Abstract
STUDY QUESTION
What are the cellular composition and single-cell transcriptomic differences between myometrium and leiomyomas as defined by single-cell RNA sequencing?
SUMMARY ANSWER
We discovered cellular heterogeneity in smooth muscle cells (SMCs), fibroblast and endothelial cell populations in both myometrium and leiomyoma tissues.
WHAT IS KNOWN ALREADY
Previous studies have shown the presence of SMCs, fibroblasts, endothelial cells and immune cells in myometrium and leiomyomas. However, there is no information on the cellular heterogeneity in these tissues and the transcriptomic differences at the single-cell level between these tissues.
STUDY DESIGN, SIZE, DURATION
We collected five leiomyoma and five myometrium samples from a total of eight patients undergoing hysterectomy. We then performed single-cell RNA sequencing to generate a cell atlas for both tissues. We utilized our single-cell sequencing data to define cell types, compare cell types by tissue type (leiomyoma versus myometrium) and determine the transcriptional changes at a single-cell resolution between leiomyomas and myometrium. Additionally, we performed MED12-variant analysis at the single-cell level to determine the genotype heterogeneity within leiomyomas.
PARTICIPANTS/MATERIALS, SETTING, METHODS
We collected five MED12-variant positive leiomyomas and five myometrium samples from a total of eight patients. We then performed single-cell RNA sequencing on freshly isolated single-cell preparations. Histopathological assessment confirmed the identity of the samples. Sanger sequencing was performed to confirm the presence of the MED12 variant in leiomyomas.
MAIN RESULTS AND ROLE OF CHANCE
Our data revealed previously unknown heterogeneity in the SMC, fibroblast cell and endothelial cell populations of myometrium and leiomyomas. We discovered the presence of two different lymphatic endothelial cell populations specific to uterine leiomyomas. We showed that both myometrium and MED12-variant leiomyomas are relatively similar in cellular composition but differ in cellular transcriptomic profiles. We found that fibroblasts influence the leiomyoma microenvironment through their interactions with endothelial cells, immune cells and SMCs. Variant analysis at the single-cell level revealed the presence of both MED12 variants as well as the wild-type MED12 allele in SMCs of leiomyomatous tissue. These results indicate genotype heterogeneity of cellular composition within leiomyomas.
LARGE SCALE DATA
The datasets are available in the NCBI Gene Expression Omnibus (GEO) using GSE162122.
LIMITATIONS, REASONS FOR CAUTION
Our study focused on MED12-variant positive leiomyomas for single-cell RNA sequencing analyses. Leiomyomas carrying other genetic rearrangements may differ in their cellular composition and transcriptomic profiles.
WIDER IMPLICATIONS FOR THE FINDINGS
Our study provides a cellular atlas for myometrium and MED12-variant positive leiomyomas as defined by single-cell RNA sequencing. Our analysis provides significant insight into the differences between myometrium and leiomyomas at the single-cell level and reveals hitherto unknown genetic heterogeneity in multiple cell types within human leiomyomas. Our results will be important for future studies into the origin and growth of human leiomyomas.
STUDY FUNDING/COMPETING INTEREST(S)
This work was supported by funding from the National Institute of Child Health and Human Development (HD098580 and HD088629). The authors declare no competing interests.
Keywords: uterine leiomyomas, single-cell RNA sequencing, MED12, cellular heterogeneity, myometrium
Introduction
Uterine leiomyomas, also known as fibroids, are benign tumors of the myometrium, affecting over 70% of women at some point during their lifetime. Approximately, 25–50% of these affected women experience severe clinical symptoms such as pelvic pain, infertility and heavy uterine bleeding (Catherino et al., 2011; Bulun, 2013; Stewart et al., 2016). These tumors can significantly affect women’s quality of life and are the single most common underlying cause of hysterectomy (Stewart, 2001). Each year, approximately 300 000 myomectomies and 200 000 hysterectomies are performed in the United States to remove either leiomyoma tumors or the whole uterus (Farquhar and Steiner, 2002; Cardozo et al., 2012). Despite the importance to women’s health, there are currently no leiomyoma-specific therapeutics. Moreover, our understanding of the origin and heterogeneity of leiomyomas continues to evolve.
Leiomyomas are considered to be monoclonal tumors of smooth muscle cells (SMCs) in the uterus (Bulun, 2013). Several studies suggest that the leiomyomas originate from a single SMC that acquires a somatic mutation (Wu et al., 2017). However, recent histological and flow cytometry studies have shown the presence of SMCs, fibroblasts, endothelial cells and immune cells in leiomyomas (Holdsworth-Carson et al., 2014a,b). Extensive genetic studies from our group and others have shown that 70% of the total leiomyoma cases are associated with genetic variants in MED12 exon 2 (Makinen et al., 2011a,b; McGuire et al., 2012; Halder et al., 2015), whereas, the MED12-variant negative leiomyomas have a highly heterogeneous genomic landscape (Mehine et al., 2013; Yatsenko et al., 2017). Studies have shown that MED12-variant negative leiomyomas are larger than leiomyomas expressing the MED12-variant allele (Rein et al., 1998). These size differences in leiomyoma are due to differences in the cell composition, rate of proliferation and accumulation of extracellular matrix (Wu et al., 2017). More recent evidence suggests that MED12-variant positive leiomyomas are composed of MED12-variant carrying SMCs and wild-type MED12 carrying fibroblasts. This study suggests that MED12-variant carrying leiomyomas form because of interactions between mutant smooth muscle and non-mutant fibroblast cells in the myometrium (Wu et al., 2017). The contribution of these cells to leiomyoma formation and molecular salient characteristics of the cellular heterogeneity in leiomyomas remains undetermined.
Here we utilized single-cell RNA sequencing to understand the underlying cellular heterogeneity in the myometrium and uterine leiomyomas. We generated a single-cell atlas for both myometrium and leiomyomas and identified previously unknown lymphatic endothelial cell (Lymphatic EC) populations in the uterine leiomyomas. We also determined the transcriptomic changes in the leiomyoma cell clusters compared to the myometrium. Based on gene ontology (GO) analysis, we identified the presence of translational and metabolic differences in the smooth muscle and fibroblast cell populations in leiomyomas compared to the myometrium. Our data shows that MED12-variant positive leiomyomas are composed of both MED12-variant positive and MED12-variant negative SMCs, suggesting that the leiomyoma origin is not monoclonal in nature.
Materials and methods
Study subjects
To resolve the cellular identity of the cells, we collected 96 573 cells from the leiomyomas and the myometrium (n = 5 patients/group). Histopathological assessment of the collected samples confirmed the identity of the samples.
Patient tissue collection and genotyping
The study was approved by the UCSF institutional review board. Uterine leiomyomas and myometrium were collected from patients undergoing hysterectomy or myomectomy due to leiomyoma presentation with informed consent. Two of the five leiomyoma samples used in the study had matched myometrium. The other three leiomyoma samples were collected from myomectomy cases where no matching myometrium was available. In cases where only myometrium sample was used in the study, the leiomyoma sample was either too small to collect enough cells to proceed for the single-cell RNA-sequencing, or the leiomyoma collected did not carry the MED12 variant. Myometrium was collected at a distance greater than 3 cm away from the tumor site by experienced pathology assistants at UCSF. All specimens were reviewed and confirmed to be either leiomyoma or myometrium by a board-certified pathologist. Fresh tissue samples were collected after the surgery, placed in ice-cold DMEM/F12, and immediately sent for further processing. A small part of each tissue was snap-frozen to perform DNA isolation for genotyping to confirm the presence of the MED12 variant as described previously by us (McGuire et al., 2012). Additionally, a part of the sample was fixed in formalin overnight for histology. Patient information is provided in Supplementary Table SI.
Preparation of single-cell suspensions from the fresh tissues
The tissue samples were collected and washed in HBSS (Sigma). Leiomyomas and myometrium were then cut into 3–4 mm pieces. These pieces were then added to 3–4 ml of digestion media containing 0.1 mg/ml Liberase (Roche, 501003280), 100 U/ml DNase I (Sigma, D4527) and 25 U/ml Dispase (Sigma, D4818) in DMEM (Life Technologies, 12634010) per gram of the tissue and mechanically dissociated using gentleMACS dissociator (Miltenyi Biotech, Germany) for 30 min at 37°C to prepare a single-cell suspension. The cell suspension was then pipetted up and down with 25, 10 and 5 ml pipettes for 1 min each and then filtered through a 70-µm filter (Corning, 431751). Debris was then removed from the cell suspension using a debris removal solution (Miltenyi Biotech, 130-109-398) as per the manufacturer’s instructions. The cells were then incubated with the red blood cell (RBC) lysis buffer (Thermo Fisher Scientific, 00-4333-57) for 5 min on ice to remove the RBCs. The cells were then resuspended in PBS containing 0.4% ultrapure bovine serum albumin (BSA) (Thermofisher Scientific, AM2616) and passed through a 70-µm cell strainer (Bel-Art, H13680-0070) to obtain a single-cell suspension.
Single-cell RNA sequencing
Freshly isolated single cells were processed through the 10× Chromium system (10× Genomics, USA) using single-cell 5′ library and Gel bead kit (10× Genomics) as per the manufacturer’s instructions. Briefly, the single-cell suspensions were partitioned into gel bead‐in‐emulsions which were used to generate the barcoded cDNA libraries. The single-cell barcoded cDNA libraries were then sequenced using an Illumina NovaSeq 6000 sequencing system (Illumina, USA). Matched myometrium and leiomyoma samples from the same patient were processed and sequenced at the same time. Wherever possible, samples from different patients were sequenced at the same time.
