Abstract
Type 1 fimbriae are surface-located adhesion organelles of Escherichia coli that are directly associated with virulence of the urinary tract. They mediate d-mannose-sensitive binding to different host surfaces by way of the minor fimbrial component FimH. Naturally occurring variants of FimH that bind strongly to terminally exposed monomannose residues have been associated with a pathogenicity-adaptive phenotype that enhances E. coli colonization of extraintestinal locations such as the urinary tract. The FimH adhesin also promotes biofilm formation in a mannose-inhibitable manner on abiotic surfaces under static growth conditions. In this study, we used random mutagenesis combined with a novel selection-enrichment technique to specifically identify mutations in the FimH adhesin that confer on E. coli the ability to form biofilms under hydrodynamic flow (HDF) conditions. We identified three FimH variants from our mutant library that could mediate an HDF biofilm formation phenotype to various degrees. This phenotype was induced by the cumulative effect of multiple changes throughout the receptor-binding region of the protein. Two of the HDF biofilm-forming FimH variants were insensitive to mannose inhibition and represent novel phenotypes not previously identified in naturally occurring isolates. Characterization of our enriched clones revealed some similarities to amino acid alterations that occur in urinary tract infection (UTI) strains. Subsequent screening of a selection of UTI FimH variants demonstrated that they too could promote biofilm formation on abiotic surfaces under HDF conditions. Interestingly, the same correlation was not observed for commensal FimH variants. FimH is a multifaceted protein prone to rapid microevolution. In addition to its previously documented roles in adherence and invasion, we have now demonstrated its function in biofilm formation on abiotic surfaces subjected to HDF conditions. The study indicates that UTI FimH variants possess adaptations that enhance biofilm formation and suggests a novel role for FimH in UTIs associated with medical implants such as catheters.
Most bacteria living in aquatic environments form sessile communities referred to as biofilms. Biofilms are compact microbial communities consisting of organisms adherent to each other and/or a target surface. They can consist of either monocultures or multispecies communities and show extraordinary resistance to mechanisms that efficiently remove free-swimming bacterial relatives (8, 9). Bacterial biofilms can establish on virtually any solid surface of inorganic or organic nature spanning a wide spectrum of environments (8). Bacterial adherence is instrumental for the successful establishment of sessile bacterial communities on mammalian tissue surfaces, notably on surfaces subjected to hydrodynamic flow (HDF) shear forces such as the urinary tract. The best-characterized group of bacterial adhesins is constituted by fimbriae (13). Type 1, or mannose-sensitive, fimbriae are found on the majority of Escherichia coli strains and are widespread among other members of the family Enterobacteriaceae (16). Interaction between type 1 fimbriae and receptor structures has in a number of studies been shown to play a key role in the colonization of various host tissues by E. coli (2, 40). Also, in certain strain backgrounds type 1 fimbriae can be regarded as virulence factors. Indeed, we and others have previously shown that the expression of type 1 fimbriae in E. coli is linked to urinary tract colonization and pathogenesis (7, 25, 36). In animal models, immunization with the type 1 fimbrial adhesin was shown to prevent urogenital mucosal infection by E. coli (20, 21).
A typical type 1-fimbriated bacterium has 200 to 500 peritrichously arranged fimbriae on its surface. A type 1 fimbria is a 7-nm-wide, approximately 1-μm-long, rod-shaped structure consisting of four different components that are added to the base of the growing organelle (22). The bulk of the structure consists of about 1,000 copies of the major subunit protein, FimA, polymerized into a right-handed helical structure, but small quantities of the minor components, FimF, FimG, and FimH are also present (14, 19). It has been shown that the receptor-recognizing element of type 1 fimbriae is the 30-kDa FimH protein (18). FimH is located at the organelle tip in a short fibrillum and perhaps additionally intercalated along the fimbrial shaft (12, 18). The FimF and FimG components seem to be required for integration of the FimH adhesin into the fimbriae (12, 14). The components of the fimbrial organelle are encoded by the chromosomally located fim gene cluster (15). In addition to the structural components, this 9.5-kb DNA segment also encodes the fimbrial biosynthesis machinery as well as regulatory elements (Fig. 1).
FIG. 1.
Schematic representation of plasmids used to display FimH variants. Plasmid pPKL115 contains the entire fim gene cluster with a translational stop linker inserted into the fimH gene (triangle). Plasmid pMAS1 is the fimH expression vector upon which all functional variants constructed in this study are based. The segment flanked by the KpnI and HincII sites was submitted to mutagenesis. Only relevant nonvector regions are shown.
By virtue of the FimH adhesin, type 1 fimbriae mediate adhesion to a variety of mannosylated glycoproteins. Additionally, a number of studies have revealed that FimH adhesins from certain clinical isolates confer binding to protein targets such as fibronectin and collagen. This specificity change was found to be due to minor variations in the amino acid sequence of FimH (27, 38). Also, the affinity of FimH variants toward mannose targets can vary due to changes in the primary structure. In about 80% of fecal E. coli isolates, the FimH adhesin is capable of binding only to trimannose receptors. In contrast, the FimH adhesins from approximately 70% of urinary tract isolates carry minor mutations (compared to the fecal isolates) that enhance their ability to recognize monomannose receptors (37). The mutant alleles confer a significantly higher tropism for the uroepithelium (36).
