the basis of these comparisons we suspect that rhamnose of 18C CPS has a beta structure. The assignment of the anomeric configuration of the rhamnose residue in 18C CPS was by chromium oxide degradation, and we have been advised by the author of that study, H. J. Jennings (personal communication), that while that method was generally used at that time, it is now known to be unreliable, and it is possible that an incorrect assignment was made. If we assume CPS2, CPS18C, and CPS23F all have a β-rhamnose moiety linked to β-d-glucose via 1-4 linkage, given the very high level of identity among the three it seems clear that all catalyze that linkage.
Assembly genes.
The pathway for assembly of bacterial polysaccharides has been best studied in gram-negative bacteria, and three pathways are known (60, 80). In the Wzy-dependent pathway, the repeat unit is synthesized on the inner face of the cytoplasmic membrane using undecaprenol pyrophosphate as the carrier and then transported across the cytoplasmic membrane by a flippase encoded by wzx before polymerization by Wzy to form O-antigen polymer. Wzx- and Wzy-like proteins were previously identified in other S. pneumoniae types based on sequence similarity and hydrophobic profiles. However, the sequence similarity even within a species is often very low for Wzx and Wzy, and distinguishing the two can be difficult because both are highly hydrophobic. In type 18C and 33F gene clusters three genes encode hydrophobic proteins with more than 10 transmembrane segments, further complicating the matter. We therefore carried out BLOCKMAKER and PSI-BLAST searches to identify Wzx and Wzy.
Each of the highly hydrophobic proteins was grouped with known or putative Wzx or Wzy proteins, and motifs were generated and used to search databases using PSI-BLAST. Only Wzx or Wzy proteins were retrieved (E values of ≤3 × 10−21 for Wzx and ≤4 × 10−26 for Wzy) after several iterations. On this basis we conclude that orf11 of type 4, orf10 of type 6B, orf9 of type 8, and orf10 of type 18C are wzx genes and that orf9 of type 4, orf9 of type 6B, orf11 of type 8, and orf11 of type 18C are wzy genes. We also identified wzx and wzy genes in all published S. pneumoniae CPS gene clusters except that for type 3 CPS, which is thought to have a similar export system to that of S. enterica O54 (36). Most confirmed earlier assignments, but the wzy genes for types 1 and 33 CPS gene clusters were not identified previously.