Pre-processing single-cell RNA-sequencing data
Cellranger v.2.1.0 single-cell software suite from 10× Genomics was used to demultiplex FastQ files, align reads to the Genome Reference Consortium Human Build 38 (hg38) transcriptome, and extract cell and UMI barcodes. Raw cell count by transcript matrices were imported into R and Seurat was used for analysis (Stuart et al., 2019). Each sample’s expression matrix was filtered to remove low-quality cells, defined as having fewer than 200 unique genes, greater than 2500 unique genes, or more than 7% mitochondrial gene expression (Ilicic et al., 2016). Samples were merged using Seurat’s integration anchors (Butler et al., 2018). Any cells with more than 1% of hemoglobin genes, HBA2, HBA1 and HBB were removed. To eliminate technical differences, the transcript counts were normalized using Seurat’s default parameters (Hafemeister and Satija, 2019).
Dimensionality reduction, clustering and differential expression analysis
Seurat (version 3.1.1) was used to cluster the merged object into subsets of cells. This workflow includes finding variable genes, running principal component analysis on variable genes, running Uniform Manifold Approximation and Projection on Principal components (UMAP) (1:20) and graphed clustering using KNN and Louvain clustering (with Seurat FindClusters resolution 0.4). Cell types were defined empirically using the expression of marker genes. Cells were partitioned into smooth muscle, fibroblasts, endothelial and immune cells based on the known signature markers. The following signature markers were used to identify the cell clusters: SMCs (MYH11, TAGLN, ACTA2, CNN1, DES, CALD1), fibroblast cells (VIM, ALDH1, CD90, FN1, DCN, OGN, MGP, COL1A1, COL1A2, COL3A1), endothelial cells (PECAM1, CD31, CDH11, VWF) and immune cells (CD3D, CD3E, FCER1G, MS4A1, CD79B, CST7, GZMB, FCGF3A, MS4A7).
Further clustering was performed for each cell type based on the expression of signature markers (with varying resolution) to delineate heterogeneity. Seurat’s Integration anchors were performed again on each subset of cells (SMC, fibroblast, endothelial and immune cells) to avoid batch effects in the cell subsets. All analysis was performed using five leiomyoma and myometrium samples unless otherwise mentioned in the Results section. All clusters heatmaps, t-distributed stochastic neighbor embedding (t-SNE), UMAP visualizations, violin plots and dot plots were produced using Seurat functions with ggplot2, heatmap and grid R packages. Differential gene expression analysis was performed using find markers in Seurat with the non-parametric Wilcoxon rank-sum test. Receptor ligand analysis was performed using Cellchat (Jin et al., 2021).
Volcano plots
Seurat’s FindMarkers with default parameters was used to determine the fold change between the MED12-positive leiomyomas and the myometrium. All data were compiled and plotted in R with ggplot2.
Immunofluorescence and RNAscope
Immunofluorescence was performed as described previously (Goad et al., 2017). Briefly, tissue sections were incubated with the primary antibodies PDPN (1:200, Cell Signaling) and NUSAP1 (1:1000, Abcam) overnight at 4°C, then incubated with secondary antibodies for 1 h at room temperature. For immunofluorescence, the images for all the samples were taken at the same exposure using a Nikon microscope (Nikon, Japan). RNAscope Fluorescent multiplex assay was performed using the RNAscope Multiplex Fluorescent v2 kit (Advanced Cell Diagnostics) as per the manufacturer’s instructions. Images were obtained using the Leica SP8 confocal microscope (Leica Biosystems, Germany).
Clonality analysis
Bam files from single-cell RNA sequencing data were uploaded to Integrative Genome Viewer (IGV) to perform variant analysis (Robinson et al., 2011, 2017). The sequencing data were analyzed for the presence of MED12 variants associated with leiomyomas in exon 2 on the X-chromosome (Makinen et al., 2011b; McGuire et al., 2012). Variants detected in the IGV analysis were confirmed by Sanger sequencing. Dsc-pileup in the freemuxlet tool with standard parameters was used to construct the barcode and variants table (Kang et al., 2018; Pique-Regi et al., 2019). The location of the MED12 variants was input into the pipeline and reads that matched the reference base and those that did not match with the reference base were counted. Variants from cells that weren’t filtered by preprocessing were used in conjunction with Seurat metadata to print UMAPs with variant information.
Statistical analysis
Statistically significant ligand–receptor interaction was found based on a permutation test on P-values obtained from a Hill function modeled based on the law of mass actions.
Results
Single-cell atlas of the human myometrium and leiomyomas
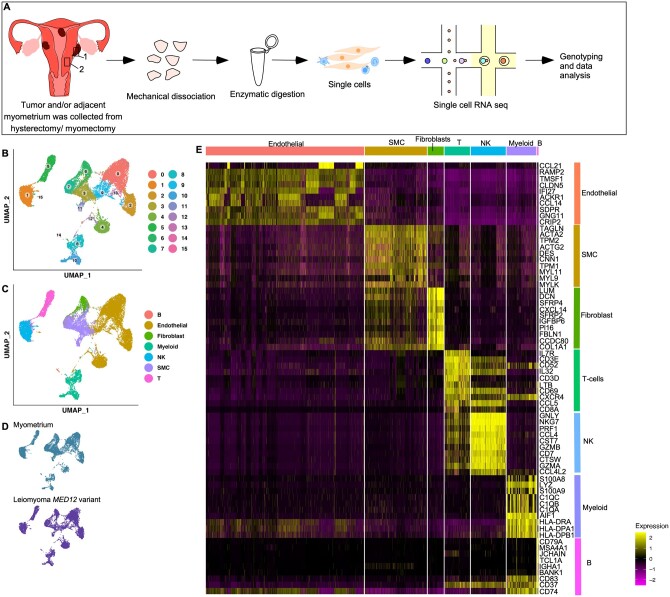
We collected a total of 96 573 cells from the myometrium and leiomyomas samples (n = 5 patients/sample) (Fig. 1A). After accounting for technical and biological variation, clustering of the 24 605 high-quality cells (10 001 from myometrium (n = 5) and 14 604 from MED12-variant positive (n = 5)) revealed the presence of 16 different clusters across known cell lineages (Fig. 1B and C). These cell clusters were highly reproducible as all clusters were represented in all patient samples (Supplementary Fig. S1A). The subpopulations included known cell types previously identified through histology and flow cytometry (Holdsworth-Carson et al., 2014b). These included: SMCs, fibroblasts, NK cells, T cells, B cells, myeloid cells and endothelial cells (Fig. 1C). The annotations were performed using differential gene expression analysis supported by known gene markers such as ACTA2, CNN1 for SMCs, VWF and PECAM for endothelial cells (Endo), PDPN Lymphatic ECs, DCN and LUM for fibroblasts (Fibro), CD3D for T cells, MS4A1 and CD79A for B cells, GNLY and NKG7 for NK cells and CD14 and S100A8 for myeloid cells (Fig. 1E and Supplementary Fig. S1C and D). All cell clusters were present in most patient samples across both the myometrium and the MED12-variant positive leiomyomas (Fig. 1D and Supplementary Fig. S1A). The contaminating endometrial cells (EPCAM+ or KRT8+) and RBCs expressing HBB, HBA1 and HBA2 were removed from the datasets. Quality control matrices for each patient sample and cell clusters are provided (Supplementary Fig. S1B and E).
Figure 1.
Single-cell atlas of myometrium and leiomyomas. (A) Overview of the dissociation protocol for both myometrium (n = 5) and leiomyomas (n = 5). (B) Clustering of 24 605 high-quality cells from MED12-variant positive leiomyomas and myometrium. (C) Cell lineages identified by the marker gene expression. (D) Annotation of the cell clusters per sample. (E) Heatmap showing at least the top nine genes used for cluster identification. Columns denote cells; rows denote genes. SMC, smooth muscle cell; NK, natural killer cells; T, T cells; B, B cells.
Characterization of the smooth muscle cells and fibroblast populations in myometrium
We first generated a UMAP from the myometrium dataset to examine the cell types within the myometrium. We identified the presence of 16 different cell type clusters. These clusters included SMCs, fibroblast cells, endothelial cells, Lymphatic ECs and immune cell populations (Supplementary Fig. S2A).
To determine if there is an additional underlying heterogeneity in the SMC population of the myometrium, we extracted all the cells expressing TAGLN, CNN1 and SMA, and clustered them at a higher resolution. These genes are known markers of SMC in the uterus and other tissues (Holdsworth-Carson et al., 2014a,b; Latif et al., 2015). Additional re-clustering of these cells revealed three different SMC clusters in the myometrium samples (Supplementary Fig. S2B and F). We removed one myometrial sample from this analysis as the number of cells contributed by this sample did not meet the minimum parameters required for batch correction using Seurat integration. All the clusters were present in the remaining four patient samples, indicating reproducibility and the absence of technical or batch effects (Supplementary Fig. S2C). Quality control matrices for SMC are provided (Supplementary Fig. S3A).
