Abstract
A major challenge to end the pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is to develop a broadly protective vaccine that elicits long-term immunity. As the key immunogen, the viral surface spike (S) protein is frequently mutated, and conserved epitopes are shielded by glycans. Here, we revealed that S protein glycosylation has site-differential effects on viral infectivity. We found that S protein generated by lung epithelial cells has glycoforms associated with increased infectivity. Compared to the fully glycosylated S protein, immunization of S protein with N-glycans trimmed to the mono-GlcNAc–decorated state (SMG) elicited stronger immune responses and better protection for human angiotensin-converting enzyme 2 (hACE2) transgenic mice against variants of concern (VOCs). In addition, a broadly neutralizing monoclonal antibody was identified from SMG-immunized mice that could neutralize wild-type SARS-CoV-2 and VOCs with subpicomolar potency. Together, these results demonstrate that removal of glycan shields to better expose the conserved sequences has the potential to be an effective and simple approach for developing a broadly protective SARS-CoV-2 vaccine.
A vaccine developed by removing glycan shields in the SARS-CoV-2 S protein conferred protection against infection with variants of concern.
A sugar-free vaccine
Vaccines that elicit broadly neutralizing antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) will be essential for curbing the ongoing coronavirus disease 2019 (COVID-19) pandemic. Here, Huang et al. evaluated the immunological impact of enzymatically removing glycan shields from the SARS-CoV-2 spike (S) protein. The authors found that immunizing hamsters or mice with S protein lacking glycan shields, termed SMG, elicited robust and broadly reactive immune responses. Vaccinated mice were protected from challenge with the alpha, gamma, and delta variants of concern. Antibodies isolated from vaccinated animals could also neutralize the omicron variant. Together, these data support further investigation into SMG as a candidate SARS-CoV-2 vaccine.
INTRODUCTION
Spike (S) protein, the main focus of vaccine development for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), contains 22 N-linked and at least 2 O-linked glycosylation sites per monomer (1–3). These sites are important for S protein folding and processing as well as for evading immune recognition by shielding specific epitopes, thus hindering the efficacy of the vaccine (2, 3). Understanding the glycosylation of S protein can uncover the role in which glycans play and guide rational vaccine design (2, 3). The glycan profiles of S protein expressed from various cell sources have been reported, revealing a conserved pattern of 8 specific N-glycosylation sites harboring at least 30% underprocessed high mannose–type and hybrid-type N-glycans, with the remaining 14 sites predominantly of the complex type (2, 4). It was shown that a trimeric S protein with its original complex type–dominant glycoform is more efficient in receptor recognition and viral entry than the high-mannose variants derived either from N-acetylglucosaminyltransferase I (GnTI)¯ human embryonic kidney (HEK) 293 (5) or alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase (MGAT1)¯ HEK293T cells (6).
In this study, we investigated the differential influences of overall and site-specific glycosylation of the S protein on SARS-CoV-2 infectivity. We further analyzed the glycosylation profile of S protein expressed from lung epithelial cells, the primary cell type infected by SARS-CoV-2, to evaluate the correlation between S protein glycosylation, protein conservation, and glycan shielding. The results led us to generate a mono-GlcNAc–decorated S protein (SMG) as a candidate vaccine immunogen to expose the conserved glycan-shielded epitopes on the protein surface. In vitro and in vivo studies of the SMG immunogen as a vaccine against SARS-CoV-2 and variants of concern (VOCs), as well as the isolation of a cross-neutralizing antibody m31A7, suggested that the vaccine design with removal of glycan shields is a simple strategy to develop a broadly reactive coronavirus disease 2019 (COVID-19) vaccine.