The FimH adhesin has been shown to be instrumental in biofilm formation by E. coli K-12 under static growth conditions (28). Type 1 fimbriae were found to be critical for the initial interaction of E. coli cells with abiotic surfaces. Furthermore, this attachment could be inhibited by d-mannosides, indicating a specific role for FimH in such interactions. The FimH adhesin is a two-domain protein, with the NH2-terminal half conferring its lectin-binding characteristics (5, 17, 21, 33, 39). In a previous study we described the construction of a FimH mutant library by PCR-induced random mutagenesis within this region of the fimH gene (34). Specific mutations that altered the ability of FimH to bind to monomannose, oligomannose, and protein targets were identified. In this work we have screened the same FimH mutant library for variants with an ability to confer biofilm formation under HDF conditions, i.e., a scenario that arguably mimics many natural environments better than static conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. E. coli K-12 strain HB101 (F′ lacIq kan) (3) was used as an intermediate host during plasmid construction work. All subsequent phenotypic analyses were performed in the E. coli Δfim strain S1918 (4). The FimH expression vector pMAS1 contains the fimH gene from E. coli K-12 strain PC31 (15) under transcriptional control of the lac promoter. In addition, the plasmid contains unique KpnI and HincII recognition sequences within the fimH gene that flank the region encoding the proposed FimH receptor-binding domain (Fig. 1). Cells were grown in Luria-Bertani (LB) broth (30) supplemented with the appropriate antibiotics.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference |
|---|---|---|
| E. coli strains | ||
| HB101 | F′ laqIq | 3 |
| KB28 | fimH gene from wild-type strain CI#10 | 37 |
| KB53 | fimH gene from wild-type strain CI#4 | 37 |
| KB91 | fimH gene from commensal strain F-18 | 37 |
| KB96 | fimH gene from wild-type strain MJ2-2 | 37 |
| S1918 | F′ laqIq ΔfimB-H::kan | 4 |
| MS72 | pMAS1 and pPKL115 in S1918 | This study |
| MS174 | pMAS58 and pPKL115 in S1918 | This study |
| MS202 | pMAS60 and pPKL115 in S1918 | This study |
| MS206 | pMAS64 and pPKL115 in S1918 | This study |
| PK799 | pPKL236 and pPKL115 in S1918 | This study |
| PK800 | pPKL238 and pPKL115 in S1918 | This study |
| PK802 | pPKL237 and pPKL115 in S1918 | This study |
| PK803 | pPKL239 and pPKL115 in S1918 | This study |
| TK130 | pTBK31 and pPKL115 in S1918 | This study |
| Plasmids | ||
| pPKL4 | Wild-type fim gene cluster | 15 |
| pPKL115 | All fim genes except fimH | 26 |
| pMAS1 | fimH gene in pUC19 | 32 |
| pMAS58 | Modified fimH gene (V28A, V56A, F71L, Y82H, T87A, V94A, W103R, S113G, V118A) | This study |
| pMAS60 | Modified fimH gene (V30L, G73E, S114R, N136Y, Q143L, V155G) | This study |
| pMAS64 | Modified fimH gene (G73E, L107F) | This study |
| pPKL236 | Modified fimH gene (W103R, S113G, V118A) | This study |
| pPKL237 | Modified fimH gene (V28A, V56A, F71L, Y82H, T87A V94A) | This study |
| pPKL238 | Modified fimH gene (S114R, N136Y, Q143L, V155G) | This study |
| pPKL239 | Modified fimH gene (V30L, G73E) | This study |
| pTBK31 | Modified fimH gene (G73E) | This study |
DNA techniques.
Plasmid DNA was isolated using a QIAprep spin plasmid kit (Qiagen). Restriction endonucleases were used as specified by the manufacturer (New England Biolabs or Pharmacia). The nucleotide sequences were determined on both DNA strands by the dideoxynucleotide chain termination method (31). Oligonucleotide primers were purchased from Gibco BRL.
Construction of the fimH mutant library.
Construction of the fimH mutant library has been described previously (34). Briefly, the 650-bp KpnI-HincII fragment of the fimH gene from pMAS1 was mutagenized by nucleotide misincorporation during suboptimal PCR conditions. Four reactions were performed, with three of the four nucleotides at a concentration of 50 mM and the other at 5 mM. Each reaction mixture also contained 7 mM MgCl2 to increase the stability of noncomplementary base pairs and 0.5 mM MnCl2 to reduce the template specificity of the polymerase. The error-prone PCR procedure was performed for 35 cycles with two primers that flank the KpnI and HindII sites of the fimH gene. The amplification products were combined, digested with KpnI and HincII, purified after agarose gel electrophoresis, and religated into similarly cut plasmid pMAS1 to construct a library of altered fimH genes. To permit expression of the corresponding FimH variants as functional constituents of type 1 fimbriae, the ligation mix was transformed into E. coli strain S1918 (Δfim) containing an auxiliary plasmid, pPKL115, which encodes the entire fim gene cluster except fimH. The transformation mixture was made up to 10 ml, grown to approximately 10 times the initial library diversity, and stored as aliquots at −80°C in 25% (vol/vol) glycerol. Analysis of 300 random transformants revealed that the mutagenesis procedure was highly successful, with approximately 60% of transformants displaying an altered yeast agglutination phenotype.
