Abstract
Carbon fixation is a critical process for our planet; however, its distribution across the bacterial and archaeal domains of life has not been comprehensively studied. Here, we performed an analysis of 52,515 metagenome-assembled genomes and discover carbon fixation pathways in 1,007 bacteria and archaea. We reveal the genomic potential for carbon fixation through the reverse tricarboxylic acid cycle in previously unrecognized archaeal and bacterial phyla (i.e. Thermoplasmatota and Elusimicrobiota) and show that the 3-hydroxypropionate bi-cycle is not, as previously thought, restricted to the phylum Chloroflexota. The data also substantially expand the phylogenetic breadth for autotrophy through the dicarboxylate/4-hydroxybutyrate cycle and the Calvin–Benson–Bassham cycle. Finally, the genomic potential for carbon fixation through the 3-hydroxypropionate/4-hydroxybutyrate cycle, previously exclusively found in Archaea, was also detected in the Bacteria. Carbon fixation thus appears to be much more widespread than previously known, and this study lays the foundation to better understand the role of archaea and bacteria in global primary production and how they contribute to microbial carbon sinks.
Keywords: carbon fixation cycles, reductive tricarboxylic acid cycle, 3-hydroxypropionate bi-cycle, Calvin–Benson–Basshan cycle, 3-hydroxypropionate/4-hydroxybutyrate
Significance Statement.
Global carbon fixation is underpinned by at least seven different biochemical pathways, of which six are exclusively found in bacteria and archaea. Here, we provide a comprehensive picture on how these carbon fixation pathways are distributed across bacterial and archaeal domains of life and discover the unexpected occurrence of certain pathways in specific phylogenetic and taxonomic groups. These findings have significant implications as they show that a much greater diversity of bacteria and archaea have the potential to contribute to carbon fixation than previously known.
Introduction
Autotrophic organisms are crucial for supporting life on Earth and are primary producers in all ecosystems. Both marine and terrestrial environments contribute equally to the rate of net primary production, and most of the global CO2 is thought to be fixed into organic matter through the Calvin–Benson–Bassham (CBB) cycle (1, 2). On land, this process is mainly carried out by eukaryotic plants, while in the oceans it is mainly driven by microscopic phytoplankton, including microalgae and cyanobacteria (3).
Although carbon fixation pathways (CFPs) other than the CBB cycle have been known for a long time, only recently has it been accepted that they also contribute significantly to global net primary production (4, 5). Besides the CBB cycle, there are six other known CFPs found in bacteria and archaea: the reverse tricarboxylic acid (rTCA) cycle, the 3-hydroxypropionate (3HP) bi-cycle, the 4-hydroxybutyrate/3-hydroxypropionate (4HB/3HP) cycle, the dicarboxylate/4-hydroxybutyrate (DC/4HB) cycle, the reductive acetyl-CoA pathway (Wood–Ljungdahl pathway—WLP), and the reductive glycine pathway (6). Some of these CFPs, such as the rTCA cycle, are found across several different bacterial and archaeal phyla, while others are thought to occur only in particular taxa (e.g. the 3HP bi-cycle in the phylum Chloroflexota) (7, 8).
Recent large-scale metagenomic studies have been able to uncover the genomic diversity of much of the bacterial and archaeal life on our planet (9–11). One of the most comprehensive studies was recently conducted by Nayfach et al., which describes 52,515 metagenome-assembled genomes (MAGs) that cover all major taxonomic groups (12). These resources now provide the opportunity to comprehensively describe the distribution of CFPs in the bacterial and archaeal domains of life.
Here, we use bioinformatics and a hand-curated analysis to describe the occurrence of CFPs in the Archaea and Bacteria. We show evidence for the previously unrecognized occurrence of the rTCA cycle in certain archaeal and bacterial phyla (Thermoplasmatota and Elusimicrobiota) as well as an expanded phylogenetic distribution of the 3HP bi-cycle, the DC/HB cycle, and CBB cycle. The HP/HB cycle, which was previously exclusively found in the Archaea, was here also detected in the Bacteria and found to be linked to photosynthesis in the genus Luminiphilus.
Results and discussion
In this study, we analyzed the distribution of five CFPs across a dataset of 52,515 MAGs that belong to the domains Archaea (5.8% of MAGs) and Bacteria (94.2% of MAGs), and contain 12,556 novel candidate species (12). CFPs were confidently identified in 1007 MAGs (Fig. 1) and 23 MAGs have more than one pathway [Table S5; see the “Material and methods” section for details on the identification process and the taxonomic classification using the Genome Taxonomy Database (GTDB) (13)]. As the WLP, the reverse oxidative TCA cycle, and the glycine reductive pathway can run in the oxidative direction with the same enzymes (i.e. in methane or acetate oxidizers) and have only reversible enzymatic reactions (14, 15), an accurate in-silico prediction is not possible and hence was not pursued here.
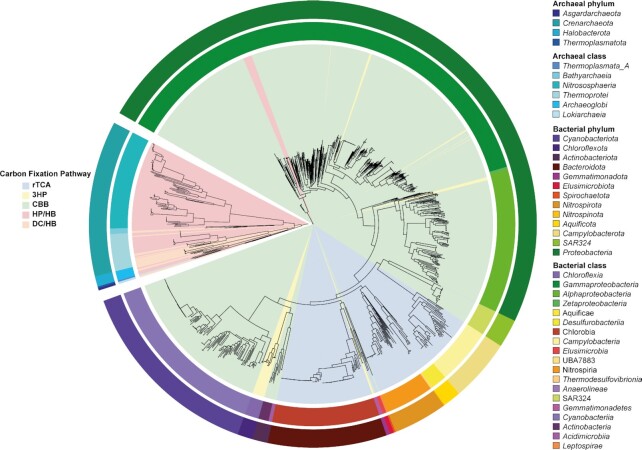
Fig. 1.
Phylogenetic tree representing the distribution of the different CFPs across the bacterial and archaeal domains of life in 1,007 MAGs. The outer ring is colored by the phylum, while the inner ring shows classes. Shading in the cycle shows the five different CFPs analyzed by the pipeline (see text for details).
Energy-generating pathways related to the oxidation of 11 different compounds as well as the biosynthesis of (bacterio-)chlorophyll were also analyzed to understand how the organisms generate reductive power for carbon fixation. No correlation was found between a specific habitat from which the MAGs were recovered and the type of CFP we detected in the MAGs (Fig. S1). Specific observations on each CFP are presented and discussed in the following sections.
Distribution of the reverse tricarboxylic acid cycle
The rTCA cycle, also known as the Arnon–Buchanan cycle, was first described in the green photosynthetic, anoxygenic bacterium Chlorobium thiosulfatophilum [phylum Bacteroidota, formerly phylum Chlorobi (16)] and most of its enzymes catalyze reversible reactions that are also used in the oxidative TCA cycle. However, the reduction of fumarate to succinate and the cleavage of citrate into acetyl-CoA and oxaloacetate reactions are considered to be irreversible (15, 17). The citrate cleavage can be catalyzed either by an ATP-dependent citrate lyase (ACL) (hereafter referred to as rTCA1, see Table 1) or by the sequential reactions of the ATP-dependent citryl-CoA synthetase (CCS) and the citryl-CoA lyase (CCL) (hereafter referred to as rTCA2, see Table 1). CO2 is fixed in the conversion of succinyl-CoA to 2-oxoglutarate as well as in the conversion of 2-oxoglutarate into isocitrate. Under low product-to-substrate ratios, the regular citrate lyase can also cleave citrate to support the rTCA (15); however, this cannot be bioinformatically distinguished from the oxidative version of the TCA and was therefore not considered in the current study. So far, this pathway has been shown to be restricted to the genus Desulfurella, but it is possible that other organisms known to express enzymes of the rTCA cycle and carrying citrate lyase genes might have the potential to use this cycle (15).
