Abstract
Hybridization outcomes vary geographically and can depend on the environment. Hybridization can also reshape biotic interactions, leading to ecological shifts. If hybrids function differently ecologically in ways that enhance or reduce fitness, and those ecological roles vary geographically, ecological factors might explain variation in hybridization outcomes. However, relatively few studies have focused on ecological traits of hybrids. We compared the feeding ecology of Catostomus fish species and hybrids by using stable isotopes (δ13C and δ15N) as a proxy for diet and habitat use, and compared two native species, an introduced species, and three interspecific hybrid crosses. We included hybrids and parental species from seven rivers where hybridization outcomes vary. Relative isotopic niches of native species varied geographically, but native species did not fully overlap in isotopic space in any river sampled, suggesting little overlap of resource use between historically sympatric species. The introduced species overlapped with one or both native species in every river, suggesting similar resource use and potential competition. Hybrids occupied intermediate, matching, or more transgressive isotopic niches, and varied within and among rivers. Ecological outcomes of hybridization varied across locations, implying that hybridization might have unpredictable, idiosyncratic ecological effects.
Keywords: Hybridization, fitness, stable isotopes, ecological interactions
Introduction
Hybridization tests reproductive isolation between diverged lineages in secondary contact (Barton and Hewitt 1989, Harrison 1990). Studies of hybridization are motivated partly by a desire for generality in our understanding of how biodiversity arises and is maintained, and increasing accessibility of large genomic datasets has led to a newfound ability to study replicate outcomes of hybridization in the wild (Narum et al. 2013, McFarlane and Pemberton 2019). However, mechanisms controlling hybridization outcomes remain unidentified in most species pairs (Gompert et al. 2017), despite evidence that hybridization outcomes often vary substantially across replicate instances of hybridization (e.g. in Ipomopsis, sculpins, mice, oaks, spruce, butterflies, toads, barn swallows, trout, and many others; Aldridge and Campbell 2009, Nolte et al. 2009, Teeter et al. 2010, Lepais and Gerber 2011, Haselhorst and Buerkle 2013, Lagache et al. 2013, Gompert et al. 2014, Arntzen et al. 2017, Scordato et al. 2017, Mandeville et al. 2019).
Hybridization outcomes are likely to be shaped by interactions between divergent parental genomes and selection on phenotypes in hybrids across different environments. Both intergenomic and intragenomic incompatibilities can influence hybridization outcomes extensively (Lindtke 2015, Moran et al. 2021). Genomic incompatibilities in hybrids typically scale with evolutionary divergence between parental species (Presgraves 2002, Bolnick and Near 2005), and in some systems hybrids between more divergent species also produce more phenotypic, functional, and ecological novelty in hybrids (Rieseberg et al. 1999, 2007, Stelkens and Seehausen 2009, Stelkens et al. 2009, Holzman and Hulsey 2017, Selz and Seehausen 2019). If genomic incompatibilities alone drive hybridization outcomes without a clear connection to ecological context, or if selection pressures act similarly in all environments, then we might expect relatively consistent outcomes of hybridization across locations, either in overall ancestry or locus-specific ancestry (e.g. Schumer et al. 2018). However, hybridization outcomes do vary in many systems, and it is likely that environment shapes hybridization in some cases. Some clear connections between environmental context and hybridization outcomes have been described (e.g. Taylor and McPhail 2000, Vines et al. 2003, Muhlfeld et al. 2017, Mandeville et al. 2019), but our understanding of the ecological context and consequences of hybridization is still largely incomplete. Not knowing how genomic outcomes link to ecological outcomes is a limitation, because ecological context can influence outcomes of hybridization both by affecting relative fitness of hybrid and parental individuals (e.g. Arnold et al. 2012) and by limiting or increasing the opportunities for hybridization (e.g. Lepais and Gerber 2011). However, few empirical studies have described variation in hybridization outcomes with precise estimates of ancestry from genomic data, and also quantified the ecological interactions that have either resulted from or shaped hybridization outcomes.
Hybridization between fish species provides examples of both environment-dependent fitness of hybrids and broader ecosystem effects of hybridization, although most of these examples come from either laboratory or mesocosm experiments rather than natural populations. For example, stickleback hybrids grow at a reduced rate and have reduced fitness in enclosures under natural conditions relative to in a laboratory setting (Hatfield and Schluter 1999). In both trout and pupfish, sympatry with hybrids has been shown to modify the fitness landscape for parental species by reducing growth, survival rate, or reproductive success of parental species through competition for resources, space, or mates (Rosenfield et al. 2004, Seiler and Keeley 2009). Phenotypes related to foraging ability can be especially important in determining outcomes of interactions between traits of hybrids and the environment. Experimental work in fish has shown that hybrids can be less successful than parental species when they have intermediate mouth morphology, because of an ecological mismatch between feeding morphology and available resources (e.g. sunfish; McGee et al. 2015) or reduced growth (e.g. sticklebacks; Arnegard et al. 2014). However, phenotypes of hybrids are hard to predict, and are not necessarily intermediate between parental species (Rieseberg et al. 1999, Stelkens et al. 2009). Instead, phenotypes of hybrid individuals might be similar to one of the parental species, or might differ from both parental species (e.g., Stelkens et al. 2009, Holzman and Hulsey 2017, Thompson et al. 2021). Hybrids can even sometimes feed more successfully on novel resources than parental species (Selz and Seehausen 2019), suggesting that under some circumstances hybrids could be better suited to their environment than parental species.
Our goal was to compare ecological interactions between multiple Catostomus fish species and their hybrid progeny across rivers in Colorado and Wyoming, incorporating sampling from multiple locations with differing hybridization outcomes, and using precise genomic ancestry estimates. The focal Catostomus species are part of a clade with extensive and highly variable interspecific hybridization between multiple species pairs (Hubbs et al. 1943, McDonald et al. 2008, Mandeville et al. 2015, 2017). Genomic outcomes of hybridization in Catostomus can vary substantially over the geographic range in which a pair of species co-occurs (Mandeville et al. 2015, 2017). Mechanisms driving this variation in hybridization outcomes remain unidentified. One hypothesis, which we explored in this study, is that geographic variation in the ecological success and survival of hybrids might cause variation in hybridization outcomes. If Catostomus hybrids were able to occupy novel ecological niches and thus gain a fitness advantage in some locations but not others, ecological variation and environment-dependent fitness of hybrids might contribute to the extreme variation we have observed in genomic outcomes of hybridization.
While some hybridization between Catostomus species is precipitated by human-caused species introductions, most notably of C. commersonii (white sucker) from east of the Continental Divide to the Upper Colorado River basin west of the Continental Divide in North America, this group of fishes likely has a long and convoluted history of hybridization (Hubbs et al. 1943, Hubbs 1955, Bangs et al. 2018), including a hypothesized allopolyploid origin (Ferris 1984). Of the three focal species in this study, C. latipinnis (flannelmouth sucker) and C. discobolus (bluehead sucker) are native to the study region and historically sympatric, while the common introduced species, white sucker, has no previous history of sympatry with the other focal species, but has been introduced to the study region within the last 150 years. Two additional species, C. catostomus (longnose sucker, introduced), and C. platyrhynchus (mountain sucker, native), were sampled at one location each, and were included in ecological analyses. Contemporary hybridization between Catostomus species is extensive; several species have hybridized with more than one congener, resulting in at least 12 different hybrid crosses between six geographically overlapping Catostomus species in the US mountain west (Mandeville et al. 2017). Previous genetic work demonstrated that species with intermediate morphology (Hubbs et al. 1943) are interspecific hybrids (McDonald et al. 2008, Mandeville et al. 2015, Bangs et al. 2018) and that hybridization varied spatially (Mandeville et al. 2017). In some locations, few crosses were sampled despite sympatry of multiple parental species; in other locations, many different crosses involving the sympatric parental species occurred. Backcrossed hybrids and later-generation recombinant hybrids were common in some locations, while other locations contained only first generation hybrids (Mandeville et al. 2015, 2017).