RESULTS
Glycosylation affects pseudovirus S protein interacting with ACE2 on several cell types
To understand the importance of glycosylation, we expressed S protein from lung epithelial cells, the primary cells for infection, and found that sialylation of S protein is required for higher avidity to the receptor (Fig. 1A). A similar pattern was also observed for HEK293T cell–generated S protein (Fig. 1B), and the avidity was also reduced for the S protein with only high-mannose glycans or in the glycoform with all N-glycans trimmed to a single N-acetylglucosamine (GlcNAc) (Fig. 1C). The impact of its glycosylation was further tested by pseudovirus infection in human angiotensin-converting enzyme 2 (hACE2)–expressing HEK293T cells, revealing a consistent trend, when the same amount of virus was applied (Fig. 1D and fig. S1). This allowed us to conclude that complex-type glycans and sialylation are functionally important for S protein–mediated infectivity. A full panel of 24 lentivirus-based pseudovirus variants (comprising the 22 N- and 2 O-glycosites) were also generated for evaluating the viral entry efficiency in five hACE2-expressing cell lines, including HEK293T, Vero-E6, and three human lung cell lines, A549, Calu-1, and Calu-3 cells (Fig. 1, E to G). These pseudoviruses were based on the S construct with C-terminal 19 amino acid deletion, which produced the highest viral titer (fig. S2). Pseudovirus production was quantified by a p24 immunoassay, and results were normalized against the titer of each mutant strain (Fig. 1F). Every N-glycosite asparagine (Asn) was substituted to glutamine (Gln) to minimize the structural influence because of their chemical similarity, and each O-glycosite threonine (Thr) or serine (Srn) was substituted to alanine (Ala). Because the mutagenesis did change the amino acids, the resulting change in infectivity would come from collective factors, including the glycosylation-related conformational shifts that affect receptor engagement and the surface abundance of S protein affected by protein expression, folding, and trafficking. Results showed that disruption of S protein glycosylation reduced infectivity, which is similar to the findings using HEK293T cells (7). A substantial reduction was observed for two mutations in the receptor binding domain (RBD), N331Q and N343Q, as well as for mutations of the two O-glycosites (T323A and S325A), despite the low occupancy of the latter (Fig. 1G) (2). In addition, deletion of N122 glycosylation in the N-terminal domain (NTD) resulted in reduced infectivity and low protein expression (Fig. 1G and fig. S3A). Two NTD mutations, N149Q and N165Q, increased infectivity in Vero-E6 and Calu-1 cells, respectively, although decreased infections were observed in other cells (Fig. 1G). Note that the glycans attached to this N165 residue are structurally proximal to the neighboring RBD in the trimeric S protein (2), and its mutation reduced ACE2 binding probably because of the conformational shift of RBD toward the “down” state (8). We identified two mutants, N801Q and N1194Q (Fig. 1F), that universally abolished virus infectivity in all five cells. The glycosite N801 is located near the fusion peptide proximal region (FPPR), and N1194 is near the center of heptad repeat 2 (HR2) helix and is the last N-glycosite preceding the transmembrane domain (Fig. 1E and fig. S3B). These mutations both caused low-yield expression (fig. S3A). The N801Q mutant was more prone to degradation, and the N1194Q mutant disrupted S protein trimerization (fig. S3, C and D), which could be part of the explanation for the reduction of infectivity by pseudoviruses carrying these mutants.
Fig. 1. S protein glycosylation impacts ACE2 receptor binding and SARS-CoV-2 infection.
(A to C) Binding avidity of ACE2 was measured for differently glycosylated S protein ectodomains (SFG; original fully glycosylated, blue; deS, nonsialylated, light blue; SHM, high-mannose, yellow; and SMG, mono-GlcNAc, red) from BEAS-2B (A), HEK293T (B), and HEK293S (GnTI¯) cells without or with Endo H digestion (C). Data of three technical replicates are shown as means ± SD, and curves fit by nonlinear regression for EC50 values. (D) Viral infectivity was measured for pseudoviruses carrying differently glycosylated S protein with the same input amount (0.3 μg/ml of p24 equivalent) colored accordingly as in (C). RLU, relative luminescence unit. Data of six technical replicates shown as means ± SD and analyzed with ordinary one-way ANOVA test followed by Tukey’s multiple comparisons test. ns, not significant; ****P < 0.0001. (E) A schematic view of SARS-CoV-2 S protein [wild type (WT)] is shown colored by domain, including N-terminal domain (NTD; 14–306; orange), receptor binding domain (RBD; 319–541; red), two subdomains (SD1/2; 542–685; yellow), fusion peptide proximal region (FPPR; 816–856; green), heptad repeat 1 (HR1; 912–984; teal), connecting domain (CD; 1063–1162; blue), heptad repeat 2 (HR2; 1163–1211; purple), and transmembrane domain (TM; 1214–1234; white). N-glycan (drawn as branches) and O-glycan (circles) sites are marked with residue number. S1 and S2 domains are shown above. (F) Viral titers are shown for pseudoviruses carrying WT S protein or mutants with glycans removed at each shown glycosite, normalized by p24 quantification, and colored accordingly as in (E). (G) Infectivity of the same panel of pseudoviruses as in (F) tested in five hACE2-expressing cell lines. Values in (F) and (G) are normalized against WT values (defined as 100%, colored in dark gray) with means ± SD of three independent experiments.
