Abstract
Background
Recurrence following abdominal surgery in Crohn disease is over 50%. The impact of genetics on postoperative recurrence is not well defined.
Methods
A literature search was conducted where inclusion required an assessment, by genotype, of postoperative recurrence. The primary endpoint was odds of surgical recurrence.
Results
Twenty-eight studies identified a total of 6715 patients. Thirteen loci were identified as modifying the risk of recurrence. NOD2 was identified as a risk factor for recurrence by multiple works (cumulative odds ratio: 1.64, P = 0.003).
Conclusions
A NOD2 risk allele is associated with recurrence following surgery in Crohn disease. Progress in this area will require standardized reporting in future works.
Keywords: Crohn disease, inflammatory bowel disease, recurrence, genetics, NOD2, CARD15
Introduction
An individual’s genetic makeup is at the root of many complex disease processes.1 Mapping of the human genome has not been the panacea that many had hoped, but advancements in the field of genomics have increased our understanding of numerous disease processes, including Crohn disease (CD).2 Several large, multinational genomic cohorts have identified specific loci associated with the disease and revealed the importance of genetic makeup on the development and phenotype of CD.2 Investigation of these genetic associations has led to significant insights into the pathogenesis of CD. The identification of NOD2, the first and most well-studied genetic loci associated with CD, highlights the importance of microbial–immune system interactions in the process of the disease, given the gene’s role in bacterial peptide recognition.3 The heterogeneity of phenotype in CD encourages a more granular approach to genetic analysis, and work has been done identifying specific loci and their association with aggressive phenotypes, and penetrating and fibrostenotic disease.
The need for surgical intervention in CD is common, with over 60% of patients requiring surgery within 10 years of diagnosis.4 Surgery is not considered curative as postoperatively up to 80% of patients experience endoscopic recurrence within 1 year of resection, and over 50% require repeat surgical intervention within a decade.5,6 These data demonstrate the importance of the problem clinically but furthering the understanding of postoperative recurrence may also be important for understanding CD in general. Surgical resection offers a microbial and inflammatory clean slate, and understanding the processes leading to recurrence in this setting may help reveal the mechanism leading to de novo disease occurrences. Given that there is a general lack of cohesive genetic information related to postoperative disease recurrence, a thorough and complete systematic review will help to identify any risk loci, and focus further work regarding the etiology and pathogenesis of postoperative recurrence.7 To date, there has been no comprehensive project aimed at identifying all loci associated with postoperative recurrence of CD. Therefore, we present the first comprehensive up-to-date systematic review and meta-analysis of the genetic factors involved in the recurrence of CD following abdominal surgery.
Materials and Methods
Search Strategy
This review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.8 The protocol was prospectively registered on the international prospective register of systematic reviews, PROSPERO (ID: CRD42017073629).
A systematic search, designed by our research librarian (TC), was performed of Medline, CINAHL, Web of Science, and Embase databases for all studies published between January 1, 1950 and February 12, 2020 examining the genetics of postoperative CD recurrence. The following key words were used, “Crohn’s disease or inflammatory bowel disease” and “genetics or genes or polymorphism or mutations or SNP” AND “recurrence” (Appendix 1). We did not limit our search to a language, nor did we exclude unpublished data. Further works were added after review of the reference lists in the publications, and manual searching of PubMed, for relevant articles missed by our search criteria. Abstracts and full-text reviews were evaluated by 2 investigators (ML and JD) in order to evaluate for inclusion and exclusion criteria. Disagreements were resolved by a third reviewer (TD).
Selection Criteria
Inclusion criteria included studies that examined postoperative recurrence of CD and defined 2 or more cohorts by specific genotypes. Both retrospective and prospective study designs were included. Exclusion criteria included studies with a lack of recurrence definition, case series or case studies, review articles, and subsets of subsequently published larger studies.
Data Extraction
Data extraction was completed independently by 2 reviewers (ML and JD) using a standard data abstraction form. The primary outcome of interest was recurrence of CD between genetic cohorts. Data collected also included year of study, study location, numbers of patients, definitions of recurrence, sex, age, tobacco use, follow-up, candidate genes and polymorphisms, genotyping method, allele frequency, and homozygosity/heterozygosity frequency. Authors of studies were contacted for missing data, and these data are presented when successfully obtained.
Risk of Bias Assessment
The Methodological Index for Non-Randomized Studies (MINORS) tool was used to assess the risk of bias for included studies and results are included in Appendix 2.9
Statistical Analysis
Descriptive categorical data are expressed as percentages and continuous data are expressed as weighted mean ± SD. Baseline differences between groups were evaluated by univariate analyses using Fisher exact test for categorical data and independent sample t test for continuous data. Meta-analysis was performed when data were available from 3 or more studies, using RevMan 5.3 software,10 and employed a random-effects model. Sensitivity analyses were planned based on the type of recurrence recorded (ie, clinical, endoscopic, or surgical) and type of surgery performed (ie, ileal resection and ileocolectomy alone). An assessment of individual data combined from each study was planned a priori when the data were available. Univariate analysis of genetic loci was performed using a log-rank test, with time to surgical recurrence or to the end of follow-up used at the time variable. Kaplan–Meier curves were created using this analysis.