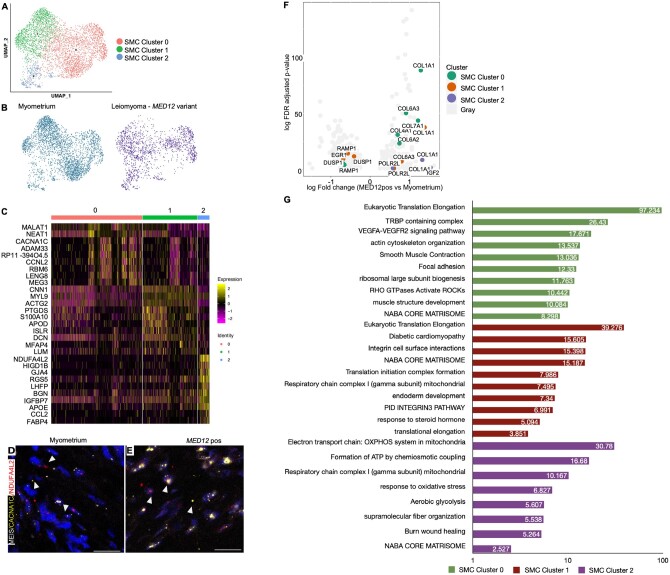
To delineate the functional role of SMCs, we performed GO enrichment analysis. Functional enrichment analysis with the biological function revealed that SMC Cluster 0 (CACNA1C+MEIS1−) was associated with collagen formation (Supplementary Table SII). SMC Cluster 1 (CACNA1C+MEIS+) expressed markers for both fibroblast cells and SMCs, indicating that these cells might be myofibroblasts. We found these cells were associated with the muscle structure development and smooth muscle contraction (Supplementary Table SII). SMC Cluster 2 was defined by the expression of NDUFA4L2. These cells showed the upregulation of genes responsible for regulating the cellular response to stimulus, translation, regulation of SMC proliferation and cellular glycolysis. A full list of the GO processes for all the clusters is provided in Supplementary Table SII. We performed in-situ hybridization to validate the biological presence of these clusters using the markers CACNA1C+MEIS1− (SMC Cluster 0), CACNA1C+MEIS+ (SMC Cluster 1) and NDUFA4L2 (SMC Cluster 2) (Fig. 2D and E). As indicated by the heatmap, we identified cells that co-expressed markers of clusters 1 and 2 in our in-situ analysis.
Figure 2.
Heterogeneity and transcriptomic changes in smooth muscle cells (SMCs) in MED12-variant positive leiomyomas compared to the myometrium. (A) Uniform Manifold Approximation and Projection on Principal (UMAP) components showing the clusters of 4650 SMCs. (B) UMAP showing that all cell clusters are present in the myometrium and MED12-variant positive leiomyomas. (C) Heatmap of the SMC clusters. The colored bar on the top represents the cluster number. Columns denote cells; rows denote genes. (D, E) In-situ images showing the validation of the SMC clusters in MED12-variant positive leiomyomas and myometrium. Scale bar is 50 µm. (F) Volcano plots showing the transcriptomic changes in the SMC clusters of MED12-variant positive cells compared to the myometrium. (G) Gene ontology (GO) analysis of DE genes showing at least 0.5-fold change in the MED12-variant positive leiomyomas as compared to the myometrium.
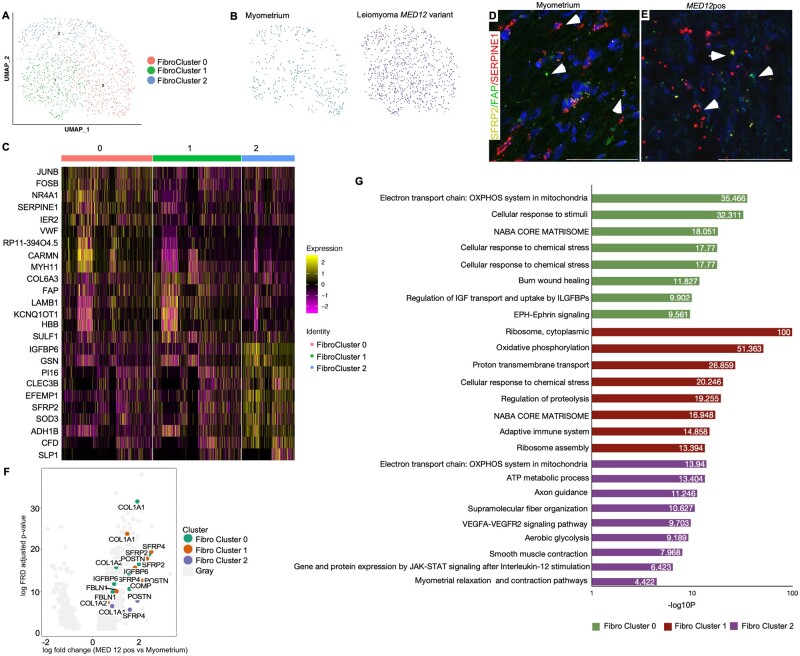
Our data analysis also identified three distinct cell subsets within fibroblast (Fibro) cell populations, present across all patient samples (Supplementary Fig. S2D, E and G). Quality control matrices for fibroblast clusters are provided (Supplementary Fig. S3B). Fibro cluster 0 identified by the expression of SERPINE1, IER2 and NR4A1 significantly resembled myofibroblast due to co-expression of SMC markers (CARMN, DES) as well as fibroblast cell markers (DCN). Interestingly, these cells expressed several markers in common with Fibro clusters 1 and 2 (Supplementary Fig. S2G). GO enrichment analysis revealed that the fibro cluster 0 was mainly involved in regulating the actin filament-based process, PI3K-Akt pathway and striated muscle contraction pathway (Supplementary Table SIII). Fibro cluster 1 (expressing FAP gene) was linked to integrin pathway and axon guidance, while fibro cluster 2 (expressing SFRP2 and PI16 genes) was enriched in terms related to the regulation of extracellular matrix organization, regulation of insulin growth factor transport and uptake by insulin growth factor binding proteins (Supplementary Table SIII). In-situ hybridization confirmed the presence of three different fibroblast cell populations (Fig. 3D and E).
Figure 3.
Intracellular heterogeneity and transcriptomic changes in fibroblast population in MED12-variant positive leiomyomas compared to the myometrium. (A) Uniform Manifold Approximation and Projection on Principal (UMAP) components showing the clusters of 1197 fibroblast cells. (B) UMAP showing that all cell clusters are present in the myometrium and MED12-variant positive leiomyomas. (C) Heatmap of the fibroblast cell clusters. The colored bar on the top represents the cluster number. Columns denote cells; rows denote genes. (D, E) In-situ images showing the validation of fibroblast populations in the myometrium and MED12-variant positive leiomyomas. Scale bar is 50 µm. (F) Volcano plots showing the transcriptomic changes in the fibroblast cell clusters of MED12-variant positive cells compared to the myometrium. (G) Gene ontology (GO) analysis of DE genes showing at least 0.5-fold change in the MED12-variant positive leiomyomas as compared to the myometrium.
Cellular heterogeneity in MED12-variant positive leiomyomas
Next, we generated a UMAP for the leiomyoma-only dataset. We performed single-cell sequencing on five leiomyomas. These leiomyomas, like the majority, about 70% of leiomyomas, carry somatic mutations in MED12 (Supplementary Table SI). Two of the leiomyoma samples had matched myometrium samples while the other three leiomyoma samples were unmatched. We identified, within the leiomyoma samples, the presence of the same 16 distinct cell clusters as identified in the myometrium samples (Supplementary Fig. S4A and C). These cell clusters included SMCs, fibroblasts, endothelial cells, Lymphatic ECs, B-cells, T-cells and myeloid cells (Supplementary Fig. S4B and C). We then isolated the smooth muscle, fibroblast, endothelial cells and immune cell populations from the integrated dataset and resolved them at a higher resolution. Again, like myometrium, our data analysis revealed the presence of three different SMC populations in the leiomyomas (Fig. 2A–C). Furthermore, all these SMC clusters were the same as those present in the myometrium (Supplementary Fig. S2B). Next, we wanted to determine if similar heterogeneity exists in the fibroblast cell population in leiomyomas. We found the presence of three distinct fibroblast subsets in both the myometrium and the leiomyomas (Fig. 3A–C). Together, these results suggest that myometrium and MED12-variant positive leiomyomas have similar cellular composition.
Remodeling of the smooth muscle and fibroblast cell populations in leiomyomas compared to the myometrium
Our analysis so far indicated similar cellular composition between myometrium and leiomyomas (Fig. 1B–D). We, therefore, examined transcriptomic changes between myometrium and leiomyoma cell clusters.
We compared the transcriptomic changes in the SMC sub-clusters derived from the MED12-variant positive leiomyomas to their counterparts in the myometrium. GO analysis revealed enrichment of genes involved in the regulation of eukaryotic translation elongation in both clusters 0 and 1 of the MED12-variant positive leiomyomas compared to clusters 0 and 1 in the myometrium (Fig. 2F and G and Supplementary Table SIV). Furthermore, we found cluster-specific transcriptional differences in leiomyoma SMC clusters compared to those of the myometrium, indicating that these cells remodel and play a different role in leiomyoma formation. SMC cluster 0 showed enrichment of GO terms responsible for actin cytoskeleton organization, smooth muscle contraction, TRBP containing complex and VEFGF-VEGFA signaling. However, SMC cluster 1 was enriched in GO terms regulating integrin cell surface interactions, NABA CORE matrisome and translational initiation complex formation. SMC cluster 2 showed enrichment of GO terms involved in the electron transport chain, regulating glycolysis and formation of ATP coupling; this indicates metabolic differences in the SMC Cluster 2 of MED12-variant leiomyomas compared to the myometrium. The full list of other transcriptomic changes in SMC clusters is provided in Supplementary Table SIV.