S protein from lung epithelial cells contains more sialylated complex-type glycans
The glycan profile analysis of S protein revealed a higher abundance of complex-type glycans (78%), and fewer hybrid-type glycans (less than 1%) for S protein were produced in the human lung epithelial cell line BEAS-2B (Fig. 2A and table S1) as compared with S protein produced in the human kidney epithelial cell line, HEK293T (61 and 23%, respectively) (Fig. 2B and table S2). Among the high mannose–type glycans, the N-linked mannose-5 glycan (man5) was the predominant type found across the HEK293T-expressed S protein, although it is only seen at site N61 from BEAS-2B cells (figs. S4 and S5). In addition, the complex-type glycans at sites N74, N149, N282, and N1194 from BEAS-2B cells were more diversely processed (multiple antennae, galactosylation, fucosylation, or sialylation) than those from HEK293T cells. In contrast, the glycans at sites N122, N331, N1098, and N1134 were less diverse (figs. S4 and S5). Furthermore, N149 and N17 harbored no core fucose in BEAS-2B. We observed an overall higher degree of sialylation on all 22 N-glycosites from BEAS-2B (53%) than that from HEK293T (35%), HEK293E (26%) (tables S1 to S3 and fig. S6), or the previously reported HEK293F cells (15%) (2). Particularly, the two N-glycosites (N331 and N343) of RBD are more sialylated in BEAS-2B (99 and 39%) than in HEK293T (49 and 15%) (tables S1 and S2). Despite the differences, the S protein from all cell types contains a non–complex-type glycan belt located around the middle section of the S2 domain (Fig. 2C and fig. S7), where the N-glycosite N801 is critical for infection (Fig. 1G), N1074 contains diverse glycans (figs. S4 to S6), and N717 is essential for S protein expression (fig. S3A).
Fig. 2. S protein glycan profiles demonstrate differences in two cell lines and correlate with sequence conservation.
(A and B) A comparison of the N-glycosylation profile of recombinant S protein expressed from BEAS-2B lung epithelial cells (A) and HEK293T kidney epithelial cells (B) is shown. Glycans are grouped and colored accordingly: complex-S (sialylated complex type; dark blue), complex (non–sialylated complex type; light blue), hybrid-S (sialylated hybrid type; dark yellow), and hybrid (nonsialylated hybrid type; light yellow), high mannose (green), and unoccupied (gray). The percentage of each group is shown for each glycosite in a pie chart, and the proportion of each glycoform (nos. 1 to 27) in a bar chart. The bar graphs represent the means ± SD of three biological replicates. Detailed structure and percentage of each glycoform can be found in tables S1 to S3 and figs. S4 to S6. Fhybrid indicates fucosylated hybrid-type glycans. (C) Glycan profiles from (A) and (B) were mapped on the 3D structure of S ectodomain (modeled from 6VSB). Glycans are colored by the highest-abundance group for BEAS-2B (left) or HEK293T (right) data as labeled (complex type, blue; hybrid type, yellow; and high mannose, green). Non–complex-type N-glycosites are labeled with residue number. (D) Mapping of relative surface accessibility (RSA) on modeled S structure protein is shown, with buried residues colored in dark yellow, glycan shielded in yellow, and exposed in light yellow. (E) Mapping of sequence variation on modeled S protein structure is shown, colored in a heatmap, with darker red indicating higher mutation rates. Several highly conserved glycan-shielded regions are highlighted. More details for (D) and (E) can be found in fig. S9 and data file S1.
Highly conserved epitopes in S protein are largely shielded by glycans
From the modeled SARS-CoV-2 S protein structure and the glycan profile from BEAS-2B cells, we conducted structural analysis of glycan coverage over protein surface areas and overlaid with multiple alignment results using 1,117,474 S protein sequences (9). It revealed several regions that were highly conserved, yet shielded by glycans, including the lower flank of RBD, the S2 stem region with the non–complex-type glycan belt, and the C-terminal part of S2 involving the connecting domain (CD) and HR2 (Fig. 2, D and E). On the primary sequence level, these regions were shown as conserved epitopes (fig. S8). Sequence conservation analysis also showed that most of the glycosite regions were highly conserved (fig. S9A), and the most conserved ones (mutation rate lower than 0.02%) included those in the NTD (N61, N122, N165, and N234), the RBD (S325, N331, and N343), the subdomain 1/2 (SD1/2) (N603 and N657), the stem region of subunit 2 (S2) (N709, N1098, N1134, N1158, N1173, and N1194), and N801 near the FPPR. Most of these regions contain around 20 to 40% of conserved surface residues (fig. S9B), and among them, a certain percentage of residues were shielded by glycans, 36% in the RBD and about 50% in other regions (fig. S9C). Although harboring no N-glycosites, the HR1 region had 69% of its conserved surface residues being covered by glycans stemmed from adjacent domains (fig. S9C). These results highlighted the importance of S protein glycosylation, both structurally and evolutionarily, leading to the thought that exposing glycan-shielded conserved regions may elicit immune responses against conserved epitopes.