Variation in wzg, wzh, wzd, and wze genes.
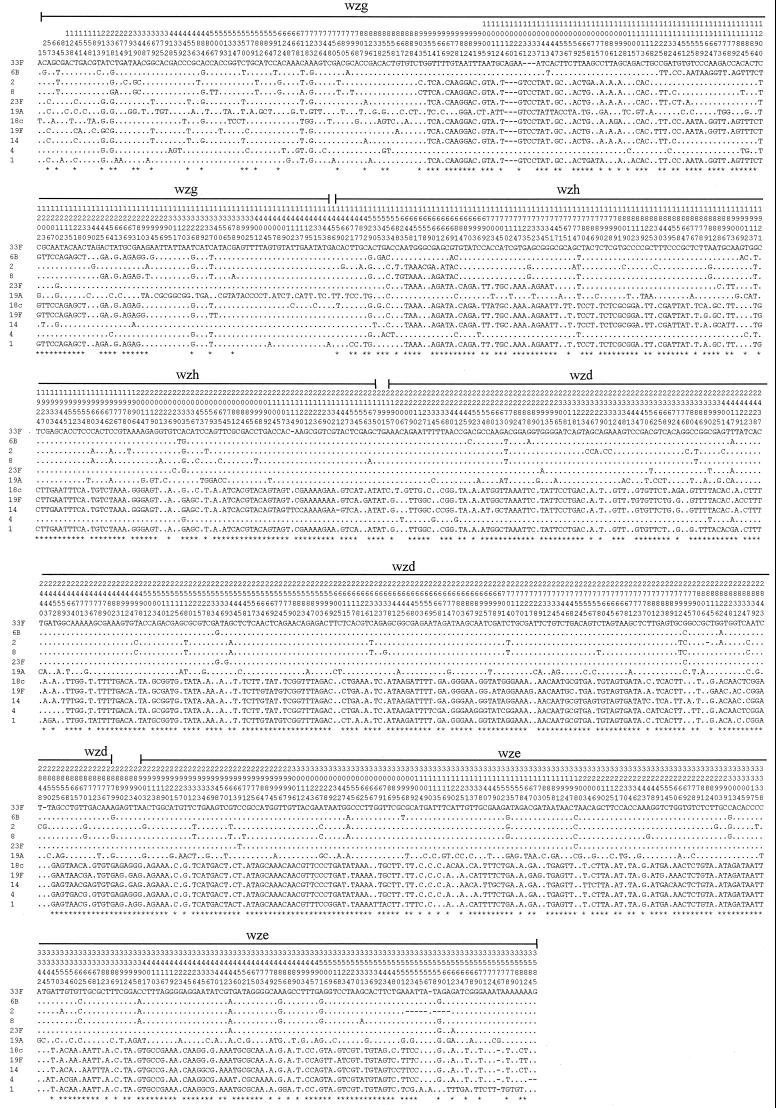
The genes at the 5′ and 3′ ends of the gene clusters, where they are common to many CPS types, show interesting patterns in the level of similarity between forms. Morona et al. (50) reported two classes of wzd and wze genes based on hybridization of DNA of CPS types 19A and 19F with DNA from strains of other CPS types and also on the sequence analysis of wzd genes. We can now compare nucleotide sequences of wzg, wzh, wzd and wze genes from eleven CPS types (Fig. 3 and 4). The comparison confirms that the four conserved genes are divided into two classes, but we now see an interesting pattern of recombination between them (Fig. 4). For the first 950 bp there is little variation, but from position 951 to the end of wze there are generally two forms, although CPS type 19A has a unique sequence to position 1529 in wzh. Types 1, 14, 18C, and 19F are in class I and types 2, 6B, 8, 19A, 23F, and 33F are in class II. CPS type 4 has a class II sequence to position 2463 and a class I sequence from that point onward. From position 951 in wzg to position 1747 in wzh there has been a lot of reassortment of the two sequence forms, with presumptive recombination events detected in wzg at positions 1118, 1125, 1146, 1153, 1245, and 1293 and in wzh at positions 1529, 1553, 1684, and 1747 (Fig. 4). The distinction between the two classes is most evident from position 1747 to the end of wze, with only type 4 showing a recombination event (position 2463) between class I and II forms in this segment. The Southern hybridization results obtained previously (38, 50, 51, 59) are generally consistent with the more detailed sequence comparison seen here.
FIG. 3.
Distribution of polymorphic sites within the sequences of wzg, wzh, wzd, and wze genes from positions 1 to 3550. The type 33F sequence is used for comparison, and only the differences are shown for the other sequences. Asterisks indicate informative sites. Gene boundaries are shown above polymorphic site numbers.
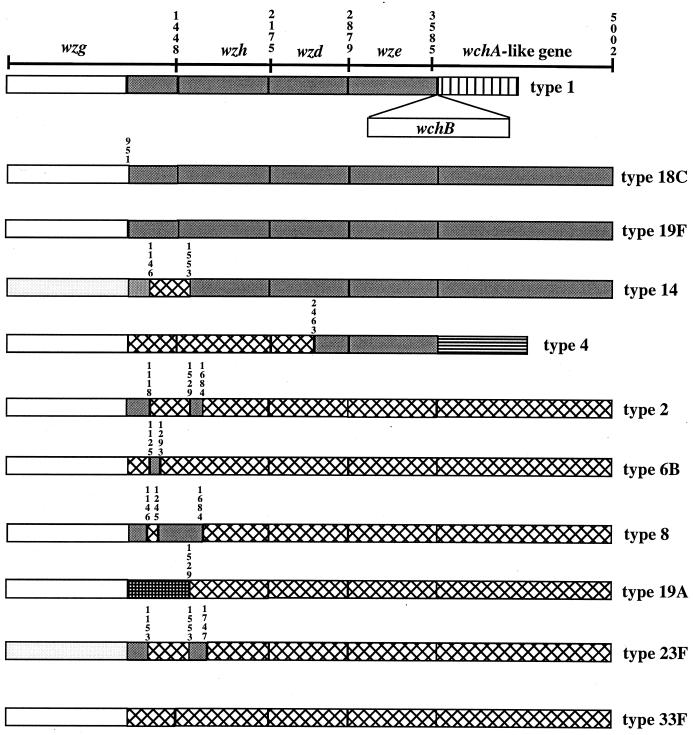
FIG. 4.