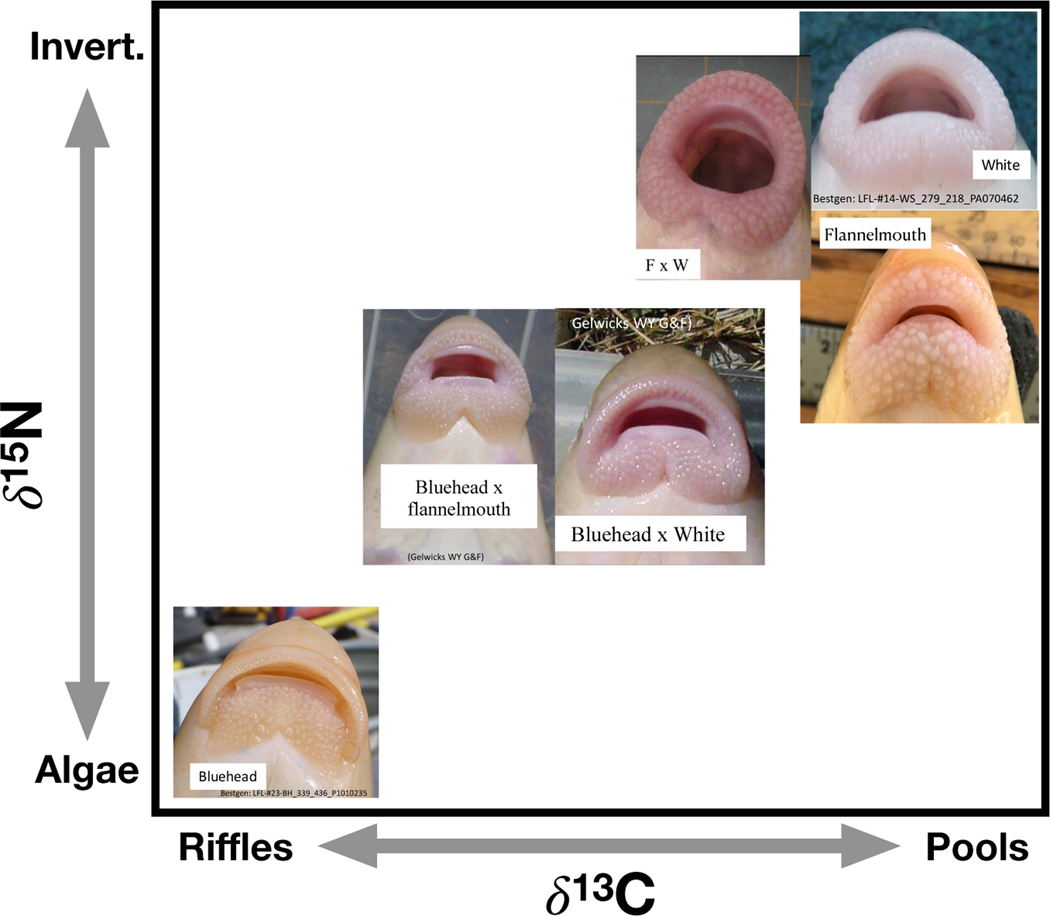
Ecological traits of these species differ. Bluehead suckers eat predominantly algae that grow in fast-moving, shallow portions of rivers (called riffles), while flannelmouth and white suckers eat more aquatic invertebrates and more often occur in deep, slow pools (Tronstad and Estes-Zumpf 2011, Cross et al. 2013, Walsworth et al. 2013). Catostomus species vary substantially in mouth morphology. Bluehead and mountain suckers have a distinctive “scraping ridge” that facilitates scraping algal biofilms from surfaces (Baxter et al. 1995, Fig. 1). In contrast, the other species have no scraping ridge, and instead have fleshy lips in varying shapes and textures (Fig. 1). Variation in dietary, behavioral, and morphological traits sets up the expectation that isotopic niche might vary by species. Nitrogen (δ15N) serves as a proxy for trophic position, and we therefore expected higher values of δ15N for fish consuming more invertebrates (Post 2002), namely flannelmouth and white suckers, than for bluehead suckers consuming more algae. Variation in carbon isotopes (δ13C) can result from intrinsic characteristics of dietary items (e.g. C3 versus C4 plants), but in rivers might also depend on patterns of CO2 exchange between water and air, source of CO2, and flow velocity over biofilms limiting mass transfer and therefore isotopic discrimination (Finlay 2001, Newsome et al. 2007). Carbon stable isotopic ratios might therefore also differ depending on how much individuals feed in riffles or pools in a river, leading to the expectation that δ13C might be lower in bluehead suckers, which favor riffles. It is also possible that more separation in δ13C could be found in rivers with more heterogeneous habitat, where there is more opportunity for divergent feeding behaviors.
Figure 1:
Hypothesized distribution of the three most widely sampled Catostomus fish species and their hybrids in isotopic space. Previous studies and mouth morphology suggested that bluehead suckers eat more algae, and feed primarily in riffles, leading to lower δ13C and δ 15N. Flannelmouth and white suckers are expected to eat more invertebrates and in pools, leading to higher δ 13C and δ 15N. Due to morphological intermediacy, bluehead×flannelmouth and bluehead × white hybrids were expected to be dietarily intermediate between parental species. Flannelmouth × white hybrids were expected to match the dietary phenotypes of their parental species. Mountain and longnose sucker were excluded from this figure because each species (and a small number of hybrids) were found in only one river each. Based on mouth morphology, mountain suckers are expected to be similar to bluehead suckers, while longnose would likely be more similar to flannelmouth and white suckers.
We used stable isotope data to identify ecological overlaps between genetically-defined parental species and hybrids (Layman et al. 2007, Newsome et al. 2007). In hybrid crosses where parental species were expected to differ in isotopic niche (e.g. bluehead×flannelmouth and bluehead×white), we expected intermediacy in diets of hybrids, based on morphology (Fig. 1) and literature on related species (Clarkson and Minckley 1988). In hybrid crosses where parental species shared a similar isotopic niche (e.g. flannelmouth×white), we expected hybrids to have similar diets to parental species (Fig. 1). Based on previous studies of Catostomus sucker ecology, we expected that species introductions and hybridization might lead to greater overlap in ecological niche (Walsworth et al. 2013). We compared body condition of parental species and hybrids as a rough estimate of foraging success and ecological suitability to available food resources. Because fertility of hybrids can vary, we consider body condition a partial fitness proxy. Ecological success of hybrids is one component of fitness, but may be independent of fertility and reproductive success in some hybrid crosses. If hybrids or introduced species were at an ecological disadvantage, we would expect to see lower body condition; however, if hybrids or introduced species were at an ecological advantage in some environments, they might be in better body condition.
Methods
We combined stable isotope and genetic analyses of 506 individual fish to identify ecological niches of Catostomus suckers with different parental species and hybrid ancestry. Sampling spanned seven different locations in the Upper Colorado River basin in Wyoming and Colorado (Table 1, Fig. 2). Fisheries biologists from state agencies collected fin tissue from each individual fish; tissue was stored in ethanol prior to use for both genetic analyses (Mandeville et al. 2015, 2017) and stable isotope analyses (this study). Ethanol preservation can distort isotopic signatures in fish tissue (Arrington and Winemiller 2002, Kelly et al. 2006), but because all fin samples were treated the same in this study, we did not correct any of the isotope ratios for preservation fractionation. Locations for isotope analysis are a subset of those used for genetic analyses, and were chosen to include rivers with a range of hybridization outcomes, based on the results of Mandeville et al. (2017).
Table 1:
Fish were sampled from seven rivers in the Upper Colorado River basin. Individuals sampled included parental species and hybrids, abbreviated in the table below. (B = blue-head, F = flannelmouth, W = white. Hybrids are represented with a × connecting parental species, i.e. flannelmouth×white = F×W).
| Location | Drainage | Individuals | Species | Hybrids (> 3 ind) |
|---|---|---|---|---|
| Big Sandy River (WY) | Green River | 61 | B, F, W | - |
| Little Sandy Creek (WY) | Green River | 80 | B, F, W | F×W |
| Escalante Creek (CO) | Gunnison | 85 | B, F, W | B×F, B×W, F×W |
| White River (CO) | White | 55 | B, F | B×F |
| Little Snake River (WY) | Yampa | 68 | F, W | B×W, F×W |
| Yampa River (CO) | Yampa | 72 | B, F, W | B×W, F×W |
| Muddy Creek (WY) | Yampa | 58 | B, W | B×W, F×W |
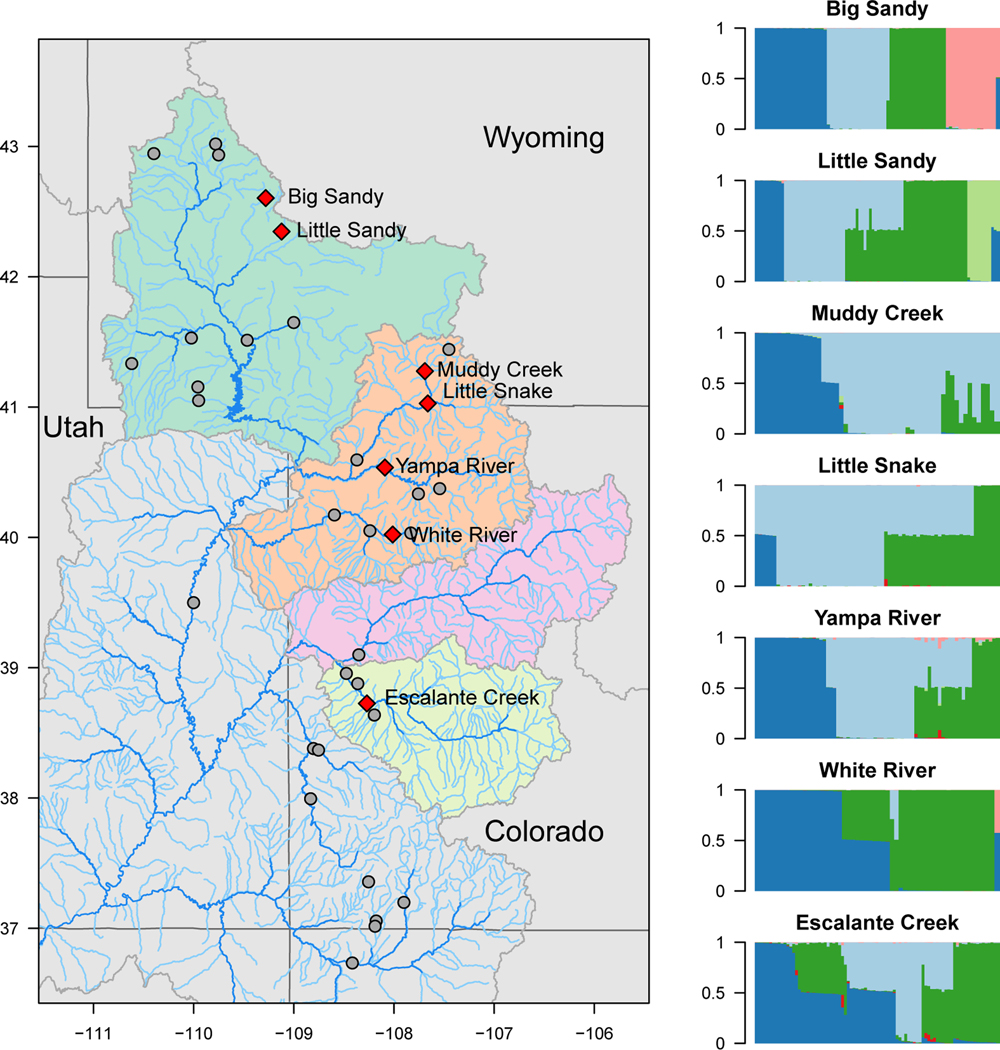
Figure 2:
Seven rivers in Wyoming and Colorado and corresponding genetics outcomes of hybridization. Gray dots on the map at left represent all sampling points from Mandeville et al. (2017); the focal rivers for this study are shown with red diamonds. Barplots at right show proportional genetic ancestry of individual fish used for stable isotope analysis (dark blue = bluehead sucker; light blue = white sucker; dark green = flannelmouth sucker; pink = longnose sucker; light green = mountain sucker). A range of hybrids between Catostomus species were present (Mandeville et al. 2017). The most common crosses were flannelmouth×white (closely related, F1 and backcrossed hybrids), and bluehead×white and bluehead×flannelmouth (more distantly related; mostly F1 hybrids).