SMG was developed as a vaccine
Our initial attempt to mutate multiple glycosites led to a markedly reduced expression of S protein (fig. S3A). Yet, when we expressed it from GnTI¯ HEK293S cells (fig. S10, A and B), we were able to produce a high-mannose glycoform S protein (SHM) with good yield and purity (fig. S10C). We then trimmed the glycans using endoglycosidase H (Endo H) to a single GlcNAc at each N-glycosite (fig. S10, A and B) (10, 11), generating a soluble trimmer mono-GlcNAc–decorated S protein, which we called SMG (Fig. 3A and fig. S10, D and E). SMG was confirmed by mass spectrometry that all N-glycosites mostly occupied with single GlcNAc (fig. S10F), and the occupancies of untreated O-glycans were too low to be detected. This modified SMG, and SHM, as well as the original fully glycosylated S protein (SFG) were mixed with aluminum hydroxide (alum) as an adjuvant and were then used to immunize BALB/c mice (n = 5) by intramuscular injection (Fig. 3B). The SFG used for comparison in this study was expressed by HEK293E cells and contained diverse glycans; this is similar to the immunogens used in many current COVID-19 vaccines that are either approved or in clinical trials, including the insect cell–expressed S protein vaccines from Sanofi and Novavax (1), the Chinese hamster ovary (CHO) cell–expressed recombinant S vaccine from Medigen (12), the adenovirus-based vaccines from AstraZeneca and Johnson & Johnson, and the mRNA vaccines from Pfizer-BioNTech and Moderna (1). Both SFG and SMG proteins demonstrate essentially the same trimeric structure in solution by negative staining analysis (fig. S10, G and H).
Fig. 3. SMG vaccination elicits stronger humoral and cellular immune responses than SFG in BALB/c mice.
(A) Structural models of SFG and SMG protein vaccine are shown (according to Fig. 2C). Blue: glycans; gray: protein. SFG was expressed by HEK293E without further modification. SMG was obtained by enzymatic digestion to truncate all N-glycans of SHM expressed by HEK293S GnTI¯ to single GlcNAc, whereas O-glycans were unmodified. (B) Immunization schedule using proteins as in (A) as immunogens in BALB/c mice (n = 5 in each experiment). SFG (blue), SHM (yellow), SMG (red), and control (gray). Alum, aluminum hydroxide. (C) Anti–S protein IgG titers of serum samples were analyzed by ELISA. (D) Neutralization titers of serum samples were measured using pseudovirus with WT S protein. (E to G) IgG subtype analysis of sera, including IgG1 (E), IgG2a (F), and the IgG2a:IgG1 ratio (G). (H to K) The percentage of Tfh in activated nonregulatory CD4 T cells (H) and the percentages of IFN-γ (I)–, IL-4 (J)–, and IL-21 (K)–expressing Tfh cells (CD4+CD19−CD44hiFoxp3−PD-1+CXCR5+) in the lymph nodes (LNs) of BALB/c mice by flow cytometry. (L) The percentage of granzyme B−producing CD8+ T cells (CD3+ B220−CD8+ CD49b−) in the LN of BALB/c mice analyzed by flow cytometry. (M) The ratio of S protein–specific B cells (CD3−CD19+S protein+) (percentage) normalized to fluorescence minus one (FMO) control staining (stained without S protein) (percentage) in the spleen is shown. (N) Kappa and lambda light chain usage is shown. (O and P) Heavy (O) and kappa (P) chain distribution of B cell repertoire analysis. Less than 5% usage is shown in white. (Q to S) Anti−S protein IgG titers (Q), pseudovirus neutralization titers (R), and authentic virus neutralization titers (S) are shown for serum isolated from BALB/c mice after three doses of indicated vaccines against SARS-CoV-2 WT (or D614G) and variants (number above each bar indicate fold of increase of SMG compared to SFG group). pNT50 represents the reciprocal dilution achieving 50% neutralization. The dotted line in bar charts represents the lower limit of detection. Data are shown as means ± SEM and analyzed by two-sided Mann-Whitney U test to compare two experimental groups, except in (N), where five samples were pooled together and a chi-squared test was used. P values shown above each bar. *P < 0.05; **P < 0.01.