Two classes of wzg, wzh, wzd, wze, and wchA-like genes. Each pattern represents a class of closely related sequences. Numbers above the patterns indicate nucleotide positions: position 1 is the first nucleotide of wzg. The wchA-like gene is downstream of wchB gene in type 1.
The pairwise difference between sequences of different classes (excluding those small segments of the same class) ranges from 25.98 to 27.62%. The average pairwise differences of strains within class I or class II (excluding those small segments of the other class) are 2.40 and 3.86%, respectively.
Variation in wchA genes.
The first transferase gene, wchA, is located immediately downstream of wze. Comparison of the nine wchA genes available revealed that they also fall into two classes, the pattern exactly following that of the preceding wze gene (Fig. 4). This is in agreement with the previous Southern hybridization data (50). wchA genes of class I average a 2.61% pairwise difference and those of class II average a 4.33% pairwise difference. Pairwise differences between genes of different classes range from 28.1 to 28.8%.
Variation in rml genes.
The four rml genes are present in types 2, 6B, 18C, 19F, 19A, and 23F CPS gene clusters as expected and also in the type 1 CPS gene cluster, although rhamnose is not present in type 1 CPS (53). The rml genes if present are always located at the 3′ end of the CPS gene cluster and have the same gene order (rmlA, rmlC, rmlB, and rmlD). As for the first four genes in the CPS gene cluster, there is a gradient in the nature of the rml gene variation. The average pairwise difference for an rml gene close to the central region is higher than that for a gene close to the end of the gene cluster. This is similar to the situation we observed for rml genes of 11 S. enterica O-antigen gene clusters, where the genes are at the 5′ end of the O-antigen gene cluster in the order rmlB, rmlD, rmlA, and rmlC. In this case, the average pairwise difference increases from rmlB to rmlC (44), that is from 5′ to 3′, the opposite of the situation in S. pneumoniae. However, we consider it very significant that in both cases the most variable gene is that adjacent to the central type-specific region, and the least variable gene is that at one end of the whole gene cluster.
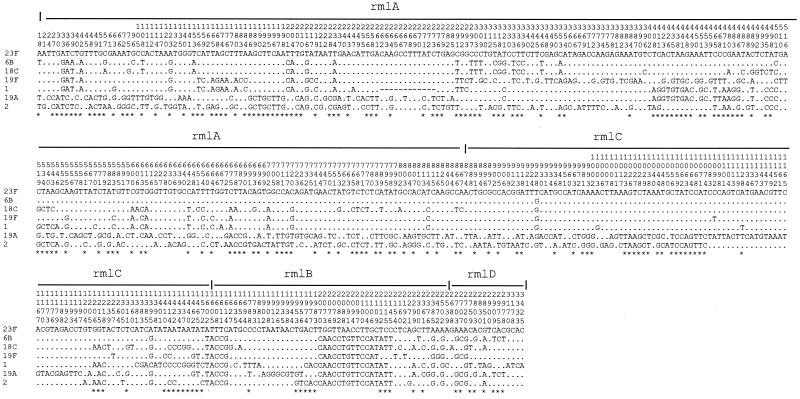
The neighbor-joining method was used to construct individual gene trees for the rml genes, and the variation in topology of the four trees suggests that recombination events have occurred in this region. For example, there is a segment in rmlA for which 6B and 18C are very similar and another where 6B and 23F are similar but different from 18C. The latter situation continues into rmlC but, in much of rmlD, 6B is similar to 19A, which for most rmlA and rmlC is quite different from 6B and 18C and indeed from most other CPS types (Fig. 5).
FIG. 5.
Distribution of polymorphic sites within rmlA, rmlC, rmlB, and rmlD. The type 23F sequence is used for comparison, and only differences are shown for the other sequences. Asterisks indicate informative sites.