Table 1.
Key enzymes, reference organisms (and accession numbers), and pathway completeness thresholds of the different CFPs.
| Autotrophic pathway | Key enzyme | Reference organism | Number of reactions in CFP | Expected completeness threshold (%) | Number of detected MAGs |
|---|---|---|---|---|---|
| rTCA1 | Citrate lyase A (ACLa)Citrate lyase B (ACLb) | Chlorobium limicola (16)(CP001097.1) | 9 | 90 | 207 |
| rTCA2 | Citryl-CoA lyase (CCL)Citryl-CoA synthetase small (CCSs)Citryl-CoA synthetase large (CCSl) | Hydrogenobacter thermophilus (18)(CP002221.1) | 12 | 73 | 10 |
| 3HP | Mesaconyl-CoA isomerase(TIGR04253) | Chloroflexus auranticus (19)(CP000909.1) | 26 | 82 | 21 |
| CBB | Phosphoribulokinase I (PRK1)Phosphoribulokinase II (PRK2) | Cereibacter(formerlyRhodobacter)sphaeroides (20)(CP001150.1) | 13 | 90 | 616 |
| HP/HB | 4-Hydroxybutyrate dehydrogenase(HBD) | Nitrosopumilus maritimus (21)(CP000866.1) | 16 | 90 | 135 |
| DC/HB | 4-Hydroxybutyrate dehydrogenase(HBD) | Ignicoccus hospitalis (22)(CP000816.1) | 14 | 75 | 18 |
Citrate is cleaved into oxalacetate (OAA) and acetyl-CoA in a one-step reaction catalyzed by ACL in rTCA1. The rTCA2 pathway is a two-step catalysis, in which citrate is first cleaved into citryl-CoA by CCS and then citryl-CoA is cleaved into OAA and acetyl-CoA.
Consistent with the literature, we found genetic support for the rTCA1/2 cycles within a wide diversity of bacterial taxa within the phyla Aquificota (genera Hydrogenobacter, Thermocrinis, Hydrogenobaculum, Persephonella, Sulfuridrogenibium, and Thermovibrio), Bacteriodota (in particular the families Chlorobiaceae and Chloroherpetonaceae), Campylobacterota (specifically the class Campylobacteria; formerly Epsilonproteobacteria), and Nitrospinota (18, 23–25) (Table S1).
In addition to these previous observed taxa, we found here that two archaeal MAGs (GEM ID 3300020171_15 and 3300027917_99) had the rTCA1 cycle with adjusted, observed completeness greater than 97% and the predicted capacity for hydrogen oxidation (Fig. 2).
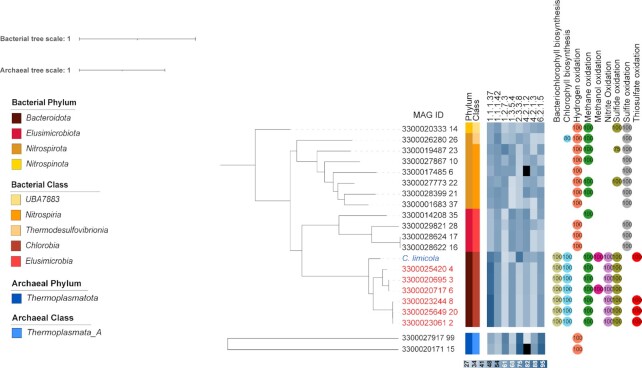
Fig. 2.
Phylogenetic tree showing the distribution of the rTCA cycle in novel bacterial and archaeal taxa and in known organisms from the family Chlorobiaceae (blue: reference genome in blue; red: MAGs assigned to the same family as the reference genome). Identity of the enzymes involved in the pathway against the UniRef database are shown in blue heat scale. 1.1.1.37: pyruvate-water dikinase; 1.1.1.42: isocitrate dehydrogenase; 1.2.7.3: 2-oxoglutarate-synthase; 1.3.5.4: phosphoenolpyruvate carboxylase; 2.3.3.8: ATP-citrate lyase; 4.2.1.2: fumarase; 4.2.1.3: aconitate hydratase; 6.2.1.5: succinyl-CoA synthetase. Numbers in circles refer to completeness of diagnostic pathways for energy generation. Black squares in the heatmap refers to missing enzymes.
The proposition of the rTCA1 cycle in the domain Archaea is not novel. Some evidence for the rTCA1 cycle’s carboxylation steps has been previously found in MAGs also belonging to the Thermoplasmatota from a hypersaline lake in the Kenyan Rift Valley; however, no solid conclusion could be made due to the predicted ACL possessing low similarity (32%) to known ACL sequences (26).
In contrast, MAGs 3300020171_15 and 3300027917_99 encoded both carboxylases required for the rTCA1 cycle as well as both subunits of the ACL (ACLa and ACLb), which had sequence similarities of 47% to 56.6% to reference sequence from C. limicola. A predicted membrane-embedded quinol: fumarate reductase as well as the predicted capacity for hydrogen-based chemolithotrophy (Fig. 2 and Tables S4 and S6) was also found in both MAGs, thus together providing the so far strongest support for the presence of rTCA1 cycle in the Thermoplasmatota.
The rTCA1 cycle was also identified in several MAGs belonging to the phylum Nitrospirota. Most of the MAGs belong to the nitrite-oxidizing order Nitrospirales, consistent with previous observation (17). However, for the first time, the complete set of enzymes for the rTCA cycle was here identified in organisms of the orders 2–01-FULL-66–17 (class Nitrospira) and the Thermodesulfovibrionales (MAG ID 3300026280_26), which all lacked the genetic capacity for nitrite-oxidation, but instead appear to rely on the oxidation of hydrogen, sulfite, and/or sulfide for energy generation. Similarly, the rTCA cycle in bacterial order UBA7883 (phylum Nitrospinota) also does not appear to rely on the oxidation of nitrite, but instead on hydrogen, sulfide, or sulfite.
We also found genes for the rTCA1 cycle in the Elusimicrobiota (4 MAGs with >97% pathway completeness), which is a novel finding for this phylum and appears to be coupled to sulfite and hydrogen oxidation (Fig. 2 and Tables S4 and S6). While members of this phylum have been reported to be facultative autotrophic, this capacity has so far only been attributed to the WLP (27).
The 3-hydroxypropionate bi-cycle beyond the Chloroflexota
The 3HP bi-cycle was first described in green, nonsulfur bacterium Chloroflexus aurantiacus (19, 28) and since then has been commonly and exclusively attributed to the phylum Chloroflexota (29, 30). However, enzymes for this pathway (e.g. 2-methylfumaryl-CoA isomerase, malonyl-CoA reductase, and propionyl-CoA synthase) have been recently detected in organisms belonging to the phyla Actinobacteriota (formerly Actinobacteria) as well as the classes Alpha- and Gammaproteobacteria within the Proteobacteria [recently renamed “Pseudomonadota (31)], raising speculation about their ability to undertake autotrophy based on the 3HP bi-cycle (32). Our analysis also detected the key enzymes for this cycle in 27 MAGs of the phylum Actinobacteriota, however the average pathway completeness was only 68.6%, which limits the support for the presence of the 3HP bi-cycle (Table S1). Instead, we propose that the detected enzymes are used to assimilate various organic substrates (i.e. glycolate, acetate, propionate, 3-hydroxypropionate, lactate, butyrate, or succinate), similar to what has been described for C. aurantiacus (33) (Supplementary Material and Fig. S2).
