Abstract
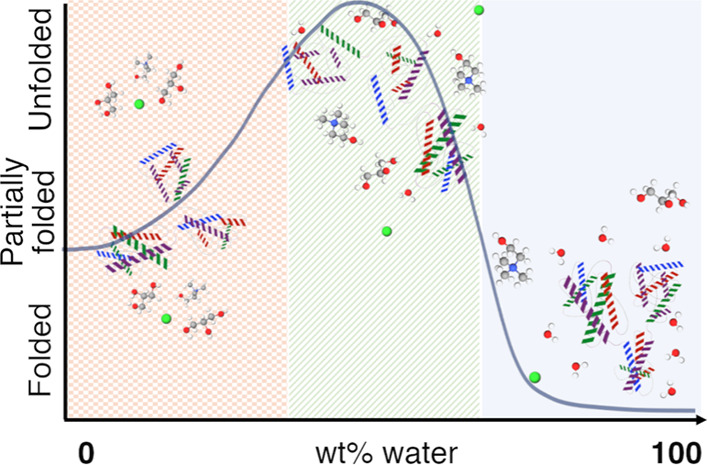
The preservation of labile biomolecules presents a major challenge in chemistry, and deep eutectic solvents (DESs) have emerged as suitable environments for this purpose. However, how the hydration of DESs impacts the behavior of proteins is often neglected. Here, we demonstrate that the amino acid environment and secondary structure of two proteins (bovine serum albumin and lysozyme) and an antibody (immunoglobulin G) in 1:2 choline chloride:glycerol and 1:2 choline chloride:urea follow a re-entrant behavior with solvent hydration. A dome-shaped transition is observed with a folded or partially folded structure at very low (<10 wt % H2O) and high (>40 wt % H2O) DES hydration, while protein unfolding increases between those regimes. Hydration also affects protein conformation and stability, as demonstrated for bovine serum albumin in hydrated 1:2 choline chloride:glycerol. In the neat DES, bovine serum albumin remains partially folded and unexpectedly undergoes unfolding and oligomerization at low water content. At intermediate hydration, the protein begins to refold and gradually retrieves the native monomer–dimer equilibrium. However, ca. 36 wt % H2O is required to recover the native folding fully. The half-denaturation temperature of the protein increases with decreasing hydration, but even the dilute DESs significantly enhance the thermal stability of bovine serum albumin. Also, protein unfolding can be reversed by rehydrating the sample to the high hydration regime, also recovering protein function. This correlation provides a new perspective to understanding protein behavior in hydrated DESs, where quantifying the DES hydration becomes imperative to identifying the folding and stability of proteins.
Introduction
Conformation and dynamics are overarching concepts in understanding protein stability, enzymatic activity, and molecular recognition.1−4 The use of proteins in their natural aqueous environment is restricted to a limited variety of conditions as proteins are marginally stable outside their native conformational window.5 The development of non-aqueous enzymology extended the range of possible solvents for protein stabilization, potentially improving protein stability and substrate specificity.6 Notably, organic solvents also allowed tailoring protein behavior, such as the selectivity of an enzymatic route toward specific products, through changes in the solvent properties.7 More recently, the alternatives for non-aqueous protein stabilization and function were further expanded by using ionic liquids as solvation environments.8 Progress has been made in understanding protein behavior in these complex solvents, where subtle changes in the solvent characteristics can lead to a wide variety of biophysical and enzymatic responses.9,10
Deep eutectic solvents (DESs) are eutectic mixtures that show a larger depression in the melting point (“deep”) than that predicted for an ideal mixing of their constituents.11 In particular, the majority of DESs discovered heretofore belong to the Type III category. Those are formed by an organic salt (typically ammonium) and a neutral compound, acting as a hydrogen bond donor, at the eutectic ratio of the mixture. Mixing these precursors leads to a highly entropic state stabilized by an extensive hydrogen bond network, often remaining liquid at room temperature.12−15 DESs are compared to ionic liquids since different precursors allow great control over the solvent physicochemical properties, such as polarity, hydrophobicity, and hydrogen potential.11,16−19 Importantly, DESs can be synthesized from bio-derived compounds, such as sugars, carboxylic acids, and ammonium salts, which bestows the system with mild character, thermal and chemical stability, sustainability, and biocompatibility.20−24 Hence, DESs offer a tailorable, suitable environment for biomolecules, probing to be milder alternatives to ionic liquids.8 Recently, particular emphasis has been placed on developing DESs as solvents for biomolecule assembly, preparation of functional biomaterials, enzymatic catalysis, and preservation of biomolecule integrity, among others.20,23,25−33
Despite the promising outlook for biotechnological applications using DESs, the inherent properties of the solvent (e.g., high viscosity, low ion mobility) often hinder further potential developments. In this sense, proteins present a challenge for incorporation into DESs as their solubilization rate is extremely slow, and their thermo-labile character precludes incorporation through heating.34−36 An accepted route to leverage these properties of DESs is adding water to the system.37,38 This approach offers an extra degree of freedom for tailoring the solvent properties, for example, by significantly reducing the viscosity of the DESs and improving the dissolution rate of recalcitrant macromolecules.38 Consequently, most investigations on proteins in DESs have employed hydrated versions of these solvents. For instance, many studies have shown that the activity of different enzymes is retained in hydrated DESs.26,36,39−42 Also, the addition of sufficient water to DESs causes proteins to retrieve their native structure.35,36,42−44 However, lower levels of solvent hydration could influence the behavior of the protein as water can either modify or disrupt the molecular interactions within the DESs.45−47 This has been widely explored for hydrated organic solvents and ionic liquids, showing that adding water can induce nonlinear responses in protein behavior and denaturation.7,48,49 Also, it has been shown that DESs can promote the crystallization of proteins under certain hydration conditions, which control the nucleation rate.50 Surprisingly, the effect of hydration is often neglected when studying the solution behavior of proteins in hydrated DESs.
To fill this knowledge gap, we present an investigation of protein behavior in hydrated DESs across a wide range of hydration levels using UV–vis absorption spectroscopy and far-UV circular dichroism (CD). The solvation environment and secondary structure of bovine serum albumin (BSA), lysozyme (Lyz), and immunoglobulin G (IgG) were studied in 1:2:n choline chloride:glycerol:water (1:2:n ChCl:Glyc:H2O), with n = 0.4 to 20 mole-to-mole ratio, that is, from 2.2 to 53.4 wt % H2O. Similarly, the behavior of Lyz in 1:2:n choline chloride:urea:water (1:2:n ChCl:Urea:H2O), with n = 0.4 to 20 mole-to-mole ratio, that is, from 2.7 to 58.1 wt % H2O, was investigated to provide a comparison of the effect caused by the DES. We decided to use the 1:2:n ChCl:Glyc:H2O and 1:2:n ChCl:Urea:H2O solvents since their physicochemical properties, molecular structure, and DES–water interactions have been well resolved, covering the full range of hydration through experimental and computational methods.12,45−47 Similarly, BSA provides a characteristic overall conformation and has been extensively characterized in the native state and under different conditions of chemical and thermal stress,35,51−55 constituting a good proxy to understanding the effect of the DES hydration on protein folding, self-association equilibrium, and stability. Thus, BSA conformation and stability were investigated in 1:2:n ChCl:Glyc:H2O using small-angle neutron scattering (SANS), fluorescence spectroscopy, and temperature-dependent CD. The behavior of the proteins in the neat DESs and in an aqueous buffer (10 mM, pH 7 sodium phosphate buffer, herein referred to as the native state) provides the baseline comparisons to the results presented here. Notably, the residual water content was measured at ca. 0.37 wt % H2O (n = 0.065) in 1:2 ChCl:Glyc and 0.24 wt % H2O (n = 0.034) in 1:2 ChCl:Urea. These are expected values of the reported remanent water content in these DESs even after extensive drying.12,45,46,56−58 Thus, we refer to these as neat DESs. The equivalences of the water content in wt % and mol % are presented in Table S1. For simplicity and due to the similarity in the hydration levels between the two DESs systems, we will use the water contents of the 1:2:n ChCl:Glyc:H2O system to label the data throughout the article unless otherwise stated.
Results
Nonlinear Structural Transitions
Initially, the solvation environment of the aromatic amino acids was investigated using UV–vis spectroscopy. To extract detailed information about the environment of the protein chromophores (e.g., solvent exposure), the second-derivative UV–vis spectra (d2Abs/dλ2) were calculated to resolve the changes in the signal of the peak centered at ca. 280 nm, from which the contributions of the tyrosine (Tyr, ca. 285 nm) and tryptophan (Trp, ca. 295 nm) sides chains can be discerned.59 Thus, the Tyr peak position (λd2AbsTyr) and the ratio between the amplitudes of the Tyr and Trp peaks (d2AbsTyr/d2AbsTrp) were determined. These data and results are presented in Figure 1. The full UV–vis absorption spectra for the different systems are presented in Figure S1. For further details on the analysis of these data, see the Supporting Information.
Figure 1.