Recombination sites and lateral transfer of CPS gene clusters.
The data on variation in 5′-terminal conserved genes (wzg-wchA) and rml genes can be compared with that in housekeeping genes. Seven housekeeping genes from 295 invasive isolates of S. pneumoniae (25) were aligned, and the Stephens test (71) used to detect recombination segments in the alleles of each gene. Both the Stephens test and visual examination indicated that the recombination frequency in the terminal CPS genes, especially wzg, wzh, rmlA, and rmlC, is much higher than in the housekeeping genes.
The high level of variation in terminal CPS genes resembles that of ddl, which is adjacent to pbpB, a gene which in some forms confers penicillin resistance. A segment of ddl sequenced from 566 S. pneumoniae isolates fell into two distinct groups (A and B) on a neighbor-joining tree (24). Group A alleles were very similar and were thought to represent the sequence of S. pneumoniae. Group B included much more divergent alleles, which differed from group A at up to 10.2% of the nucleotide sites. All group B strains were penicillin resistant, and the high level of variation was ascribed to a hitchhiking effect whereby interspecies recombinational exchanges involving pbpB, selected by penicillin resistance, often extended into, or through, the ddl gene. The high level of sequence variation in the 5′ end conserved genes (Fig. 3), and the rml gene set (Fig. 5) may be driven in the same way by natural selection for the transfer of CPS genes rather than representing random genetic drift. There is a well-documented example (15) in which eight CPS type 19F variants of the major Spanish multiresistant CPS type 23F clone were found to have large recombinational replacements, including CPS genes. In two cases, one of the crossover points was within an rml gene, rmlC in one case and rmlB in the other.
The maximum pairwise difference observed within rmlA, between type 19A or type 2 and other CPS types (from 14.25 to 20.11%), is within the range for interspecies variation. For the 5′-end conserved genes, the maximum pairwise difference within wzd is 30.12%.
The wide distribution of rml genes could make them useful in studying the relationships of bacterial polysaccharide gene clusters, and this has been attempted for O-antigen gene clusters of S. enterica (44), for which it appears that the two ends of the rml gene sets have different evolutionary histories. The correlation of 5′-end sequence variation with subspecies indicates that this part was in S. enterica prior to subspeciation and diverged with the subspecies. The 3′ end sequence is O-antigen specific and may represent the O-antigen clusters as first acquired by S. enterica by lateral transfer from other species, the high level of sequence variation indicating the divergent sources for these gene clusters (44). In the case of S. pneumoniae we do not have a subspecies structure, and we cannot draw any specific conclusion on the sources of rml genes. However, on the basis of the data presented above we suggest a similar evolutionary history.
The analysis of genes at the two ends of the CPS clusters also suggests that inter- and intraspecies transfer driven by selection for antigenic variations might have contributed to the distribution of the various S. pneumoniae CPS structures.
Concluding comments.
Now that the number of CPS gene clusters sequenced has reached 14, we begin to see patterns emerging. There are enough sequences of both the wzg, wzh, wzd, and wze and the rmlACBD sets of genes to see the crossover events and to start a detailed analysis of the role of these sets of genes in facilitating transfer of CPS gene clusters within S. pneumoniae and perhaps between Streptococcus species. It is probably no coincidence that the rather common rml genes are at one end of both the S. pneumoniae capsule and the E. coli-S. enterica O-antigen gene clusters. This not only facilitates lateral transfer but also impedes disruption of a gene cluster by recombination between genes common to two clusters. However, even this role has an exception, and type 18C is the first CPS cluster to have a gene distal to the rml genes.
It is clear that, as in the E. coli and S. enterica O-antigen gene clusters, wzx and wzy genes are extremely divergent such that motif searches are needed to identify them convincingly.
Allocating pathway genes is becoming easier. For rmlA-D, ugd, gct, and mnaA, it is possible to find a good level of similarities (at amino acid level) to genes of known function. As for l-Fuc2NAc, the number of sequenced gene clusters has grown to enable the gene or genes of the pathways involved to be identified with reasonable confidence, even if they cannot be allocated to specific steps until the pathway is established.