As for the Proteobacteria, we discovered MAGs with adjusted pathway completeness above the required threshold within the families Ga0077523 and Burkholderiaceae (both class Gammaproteobacteria, order Burkholderiales), both of which have also the predicted capacity of photosynthesis and oxidation of various inorganic electron donors (Fig. 3).
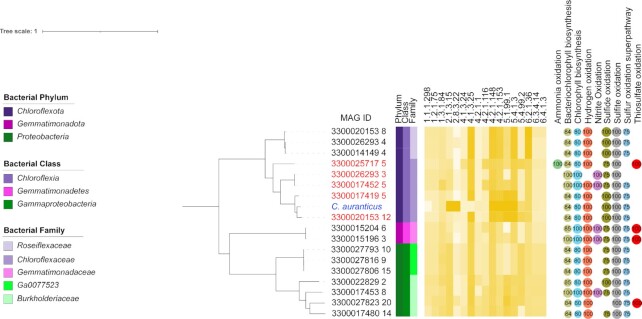
Fig. 3.
Phylogenetic tree showing the distribution of the 3HP cycle in novel bacterial taxa and in known organisms from the family Chloroflexaceae (blue: reference genome; red: MAGs assigned to the same family as the reference genome). Identity of the enzymes involved in the pathway against the UniRef database are shown in yellow heat scale. 1.1.1.298: 3-hydroxypropionate: NADP + oxidoreductase; 1.2.1.75: malonate semialdehyde: NADP + oxidoreductase (malonate semialdehyde-forming); 1.3.1.84: acrylyl-CoA reductase; 2.1.3.15: acetyl-CoA carboxyltransferase; 2.8.3.22: succinyl CoA: L-malate CoA transferase; 4.1.3.24: beta-methylmalyl CoA lyase; 4.1.3.25: S,-citramalyl-CoA lyase; 4.2.1.1: carbonic anhydrase; 4.2.1.116: 3-hydroxypropionyl-CoA dehydratase; 4.2.1.148: beta-methylmalyl-CoA dehydratase; 4.2.1.153: mesaconyl-C4-CoA hydratase; 5.1.99.1: methylmalonyl-CoA epimerase; 5.4.1.3: mesaconyl-C1-CoA-C4-CoA transferase; 5.4.99.2: methylmalonyl-CoA mutase; 6.2.1.36: 3-hydroxypropionate: CoA ligase; 6.3.4.14: biotin carboxylase; 6.4.1.3: propionyl-CoA carboxylase. Numbers in circles refer to completeness of diagnostic pathways for energy generation. Black squares in the heatmap refers to missing enzymes. The tree was rooted with a MAG from the Actinobacteriota, which is not shown here.
The three MAGs of the family Ga0077523 belong to the same genus (Ga0077523) and contain all genes for the glyoxylate salvage in the same operon, a characteristic conserved among organisms known to fix carbon through the 3HP bi-cycle (Supplementary Material) (33). Similarly, the four Burkholderiaceae MAGs have the genomic potential do fix carbon through the 3HP bi-cycle and display a similar organization of the genes related to the glyoxylate salvage (Supplementary Material).
We also detected all genes required for the 3HP bi-cycle for the first time in the phylum Gemmatimonadota (two MAGs assigned to the order Gemmatimonadales), whose members are usually found in plant rhizospheres where they thrive in carbon- and nitrogen-depleted environments (34, 35). Gemmatimonadota have also recently been found in hadal trenches, where they are considered to play an important role in organic carbon turnover (36). The arrangement of genes for the glyoxylate salvage pathway were also similar to those the other MAGs or reference organism. (Supplementary Material and Fig. S3). The predicted capacity for autotrophy in these Gemmatimonadales might be linked to the oxidation of various inorganic electron donors (Fig. 3 and Tables S4 and S6).
An expanded range of the CBB cycle
The CBB cycle is present in all eukaryotic plants and cyanobacteria and widespread in several chemoautotrophic bacteria and purple phototrophic bacteria (37–39). Consistent with the previous literature, we detected the genetic potential for the CBB in members of the phyla Actinobacteriota, Cyanobacteriota, Chloroflexota, CPR SAR324 (a former clade within the Deltaproteobacteria), Gemmatimonadota, Leptospirae, Proteobacteria, Methylomirabilota and Verrucomicrobiota and specific orders and families within them (40–44) (Supplementary Material and Table S1).
Despite its widespread and extensively studied distribution, we found genomic evidence for the CBB cycle in many new taxa and these includes members of the order Microtrichales (phylum Actinobacteriota, class Acidimicrobiia) and Propionibacteriales (phylum Actinobacteriota, class Actinobacteria) (Fig. 4). These MAGs have the genetic potential to perform the sedoheptulose-1,7-bisphosphate (SBPase) variant of the cycle, where erythrose 4-phosphate and dihydroxyacetone phosphate are condensed into sedoheptulose 1,7-biphosphate by an aldolase (45). We also found here first evidence for its presence in members of the families Beijerinckiaceae, Methyloligellaceae, Pseudoxanthobacteraceae, and UBA4765, which all belong to the order Rhizobiales that has recently been shown to contain autotrophic organisms (46).
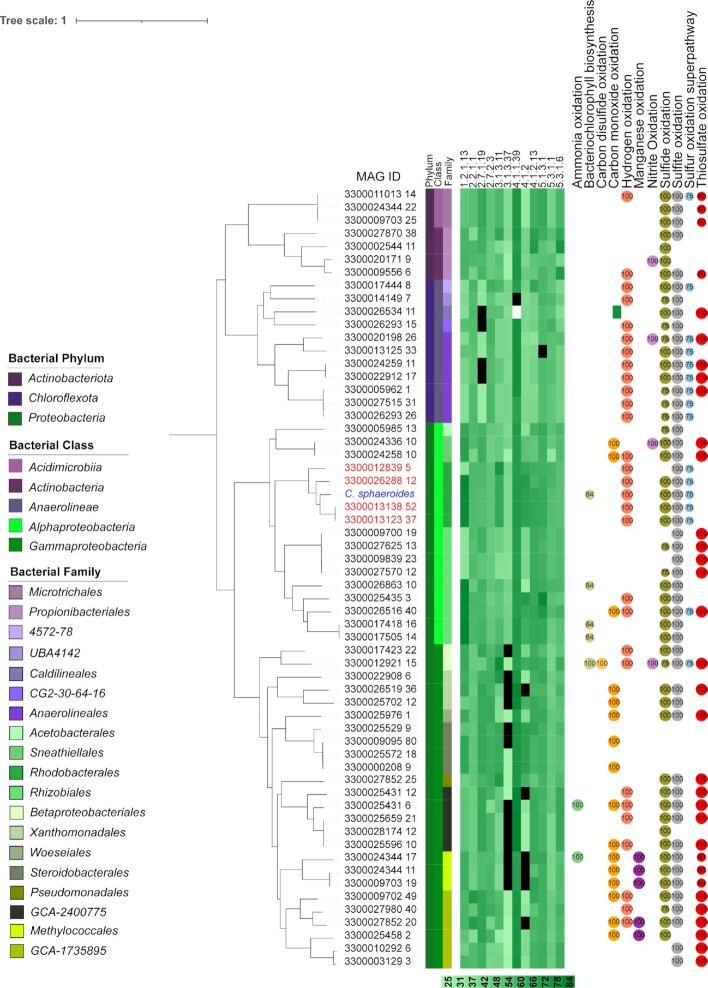
Fig. 4.