Included studies were then tested for heterogeneity using the chi-square test with significance set at P < 0.10 and the amount of heterogeneity was quantified by the I2 statistic as follows: (1) low 25%, (2) moderate 50%, and (3) high 75%.11
Results
Study Selection
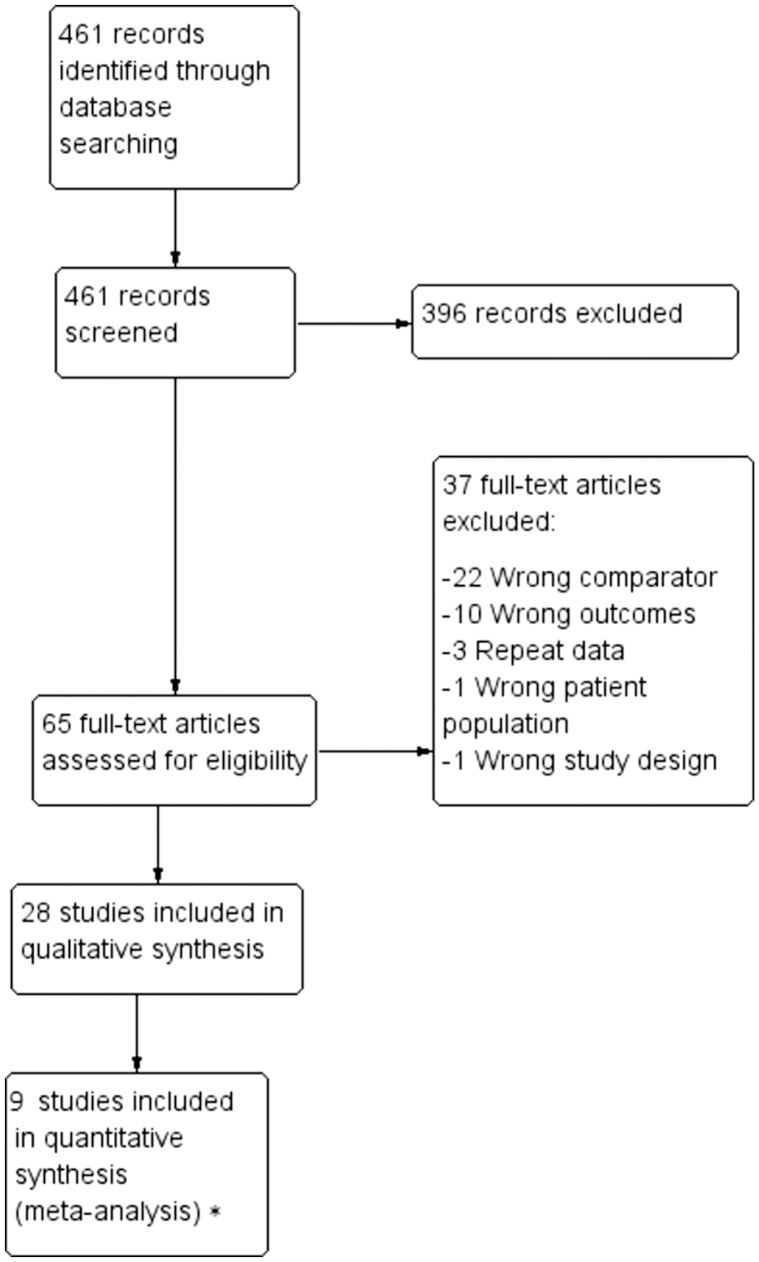
A comprehensive literature search yielded 461 potentially relevant articles (Fig. 1). After title and abstract screening 65 articles remained for full-text review. From these, 28 studies were eligible for inclusion (Table 1). Five of the included studies were published as abstracts only. Both retrospective and prospective studies were identified and included. Attempt to contact all corresponding and first authors were made, 19 of which were unsuccessful and received no response. Four investigators responded but did not possess the necessary data. Five authors were able to provide the requested information.
Figure 1.
PRISMA flow diagram. *Nine studies were appropriate for meta-analysis based on the presence of a risk allele at NOD2. Five studies were appropriate for meta-analysis based upon the presence of separate NOD2 subtypes. Eight studies were appropriate for meta-analysis based on the presence of a risk allele at NOD2 specifically for surgical recurrences. Four studies were appropriate for meta-analysis based on the presence of a risk allele at NOD2 specifically for surgical recurrences after small bowel or ileocolonic resection. Three studies were appropriate for a time-to-event analysis based on the presence of a risk allele at NOD2.
Table 1.
Study Characteristics12–39
| Author | Year | Location | Clinical Data Source | Journal | n | Genotyping Method | Genes Examined | No. Loci Examined | Abstract or Full Article |
|---|---|---|---|---|---|---|---|---|---|
| Ahmad | 2002 | United Kingdom | Prospective database | Gastroenterology | 158 | PCR sequence-specific primers | NOD2 | 3 | Article |
| Alvarez-Lobos | 2005 | Spain | Prospective database | Annals of Surgery | 170 | PCR sequence-specific primers | NOD2 | 3 | Article |
| Bhullar | 2014 | Australia | Retrospective | World Journal of Gastroenterology | 30 | dHPLC and direct sequencing | NOD2 | 4 | Article |
| Buning | 2004 | Germany | Retrospective | Alimentary Pharmacology and Therapeutics | 180 | PCR sequence-specific primers | NOD2 | 3 | Article |
| Burke | 2013 | Ireland | Prospective database | Annals of Surgery | 147 | SNP Assay | SMAD3 | 5 | Article |
| CTGF | |||||||||
| Chen | 2011 | NR | NR | — | 285 | SNP Assay | NOD2 | 4 | Abstract |
| ATG16L1 | |||||||||
| Fowler | 2014 | United States | Prospective database | Journal of Crohn’s and Colitis | 194 | Sequenom genotyping platform | NOD2, CARD9, ITLN1, ATG16L1, IRGM, LRRK2-MUC19, IL23R, JAK2, STAT3, TYK2, SMAD2, TNFSF15 | 25 | Article |
| Gathungu | 2018 | United States | Prospective database | World Journal of Gastroenterology | 412 | Illumina Golden Gate custom Immunochip array | NOD2, ATG16L1, IRGM, CARD9, XBP1, ORMDL, Others NR | NR | Article |
| Gerich | 2013 | NR | NR | — | 589 | 200k Immunochip platform | Multiple | NR | Abstract |
| Germain | 2016 | France | Prospective database | Surgery | 280 | SNPlex technology | CARD8 | 200 | Article |
| NAT2, Others NR | |||||||||
| Ghaly | 2016 | Australia | Prospective database | Inflammatory Bowel Disease | 309 | Taqman PCR | VDBP | 1 | Article |
| Laffin | 2018 | Canada | Prospective database | Inflammatory Bowel Disease | 191 | Illumina Goldengate assay platform | SMAD3, IL10RB, IL15RA, BACH2, IL12B, IL18RAP, IFNGR2, JAK2 | 8 | Article |
| Li | 2019 | United States | Prospective | PLoS One | 106 | Illumina Immunochip or Taqman Genotyping Assays | NOD2, ATG16L1, IRGM, CARD9, XBP1, ORMDL3 | 6 | Article |
| Liu | 2014 | United States | Retrospective | — | 178 | NR | NOD2, ATG16L1, Others NR | NR | Abstract |
| Liu | 2017 | United States | Retrospective | — | 98 | Japonica array | Multiple | 659,253 | Article |
| Maconi | 2009 | Italy | Prospective database | The American Journal of Gastroenterology | 253 | NR | NOD2 | 3 | Article |
| Martinek | 2015 | NR | Retrospective | Rozhledy v chirurgii | 76 | Primer-specific PCR | NOD2 | 3 | Article |
| Meijer | 2009 | Netherlands | Retrospective | Inflammatory Bowel Disease | 87 | PCR-RFLP and tetra primer ARMS PCR | MMP-TIMP | 1 | Article |
| Meresse | 2002 | NR | NR | Gut | 79 | Primer-specific PCR | IL10 promoter | 1 | Article |
| Naito | 2016 | Japan | Retrospective | — | 113 | Japonica array | Multiple | 659,253 | Abstract |
| Onnie | 2007 | United Kingdom | NR | European Journal of Gastroenterology & Hepatology | 630 | Taqman PCR | NOD2, IBD5 | 4 | Article |
| Potdar | 2019 | United States | Prospective database | Journal of Crohn’s and Colitis | 139 | Illumina Immuno-BeadChip | Multiple | NR | Article |
| Renda | 2008 | Italy | Prospective | American Journal of Gastroenterology | 182 | Allele-specific reverse dot-blot hybridization | NOD2 | 3 | Article |
| Sehgal | 2012 | United States | Retrospective | Diseases of the Colon & Rectum | 66 | DNA microarray | Multiple | 66 | Article |
| Seiderer | 2006 | Germany | Retrospective | Scandinavian Journal of Gastroenterology | 303 | Primer-specific PCR | NOD2 | 3 | Article |
| Siegel | 2011 | NR | Prospective | — | 376 | NR | Multiple | NR | Abstract |
| Van Dussen | 2014 | United States | Retrospective | Gastroenterology | 178 | Human OmniQuad SNP genotyping arrays and Immunochip | NOD2, Others NR | NR | Article |
| Yang | 2014 | Korea | Prospective database | Journal of Crohn’s and Colitis | 906 | Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry-based system | TNFSF15 | 5 | Article |
dHPLC, denaturing high-performance liquid chromatography; NR, not reported; PCR, polymerase chain reaction; SNP, single nucleotide polymorphism.