Next, we decided to compare the transcriptional changes between the MED12-variant positive leiomyoma fibroblast subclusters and the myometrium fibroblast subclusters. We found that both clusters 0 and 1 regulate common biological pathways in leiomyomas (Supplementary Table SV). We found enrichment of genes involved in the cellular response to stimuli and EPH-EPHRIN signaling, specifically in the MED12-variant Fibro Cluster 0 compared to the myometrium (Fig. 3G). MED12-variant positive Fibro Cluster 1 showed enrichment of ribosome, cytoplasmic, oxidative phosphorylation and ribosome assembly, indicating a difference in translation activity in MED12-variant positive Fibro Cluster 1 compared to the same in myometrium (Fig. 3F and G). Fibroblast cluster 2 seemed to be regulated by WNT signaling in myometrium and MED12-variant positive leiomyomas (Fig. 3C–E). This cell cluster showed enrichment in genes responsible for response to wound healing, the ATP metabolic process, glycolysis, smooth muscle contraction, JAK-STAT signaling, and myometrium relaxation and contraction pathway compared to the Fibro cluster 2 in the myometrium (Fig. 3G and Supplementary Table SV). Collectively, these data suggest that SMCs and fibroblast cells remodel differently in leiomyomas compared to the myometrium.
Lymphatic ECs are present in uterine leiomyomas
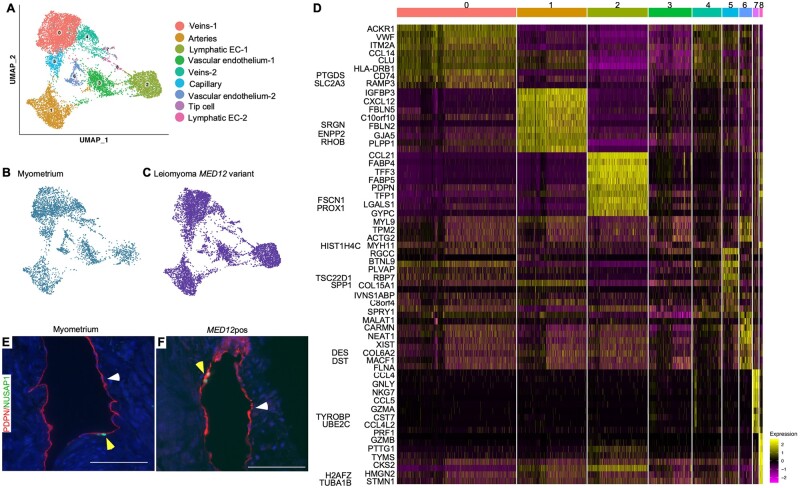
Unsupervised clustering of endothelial cells revealed the presence of seven clusters of endothelial cells in both the myometrium and the leiomyoma samples (Fig. 4A–D). Based on the differential gene expression, we identified these endothelial cell clusters as veins (Cluster 0 and 4; ACKR1+), arteries (Cluster 1; GJA5), Lymphatic ECs (Cluster 2 and 8; PDPN), capillaries (Cluster 5; RGCC), tip cells (Cluster 7; CXCR4) and vascular endothelial cells (Cluster 3 and 6; MYH11, FLNA). We observed that endo clusters 2 and 8, identified as Lymphatic EC clusters, were present in both the myometrium and the MED12-variant positive leiomyomas (Fig. 4E and F). It is noteworthy that Lymphatic ECs have previously been discovered in myometrium but not in leiomyomas. Lymphatic ECs are known to enable immune cell infiltration in tissue (Lucas and Tamburini, 2019). Our GO analysis revealed upregulation of genes involved in response to cytokines in lymphatic endothelial cells 1 (Lymphatic EC-1) in the MED12-variant positive leiomyomas compared to the Lymphatic EC-1 in the myometrium. However, the leiomyoma lymphatic endothelial cells 2 (Lymphatic EC-2) showed upregulation of the complement and coagulation cascade when compared to the Lymphatic EC-2 in the myometrium (Supplementary Table SVI). These data indicate that Lymphatic ECs might be involved in regulating immune response. Additionally, the GO analysis highlighted differences in biological processes in endothelial cells between the MED12-variant positive leiomyoma and the myometrium. We found an upregulation of genes involved in leukocyte migration, regulation of cytokine production and regulation of inflammatory response within leiomyomas as compared to the myometrium (Supplementary Table SVI).
Figure 4.
Lymphatic ECs are present in leiomyomas. (A) Uniform Manifold Approximation and Projection on Principal (UMAP) components showing the clusters of 10 448 endothelial cells. (B, C) UMAP showing the cluster annotation per condition: myometrium and MED12-variant positive leiomyomas. (D) Heatmap of endothelial cell clusters. The colored bar on the top represents cluster number. Columns denote cells; rows denote genes. (E, F) Immunostaining for PDPN (white arrowheads) and NUSAP1 (yellow arrowheads) showing the presence of Lymphatic EC clusters in normal myometrium (E) and MED12-variant positive leiomyomas (F). Scale bar is 100 µm.
Immune cell infiltration in MED12-variant positive leiomyomas and myometrium
We also analyzed the immune profile of the myometrium samples and compared it to the leiomyoma samples. Our data showed the presence of natural killer (NK) cells, B-cells, dendritic cells, T-cells, CD8+ve T-cells, monocytes, CD14 monocytes, FCER3A+ monocytes and macrophages in both the myometrium and the MED12-variant positive leiomyomas (Supplementary Fig. 5A–C). These results are in agreement with the previously reported immune cell populations in the myometrium (Siewiera and Erlebacher, 2020). GO analysis for the biological processes showed enrichment of terms associated with the adaptive immune response, antigen processing and presentation, and regulation of leukocyte activation in the MED12-variant leiomyomas when compared to the myometrium (Supplementary Table SVII). These data indicate preferential activation of adaptive immune response in the MED12-variant leiomyomas as compared to the myometrium.
Fibroblast populations drive cell–cell communication in leiomyomas
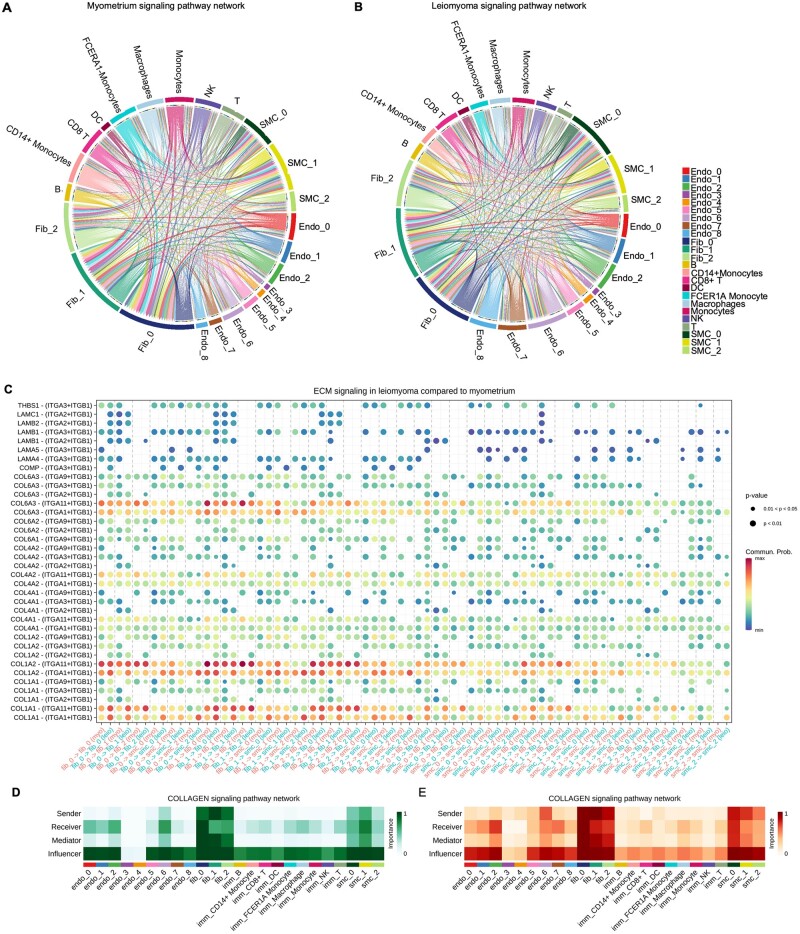
We performed cell–cell communication analysis using CellChat, a manually curated repository of receptors and ligands (Jin et al., 2021), to predict the cross talk between different cell types in leiomyomas and myometrium. Analysis of overall signaling pathways revealed interactions between SMCs, fibroblasts, endothelial cells and immune cells. We found differential expression of 2397 ligands and 1057 receptors in the leiomyoma samples and 2407 ligands and 1079 receptors in the myometrium samples (Supplementary Fig. S6A). Out of these, 564 receptor–ligand pair interactions were significantly upregulated in the leiomyomas and 98 receptor–ligand pair interactions were significantly downregulated in the leiomyomas (log fold change ≤ 0.5) compared to the myometrium. We found that the number of interactions in both SMCs and fibroblast populations increased in the leiomyomas compared to the myometrium (Supplementary Fig. S6B and C). However, our data suggested that the fibroblast populations are major drivers of the signaling in the leiomyomas. We found that fibroblasts were involved in a total number of 172 596 interactions in leiomyomas compared to 145 818 total number of interactions in myometrium (P-value < 0.05; Supplementary Fig. S6C), while SMCs were involved in a total of 119 081 interactions in leiomyomas compared to 107 148 total number of interactions in myometrium (Supplementary Fig. S6B). We predict that fibroblasts interact with immune cells and endothelial cells and might play a role in immune cell infiltration (Fig. 5A and B).