Phylogenetic tree showing the distribution of the CBB cycle in novel bacterial taxa and in known organisms from the family Rhodobacteraceae (blue: reference genome; red: MAGs assigned to the same family as the reference genome Identity of the enzymes involved in the pathway against the UniRef database are shown in green heat scale). 1.2.1.13: glyceraldehyde-3-phosphate dehydrogenase; 2.2.1.1: D-fructose 6-phosphate: D-glyceraldehyde-3-phosphate glycolaldehydetransferase; 2.7.1.19: phosphoribulokinase; 2.7.2.3: phosphoglycerate kinase; 3.1.3.11: fructose 1,6-bisphosphatase; 3.1.3.37: sedoheptulose bisphosphatase; 4.1.1.39: ribulose bisphosphate carboxylase; 4.1.2: sedoheptulose-1,7-bisphosphate aldolase, 4.1.2.13: fructose-bisphosphate aldolase; 5.1.3.1: pentose-5-phosphate 3-epimerase; 5.3.1.1: triosephosphate isomerase; 5.3.1.6: ribose 5-phosphate isomerase. Numbers in circles refer to completeness of diagnostic pathways for energy generation. Black squares in the heatmap refers to missing enzymes. The tree was rooted with a MAG from the Gemmatimonadota, which is not shown here.
Novel is also the finding of a complete set of CBB genes in the family Acetobacteraceae (order Acetobacterales, class Alphaproteobacteria). It has been previously reported that organisms in this family encode most of the enzymes required for the CBB, but no genetic evidence for a phosporibulokinase (PRK) was found (47). In contrast, we detected in MAG 3300005985_13 of this family a protein sequence for PRK I with a sequence similarity of 69.4% to the reference PRK I from Cereibacter sphaeroides. For the first time, we were also able to identify the genes for a complete CBB cycle in the order Sneathiellales, which have so far only been described as chemoheterotrophic (48). Both MAGs (3300024336_10 and 3300024258_10) identified are unassigned at the family and genus level, which limits further comparison with the literature, but they have the predicted capacity for chemolithotrophy via the oxidation of carbon monoxide, hydrogen, nitrite, sulfide, sulfite and/or thiosulfate.
A full set of genes for the CBB cycle was also found in two MAGs of the family Marinicellaceae and one MAG assigned to the family Rhodanobacteraceae (both order Xanthomonadales, class Gammaproteobacteria). Xanthomonadales are primarily chemoorganoheterotrophs, although evidence for chemolithotrophy based on sulfur oxidation has been found (49). We were able to detect genes encoding for the oxidation of carbon monoxide, sulfide, sulfite, and/or thiosulfate in the Marinicellaceae MAGs (3300025702_12 and 3300026519_36), while for the Rhodanobacteraceae MAG (3300022908_6), no pathways for lithotrophy or photosynthesis could be found.
The order GCA-1,735,895 (class Gammaproteobacteria) has been previously inferred to be mixotrophic, but no CFPs were identified (50). Our findings indicate that its autotrophic growth is based on the CBB cycle as well as the potential to generate energy through the oxidation of carbon monoxide, hydrogen, manganese, nitrite, sulfide, sulfite, or thiosulfate (Fig. 4 show six representative MAGs and for the remainder see Tables S1, S4, and S6). Similarly, organisms assigned to the order GCA-2,400,775 have been predicted to be chemolithotrophic, even though no CFP had been proposed (51). In our analysis, 96 MAGs assigned to this taxon have genes for the CBB cycle and have the predicted capacity to oxidise ammonia, carbon monoxide, hydrogen, sulfide, sulfite, or thiosulfate (Fig. 4 show five representative MAGs and for the remainder see Tables S1, S4, and S6). We also found all genes for the CBB cycle in organisms assigned to the family IMCC2047 (order Pseudomonadales), which has genes for the oxidation of sulfide, sulfite, and thiosulfate. Within the order Betaproteobacteriales, we also detected genomic evidence for the CBB cycle in the family Ga0077523 (MAG 3300012921_15). This MAGs is predicted to be capable of oxidizing carbon disulfide, hydrogen, nitrite, sulfide, sulfite, sulfur, or thiosulfate, or synthetize bacteriochlorophyll.
Intriguingly, we also found all genes for the CBB cycle with the exception of sedoheptulose bisphosphatase (SBPase) in MAG 3300017423_22 from the order Betaproteobacteriales (family Methylophilaceae). However, fructose bisphosphatase has been reported to be a bifunctional enzyme that can act as SBPase in some organisms (45, 52). This family is known to be strictly methylotrophic and assimilates methylated compounds through the ribulose monophosphate cycle (RuMP), which shares its reduction steps with the CBB cycle (53), but does not utilize PRK and ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (45). This MAGs also lacks genes for methanol oxidation, but instead genes encoding for enzyme to oxidize hydrogen, sulfite, and sulfide. In addition, we detected in this MAG genes encoding for hexulose-6-phosphate synthase (HPS), hexulose-6-phosphate isomerase (PHI), methylene-tetrahydromethanopterin dehydrogenase, and methylene-H4MPT cyclohydrolase, which are part of the recently described reductive hexulose-phosphate (RHP) pathway (53). This pathway is different from the CBB in that PHI isomerizes fructose 6-phosphate to hexulose 6-phosphate, which is then cleaved by HPS into formaldehyde and ribulose 5-phosphate. This pathway has been proposed for an archaeon (53) and our genomic evidence suggest that it might also be operational in the bacterial family Methylophilaceae. This proposition for autotrophy is also supported by the fact that the majority of the genes for the RHP pathway are collocated in the same genomic region of MAG 3300017423_22 (Fig. S4).
Growth supported by the CBB cycle has been previously described for members of the methanotrophic order Methylococcales, but only for the family Methylococcaceae (54). We found all genes for the CBB in three MAGs assigned to the family Methylomonadaceae of that order. This family was thought to be strictly heterotrophic (55), but our results rather support the notion that they can also grow autotrophically, possibly generating energy from the oxidation of ammonia, carbon monoxide, manganese, sulfide, sulfite, or thiosulfate. Growth using the CBB cycle has also been predicted for members of the family Woeseiaceae of the order Steroidobacterales (56). Here, we expand the phylogenetic breadth of the CBB within this order by discovering also all necessary genes for the CBB cycle in MAGs of the family Steroidobacteraceae and we show that MAGs 3300009095_80 and 3300000208_9 have the potential to oxidize carbon monoxide.
Recently, a new form I’ of RuBisCO was described in the phylum Chloroflexota, which shares a high similarity with RuBisCO form I, but does not depend on the small subunit to be active (57). Form I’ was found in MAGs belonging to the order Anaerolineales (class Anaerolineae, phylum Chloroflexota) alongside with several other enzymes of the CBB cycle, such as PRK, glyceraldehyde-3-phosphate dehydrogenase (GAPD), transaldolase (TAL), and FBPase, suggesting autotrophic growth. Our results suggest that this mechanism might also span in the Caldilineales (class Anaerolineae), as one MAGs in that order have genes for PRK, GDAP, FBPase, and TAL and also only encode for the large subunit of RuBisCO, which has 78.87% sequence similarity to the form I’ reference sequence (PDB: 6URA_A). Similar results were found for MAGs in the placeholder orders CG2-30-64–16, 4572–78, and UBA4142 of the Anaerolineae, suggesting that RuBisCO form I’ might be a synapomorphy for the class, not only for the order.
The expanded distribution of the dicarboxylate/4-hydroxybutyrate cycle in the Archaea
The dicarboxylate/4-hydroxybutyrate (DC/HB) cycle was initially described in Ignicoccus hospitalis, an hydrogen-oxidizing and sulfur-reducing archaeon (58). The DC/HB cycle’s conversion of succinyl-CoA to acetyl-CoA uses the same enzymes as the HP/HB cycles, but the regeneration of the former involves the carboxylation of acetyl-CoA into pyruvate catalyzed by an oxygen-sensitive, ferredoxin-dependent pyruvate synthase. This cycle appeared to be restricted to anaerobic archaea of the orders Sulfolobales and Thermoproteales (phylum Thermoproteota, class Thermoproteia) (59, 60), but our analysis substantially expands its range within the archaeal domain (Fig. 5 and Table S1).