ACKNOWLEDGMENTS
We thank James C. Paton for kindly supplying S. pneumoniae strains. We also thank Bernard Henrissat for helping us allocate the putative glycosyltransferase genes by searching their classification database.
This study was supported by the Australian Research Council.
REFERENCES
- 1.Altschul A F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3398–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrianopoulos K, Wang L, Reeves P R. Identification of the fucose synthetase gene in the colanic acid gene cluster of Escherichia coli K-12. J Bacteriol. 1998;180:998–1001. doi: 10.1128/jb.180.4.998-1001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrecubieta C, Carcia E, Lopez R. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene. 1995;167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 5.Arrecubieta C, Garcia E, Lopez R. Demonstration of UDP-glucose dehydrogenase activity in cell extracts of Escherichia coli expressing the pneumococcal cap3A gene required for the synthesis of type 3 capsular polysaccharide. J Bacteriol. 1996;178:2971–2974. doi: 10.1128/jb.178.10.2971-2974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrecubieta C, Lopez R, Garcia E. Type 3-specific synthase of Streptococcus pneumoniae (cap3B) directs type 3 polysaccharide biosynthesis in Escherichia coli and in pneumococcal strains of different serotypes. J Exp Med. 1996;184:449–455. doi: 10.1084/jem.184.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Austrain R. Pneumococcus: the first one hundred years. Rev Infect Dis. 1981;3:183–189. doi: 10.1093/clinids/3.2.183. [DOI] [PubMed] [Google Scholar]
- 8.Austrain R, Bernheimer H P, Smith E E B, Mills G T. Simultaneous production of two capsular polysaccharides by pneumococcus. II. The genetics and biochemical bases of binary capsulation. J Exp Med. 1959;110:585–602. doi: 10.1084/jem.110.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 10.Boyd A C. Turbo cloning: a fast, efficient method for cloning PCR products and other blunt-ended DNA fragments into plasmid. Nucleic Acids Res. 1993;21:817–821. doi: 10.1093/nar/21.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown P K, Romana L K, Reeves P R. Molecular analysis of the rfb gene cluster of Salmonella serovar Muenchen (strain M67): genetic basis of the polymorphism between groups C2 and B. Mol Microbiol. 1992;6:1385–1394. doi: 10.1111/j.1365-2958.1992.tb00859.x. [DOI] [PubMed] [Google Scholar]
- 12.Campbell J A, Davies G J, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffey T J, Daniels M, Enright M C, Spratt B G. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology. 1999;145:2023–2031. doi: 10.1099/13500872-145-8-2023. [DOI] [PubMed] [Google Scholar]
- 14.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal gene transfer of multiple penicillin-binding protein genes and capsular biosynthetic genes in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 15.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 16.Coffey T J, Enright M C, Daniels M, Wilkinson P, Berron S, Fenoll A, Spratt B G. Serotype 19A variants of the Spanish serotype 23F multiresistant clone of Streptococcus pneumoniae. Microb Drug Resist. 1998;4:51–55. doi: 10.1089/mdr.1998.4.51. [DOI] [PubMed] [Google Scholar]
- 17.Comstock L E, Coyne M J, Tzianbos A O, Kasper D L. Interstrain variation of the polysaccharide B biosynthesis locus of Bacteroides fragilis: charaterization of the region from strain 638R. J Bacteriol. 1999;181:6192–6196. doi: 10.1128/jb.181.19.6192-6196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean C R, Franklund C V, Retief J D, Coyne M J, Jr, Hatano K, Evans D J, Pier G B, Goldberg J B. Characterization of the serotype O11 O-antigen locus of Pseudomonas aeruginosa PA103. J Bacteriol. 1999;181:4275–4284. doi: 10.1128/jb.181.14.4275-4284.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demerec M, Adelberg E, Clark A, Hartman P. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966;54:61–74. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dillard J P, Vandersea M W, Yother J. Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med. 1995;181:973–983. doi: 10.1084/jem.181.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drummelsmith J, Whitfield C. Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli. Mol Microbiol. 1999;31:1321–1332. doi: 10.1046/j.1365-2958.1999.01277.x. [DOI] [PubMed] [Google Scholar]