Quality Assessment of Included Studies
Studies were assessed for bias and methodological quality using the MINORS criteria (Appendix 2).9 None of the included articles met the global ideal score for noncomparative studies with 9 studies having a score less than 10. This was due to limitations with the inclusion of consecutive patients, the prospective collection of data, and reporting of follow-up periods. Furthermore, none of the included studies performed a prospective calculation of sample sizes.
Patient Characteristics
A total of 6715 patients were included from North America, Europe, Asia, and Australia (Table 1). The weighted mean age was 28.8 years at diagnosis or time of surgery and weighted sex was 52.9% male. The rates of active smokers ranged from 12.9% to 45.6%. The most common index surgical procedures included were primarily ileocolectomy or ileal resection (10 studies), and unspecified bowel resection (9 studies) (Table 2). Index surgical procedure was not reported in 2 studies. Median follow-up ranged from 3.4 to 16.2 years postoperatively. Postoperative recurrence occurred in 36.3% of patients. The majority of studies defined recurrence as surgical (67.9%), with fewer studies using clinical (17.9%), endoscopic (10.7%), and radiologic (7.1%) definitions. Three studies12,13,28 defined endoscopic recurrence as a Rutgeerts score of i2 or more. Four studies did not report on their definition of recurrence.
Table 2.
Study Outcomes
| Study | Genetic Loci | Associated Gene | Gene Carriers | Index Surgical Procedures Included | Definition of Recurrence | Total Recurrence | Recurrence With SNP | Recurrence Without SNP | P |
|---|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | ||||||
| Ahmad 2002 | G908R, L1007fs, R702W | NOD2 | NR | ICR for stenotic disease | Surgical | 66 (41.8%) | NR | NR | NR |
| Alvarez-Lobos 2005 | G908R, L1007fs, R702W | NOD2 | 28 (40.0) | Any bowel resection or strictureplasty | Clinical and surgical | 23 (32.9%) | 14 (50.0) | 9 (21.4) | 0.019 |
| Bhullar 2014 | G908R, L1007fs, R702W, P268S | NOD2 | 8 (26.7) | ICR, small bowel resection and strictureplasty | Surgical | 16 (53.3) | 6 (75.0) | 10 (45.5) | 0.226 |
| Buning 2004 | G908R, L1007fs, R702W | NOD2 | 29 (34.5) | ICR | Surgical | 14 (27.5) | 12 (41.4) | 2 (9.1) | <0.001 |
| Burke 2013 | rs9402373 (GC) | CTGF | 43 (29.7) | ICR | Surgical | 32 (22.1) | 26 (25.5) | 6 (14.0) | 0.187 |
| rs17293632 (TC) | SMAD3 | 50 (36.8) | 32 (22.1) | 17 (19.8) | 15 (30.0) | 0.210 | |||
| Chen 2011 | G908R, L1007fs, R702W | NOD2 | 117 (41.1) | ICR | Surgical | NR | NR | NR | NR |
| T300A | ATG16L1 | 234 (82.1) | NR | NR | NR | NR | |||
| Fowler 2014 | Multiple | Multiple | NR | Any bowel resection | Surgical | 69 (35.6) | NR | NR | NR |
| Gathungu 2018 | rs2066843, rs2066844, rs2066845, rs2076756 and rs2066847, rs2241880, rs13361189 | NOD2 | NR | ICR or isolated ileal resection | Surgical | NR | NR | NR | NR |
| CARD9 | |||||||||
| IRGM | |||||||||
| ATG16L1 | |||||||||
| Gerich 2013 | NR | IRF8 | NR | Any bowel resection | Surgical | 210 (35.7) | NR | NR | 0.015 |
| LSP1/TNNI2 | 0.04 | ||||||||
| 0.04 | |||||||||
| PTGER4 | 0.04 | ||||||||
| DAP | <0.001 | ||||||||
| FAM49B | <0.001 | ||||||||
| PELI3 | <0.001 | ||||||||
| CHL1 | <0.001 | ||||||||
| PARVB | <0.001 | ||||||||
| STK24 | |||||||||
| Germain 2016 | Multiple | Multiple | NR | Any bowel resection | Surgical | 38 (27.7) | NR | NR | NR |
| Ghaly 2016 | rs7041 | VDBP | NR | NR | Clinical | 80 (79.2) | NR | NR | NR |
| Laffin 2018 | rs1847472 | BACH2 | 88 (46.1) | Any bowel resection | Surgical | 87 (45.5) | 47 (53.4) | 40 (38.8) | 0.04 |
| rs17293632 | SMAD3 | ||||||||
| rs2284553 | IL10RB | ||||||||
| rs12722515 | IL15RA | ||||||||
| rs6871626 | IL12B | ||||||||
| rs917997 | IL18RAP, IL18R1 | ||||||||
| rs2284553 | IFNGR2 | ||||||||
| rs10758669 | JAK2 | ||||||||