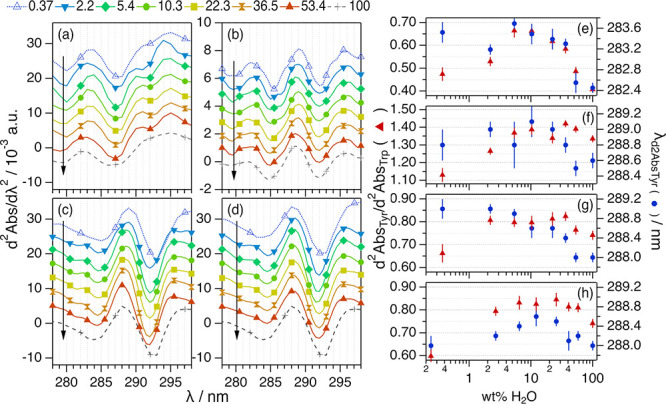
Second-derivative UV–vis spectra for (a) 100 μM BSA, (b) 20.4 μM IgG, and (c) 206 μM Lyz in 1:2:n ChCl:Glyc:H2O; (d) 224 μM Lyz in 1:2 ChCl:Urea:H2O at different DES hydrations expressed in wt % H2O in the solvent. The data and results for the proteins in aqueous buffer (100 wt % H2O) are presented for comparison. Data have been offset for clarity by (a,c,d) +0 (100 wt % H2O) +0.004, +0.008, +0.012, +0.016, +0.020, +0.024, and +0.028 (0.37 wt % H2O) and (b) +0 (100 wt % H2O), +0.001, +0.002, +0.003, +0.004, +0.005, +0.006, and +0.007 (0.37 wt % H2O). Hydration increases in the direction of the arrows. (e–h) present the results derived from the analysis of the spectroscopy data of the proteins, λd2AbsTyr and d2AbsTyr/d2AbsTrp, in DES at different hydration levels. Where not seen, error bars are within the markers.
In neat and hydrated DESs, the proteins present the characteristic protein absorption spectrum with a peak centered at ca. 280 nm (Figure S1), similar in shape and intensity to the aqueous buffer, indicating the capacity of the neat and hydrated DESs to solubilize the proteins used in this study. The second-derivative UV–vis spectrum (d2Abs/dλ2) enables accessing the changes occurring in the molecular environment of the protein chromophores with the advantage of increased resolution compared to the relatively featureless UV–vis spectrum.59 From the results, it is observed that the features in the second-derivative UV–vis varied when changing the level of DES hydration; this qualitatively confirms changes in the amino acid environment and that the extent of those depends on the water content. The characteristic parameters of second-derivative UV–vis spectra are very sensitive to changes in the Tyr environment (peak at ca. 287 nm) and, to a minor extent, in the Trp residue (peak at ca. 295 nm), where an increase in d2AbsTyr/d2AbsTrp and a bathochromic shift in λd2AbsTyr relate to the exposure of the aromatic residues to a more polar environment.59
Generally, a dome-shaped transition is observed in the λd2AbsTyr and d2AbsTyr/d2AbsTrp for all systems investigated here (see Figure 1e–h). Three regimes can be differentiated: (i) At low hydration levels, the characteristic parameters gradually increase to their maxima around 4–8 wt % H2O; (ii) when the water content is increased between ca. 10 and ca. 40 wt % H2O, the value of d2AbsTyr/d2AbsTrp gradually decreases and a hypsochromic shift in λd2AbsTyr is observed; and (iii) above ca. 40 wt % H2O, the spectral parameters become similar to those in native conditions. Interestingly, the λd2AbsTyr and d2AbsTyr/d2AbsTrp in neat DESs reach similar magnitudes to those in aqueous buffer in some cases.
These transitions in the UV–vis spectral features are commonly associated with conformational changes in the protein, correlated to the exposure of buried aromatic residues that arises from a change in the amino acid milieu.59 To study the origin of those variations, the secondary structure of the proteins was investigated using far-UV CD measurements. The resulting CD spectra were analyzed to determine the population of each secondary structure motif in the protein using the software BeStSel in the wavelength range between 200 and 250 nm.60 The results are presented in Figure 2.
Figure 2.
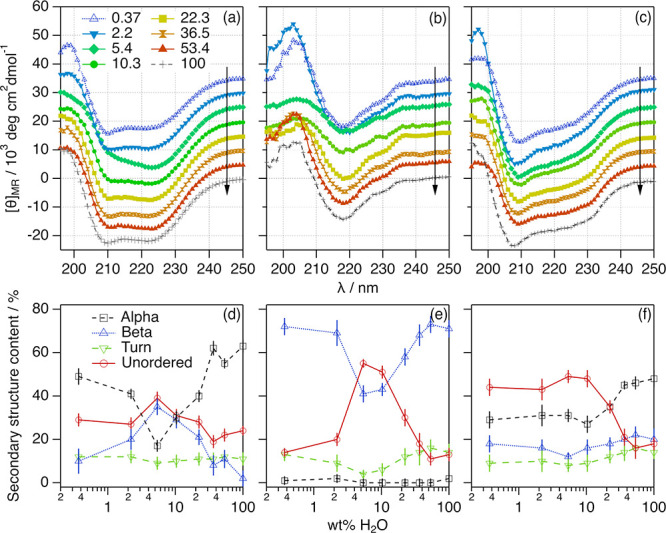
Mean residue ellipticity of (a) 59 μM BSA, (b) 20.4 μM IgG, and (c) 206 μM Lyz in ChCl:Glyc:H2O at different hydration levels. The data and results for the proteins in aqueous buffer (100 wt % H2O) are presented for comparison. Data have been offset for clarity by +0 (100 wt % H2O), +5 × 103, +10 × 103, +15 × 103, +20 × 103, +25 × 103, +30 × 103, and +35 × 103 (0.37 wt % H2O). Hydration increases in the direction of the arrows. Hydration-dependent changes in the content of secondary structure motifs of (d) BSA, (e) IgG, and (f) Lyz were determined from the far-UV CD spectra. These values correspond to the average fitted values from three independent CD spectra. Where not seen, error bars are within the markers.
The proteins investigated provide a detailed comparison of the impact of DES hydration on specific folding motifs. In the native state, BSA is predominantly populated by α-helices with no β-sheet content,61 IgG folds into β-sheets with marginal amounts of α-helices,62 and Lyz contains populations of both ordered motifs (Tables S3–S5).63 Our results confirm that the hydration of the DESs induces changes in the secondary structure of the proteins, as observed in the spectral features of the CD data. Notably, the deconvolution of the CD spectra shows a general trend for the three proteins in 1:2:n ChCl:Glyc:H2O, where several transitions can be observed at different hydration regimes: (i) at low hydration (below ca. 8 wt % H2O), the proteins lose a certain amount of the main ordered secondary structure motif and the content of unordered structure increases; (ii) when water is increased within an intermediate hydration regime (between ca. 10 and ca. 40 wt % H2O), the ordered secondary structure of the proteins is gradually retrieved and the unordered structure decreases; and (iii) at high hydration (above ca. 40 wt % H2O), the native secondary structure is retrieved (within the error).
Interestingly, hydration affects differently to each protein and changes are particularly pronounced for BSA and IgG. In the case of BSA, a relative decrease of 0.78-fold in the α-helix content is observed in 1:2 ChCl:Glyc, accompanied by a subtle increase in the β-sheet and unordered content compared to those in the native state. At 5.4 wt % H2O, the protein loses 0.27-fold of the α-helix content, and the β-sheet and unordered contents increase by 17-fold and 2-fold, respectively. Finally, a similar secondary structure (within the error) to the BSA native state is observed above 36.5 wt % H2O. This suggests that DES hydration induces the unfolding of the secondary structure of BSA, with a characteristic α-helix-to-β-sheet transition, as previously observed upon albumin unfolding.64 For IgG, similar transitions are observed but affect the secondary structure motifs differently. In this case, the secondary structure population of the antibody is practically the same in 1:2 ChCl:Glyc as in aqueous buffer. However, the addition of water in the low hydration regime (5.4 wt % H2O) induces a 0.58-fold decrease in the population of β-sheets and a 4.2-fold increase in the content of unordered structures. This unfolding is again gradually reverted with increasing hydration. Changes for Lyz follow the same trends, although they are less pronounced. The low hydration unfolding is associated with a 0.6-fold decrease of the α-helix and β-sheet content and a 2.4-fold increase in the amount of unordered secondary structure. Also, the content of turn motifs in the proteins (ca. 10% of the total) hardly changes upon hydration.
It should be noted that the strong optical absorption of 1:2 ChCl:Urea in the far-UV region does not allow measuring the CD signal below 210 nm, even when using the narrow path cuvette (0.1 mm). Therefore, no quantitative estimation of the secondary structure of Lyz in 1:2:n ChCl:Urea:H2O can be performed with the available data at low hydration (Figure S2). However, some meager analysis was performed for the system at high hydration (Table S6), where the solvent is known to behave as an aqueous solution of the DES components.45 Our results confirm that the urea-based DES at high hydration (>40 wt % H2O) impacts protein structure to a larger extent than the glycerol analogues. This results in a decrease of the ordered secondary structure with decreasing water content in this hydration regime (e.g., 0.5-fold decrease in the α-helix content in 40.9 wt % H2O compared to the native secondary structure). The interaction of freely diffusing urea, a known potent denaturant, with Lyz is possibly the origin of this structural change; it has been shown that the disruption of the DES structure upon hydration enables specific interactions between the protein and the DES constituents.65,66 Still, it is observed that the native structure of the protein changes with hydration in 1:2:n ChCl:Urea:H2O and, considering the results from the UV–vis characterization of the system, these are likely to be non-monotonic.
Thus, a general mechanism seems to govern the amino acid environment and local folding of the proteins in hydrated DESs. Three particular regimes are observed with increasing hydration: (i) a low hydration regime, where water contributes to disrupting the structure of the protein, (ii) an intermediate hydration regime, where the protein gradually retrieves its native local fold, and (iii) a high hydration regime, where the native structure of the proteins is fully regained despite the high remanent amount of DES in the solvent.
Protein Conformational Landscape
To relate the observed re-entrant changes above to the protein’s hierarchical structure, SANS experiments were performed on 55 μM BSA (3.66 mg/mL, average concentration) to study the effect of 1:2:n ChCl:Glyc:H2O at different levels of hydration. Deuterium-labeled solvents were required to gain contrast in SANS experiments (1:2:n d9-ChCl:d8-Glyc:D2O). Previous investigations have shown that solvent deuteration has a minimal impact on the behavior of this protein in DESs.35 Also, SANS does not cause any radiation damage to the sample. Thus, we preferred this method to other alternatives, such as synchrotron-radiation small-angle X-ray scattering, which is known to cause beam damage due to the high electron density of the DESs.67 The data, models, and pair-distance distribution functions (p(r)) of the scatterer are presented in Figure 3.