Construction of defined fimH mutations.
Specific amino acid substitutions from the mutant fimH genes were introduced into the wild-type fimH sequence by overlapping PCR. The following primers were used: ms1 (5′-GTGATAAGCTTCACCATACCTACAGC), ms2 (5′-GCTCGAATTCCAGCATTAGCAATGTCC), 137 (5′-ATAATCGAGAACGGATAAGC), and 138 (5′-GCTTATCCGTTCTCGAATTAT). Each construct was sequenced to ensure fidelity of the PCR. Plasmids containing these chimeric fimH genes were introduced into S1918(pPKL115) and tested for yeast agglutination and biofilm formation.
Biofilm screening assay.
An aliquot of the fimH mutant library was grown overnight in LB and diluted to approximately 108 cells ml−1 in phosphate-buffered saline (PBS), and 100-μl aliquots were placed in 10 wells of a polystyrene microtiter plate. The plate was incubated at 37°C for 1 h with shaking and washed with 10 times with PBS; any remaining bound cells were then resuspended in 100 μl of LB. Following growth overnight at 37°C with shaking the cells were diluted to approximately 108 cells ml−1, and the procedure was carried out three more times. After the last incubation, the cultures were streaked for single colonies.
Assays for biofilm formation under hydrodynamic growth conditions were performed as previously described (10), with some modifications. Briefly, early-exponential-phase cultures (optical density at 600 nm [OD600] = 0.2) were incubated in 100-μl volumes in a microtiter plate well for 16 h with vigorous shaking. Unbound cells were removed by inversion of the microtiter plate and tapping on absorbent paper. Adhered cells were subsequently stained by the addition of 200 μl of 0.1% crystal violet. The stain was removed by thorough washing with PBS, and the wells were allowed to dry. The crystal violet was solubilized by the addition of 200 μl of ethanol-acetone (80:20, wt/wt), and A600 was determined.
Agglutination of yeast cells.
The capacity of bacteria to express a d-mannose-binding phenotype was assayed by their ability to agglutinate yeast (Saccharomyces cerevisiae) cells on glass slides. Aliquots of washed bacterial suspensions at OD550 = 0.5 and 5% yeast cells were mixed, and the time agglutination occurred was measured. Furthermore, clones which did not cause any agglutination under these conditions were also tested at OD550 = 20 and/or low temperature but still did not react.
RESULTS
Identification of FimH variants capable of HDF biofilm formation.
In E. coli K-12, type 1 fimbriation was shown to be critical for biofilm formation on abiotic surfaces under static growth conditions; this property could be abolished by addition of methyl-α-d-mannopyranoside, indicative of FimH involvement (Fig. 2A) (28). However, under HDF conditions, the wild-type (wt) FimH adhesin was unable to confer biofilm formation (Fig. 2B). We hypothesized that variants of the FimH adhesin that would induce biofilm formation under HDF conditions could be selected for. To identify such variants we screened our FimH mutant library, which consists of a pool of fimH genes with PCR-introduced random mutations within the receptor-binding region of the FimH adhesin. More specifically, the mutagenesis was targeted to the region encompassing amino acids 8 to 225 of the mature FimH protein (34). An aliquot of the FimH mutant library (approximately 108 cells ml−1) was incubated in a polystyrene microtiter plate and subjected to the selection-enrichment procedure described in Materials and Methods.
FIG. 2.
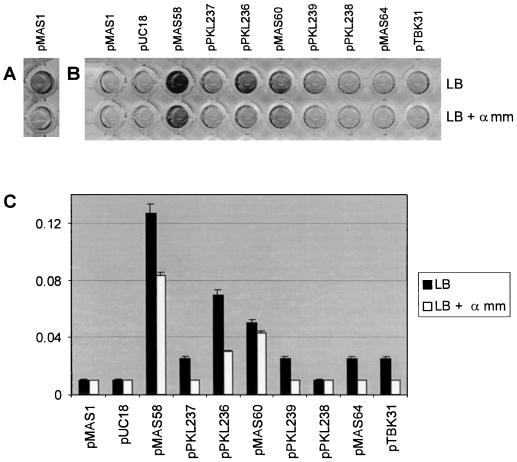
Biofilm formation by E. coli cells expressing FimH variants. Each FimH variant is indicated by its expression plasmid. (A) Biofilm formation on a microtiter plate surface by cells expressing the K-12 fimH allele when grown in LB under static conditions. (B) Biofilm formation on a microtiter plate surface by cells expressing FimH variants when grown in LB under HDF conditions. Note that no biofilm is produced by the K-12 fimH allele (pMAS1) under these conditions. (C) Quantification of biofilm formation as determined by the amount of crystal violet staining for each of the FimH variants. All experiments were performed in the presence and absence of methyl-α-d-mannopyranoside (α mm). Results are expressed as the average of four independent experiments (± standard deviation).