Figure 5.
Cell–cell communication in myometrium and leiomyomas. (A) Chord diagram showing the cell–cell interactions in myometrium. (B) Chord diagram showing cell–cell communication in MED12-variant positive leiomyomas. The perimeter of the chord diagram is color-coded by cellular subtype (colored outer arcs). The inner connecting lines show the interactions between target and receiver cell types. The connecting lines are color coded consistently with cellular subtypes. The thickness of each edge (edge weight) is proportional to the interaction strength. A thicker edge line indicates a stronger signal. (C) Dot plot showing the upregulated ECM signaling pathway receptor–ligand interactions in subcellular clusters. Color indicates the communication probability. Dot size indicates the P-value. (D, E) Heatmaps show the relative importance of each cell group based on the computed four network centrality measures of ECM signaling network in (D) myometrium and (E) leiomyoma.
Extracellular matrix deposition is a characteristic feature of leiomyomas. As expected, we found an increase in autocrine and paracrine collagen signaling from SMCs and fibroblasts (e.g., COL1A1-ITGA1+ITGB1, COL6A3-ITGA1+ITGB1, COMP-ITGA3+ITGB1) (Fig. 5C–E). These results indicate the participation of both SMCs and fibroblast populations in the collagen signaling network in the MED12-variant positive leiomyomas (Fig. 5D and E). We observed upregulation of SEMA3 signaling (SEMA3C-PLXNA1, SEMA3B-NRP1-PLXNA4) in the leiomyomas compared to the myometrium (Supplementary Fig. S6D–G). SEMA3C was predominantly expressed by the SMC clusters and the fibroblast cell clusters. Semaphorins are known to participate in wound healing and actin remodeling and are commonly associated with cells’ anchorage to the extracellular matrix (Jackson and Eickholt, 2009). The receptor for semaphorins, NRP1/2-PLXND1/2/4, was observed in cell types including SMCs, fibroblasts and endothelial cells. These data further support that both SMCs and fibroblasts contribute to actin remodeling in leiomyomas.
Furthermore, we also observed increased interactions between fibroblasts and immune cells in the MED12-variant positive leiomyomas (193 757 interactions) compared to the myometrium (162 762, P-value < 0.05; Supplementary Fig. S7A). Fibroblast and immune cell populations showed upregulation of JAG_NOTCH through interactions among JAG1/2 and NOTCH1/2/4 signaling components and the mTOR signaling pathway through the expression of ROCK2, PRSS23, OSBPL8, RPL38, and PDPK1 (Supplementary Fig. S7B).
SMCs are the major contributor of extracellular matrix secretion in MED12-variant positive leiomyomas
Leiomyomas are characterized by the presence of excess extracellular matrix. Both SMCs and fibroblast cell populations secrete collagen in MED12-variant positive leiomyomas (Wu and DeMayo, 2017). Our data suggest the presence of intracellular heterogeneity in both SMCs and fibroblasts. Using our data, we wanted to investigate which specific cellular populations are associated with collagen secretion. Therefore, we compared the expression of collagen genes associated with the leiomyomas in both the smooth muscle and the fibroblast cell clusters. In agreement with previous studies, we found upregulation of collagen genes in both smooth muscle and fibroblast cell clusters in the MED12-variant positive leiomyomas compared to the myometrium (Supplementary Fig. S8A and B). All three SMC clusters showed increased average expression of collagen genes compared to the myometrium (Supplementary Fig. S8A). In contrast, in fibroblast cell populations, we observed increased average expression of COL1A1, COL1A2 and COL3A1 in the MED12-variant positive leiomyomas compared to the myometrium. However, there was decreased average expression of COL4A1, COL5A2, COL6A2, COL6A3, COL7A1, COL12A1, COL16A1, FN1 and ADAM19 compared with the myometrium (Supplementary Fig. S8B). These results indicate that both SMCs and fibroblast cells contribute to extracellular matrix secretion in MED12-variant positive leiomyomas.
MED12-positive leiomyomas show reduced expression of hormone receptors
Uterine leiomyomas are dependent on ovarian steroid hormones for their growth (Kim and Sefton, 2012). A recent study utilizing the patient-derived tumor xenograft has shown that the fibroblast population in MED12-variant positive tumors are estrogen-responsive, while progesterone does not affect fibroblasts (Wu and DeMayo, 2017). Using our data, we wanted to identify cellular subcluster-specific changes in estrogen and progesterone receptor expression. We found that 15–20% of cells in the leiomyoma SMC cluster 0 expressed progesterone receptor compared to 10% of the cells in the myometrium SMC cluster 0 (Supplementary Fig. S8C). The average expression of progesterone receptors was reduced to 10% of cells in the SMC cluster 1 in the MED12-variant leiomyomas compared to 20% of the cells in SMC cluster 1 in the myometrium. We observed a decrease in the average expression of estrogen receptors in the SMCs in the MED12-variant leiomyomas compared to the SMCs in the myometrium (Supplementary Fig. S8C). In contrast, we observed a decrease in the average expression of both estrogen and progesterone receptors in all fibroblast clusters in the MED12-variant positive leiomyomas compared with the myometrium (Supplementary Fig. S8D). These data show overall reduced expression of hormone receptors in the MED12-variant leiomyomas compared to the myometrium.
MED12-variant effects on leiomyoma signaling pathways
The Wnt signaling pathway has been implicated in leiomyoma formation. Deletion of Wnt/β-catenin in mouse myometrium leads to the conversion of smooth muscle into adipocytes (Arango et al., 2005), while overexpression of Wnt/β-catenin in uterine mesenchyme results in the formation of leiomyoma-like lesions in the mouse uterus (Tanwar et al., 2009). It has been suggested that Wnt/β-catenin signaling is involved in regulating the leiomyoma stem cell population (Reya and Clevers, 2005). We wanted to investigate whether the MED12-variant affects the WNT signaling pathway. We found a decrease in expression of CTNNB1 in both SMCs and fibroblasts in the MED12-variant positive leiomyomas as compared to the myometrium (Supplementary Fig. S8E and F). We also found upregulation of SFRP2 in all fibroblast populations in the MED12-variant positive leiomyomas (Supplementary Fig. S8F). SFRP2 is a known negative regulator of the canonical WNT signaling pathway (Ladher et al., 2000).
Another known pathway associated with leiomyoma formation is the mTOR/PI3K signaling pathways in leiomyoma formation. Loss of REST is known to activate the mTOR/PI3K pathway leading to the proliferation of leiomyoma cells (Varghese et al., 2013). We found relatively decreased expression of REST in both smooth muscle and fibroblast cell clusters in the MED12-variant positive leiomyomas (Supplementary Fig. S8E and F). Interestingly, we also observed increased average expression of mTOR, TSC1 and TSC2 in only SMC cluster 0 in the MED12-variant leiomyomas compared to myometrium (Supplementary Fig. S8E). These data indicate SMC-cluster specific activation of the mTOR pathway in MED12-variant leiomyomas, which may be contributing to the cell proliferation of these cells in MED12-variant positive leiomyomas.
MED12-variant positive leiomyoma cell types are genetically heterogeneous
Our single-cell RNA sequencing data indicates the presence of cellular heterogeneity in the SMC populations, fibroblast cell populations and endothelial cell populations in the leiomyomas and the myometrium (Fig. 1B and C). If leiomyomas are purely monoclonal and derive from a ‘stem cell’, which gives rise to all the cell types within the leiomyoma, then all the cell types identified in leiomyomas would be expected to have MED12-mutant variant positive genotype. Single-cell RNA sequencing has an advantage over the previously used techniques to determine the clonality of the tumor because of the ability to capture variants in individual cells and interrogate variants at a higher resolution.