Figure 3.
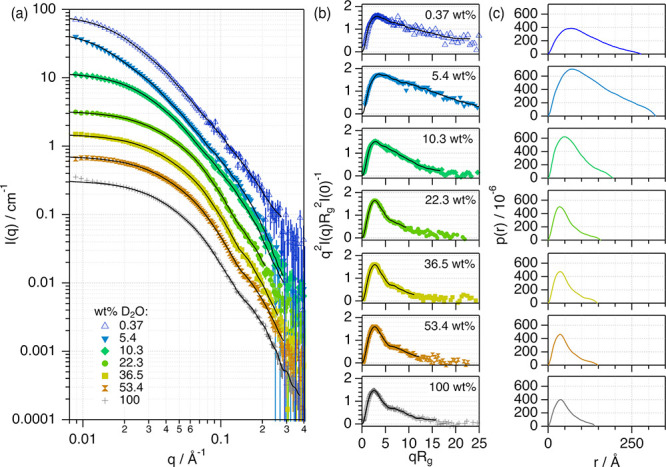
(a) SANS data and models of 55 μM BSA in 1:2:n d9-ChCl:d8-Glyc:D2O at different water contents in the solvent expressed in wt % D2O. (b) Normalized Kratky representation of the SANS data and models and (c) pair-distance distribution functions of the protein in different solvents. The data and results for BSA aqueous buffer (100 wt % D2O) are presented for comparison. The models obtained from the indirect Fourier transform analysis are presented as solid black lines. The legend for the data is presented in panels (a,b). Data and models in (a) have been offset for clarity by ×1 (100 wt % D2O), ×2, ×4, ×8, ×16, ×32, and ×64 (0.37 wt % D2O). Where not seen, error bars are within the markers.
Our SANS data show that the signal from BSA significantly changes when the water content varies in the DES. These differences are highlighted in the normalized Kratky plots, which represent the normalized scattering intensity, q2I(q)Rg2I(0)−1, against qRg, where I(q) is the scattered intensity, I(0) is the extrapolated scattering intensity at angle zero, q is the momentum transfer vector, and Rg is the radius of gyration of the scatterer. In this plot, the decay at high qRg relates to the folding state of the protein (see Figure 3b).68 The bell shape of the Kratky plot for BSA in 1:2 ChCl:Glyc shows that the decay at high qRg (>5) is less pronounced in neat DES than in the aqueous buffer. This confirms that the protein changes conformation but retains a certain degree of globularity in the DES compared to aqueous buffer.35 In the hydrated DESs, different regimes are observed: (i) at a low water content (5.4 wt %), the protein swells as reflected by the change in the negative slope at high qRg; (ii) when the water content is increased to 10.3 wt % D2O, the slope of the Kratky representation suggests that the protein folds into a more compact conformation compared to that at low DES hydration; and (iii) above 36.5 wt % D2O, the shape of the Kratky plot is similar to that of the protein in the native state.
The indirect Fourier transform method was used to calculate the p(r), representing a histogram of the distances between two points within the scatterer, to extract detailed structural information.69−71 From the p(r), the characteristic structural parameters of BSA were calculated: the maximum dimension of the protein, Dmax, the folded state of the monomer parameterized as the position of the first peak in the p(r), r1, and the protein self-association parameterized as the aggregation number, Nagg (Figure 4). Details on the data analysis and the determination of these parameters are presented in the Supporting Information.
Figure 4.
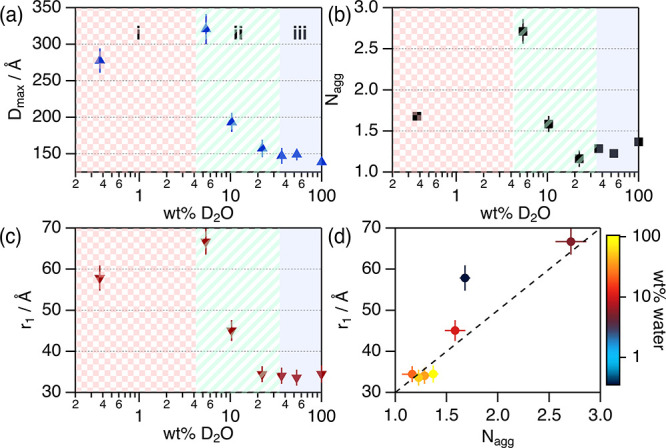
Parameters derived from the analysis of the SANS data of 55 μM BSA in 1:2:n d9-ChCl:d8-Glyc:D2O at different water contents: (a) Dmax, (b) Nagg, and (c) r1 as a function of wt % D2O. (d) Parametric plot of the variation of r1 vs Nagg at different levels of solvent hydration. The water content in (d) is represented following the color scale. The dashed line is a guide for the eye of the trend described by the results in hydrated DES.
In the native state, the conformation of BSA in solution is a mixture of folded (globular) monomers and dimers in a dynamic equilibrium.52,72 Our characterization of BSA in aqueous buffer agrees with the previously reported conformation of this protein in the native state, confirming that the selected buffer does not affect the structure of the protein. BSA’s calculated p(r) in the different solvents confirms that the neat DES promotes conformational changes (see Figure 4a). We observe that the Dmax of BSA in 1:2 ChCl:Glyc (280 ± 17 Å) doubles that of the protein in aqueous buffer (138 ± 5 Å).73 The size change confirms a shift in the native BSA self-association (monomer–dimer equilibrium) toward a higher population of oligomers in the neat DES, potentially a monomer–dimer–tetramer equilibrium.72 This behavior has been previously reported for aqueous solutions of BSA at high ionic strength, where the protein self-associates into tetramers (maximum diameter ca. 300 Å) due to an increase in protein–protein attraction.53,54 The r1 value increases when BSA is solvated in 1:2 ChCl:Glyc (57.8 ± 3.1 Å) compared to that in the aqueous buffer (34.5 ± 2.1 Å). This larger value, together with the shape of the Kratky plot, suggests that the BSA monomer is partially folded in the neat DES compared to the fully folded native folded state.
The addition of water to 1:2 ChCl:Glyc induces changes in the conformational state of BSA in both the folding state and self-association, even at low water content. Importantly, these changes are non-monotonic: (i) An increase in Dmax, r1, and Nagg is observed at 5.4 wt % D2O compared to the neat DES, showing that BSA is further unfolded and increases self-association. (ii) At 10.3 wt % D2O, the protein becomes more compact and the self-association decreases but remains in a partially folded, self-associated state. (iii) At 36.5 wt % D2O and above, the protein retrieves its native folding and self-association equilibrium. From the results presented in the parametric plot (Figure 4d), we observe that the protein populates different conformational states depending on the water content: there is a linear correlation between the monomer structure (r1) and self-association (Nagg) in hydrated DES; however, the protein does not follow these conformational features in neat DES. Therefore, water must play a significant role in the behavior of the protein, possibly connected to the changes in the nanoscale organization of the solvent.45−47
BSA has two Trp residues buried at the hydrophobic pockets of domains I and II.74 As such, intrinsic Trp fluorescence spectroscopy can be used to determine the degree of solvent accessibility to the hydrophobic core associated with the conformational changes. From the experimental data, the center of spectral mass (CSM) of the Trp emission spectra was determined at each hydration level. The CSM accounts for the position and intensity distribution across the emission peak; thus, it can be used to identify changes in the environment of the Trp residue. These results are shown in Figure 5.
Figure 5.
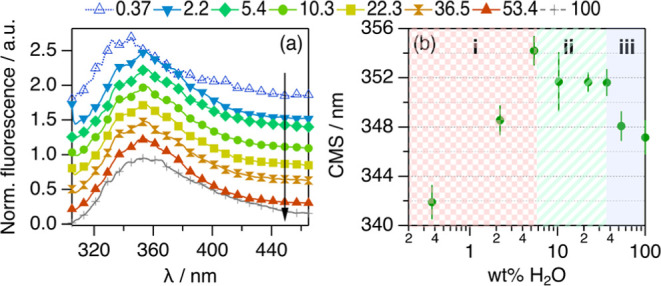
(a) Trp emission spectra from 10 μM BSA in 1:2:n ChCl:Glyc:H2O at different hydration levels expressed in wt % H2O in the solvent. The data and results for BSA aqueous buffer (100 wt % H2O) are presented for comparison. Data in (a) have been offset for clarity by +0 (100 wt % H2O), +0.25, +0.50, +0.75, +1.00, +1.25, +1.50, and +1.75 (0.37 wt % H2O). Hydration increases in the direction of the arrows. (b) CSM results derived from the analysis of the Trp emission spectra between 305 and 380 nm.
The Trp emission spectra and CSM show the same characteristic transitions observed in the UV–vis results (see Figure 5b): (i) a bathochromic shift is observed at low water content, (ii) this is followed by a hypsochromic shift when hydration is increased within intermediate water contents, and (iii) the CSM plateaus at high hydration levels at a similar value to that in the native state. These changes could be attributed to variations in the polarity of Trp’s solvation milieu or specific Trp–DES interactions. For instance, it has been shown that the choline cation interacts with the exposed Trp residues of Lyz in choline-based DESs.50,65 However, previous investigations showed that the emission wavelength of L-Trp in 1:2 ChCl:Glyc increases, at least, by 2 nm compared to that in aqueous buffer.75 Here, it is observed that the change in the emission of the Trp residues of BSA in 1:2 ChCl:Glyc is opposite to that of the isolated amino acid. As most of these residues are buried in the hydrophobic core of the protein in the native state,51,55,74 these changes cannot be solely attributed to solvent exposure and must relate to conformational transitions occurring in the protein, where the Trp side chains are relocated inside the protein envelope. In contrast, the bathochromic shifts observed at low hydration potentially relate to an increase in the solvent exposure of Trp and a more pronounced unfolding. Similarly, the hypsochromic shifts observed with further increasing hydration are associated with the refolding of BSA. Thus, these changes agree with the conformational transitions determined from the SANS data.