Fifty colonies enriched by this procedure were randomly selected and tested for biofilm formation under HDF conditions on a polystyrene surface. This involved growth of each clone in a microtiter plate for 16 h with vigorous shaking, removal of unbound cells, and subsequent staining of bound cells with 0.1% crystal violet. Cells attached to the abiotic surface could be visualized, as they were stained purple with crystal violet. Thirty (60%) of the fifty colonies were observed to form a biofilm under these conditions. To ensure that the observed biofilm-forming phenotype was indeed the result of specific alteration of the fimH gene, each of the 30 fimH-encoding plasmids was purified and retransformed into S1918(pPKL115). The new recombinant clones displayed the same biofilm-forming phenotype as the original isolates, indicating that this phenotype was indeed plasmid encoded.
Characterization of biofilm forming mutants.
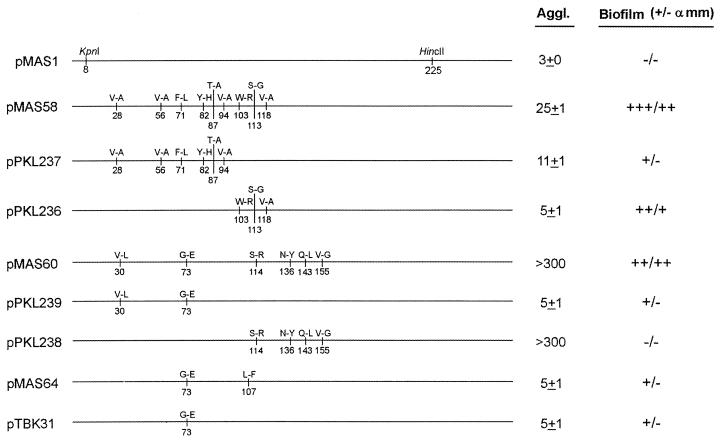
The nucleotide sequences of the fimH genes from the 30 HDF biofilm-forming clones were determined. The sequences revealed that there were indeed only three different clone types; the three plasmids were referred to as pMAS58, pMAS60, and pMAS64. Plasmid pMAS58 was found in 24 of the 30 clones, plasmid pMAS60 was found in 5 of the clones, and plasmid pMAS64 was present in 1 clone. All of the amino acid changes were located within the first 155 amino acids of the mature FimH protein (Fig. 3). Two of the FimH variants (pMAS58 and pMAS60) contained multiple amino acid changes, while the FimH of plasmid pMAS64 had only two amino acid changes. The abilities of the clones to form a biofilm under HDF conditions differed (Fig. 2). The FimH variant encoded on pMAS58 conferred the highest level of biofilm formation, being about 2.5- and 6.5-fold better than pMAS60 and pMAS64, respectively. This feature was reflected in the enrichment frequency (80% of all isolated biofilm-forming clones).
FIG. 3.
Depiction of mutations introduced into each of the FimH variants, along with a summary of their agglutination (Aggl.) and biofilm phenotypes. α mm, methyl-α-d-mannopyranoside.
Inhibition by soluble inhibitors.
Under natural conditions bacterial adhesion to eucaryotic cells is a function of the ability of the adhesin to interact with the cognate receptor on the cell surface but also depends on the sensitivity of the adhesin to soluble inhibitory compounds that often bathe the cellular target. Since the mutations in all three clones fall within the d-mannose receptor recognition domain of FimH, we speculated that the sensitivity to soluble inhibitors might be affected. Therefore, we determined the sensitivity of the FimH variants to biofilm formation under HDF conditions in the presence of methyl-α-d-mannopyranoside. Interestingly, the biofilm-forming abilities of strains harboring pMAS58 and pMAS60 were only partially (35 and 10%, respectively) reduced under these conditions (Fig. 2). In previously described experiments (28), FimH-mediated biofilm formation under static growth conditions was abolished by d-mannose. Thus, the pMAS58 and pMAS60 FimH variants possess novel phenotypes that have not been previously identified. In contrast to these results, the weaker biofilm-forming FimH variant encoded by pMAS64 could not form a biofilm in the presence of methyl-α-d-mannopyranoside.
Detailed analysis of the amino acid changes in FimH.