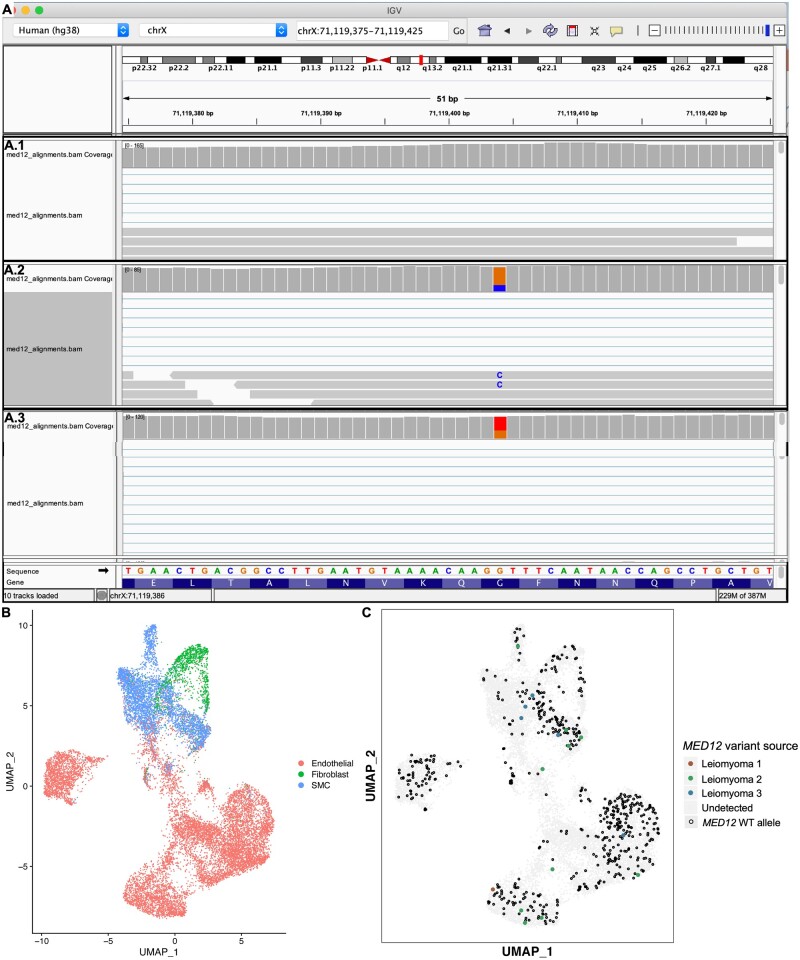
We performed variant analysis of the single-cell RNA sequencing datasets of the MED12-variant positive leiomyomas using Integrative Genomics Viewer (IGV). We exploited the MED12-variant location on the X-chromosome, subject to X-chromosome inactivation, as a marker to interrogate clonality. IGV analysis revealed expression of both the mutant variants (G44T, G44C) and the wild-type MED12 allele in MED12-variant positive leiomyomas (Fig. 6A.2–3). As expected, we did not observe the presence of the MED12-variant allele in the affected myometrium sample at the single-cell level (Fig. 6A.1). To determine which cells, express the MED12 variant, we generated a UMAP showing SMCs, fibroblasts, and endothelial cells from three MED12-positive fibroid samples (Fig. 6B). We generated a UMAP from only these cells to determine whether the expression of wild-type MED12 is a result of immune cell infiltration in the tumor. We then identified the MED12 variants in the single-cell RNA sequencing data by utilizing the dsc-pileup software developed from a previously published tool, demuxlet (Kang et al., 2018; Pique-Regi et al., 2019). We plotted the MED12 variants in the UMAP generated using the SMC, fibroblast and endothelial cell populations of the MED12-variant carrying leiomyomas. We found expression of MED12 variants (MED12 G > C 27% and MED12 wild-type allele 73%; MED12 G > T 61%, and MED12 wild-type allele 39%) in IGV analysis (Fig. 6A). The expression of the MED12-variant allele and wild-type allele varied from patient to patient with some patients expressing roughly 80% of the MED12-variant allele while others expressed only 20% of the MED12-variant allele within leiomyomas (Fig. 6A). Upon plotting the wild-type MED12 allele of these cells, we found the presence of the wild-type MED12 in all SMCs, fibroblast cells and endothelial cell clusters (Fig. 6C). Interestingly, we did not find MED12-variant allele-carrying cells in the fibroblast clusters (Fig. 6C). Our data indicate that MED12-variant positive leiomyomas are composed of genetically heterogeneous cell populations.
Figure 6.
MED12-variant positive uterine leiomyomas are not monoclonal. (A) Integrative Genome Viewer (IGV) analysis shows the presence of only wild-type MED12 allele in the myometrium (A.1) and the presence of both the variant and wild-type codon at c.131 in the MED12-variant positive leiomyomas (A.2 and A.3) in single-cell RNA sequencing data. The column med12 Bam alignment coverage shows the summary of the variant and wild-type allele detected at the c.131 location on MED12. The column MED12 alignment shows a snapshot of a small part of the data alignment track used to visualize individual MED12 aligned reads. (B) Uniform Manifold Approximation and Projection on Principal (UMAP) components of the mesenchymal cell populations in the leiomyomas. (C) UMAP showing the presence of the mutant MED12 variant (colored dots) and the wild-type MED12 (black dots) in SMCs, fibroblasts and endothelial cells.
Discussion
Uterine leiomyomas are thought to originate from SMCs in the myometrium. However, recent studies have reported the presence of fibroblasts and endothelial cells in uterine leiomyomas in addition to SMC populations (Holdsworth-Carson et al., 2014a; Wu et al., 2017), thus suggesting the presence of cellular heterogeneity in leiomyomas. We applied the single-cell RNA sequencing technique to create a cell atlas for myometrium and leiomyoma samples. Our data shows that the myometrium is composed of at least 16 different cell types, including different subtypes of SMCs, fibroblasts, vascular endothelial cells, lymphatic ECs, and immune cells. Both the myometrium and the leiomyomas share similar cell composition but show significant transcriptomic differences in smooth muscle and fibroblast populations.
Previous studies have established the differences in the transcriptomic profile between myometrium and leiomyomas (Kampjarvi et al., 2016; Moyo et al., 2020). These studies have consistently identified differential expression of genes regulating ECM metabolism (COL12A1, COL6A3, FN1, ADAM19), hormone responsiveness (PRLHR, EGFR, CYP26A1, EGR1) and muscle physiology (RGS2, CACNA1C, MYH2) (Kampjarvi et al., 2016; Moyo et al., 2020). We analyzed the transcriptional changes in leiomyomas and myometrium at the single-cell resolution. Like previous reports, we found upregulation of genes involved in collagen fibril organization and actin filament fragmentation (COL1A1, COL1A2, COL6A1, COL6A3, CFL1, DSTN), hormone-responsive genes (FOS, RHOA, IGFBP7), muscle structure development and contraction (EGR1, DMD, PDLIM1, SYNE1, SORBS1, KCNMA1) in leiomyomas compared to the myometrium. When comparing the transcriptomic changes at the single-cell level, we found that there were cell-cluster specific differences between the MED12-variant positive leiomyomas and the myometrium. We found that smooth muscle and fibroblast cells show upregulation of genes associated with the ECM metabolism. Both fibroblasts and SMCs influence ECM signaling in MED12-variant positive leiomyomas. Our results agree with a previous report that shows that both SMCs and fibroblasts are responsible for ECM production in MED12-variant positive leiomyomas (Wu et al., 2017).
Previous studies using human tissue and transgenic mouse models have implicated the role of Wnt/β-catenin, and mTOR signaling pathways in leiomyoma pathology (Tanwar et al., 2009; Prizant et al., 2013; Varghese et al., 2013; Commandeur et al., 2015; Kampjarvi et al., 2016). In agreement with these studies, our human single-cell RNA sequencing data show dysregulation of WNT signaling and mTOR signaling pathway components in different cell clusters of smooth muscle and fibroblast cell populations in MED12-variant positive leiomyomas. However, we found a smooth muscle or fibroblast cell cluster can have dysregulation of multiple signaling pathways in MED12-variant positive leiomyomas. It has been previously shown that overexpression of Wnt/β-catenin signaling in mesenchyme leads to leiomyoma-like tumors (Tanwar et al., 2009). However, these studies have not analyzed the relationship between MED12 mutations and Wnt signaling. We found negative regulation of Wnt/β-catenin signaling in both smooth muscle and fibroblast cells in MED12-variant positive leiomyomas. Interestingly, we found loss of REST and an increase in mTOR components in both smooth muscle and fibroblast populations in MED12-variant positive leiomyomas. Loss of REST is known to activate the mTOR/PI3K signaling pathway resulting in the proliferation of leiomyoma cells (Varghese et al., 2013). These results indicate that a specific signaling pathway might not be responsible for leiomyoma formation. We have previously shown that a common nonsynonymous MED12 exon 2 variant (c.131G>A) causes leiomyomagenesis in a mouse model (Mittal et al., 2015). Using our MED12 mouse model, we found dysregulation of the Wnt signaling pathway, Ras signaling and mTOR signaling pathways in leiomyomas (Mittal et al., 2015). The involvement of multiple pathways is likely due to MED12, which plays a known role as part of the mediator complex that regulates global RNA polymerase II-dependent transcription. Thus, the MED12 variant is likely to have a global effect (Makinen et al., 2011a,b).
Uterine leiomyomas are hormone-responsive in nature (Kim and Sefton, 2012). Previous studies have shown that leiomyomas increase in size with the presence of estrogen and progesterone (Bulun, 2013; Mittal et al., 2015). Leiomyomas with HMGA2 rearrangements and MED12 mutations show increased proliferation of SMCs in the presence of estrogen and progesterone, whereas the fibroblast population proliferates solely in response to estrogen (Wu et al., 2017). Our study shows an increase in the percentage of cells expressing estrogen and progesterone receptors in only one of the SMC clusters in MED12-variant positive leiomyomas compared to the myometrium. However, the other SMC clusters showed a decrease in the average expression of both estrogen and progesterone receptors in the leiomyomas. We also found reduced expression of estrogen and progesterone receptors in fibroblast cell clusters. It is possible that this one smooth muscle cluster is responsible for interacting with all other SMC clusters and fibroblast clusters in a paracrine manner to drive the leiomyoma growth. The expression of the estrogen and progesterone hormone pathway receptors and ligands were not statistically significant in our data to perform the CellChat analysis. Further mechanistic studies are warranted to understand these cellular interactions in response to ovarian hormones.