Notably, the boundaries of each defined region (i, ii, and iii) overlap between the different techniques (UV–vis, CD, SANS, and fluorescence). This confirms that protein concentration in the dilute regime and isotope substitution in the solvent have negligible effects on the behavior of the protein.
Protein Thermal Stability
Considering the conformational transitions of BSA, temperature-dependent CD measurements were performed to determine whether solvent hydration affects the thermal stability of the protein (see Figure S4). Increasing the temperature from 20 to 120 °C, the spectra showed a progressive reduction of the negative intensity at 208 and 222 nm. This change is associated with the loss of the defined secondary structure, α-helix and β-sheet, and the concomitant increase in the unordered content. To rigorously quantify the denaturation transition, the equilibrium fraction denatured (fD) of BSA at each temperature was calculated from the variation of the spectral signal at 222 nm (see the Supporting Information for details on these calculations). The fD as a function of temperature for each system is presented in Figure 6.
Figure 6.
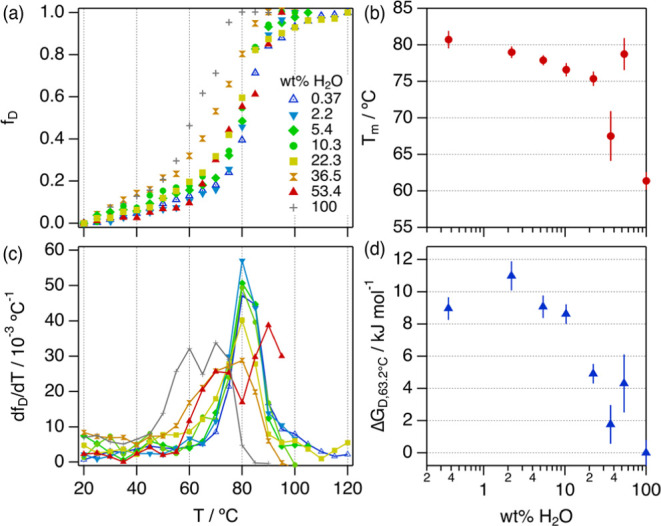
(a) Equilibrium fraction denatured and (c) first derivative of the fraction denatured for BSA in 1:2:n ChCl:Glyc:H2O at different hydration levels as a function of the temperature. The legend for the data is presented in (a). (b) Half denaturation temperature and (d) free energy of denaturation for BSA in neat and hydrated DESs at 63.2 °C. Error bars represent the standard deviation to the observed mean from three repeats performed on independent samples. Where not seen, error bars are within the markers.
The fD gradually increases when the sample temperature increases with a sharp transition at the thermal denaturation of the protein. Notably, the temperature at which the transition occurs is different for each hydration level, as observed in the first derivative of the fraction denatured (see Figure 6b). The latest transition occurs for BSA in neat ChCl:Glyc, and the earliest transition is observed for BSA in aqueous buffer. The solvents with hydrations at and below 22.3 wt % H2O show a sharp sigmoidal transition in fD and a single peak in the dfD/dT. This trend confirms that the thermal denaturation process at low hydration essentially follows a two-state unfolding mechanism.2,76,77 On the contrary, it is observed that BSA in aqueous buffer (100% H2O) and hydrated DES with 53.4 wt % H2O shows two peaks in the first derivative, potentially associated with a transition involving unfolding intermediates. More complex is the broad transition observed for BSA in the solvent containing 36.5 wt % H2O, which can be attributed to a multiple-state denaturation mechanism.78
Using a two-state transition model, the characteristic thermodynamic parameters associated with the unfolding process, that is, the half-denaturation temperature (Tm) and the free energy of denaturation at 63.2 °C (ΔGD), were determined (see Figures 6b,d and S5).76,79 For the system at high water content (36.5, 53.4, and 100 wt % H2O), the system was approximated as a two-state transition, provided that the shape of the fD does not show any sharp features in the transition region that can be attributed to the accumulation of unfolding intermediates around the Tm (e.g., molten globules and aggregates).76Tm is the highest in neat 1:2 ChCl:Glyc with a 31.5% ± 0.4% increase relative to Tm in aqueous buffer. When the DES is hydrated, Tm gradually decreases with increasing hydration between 2.2 and 36.5 wt % H2O. Surprisingly, this trend is reversed at 53.4 wt % H2O as the Tm increases by 28.3% ± 1.9% compared to the aqueous buffer. The stabilizing effect promoted by the presence of the DESs is also reflected in ΔGD. BSA in hydrated DESs shows a higher ΔGD value than that in aqueous buffer (ΔGD,63.2°C = 0 kJ/mol) in all cases, indicative of increased stability. In particular, the solvents with the lowest level of hydration, that is, between 0.37 and 10.3 wt % H2O, show the largest gain in stability with a ΔGD of ca. 9 kJ/mol. At higher hydration, the variation becomes again non-monotonic as the hydrated DES with 53.4 wt % H2O shows a higher ΔGD than that with 36.5 wt % H2O.
These results prove that the neat and hydrated DESs induce a stabilizing effect against the thermal denaturation of BSA. Two different stabilization pathways are observed. (1) At low DES hydration (<22.3 wt % H2O), BSA unfolding follows a two-state mechanism, where the protein evolves from the partially folded conformation to a denature state upon heating. Importantly, BSA shows the highest stability in the conditions investigated here in this hydration regime. (2) At high DES hydration (>36.5 wt % H2O), BSA follows a non-two-state transition. This is similar to BSA in aqueous buffer, where protein aggregation upon heating restricts the refolding of early unfolded protein and generates denaturation intermediates.78 Interestingly, the thermal stabilization at 53.4 wt % H2O potentially arises from an indirect protection mechanism as the conformation and solvation of the protein are very similar to that in the native state. However, this demonstrates that even dilute 1:2 ChCl:Glyc is remarkably capable of protecting the protein against thermal unfolding.
Conformational and Functional Recovery
To test whether the unfolding observed at low hydration, that is, regime i, led to the formation of trapped unfolded states or unfunctional aggregates, we study the reversibility of the system upon rehydration to regime iii. After stabilizing Lyz in the hydrated 1:2:1 ChCl:Glyc:H2O (5.4 wt % H2O) for 24 h and 1:2:1 ChCl:Urea:H2O (6.5 wt % H2O), where the protein remains unfolded (see Figure 1), water was added to the sample to reach 53.4 wt % H2O and 58.1 wt % H2O for the glycerol-based and urea-based DESs, respectively. At this degree of hydration, the protein is expected to regain its folded structure and activity if the unfolding at low hydration is reversible. These samples are labeled as rehydrated from hereon. Far-UV CD were used to study the secondary structure of Lyz, and activity assays were performed to study Lyz function under the different conditions.33 The results from the characterization are shown in Figure 7.
Figure 7.
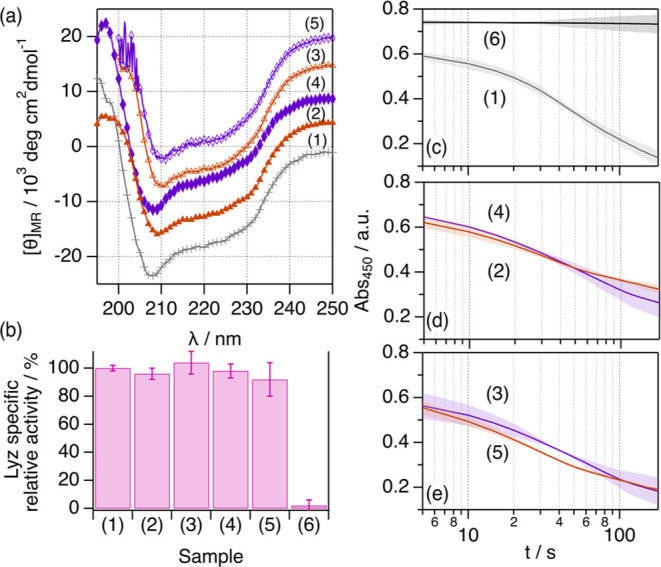
(a) Mean residue ellipticity for 107 μM Lyz in (1) aqueous buffer, (2) 1:2:20 ChCl:Glyc:H2O with 53.4 wt % H2O, (3) rehydrated 1:2:20 ChCl:Glyc:H2O to 53.4 wt % H2O, (4) 1:2:20 ChCl:Urea:H2O with 58.1 wt % H2O, and (5) rehydrated 1:2:20 ChCl:Urea:H2O to 58.1 wt % H2O. Data have been offset for clarity by +0 (aqueous buffer), +5 × 103, +10 × 103, +15 × 103, and +20 × 103 (rehydrated 1:2:20 ChCl:Urea:H2O). (b) Specific relative activities of Lyz in the different samples. Sample (6) corresponds to the data recorded for a Micrococcus lysodeikticus suspension in the absence of Lyz. The sample activities (IU mg–1 of Lyz) were normalized to the activity of Lyz in aqueous buffer. (c–e) Change in the absorbance at 450 nm of a Micrococcus lysodeikticus cell suspension upon the addition of Lyz for the spectrophotometric determination of Lyz activity in the different samples. Error bars represent the standard deviation to the observed mean from three repeats.