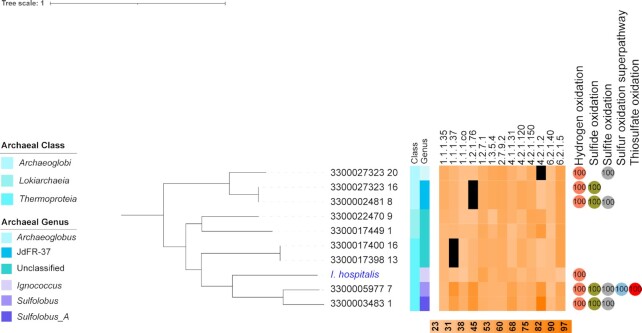
Fig. 5.
Phylogenetic tree showing the distribution of the DC/HB cycle in novel archaeal taxa and the reference genome of I. hospitalis. Identity of the enzymes involved in the pathway against the UniRef database are shown in orange heat scale. 1.1.1.35: S-3-hydroxybutyryl-CoA dehydrogenase; 1.1.1.37: malate dehydrogenase; 1.1.1.co: succinate semialdehyde reductase; 1.2.1.76: succinyl-CoA reductase; 1.2.7.1: pyruvate synthase (ferredoxin-dependent); 1.3.5.4: fumarate reductase; 2.7.9.2: phosphoenolpyruvate synthetase; 4.1.1.31: phosphoenolpyruvate carboxylase; 4.2.1.120: 4-hydroxybutyryl-CoA dehydratase; 4.2.1.150: S,-3-hydroxybutanoyl-CoA dehydrogenase; 4.2.1.2: fumarase; 6.2.1.40: 4-hydroxybutyryl-CoA synthetase; 6.2.1.5: succinyl-CoA synthetase. Numbers in circles refer to completeness of diagnostic pathways for energy generation. Black squares in the heatmap refers to missing enzymes. The tree was rooted with a MAG from the Methanobacteriota, which is not shown here.
Two MAGs assigned to the class Lokiarchaeia (phylum Asgardarchaeota) contained all genes for the DC/HB cycle. Member of the Asgardarchaeota have been suggested to be capable of growing autotrophically, through the WLP or even through reactions mediated by a type III-like Rubisco (Bulzu, P et. al, 2019) (61, 62), but also, as we show here, possibly through the DC/HB cycle. Further work is required to determine which CFPs are used by this metabolically diverse class and phylum.
And finally, we find, for the first time, genomic evidence for the DC/HB cycle in the genus Sulfolobus (Fig. 5), whose members have been reported to be microaerophilic thermophiles that can oxidise reduced sulfur compounds (Fig. 6) (63). Such growth conditions might allow oxygen-sensitive enzymes of the DC/HB cyle, such as the ferredoxin-dependent pyruvate synthase, to be active (22). To the best of our knowledge, Sulfolobus has so far been thought to grow autotrophically exclusively through the HP/HB cycle (60, 21, 64). Our genomic evidence suggests that it can also grow through the DC/HB cycle, because of the presence of all the enzymes required for the regeneration of acetyl-CoA from succinyl-CoA (Fig. 5).
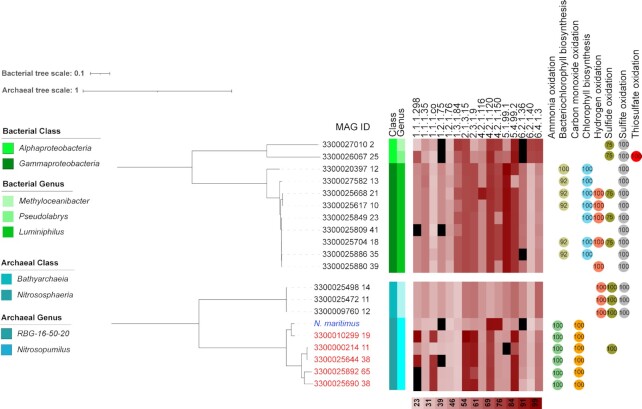
Fig. 6.
Phylogenetic tree showing the distribution of the HP/HB cycle in novel bacterial and archaeal taxa and in known organisms from the family Nitrosopumilaceae (blue: reference genome; red: MAGs assigned to the same family as the reference genome). Identity of the enzymes involved in the pathway against the UniRef database are shown in red heat scale. 1.1.1.298: malonate semialdehyde reductase; 1.1.1.35: S,-3-hydroxybutyryl-CoA dehydrogenase; 1.1.1.co: succinate semialdehyde reductase; 1.2.1.75: malonyl-coenzyme A reductase; 1.2.1.76: succinyl-CoA reductase; 1.3.1.84: acrylyl-CoA reductase; 2.1.3.15: acetyl-CoA carboxytransferase; 2.3.1.9: acetoacetyl-CoA beta-ketothiolase; 4.2.1.116: 3-hydroxypropionyl-CoA dehydratase; 4.2.1.120: 4-hydroxybutyryl-CoA dehydratase; 4.2.1.150: S,-3-hydroxybutanoyl-CoA dehydrogenase; 5.1.99.1: methylmalonyl-CoA epimerase; 5.4.99.2: methylmalonyl-CoA mutase; 6.2.1.36: 3-hydroxypropionyl-CoA synthetase; 6.2.1.40: 4-hydroxybutyryl-CoA synthetase; 6.4.1.3: propionyl CoA carboxylase. Numbers in circles refer to completeness of diagnostic pathways for energy generation. Black squares in the heatmap refers to missing enzymes.
The 3-hydroxypropionate/4-hydroxybutyrate cycle in the Archaea and Bacteria
The HP/HB cycle was first discovered in the thermoacidophilic, iron-oxidizing archaeon Metallosphaera sedula (65) and has been more recently described in detail for the ammonium-oxidizing archaeaon Nitrosopumilus maritimus (66, 67). So far, this cycle has only been reported for the Archaea. The 4-hydroxybutyrate dehydratase (HBD) is commonly considered a key enzyme (65) as it appears in the Archaea to be only involved in the HP/HB cycle. In the Bacteria, however, HBD is part of different metabolic pathways (i.e succinate fermentation and 4-aminobutanoate degradation) (68).
As expected, we found ample of genomic evidence for this cycle in MAGs belonging to the archaeal phylum Thermoproteota as well as the classes Nitrososphaeria (order Nitrososphaerales) and Thermoprotei (order Sulfolobales) (Table S1). We also found evidence for this cycle in the genus Archaeoglobus (phylum Halobacteriota, order Archaeoglobales) (Table S1). However, it has been previously experimentally shown that Archaeoglobus lithotrophicus fixes carbon solely through the WLP, even though predicted homologs for HBD were found in its genome (69). It is therefore unclear what role the HP/HB cycle enzymes have in this genus.
Novel here is the discovery of the HP/HB cycle in three MAGs of the family RBG-16–50–20 within the class Bathyarchaeia. These MAGs have the genomic potential to encode all the enzymes required for carbon fixation through the 3-hydroxypropionate/4-hydroxybutyrate cycle and this process seems to be supported by oxidation of hydrogen, sulfide, and/or sulfite.