Lymphatic ECs were previously reported in myometrium but not in leiomyomas (Koukourakis et al., 2005; Red-Horse, 2008). In this study, we found the presence of two distinct populations of Lymphatic ECs in both myometrium and MED12-variant positive leiomyomas. In lymph nodes, the balance between pro-lymphangiogenesis and anti-lymphangiogenesis is maintained by inflammatory stimulus (Kim et al., 2014). Our data suggests that the second Lymphatic EC population detected in leiomyomas might be in response to inflammation. The Lymphatic ECs are known to facilitate the recruitment of immune cells to the tissue (Lucas and Tamburini, 2019).
We did not observe any difference in the immune cell populations between myometrium and MED12-variant positive leiomyomas. In our current analysis, we did not identify neutrophils and mast cells in either myometrium or leiomyomas. These cells were previously reported in myometrium by immunohistochemistry (Fox and Abell, 1965; Thomson et al., 1999; Siewiera and Erlebacher, 2020). The reason we did not identify these cells in our data may be due to neutrophils being sparsely present in the myometrium before the onset of labor. In addition, neutrophils are sensitive to degradation after sample collection (Thomson et al., 1999), whereas mast cells are mostly present on the luminal side of the myometrium (Mori et al., 1997) and may have been missed during our collection. We did observe transcriptomic differences in the immune cell populations of leiomyoma compared with myometrium, indicating the activation of adaptive immune response in MED12-variant leiomyomas.
Based on studies utilizing the HUMARA assay and the inactivation of glucose-6-phosphate dehydrogenase isoform expression, uterine leiomyomas are widely accepted to be monoclonal in nature (Linder and Gartler, 1965; Mashal et al., 1994). These assays are widely used to study the clonality of different tumors and tissues (Kopp et al., 1997). However, recent studies have questioned the accuracy of these assays which are partly affected by inconsistent methylation of X-chromosome genes (Carrel and Willard, 2005; Swierczek et al., 2012). MED12 mutation is present in 70% of leiomyomas (McGuire et al., 2012). MED12 is located on the X-chromosome and, because of X chromosome inactivation, either the variant or the wild-type MED12 allele will be expressed (Ercan, 2015). It has previously been shown that MED12-variant positive leiomyomas express the variant allele (Makinen et al., 2011b; McGuire et al., 2012). If leiomyomas were monoclonal and originated from a single MED12-variant carrying stem cells, then all the cells present in a leiomyoma would be expected to carry the same MED12 variant. Our analysis of the data, exploiting the unique genotype of MED12-variant positive leiomyomas, shows that uterine leiomyomas’ cellular moiety is heterogeneous by genotype. We found that MED12-variant positive leiomyomas were composed of a mixture of cells expressing both MED12-variant allele as well as MED12 wild-type allele. Other studies using alternative techniques support our findings. FACS sorting study of leiomyomas into SMCs and fibroblast populations revealed that MED12-variant positive leiomyomas are composed of SMCs carrying MED12-variant allele whereas the fibroblast cells in leiomyomas do not carry the MED12 variant (Wu et al., 2017). It was concluded that the leiomyoma causative mutations are present in SMCs only. However, we found that leiomyomas are a mixture of cells expressing both the MED12-variant and wild-type allele in multiple cell types examined, which included SMCs and endothelial cells. Interestingly, like previous observations, we did not find any MED12-variant carrying cells in the fibroblast cell population. Although our results show that leiomyoma cellular moiety is heterogenous by genotype, the early origin of leiomyomas remains unclear. It is still possible that “a Med12-variant positive stem cell” initiates events that lead to the recruitment of surrounding, wild-type cells to support leiomyoma growth. These findings are of particular significance because all the experimental design for understanding leiomyoma biology assumes that leiomyomas are monoclonal in origin. Previous studies have shown that the fibroblast cell population overtakes the MED12-mutant cells in the primary cell culture of leiomyomas (Bloch et al., 2017). Our research findings are consistent with the interpretation that fibroblasts are the major drivers of cell–cell communication in leiomyomas.
There is a chance that we might have lost a few cellular subtypes in our analysis due to filtering out low-quality cells. The presence of extracellular matrix and a bone-like density makes it hard to acquire good quality single-cell suspension from leiomyomas in a time-limited manner critical for the success of single-cell RNA sequencing experiments. Despite using fresh samples in our experiment, we still had to remove a significant number of dead cells prior to sequencing and analysis. Future studies and developing methodologies to work around hypocellular and hard tissues like leiomyomas will likely add to our current findings.
In conclusion, the single-cell atlas of human myometrium and leiomyomas shows the presence of intracellular heterogeneity in smooth muscle and fibroblast cells. We found the presence of Lymphatic ECs in leiomyomas. We found that both myometrium and leiomyomas have similar cellular composition. The main difference between the MED12-variant leiomyomas and myometrium are transcriptomic changes that may account for hormone responsiveness and ECM accumulation. Moreover, our studies show that leiomyoma cell moiety is genetically heterogenous, which should be a major consideration when designing future experiments and therapeutics against leiomyomas.
Supplementary Material
Acknowledgements
We thank Dr Rohit K. Gupta for helping us with the sample preparation protocol, Meghana Sukhthankar for helping with the sample preparation and Dr Katja Rust and Tania Moody for discussions about the data analysis. We thank all of the Rajkovic lab members for the critical reading of the document.
Authors’ roles
J.G. and A.R. conceived the study. J.G. designed and performed the experiments, data analysis and interpretation. A.R. supervised the study, designed experiments and performed the data analysis. J.R. and M.Z. performed the computational analysis. J.J.W. helped with the pathological examination of the tissues used in the study. M.T., Y.D., S.B. and D.C. helped with the data analysis. J.G. and A.R. wrote the manuscript with input from all of the authors.
Funding
This work was supported by funding from the National Institute of Child Health and Human Development (5P50HD098580).
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Jyoti Goad, Department of Pathology, University of California, San Francisco, CA, USA; Department of Obstetrics, Gynecology and Reproductive Sciences, University of California, San Francisco, CA, USA.
Joshua Rudolph, Department of Medicine, Lung Biology Center, University of California, San Francisco, CA, USA.
Mehrdad Zandigohar, Department of Biomedical Engineering, University of Illinois at Chicago, Chicago, IL, USA.
Matthew Tae, Department of Pathology, University of California, San Francisco, CA, USA.
Yang Dai, Department of Biomedical Engineering, University of Illinois at Chicago, Chicago, IL, USA.
Jian-Jun Wei, Department of Pathology, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA.
Serdar E Bulun, Division of Reproductive Sciences in Medicine, Department of Obstetrics and Gynecology, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA.
Debabrata Chakravarti, Division of Reproductive Sciences in Medicine, Department of Obstetrics and Gynecology, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA.
Aleksandar Rajkovic, Department of Pathology, University of California, San Francisco, CA, USA; Department of Obstetrics, Gynecology and Reproductive Sciences, University of California, San Francisco, CA, USA; Institute of Human Genetics, University of California, San Francisco, CA, USA.
Data Availability
The datasets generated and analyzed in the study are available in the NCBI Gene Expression Omnibus (GEO) (GSE162122) and Sequence Read Archive (SRA) (SRP294127). Code is available at: https://github.com/JyotiGoad/Single_Cell_Atlas_Of_Leiomyoma. All custom scripts can be accessed upon request to the corresponding author.