The estimation of the secondary structure content in the rehydrated sample shows a recovery of the local folding of Lyz, with similar populations within the error to those of the sample at high hydration (Table S8). Again, the sample containing the rehydrated urea-based DES shows a slight drop in the population of ordered structures, as seen in the hydrated 1:2:20 ChCl:Urea:H2O. Also, the addition of Lyz to the cell suspension prompts a rapid decrease in the absorbed intensity in all cases, associated with the bactericidal action of the protein. Note that the absorbed intensity of the cell suspension does not decrease in the absence of Lyz. From the initial rate of the lysis, the activity of the enzyme in the rehydrated DESs was calculated and compared to that of the systems at high hydration and to that of the enzyme in aqueous buffer. Our results show that the normalized activity values in the rehydrated samples are similar to those of Lyz in DESs at high hydration and in native conditions. Thus, the enzyme is fully active in these samples. This confirms that the unfolding observed at low water content can be reversed through the addition of water, that is, rehydration, accompanied by the recovery of the protein function.
Discussion
The re-entrant character of protein behavior suggests a complex interplay between the DES components and the protein. As the addition of water should increase the solvophobic effect and, thus, the folding tendency of the protein, an alternative mechanism is required to explain the results gathered here, where hydration cannot solely explain the behavior of the proteins.9 From these results, we propose a mechanism for protein behavior in neat and hydrated DESs.
In neat DESs, the protein undergoes small changes in the secondary structure, which alter the overall conformation of the protein to a partially folded state compared to the native folding. Molecular dynamics (MD) simulations showed that the changes are attributed to forming of a solvation matrix stabilized by hydrogen bond interactions between the protein residues and the DES constituents.51 These solvation and stabilization mechanisms were previously observed for several proteins, such as α-chymotrypsin, lysozyme, and subtilisin in glycerol-based DESs.35,36,42,80 A similar protective effect has been reported in glycerol, which also enhances protein stability.81 However, our results show that glycerol alone does not cause significant conformational changes in BSA (Figure S3, Table S7). Thus, the changes observed for the proteins in DESs must come from contributions of the ionic species in the solvent, that is, choline chloride. Furthermore, the solubilization of antibodies in choline-based ionic liquids was shown to induce conformational transitions, which could enhance the stability under certain conditions.82,83 This solubilization mechanism hinders fluctuations in the protein structure,51,66 and could also contribute to the enhanced thermal stability. These arrested dynamics could reduce the denaturation tendency despite the partially folded conformation of the protein.
Upon DES hydration, we observe a re-entrant behavior of the folding and solvation environment of the protein. We hypothesize that these changes are connected to the non-monotonic variations in the solvent nanostructure upon solvation,45−47 possibly involving non-specific (preferential solvation) and protein–DES-specific interactions, as previously observed using MD simulations of proteins in neat and diluted DESs.51,65 At low hydration levels (i, < ca. 10 wt % water) in 1:2 ChCl:Glyc, water becomes part of the DES network by displacing Cl– anions. The resulting solvent arrangement could be more prone to interact with the proteins through hydrogen bonds and electrostatic interactions, affecting their electrostatic landscape and promoting further loss of folding compared to the more charge-balanced neat DESs. Concomitantly, the pronounced exposure of hydrophobic residues increases patchy attractive interactions and promotes the formation of more transient oligomers at low hydration levels.52,84 When the water content is increased between 10 and 40 wt % (ii), water molecules dissociate the DES components into smaller clusters and fill interstitial spaces. This potentially weakens the interaction between the protein and the DES components, and water starts to solvate the protein preferentially, as observed for other small solutes such as thiocyanate.85 The weakened DES–protein paired interaction results in the gradual refolding of the protein in this solvation regime. At high hydration levels (iii, > ca. 40 wt % water), the DES components are fully dissociated and freely diffusing. As a result, we observe that the effects of the DES on the behavior of the protein vanish, and water preferentially solvates the proteins.85 This could parallel the observations of specific solvation in glycerol/water mixtures, where water preferentially solvates the protein above 40 wt % water.86 It is also important to keep in mind that the dissociated species in regime iii could effect protein behavior,45 as shown for the urea-based hydrated DES.
Unlike the effect of chemical denaturants, which reduce the stability of proteins in connection to the loss of a folded conformation,55 the glycerol-based DES and DES–water mixtures enhance BSA’s stability in a partially folded state. This behavior has been reported for molten globules in aqueous buffers, ionic liquids, and chemically engineered proteins solubilized in different media.10,77,79,87−89 However, the enhanced stability is not a general case in neoteric solvents as hydrated ionic liquids reduce the stability of the green fluoresce protein.10 These effects could be intrinsically related to the properties of the cybotactic region, as the high viscosity of a choline-based DES was shown to control the folding of DNA and the stabilization of kinetically trapped non-native folds.90 This is a similar mechanism to that observed in organic solvents, where the solubilization of proteins in anhydrous organic media significantly enhances the stability of proteins due to the restrictions in protein conformational mobility.6,7,48 However, adding water to the system facilitates conformational changes in the protein, as it happens in the DESs, which gradually decreases this stability enhancement. The addition of water to the hydrated systems can also be used to shift the conformational state of the protein. The unfolded proteins at low hydration can be refolded via rehydration, also leading to recovery of the protein function at the water content. Notably, the protein does not show any signs of aggregation at any hydration level, possibly attributed to the presence of electrostatic colloidal repulsion in the neat and hydrated DESs despite the high ionic strength of the solvent.91
Therefore, the behavior of these proteins in neat and hydrated DESs parallels that of other proteins in organic solvents and ionic liquids, where specific solvent–protein interactions and the formation of segregated solvent domains strongly correlate to the differences observed in protein conformation, stability, and function.9,10,48,49 DESs appear as promising candidates for enzymatic catalysis,92 biomolecule preservation,33 protein crystallization,50 and drug delivery,83 among others; these results will help in the rational development of pre-design solvents for optimized performance. In particular, the stabilization of antibodies, which is of great importance in pharmaceutical technology, makes DESs a potential alternative for the protection of these labile biomolecules in solution.83 In addition, future investigations involving MD simulations will help to reveal the molecular origin of the protein–solvent interactions, helping to boost those that enhance protein performance and stability.38 Notably, further developments are required to reliably incorporate biomolecules into DES-containing simulation boxes, where slight inaccuracies in the highly complex energy landscape of the solvent could override the fine energy balance that defines protein folding in these systems.
Conclusions
In summary, we demonstrate that two model proteins and an antibody undergo several transitions with the hydration of 1:2 ChCl:Glyc and 1:2 ChCl:Urea following an overarching mechanism. These changes are non-monotonic and follow a re-entrant fashion: a folded or partially folded conformation is observed in neat DES and adding small amounts of water to the system promotes further unfolding. This trend is reversed with higher hydration as the native fold is gradually retrieved above the threshold of ca. 10 wt % water in the DESs. However, it takes ca. 40 wt % water to completely retrieve the native protein folding and solvation environment in glycerol-based hydrated DES. In contrast, the urea-based DES at high hydration affects protein structure but still to a lower extent than this solvent at low hydration. This is potentially attributed to the presence of freely diffusing urea acting as a protein denaturant.
We also show that the changes induced by hydration affect the higher structure of proteins, as shown for BSA. Consequently, changes in the secondary structure correlate to transitions in the folded state and self-association of the protein. We also report that the solubilization of the protein in neat and hydrated 1:2 ChCl:Glyc prompts an increase in the thermal stability of BSA. The protein shows the highest stability at low hydration (<10 wt % water), but even the DESs in dilute conditions show a remarkable stabilizing effect against thermal denaturation. One of the interesting aspects of the conformational landscape in hydrated DESs is that transitions can be achieved with variations in the water content, for example, rehydration, leading to a fine control over the protein behavior and the stabilization of folding intermediates.
These results confirm that the conformation of the protein is strongly correlated to the hydration of DESs, where the changes in protein conformation, local folding, and solvation environment co-occur. In particular, we hypothesize that such transitions are interconnected in a way that the solvation environment of the protein, defined by non-specific preferential solvation and specific interactions between the protein and the solvent (either the DES components or water), controls the overall folding of the proteins. As the nanoscopic changes in DESs upon hydration seem to be ruled by a general mechanism, where water has a nonlinear de-structuring effect on the DES,45−47 the transitions reported here can potentially be extrapolated to other protein systems. Therefore, quantifying the water content of the system becomes imperative as unexpected changes in protein behavior may occur at different hydration levels or upon water adsorption due to the hygroscopic nature of DES.
Experimental Section
1:2 choline chloride:glycerol, its deuterated analogue and 1:2 choline chloride:urea were prepared by mixing the components and heating at 60 °C under an argon atmosphere until a colorless, transparent liquid was formed. The proteins were incorporated into the neat DESs using a freeze-drying approach on an Epsilon 2-6D LSCplus from Martin Christ. The residual water content in the neat DESs was determined using Karl–Fischer titration and found to be 0.37 ± 0.10 and 0.24 ± 0.08 wt % water. Samples containing protein in hydrated DESs were prepared by mixing an aqueous protein stock solution with the DES and water or D2O to the required protein concentration and water content. Protein concentration was determined using an ND-1000 Spectrophotometer (Saveen Werner).
SANS experiments were performed on D22 at Institut Laue-Langevin under experiment number 8-03-1049.93 Reduced data were analyzed using the indirect Fourier transform method implemented in the GNOM software.71 CD measurements were performed on a Jasco J-715, and spectra were analyzed using BeStSel.60 Temperature-dependent CD data were collected using a JASCO J-1100 equipped with a Peltier sample stage. UV–vis spectroscopy measurements were performed on a Varian Cary 50 UV–Vis Spectrometer and a JASCO V-770 UV–vis/NIR Spectrophotometer. Fluorescence spectroscopy measurements were performed on a Cary Eclipse fluorescence spectrometer. Data are openly available at DOI:10.5281/zenodo.6341232 and 10.5291/ILL-DATA.8-03-1049.93
Acknowledgments
A.S.F. acknowledges the Spanish Ministerio de Universidades for the awarded Maria Zambrano fellowship. Also, the research in this study was performed with financial support from Vinnova—Swedish Governmental Agency for Innovation Systems within the NextBioForm Competence Centre and from The Crafoord Foundation (grant #20190750). The authors thank the Institute Laue-Langevin for the awarded beamtime (8-03-1049).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c11190.