| Li 2019 | rs2066847, rs2066884, rs2066845, rs5743289 | NOD2 | 37 (34.9) | ICR, total abdominal colectomy, right hemicolectomy | Endoscopic | 27 (44.2)* | NR | NR | NR |
| NR | NR | NR | |||||||
| rs2241880 | ATG16L1 | 91 (85.8) | NR | NR | NR | ||||
| rs13361189 | IRGM | 30 (28.3) | NR | NR | NR | ||||
| rs10870077 | CARD9 | 101 (95.3) | NR | NR | NR | ||||
| rs35873774 | XBP1 | 14 (13.2) | NR | NR | NR | ||||
| rs2872507 | ORMDL3 | 79 (74.5) | |||||||
| Liu 2014 | NR | NOD2 | NR | Any bowel resection | NR | NR | NR | NR | NR |
| T300A | ATG16L1 | ||||||||
| NR | Others NR | ||||||||
| Liu 2017 | NR | Multiple | NR | Ileal resection | Endoscopic and radiologic | NR | NR | NR | NR |
| Maconi 2009 | G908R, L1007fs, R702W | NOD2 | 91 (36.0) | Any bowel resection or strictureplasty | Surgical | 89 (35.2) | 31 (34.1) | 58 (35.8) | 0.891 |
| Martinek 2015 | G908R, L1007fs, R702W | NOD2 | 24 | Any bowel resection or strictureplasty | Surgical | 25 (48.1) | 10 (41.7) | 15 (28.8) | 0.471 |
| Meijer 2009 | 372 T/C | TIMP-1 | 60 (69.8) | ICR, small bowel resection, subtotal colectomy | Clinical, endoscopic, radiologic and surgical | NR | NR | NR | NR |
| Meresse 2002 | IL-10 | G7-8 and G10-13 | 17 (47.2) | ICR | Endoscopic | 19 (52.8) | 7 (41.2) | 12 (63.2) | 0.316 |
| Naito 2016 | Multiple | Multiple | NR | Ileal resection | NR | NR | NR | NR | NR |
| Onnie 2007 | G908R, L1007fs, R702W | NOD2 | 90 (24.9) | Any bowel resection | Surgical | 111 (30.7) | 37 (41.1) | 74 (27.3) | 0.017 |
| IGR2063 | IBD5 | 111 (30.7) | 58 (35.2) | 53 (27.0) | 0.109 | ||||
| Potdar 2019 | Multiple | Multiple | 54 (38.9) | Small bowel resection | Clinical and surgical | NR | NR | NR | NR |
| Renda 2008 | R702W | NOD2 | 35 (31.8) | Any bowel resection | Surgical | 32 (29.1) | 12 (34.3) | 20 (26.7) | 0.500 |
| Sehgal 2012 | Multiple | Multiple | NR | ICR | Surgical | NR | NR | NR | NR |
| Seiderer 2006 | G908R, L1007FS, R702W | NOD2 | NR | NR | NR | NR | NR | NR | NR |
| Siegel 2011 | NR | Multiple | NR | Any bowel resection | Clinical | NR | NR | NR | NR |
| VanDussen 2014 | G908R, L1007fs, R702W | NOD2 | 39 (41.1) | ICR or ileal resection | NR | 50 (52.6) | 21 (53.8) | 29 (51.8) | 1.000 |
| T300A | ATG16L1 | 42 (44.2) | 50 (52.6) | 24 (57.1) | 26 (49.1) | 0.536 | |||
| Yang 2014 | Multiple | TNFSF15 | NR | Any bowel resection or intestinal bypass | Surgical | 87 (25.9) | NR | NR | NR |
Only most significant results reported from each study. P values calculated using Fisher exact test.
*Only 61 of 106 patients were assessed endoscopically for recurrence.
ICR, ileocolonic resection; NR, not reported; SNP, single nucleotide polymorphism.
Genetic Data
The total number of loci interrogated across all included studies was over 650,000. Available data are summarized in Table 2. In total, complete data were obtained from 13 out of 28 studies. NOD2 was analyzed in 14 studies, while other genes examined in single studies included BACH2, CTGF, CARD8, SMAD3, ATG16L1, TIMP-1, IL-10, VDBP, IBD5, and TNFSF15. Only 3 loci were appropriate for meta-analysis, all belonging to NOD2. These loci were G908R, L1007fs, R702W and were analyzed in a combined manner, as well as by subtype.
Genes Associated With Postoperative Recurrence
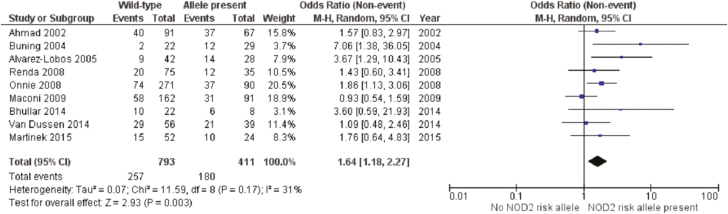
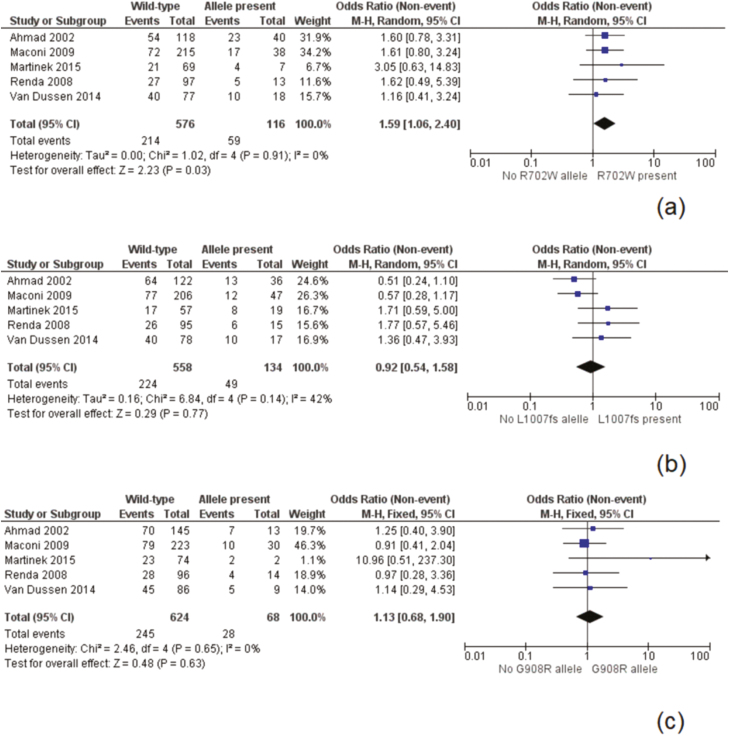
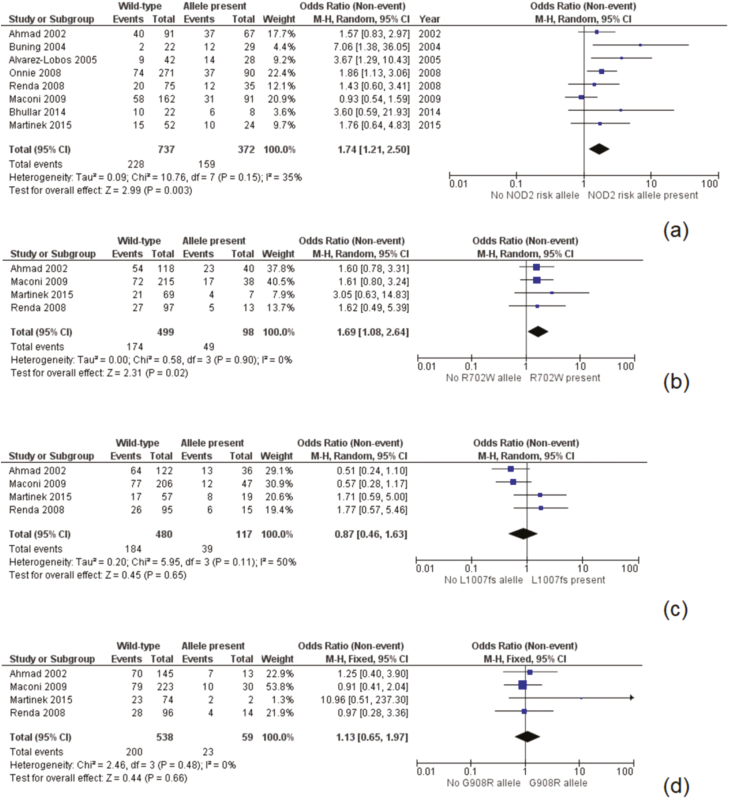
NOD2 was the identified loci with the most available data, as 9 studies presented data suitable for meta-analysis at this risk allele14–20 (Fig. 2). These studies included 1204 patients of which 411 (34.1%) had a NOD2 mutation. The overall recurrence rate was 43.8% in the NOD2 group compared to 32.4% for wild-type patients. A meta-analysis found the presence of a risk allele at NOD2 to be a significant risk factor for postoperative recurrence [odds ratio (OR) 1.64, 95% confidence interval (CI) 1.18–2.27, P = 0.003]. There was moderate heterogeneity in the included studies (I2 = 31%, P = 0.17). A separate meta-analysis was performed