References
- Arango NA, Szotek PP, Manganaro TF, Oliva E, Donahoe PK, Teixeira J.. Conditional deletion of beta-catenin in the mesenchyme of the developing mouse uterus results in a switch to adipogenesis in the myometrium. Dev Biol 2005;288:276–283. [DOI] [PubMed] [Google Scholar]
- Bloch J, Holzmann C, Koczan D, Helmke BM, Bullerdiek J.. Factors affecting the loss of MED12-mutated leiomyoma cells during in vitro growth. Oncotarget 2017;8:34762–34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulun SE. Uterine fibroids. N Engl J Med 2013;369:1344–1355. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo ER, Clark AD, Banks NK, Henne MB, Stegmann BJ, Segars JH.. The estimated annual cost of uterine leiomyomata in the United States. Am J Obstet Gynecol 2012;206:211.e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF.. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 2005;434:400–404. [DOI] [PubMed] [Google Scholar]
- Catherino WH, Parrott E, Segars J.. Proceedings from theNational Institute of Child Health and Human Development conference on the Uterine Fibroid Research Update Workshop. Fertil Steril 2011;95:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commandeur AE, Styer AK, Teixeira JM.. Epidemiological and genetic clues for molecular mechanisms involved in uterine leiomyoma development and growth. Hum Reprod Update 2015;21:593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan S. Mechanisms of x chromosome dosage compensation. J Genomics 2015;3:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar CM, Steiner CA.. Hysterectomy rates in the United States 1990–1997. Obstet Gynecol 2002;99:229–234. [DOI] [PubMed] [Google Scholar]
- Fox JE, Abell MR.. Mast cells in uterine myometrium and leiomyomatous neoplasms. Am J Obstet Gynecol 1965;91:413–418. [DOI] [PubMed] [Google Scholar]
- Goad J, Ko YA, Syed SM, Crossingham YJ, Tanwar PS.. Data on the mRNA expression by in situ hybridization of Wnt signaling pathway members in the mouse uterus. Data Brief 2017;12:208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, Satija R.. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 2019;20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder SK, Laknaur A, Miller J, Layman LC, Diamond M, Al-Hendy A.. Novel MED12 gene somatic mutations in women from the Southern United States with symptomatic uterine fibroids. Mol Genet Genomics 2015;290:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth-Carson SJ, Zaitseva M, Girling JE, Vollenhoven BJ, Rogers PA.. Common fibroid-associated genes are differentially expressed in phenotypically dissimilar cell populations isolated from within human fibroids and myometrium. Reproduction 2014a;147:683–692. [DOI] [PubMed] [Google Scholar]
- Holdsworth-Carson SJ, Zaitseva M, Vollenhoven BJ, Rogers PA.. Clonality of smooth muscle and fibroblast cell populations isolated from human fibroid and myometrial tissues. Mol Hum Reprod 2014b;20:250–259. [DOI] [PubMed] [Google Scholar]
- Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, Teichmann SA.. Classification of low quality cells from single-cell RNA-seq data. Genome Biol 2016;17:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RE, Eickholt BJ.. Semaphorin signalling. Curr Biol 2009;19:R504–R507. [DOI] [PubMed] [Google Scholar]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q.. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampjarvi K, Makinen N, Mehine M, Valipakka S, Uimari O, Pitkanen E, Heinonen HR, Heikkinen T, Tolvanen J, Ahtikoski A. et al. MED12 mutations and FH inactivation are mutually exclusive in uterine leiomyomas. Br J Cancer 2016;114:1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 2018;36:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kataru RP, Koh GY.. Inflammation-associated lymphangiogenesis: a double-edged sword? J Clin Invest 2014;124:936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Sefton EC.. The role of progesterone signaling in the pathogenesis of uterine leiomyoma. Mol Cell Endocrinol 2012;358:223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp P, Jaggi R, Tobler A, Borisch B, Oestreicher M, Sabacan L, Jameson JL, Fey MF.. Clonal X-inactivation analysis of human tumours using the human androgen receptor gene (HUMARA) polymorphism: a non-radioactive and semiquantitative strategy applicable to fresh and archival tissue. Mol Cell Probes 1997;11:217–228. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Sivridis E, Simopoulos C, Gatter KC, Harris AL, Jackson DG.. LYVE-1 immunohistochemical assessment of lymphangiogenesis in endometrial and lung cancer. J Clin Pathol 2005;58:202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladher RK, Church VL, Allen S, Robson L, Abdelfattah A, Brown NA, Hattersley G, Rosen V, Luyten FP, Dale L. et al. Cloning and expression of the Wnt antagonists Sfrp-2 and Frzb during chick development. Dev Biol 2000;218:183–198. [DOI] [PubMed] [Google Scholar]
- Latif N, Sarathchandra P, Chester AH, Yacoub MH.. Expression of smooth muscle cell markers and co-activators in calcified aortic valves. Eur Heart J 2015;36:1335–1345. [DOI] [PubMed] [Google Scholar]
- Linder D, Gartler SM.. Glucose-6-phosphate dehydrogenase mosaicism: utilization as a cell marker in the study of leiomyomas. Science 1965;150:67–69. [DOI] [PubMed] [Google Scholar]
- Lucas ED, Tamburini BAJ.. Lymph node lymphatic endothelial cell expansion and contraction and the programming of the immune response. Front Immunol 2019;10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen N, Heinonen HR, Moore S, Tomlinson IP, van der Spuy ZM, Aaltonen LA. MED12 exon 2 mutations are common in uterine leiomyomas from South African patients. Oncotarget 2011a;2:966–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, Gentile M, Yan J, Enge M, Taipale M. et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011b;334:252–255. [DOI] [PubMed] [Google Scholar]
- Mashal RD, Fejzo ML, Friedman AJ, Mitchner N, Nowak RA, Rein MS, Morton CC, Sklar J.. Analysis of androgen receptor DNA reveals the independent clonal origins of uterine leiomyomata and the secondary nature of cytogenetic aberrations in the development of leiomyomata. Genes Chromosomes Cancer 1994;11:1–6. [DOI] [PubMed] [Google Scholar]
- McGuire MM, Yatsenko A, Hoffner L, Jones M, Surti U, Rajkovic A.. Whole exome sequencing in a random sample of North American women with leiomyomas identifies MED12 mutations in majority of uterine leiomyomas. PLoS One 2012;7:e33251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehine M, Kaasinen E, Makinen N, Katainen R, Kampjarvi K, Pitkanen E, Heinonen HR, Butzow R, Kilpivaara O, Kuosmanen A. et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med 2013;369:43–53. [DOI] [PubMed] [Google Scholar]
- Mittal P, Shin YH, Yatsenko SA, Castro CA, Surti U, Rajkovic A. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J Clin Invest 2015;125:3280–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori A, Zhai YL, Toki T, Nikaido T, Fujii S.. Distribution and heterogeneity of mast cells in the human uterus. Hum Reprod 1997;12:368–372. [DOI] [PubMed] [Google Scholar]
- Moyo MB, Parker JB, Chakravarti D.. Altered chromatin landscape and enhancer engagement underlie transcriptional dysregulation in MED12 mutant uterine leiomyomas. Nat Commun 2020;11:1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R, Romero R, Tarca AL, Sendler ED, Xu Y, Garcia-Flores V, Leng Y, Luca F, Hassan SS, Gomez-Lopez N.. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 2019;8:e52004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prizant H, Sen A, Light A, Cho SN, DeMayo FJ, Lydon JP, Hammes SR.. Uterine-specific loss of Tsc2 leads to myometrial tumors in both the uterus and lungs. Mol Endocrinol 2013;27:1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K. Lymphatic vessel dynamics in the uterine wall. Placenta 2008;29(Suppl A):S55–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein MS, Powell WL, Walters FC, Weremowicz S, Cantor RM, Barbieri RL, Morton CC.. Cytogenetic abnormalities in uterine myomas are associated with myoma size. Mol Hum Reprod 1998;4:83–86. [DOI] [PubMed] [Google Scholar]
- Reya T, , Clevers H.. Wnt signalling in stem cells and cancer. Nature 2005;434:843–850. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Wenger AM, Zehir A, Mesirov JP.. Variant review with the integrative genomics viewer. Cancer Res 2017;77:e31–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP.. Integrative genomics viewer. Nat Biotechnol 2011;29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewiera J, Erlebacher A.. Myometrial leukocytes. Curr Opin Physiol 2020;13:6–13. [Google Scholar]
- Stewart EA. Uterine fibroids. Lancet 2001;357:293–298. [DOI] [PubMed] [Google Scholar]
- Stewart EA, Laughlin-Tommaso SK, Catherino WH, Lalitkumar S, Gupta D, Vollenhoven B.. Uterine fibroids. Nat Rev Dis Primers 2016;2:16043. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R.. Comprehensive integration of single-cell data. Cell 2019;177:1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczek SI, Piterkova L, Jelinek J, Agarwal N, Hammoud S, Wilson A, Hickman K, Parker CJ, Cairns BR, Cairns B. et al. Methylation of AR locus does not always reflect X chromosome inactivation state. Blood 2012;119:e100–e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanwar PS, Lee HJ, Zhang L, Zukerberg LR, Taketo MM, Rueda BR, Teixeira JM.. Constitutive activation of Beta-catenin in uterine stroma and smooth muscle leads to the development of mesenchymal tumors in mice. Biol Reprod 2009;81:545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AJ, Telfer JF, Young A, Campbell S, Stewart CJ, Cameron IT, Greer IA, Norman JE.. Leukocytes infiltrate the myometrium during human parturition: further evidence that labour is an inflammatory process. Hum Reprod 1999;14:229–236. [PubMed] [Google Scholar]
- Varghese BV, Koohestani F, McWilliams M, Colvin A, Gunewardena S, Kinsey WH, Nowak RA, Nothnick WB, Chennathukuzhi VM.. Loss of the repressor REST in uterine fibroids promotes aberrant G protein-coupled receptor 10 expression and activates mammalian target of rapamycin pathway. Proc Natl Acad Sci USA 2013;110:2187–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SP, DeMayo FJ.. Progesterone receptor signaling in uterine myometrial physiology and preterm birth. Curr Top Dev Biol 2017;125:171–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Serna VA, Thomas J, Qiang W, Blumenfeld ML, Kurita T.. Subtype-specific tumor-associated fibroblasts contribute to the pathogenesis of uterine leiomyoma. Cancer Res 2017;77:6891–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko SA, Mittal P, Wood-Trageser MA, Jones MW, Surti U, Edwards RP, Sood AK, Rajkovic A.. Highly heterogeneous genomic landscape of uterine leiomyomas by whole exome sequencing and genome-wide arrays. Fertil Steril 2017;107:457–466.e459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed in the study are available in the NCBI Gene Expression Omnibus (GEO) (GSE162122) and Sequence Read Archive (SRA) (SRP294127). Code is available at: https://github.com/JyotiGoad/Single_Cell_Atlas_Of_Leiomyoma. All custom scripts can be accessed upon request to the corresponding author.