Materials and methods, UV–vis data of the proteins in the different solvents, populations of secondary structure motifs in hydrated 1:2:n ChCl:Glyc:H2O, circular dichroism of Lyz in 1:2:n ChCl:Urea:H2O, BSA conformation in glycerol with 10 wt % water, temperature-dependent CD spectra, and thermodynamic analysis of the temperature-dependent scans (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Branden C. I.; Tooze J., Folding and Flexibility. Introduction to Protein Structure, 2nd Edition ed.; Garland Science: 2012; pp 89–119. [Google Scholar]
- Rader A. J.; Hespenheide B. M.; Kuhn L. A.; Thorpe M. F. Protein unfolding: rigidity lost. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 3540–3545. 10.1073/pnas.062492699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A. Dominant forces in protein folding. Biochemistry 1990, 29, 7133–7155. 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Babine R. E.; Bender S. L. Molecular Recognition of Protein–Ligand Complexes: Applications to Drug Design. Chem. Rev. 1997, 97, 1359–1472. 10.1021/cr960370z. [DOI] [PubMed] [Google Scholar]
- Goldenzweig A.; Fleishman S. J. Principles of Protein Stability and Their Application in Computational Design. Annu. Rev. Biochem. 2018, 87, 105–129. 10.1146/annurev-biochem-062917-012102. [DOI] [PubMed] [Google Scholar]
- Zaks A.; Klibanov A. M. Enzymatic Catalysis in Organic Media at 100°C. Science 1984, 224, 1249–1251. 10.1126/science.6729453. [DOI] [PubMed] [Google Scholar]
- Klibanov A. M. Improving enzymes by using them in organic solvents. Nature 2001, 409, 241–246. 10.1038/35051719. [DOI] [PubMed] [Google Scholar]
- Schindl A.; Hagen M. L.; Muzammal S.; Gunasekera H. A. D.; Croft A. K. Proteins in Ionic Liquids: Reactions, Applications, and Futures. Front. Chem. 2019, 7, 347. 10.3389/fchem.2019.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijaya E. C.; Separovic F.; Drummond C. J.; Greaves T. L. Activity and conformation of lysozyme in molecular solvents, protic ionic liquids (PILs) and salt-water systems. Phys. Chem. Chem. Phys. 2016, 18, 25926–25936. 10.1039/c6cp03334b. [DOI] [PubMed] [Google Scholar]
- Bui-Le L.; Clarke C. J.; Bröhl A.; Brogan A. P. S.; Arpino J. A. J.; Polizzi K. M.; Hallett J. P. Revealing the complexity of ionic liquid-protein interactions through a multi-technique investigation. Commun. Chem. 2020, 3, 55. 10.1038/s42004-020-0302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B. B.; Spittle S.; Chen B.; Poe D.; Zhang Y.; Klein J. M.; Horton A.; Adhikari L.; Zelovich T.; Doherty B. W.; Gurkan B.; Maginn E. J.; Ragauskas A.; Dadmun M.; Zawodzinski T. A.; Baker G. A.; Tuckerman M. E.; Savinell R. F.; Sangoro J. R. Deep Eutectic Solvents: A Review of Fundamentals and Applications. Chem. Rev. 2021, 121, 1232–1285. 10.1021/acs.chemrev.0c00385. [DOI] [PubMed] [Google Scholar]
- Hammond O. S.; Bowron D. T.; Edler K. J. Liquid structure of the choline chloride-urea deep eutectic solvent (reline) from neutron diffraction and atomistic modelling. Green Chem. 2016, 18, 2736–2744. 10.1039/c5gc02914g. [DOI] [Google Scholar]
- Hammond O. S.; Bowron D. T.; Jackson A. J.; Arnold T.; Sanchez-Fernandez A.; Tsapatsaris N.; Garcia Sakai V.; Edler K. J. Resilience of Malic Acid Natural Deep Eutectic Solvent Nanostructure to Solidification and Hydration. J. Phys. Chem. B 2017, 121, 7473–7483. 10.1021/acs.jpcb.7b05454. [DOI] [PubMed] [Google Scholar]
- Turner A. H.; Holbrey J. D. Investigation of glycerol hydrogen-bonding networks in choline chloride/glycerol eutectic-forming liquids using neutron diffraction. Phys. Chem. Chem. Phys. 2019, 21, 21782–21789. 10.1039/c9cp04343h. [DOI] [PubMed] [Google Scholar]
- Faraone A.; Wagle D. V.; Baker G. A.; Novak E. C.; Ohl M.; Reuter D.; Lunkenheimer P.; Loidl A.; Mamontov E. Glycerol Hydrogen-Bonding Network Dominates Structure and Collective Dynamics in a Deep Eutectic Solvent. J. Phys. Chem. B 2018, 122, 1261–1267. 10.1021/acs.jpcb.7b11224. [DOI] [PubMed] [Google Scholar]
- Pandey A.; Rai R.; Pal M.; Pandey S. How polar are choline chloride-based deep eutectic solvents?. Phys. Chem. Chem. Phys. 2014, 16, 1559–1568. 10.1039/c3cp53456a. [DOI] [PubMed] [Google Scholar]
- van Osch D. J. G. P.; Dietz C. H. J. T.; Warrag S. E. E.; Kroon M. C. The Curious Case of Hydrophobic Deep Eutectic Solvents: A Story on the Discovery, Design, and Applications. ACS Sustain. Chem. Eng. 2020, 8, 10591–10612. 10.1021/acssuschemeng.0c00559. [DOI] [Google Scholar]
- Martins M. A. R.; Crespo E. A.; Pontes P. V. A.; Silva L. P.; Bülow M.; Maximo G. J.; Batista E. A. C.; Held C.; Pinho S. P.; Coutinho J. A. P. Tunable Hydrophobic Eutectic Solvents Based on Terpenes and Monocarboxylic Acids. ACS Sustain. Chem. Eng. 2018, 6, 8836–8846. 10.1021/acssuschemeng.8b01203. [DOI] [Google Scholar]
- Wu J.; Yin T. Amphiphilic Deep Eutectic Solvent Based on Lidocaine and Lauric Acid: Formation of Microemulsion and Gel. Langmuir 2022, 38, 1170–1177. 10.1021/acs.langmuir.1c02837. [DOI] [PubMed] [Google Scholar]
- Álvarez M. S.; Zhang Y. Sketching neoteric solvents for boosting drugs bioavailability. J. Controlled Release 2019, 311-312, 225–232. 10.1016/j.jconrel.2019.09.008. [DOI] [PubMed] [Google Scholar]
- Dai Y.; van Spronsen J.; Witkamp G. J.; Verpoorte R.; Choi Y. H. Natural deep eutectic solvents as new potential media for green technology. Anal. Chim. Acta 2013, 766, 61–68. 10.1016/j.aca.2012.12.019. [DOI] [PubMed] [Google Scholar]
- Macário I. P. E.; Oliveira H.; Menezes A. C.; Ventura S. P. M.; Pereira J. L.; Gonçalves A. M. M.; Coutinho J. A. P.; Gonçalves F. J. M. Cytotoxicity profiling of deep eutectic solvents to human skin cells. Sci. Rep. 2019, 9, 3932. 10.1038/s41598-019-39910-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva A.; Craveiro R.; Aroso I.; Martins M.; Reis R. L.; Duarte A. R. C. Natural Deep Eutectic Solvents - Solvents for the 21st Century. ACS Sustain. Chem. Eng. 2014, 2, 1063–1071. 10.1021/sc500096j. [DOI] [Google Scholar]
- Florindo C.; Branco L. C.; Marrucho I. M. Quest for Green-Solvent Design: From Hydrophilic to Hydrophobic (Deep) Eutectic Solvents. ChemSusChem 2019, 12, 1549–1559. 10.1002/cssc.201900147. [DOI] [PubMed] [Google Scholar]
- Anselmo A. C.; Gokarn Y.; Mitragotri S. Non-invasive delivery strategies for biologics. Nat. Rev. Drug Discovery 2019, 18, 19–40. 10.1038/nrd.2018.183. [DOI] [PubMed] [Google Scholar]
- Kist J. A.; Zhao H.; Mitchell-Koch K. R.; Baker G. A. The study and application of biomolecules in deep eutectic solvents. J. Mater. Chem. B 2021, 9, 536–566. 10.1039/d0tb01656j. [DOI] [PubMed] [Google Scholar]
- Gorke J. T.; Srienc F.; Kazlauskas R. J. Hydrolase-catalyzed biotransformations in deep eutectic solvents. Chem. Commun. 2008, 1235–1237. 10.1039/b716317g. [DOI] [PubMed] [Google Scholar]
- Gorke J. T.; Srienc F.; Kazlauskas R. J.. Deep Eutectic Solvents forCandida antarcticaLipase B-Catalyzed Reactions. Ionic Liquid Applications: Pharmaceuticals, Therapeutics, and Biotechnology; American Chemical Society, 2010; Vol. 1038, pp 169–180. [Google Scholar]
- Gorke J.; Srienc F.; Kazlauskas R. Toward advanced ionic liquids. Polar, enzyme-friendly solvents for biocatalysis. Biotechnol. Bioprocess Eng. 2010, 15, 40–53. 10.1007/s12257-009-3079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamajanov I.; Engelhart A. E.; Bean H. D.; Hud N. V. DNA and RNA in anhydrous media: duplex, triplex, and G-quadruplex secondary structures in a deep eutectic solvent. Angew. Chem., Int. Ed. Engl. 2010, 49, 6310–6314. 10.1002/anie.201001561. [DOI] [PubMed] [Google Scholar]
- Gállego I.; Grover M. A.; Hud N. V. Folding and imaging of DNA nanostructures in anhydrous and hydrated deep-eutectic solvents. Angew. Chem., Int. Ed. Engl. 2015, 54, 6765–6769. 10.1002/anie.201412354. [DOI] [PubMed] [Google Scholar]