In an attempt to define the amino acid changes in FimH responsible for biofilm formation under HDF conditions, we used overlapping PCR to split the mutations (Fig. 3). The fimH gene in the predominant plasmid, pMAS58, contained nine amino acid-altering mutations. The split derivative plasmids pPKL236 and pPKL237 both turned out to be weaker HDF biofilm formers than the parent. This suggests that the inherent biofilm formation phenotype of the pMAS58 FimH variant is the cumulative result of several mutations within different regions of the receptor-binding domain. In contrast, the HDF biofilm-forming phenotype of plasmids pMAS60 (six changes) and pMAS64 (two changes) could be at least partly attributed to a mutation, G73E, found in both FimH variants. Apart from the presence of this mutation in both clones, a significant biofilm-forming potential was present in one of the split clones of pMAS60, i.e., pPKL239, harboring this mutation, whereas the other split clone, pPKL238, exhibited no HDF biofilm formation (Fig. 2). This led us to focus on the G73E change. Indeed, when the G73E mutation was uniquely introduced into the wt fimH, resulting in plasmid pTBK31 (Fig. 3), an HDF biofilm-forming phenotype was observed (Fig. 2). Taken together, the data demonstrate that a single amino acid substitution can enhance the binding repertoire of the FimH adhesin and permit adhesion to and biofilm formation on an abiotic surface under HDF conditions.
Agglutination phenotypes of FimH variants.
The classical way to monitor type 1 fimbria-mediated adhesion to eucaryotic cells is agglutination of erythrocytes or yeast cells. Yeast cell agglutination is the most conserved binding property among natural E. coli isolates and therefore was used to evaluate the receptor binding exhibited by the biofilm-forming FimH variants. The pMAS58 FimH conferred a weak agglutination phenotype. Splitting of the mutations in this variant resulted in an increased ability to agglutinate yeast cells in both progeny clones (Fig. 3). In contrast, the pMAS60 FimH did not confer an ability to agglutinate yeast cells. Furthermore, analysis of the split clones revealed that some combination of the S114R, N136Y, Q143L, and V155G mutations was responsible for abolishing the mannose-binding ability of FimH. In this respect it is important to note that mutagenesis of position 136 was previously shown to abolish agglutination (33). The pMAS64 FimH also exhibited a reduced capacity to agglutinate yeast cells.
Biofilm formation induced by wt fimH alleles.
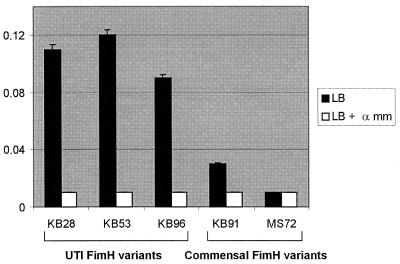
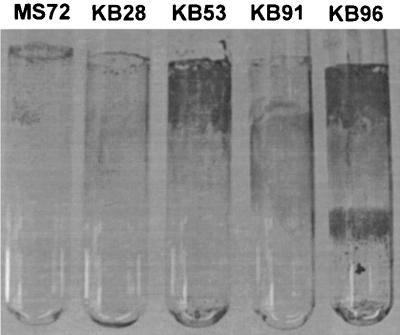
We identified the G73E mutation of the pMAS60 FimH variant as a functional alteration involved in biofilm formation under HDF conditions. Interestingly, the same mutation has also been identified in the pathogenicity-adapted FimH variant of a wt urinary tract isolate (38). The mutation in this variant imparts a higher affinity for monomannose targets and thus provides an adaptive advantage for the bacterial colonization of the urinary tract (34, 36). We speculated that there might exist a correlation between UTI FimH pathoadaptive phenotypes and HDF biofilm formation potential. To test this, we examined a selection of clones expressing previously characterized wt fimH alleles from urinary tract isolates (37) for the ability to form biofilms on abiotic surfaces. Indeed, all three of the strains tested turned out to be very good HDF biofilm formers. Furthermore, unlike our enriched clones, this biofilm formation phenotype could be inhibited by mannose (Fig. 4). Two of these strains (KB53 and KB96) also formed striking biofilms on glass surfaces under HDF conditions (Fig. 5).
FIG. 4.
Biofilm formation under HDF conditions on a microtiter plate surface by recombinant E. coli cells expressing fimH alleles from UTI isolates (KB28, KB53, and KB96) and commensal isolates (KB91 and MS72). Results are expressed as the average of four independent experiments (± standard deviation). α mm, methyl-α-d-mannopyranoside.
FIG. 5.
Biofilm formation on a glass surface by recombinant E. coli cells expressing fimH alleles from UTI isolates (KB28, KB53, and KB96) and commensal isolates (MS72 and KB91). Cultures were grown overnight in LB under HDF conditions.
To explore this correlation further, we examined the biofilm formation potential of FimH variants isolated from commensal strains. To this end, we used the FimH alleles from E. coli K-12 strain PC31 and the wt strain F-18, both of which were originally isolated from normal human fecal flora (1, 6). The specific amino acid changes in these FimH alleles (15, 37) and their adhesion to mannose substrates (35, 37) have been documented. Compared to the UTI FimH variants, the commensal types displayed a significantly reduced ability to form HDF biofilms on abiotic surfaces (Fig. 4). This is the first report to document a correlation between HDF biofilm formation on abiotic surfaces and UTI pathogenicity-adapted FimH phenotypes.