Genomic ancestry of hybrid and parental individuals
We used results of genomic analyses from Mandeville et al. (2017) to identify ancestry of individual fish (Fig. 2). Genomic analyses of hybridization for the individuals in this study used genotyping-by-sequencing data (Parchman et al. 2012). Specifically, we relied on results from a genetic clustering algorithm, entropy (Gompert et al. 2014, Shastry et al. 2021), based on 11,221 single nucleotide polymorphisms (SNPs) to identify parental species and hybrids genetically. This analysis produced precise estimates of each individual fish’s ancestry, including proportion of ancestry from each parental species in hybrids. Estimates of interspecific ancestry from entropy also revealed extent of hybridization and backcrossing, so we know that a substantial proportion of hybrids in this study were F1 (first generation) hybrids or BC1 (backcrosses resulting from mating between an F1 hybrid and a parental species). We used both categorical designations of ancestry (species or hybrid cross) and a continuously valued measure of ancestry (proportion of ancestry) as covariates for stable isotopic ratios and body condition of hybrids in this study. We also quantified extent of backcrossing in flannelmouth×white hybrids (the most geographically widespread cross) in each river using the 95% quantiles of the distribution of proportion of ancestry (q) across hybrid More extensive discussion of genomic outcomes of hybridization in Catostomus fishes can be found in Mandeville et al. (2017).
Stable isotope analyses of ecological niche
To obtain stable isotope data, we prepared samples of fin tissue for analysis of δ13C and δ15N by first desiccating them in a drying oven (60 °C) for 2–3 days. We chose to use fin tissue because it can be sampled non-lethally and has a relatively slow turnover time, on the order of several months (comparable to muscle, a more frequently used tissue; Jardine Thomas and Crowther 2015). Recent work confirms that δ13C and δ15N are highly correlated in muscle and fin in C. commersonii and C. catostomus (Maitland and Rahel 2021), confirming that use of fin is appropriate for these study species. Dried fin samples were analyzed for δ13C and δ15N ratios the University of Wyoming Stable Isotope Facility (UW-SIF) via high temperature oxidation in a Costech 4010 elemental analyzer interfaced to a Finnigan DeltaPlus XP mass spectrometer.
For each species or hybrid cross in each river, as defined by estimates of genetic ancestry (Mandeville et al. 2017), we estimated an ellipse in dual isotopic space to quantify the relative position of δ13C and δ15N values and variation in those values within a group (Layman et al. 2007). We used the maximum likelihood standard.ellipse function in the siar package in R to fit the standard ellipses (Parnell et al. 2010, Jackson et al. 2011). This model assumes that bivariate isotopic data (isotopic ratios of δ13C and δ15N) for a group of individuals is best fit by a multivariate normal distribution, and uses sample means and a covariance matrix to estimate parameters of a standard ellipse containing 40% of the data (Jackson et al. 2011), which is analogous to a standard deviation for a univariate normal distribution. Notably, an ellipse of this size is generally consistent in area across variable sample sizes, while approaches enclosing a larger of the proportion are more sensitive to sample size variation and individual outliers. To account for potential underestimation of ellipse size at low sample sizes, an additional correction of (n-1)/(n-2) is applied to produce SEAc, a standard ellipse that is unbiased with respect to sample size. We used the SEAc version of the standard ellipse for all analyses, because simulations suggest that it is less sensitive to sample size variation and small sample size (Jackson et al. 2011), and our sample sizes varied across rivers and species sampled (Table 1). We quantified isotopic similarity of parental species and hybrids within each river using the overlap function in siar to calculate the overlap of standard ellipses (Jackson et al. 2011). We then compared area of isotopic overlap in each pair of species or hybrids across rivers to understand how consistent relative positions of species are in isotopic space. We also examined the relationship between isotopic ratios and a continuous metric of genetic ancestry, using estimates of q (proportion of ancestry) from Mandeville et al. 2017 rather than categorical binning of individuals into species or hybrid crosses.
Fin tissue was originally sampled for genetic analysis, and because of this focus no food web baseline samples for isotope analysis were collected at the time of sampling. We decided that a post-hoc sampling of the food web would not accurately represent conditions when fish were originally sampled, as food webs vary seasonally and year-to-year (Cross et al. 2013). Rivers likely varied in baseline δ13C and δ15N ratios, as well as the food resources available, making comparisons of absolute values of δ13C and δ15N across rivers inappropriate. We addressed these constraints by calculating overlaps in isotopic space only within a river, not across rivers, and considering relative positions of species and hybrids in isotopic space within a river rather than absolute values of δ13C and δ15N.
Clustering of individuals in isotopic space without respect to ancestry
Species and hybrid crosses are commonly considered as “types” in that conspecific individuals are assumed to be more similar to one another than to heterospecific individuals. However, individual variation may lead to differences among individuals of the same species, and it is unclear to what extent we should expect conspecific homogeneity in ecological niche. To assess empirical evidence for isotopic niche clustering in Catostomus species and hybrid crosses, we also assessed the distribution of individuals in isotopic space using a hierarchical Bayesian model, isoclust. The model is written in JAGS and implemented through R (code included in Supplement). This model identifies the best-supported number of distinct clusters in isotopic space for each river with no prior information about ancestry of individuals, thus quantifying how clustered individuals are in resource use without assuming that individuals of a species are similar to one another. We sequentially fit models for 1 to 6 clusters in isotopic space, where each individual is assigned categorically to a cluster, and identified the best fit of model to data using a penalized measure of model deviance (pD). This approach complements our more conventional analyses where we quantified resource use by binning individuals a priori by species. By identifying statistically how many clusters are represented in isotope space, we can better quantify distinctiveness and overlap across species and hybrid categories, rather than simply describing clusters qualitatively based on the assumption that individuals with similar ancestry will be ecologically similar (i.e., grouping by ancestry as in Fig. 3).
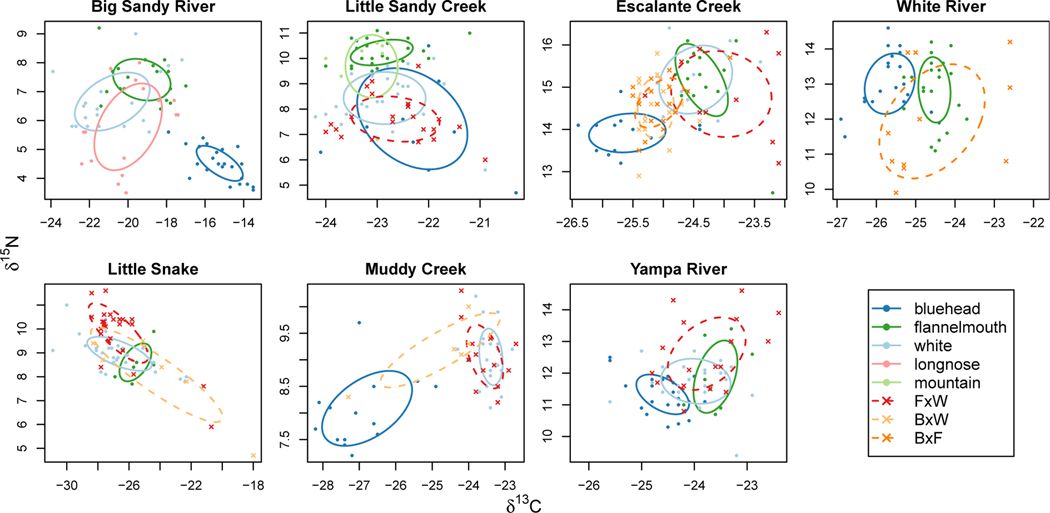
Figure 3:
Standard ellipses summarize stable isotopic ratios (δ13C and δ15N; plots oriented the same as Fig. 1) by encompassing approximately 40% of individual fish within each of five species and three hybrid crosses. Ellipses were estimated using a maximum likelihood and a bivariate normal distribution, implemented in SIAR (Jackson et al. 2011). Species ellipses are solid lines; hybrid ellipses are dashed lines. Ecological relationships between parental species varied across rivers, but the two native parental species (bluehead and flannelmouth) did not overlap in isotopic space in any sampled locations. Hybrids among these three parental species had matching, intermediate, or transgressive isotopic ratios relative to parental species, and are shown more clearly in Fig. 4.