- Svigelj R.; Dossi N.; Toniolo R.; Miranda-Castro R.; de-los-Santos-Álvarez N.; Lobo-Castañón M. J. Selection of Anti-gluten DNA Aptamers in a Deep Eutectic Solvent. Angew. Chem., Int. Ed. Engl. 2018, 57, 12850–12854. 10.1002/anie.201804860. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fernandez A.; Prevost S.; Wahlgren M. Deep eutectic solvents for the preservation of concentrated proteins: the case of lysozyme in 1 : 2 choline chloride : glycerol. Green Chem. 2022, 24, 4437–4442. 10.1039/d1gc04378a. [DOI] [Google Scholar]
- Sanchez-Fernandez A.; Jackson A. J.. Proteins in deep eutectic solvents: Structure, dynamics and interactions with the solvent. In Eutectic Solvents and Stress in Plants; Academic Press, 2021, pp 69–94. 10.1016/bs.abr.2020.09.003 [DOI] [Google Scholar]
- Sanchez-Fernandez A.; Edler K. J.; Arnold T.; Alba Venero D.; Jackson A. J. Protein conformation in pure and hydrated deep eutectic solvents. Phys. Chem. Chem. Phys. 2017, 19, 8667–8670. 10.1039/c7cp00459a. [DOI] [PubMed] [Google Scholar]
- Esquembre R.; Sanz J. M.; Wall J. G.; del Monte F.; Mateo C. R.; Ferrer M. L. Thermal unfolding and refolding of lysozyme in deep eutectic solvents and their aqueous dilutions. Phys. Chem. Chem. Phys. 2013, 15, 11248–11256. 10.1039/c3cp44299c. [DOI] [PubMed] [Google Scholar]
- Dai Y.; Witkamp G. J.; Verpoorte R.; Choi Y. H. Tailoring properties of natural deep eutectic solvents with water to facilitate their applications. Food Chem. 2015, 187, 14–19. 10.1016/j.foodchem.2015.03.123. [DOI] [PubMed] [Google Scholar]
- Ma C.; Laaksonen A.; Liu C.; Lu X.; Ji X. The peculiar effect of water on ionic liquids and deep eutectic solvents. Chem. Soc. Rev. 2018, 47, 8685–8720. 10.1039/c8cs00325d. [DOI] [PubMed] [Google Scholar]
- Wu B.-P.; Wen Q.; Xu H.; Yang Z. Insights into the impact of deep eutectic solvents on horseradish peroxidase: Activity, stability and structure. J. Mol. Catal. B: Enzym. 2014, 101, 101–107. 10.1016/j.molcatb.2014.01.001. [DOI] [Google Scholar]
- Milano F.; Giotta L.; Guascito M. R.; Agostiano A.; Sblendorio S.; Valli L.; Perna F. M.; Cicco L.; Trotta M.; Capriati V. Functional Enzymes in Nonaqueous Environment: The Case of Photosynthetic Reaction Centers in Deep Eutectic Solvents. ACS Sustain. Chem. Eng. 2017, 5, 7768–7776. 10.1021/acssuschemeng.7b01270. [DOI] [Google Scholar]
- Toledo M. L.; Pereira M. M.; Freire M. G.; Silva J. P. A.; Coutinho J. A. P.; Tavares A. P. M. Laccase Activation in Deep Eutectic Solvents. ACS Sustain. Chem. Eng. 2019, 7, 11806–11814. 10.1021/acssuschemeng.9b02179. [DOI] [Google Scholar]
- Yadav N.; Bhakuni K.; Bisht M.; Bahadur I.; Venkatesu P. Expanding the Potential Role of Deep Eutectic Solvents toward Facilitating the Structural and Thermal Stability of α-Chymotrypsin. ACS Sustain. Chem. Eng. 2020, 8, 10151–10160. 10.1021/acssuschemeng.0c02213. [DOI] [Google Scholar]
- Xin R.; Qi S.; Zeng C.; Khan F. I.; Yang B.; Wang Y. A functional natural deep eutectic solvent based on trehalose: Structural and physicochemical properties. Food Chem. 2017, 217, 560–567. 10.1016/j.foodchem.2016.09.012. [DOI] [PubMed] [Google Scholar]
- Hossain S. S.; Paul S.; Samanta A. Structural Stability and Conformational Dynamics of Cytochrome c in Hydrated Deep Eutectic Solvents. J. Phys. Chem. B 2021, 125, 5757–5765. 10.1021/acs.jpcb.1c01975. [DOI] [PubMed] [Google Scholar]
- Hammond O. S.; Bowron D. T.; Edler K. J. The Effect of Water upon Deep Eutectic Solvent Nanostructure: An Unusual Transition from Ionic Mixture to Aqueous Solution. Angew. Chem., Int. Ed. Engl. 2017, 56, 9782–9785. 10.1002/anie.201702486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A. S. D.; Craveiro R.; Duarte A. R.; Barreiros S.; Cabrita E. J.; Paiva A. Effect of water on the structure and dynamics of choline chloride/glycerol eutectic systems. J. Mol. Liq. 2021, 342, 117463. 10.1016/j.molliq.2021.117463. [DOI] [Google Scholar]
- Weng L.; Toner M. Janus-faced role of water in defining nanostructure of choline chloride/glycerol deep eutectic solvent. Phys. Chem. Chem. Phys. 2018, 20, 22455–22462. 10.1039/c8cp03882a. [DOI] [PubMed] [Google Scholar]
- Griebenow K.; Klibanov A. M. On Protein Denaturation in Aqueous–Organic Mixtures but Not in Pure Organic Solvents. J. Am. Chem. Soc. 1996, 118, 11695–11700. 10.1021/ja961869d. [DOI] [Google Scholar]
- Takekiyo T.; Yamazaki K.; Yamaguchi E.; Abe H.; Yoshimura Y. High ionic liquid concentration-induced structural change of protein in aqueous solution: a case study of lysozyme. J. Phys. Chem. B 2012, 116, 11092–11097. 10.1021/jp3057064. [DOI] [PubMed] [Google Scholar]
- Belviso B. D.; Perna F. M.; Carrozzini B.; Trotta M.; Capriati V.; Caliandro R. Introducing Protein Crystallization in Hydrated Deep Eutectic Solvents. ACS Sustain. Chem. Eng. 2021, 9, 8435–8449. 10.1021/acssuschemeng.1c01230. [DOI] [Google Scholar]
- Kumari M.; Kumari P.; Kashyap H. K. Structural adaptations in the bovine serum albumin protein in archetypal deep eutectic solvent reline and its aqueous mixtures. Phys. Chem. Chem. Phys. 2022, 24, 5627–5637. 10.1039/d1cp05829k. [DOI] [PubMed] [Google Scholar]
- Molodenskiy D.; Shirshin E.; Tikhonova T.; Gruzinov A.; Peters G.; Spinozzi F. Thermally induced conformational changes and protein-protein interactions of bovine serum albumin in aqueous solution under different pH and ionic strengths as revealed by SAXS measurements. Phys. Chem. Chem. Phys. 2017, 19, 17143–17155. 10.1039/c6cp08809k. [DOI] [PubMed] [Google Scholar]
- Barbosa L. R.; Ortore M. G.; Spinozzi F.; Mariani P.; Bernstorff S.; Itri R. The importance of protein-protein interactions on the pH-induced conformational changes of bovine serum albumin: a small-angle X-ray scattering study. Biophys. J. 2010, 98, 147–157. 10.1016/j.bpj.2009.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock J. J.; Brancaleon L. Bovine serum albumin oligomers in the E- and B-forms at low protein concentration and ionic strength. Int. J. Biol. Macromol. 2013, 53, 42–53. 10.1016/j.ijbiomac.2012.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S.; Pyne P.; Samanta N.; Ebbinghaus S.; Mitra R. K. Thermal stability modulation of the native and chemically-unfolded state of bovine serum albumin by amino acids. Phys. Chem. Chem. Phys. 2019, 22, 179–188. 10.1039/c9cp04887a. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Yu D.; Chen W.; Fu L.; Mu T. Water absorption by deep eutectic solvents. Phys. Chem. Chem. Phys. 2019, 21, 2601–2610. 10.1039/c8cp07383j. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fernandez A.; Arnold T.; Jackson A. J.; Fussell S. L.; Heenan R. K.; Campbell R. A.; Edler K. J. Micellization of alkyltrimethylammonium bromide surfactants in choline chloride:glycerol deep eutectic solvent. Phys. Chem. Chem. Phys. 2016, 18, 33240–33249. 10.1039/c6cp06053f. [DOI] [PubMed] [Google Scholar]
- Bryant S. J.; Atkin R.; Warr G. G. Effect of Deep Eutectic Solvent Nanostructure on Phospholipid Bilayer Phases. Langmuir 2017, 33, 6878–6884. 10.1021/acs.langmuir.7b01561. [DOI] [PubMed] [Google Scholar]
- Ragone R.; Colonna G.; Balestrieri C.; Servillo L.; Irace G. Determination of tyrosine exposure in proteins by second-derivative spectroscopy. Biochemistry 1984, 23, 1871–1875. 10.1021/bi00303a044. [DOI] [PubMed] [Google Scholar]
- Micsonai A.; Wien F.; Kernya L.; Lee Y. H.; Goto Y.; Réfrégiers M.; Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, E3095–E3103. 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güler G.; Vorob’ev M. M.; Vogel V.; Mäntele W. Proteolytically-induced changes of secondary structural protein conformation of bovine serum albumin monitored by Fourier transform infrared (FT-IR) and UV-circular dichroism spectroscopy. Spectrochim. Acta, Part A 2016, 161, 8–18. 10.1016/j.saa.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Kanthe A.; Ilott A.; Krause M.; Zheng S.; Li J.; Bu W.; Bera M. K.; Lin B.; Maldarelli C.; Tu R. S. No ordinary proteins: Adsorption and molecular orientation of monoclonal antibodies. Sci. Adv. 2021, 7, eabg2873 10.1126/sciadv.abg2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng L.; Wang J.; Huang M.; Xu Q.; Ma M. The changes of secondary structures and properties of lysozyme along with the egg storage. Int. J. Biol. Macromol. 2016, 92, 600–606. 10.1016/j.ijbiomac.2016.07.068. [DOI] [PubMed] [Google Scholar]