DISCUSSION
In natural aquatic environments surfaces are often exposed to HDF conditions. The urinary tract is such an example, and bacteria infecting the urinary tract are faced with brutal HDF shear forces. An obvious way for bacteria to counter this problem is to express efficient adhesions, enabling them to attach and eventually form sessile communities or biofilms. More than 50% of all microbial infections have now been associated with the formation of biofilms (9). In the urinary tract, biofilm formation is associated with chronic cystitis and infections related to medical implants such as catheters (24). The majority (80%) of urinary tract infections in humans are caused by E. coli. UTIs are a huge burden on our health care system; it is estimated that between 50 and 80% of women will experience at least one UTI infection at some time (11).
Uropathogenic E. coli express a number of different adhesive organelles including type 1, P, S, and F1C fimbriae. It is now well established that type 1 fimbriae are required for E. coli virulence of the urinary tract and that the FimH adhesin is instrumental in this regard (7, 25, 36). More recent data indicate that FimH can directly trigger host cell signaling cascades that lead to bacterial internalization (23). Type 1 fimbriae have also been described as key factors in biofilm establishment on abiotic surfaces (29). More specifically, the FimH adhesin was shown to be required for adherence to abiotic surfaces under static growth conditions. In this study, we used a novel random mutagenesis approach followed by a selection-enrichment procedure to specifically identify FimH variants capable of mediating biofilm formation on abiotic surfaces. Furthermore, to more realistically mimic real-life scenarios, our selection procedure was performed under HDF conditions.
We have previously demonstrated that the PCR mutagenesis approach used to construct our FimH mutant library was highly successful for the introduction of a limited number of random structural alterations in the FimH primary sequence. Nucleotide sequences were distributed randomly along the target sector, and the observed amino acid changes were diverse in nature (34). Therefore, it is intriguing that although the targeted region encompassed codons 8 to 225, no changes resulting in amino acid alterations were identified in the fimH alleles of the three clones enriched for HDF biofilm formation downstream of codon 155. For the sake of completeness, it should be noted that the fimH allele of plasmid pMAS60 contained two nucleotide changes downstream of codon 155 that did not alter the amino acid sequence. Thus, it appears that FimH-mediated biofilm formation as defined by our selection procedure is specifically linked to alterations in the N-terminal receptor-binding domain of the protein.
We identified three FimH variants that could mediate biofilm formation on abiotic surfaces subjected to HDF conditions. Two of these HDF biofilm library clones (pMAS58 and pMAS60) were partly immune to mannose inhibition. The strongest biofilm former identified in our selections was that encoded by the most predominantly enriched plasmid, pMAS58. From our split clones it is clear that some combination of the W103R, S113G, and V118A alterations is critical to impart this phenotype. In the case of plasmid pMAS60, biofilm formation was also associated with multiple amino acid changes. Our data indicates that the G73E mutation is required in combination with other changes to produce this phenotype. This mutation was also present in an independently isolated clone (pMAS64), providing additional evidence that it has a functional impact on the HDF biofilm formation phenotype. Furthermore, when the G73E change was introduced separately into the parental K-12 FimH background (i.e., plasmid pTBK31), enhanced biofilm formation was observed. When we compared the mutations in our enriched clones with those already documented in UTI FimH variants, some striking similarities were noted. First, the G73E mutation has been identified as the functional change in a naturally occurring FimH variant (38). In the case of the FimH encoded by the predominant plasmid pMAS58, the functional changes associated with biofilm formation partially overlap with a deletion of four amino acids (117 to 120) identified in another naturally occurring UTI FimH variant (38). These similarities between our own in vitro-selected clones and naturally occurring pathogenicity-adapted isolates prompted us to investigate biofilm formation mediated by naturally occurring FimH variants.
We examined three UTI FimH variants for their role in biofilm formation on abiotic surfaces subjected to HDF conditions. These variants have been well characterized with regard to their mannose-binding properties, and all contain minor amino acid changes in their amino-terminal domains that result in altered receptor specificity (35, 38). Furthermore, all three recombinant strains carrying the wt fimH alleles are similar with regard to level of fimbriation and relative levels of FimH incorporation, and importantly they are identical to the respective parental clinical isolates with regard to FimH adhesive phenotypes (37). We observed that these structural changes also impart an ability to form HDF biofilms on abiotic surfaces. It is intriguing to speculate on this correlation and the implications for pathogenesis. The ability to form biofilms is clearly a well-defined virulence trait among bacteria infecting the urinary tract (9). Thus, in addition to adhesion to mannose-containing substrates, FimH may be critical for the formation of biofilms in a hydrodynamic environment such as the urinary tract. Furthermore, the data implicate FimH as a potential mediator of biofilm formation on abiotic medical devices such as catheters.