Unexpectedly, we found genomic evidence of the HP/HB cycle in the Bacteria. Eight MAGs belonging to the genus Luminiphilus (phylum Proteobacteria, class Gammaproteobacteria, order Pseudomonadales, family Halieaceae) encoded all the enzymes for this cycle (Fig. 6 and Table S1). We have also detected genes for the synthesis of (bacterio-) chlorophyll in most of these MAGs. Members of the genus Luminiphilus have previously been described to use organic carbon sources and have a full photosynthetic apparatus but lack PRK or RuBisCO genes. They were therefore thought to be photoheterotrophic organisms (70), however, our observation shows that they are instead likely capable of autotrophic growth through the HP/HB cycle, linking this cycle for the first time also to photosynthesis.
Our observation represents only the second time that the HP/HB cycle has been proposed to exist in the bacterial domain of life, with previous work suggesting its presence in the candidate phylum SBR1093 and in some organisms within the Bradyrhizobiaceae of the order Rhizobiales (71). Our results showed that the MAGs 3300026067_25 (genus Pseudolabrys) and 3300027010_2 (genus Methyloceanibacter) within this order have the potential to fix carbon through this cycle (88% completeness). Further inspection showed this to be due to missing genes for the malonyl-CoA reductase and the 3-hydroxypropionyl-CoA synthetase. Distant homologs, however, may exist for members of both families as manual analysis revealed proteins sharing 30.5% identity across their full lengths with a predicted malonyl-CoA reductase (UniRef cluster UniRef50_A0A0P8CE70) and 28.5% identity with a predicted 3-hydroxypropionyl-CoA synthetase (UniRef cluster UniRef50_A5UY60) for the Pseudolabrys MAG. For the MAGs classified as Methyloceanibacter, we also found protein sequences sharing 30.1% identity across their entire length with predicted malonyl-CoA reductase and 28.5% identity with 92.2% coverage with a predicted 3-hydroxypropionyl-CoA synthetase sequence of corresponding UniRef clusters. Altogether, this suggests the potential for the HP/HB cycle in these taxa, which also have the predicted capacity of sulfur-based chemolithotrophy (Fig. 6).
Together, these results show that the HP/HB cycle occurs not only in the archaeal domain, but also within the Bacteria. Furthermore, the data support the notion that this cycle is not only restricted to chemolithotrophic organisms, but can also be found in phototrophs.
Conclusion
Our analysis has revealed that CFPs are much more widespread in the archaeal and bacterial domains of life than previously anticipated. Evolutionarily, this is likely underpinned by complex patterns of horizontal gene transfer and gene loss (7, 43). Future work could monitor the CFP expression for these new taxa found here to better understand their role in global primary production and define how they contribute to microbial carbon sinks in various ecosystems.
Material and methods
The nucleotide and predicted protein sequences of 52,515 MAGs were obtained from the GEM catalogue, available at JGI Genome Portal with the identifier DOI: 10.1038/s41587-020-0718-6 (12). These MAGs were generated from 3,170 metagenomic samples obtained from a wide range of habitats (including freshwater, marine water, soil, and thermal springs; see Fig. S1 and Table S1).
For each CFP, we defined key enzymes, which are unique to the specific pathway, and built hidden Markov models (HMMs) using the HMMER suite version 3.3 (Table 1). For the models, we only selected sequences that were experimentally proven to catalyze the specific reaction in order to prevent issues related to “genome annotation rot” (72). When only one experimentally validated sequence was available, we did BLASTP searches to select enzymes with an identity higher than 90% from different organisms. HMMsearch results are available in Table S3. For the CBB, we decided to use phosphoribulokinase (PRK) as the key enzyme for our initial searches instead of RuBisCO as the latter has five different isoforms, six different clades, and different subunits for isoforms, which would have complicated HMM building (57). Accession numbers and seed alignments can be found in Table S2. An HMM for the mesaconyl-CoA isomerase (TIGR04253) was previously available (http://tigrfams.jcvi.org/cgi-bin/index.cgi) (73).
All MAGs with a positive hit (E-value < 10−99) for the key enzymes had the completeness of their CFPs analyzed using GapSeq version 1.1 with the UniProt database version UniRef 2021_01 (74). UniRef stores protein sequence clusters at different resolutions that account for sequence similarities within each cluster, which allow to infer homology between proteins. GapSeq combines searches against protein reference sequence databases, such as UniProt, with pathway and reaction databases (ModelSEED, KEGG, and MetaCyc) in order to infer the presence and completeness of metabolic pathway in a genome. Carboxylation reactions were also considered key reactions for all GapSeq analyses. GapSeq parameters were set to use unreviewed protein sequences only when reviewed sequences were not available for a particular reaction or enzyme function, and a bit score higher than 100 was required (Table S1).
Thresholds for pathway completeness were defined using the complete genomes of well-studied model organisms for each particular CFP (Table 1). Pathway completeness for the model organisms was analyzed by GapSeq. The expected completeness thresholds were set 10% lower than the reference genome values obtained by GapSeq in a conservative measure to avoid false negatives. The observed CFP completeness for the MAGs was then divided by the genome completeness, which was estimated using the CheckM algorithm (75) with lineage-specific marker genes, to obtain the adjusted, observed completeness. A particular CFP was considered to be present if a MAG is adjusted, observed CFP completeness was above the expected completeness threshold. MAGs with estimated genome completeness lower than 75% were generally not considered. MAGs 3300013125_33 and 3300017423_22 were the exception as we manually checked them for the form I RuBisCO.
In most cases, multiple, phylogenetically closely related MAGs with the same predicted CFP were found and this gave confidence that the pathway’s presence in a specific taxon or clade is not due to contaminations. However, in some cases, only one MAG with a particular CFP was recovered for a specific taxon. In those cases, genomic regions flanking the CFP genes were checked for potential mis-assemblies using GC content and taxonomic assignment of neighbouring genes.
Taxonomy was assigned to MAGs using the GTDB version 95 and GTDB-Tk toolkit version 94 (76). Mapping of taxonomic names in GTDB to those used in the taxonomy of the National Center for Biotechnology Information (NCBI) can be found at http://gtdb.ecogenomic.org. Unrooted bacterial and archaeal phylogenetic trees based on 120 phylogenetic marker genes for bacteria and 122 marker genes for archaea were generated by GTDB-Tk and visualized using iTOL (77).
Energy-generating pathways were inferred using GapSeq outputs focusing on 34 diagnostic pathways in the MetaCyc database, which are involved in the oxidation of 11 different inorganic compounds. In addition, we determined the potential for photosynthesis through the MetaCyc superpathways for chlorophyll and bacteriochlorophyll syntheses. Completeness of the pathways was determined without considering MAG completeness and values of >75% are reported for the most complete diagnostic pathways for each compound. The percentage shown in the dot plot refers to the most complete oxidation pathway for a specific compound (e.g. if four different sulfide oxidation pathways were available to be analyzed, then the percentage in the plot represents the percentage of the most complete pathway). A list of the analyzed pathways and their pathway completeness as well as all genes found are available in Tables S4 and S6.
Supplementary Material
ACKNOWLEDGEMENTS
This research includes computations using the computational cluster Katana supported by Research Technology Services at UNSW Sydney.
Notes
Competing Interest: The authors declare no competing interests.
Contributor Information
Alessandro N Garritano, Centre for Marine Science and Innovation, School of Biological, Earth and Environmental Sciences, Faculty of Science, The University of New South Wales, Kensington, NSW 2052, Australia.
Weizhi Song, Centre for Marine Science and Innovation, School of Biological, Earth and Environmental Sciences, Faculty of Science, The University of New South Wales, Kensington, NSW 2052, Australia.
Torsten Thomas, Centre for Marine Science and Innovation, School of Biological, Earth and Environmental Sciences, Faculty of Science, The University of New South Wales, Kensington, NSW 2052, Australia.