SMG vaccine elicited better immune response using different antibody subclasses
Mice immunized with SMG induced superior humoral immune response after second immunization as compared with SFG, with a 1.44-fold significantly higher immunoglobulin G (IgG) titer against S protein (end point titer: SFG, 39,408 ± 1,619; SMG, 56,957 ± 5,091; P = 0.0079) (Fig. 3C) and 3.6-fold stronger antibody neutralization potency based on the inhibition of SARS-CoV-2 pseudovirus infection (reciprocal half maximal neutralization titer pNT50: SFG, 1346 ± 285; SMG, 4791 ± 767; P = 0.0159) (Fig. 3D), whereas SHM-immunized group shows similar anti–S IgG titers (39,086 ± 11,654) and no difference in pNT50 titer compared with the SFG group. The analysis of IgG subtype titer and interferon-γ (IFN-γ) or interleukin-4 (IL-4) production by T follicular helper (Tfh) cells revealed that SMG vaccine induced more IgG2a, which is the marker for T helper 1 cell (TH1) lymphocytes in BALB/c mice, a more balanced TH1/TH2 response, and more IFN-γ–expressing Tfh cells compared with the SFG- and SHM-vaccinated groups (Fig. 3, E to J). Furthermore, the SMG vaccine induced higher frequency of IL-21+ Tfh cells (Fig. 3K) and an elevated frequency of granzyme B–producing CD8+ T cells (Fig. 3L). These data indicated that a more potent humoral and cellular adaptive immune response was elicited by SMG, as compared with that induced by SFG. We then examined the frequency of S protein–specific B cells (CD3−CD19+S+) from the spleen of mice immunized after the third dose of SFG or SMG (Fig. 3A) and found that mice immunized with SMG generated more S protein–specific B cells (Fig. 3M and fig. S11, A and B). The B cell repertoire analysis from SFG- and SMG-immunized mice (n = 5) indicated that more lambda light chain genes were used in the SMG group compared with that in the SFG group (SFG, 1.92%; SMG, 9.68%) (Fig. 3N). In addition, antibodies derived from several specific loci of the Ig heavy chain variable region (IGHV) (Fig. 3O and fig. S11C) and the Ig kappa chain variable region (IGKV) genes (Fig. 3P and fig. S11D) were overrepresented in the SMG group than in the SFG group, especially the IGHV1-18 gene (Fig. 3O). This finding suggested that B cell epitopes may be processed differently in these two groups, and it remains to be further explored whether and why this difference is immunologically beneficial. In addition, the three-dose SMG vaccination elicited higher end point titer IgG than the two-dose vaccination against wild-type (WT) S protein (end point titer: SFG, 208,911 ± 50,092; SMG, 376,410 ± 80,873). We observed differences between the SMG and SFG groups in serum IgG-binding curves measured by enzyme-linked immunosorbent assay (ELISA) against the S protein from SARS-CoV-2 VOCs (13), including alpha (B.1.1.7; P = 0.0488), beta (B.1.351; P = 0.0010), gamma (P.1; P = 0.0068), and delta (B.1.617.2; P = 0.0068) (fig. S12A), but no statistical differences in end point titers analysis (Fig. 3Q). Differences in neutralizing antibody responses against VOCs by pseudovirus neutralization curve, including alpha (P = 0.0156), beta (P = 0.0156), and delta (P = 0.0078) (fig. S12B), were also observed, but no differences were observed in the pNT50 titer values for pseudovirus or authentic virus neutralization (Fig. 3, R and S; and fig. S12, C and D), as compared with SFG. These VOCs are of particular concern because they have shown resistance to several therapeutic antibodies and convalescent serum and because they have exhibited reduced protection by many approved vaccines (14–18).