FimH variants from E. coli strains of human fecal origin differ in receptor specificity from those from UTI isolates. Indeed, FimH variants from commensal isolates primarily recognize oligomannose-like receptors, while those from UTI isolates preferentially bind monomannose-like receptors and provide an adaptive advantage for bacterial colonization of the urinary tract (36). We examined HDF biofilm formation mediated by FimH variants from two commensal strains isolated from human fecal flora (E. coli K-12 strain PC31 and the wt strain F-18). Contrary to the FimH variants of UTI origin, the FimH variants from both of the commensal strains did not promote significant biofilm formation on abiotic surfaces under HDF conditions. Taken together, these findings suggest that the UTI FimH variants may have a greater capacity for HDF biofilm formation. At this stage one can only speculate on the role such FimH variants might play in establishing biofilms in the urinary tract, especially in catheterized patients. Although there might not be a complete correlation between HDF biofilm-forming FimH variants and pathogenicity, it may be that the biofilm assay described here could be used as a crude, yet simple and inexpensive means for differentiation of clinical isolates to indicate potential UTI pathogens.
Complicated UTIs and bacterial biofilms on the surface of catheters are often refractory to treatment with antibiotics. Our current knowledge suggests a multifunctional role for FimH in virulence traits that encompass adhesion, biofilm formation, and invasion. Our proposed role for FimH in biofilm formation on surfaces subjected to HDF conditions may lead to a better understanding in the design of treatment regimens to prevent UTIs. We have shown that the pathogenicity-adaptive phenotype that specifies altered affinity for different FimH alleles toward monomannose and trimannose can now be extended to include adherence to abiotic surfaces under HDF conditions. This novel phenotype has not previously been observed in naturally occurring FimH variants. Natural UTI FimH variants are probably functional compromises selected for their abilities to target and bind to mannose receptors and to form HDF biofilm. It is possible that the specific FimH isotypes enriched in our assays represent adhesive phenotypes selected against in the urinary tract. A highly intriguing aspect of this work is the observation that UTI FimH alleles possess greater biofilm formation potential than their commensal FimH counterparts.
ACKNOWLEDGMENTS
We thank Birthe Jul Jørgensen for expert technical assistance and David Hasty (University of Tennessee) and Evgeni Sokurenko (University of Washington) for providing strains.
This work was supported by the Danish Medical Research Council (grant 9802358).
REFERENCES
- 1.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 2.Bloch C A, Stocker B A D, Orndorff P E. A key role for type 1 pili in enterobacterial communicability. Mol Microbiol. 1992;6:697–701. doi: 10.1111/j.1365-2958.1992.tb01518.x. [DOI] [PubMed] [Google Scholar]
- 3.Boyer H W, Roulland-Dussoix D. A complementary analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 4.Brown S. Engineering iron oxide-adhesion mutants of the Escherichia coli phage lambda receptor. Proc Natl Acad Sci USA. 1992;89:8651–8655. doi: 10.1073/pnas.89.18.8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinker J, Hultgren S J, Knight S D. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 6.Cohen P S, Arruda J C, Williams T J, Laux D C. Adhesion of a human fecal Escherichia coli strain to mouse colonic mucus. Infect Immun. 1985;48:139–145. doi: 10.1128/iai.48.1.139-145.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connell H, Agace W, Klemm P, Schembri M, Måarild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerson J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 9.Costerson J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 10.Genevaux P, Muller S, Bauda P. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol Lett. 1996;142:27–30. doi: 10.1111/j.1574-6968.1996.tb08402.x. [DOI] [PubMed] [Google Scholar]
- 11.Gupta K, Stamm W E. Pathogenesis and management of recurrent urinary tract infections in women. World J Urol. 1999;17:415–420. doi: 10.1007/s003450050168. [DOI] [PubMed] [Google Scholar]
- 12.Jones C H, Pinkner J S, Roth R, Heuser J, Nicholes A V, Abraham S N, Hultgren S J. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klemm P. Fimbriae: adhesion, genetics, biogenesis and vaccines. Boca Raton, Fla: CRC Press, Inc.; 1994. [Google Scholar]
- 14.Klemm P, Christiansen G. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1987;208:439–445. doi: 10.1007/BF00328136. [DOI] [PubMed] [Google Scholar]
- 15.Klemm P, Jørgensen B J, van Die I, de Ree H, Bergmans H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli. Mol Gen Genet. 1985;199:410–414. doi: 10.1007/BF00330751. [DOI] [PubMed] [Google Scholar]
- 16.Klemm P, Krogfelt K A. Type 1 fimbriae of Escherichia coli. In: Klemm P, editor. Fimbriae, adhesion, genetics, biogenesis and vaccines. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 9–26. [Google Scholar]