- 23.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 24.Enright M C, Spratt B G. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol Biol Evol. 1999;16:1687–1695. doi: 10.1093/oxfordjournals.molbev.a026082. [DOI] [PubMed] [Google Scholar]
- 25.Enright M C, Spratt B G. Multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 26.Fournier J M, Vann W F, Karakawa W W. Purification and characterization of Staphylococcus aureus type 8 capsular polysaccharide. Infect Immun. 1994;45:87–93. doi: 10.1128/iai.45.1.87-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frosch M, Weisgerber C, Meyer T F. Molecular characterisation and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis group B. Proc Natl Acad Sci USA. 1989;86:1669–1673. doi: 10.1073/pnas.86.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia E, Lopez R. Molecular biology of the capsular genes of Streptococcus pneumoniae. FEMS Microbiol Lett. 1997;149:1–10. doi: 10.1111/j.1574-6968.1997.tb10300.x. [DOI] [PubMed] [Google Scholar]
- 29.Gordon D, Abajian C, Green P. CONSED—a graphical tool for sequence finishing. PCR Methods Appl. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 30.Henikoff S, Henikoff J G, Alford W J, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- 31.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansson P E, Lindberg B, Anderson M, Lindquist U, Henrichsen J. Structural studies of the capsular polysaccharide from Streptococcus pneumoniae type 2, a reinvestigation. Carbohydr Res. 1988;182:111–117. doi: 10.1016/0008-6215(88)84095-3. [DOI] [PubMed] [Google Scholar]
- 33.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar Typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 34.Jones C, Currie F, Forster M J. N.m.r. and conformational analysis of the capsular polysaccharide from Streptococcus pneumoniae type 4. Carbohydr Res. 1991;221:95–121. doi: 10.1016/0008-6215(91)80051-n. [DOI] [PubMed] [Google Scholar]
- 35.Jones J K, Perry M B. The structure of the type VIII pneumococcus specific polysaccharide. J Am Chem Soc. 1957;79:2787–2793. [Google Scholar]
- 36.Keenleyside W J, Whitefield C. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J Biol Chem. 1996;271:28581–28592. doi: 10.1074/jbc.271.45.28581. [DOI] [PubMed] [Google Scholar]
- 37.Knirel Y A, Vinogradov E V, Kocharova N A, Paramonov N A, Kochetkov N K, Dmitriev B A, Stanislavsky E S, Lanyi B. The structure of O-specific polysaccharides and serological classification of Pseudomonas aeruginosa (a review) Acta Microbiol Hungar. 1988;35:3–24. [PubMed] [Google Scholar]
- 38.Kolkman M A B, van der Zeijst B A M, Nuijten P J M. Diversity of capsular polysaccharide synthesis gene clusters in Streptococcus pneumoniae. J Biochem. 1998;123:937–945. doi: 10.1093/oxfordjournals.jbchem.a022028. [DOI] [PubMed] [Google Scholar]
- 39.Kolkman M A B, van der Zeijst B A M, Nuijten P J M. Functional analysis of glycosyltransferases encoded by the capsular polysacharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J Biol Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 40.Kroll J S, Moxon E R. Capsulation in distantly related strains of Haemophilus influenzae type b: genetic drift and gene transfer at the capsulation locus. J Bacteriol. 1990;172:1347–1379. doi: 10.1128/jb.172.3.1374-1379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kroll J S, Zamze S, Loynds B, Moxon E R. Common organization of chromosomal loci for production of different capsular polysaccharides in Haemophilus influenzae. J Bacteriol. 1989;174:3343–3347. doi: 10.1128/jb.171.6.3343-3347.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lazarevic V, Margot P, Soldo B, Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J Gen Microbiol. 1992;138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- 43.Lee J C, Lee C Y. Capsular polysaccharides of Staphylococcus aureus. In: Goldberg J B, editor. Genetics of bacterial polysaccharide. Boca Raton, Fla: CRC Press; 1999. pp. 185–205. [Google Scholar]