on prominent variants (R702W, G908R, and L1007fs) at the NOD2 loci with data included from 5 studies (Fig. 3).17,19–22 The R702W SNP was found to be significantly associated with recurrence (OR 1.59, 95% CI 1.06–2.40, P = 0.02), with a low degree of heterogeneity (I2 = 0%, P = 0.91) (Fig. 3). The mutations L1007fs and G908R were not significantly associated with postoperative recurrence. Two additional sensitivity analyses were performed to similar results, examining first only studies including surgical recurrence (Fig. 4), and second, examining only studies including surgical recurrence following small bowel or ileocolonic resection (Supplementary Material 1).
Figure 2.
Postoperative recurrence by NOD2 risk allele.
Figure 3.
Postoperative recurrence by NOD2 subtype: Panel A, R702W subtype; Panel B, L1007fs subtype; Panel C, G908W subtype.
Figure 4.
Postoperative surgical recurrence by NOD2 and subtype: Panel A, grouped NOD2 risk alleles; Panel B, R702W subtype; Panel C, L1007fs subtype; Panel D, G908W subtype.
Data on time to recurrence for individual subjects were obtained for 3 works, containing 521 subjects.17,19,21 No differences in survival were noted for NOD2 risk alleles combined, or individually (Supplementary Material 2).
Several genes were analyzed by other papers, none of which were appropriate for meta-analysis (Table 3). Genetic factors that were found to be associated (ie, P < 0.05) with postoperative recurrence in single work included BACH2 homozygosity (hazard ratio [HR]: 1.54),34 CARD8 homozygosity (OR: 7.56),40 TNFSF15 (with the development of strictures) (HR: 1.71),23 and IRGM (HR: not reported),24 IRF8 (HR: 0.6), LSP1/TNNI2 (HR: 1.4), PTGER4 (HR: 1.3), DAP (HR: 1.4), FAM49B (HR: 3.2), PELI3 (HR: 1.8), CHL1 (HR: 2.6), PARVB (HR: 0.5), and STK24 (HR: 1.7), though many of these findings were either not analyzed in a multivariable manner or corrected for multiple testing.
Table 3.
Summary of Genes Other Than NOD2 With a Significant Impact on Postoperative Recurrence of CD
| Study | Identified Genetic Factor Associated With Recurrence | Statistical Analysis | Definition of Recurrence | Change in Risk | P |
|---|---|---|---|---|---|
| Gerich 2013 | IRF8 | Univariate survival analysis. | Repeat surgery | HR: 0.6 | 0.015 |
| LSP1/TNNI2 | No correction for multiple testing. | HR: 1.4 | 0.04 | ||
| PTGER4 | HR: 1.3 | 0.04 | |||
| DAP | HR: 1.4 | 0.04 | |||
| FAM49B | HR: 3.2 | <0.001 | |||
| PELI3 | HR: 1.8 | <0.001 | |||
| CHL1 | HR: 2.6 | <0.001 | |||
| PARVB | HR: 0.5 | <0.001 | |||
| STK24 | HR: 1.7 | <0.001 | |||
| Germain 2016 | Homozygosity for the variant at CARD8 snp rs2043211 | Multivariable survival analysis. | Repeat surgery | OR: 7.56 | 0.04 |
| No correction for multiple testing. | |||||
| Laffin 2018 | Homozygosity for the variant at BACH2 snp rs1847472 | Multivariable survival analysis. | Repeat surgery | HR: 1.54 | <0.05 |
| No correction for multiple testing. | |||||
| Sehgal 2012 | Presence of the variant at IRGM rs4958847 | Multivariable survival analysis. | Repeat surgery | NR | 0.007 |
| Corrected for multiple testing. | |||||
| Yang 2014 | Homozygosity for the variant at TNFSF15 snp rs6478108 | Multivariable survival analysis. | Development of stricture | HR: 1.7 | 0.005 |
| No correction for multiple testing. |
NR, not reported.
Discussion
This work represents the first systematic review comprehensive for all known genetic loci associated with the postoperative recurrence of CD. It is also the first meta-analysis to find an association between any specific gene, namely NOD2, and disease recurrence. This effect was impressive, with an OR of 1.64 for postoperative recurrence in the NOD2 risk allele cohort. The inability to perform additional meta-analysis on the numerous other studied highlights a major issue with the study of postoperative recurrence in CD, namely the lack of standardized reporting and data transparency.