- 17.Knudsen T B, Klemm P. Probing the receptor recognition site of the FimH adhesin by fimbriae-displayed FimH-FocH hybrids. Microbiology. 1998;144:1919–1929. doi: 10.1099/00221287-144-7-1919. [DOI] [PubMed] [Google Scholar]
- 18.Krogfelt K A, Bergmans H, Klemm P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun. 1990;58:1995–1998. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogfelt K A, Klemm P. Investigation of minor components of Escherichia coli type 1 fimbriae: protein chemical and immunological aspects. Microb Pathog. 1988;4:231–238. doi: 10.1016/0882-4010(88)90073-3. [DOI] [PubMed] [Google Scholar]
- 20.Langermann S, Mollby R, Burlein J E, Palaszynski S R, Auguste C G, DeFusco A, Strouse R, Schenerman M A, Hultgren S J, Pinkner J S, Winberg J, Guldevall L, Soderhall M, Ishikawa K, Normark S, Koenig S. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J Infect Dis. 2000;181:774–778. doi: 10.1086/315258. [DOI] [PubMed] [Google Scholar]
- 21.Langermann S, Palaszynsky S, Barnhart M, Auguste G, Pinkner J S, Burlein J, Barren P, Koenig S, Leath S, Jones C H, Hultgren S J. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 22.Lowe M A, Holt S C, Eisenstein B I. Immunoelectron microscopic analysis of elongation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1987;169:157–163. doi: 10.1128/jb.169.1.157-163.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez J J, Mulvey M A, Schilling J D, Pinkner J S, Hultgren S J. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris N S, Stickler D J, McLean R J C. The development of bacterial biofilms on indwelling urethral catheters. World J Urol. 1999;17:345–350. doi: 10.1007/s003450050159. [DOI] [PubMed] [Google Scholar]
- 25.Mulvey M A, Lopez-Boado Y S, Wilson C L, Roth R, Parks W C, Heuser J, Hultgren S J. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 26.Pallesen L, Poulsen L K, Christiansen G, Klemm P. Chimeric FimH adhesin of type 1 fimbriae: a bacterial display system for heterologous sequences. Microbiology. 1995;141:2839–2848. doi: 10.1099/13500872-141-11-2839. [DOI] [PubMed] [Google Scholar]
- 27.Pouttu R, Puustinen T, Virkola R, Hacker J, Klemm P, Korhonen T K. Amino acid residue Ala-62 in the FimH fimbrial adhesin is critical for the adhesiveness meningitis-associated Escherichia coli to collagens. Mol Microbiol. 1999;31:1747–1757. doi: 10.1046/j.1365-2958.1999.01311.x. [DOI] [PubMed] [Google Scholar]
- 28.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 29.Pratt L A, Kolter R. Genetic analyses of bacterial biofilm formation. Curr Opin Microbiol. 1999;2:598–603. doi: 10.1016/s1369-5274(99)00028-4. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schembri M A, Klemm P. Heterobinary adhesins based on the Escherichia coli FimH fimbrial protein. Appl Environ Microbiol. 1998;64:1628–1633. doi: 10.1128/aem.64.5.1628-1633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schembri M A, Pallesen L, Connell H, Hasty D L, Klemm P. Linker insertion analysis of the FimH adhesin of type 1 fimbriae in an Escherichia coli fimH-null background. FEMS Microbiol Lett. 1996;137:257–263. doi: 10.1111/j.1574-6968.1996.tb08115.x. [DOI] [PubMed] [Google Scholar]
- 34.Schembri M A, Sokurenko E V, Klemm P. Functional flexibility of the FimH adhesin: insights from a random mutant library. Infect Immun. 2000;68:2638–2646. doi: 10.1128/iai.68.5.2638-2646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sokurenko E V, Chesnokova V, Doyle R J, Hasty D L. Diversity of Escherichia coli type 1 fimbrial lectin. J Biol Chem. 1997;272:17880–17886. doi: 10.1074/jbc.272.28.17880. [DOI] [PubMed] [Google Scholar]
- 36.Sokurenko E V, Chesnokova V, Dykhuizen D E, Ofek I, Wu X-R, Krogfelt K A, Struve C, Schembri M A, Hasty D L. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci USA. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokurenko E V, Courtney H S, Maslow J, Sitonen A, Hasty D L. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J Bacteriol. 1995;177:3680–3686. doi: 10.1128/jb.177.13.3680-3686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokurenko E V, Courtney H S, Ohman D E, Klemm P, Hasty D L. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J Bacteriol. 1994;176:748–755. doi: 10.1128/jb.176.3.748-755.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thankavel K, Madison B, Ikeda T, Malavia R, Shah A H, Arumugam P M, Abraham S N. Localization of a domain in the FimH adhesin of Escherichia coli type 1 fimbriae capable of receptor recognition and use of a domain-specific antibody to confer protection against experimental urinary tract infection. J Clin Investig. 1997;100:1123–1136. doi: 10.1172/JCI119623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto T, Fujita K, Yokota T. Adherence characteristics to human intestinal mucosa of Escherichia coli isolated from patients with diarrhea or urinary tract infections. J Infect Dis. 1990;162:896–908. doi: 10.1093/infdis/162.4.896. [DOI] [PubMed] [Google Scholar]