Body condition as a measure of ecological success
We calculated body condition using length and weight data for 255 individual fish that had these data available (Bolger and Connolly 1989, Jakob et al. 1996). Body condition is sometimes used as a fitness proxy, but because hybrids can vary in fertility independent of their body condition, we interpret body condition as ecological success but do not fully equate ecological success with overall fitness. We used two methods, Fulton’s condition factor (K; Bolger and Connolly 1989) and relative weight (Wr; Wege and Anderson 1978). We also modeled body condition as a function of several predictor variables, including isotopic ratios (both carbon and nitrogen), as well as proportional ancestry from white, flannelmouth, and bluehead suckers.
Results
Isotopic niche differentiation of native species and niche overlap with introduced species
Sympatric native species (bluehead and flannelmouth) did not overlap in isotopic space, suggesting little ecological overlap in any of the five rivers where both species were sampled (Fig. 3). In two rivers (Big Sandy River and Escalante Creek), these two species were substantially separated in dual isotope plots, whereas in the other three rivers, standard ellipses for these species were directly adjacent but not overlapping (Fig. 3). Flannelmouth suckers were more enriched in nitrogen than bluehead suckers in four of five rivers where both species were sampled, mostly consistent with our expectation that flannelmouth suckers typically feed on a higher trophic level, but with some variation. In one river (Little Sandy Creek), flannelmouth suckers were differentiated from blueheads only in δ15N, not in δ13C. In another location (White River), flannelmouth and bluehead suckers were differentiated only in δ13C, not in δ15N.
In contrast to the ecological niche differentiation between native flannelmouth and blue-head suckers, white suckers, a widespread introduced species, overlapped in isotopic niche with at least one of the native species in each river (Fig. 3). In four locations (Big Sandy River, Escalante Creek, Little Snake River, and Muddy Creek), white suckers overlapped with flannelmouth suckers in isotopic space, and Little Sandy Creek, white suckers overlapped with bluehead suckers. In Yampa River, white suckers overlapped with both parental species. The variation in overlap between native and introduced species suggests different ecological effects of the white sucker introduction in different locations.
Matching, intermediate, and transgressive isotopic ratios in hybrids
Relative isotopic niches of hybrids and parental species varied, reflecting likely variation in ecological niches as well. In Escalante Creek, bluehead × white and bluehead × flannelmouth hybrids had intermediate isotopic ratios between parental species for both carbon and nitrogen (Fig. 3, Fig. 4), and also slightly overlapped with both parental species. In another river (White River), bluehead×flannelmouth hybrids overlapped with parental species in carbon isotopic ratio, but had extremely variable nitrogen ratios that were sometimes much lower than either parental species. Flannelmouth×white hybrids overlapped substantially in dual isotope plots with both white and flannelmouth suckers, suggesting that some hybrid individuals might use similar food resources to parental species. However, in each river sampled, a subset of flannelmouth × white hybrid individuals also had transgressive isotopic ratios (Fig. 4), or isotopic ratios that substantially exceeded the range of either parental species and were not intermediate between parental species. Location of transgressive hybrids relative to parental species varied substantially, suggesting a variety of ecological niches. In Escalante Creek, these transgressive hybrids were more enriched in carbon than parental species, suggesting that they might use a different carbon source, consistent with foraging in a different location. In the Yampa and Little Snake River, transgressive hybrids were more enriched in nitrogen, suggesting that they might have foraged on a higher trophic level. In Big Sandy River, transgressive hybrids had lower carbon isotopic ratios, suggesting foraging on a lower trophic level.
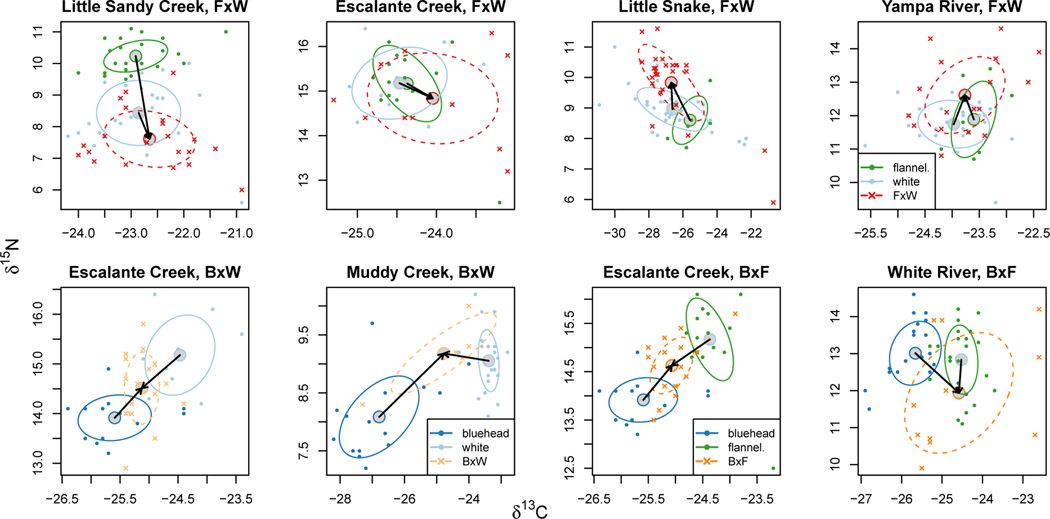
Figure 4:
Subset plots show pairwise relationships of parental species and hybrids. Means for genetically-defined species and hybrid crosses are shown with gray points; individual fish are denoted by points color-coded by species or hybrid cross. Flannelmouth×white sucker hybrids (top row) displayed matching and transgressive isotopic ratios relative to parental species, as shown by arrows from the mean values for parental species to the mean value for hybrids. Bluehead×white sucker hybrids (bottom row, left) were intermediate between isotopic values for the two parental species. Bluehead×flannelmouth sucker hybrids (bottom row, right) were intermediate between values for the two parental species in Escalante Creek, but individuals were a mixture of matching and transgressive relative to parental species in the White River.
In addition to considering hybrid individuals of each cross as a group with similar attributes, we also treated ancestry as a continuous correlate of isotopic ratios (Fig. S1, Fig. S2). Hybrids between bluehead and white sucker were mostly intermediate genetically and were first-generation hybrids; as seen using estimated standard ellipses (Fig. 4), these hybrids were usually intermediate in isotopic ratio as well. Hybrids between flannelmouth and white sucker represented more of a continuum in ancestry between the two parental species (specifically, more backcrossed hybrids; see also Fig. 2), but no clear patterns of clinal variation in isotopic ratios were evident (Fig. S1, Fig. S2). We explored modeling isotopic ratios as a function of a suite of predictor variables, including differentiation (FST) between parental species in each river (Table S1), proportional ancestry from each species, ecological differentiation between parental species, and overlap of parental species. However, only four replicate localities had complete data, which was not a sufficient level of replication to build linear models with multiple predictor variables. We therefore tested for relationships between individual predictors and isotopic signature, but no significant pairwise correlations between isotopic signatures and predictor variables were identified. Additional description of these analyses may be found in the Supplement.
Clustering of individuals in isotopic space and ecological niche overlap
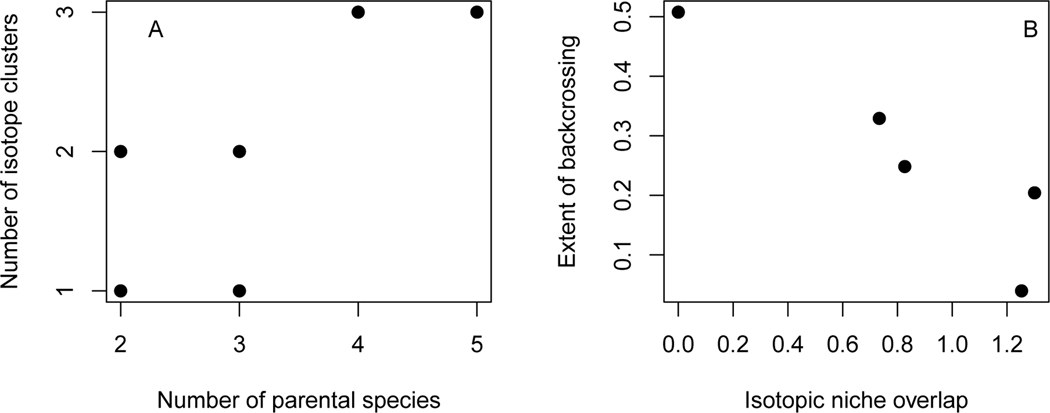
Statistically supported clusters of ecologically similar individuals did not correspond to species or hybrid crosses, nor did all individuals of a species fit into the same cluster. Models with 1–3 clusters best fit the isotopic ratio data for individual fish in each river (Fig. S3). The number of statistically supported clusters (1–3) weakly correlated with the number of species (2–5) sampled in each river (p = 0.046; Pearson correlation 0.76), but in each location, fewer clusters were identified in isotopic space than there were species present (Fig. 5A), suggesting that species do not form distinct clusters in isotope space but rather occupy continuous, overlapping isotopic niches. There was no relationship between number of genetic categories (species plus hybrid crosses) and the number of statistically defined clusters in isotope space (Fig. S4), implying that hybrids do not occupy wholly distinct isotopic niche space from parental species. Additionally, with the exception of bluehead suckers in Big Sandy River and Muddy Creek, clusters defined by our model did not correspond to species (Fig. S3). The extent of backcrossing in flannelmouth×white sucker hybridization correlated negatively with the overlap in isotopic niche area in a river; there was less backcrossing of hybrids to parental species in rivers with more ecological overlap between parental species, and more backcrossing in rivers with less ecological overlap between flannelmouth and white suckers (Fig. 5B; p = 0.026; Pearson correlation −0.92).