Funding
This work was supported by an R&D project registered as ANP 21005-4, “PROBIO-DEEP—Survey of potential impacts caused by oil and gas exploration on deep-sea marine holobionts and selection of potential bioindicators and bioremediation processes for these ecosystems” (UFRJ/Shell Brasil/ANP), sponsored by Shell Brasil under the ANP R&D levy as “Compromisso de Investimentos com Pesquisa e Desenvolvimento.”
Authors' Contributions
A.N.G.: data analysis, development of figures, bioinformatical pipeline development, and manuscript drafts. W.S.: bioinformatical pipeline development. T.T.: data analysis, bioinformatical pipeline development, and manuscript revision.
Data Availability
Download for the 52,515 MAGs and their amino acids files is available at https://genome.jgi.doe.gov/GEMs and https://portal.nersc.gov/GEM. HMM models used are available at https://github.com/alegarritano/HMM_CFP.
References
- 1. Ward LM, Shih PM. 2019. The evolution and productivity of carbon fixation pathways in response to changes in oxygen concentration over geological time. Free Radic Biol Med. 140:188–199. [DOI] [PubMed] [Google Scholar]
- 2. Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 281:237–240. [DOI] [PubMed] [Google Scholar]
- 3. Olajire AA, Essien JP. 2014. Aerobic degradation of petroleum components by microbial consortia. J Pet Environ Biotechnol. 5:195. [Google Scholar]
- 4. Baltar F, et al. 2010. Significance of non-sinking particulate organic carbon and dark CO2 fixation to heterotrophic carbon demand in the mesopelagic Northeast Atlantic. Geophys Res Lett. 37:L09602. [Google Scholar]
- 5. Herndl GJ, et al. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 71:2303–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sánchez-Andrea I, et al. 2020. The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Nat Commun. 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Becerra A, Rivas M, García-Ferris C, Lazcano A, Peretó J. 2014. A phylogenetic approach to the early evolution of autotrophy: the case of the reverse TCA and the reductive acetyl-CoA pathways. Int Microbiol. 17:91–97. [DOI] [PubMed] [Google Scholar]
- 8. Hügler M, Huber H, Stetter KO, Fuchs G. 2003. Autotrophic CO2 fixation pathways in archaea (Crenarchaeota). Arch Microbiol. 179:160–173. [DOI] [PubMed] [Google Scholar]
- 9. Youngblut ND, et al. 2020. Large scale metagenome assembly reveals novel animal-associated microbial genomes, biosynthetic gene clusters, and other genetic diversity. mSystems. 5:e01045–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai R, Zhang J, Liu R, Sun C. 2021. Metagenomic insights into the metabolic and ecological functions of abundant deep-sea hydrothermal vent DPANN archaea. Appl Environ Microbiol. 87:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He C, et al. 2021. Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems. Nat Microbiol. 6:354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nayfach S, et al. 2020. A genomic catalog of Earth’s microbiomes. Nat Biotechnol. 39:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2020. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 36:1925–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kadnikov VV, Mardanov AV, Beletsky AV, Karnachuk OV, Ravin NV. 2019. Genome of the candidate phylum Aminicenantes bacterium from a deep subsurface thermal aquifer revealed its fermentative saccharolytic lifestyle. Extremophiles. 23:189–200. [DOI] [PubMed] [Google Scholar]
- 15. Mall A, et al. 2018. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science. 359:563–567. [DOI] [PubMed] [Google Scholar]
- 16. Evans MC, Buchanan BB, Arnon DI. 1966. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc Natl Acad Sci. 55:928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hügler M, Sievert SM. 2011. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev Mar Sci. 3:261–289. [DOI] [PubMed] [Google Scholar]
- 18. Hügler M, Huber H, Molyneaux SJ, Vetriani C, Sievert SM. 2007. Autotrophic CO2 fixation via the reductive tricarboxylic acid cycle in different lineages within the phylum Aquificae: evidence for two ways of citrate cleavage. Environ Microbiol. 9:81–92. [DOI] [PubMed] [Google Scholar]
- 19. Holo H. 1989. Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch Microbiol. 151:252–256. [Google Scholar]
- 20. Qian Y, Tabita FR. 1996. A global signal transduction system regulates aerobic and anaerobic CO2 fixation in Rhodobacter sphaeroides. J Bacteriol. 178:12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu L, et al. 2021. Convergent evolution of a promiscuous 3-hydroxypropionyl-CoA dehydratase/crotonyl-CoA hydratase in Crenarchaeota and Thaumarchaeota. mSphere. 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berg IA, Ramos-Vera WH, Petri A, Huber H, Fuchs G. 2010. Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiology. 156:256–269. [DOI] [PubMed] [Google Scholar]
- 23. Waite DW, et al. 2017. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol. 8:682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lücker S, et al. 2010. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci. 107:13479–13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park S-J, et al. 2020. Expanded diversity and metabolic versatility of marine nitrite-oxidizing bacteria revealed by cultivation- and genomics-based approaches. Appl Environ Microbiol. 86:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mwirichia R, et al. 2016. Metabolic traits of an uncultured archaeal lineage-MSBL1-from brine pools of the Red Sea. Sci Rep. 6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Méheust R, et al. 2020. Groundwater Elusimicrobia are metabolically diverse compared to gut microbiome Elusimicrobia and some have a novel nitrogenase paralog. ISME J. 14:2907–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holo H, Sirevåg R. 1986. Autotrophic growth and CO2 fixation of Chloroflexus aurantiacus. Arch Microbiol. 145:173–180. [Google Scholar]
- 29. Lannes R, Olsson-Francis K, Lopez P, Bapteste E. 2019. Carbon fixation bymarine ultrasmall prokaryotes. Genome Biol Evol. 11:1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gayathri R, et al. 2020. A review on biological carbon sequestration: a sustainable solution for a cleaner air environment, less pollution and lower health risks. J King Saud Univ - Sci. 33:101282. [Google Scholar]
- 31. Oren A, Garrity GM. 2021. Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol. 71(10):005056. [DOI] [PubMed] [Google Scholar]
- 32. Ruiz-Fernández P, Ramírez-Flandes S, Rodríguez-León E, Ulloa O. 2020. Autotrophic carbon fixation pathways along the redox gradient in oxygen-depleted oceanic waters. Environ Microbiol Rep. 12:334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zarzycki J, Fuchs G. 2011. Coassimilation of organic substrates via the autotrophic 3-hydroxypropionate bi-cycle in Chloroflexus aurantiacus. Appl Environ Microbiol. 77:6181–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang H, et al. 2003. Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int J Syst Evol Microbiol. 53:1155–1163. [DOI] [PubMed] [Google Scholar]
- 35. Chee-Sanford JC, Connor L, Krichels A, Yang WH, Sanford RA. 2020. Hierarchical detection of diverse Clade II (atypical) nosZ genes using new primer sets for classical- and multiplex PCR array applications. J Microbiol Methods. 172:105908. [DOI] [PubMed] [Google Scholar]
- 36. Liu R, et al. 2020. Bulk and active sediment prokaryotic communities in the Mariana and Mussau Trenches. Front Microbiol. 11:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Calvin M, Benson AA. 1948. The path of carbon in photosynthesis. Science. 107:476–480. [DOI] [PubMed] [Google Scholar]
- 38. Bassham JA, et al. 1954. The path of carbon in photosynthesis. XXI. The cyclic regeneration of carbon dioxide acceptor. J Am Chem Soc. 76:1760–1770. [Google Scholar]