- 44.Li Q, Reeves P R. Genetic variation of dTDP-l-rhamnose pathway genes in Salmonella enterica. Microbiology. 2000;146:2291–2307. doi: 10.1099/00221287-146-9-2291. [DOI] [PubMed] [Google Scholar]
- 45.Liu D, Verma N K, Romana L K, Reeves P R. Relationships among the rfb regions of Salmonella serovars A, B, and D. J Bacteriol. 1991;173:4814–4819. doi: 10.1128/jb.173.15.4814-4819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llull D, Munoz R, Lopez R, Garcia E. A single gene (tts) located outside the cap locus directs the formation of Streptococcus pneumoniae type 37 capsular polysaccharide: type 37 pneumococci are natural, genetically binary strains. J Exp Med. 1999;190:241–251. doi: 10.1084/jem.190.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lugowski C, Jennings H J. Structural determination of the capsular polysaccharide of Streptococcus pneumoniae type 18C. Carbohydr Res. 1984;131:119–129. doi: 10.1016/0008-6215(84)85409-9. [DOI] [PubMed] [Google Scholar]
- 48.Manca M C, Weintraub A, Widmalm G. Structural studies of the Escherichia coli O26 O-antigen polysaccharide. Carbohydr Res. 1996;281:155–160. doi: 10.1016/0008-6215(95)00333-9. [DOI] [PubMed] [Google Scholar]
- 49.Moreau M, Richards J C, Fournier J M, Byrd R A, Karakawa W W, Vann W F. Structure of the type 5 capsular polysaccharide of Staphylococcus aureus. Carbohydr Res. 1990;201:285–297. doi: 10.1016/0008-6215(90)84244-o. [DOI] [PubMed] [Google Scholar]
- 50.Morona J K, Morona R, Paton J C. Analysis of the 5′ portion of the type 19A capsule locus identifies two classes of cpsC, cpsD, and cpsE genes in Streptococcus pneumoniae. J Bacteriol. 1999;181:3599–3605. doi: 10.1128/jb.181.11.3599-3605.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morona J K, Morona R M, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 52.Munoz R, Mollerach M, Lopez R, Garcia E. Characterization of the type 8 capsular gene cluster of Streptococcus pneumoniae. J Bacteriol. 1999;181:6214–6219. doi: 10.1128/jb.181.19.6214-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munoz R, Mollerach M, Lopez R, Garcia E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 54.Nassau P M, Martin S L, Brown R E, Weston A, Monsey D, R M M, Duncan K. Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J Bacteriol. 1996;178:1047–1052. doi: 10.1128/jb.178.4.1047-1052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;1998:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 56.Park Y S, Sweitzer T D, Dixon J E, Kent C. Expression, purification, and characterization of CTP: glycerol-3-phosphate cytidylytransferase from Bacillus subtilis. J Biol Chem. 1993;268:16648–16654. [PubMed] [Google Scholar]
- 57.Paulsen I T, Beness A M, Saier M H., Jr Computer-based analyses of the protein constituents of transport systems catalyzing export of complex carbohydrates in bacteria. Microbiology. 1997;143:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- 58.Philips L R, Nishmura O, Fraser B A. The structure of the repeating oligosaccharide unit of the pneumococcal capsular polysaccharide type 18C. Carbohydr Res. 1983;121:243–255. doi: 10.1016/0008-6215(83)84021-x. [DOI] [PubMed] [Google Scholar]
- 59.Ramirez M, Tomasz A. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J Bacteriol. 1998;180:5273–5278. doi: 10.1128/jb.180.19.5273-5278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reeves P R. Biosynthesis and assembly of lipopolysaccharide. In: Neuberger A, van Deenen L L M, editors. Bacterial cell wall. Vol. 27. Amsterdam, The Netherlands: Elsevier Science Publishers; 1994. pp. 281–314. [Google Scholar]
- 61.Reeves P R, Farnell L, Lan R. MULTICOMP: a program for preparing sequence data for phylogenetic analysis. CABIOS. 1994;10:281–284. doi: 10.1093/bioinformatics/10.3.281. [DOI] [PubMed] [Google Scholar]