- Mitra R. K.; Sinha S. S.; Pal S. K. Hydration in protein folding: thermal unfolding/refolding of human serum albumin. Langmuir 2007, 23, 10224–10229. 10.1021/la7014447. [DOI] [PubMed] [Google Scholar]
- Kumari P.; Kumari M.; Kashyap H. K. How Pure and Hydrated Reline Deep Eutectic Solvents Affect the Conformation and Stability of Lysozyme: Insights from Atomistic Molecular Dynamics Simulations. J. Phys. Chem. B 2020, 124, 11919–11927. 10.1021/acs.jpcb.0c09873. [DOI] [PubMed] [Google Scholar]
- Monhemi H.; Housaindokht M. R.; Moosavi-Movahedi A. A.; Bozorgmehr M. R. How a protein can remain stable in a solvent with high content of urea: insights from molecular dynamics simulation of Candida antarctica lipase B in urea : choline chloride deep eutectic solvent. Phys. Chem. Chem. Phys. 2014, 16, 14882–14893. 10.1039/c4cp00503a. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fernandez A.; Edler K. J.; Arnold T.; Heenan R. K.; Porcar L.; Terrill N. J.; Terry A. E.; Jackson A. J. Micelle structure in a deep eutectic solvent: a small-angle scattering study. Phys. Chem. Chem. Phys. 2016, 18, 14063–14073. 10.1039/c6cp01757f. [DOI] [PubMed] [Google Scholar]
- Rambo R. P.; Tainer J. A. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 2011, 95, 559–571. 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatter O. The interpretation of real-space information from small-angle scattering experiments. J. Appl. Crystallogr. 1979, 12, 166–175. 10.1107/s0021889879012139. [DOI] [Google Scholar]
- Svergun D. I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992, 25, 495–503. 10.1107/s0021889892001663. [DOI] [Google Scholar]
- Petoukhov M. V.; Franke D.; Shkumatov A. V.; Tria G.; Kikhney A. G.; Gajda M.; Gorba C.; Mertens H. D.; Konarev P. V.; Svergun D. I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012, 45, 342–350. 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameseder F.; Radulescu A.; Holderer O.; Falus P.; Richter D.; Stadler A. M. Relevance of Internal Friction and Structural Constraints for the Dynamics of Denatured Bovine Serum Albumin. J. Phys. Chem. Lett. 2018, 9, 2469–2473. 10.1021/acs.jpclett.8b00825. [DOI] [PubMed] [Google Scholar]
- Bucciarelli S.; Midtgaard S. R.; Nors Pedersen M.; Skou S.; Arleth L.; Vestergaard B. Size-exclusion chromatography small-angle X-ray scattering of water soluble proteins on a laboratory instrument. J. Appl. Crystallogr. 2018, 51, 1623–1632. 10.1107/S1600576718014462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujacz A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68, 1278–1289. 10.1107/s0907444912027047. [DOI] [PubMed] [Google Scholar]
- Kadyan A.; Juneja S.; Pandey S. Photophysical Behavior and Fluorescence Quenching of l-Tryptophan in Choline Chloride-Based Deep Eutectic Solvents. J. Phys. Chem. B 2019, 123, 7578–7587. 10.1021/acs.jpcb.9b04659. [DOI] [PubMed] [Google Scholar]
- Dicko C.; Kasoju N.; Hawkins N.; Vollrath F. Differential scanning fluorimetry illuminates silk feedstock stability and processability. Soft Matter 2016, 12, 255–262. 10.1039/c5sm02036k. [DOI] [PubMed] [Google Scholar]
- Baker S. N.; McCleskey T. M.; Pandey S.; Baker G. A. Fluorescence studies of protein thermostability in ionic liquidsElectronic supplementary information (ESI) available: synthesis of [C4mpy][Tf2N]. See http://www.rsc.org/suppdata/cc/b4/b401304m/. Chem. Commun. 2004, 940–941. 10.1039/b401304m. [DOI] [PubMed] [Google Scholar]
- Luo J. J.; Wu F. G.; Yu J. S.; Wang R.; Yu Z. W. Denaturation behaviors of two-state and non-two-state proteins examined by an interruption-incubation protocol. J. Phys. Chem. B 2011, 115, 8901–8909. 10.1021/jp200296v. [DOI] [PubMed] [Google Scholar]
- Nishii I.; Kataoka M.; Goto Y. Thermodynamic stability of the molten globule states of apomyoglobin. J. Mol. Biol. 1995, 250, 223–238. 10.1006/jmbi.1995.0373. [DOI] [PubMed] [Google Scholar]
- Zhao H.; Baker G. A.; Holmes S. Protease activation in glycerol-based deep eutectic solvents. J. Mol. Catal. B: Enzym. 2011, 72, 163–167. 10.1016/j.molcatb.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm I.; Sanchez-Fernandez A.; Choi J.; Lang C.; Schagerlöf J.; Wahlgren H.; Nilsson M.; Nilsson L. The Impact of Glycerol on an Affibody Conformation and Its Correlation to Chemical Degradation. Pharmaceutics 2021, 13, 1853. 10.3390/pharmaceutics13111853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmool T. A.; Martin L. K.; Bui-Le L.; Moya-Ramirez I.; Kotidis P.; Matthews R. P.; Venter G. A.; Kontoravdi C.; Polizzi K. M.; Hallett J. P. An experimental approach probing the conformational transitions and energy landscape of antibodies: a glimmer of hope for reviving lost therapeutic candidates using ionic liquid. Chem. Sci. 2021, 12, 9528–9545. 10.1039/d1sc02520a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angsantikul P.; Peng K.; Curreri A. M.; Chua Y.; Chen K. Z.; Ehondor J.; Mitragotri S. Ionic Liquids and Deep Eutectic Solvents for Enhanced Delivery of Antibodies in the Gastrointestinal Tract. Adv. Funct. Mater. 2021, 31, 2002912. 10.1002/adfm.202002912. [DOI] [Google Scholar]
- Li W.; Persson B. A.; Morin M.; Behrens M. A.; Lund M.; Zackrisson Oskolkova M. Charge-induced patchy attractions between proteins. J. Phys. Chem. B 2015, 119, 503–508. 10.1021/jp512027j. [DOI] [PubMed] [Google Scholar]
- Sakpal S. S.; Deshmukh S. H.; Chatterjee S.; Ghosh D.; Bagchi S. Transition of a Deep Eutectic Solution to Aqueous Solution: A Dynamical Perspective of the Dissolved Solute. J. Phys. Chem. Lett. 2021, 12, 8784–8789. 10.1021/acs.jpclett.1c02118. [DOI] [PubMed] [Google Scholar]
- Hirai M.; Ajito S.; Sugiyama M.; Iwase H.; Takata S. I.; Shimizu N.; Igarashi N.; Martel A.; Porcar L. Direct Evidence for the Effect of Glycerol on Protein Hydration and Thermal Structural Transition. Biophys. J. 2018, 115, 313–327. 10.1016/j.bpj.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogan A. P. S.; Bui-Le L.; Hallett J. P. Non-aqueous homogenous biocatalytic conversion of polysaccharides in ionic liquids using chemically modified glucosidase. Nat. Chem. 2018, 10, 859–865. 10.1038/s41557-018-0088-6. [DOI] [PubMed] [Google Scholar]
- Brogan A. P.; Hallett J. P. Solubilizing and Stabilizing Proteins in Anhydrous Ionic Liquids through Formation of Protein-Polymer Surfactant Nanoconstructs. J. Am. Chem. Soc. 2016, 138, 4494–4501. 10.1021/jacs.5b13425. [DOI] [PubMed] [Google Scholar]
- Brogan A. P. S.; Siligardi G.; Hussain R.; Perriman A. W.; Mann S. Hyper-thermal stability and unprecedented re-folding of solvent-free liquid myoglobin. Chem. Sci. 2012, 3, 1839–1846. 10.1039/c2sc20143g. [DOI] [Google Scholar]
- Lannan F. M.; Mamajanov I.; Hud N. V. Human telomere sequence DNA in water-free and high-viscosity solvents: G-quadruplex folding governed by Kramers rate theory. J. Am. Chem. Soc. 2012, 134, 15324–15330. 10.1021/ja303499m. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fernandez A.; Jackson A. J.; Prévost S. F.; Doutch J. J.; Edler K. J. Long-Range Electrostatic Colloidal Interactions and Specific Ion Effects in Deep Eutectic Solvents. J. Am. Chem. Soc. 2021, 143, 14158–14168. 10.1021/jacs.1c04781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav N.; Venkatesu P. Current understanding and insights towards protein stabilization and activation in deep eutectic solvents as sustainable solvent media. Phys. Chem. Chem. Phys. 2022, 24, 13474–13509. 10.1039/d2cp00084a. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fernandez A.; Jackson A. J.; Leung A. E.; Prevost S.. Protein Conformation and Self-Association in Deep Eutectic Solvents with Tailored Hydrophobicity; Laue-Langevin, 2021; Vol. 1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