CD is a chronic inflammatory bowel disease that follows a relapsing and remitting course. Longstanding, active inflammation results in progression of disease and development of penetrating or stricturing complications, often requiring surgical management. Both the incidence and prevalence of CD are rising in Western and newly industrialized countries,41 especially in younger patients.42 Despite advances in medical therapy, postoperative recurrence of CD still occurs endoscopically in 85%–100% and clinically in 34%–86% of patients within 3 years of surgery.43 It is estimated that up to 60% of patients will require a second major abdominal surgery 20 years from their initial surgery.44 Postoperative recurrence of Crohn, especially in younger individuals, can predispose patients to multiple surgeries over their lifetime, resulting in significantly lower health-related quality of life, increased medical care costs, and lower earnings.45 Thus, identifying factors leading to postoperative CD recurrence is paramount in both understanding and treating CD.
If high-risk loci sufficiently predict recurrence, they may 1 day help guide clinical postoperative therapy choices. Thiopurines and anti-TNF therapies have been identified as effective for the prevention of postoperative recurrence, though the data here do not demonstrate a clear relationship between time of publication and rate of recurrence. The use of postsurgical medical therapies results in serious adverse events in over 15% of enrolled subjects. Therefore, to balance the risk and benefits of postoperative therapy, clinicians must be able to identify patients at a high risk of recurrence. A number of high-risk clinical features have been identified that predict postoperative recurrence including a younger patient (age <30 years old), disease duration greater than 10 years, tobacco usage, previous surgery, and family history.46,47 Less well understood are the nonclinical, patient-specific factors that predispose patients to postoperative recurrence such a patient’s individual microbiome, genetic makeup, and the complex interplay between these factors and the environmental exposures that drive inflammation in CD. The gut microbiome has long been thought to play a causative role in the high rates of CD recurrence following surgical resection.48 Several studies48–51 have shown that the mucosal microbial composition in CD patients at the time of surgery is predictive of future disease relapse.52 Work in this area is ongoing and may lead to identification a microbial signature predicting recurrence, which could assist clinicians in predicting recurrence, or altering the microbiome in a way that prevents disease progression. The addition of genetic information to these other factors, may improve the utility of any method of disease prognostication.
The clinical importance of postoperative recurrence of CD is well established, and an understanding of the pathophysiology of this disease process may play a role in the broader context of CD. Our understanding of the inciting events in the development of CD remains underdeveloped. This is in large part due to the difficulty in surveying and identifying patients at risk of disease development over the years they are most at risk, as well as the tendency for the disease to progress for many years prior to presentation to medical personnel. The postoperative period offers a unique opportunity to surveil patients with a high risk of disease development and progression. The bowel, postresection, should offer a relative clean slate in terms of inflammation, disease activity, and microbial composition. Therefore, factors identified to be associated with the development of recurrent disease, free from the confounding influence of an established inflammatory milieu and entrenched microbial communities, may also play a disproportionally large role in the development of de novo disease. The specific genotypes implicated in postoperative recurrence may be worth investigation as loci of interest in terms of initial disease development.
The NOD2 gene was first linked to genetic predisposition to developing CD in 2001.53,54 NOD2, located within the IBD1 susceptible region on chromosome 16, allows intracellular recognition of gut microbes through detection of released peptidoglycans55 and is important in regulating innate and adaptive immunity in the gut through NF-κB.56 Autophagic pathways that act through both NOD2 and ATG16L1 can also be impaired with comutations in both genes.57 Since the association was first identified, NOD2 mutation has been shown not only to increase the susceptibility to developing CD, but also predict younger age of disease onset, more severe stricturing and penetrating phenotype, and need for surgical intervention.58 A previous systematic review and meta-analysis demonstrated no association between NOD2 and postoperative recurrence.7 However, in our study, which is the first comprehensive review of all genetics involved in postoperative CD, we found that there was a strong correlation of presence of NOD2 and recurrence of disease. We also identified that the G908R genotype was significantly associated with disease recurrence, while the other common 2 genotypes were not. This phenomenon has not been described prior to this work. The exact functional impact of G908R is not known. The lack of association seen between the L1007fs mutation and postoperative recurrence may be related to the recent discovery that those patients with a L100fs variant are less likely to smoke,59 as smoking is the most well-established risk factor in the recurrence of CD following surgery. The G908R mutation is not protective from smoking, and therefore those individuals with the G908R mutation may be more likely to smoke, increasing their risk compared to the L100fs cohort.
In addition to NOD2, 7 additional genes have been implicated in postoperative recurrence, namely BACH2, CARD8, TNFSF15, IRGM, IRF8, LSP1/TNNI2, DAP, PTGER4, PELI3, CHL1, PARVB, and STK24. However, these genes have only been found in individual cohort studies, and their discovery is not unified by 1 biologic process or mechanism. The heterogeneity of included index surgical procedures and indications makes meta-analysis more challenging. This work employed multiple sensitivity analyses to address these concerns, including focusing on specific index surgical procedures and types of recurrence. The findings regarding the role of NOD2 in recurrence postoperatively were robust across these comparisons. Additionally, some studies employed multi-SNP arrays, leading to a high risk of false discovery due to the measurement of up to hundreds of thousands of gene loci.60 Finally, the defined endpoint of recurrence varied between studies. Attempts to replicate these exploratory studies have not been published and the lack of consistency in loci examined across published works greatly limits the utility of any meta-analysis. Further, the reporting on the rates of recurrence for the vast majority of interrogated genetic loci in the identified works was incomplete. This prevented our study from including data in our analysis and certainly skews our findings based on reporting bias. These concerns highlight many established issues in genetics research. For the genetics of any disease to be studied in coordinated manner, a framework must be established, clearly identifying important data that must be reported. In general, for genetic studies, a number of factors should be documented, including geographic area in which the study takes place, population age, population gender, loci studied, alleles analyzed, method of genotyping and the minor allele and homozygote frequency in the population. Specific disease factors must be described as well, and in the case of postoperative recurrence of CD, type of intraabdominal surgery performed (specifically resection vs strictureplasty), method of assessing disease recurrence, and prevalence of smoking in the cohort. We also encourage the reporting of deidentified individual patient data, including time to recurrence, Montreal disease classification, and postoperative medical therapy, as this information will add granularity to future study. Here, we include a standardized reporting (Supplementary Materials 4 and 5) form for genetic associations in postoperative recurrence of CD, which represents the minimum we suggest be reported in future published works on this topic.
The presence of the NOD2 risk allele increased the odds of surgical recurrence following intestinal resection in CD. Other genes identified as associated with recurrence were BACH2, CARD8, TIMP-1, TNFSF15, and IRGM, IRF8, LSP1/TNNI2, DAP, PTGER4, PELI3, CHL1, PARVB, and STK24, however these were only represented in single studies and future research must be conducted in the context of a standardized framework examining the genetics of postoperative recurrence in CD.