SMG vaccine provided superior protection against SARS-CoV-2 and variants in vivo
To evaluate the in vivo protective efficacy of SMG vaccine against SARS-CoV-2, we first carried out WT SARS-CoV-2 challenge in Syrian hamsters vaccinated with SMG or SFG (Fig. 4A). SMG-vaccinated hamsters (n = 5) showed less reduction in body weight as compared with the SFG and phosphate-buffered saline (PBS) groups (Fig. 4B), whereas similar virus titer reductions were observed in the lungs of both SFG- and SMG-vaccinated hamsters (Fig. 4C). In addition, according to histopathological staining and anti–nucleocapsid (N) protein immunostaining data, fewer lesions were observed in the lungs of immunized hamsters (Fig. 4D and fig. S13). Because hamsters only showed mild to moderate sickness upon SARS-CoV-2 infection, we then used severe disease models, the highly susceptible CAG-hACE2 (19) or K18-hACE2 (20) transgenic mice (Fig. 4E). The analysis of anti–S IgG binding titer, neutralizing titers, anti–S subtype IgG, and IgG2c:IgG1 ratio (Fig. 4, F to I) in CAG-hACE2 mice all showed similar results to the BALB/c mice (Fig. 3, C to G). Following challenge with WT SARS-CoV-2 intranasally, virus was not detectable in the lungs of both SFG- and SMG-vaccinated CAG-hACE2 mice (n = 3) by anti-N staining at 7 days postinfection (dpi) (Fig. 4J) or median tissue culture infectious dose (TCID50) assay at 4 dpi (Fig. 4K), whereas a viral titer of over 1,000 TCID50 was observed in the control group (Fig. 4K). The SMG group (n = 4) exhibited better (75%) survival rate than the SFG (50%) at 14 dpi (Fig. 4, L and M). We then evaluated the degree of protection conferred by SMG vaccination against challenge with the alpha variant in CAG-hACE2 mice (n = 5). We found that SMG vaccination provided 100% survival rate until 14 dpi (Fig. 4, N and O). SMG-vaccinated mice also showed a 60% survival rate in the gamma variant challenge in CAG-hACE2 mice (n = 5) (Fig. 4, P and Q) and a 75% survival rate in the delta variant challenge in K18-hACE2 mice (n = 4) (Fig. 4, R and S), whereas less than 50% SFG-vaccinated mice survived until 14 dpi in the gamma and delta variant challenge (Fig. 4, Q to S). The improved in vivo protection conferred by SMG vaccination provides further evidence that removal of glycan shields from an immunogen is an advantageous strategy to elicit superior immune response.
Fig. 4. SMG vaccination provides enhanced protection against SARS-CoV-2 infection in vivo.
(A) The immunization schedule for Syrian hamsters is shown. SFG (blue), SMG (red), and control (gray). (B) Weight change was measured in Syrian hamsters after WT SARS-CoV-2 challenge. (C) Lung virus titers of challenged hamsters are shown. The dashed line indicates the lower limit of detection. (D) Representative images shown histopathology, immunohistochemistry, and immunofluorescence of the lungs from an infected hamster (3 dpi). First row: Hematoxylin and eosin (H&E) staining; scale bar, 50 μm. Second row: Immunohistochemistry (IHC) staining; scale bar, 50 μm. Third row: Immunofluorescence (IF) staining; scale bar, 100 μm. SARS-CoV-2 N-specific polyclonal antibodies were used for virus detection as brown dots in IHC and green dots in IF staining. Blue: 4,6-diamidino-2-phenylindole (DAPI). (E) The immunization schedule for CAG-hACE2 or K18-hACE2 transgenic mice is shown. (F to I) Anti−S IgG titers (F), SARS-CoV-2 WT microneutralization titers (G), and subtype IgG analysis, including IgG1, IgG2c (H), and IgG2c:IgG1 ratio (I), are shown for serum samples collected from immunized CAG-hACE2 transgenic mice (n = 7). (J) Representative histopathology, immunohistochemistry, and immunofluorescence of the infected mouse lungs (7 dpi) are shown. Scale bars are the same as in (D). (K) Lung virus titers of the infected CAG-hACE2 mice (n = 3). The dashed line indicates the lower limit of detection. (L and M) Weight change (L) and survival analysis (M) are shown for WT-SARS-CoV-2–challenged CAG-hACE2 transgenic mice (n = 4). (N and O) Weight change (N) and survival analysis (O) are shown for SARS-CoV-2 alpha variant–challenged CAG-hACE2 transgenic mice (n = 5). (P and Q) Weight change (P) and survival analysis (Q) are shown for SARS-CoV-2 gamma variant–challenged CAG-hACE2 transgenic mice (n = 5). (R and S) Weight change (R) and survival analysis (S) are shown for SARS-CoV-2 delta variant–challenged K18-hACE2 transgenic mice (n = 4). Data are shown as means ± SEM and analyzed by two-sided Mann-Whitney U tests to compare two experimental groups. ns, not significant; *P < 0.05.