- 62.Reeves P R, Hobbs M, Valvano M, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C, Rick P. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 63.Reisner A H, Bucholtz C A, Smelt J, McNeil S. Proceedings of the Twenty-Sixth Annual Hawaii International Conference on Systems Science. 1993. Australia's National Genomic Information Service; pp. 595–602. [Google Scholar]
- 64.Richards J C, Perry M B. Structure of the specific capsular polysaccharide of Streptococcus pneumoniae type 23F (American type 23) Biochem Cell Biol. 1988;66:758–771. doi: 10.1139/o88-087. [DOI] [PubMed] [Google Scholar]
- 65.Robbins J B, Austrian R, Lee C J, Rastogi S C, Schiffman G, Henrichsen J, Mäkelä P H, Broome C V, Fracklam R R, Tiesjema R H, Parke J C J. Considerations for formulating the second-generation pneumococcal capsular polysaccharide vaccine with emphasis on the cross-reactive types within groups. J Infect Dis. 1983;148:1136–1159. doi: 10.1093/infdis/148.6.1136. [DOI] [PubMed] [Google Scholar]
- 66.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 67.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogentic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 68.Sau S, Bhasin N, Wann E R, Lee J C, Foster T J, Lee C Y. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology. 1997;143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 69.Scott J A, Hall A J, Dagan R, Dixon J M, Eykyn S J, Fenoll A, Hortal M, Jette L P, Jorgensen J H, Lamothe F, Latorre C, Macfarlane J T, Shlaes D M, Smart L E, Taunay A. Serogroup-specific epidemiology of Streptococcus pneumoniae: associations with age, sex, and geography in 7,000 episodes of invasive disease. Clin Infect Dis. 1996;22:973–981. doi: 10.1093/clinids/22.6.973. [DOI] [PubMed] [Google Scholar]
- 70.Shibaev V N. Biosynthesis of bacterial polysaccharide chains composed of repeating units. Adv Carbohydr Chem Biochem. 1986;44:277–339. doi: 10.1016/s0065-2318(08)60080-3. [DOI] [PubMed] [Google Scholar]
- 71.Stephens J C. Statistical methods of DNA sequence analysis: detection of intragenic recombination or gene conversion. Mol Biol Evol. 1985;2:539–556. doi: 10.1093/oxfordjournals.molbev.a040371. [DOI] [PubMed] [Google Scholar]
- 72.Stevenson G, Andrianopoulos K, Hobbs H, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Utterback T R, McDonald L A, Fuldner R A. A reliable, efficient protocol for 96-well plasmid DNA miniprep with rapid DNA quantification for high-throughput automated DNA sequencing. Genome Sci Technol. 1995;1:1–8. [Google Scholar]
- 74.van Dam J E G, Breg J, Komen R, Kamerling J P, Vliegenthart J F G. Isolation and structural studies of phosphate-containig oligosaccharides from alkaline and acid hydrolysates of Streptococcus pneumoniae type 6B capsular polysaccharide. Carbohydr Res. 1989;187:267–286. doi: 10.1016/0008-6215(89)80008-4. [DOI] [PubMed] [Google Scholar]
- 75.van Kranenburg R, van Swam I I, Marugg J D, Kleerebezem M, de Vos W M. Exopolysaccharide biosynthesis in Lactococcus lactis NIZO B40: functional analysis of the glycosyltransferase genes involved in synthesis of the polysaccharide backbone. J Bacteriol. 1999;181:338–340. doi: 10.1128/jb.181.1.338-340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Kranenburg R, Vos H R, van Swam I I, Kleerebezem M, de Vos W M. Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: complementation, expression, and diversity. J Bacteriol. 1999;181:6347–6353. doi: 10.1128/jb.181.20.6347-6353.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, Liu D, Reeves P R. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalysing the first step of O antigen synthesis. J Bacteriol. 1996;178:2598–2604. doi: 10.1128/jb.178.9.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, Reeves P R. Organization of Escherichia coli O157 O-antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, Romana L K, Reeves P R. Molecular analysis of a Salmonella enterica group E1 rfb gene cluster: O antigen and the genetic basis of the major polymorphism. Genetics. 1992;130:429–443. doi: 10.1093/genetics/130.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]