Supplementary Material
Conference Presentation: Canadian Surgery Forum 2019, Montreal, Quebec, Canada.
Funding: This work was supported by the Digestive Health Strategic Clinical Network Systematic Review Grant from Alberta Health Services.
Conflict of Interest: The authors have no conflicts of interest to declare.
Data Availability
No new data were created for this study.
References
- 1. Lees CW, Barrett JC, Parkes M, et al. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. [DOI] [PubMed] [Google Scholar]
- 2. Mirkov MU, Verstockt B, Cleynen I. Genetics of inflammatory bowel disease: beyond NOD2. Lancet Gastroenterol Hepatol. 2017;2:224–234. [DOI] [PubMed] [Google Scholar]
- 3. Sidiq T, Yoshihama S, Downs I, et al. Nod2: a critical regulator of ileal microbiota and Crohn’s disease. Front Immunol. 2016;7:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burisch J, Jess T, Martinato M, et al. ; ECCO-EpiCom . The burden of inflammatory bowel disease in Europe. J Crohns Colitis. 2013;7:322–337. [DOI] [PubMed] [Google Scholar]
- 5. Rutgeerts P. Strategies in the prevention of post-operative recurrence in Crohn’s disease. Best Pract Res Clin Gastroenterol. 2003;17:63–73. [DOI] [PubMed] [Google Scholar]
- 6. Landsend E, Johnson E, Johannessen HO, et al. Long-term outcome after intestinal resection for Crohn’s disease. Scand J Gastroenterol. 2006;41:1204–1208. [DOI] [PubMed] [Google Scholar]
- 7. Solon JG, Burke JP, Walsh SR, et al. The effect of NOD2 polymorphism on postsurgical recurrence in Crohn’s disease: a systematic review and meta-analysis of available literature. Inflamm Bowel Dis. 2013;19:1099–1105. [DOI] [PubMed] [Google Scholar]
- 8. Moher D, Liberati A, Tetzlaff J, et al. ; PRISMA Group . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–269, W64. [DOI] [PubMed] [Google Scholar]
- 9. Slim K, Nini E, Forestier D, et al. Methodological index for non-randomized studies (minors): development and validation of a new instrument. ANZ J Surg. 2003;73:712–716. [DOI] [PubMed] [Google Scholar]
- 10. Collaboration C. Review Manager (Version 5.3) [Computer Software]. Copenhagen, Denmark: Nord Cochrane Cent; 2014. [Google Scholar]
- 11. Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li E, Zhang Y, Tian X, et al. Influence of Crohn’s disease related polymorphisms in innate immune function on ileal microbiome. PLoS One. 2019;14:e0213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meresse B, Rutgeerts P, Malchow H, et al. Low ileal interleukin 10 concentrations are predictive of endoscopic recurrence in patients with Crohn’s disease. Gut. 2002;50:25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alvarez-Lobos M, Arostegui JI, Sans M, et al. Crohn’s disease patients carrying Nod2/CARD15 gene variants have an increased and early need for first surgery due to stricturing disease and higher rate of surgical recurrence. Ann Surg. 2005;242:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bhullar M, Macrae F, Brown G, et al. Prediction of Crohn’s disease aggression through NOD2/CARD15 gene sequencing in an Australian cohort. World J Gastroenterol. 2014;20:5008–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Büning C, Genschel J, Bühner S, et al. Mutations in the NOD2/CARD15 gene in Crohn’s disease are associated with ileocecal resection and are a risk factor for reoperation. Aliment Pharmacol Ther. 2004;19:1073–1078. [DOI] [PubMed] [Google Scholar]
- 17. Maconi G, Colombo E, Sampietro GM, et al. CARD15 gene variants and risk of reoperation in Crohn’s disease patients. Am J Gastroenterol. 2009;104:2483–2491. [DOI] [PubMed] [Google Scholar]
- 18. Onnie CM, Fisher SA, Prescott NJ, et al. Diverse effects of the CARD15 and IBD5 loci on clinical phenotype in 630 patients with Crohn’s disease. Eur J Gastroenterol Hepatol. 2008;20:37–45. [DOI] [PubMed] [Google Scholar]
- 19. Renda MC, Orlando A, Civitavecchia G, et al. The role of CARD15 mutations and smoking in the course of Crohn’s disease in a Mediterranean area. Am J Gastroenterol. 2008;103:649–655. [DOI] [PubMed] [Google Scholar]
- 20. VanDussen KL, Liu TC, Li D, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology. 2014;146:200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahmad T, Armuzzi A, Bunce M, et al. The molecular classification of the clinical manifestations of Crohn’s disease. Gastroenterology. 2002;122:854–866. [DOI] [PubMed] [Google Scholar]
- 22. Martinek L, Kupka T, Simova J, et al. Original article: NOD2/CARD15 mutations and the risk of reoperation in patients with Crohn’ s disease. Rozhl. Chir. 2015;94:242–46. [PubMed] [Google Scholar]
- 23. Yang DH, Yang SK, Song K, et al. TNFSF15 is an independent predictor for the development of Crohn’s disease-related complications in Koreans. J Crohns Colitis. 2014;8:1315–1326. [DOI] [PubMed] [Google Scholar]
- 24. Sehgal R, Berg A, Polinski JI, et al. Mutations in IRGM are associated with more frequent need for surgery in patients with ileocolonic Crohn’s disease. Dis Colon Rectum. 2012;55:115–121. [DOI] [PubMed] [Google Scholar]
- 25. Ta-Chiang L, Feng G,Thaddeus S. P-070 an integrated risk stratification system incorporating paneth cell phenotype and clinical parameter predicts outcome in post-operative Crohn’s disease patients. Inflamm Bowel Dis. 2014;20.