A broadly neutralizing antibody was isolated from B cells of mice immunized with SMG
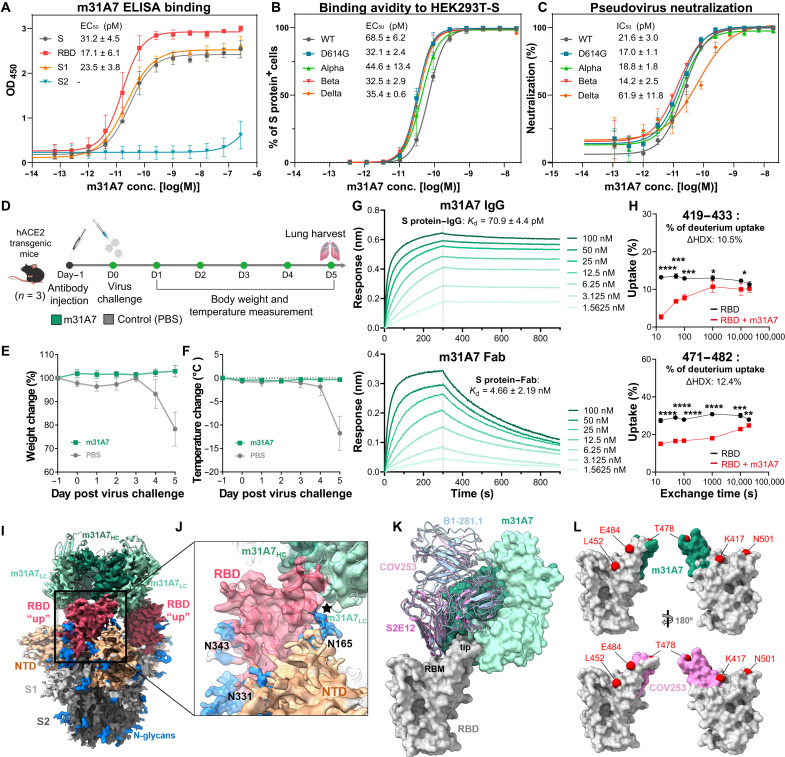
The sorting of S protein–specific B cells from SMG-immunized mice led to the identification of a monoclonal antibody (mAb) m31A7 from the IGHV1-18 amplified clones, a subset that is uniquely abundant in SMG-immunized B cell repertoire (Fig. 3O and fig. S11B). This mAb interacts with the full-length S protein, S1, and RBD, but not S2 (Fig. 5A), and binds to HEK293T cells that express the S protein from different SARS-CoV-2 variants (Fig. 5B). In addition, m31A7 was shown to neutralize various pseudovirus variants (WT, D614G, alpha, beta, and delta) at subpicomolar half-maximal inhibitory concentration (IC50) up to 1000-fold higher than the reported human mAb EY6A (Fig. 5C and fig. S14) (21). A prophylactic study also demonstrated good in vivo efficacy of m31A7 in K18-hACE2 mice (n = 3) challenged with WT SARS-CoV-2 (Fig. 5D). Prophylactically treated mice maintained both body weight and temperature (Fig. 5, E and F). Biolayer interferometry (BLI) analysis was used to measure the dissociation constants of m31A7 and its Fab binding to S protein at 70.9 pM and 4.66 nM, respectively (Fig. 5G). Epitope mapping by hydrogen-deuterium exchange mass spectrometry (HDX-MS) revealed its potential binding regions on RBD (Fig. 5H and fig. S15), which overlapped with the observed epitope in the crystal structure of RBD in complex with m31A7-Fab (fig. S16). The cryo–electron microscopy (EM) structure further clarified the binding of m31A7 only to RBD in the “up” state (Fig. 5I), with the N165-glycan from neighboring NTD in the vicinity of the RBD-m31A7 interface (Fig. 5J). The footprint of m31A7 on the RBD is similar to that of human VH1-58 class mAbs (Fig. 5K) (14, 22, 23), but it approaches the RBD from a different angle, with shifted local contact areas on the tip loop, bypassing most of key mutated residues of VOCs such as E484 and K417, but not T478 (Fig. 5L) (22, 23). The detailed RBD-m31A7 interface and the inhibitory mechanism of this SMG-elicited mAb are under further investigation.
Fig. 5. Functional, prophylactic, and structural characterization of antibody m31A7 elicited by SMG vaccination indicates cross-neutralizing capacity.