- 39. Biel K, Fomina I. 2015. Benson–Bassham–Calvin cycle contribution to the organic life on our planet. Photosynthetica. 53:161–167. [Google Scholar]
- 40. Rasigraf O, Kool DM, Jetten MSM, Sinninghe Damsté JS, Ettwig KF. 2014. Autotrophic carbon dioxide fixation via the Calvin–Benson–Bassham cycle by the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera”. Appl Environ Microbiol. 80:2451–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grostern A, Alvarez-Cohen L. 2013. RubisCO-based CO2 fixation and C1 metabolism in the actinobacterium Pseudonocardia dioxanivorans CB1190. Environ Microbiol. 15:3040–3053. [DOI] [PubMed] [Google Scholar]
- 42. Severino R, et al. 2019. High-quality draft genome sequence of Gaiella occulta isolated from a 150 meter deep mineral water borehole and comparison with the genome sequences of other deep-branching lineages of the phylum Actinobacteria. Microbiologyopen. 8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ward LM, Hemp J, Shih PM, McGlynn SE, Fischer WW. 2018. Evolution of phototrophy in the Chloroflexi phylum driven by horizontal gene transfer. Front Microbiol. 9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zorz JK, et al. 2019. A shared core microbiome in soda lakes separated by large distances. Nat Commun. 10:4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stolzenberger J, Lindner SN, Persicke M, Brautaset T, Wendisch VF. 2013. Characterization of fructose 1,6-bisphosphatase and sedoheptulose 1,7-bisphosphatase from the facultative ribulose monophosphate cycle methylotroph Bacillus methanolicus. J Bacteriol. 195:5112–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tong H, et al. 2021. Microaerophilic oxidation of Fe(II) coupled with simultaneous carbon fixation and As(III) oxidation and sequestration in karstic paddy soil. Environ Sci Technol. 55:3634–3644. [DOI] [PubMed] [Google Scholar]
- 47. Noh HJ, et al. 2020. Lichenicola cladoniae gen. nov., sp. nov., a member of the family Acetobacteraceae isolated from an Antarctic lichen. Int J Syst Evol Microbiol. 70:5918–5925. [DOI] [PubMed] [Google Scholar]
- 48. Jordan EM, et al. 2007. Sneathiella chinensis gen. nov., sp. nov., a novel marine alphaproteobacterium isolated from coastal sediment in Qingdao, China. Int J Syst Evol Microbiol. 57:114–121. [DOI] [PubMed] [Google Scholar]
- 49. Li X, Kappler U, Jiang G, Bond PL. 2017. The ecology of acidophilic microorganisms in the corroding concrete sewer environment. Front. Microbiol. 8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen YJ, et al. 2021. Metabolic flexibility allows generalist bacteria to become dominant in a frequently disturbed ecosystem. The ISME journal. 15:2986–3004.. 10.1101/2020.02.12.945220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cabello-Yeves PJ, et al. 2020. Microbiome of the Black Sea water column analyzed by genome centric metagenomics. Environmental Microbiome. 16:1–15.. 10.1101/2020.10.30.362129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yan C, Xu X. 2008. Bifunctional enzyme FBPase/SBPase is essential for photoautotrophic growth in cyanobacterium Synechocystis sp. PCC 6803. Prog Nat Sci. 18:149–153. [Google Scholar]
- 53. Kono T, et al. 2017. A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea. Nat Commun. 8:14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Baxter NJ, et al. 2002. The ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of Methylococcus capsulatus (Bath). Arch Microbiol. 177:279–289. [DOI] [PubMed] [Google Scholar]
- 55. Islam T, Larsen Ø, Birkeland NK. 2020. A novel cold-adapted Methylovulum species, with a high C16:1ω5c content, isolated from an Arctic thermal spring in Spitsbergen. Microbes Environ. 35:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mußmann M, Pjevac P, Krüger K, Dyksma S. 2017. Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J. 11:1276–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Banda DM, et al. 2020. Novel bacterial clade reveals origin of form I Rubisco. Nature Plants. 6:1158–1166. [DOI] [PubMed] [Google Scholar]
- 58. Huber H, et al. 2008. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc Natl Acad Sci. 105:7851–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hawkins AS, Han Y, Bennett RK, Adams MWW, Kelly RM. 2013. Role of 4-hydroxy buty rate-CoA synthetase in the CO2 fixation cycle in thermoacidophilic archaea. J Biol Chem. 288:4012–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu L, Huber H, Berg IA. 2020. Enzymes catalyzing crotonyl-CoA conversion to acetoacetyl-CoA during the autotrophic CO2 fixation in Metallosphaera sedula. Front Microbiol. 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martin WF, Neukirchen S, Zimorski V, Gould SB, Sousa FL. 2016. Energy for two: new archaeal lineages and the origin of mitochondria. Bioessays. 38:850–856. [DOI] [PubMed] [Google Scholar]
- 62. Macleod F, Kindler GS, Wong HL, Chen R, Burns BP. 2019. Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol. 5:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schönheit P, Schäfer T. 1995. Metabolism of hyperthermophiles. World J Microbiol Biotechnol. 11:26–57. [DOI] [PubMed] [Google Scholar]
- 64. Schocke L, Bräsen C, Siebers B. 2019. Thermoacidophilic Sulfolobus species as source for extremozymes and as novel archaeal platform organisms. Curr Opin Biotechnol. 59:71–77. [DOI] [PubMed] [Google Scholar]
- 65. Berg IA, Kockelkorn D, Buckel W, Fuchs G. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in archaea. Science. 318:1782–1786. [DOI] [PubMed] [Google Scholar]
- 66. Destan E, et al. 2021. Structural insights into bifunctional thaumarchaeal crotonyl-CoA hydratase and 3-hydroxypropionyl-CoA dehydratase from Nitrosopumilus maritimus. Sci Rep. 11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu L, Schubert DM, Könneke M, Berg IA. 2021. (S)-3-hydroxybutyryl-CoA dehydrogenase from the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Nitrosopumilus maritimus. Front Microbiol. 12:712030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Müh U, Çinkaya I, Albracht SPJ, Buckel W. 1996. 4-hydroxybutyryl-CoA dehydratase from Clostridium aminobutyricum: characterization of FAD and iron-sulfur clusters involved in an overall non-redox reaction. Biochemistry. 35:11710–11718. [DOI] [PubMed] [Google Scholar]
- 69. Estelmann S, et al. 2011. Carbon dioxide fixation in “Archaeoglobus lithotrophicus”: are there multiple autotrophic pathways?. FEMS Microbiol Lett. 319:65–72. [DOI] [PubMed] [Google Scholar]
- 70. Spring S, Riedel T. 2013. Mixotrophic growth of bacteriochlorophyll a-containing members of the OM60/NOR5 clade of marine gammaproteobacteria is carbon-starvation independent and correlates with the type of carbon source and oxygen availability. BMC Microbiol. 13:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang Z, Guo F, Liu L, Zhang T. 2014. Evidence of carbon fixation pathway in a bacterium from candidate phylum SBR1093 revealed with genomic analysis. PLoS One. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Furnham N, Garavelli JS, Apweiler R, Thornton JM. 2009. Missing in action: enzyme functional annotations in biological databases. Nat Chem Biol. 5:521–525. [DOI] [PubMed] [Google Scholar]
- 73. Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zimmermann J, Kaleta C, Waschina S. 2020. gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models. Genome Biol. 22:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Parks DH, et al. 2020. A complete domain-to-species taxonomy for bacteria and archaea. Nat Biotechnol. 38:1079–1086. [DOI] [PubMed] [Google Scholar]
- 77. Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Download for the 52,515 MAGs and their amino acids files is available at https://genome.jgi.doe.gov/GEMs and https://portal.nersc.gov/GEM. HMM models used are available at https://github.com/alegarritano/HMM_CFP.