Figure 5:
A) Number of clusters of ecologically similar individuals (identified by a hierarchical Bayesian model) correlated with the number of parental species sampled in a river (Pearson correlation 0.764, p < 0.05). B) Extent of backcrossing in flannelmouth×white hybrids negatively correlated with isotopic niche area overlap in 6 river locations (Pearson correlation −0.922, p < 0.05).
Body condition suggests little variation in ecological success
Body condition of suckers varied among rivers but not in any particularly predictive pattern (Fig. S5, Fig. S6). Using Fulton’s condition factor, in some locations white suckers and hybrids had higher body condition than native bluehead and flannelmouth suckers (Fig. S6). Relative body condition of species differed across locations; in some locations (Escalante Creek, White River) body condition for bluehead and flannelmouth suckers were not significantly different, while in one location (Yampa River) flannelmouth suckers had better body condition than bluehead suckers (Welch Two Sample t-test, p < 0.05). These results have some caveats, notably that it is difficult to compare Fulton’s condition factor across species with slightly different body shapes. We also used relative weight, Wr, to compare body condition in flannelmouth suckers and white suckers, the two species with existing Wr equations, in three locations where adequate numbers of both species were sampled. Estimates of Wr (Fig. S6) contradicted findings using Fulton’s condition factor, and showed that flannelmouth suckers had better condition than white suckers in one location (Escalante Creek; Welch Two Sample t-test, p < 0.05), but in similar condition in the other two rivers where this both species were present. Wr was similar for flannelmouth suckers and white suckers in the Little Snake River and Yampa River. Taken together, the two metrics of body condition suggest similar ecological success across ancestry classes, with no clear patterns of differential ecological success and no clear implications for the relative fitness of hybrids and parental species. Across all species, Wr and Fulton’s condition factor were also positively correlated with δ13C (Fig. S7; Pearson correlation = 0.49 and 0.26 respectively, p<0.05 for both). This finding suggests a potential relationship between resource availability and body condition (and by extension fitness), but it would require more detailed environmental data to gain a mechanistic understanding.
Discussion
Relative ecological niche of parental species and hybrid suckers varied across locations, suggesting that ecological outcomes of hybridization varied geographically. Within each genetic category, there was also substantial variation among individuals, especially in hybrids. Importantly, hybrids did not conform neatly to our hypotheses about intermediate versus matching phenotypes relative to parental species (Fig. 1). Transgressive isotopic ratios were common in flannelmouth × white hybrids (Fig. 4). The parental species had similar isotopic niches in most rivers, but flannelmouthxwhite hybrids were transgressive in different directions in different locations. In contrast, hybrids between parental species with divergent phenotypes did tend to be more intermediate, but relative isotopic niche and degree of variation among hybrids varied geographically (Fig. 4). It is likely that hybridization leads to ecological displacement, and that availability of ecological niches could constrain hybridization under some circumstances. We suggest that there is unlikely to be a consistent ecological outcome of hybridization for the same species pair or suite of species hybridizing in multiple locations. To truly understand ecological effects and environmental dependence of hybridization, we will need to prioritize studies that address both ecological and genomic outcomes of hybridization across locations, including quantifying relative fitness of parental species and hybrids.
Native species did not overlap in isotopic niche, but introduced species and hybrids overlapped substantially
Overlap among species in isotopic space varied across rivers, as did relative positions in isotopic space. However, one consistent trend was that bluehead and flannelmouth suckers, the two native parental species, did not overlap substantially in isotopic space (Fig. 3). In three of five rivers with >1 statistically supported cluster, individuals from these two species were also largely but not entirely assigned to different clusters in our hierarchical Bayesian clustering algorithm (Fig. S3). These results suggest that bluehead and flannelmouth sucker diets generally did not substantially overlap where the species co-occurred. Both species are historically native to the study rivers, and it is possible that non-overlapping isotopic niches (and diets) have evolved to avoid competition, consistent with ecological character displacement in response to competition (Connell 1980, Robinson and Wilson 1994), which can contribute to evolutionary origins and maintenance of species diversity (Schluter 2000, Seehausen 2007).
In contrast, introduced white suckers overlapped in isotopic niche space with flannelmouth, bluehead, or with both species, suggesting some degree of shared resource use between white sucker and one or both native species in each river. Overlap of white suckers with bluehead suckers was unexpected because we anticipated white suckers would be more similar ecologically to flannelmouth than to bluehead suckers, due to mouth morphology (Fig. 1) and previous work showing that white suckers preferentially eat invertebrates rather than algae (Baxter et al. 1995, Cross et al. 2013, Walsworth et al. 2013). These results suggest that different native species might face competition from introduced white suckers in different rivers. Variable position of white suckers relative to the other species in this study also suggests that the resource use of white suckers might be flexible or opportunistic (as in Corse et al. 2009). If this pattern were true, then ecological flexibility and generalist traits of white suckers might have facilitated successful invasion of the Upper Colorado River basin, and might explain to some extent the success of hybrid individuals with white sucker ancestry.
Intermediate, matching, and transgressive phenotypes in hybrids
Hybrids also occupied different isotopic niches relative to parental species in different locations (Fig. 3). Hybridization of bluehead suckers with both flannelmouth and white suckers produced hybrids that were on average intermediate in isotopic ratios relative to parental species, as anticipated based on predictions of intermediate mouth morphology, although there was substantial variation across individuals (Fig. 1, Fig. 4). Some individual bluehead × white and bluehead × flannelmouth sucker hybrids also matched or were more similar to parental phenotypes in isotopic space, suggesting ecological similarity to parental species, consistent with the idea that in some hybrid crosses, traits of one parental species are dominant even in F1 hybrids (Thompson et al. 2021). Bluehead×white and bluehead × flannelmouth hybrids typically have intermediate mouth morphology, including a reduced scraping ridge (Hubbs et al. 1943, Hubbs 1955, Quist et al. 2009), and these traits likely drive intermediacy in diet, similar other fish species. Both stickleback and centrarchid fish hybrids use intermediate food sources when they have intermediate feeding morphology (Arnegard et al. 2014, McGee et al. 2015), with variable ecological success depending on the quantity and quality of food resources available. A parental-like feeding phenotype might be advantageous to hybrid individuals if parental phenotypes are optimal for feeding on available resources (as in Arnegard et al. 2014), but also might lead to competition between the parental species and their hybrid offspring if preferred resources are limited (as in Seiler and Keeley 2009). If overlap in isotopic space does correspond to consumption of similar food resources in these rivers, competition between hybrids and parental species might occur. Competition might therefore be a serious concern for conservation of native Catostomus species in locations where hybridization is ongoing and resources are limited. We currently do not know to what extent fish production in these rivers is food limited (sensu Cross et al. 2013), so we cannot infer how strong selective pressures associated with competition are, but matching ecological phenotypes suggests that competition is possible.
We observed transgressive isotopic ratios in a subset of flannelmouth× white hybrid individuals (Fig. 4). These individuals exceeded values for any other Catostomus species in these rivers, including parental species of the hybrids, potentially pointing to novel food or habitat use. Hybrids were transgressive in different directions in different rivers (Fig. 4). Without additional data on stomach contents or isotopic baselines, we cannot pinpoint specific causes for these transgressive isotopic values, but we highlight some potential explanations. Hybrids with transgressive δ15N probably feed on a higher trophic level, potentially eating more invertebrates than flannelmouth or white suckers, or consuming some other resource like larval fish. Transgressive values for δ15N could also correspond to use of a different portion of the river (e.g., a deep pool where denitrification is occurring), but nitrogen isotope values are more commonly interpreted as a rough indicator of trophic position (Post 2002). Transgressive values for δ13C likely correspond to use of different carbon resources, potentially in a different part of the river (Finlay 2001), consistent with possible divergence in habitat use. One caveat associated with interpretation of δ13C separation in isotope space more generally, however, is that habitat heterogeneity is required for divergence in δ13C in association with different foraging behavior to emerge. More geomorphic and hydraulic data would be required to better assess how habitat heterogeneity would shape isotopic ratios, and to understand trends in δ13C across rivers.