- 26. Chen C, Zhang T, Li E, et al. The effects of NOD2 genotype, smoking and immunomodulators on postoperative recurrence of ileal Crohn’s disease Chien-Huan. Gastroenterology. 2011;6:s29. [Google Scholar]
- 27. Fowler SA, Ananthakrishnan AN, Gardet A, et al. SMAD3 gene variant is a risk factor for recurrent surgery in patients with Crohn’s disease. J Crohns Colitis. 2014;8:845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu TC, Naito T, Liu Z, et al. LRRK2 but not ATG16L1 is associated with Paneth cell defect in Japanese Crohn’s disease patients. JCI Insight. 2017;2:e91917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siegel CA, Fleshner P, Siegel LS, et al. Predicting Crohn’s disease post-operative recurrence using clinical, endoscopic, serologic and genetic factors. Gastroenterology. 2011;140:S-153. [Google Scholar]
- 30. Naito T, Liu T-C, Kakuta Y, et al. 321 Paneth cell phenotype is associated with novel genetic determinants and clinical outcome in Japanese Crohn’s disease patients. Gastroenterology. 2016;150:S75. [Google Scholar]
- 31. Seiderer J, Schnitzler F, Brand S, et al. Homozygosity for the CARD15 frameshift mutation 1007fs is predictive of early onset of Crohn’s disease with ileal stenosis, entero-enteral fistulas, and frequent need for surgical intervention with high risk of re-stenosis. Scand J Gastroenterol. 2006;41:1421–1432. [DOI] [PubMed] [Google Scholar]
- 32. Potdar AA, Li D, Haritunians T, et al. Ileal gene expression data from Crohn’s disease small bowel resections indicate distinct clinical subgroups. J Crohns Colitis. 2019;13:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Germain A, Guéant RM, Chamaillard M, et al. NOD2 gene variant is a risk factor for postoperative complications in patients with Crohn’s disease: a genetic association study. Surgery. 2016;160:74–80. [DOI] [PubMed] [Google Scholar]
- 34. Laffin MR, Fedorak RN, Wine E, et al. A BACH2 gene variant is associated with postoperative recurrence of Crohn’s disease. J Am Coll Surg. 2018;226:902–908. [DOI] [PubMed] [Google Scholar]
- 35. Gathungu G, Zhang Y, Tian X, et al. Impaired granulocyte-macrophage colony-stimulating factor bioactivity accelerates surgical recurrence in ileal Crohn’s disease. World J Gastroenterol. 2018;24:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gerich ME, Fleshner P, Panikkath D, et al. Mo1299 genotype and post-operative immunosuppression impact surgical recurrence in Crohn’s disease. Gastroenterology. 2013;144:S-630. [Google Scholar]
- 37. Ghaly S, Murray K, Baird A, et al. High vitamin D-binding protein concentration, low albumin, and mode of remission predict relapse in Crohn’s disease. Inflamm Bowel Dis. 2016;22:2456–2464. [DOI] [PubMed] [Google Scholar]
- 38. Burke JP, O’Connell RM, Lennon G, et al. The influence of CTGF single-nucleotide polymorphisms on outcomes in Crohn’s disease. Ann Surg. 2013;258:767–773; discussion 773. [DOI] [PubMed] [Google Scholar]
- 39. Meijer MJ, Mieremet-Ooms MA, Sier CF, et al. Matrix metalloproteinases and their tissue inhibitors as prognostic indicators for diagnostic and surgical recurrence in Crohn’s disease. Inflamm Bowel Dis. 2009;15:84–92. [DOI] [PubMed] [Google Scholar]
- 40. Germain A, Guéant RM, Chamaillard M, et al. CARD8 gene variant is a risk factor for recurrent surgery in patients with Crohn’s disease. Dig Liver Dis. 2015;47:938–942. [DOI] [PubMed] [Google Scholar]
- 41. Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- 42. Benchimol EI, Fortinsky KJ, Gozdyra P, et al. Epidemiology of pediatric inflammatory bowel disease: a systematic review of international trends. Inflamm Bowel Dis. 2011;17:423–439. [DOI] [PubMed] [Google Scholar]
- 43. Buisson A, Chevaux JB, Allen PB, et al. Review article: the natural history of postoperative Crohn’s disease recurrence. Aliment Pharmacol Ther. 2012;35:625–633. [DOI] [PubMed] [Google Scholar]
- 44. Peyrin-Biroulet L, Harmsen WS, Tremaine WJ, et al. Surgery in a population-based cohort of Crohn’s disease from Olmsted County, Minnesota (1970–2004). Am J Gastroenterol. 2012;107:1693–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ganz ML, Sugarman R, Wang R, et al. The economic and health-related impact of Crohn’s disease in the united states: evidence from a nationally representative survey. Inflamm Bowel Dis. 2016;22:1032–1041. [DOI] [PubMed] [Google Scholar]
- 46. Regueiro M, Velayos F, Greer JB, et al. American gastroenterological association institute technical review on the management of Crohn’s disease after surgical resection. Gastroenterology. 2017;152:277–295.e3. [DOI] [PubMed] [Google Scholar]
- 47. Unkart JT, Anderson L, Li E, et al. Risk factors for surgical recurrence after ileocolic resection of Crohn’s disease. Dis Colon Rectum. 2008;51:1211–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wright EK, Kamm MA, Wagner J, et al. Microbial factors associated with postoperative Crohn’s disease recurrence. J Crohns Colitis. 2017;11:191–203. [DOI] [PubMed] [Google Scholar]
- 49. Mondot S, Lepage P, Seksik P, et al. Structural robustness of the gut mucosal microbiota is associated with Crohn’s disease remission after surgery. Gut. 2016;65:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Cruz P, Kang S, Wagner J, et al. Association between specific mucosa‐associated microbiota in Crohn’s disease at the time of resection and subsequent disease recurrence: A pilot study. J Gastroenterol Hepatol. 2015;30:268–278. [DOI] [PubMed] [Google Scholar]
- 51. Dey N, Soergel DA, Repo S, et al. Association of gut microbiota with post-operative clinical course in Crohn’s disease. BMC Gastroenterol. 2013;13:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Laffin MR, Perry T, Park H, et al. Endospore forming bacteria may be associated with maintenance of surgically-induced remission in Crohn’s disease. Sci Rep. 2018;8:9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hampe J, Cuthbert A, Croucher PJ, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet. 2001;357:1925–1928. [DOI] [PubMed] [Google Scholar]
- 54. Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. [DOI] [PubMed] [Google Scholar]
- 55. Girardin SE, Boneca IG, Viala J, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. [DOI] [PubMed] [Google Scholar]
- 56. Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. [DOI] [PubMed] [Google Scholar]
- 57. Plantinga TS, Crisan TO, Oosting M, et al. Crohn’s disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011;60:1229–1235. [DOI] [PubMed] [Google Scholar]
- 58. Adler J, Rangwalla SC, Dwamena BA, et al. The prognostic power of the NOD2 genotype for complicated Crohn’s disease: a meta-analysis. Am J Gastroenterol. 2011;106:699–712. [DOI] [PubMed] [Google Scholar]
- 59. Kuenzig ME, Yim J, Coward S, et al. The NOD2-smoking interaction in Crohn’s disease is likely specific to the 1007fs mutation and may be explained by age at diagnosis: a meta-analysis and case-only study. Ebiomedicine. 2017;21:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brzyski D, Peterson CB, Sobczyk P, et al. Controlling the rate of GWAS false discoveries. Genetics. 2017;205:61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were created for this study.