Novel phenotypes and resource use in hybrids are well-documented in other organisms (e.g. sunflowers, butterflies, and cichlids; Rieseberget al. 1999, Gompert et al. 2006, Stelkens and Seehausen 2009), and have promoted evolutionary diversification in diverse groups of organisms (e.g. Rieseberg et al. 2003, Marques et al. 2019, Meier et al. 2019). Some transgressive hybrids might be better able to use novel resources than parental species, and might therefore be better poised to take advantage of ecological opportunity (Selz and Seehausen 2019). In cichlid fishes, likelihood of transgressive phenotypes is correlated with evolutionary distance (Stelkens et al. 2009) in some cases, but in contrast, trophic morphology of hybrids can be more transgressive in closely related hybridizing species pairs (Holzman and Hulsey 2017). In Catostomus fishes the transgressive hybrids resulted from hybridization between closely related and ecologically similar taxa (flannelmouth and white suckers), while more evolutionarily distant crosses (bluehead × white and bluehead × flannelmouth) produced intermediate or matching hybrids. Transgressive phenotypes in hybrids between closely related and ecologically similar species (flannelmouth × white) suggest that ecological competition and character displacement could help drive this pattern (Connell 1980, Robinson and Wilson 1994).
Food web effects of hybridization
Ecological outcomes of hybridization varied geographically, but some trends were apparent from our comparison of ecological and genomic outcomes of hybridization. In locations where flannelmouth and white sucker (the two most widespread parental species), overlapped more in isotopic space, there were fewer backcrossed hybrids and hybridization was mostly constrained to first generation hybrids (Fig. 5B). This pattern might reflect filtering of hybridization outcomes by environmental conditions and available niche space, if backcrossed hybrids are unable to compete in locations where parental species have greater overlap and therefore those hybrids do not survive to be sampled as adults. However, the causality could run the other direction: hybridization might entirely reshape food webs, promoting greater niche overlap. Non-native species commonly alter food web dynamics in freshwater fish communities, including isotopic niche (Sagouis et al. 2015, Rogosch and Olden 2020), but the role of hybrids is less well documented. One study of Catostomus ecology that used stable isotopes, Walsworth et al. (2013), showed that introduction of white suckers and other non-native species into tributaries of the Upper Colorado River basin could increase food chain length and increase niche crowding, consistent with our findings here. However, Walsworth et al. 2013 excluded hybrids, which make up a large fraction of the Catostomus fish present in many parts of the Upper Colorado River basin (Gill et al. 2007). In our study, although the number of identified distinct clusters in isotopic space was correlated with number of parental species (Fig. 5A), the same relationship did not hold for number of clusters and number of ancestry classes, i.e., parental species plus hybrid crosses. This finding suggests that hybrids were not likely to be accessing different resources or occupying distinct available niches, except perhaps in the case of individuals with transgressive phenotypes, but rather were overlapping with ecological resource use of parental species, with the potential for competition.
It is unclear what fitness consequences these interactions between parental species and hybrids might have. Attempts to use body condition as a measure of ecological success and proxy for fitness produced equivocal results, because different measures of body condition produced opposing patterns (Fig. S5, Fig. S6). These contradictory results illustrate some of the difficulties associated with estimating body condition (Peig and Green 2010), and suggest caution is warranted in attaching too much weight to body condition as a component of fitness. However, based on the lack of clear signal in our two metrics of body condition, it is likely that hybrids and parental species are in similar body condition, indicating that no particular species or hybrid cross is limited by fit to ecological resources.
More empirical studies of fitness in hybrid individuals, including Catostomus hybrids, will be needed to develop a mechanistic understanding of hybridization. Clarifying the fitness of hybrids is essential because relative fitness of parental species and hybrids has a profound effect on how hybridization influences the evolution of species (reviewed in Arnold and Hodges 1995, Arnold et al. 2012). In some instances of interspecific hybridization, hybrids survive at higher rates than either parental species (as in tiger salamanders, Fitzpatrick and Shaffer 2007). However, it is important to distinguish between the well-being of individuals—i.e., survival, growth, and foraging success—and reproductive success. Survival and growth cannot be equated with fitness if hybrids cannot reproduce, and there are many examples of hybrid sterility (e.g. Sweigart et al. 2006, Good et al. 2008), but also of viable, fertile offspring between quite divergent lineages. Generally, viability and fertility of hybrids is negatively correlated with genetic divergence (Bolnick and Near 2005, Stelkens et al. 2010), with additional genomic incompatibilities more likely with increased evolutionary distance (Moran et al. 2021). If genomic incompatibilities make some hybrid crosses inviable, fitness related to ecological success might be less important for these species pairs. There are frequently fitness differences between first-generation hybrids and later generation hybrids (i.e. heterosis in F1 hybrids). For example, in native westslope cutthroat trout (Oncorhynchus clarkii lewisi) and introduced rainbow trout (Oncorhynchus mykiss) hybrids, F\ hybrids had relatively high fitness, including high reproductive success, but later-generation hybrids had markedly lower fitness (Muhlfeld et al. 2009). Studies of fitness in wild populations are logistically challenging, and have not yet been completed in Catostomus suckers, but would substantially improve our understanding of the causes and consequences of interspecific hybridization.
Conservation implications
Bluehead and flannelmouth suckers remain the target of extensive management and conservation efforts (Gelwicks et al. 2009, Senecal et al. 2010). Previous work on genomic outcomes of hybridization suggested that hybridization might act as a demographic sink due to reduced conspecific reproduction and apparent absence of hybrids beyond the F1 generation in some crosses (Mandeville et al. 2015, 2017). We can also now infer some potential ecological effects of hybridization. Hybrids and non-native parental species overlapped substantially in isotopic space with native parental species, which suggests that hybrids and introduced species occupied similar ecological niches to native species, and therefore might compete for resources. Additionally, in some rivers hybrids and non-native species had higher body condition than native species by one metric, suggesting that hybrids and introduced species might sometimes exploit resources better than parental species. When combined, these lines of evidence suggest additional potential obstacles to conservation of native species: hybrids and introduced species are likely to compete with native species, but hybrids do not contribute substantially to the gene pool. Thus, hybridization might sometimes be both a demographic sink and a resource sink. However, the variability uncovered by both this study and genetic studies of hybridization suggest that these dynamics might vary substantially by river, making outcomes of hybridization more complicated to predict. If we had studied these ecological dynamics in a single river, our conclusions would have been incorrect; it is critical to recognize that ecological outcomes of hybridization, like genomic outcomes, may vary substantially and might require different conservation strategies in different locations.
Conclusion
In this study, we showed that ecological niches occupied by hybrid individuals varied within and across rivers. Across multiple locations, hybrids of similar ancestry had variable feeding ecology, either matching parental species isotopic ratios, intermediate between parental species, or displaying a transgressive isotopic ratio beyond the range of either parental species. This variation might have arisen due to greater plasticity in hybrid feeding phenotypes, or might suggest that postzygotic selection constrained hybrids to the range of phenotypes that would be ecologically successful in a given location. Additionally, differing genetic composition of hybrids, either due to selection on recombinant hybrid genotypes or associated with locus-specific genotypes at functionally important loci, could contribute to variation in ecological outcomes of hybridization. We also suggest that overall food web structure might either influence hybridization, or be altered by the presence of hybrids and non-native species. Together, these results make a compelling case for more study of how ecological interactions can constrain or facilitate hybridization, and how hybridization might alter ecological function of communities and modify fitness landscapes for hybridizing species.
Supplementary Material
Acknowledgments
Isotope analysis was funded by the Biodiversity Institute at the University of Wyoming (Biodiversity Research Grant to EGM). Genomic data were generated with funding through the Wyoming Game and Fish Department (State Wildlife Grant #001793 and Bureau of Land Management Cooperative Agreement 12AC20048) and Colorado Parks and Wildlife (Species Conservation Trust Fund project SCTF001C). EGM was supported in part by NIH INBRE funding to the University of Wyoming (NCRR P20RR016474/NIGMS P20GM103432) and by the UW Biodiversity Institute. Computing was accomplished with an allocation from from the University of Wyoming’s Advanced Research Computing Center, on its Mount Moran IBM System X cluster (http://n2t.net/ark:/85786/m4159c) and Teton Intel x86_64 cluster (https://doi.org/10.15786/M2FY47) and through a RRG Allocation on Compute Canada’s Graham cluster to E.G. Mandeville. We thank the Wyoming Game and Fish Department and Colorado Parks and Wildlife for providing samples and supporting our work on Catostomus hybridization, most notably Mark Smith, Kevin Gelwicks, Bobby Compton, and Kevin Thompson. We also thank the staff of the Stable Isotope Facility at the University of Wyoming, especially Chandelle MacDonald, for their assistance and expert advice. We thank Kevin Bestgen, Kevin Gelwicks, Kevin Thompson, Zachary Hooley-Underwood, and Gwen Harris for permission to use the photos in Fig. 1. This manuscript was improved by comments from the Walters lab at the University of Wyoming, the Mandeville lab at the University of Guelph, and feedback from anonymous reviewers.
Data accessibility
Isotope data and genetic summary statistics presented in this paper are available on Dryad at https://doi.org/10.5061/dryad.0cfxpnw1f. R scripts used in analysis are available on Zenodo at https://doi.org/10.5281/zenodo.6958091. Genetic data associated with this paper are archived in connection with the publication of Mandeville et al. 2017.
References
- Aldridge G, and Campbell DR 2009. Genetic and morphological patterns show variation in frequency of hybrids between Ipomopsis (Polemoniaceae) zones of sympatry. Heredity 102:257–265. [DOI] [PubMed] [Google Scholar]
- Anderson R, and Neumann RM 1996. Length, weight, and associated structural indices. Pages 447–482 in Fisheries Techniques, 2nd Edition. American Fisheries Society. [Google Scholar]
- Arnegard ME, McGee MD, Matthews B, Marchinko KB, Conte GL, Kabir S, Bedford N, Bergek S, Chan YF, Jones FC, Kingsley DM, Peichel CL, and Schluter D. 2014. Genetics of ecological divergence during speciation. Nature 511:307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML, Ballerini ES, and Brothers AN 2012. Hybrid fitness, adaptation and evolutionary diversification: lessons learned from Louisiana Irises. Heredity 108:159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML, and Hodges SA 1995. Are natural hybrids fit or unfit relative to their parents? Trends in Ecology & Evolution 10:67–71. [DOI] [PubMed] [Google Scholar]
- Arntzen JW, de Vries W, Canestrelli D, and Martinez-Solano I. 2017. Hybrid zone formation and contrasting outcomes of secondary contact over transects in common toads. Molecular Ecology 26:5663–5675. [DOI] [PubMed] [Google Scholar]
- Arrington DA, and Winemiller KO 2002. Preservation Effects on Stable Isotope Analysis of Fish Muscle 131:337–342. [Google Scholar]
- Bangs MR, Douglas MR, Mussmann SM, and Douglas ME 2018. Unraveling historical introgression and resolving phylogenetic discord within Catostomus (Osteichthys: Catostomidae). BMC Evolutionary Biology 18:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, and Hewitt GM 1989. Adaptation, speciation and hybrid zones. Nature 341:497–503. [DOI] [PubMed] [Google Scholar]
- Baxter GT, Stone MD, and Parker L. 1995. Fishes of Wyoming. Wyoming Game and Fish Department Cheyenne. [Google Scholar]
- Bister TJ, Willis DW, Brown ML, Jordan SM, Neumann RM, Quist MC, and Guy CS 2000. Proposed Standard Weight (W s ) Equations and Standard Length Categories for 18 Warmwater Nongame and Riverine Fish Species. North American Journal of Fisheries Management 20:570–574. [Google Scholar]
- Bolger T, and Connolly PL 1989. The selection of suitable indices for the measurement and analysis of fish condition. Journal of Fish Biology 34:171–182. [Google Scholar]
- Bolnick DI, and Near TJ 2005. Tempo of hybrid inviability in centrarchid fishes (Teleostei: Centrarchidae). Evolution 59:1754–1767. [PubMed] [Google Scholar]
- Clarkson RW, and Minckley WL 1988. Morphology and Foods of Arizona Catosto- mid Fishes: Catostomus insignis, Pantosteus clarki, and Their Putative Hybrids. Copeia 1988:422. [Google Scholar]
- Connell JH 1980. Diversity and the Coevolution of Competitors, or the Ghost of Competition Past. Oikos 35:131–138. [Google Scholar]
- Corse E, Costedoat C, Pech N, Chappaz R, Grey J, and Gilles A. 2009. Trade-off between morphological convergence and opportunistic diet behavior in fish hybrid zone. Frontiers in zoology 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross WF, Baxter CV, Rosi-Marshall EJ, Hall RO, Kennedy TA, Donner KC, Kelly HAW, Seegert SEZ, Behn KE, and Yard MD 2013. Food-web dynamics in a large river discontinuum. Ecological Monographs 83:311–337. [Google Scholar]
- Didenko AV, Bonar SA, and Matter WJ 2004. Standard Weight (W s ) Equations for Four Rare Desert Fishes. North American Journal of Fisheries Management 24:697–703. [Google Scholar]
- Ferris SD 1984. Tetraploidy and the evolution of the catostomid fishes. Pages 55–93 in Evolutionary genetics of fishes. Springer, Boston. [Google Scholar]
- Finlay JC 2001. Stable-carbon-isotope ratios of river biota: Implications for energy flow in lotic food webs. Ecology 82:1052–1064. [Google Scholar]
- Fitzpatrick BM, and Shaffer HB 2007. Hybrid vigor between native and introduced salamanders raises new challenges for conservation. Proceedings of National Academy of Sciences 104:15793–15798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelwicks KR, Gill CJ, Kern AI, and Keith R. 2009. Current Status of Roundtail Chub, Flannelmouth Sucker and Bluehead Sucker in the Green River Drainage of Wyoming. Wyoming Game and Fish Department, Fish Division. [Google Scholar]
- Gill CJ, Gelwicks KR, and Keith RM 2007. Current distribution of bluehead sucker, flannelmouth sucker, and roundtail chub in seven subdrainages of the Green River, Wyoming. Page 121 in American Fisheries Society Symposium. Vol. 53. American Fisheries Society. [Google Scholar]
- Gompert Z, Fordyce JA, Forister ML, Shapiro AM, and Nice CC 2006. Homoploid hybrid speciation in an extreme habitat. Science 314:1923–1925. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Lucas LK, Buerkle CA, Forister ML, Fordyce JA, and Nice CC 2014. Admixture and the organization of genetic diversity in a butterfly species complex revealed through common and rare genetic variants. Molecular Ecology 23:4555–4573. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Mandeville EG, and Buerkle CA 2017. Analysis of Population Genomic Data from Hybrid Zones. Annual Review of Ecology Evolution and Systematics 48:227–229. [Google Scholar]
- Good JM, Handel MA, and Nachman MW 2008. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution 62:50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG 1990. Hybrid zones: windows on evolutionary process. Oxford Surveys in Evolutionary Biology 7:69–128. [Google Scholar]
- Haselhorst MSH, and Buerkle CA 2013. Population genetic structure of Picea engelmannii, P. glauca and their previously unrecognized hybrids in the central Rocky Mountains. Tree Genetics & Genomes 9:669–681. [Google Scholar]
- Hatfield T, and Schluter D. 1999. Ecological speciation in sticklebacks: environment-dependent hybrid fitness. Evolution pages 866–873. [DOI] [PubMed] [Google Scholar]
- Holzman R, and Hulsey CD 2017. Mechanical Transgressive Segregation and the Rapid Origin of Trophic Novelty. Scientific Reports 7:40306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbs CL 1955. Hybridization between fish species in nature. Systematic zoology pages 1–20. [Google Scholar]
- Hubbs CL, Hubbs LC, and Johnson RE 1943. Hybridization in Nature Between Species of Catostomid Fishes. University of Michigan Press. [Google Scholar]
- Jackson AL, Inger R, Parnell AC, and Bearhop S. 2011. Comparing isotopic niche widths among and within communities: SIBER-Stable Isotope Bayesian Ellipses in R. Journal of Animal Ecology 80:595–602. [DOI] [PubMed] [Google Scholar]
- Jakob EM, Marshall SD, and Uetz GW 1996. Estimating Fitness : A Comparison of Body Condition Indices. Oikos 77:61–67. [Google Scholar]
- Jardine TD 2011. A non-lethal sampling method for stable carbon and nitrogen isotope studies of tropical fishes. Marine & freshwater research 62:83–90. [Google Scholar]
- Kelly B, Dempson JB, and Power M. 2006. The effects of preservation on fish tissue stable isotope signatures. Journal of Fish Biology 69:1595–1611. [Google Scholar]
- Lagache L, Klein EK, Guichoux E, and Petit RJ 2013. Fine-scale environmental control of hybridization in oaks. Molecular Ecology 22:423–436. [DOI] [PubMed] [Google Scholar]
- Layman CA, Arrington DA, Montana CG, and Post DM 2007. Can stable isotope ratios provide for community-wide measures of trophic structure? Ecology 88:42–48. [DOI] [PubMed] [Google Scholar]
- Lepais O, and Gerber S. 2011. Reproductive patterns shape introgression dynamics and species succession within the European white oak species complex. Evolution 65:156–170. [DOI] [PubMed] [Google Scholar]
- Lindtke D. 2015. The genetic architecture of hybrid incompatibilities and their effect on barriers to introgression in secondary contact Evolution 69:1987–2004. [DOI] [PubMed] [Google Scholar]
- Maitland BM, and Rahel FJ 2021. Nonlethal Fin Sampling of North American Freshwater Fishes for Food Web Studies Using Stable Isotopes. North American Journal of Fisheries Management 41:410–420. [Google Scholar]
- Mandeville E, Parchman T, Song S, Thompson K, Compton R, Gelwicks K, and Buerkle C. 2017. Inconsistent reproductive isolation revealed by interactions between Catostomus fish species. Evolution Letters 1:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandeville EG, Parchman TL, McDonald DB, and Buerkle CA 2015. Highly variable reproductive isolation among pairs of Catostomus species. Molecular Ecology 24:1856–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandeville EG, Walters AW, Nordberg BJ, Higgins KH, Burckhardt JC, and Wagner CE 2019. Variable hybridization outcomes in trout are predicted by historical fish stocking and environmental context. Molecular Ecology 28:3738–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques DA, Lucek K, Sousa VC, Excoffier L, and Seehausen O. 2019. Admixture between old lineages facilitated contemporary ecological speciation in Lake Constance stickleback. Nature Communications 10:4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald DB, Parchman TL, Bower MR, Hubert WA, and Rahel FJ 2008. An introduced and a native vertebrate hybridize to form a genetic bridge to a second native species. Proceedings of National Academy of Sciences 105:10837–10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane SE, and Pemberton JM 2019. Detecting the True Extent of Introgression during Anthropogenic Hybridization. Trends in Ecology and Evolution 34:315–326. [DOI] [PubMed] [Google Scholar]
- McGee MD, Reustle JW, Oufiero CE, and Wainwright PC 2015. Intermediate Kinematics Produce Inferior Feeding Performance in a Classic Case of Natural Hybridization. The American Naturalist 186:807–814. [DOI] [PubMed] [Google Scholar]
- Meier JI, Stelkens RB, Joyce DA, Mwaiko S, Phiri N, Schliewen UK, Selz OM, Wagner CE, Katongo C, and Seehausen O. 2019. The coincidence of ecological opportunity with hybridization explains rapid adaptive radiation in Lake Mweru cichlid fishes. Nature Communications 10:5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran BM, Payne C, Langdon Q, Powell DL, Brandvain Y, and Schumer M. 2021. The genomic consequences of hybridization. eLife 10:e69016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlfeld CC, Kalinowski ST, McMahon TE, Taper ML, Painter S, Leary RF, and Allendorf FW 2009. Hybridization rapidly reduces fitness of a native trout in the wild. Biology Letters 5:328–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlfeld CC, Kovach RP, Al-Chokhachy R, Amish SJ, Kershner JL, Leary RF, Lowe WH, Luikart G, Matson P, Schmetterling DA, Shepard BB, Westley PAH, Whited D, Whiteley A, and Allendorf FW 2017. Legacy introductions and climatic variation explain spatiotemporal patterns of invasive hybridization in a native trout. Global Change Biology 23:4663–4674. [DOI] [PubMed] [Google Scholar]
- Narum SR, Buerkle CA, Davey JW, Miller MR, and Hohenlohe PA 2013. Genotyping-by-sequencing in ecological and conservation genomics. Molecular Ecology 22:2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome SD, Del Rio CM, Bearhop S, and Phillips DL 2007. A niche for isotopic ecology. Frontiers in Ecology and the Environment 5:429–436. [Google Scholar]
- Nolte AW, Gompert Z, and Buerkle CA 2009. Variable patterns of introgression in two sculpin hybrid zones suggest that genomic isolation differs among populations. Molecular Ecology 18:2615–2627. [DOI] [PubMed] [Google Scholar]
- Parchman TL, Gompert Z, Mudge J, Schilkey F, Benkman CW, and Buerkle CA Genome-wide association genetics of an adaptive trait in lodgepole pine. Molecular Ecology 21:2991–3005. [DOI] [PubMed] [Google Scholar]
- Parnell AC, Inger R, Bearhop S, and Jackson AL 2010. Source partitioning using stable isotopes: Coping with too much variation. PLoS ONE 5:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peig J, and Green AJ 2010. The paradigm of body condition: A critical reappraisal of current methods based on mass and length. Functional Ecology 24:1323–1332. [Google Scholar]
- Post DM 2002. The long and short of food-chain length. Trends in Ecology and Evolution 17:269–277. [Google Scholar]
- Presgraves DC 2002. Patterns of postzygotic isolation in {L}epidoptera. Evolution 56:1168–1183. [DOI] [PubMed] [Google Scholar]
- Quist MC, Bower MR, Hubert WA, Parchman TL, and McDonald DB 2009. Morphometric and Meristic Differences among Bluehead Suckers, Flannelmouth Suckers, White Suckers, and Their Hybrids: Tools for the Management of Native Species in the Upper Colorado River Basin. North American Journal of Fisheries Management 29:460–467. [Google Scholar]
- Rieseberg LH, Archer M. a., and Wayne RK 1999. Transgressive segregation, adaptation and speciation. Heredity 83:363–372. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, and Lexer C. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Kim S-C, Randell RA, Whitney KD, Gross BL, Lexer C, and Clay K. 2007. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129:149–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BW, and Wilson DS 1994. Character Release and Displacement in Fishes: A Neglected Literature The American Naturalist 144:596–627. [Google Scholar]
- Rogosch JS, and Olden JD 2020. Invaders induce coordinated isotopic niche shifts in native fish species. Canadian Journal of Fisheries and Aquatic Sciences 77:1348–1358. [Google Scholar]
- Rosenfield JA, Nolasco S, Lindauer S, Sandoval C, and Kodric-Brown A. 2004. The role of hybrid vigor in the replacement of pecos pupfish by its hybrids with sheepshead minnow. Conservation Biology 18:1589–1598. [Google Scholar]
- Sagouis A, Cucherousset J, Villeger S, Santoul F, and Bouletreau S. 2015. Non-native species modify the isotopic structure of freshwater fish communities across the globe. Ecography 38:979–985. [Google Scholar]
- Schluter D. 2000. Ecological Character Displacement in Adaptive Radiation. The American Naturalist 156:S4–S16. [Google Scholar]
- Schumer M, Xu C, Powell DL, Durvasula A, Skov L, Holland C, Blazier JC, and Sankararaman S. 2018. Natural selection interacts with recombination to shape the evolution ofhybrid genomes. Science 660:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scordato ESC, Wilkins MR, Semenov G, Rubtsov AS, Kane NC, and Safran RJ 2017. Genomic variation across two barn swallow hybrid zones reveals traits associated with divergence in sympatry and allopatry. Molecular Ecology 26:5676–5691. [DOI] [PubMed] [Google Scholar]
- Seehausen O. 2007. Evolution and ecological theory: Chance, historical contingency and ecological determinism jointly determine the rate of adaptive radiation. Heredity 99:361–363. [DOI] [PubMed] [Google Scholar]
- Seiler SM, and Keeley ER 2009. Competition between native and introduced salmonid fishes: cutthroat trout have lower growth rate in the presence of cutthroat-rainbow trout hybrids. Canadian Journal of Fisheries and Aquatic Sciences 66:133–141. [Google Scholar]
- Selz OM, and Seehausen O. 2019. Interspecific hybridization can generate functional novelty in cichlid fish. Proceedings of the Royal Society B: Biological Sciences 286:20191621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecal AC, Gelwicks KR, Cavalli PA, and Keith RM 2010. WGFD Short-term Plan for the Three Species in the Green River Drainage of Wyoming; 2009–2014. Wyoming Game and Fish Department, Fish Division. [Google Scholar]
- Shastry V, Adams PE, Lindtke D, Mandeville EG, Parchman TL, Gompert Z, and Buerkle CA 2021. Model-based genotype and ancestry estimation for potential hybrids with mixed-ploidy. Molecular Ecology Resources 21:1434–1451. [DOI] [PubMed] [Google Scholar]
- Stelkens R, Schmid C, Selz O, and Seehausen O. 2009. Phenotypic novelty in experimental hybrids is predicted by the genetic distance between species of cichlid fish. BMC Evolutionary Biology 9:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelkens R, and Seehausen O. 2009. Genetic distance between species predicts novel trait expression in their hybrids. Evolution 63:884–897. [DOI] [PubMed] [Google Scholar]
- Stelkens RB, Young KA, and Seehausen O. 2010. The accumulation of reproductive incompatibilities in African cichlid fish. Evolution 64:617–633. [DOI] [PubMed] [Google Scholar]
- Sweigart AL, Fishman L, and Willis JH 2006. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics 172:2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB, and McPhail JD 2000. Historical contingency and ecological determinism interact to prime speciation in sticklebacks, Gasterosteus. Proceedings of the Royal Society B: Biological Sciences 267:2375–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Thibodeau LM, Gompert Z, Buerkle CA, Nachman MW, and Tucker PK 2010. The variable genomic architecture of isolation between hybridizing species of house mouse. Evolution 64:472–485. [DOI] [PubMed] [Google Scholar]
- Thomas SM, and Crowther TW 2015. Predicting rates of isotopic turnover across the animal kingdom: a synthesis of existing data. Journal of Animal Ecology 84:861–870. [DOI] [PubMed] [Google Scholar]
- Thompson KA, Urquhart-Cronish M, Whitney KD, Rieseberg LH, and Schluter D. 2021. Patterns, Predictors, and Consequences of Dominance in Hybrids. American Naturalist 197:E72–E88. [DOI] [PubMed] [Google Scholar]
- Tronstad LM, and Estes-Zumpf W. 2011. Is the Muddy Creek food web affected by coalbed natural gas inputs? Tech. rep., Report prepared by the Wyoming Natural Diversity Database for the Water Research Program, Laramie, Wyoming. [Google Scholar]
- Vines TH, Kohler SC, Thiel A, Ghira I, Sands TR, MacCallum CJ, Barton NH, and Nurnberger B. 2003. The maintenance of reproductive isolation in a mosaic hybrid zone between the fire-bellied toads Bombina bombina and B. variegata. Evolution 57:1876–1888. [DOI] [PubMed] [Google Scholar]
- Walsworth TE, Budy P, and Thiede GP 2013. Longer food chains and crowded niche space: effects of multiple invaders on desert stream food web structure. Ecology of Freshwater Fish 22:439–452. [Google Scholar]
- Wege GJ, and Anderson RO 1978. Relative weight (Wr): a new index of condition for largemouth bass. New approaches to the management of small impoundments. American Fisheries Society, North Central Division, Special Publication 5:79–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Isotope data and genetic summary statistics presented in this paper are available on Dryad at https://doi.org/10.5061/dryad.0cfxpnw1f. R scripts used in analysis are available on Zenodo at https://doi.org/10.5281/zenodo.6958091. Genetic data associated with this paper are archived in connection with the publication of Mandeville et al. 2017.