(A) ELISA binding of m31A7 to S1, S2, RBD, or the entire S ectodomain. (B) Flow cytometry analysis of m31A7 binding to HEK293T cells expressing S protein of SARS-CoV-2 WT and variants. (C) Neutralization activity of m31A7 against pseudoviruses carrying WT or variant S proteins. Data of three technical replicates for (A), (B), and (C) are shown as means ± SD and curves fit by nonlinear regression for EC50 values. (D) Antibody injection and challenge schedule for K18-hACE2 transgenic mice (n = 3) is shown. (E and F) Weight change (E) and body temperature change (F) are shown for mice treated with m31A7 or PBS. Data are presented as means ± SEM. (G) Binding kinetics of m31A7 IgG and Fab to S protein are presented, with dissociation constants (Kd) shown above. (H) Epitope mapping by HDX-MS of m31A7 is shown in a time course revealing two peptide candidates, 419–433 and 471–482, with greater than 10% ΔHDX at 15 s. Data are shown as means ± SD and analyzed by multiple t tests at each time point. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (I) The cryo-EM map fitted with m31A7-Fab/S protein complex structure is shown. Heavy chain, dark green; light chain, light green; RBD, red; NTD, orange; the rest of S1, light gray; S2, dark gray; and N-glycans, blue. (J) An enlarged view of RBD-m31A7 interface is shown. The star marks the vicinity between m31A7 light chain and N165-glycan. (K) Superimposition of previously reported mAbs S2E12 (magenta), COV253 (pink), and B1-182.1 (light blue) (PDB 7BEN, 7K4N, and 7MLZ) onto the m31A7-bound RBD (gray). The receptor binding motif and RBD tip are highlighted. (L) A footprint comparison of COV253 (pink) and m31A7 (green) on RBD (gray) shows similarity, with residues of VOCs labeled and drawn as red spheres.
DISCUSSION
Two years into the current global pandemic, a next-generation, broadly protective COVID-19 vaccine remains urgently needed. Although numerous studies have been published since the outbreak evaluating S protein glycosylation as well as different strategies for vaccine development, our work on a glycoengineered vaccine combines these two fields. The clinical translation of the current findings is promising, as protein-based vaccine platforms have been widely accepted with proven efficacy (24). Furthermore, the enzymatic digestion process to remove unwanted glycans is straightforward and could be readily incorporated into current manufacturing procedures. In a parallel effort, a glycoengineered mRNA vaccine recently developed by our group also showed impressive results, supporting our design concept that uncovering glycan-shielded epitopes led to a better vaccine with improved cross-protection (25).
A major concern at the present time is the inevitable emergence of new SARS-CoV-2 variants that carry immune-escape mutations (14, 26). Not a single N-glycosite mutation has been observed in any SARS-CoV-2 VOC identified so far except for the moderately conserved N17 site; this is true even for the omicron variant, which has more than 30 mutations in the S protein (27), again highlighting the importance of S protein glycosylation during viral evolution. In addition, an interesting comparison can be made between our SMG vaccine and other RBD-based vaccines, because S protein RBD, in its isolated form, is also barely covered by glycans (with the major coverage from N343). Nonetheless, the RBD contains several variation hotspots that are under stringent selective pressure, whereas our SMG vaccine targets the entire S protein ectodomain, stimulating the elicitation of both RBD and non–RBD-neutralizing antibodies that are critical for cross protection. Moreover, the improved CD4+ and CD8+ T cell responses elicited by SMG vaccination represent an additional advantage for this protein-based vaccine, especially given the increasing evidence that T cell immunity plays a central role against SARS-CoV-2 infection (28).
A matter that may be raised about the glycan removal design is that antibodies elicited by the SMG vaccine may fail to bind to the original glycosylated immunogen because of marked changes at the protein surface. This may have been the case in previous studies of HIV (2, 29), because neutralizing antibodies that target Env protein gp120 (with 480 residues per protomer and about 25 N-glycosites) are facing a heavily glycosylated surface with few accessible protein epitopes. However, SMG vaccines have been shown to be advantageous for both influenza virus and SARS-CoV-2, probably because influenza hemagglutinin (H1N1 HA; with 560 residues per protomer and about 6 to 8 N-glycosites) (10) and S protein (with 1270 residues per protomer and 22 N-glycosites) are both low to moderately glycosylated. In addition, these glycan chains are likely flexible enough to allow antibody binding even to the glycan-shielded, conserved epitopes.
We also recognized that there are some limitations of our present study. First, our experiments analyzing the impact of glycosites of S protein on interacting with ACE2 and the glycan profiling of S protein were done in human cell lines. It may be closer to the real scenario if primary lung epithelial cells, lung organoids, or other primary tissues expressing ACE2 can be used. Second, a comprehensive analysis of the differential T cell repertoire remains to be identified, particularly of the CD8 T cell repertoire elicited by SFG and SMG vaccination. It will reveal further information on whether and how glycans on S protein may affect antigen presentation. Third, although we have portrayed the differential IGHV, IGKV, and IGLV locus usage by SFG and SMG vaccination, the importance of those differentially elicited B cell repertoires contributing to the potent protection by SMG vaccination deserved further study. Other mAbs, particularly those belonging to the IGHV1-18 family, are worthwhile to be cloned and investigated. Last, the strategy has only been validated in animal models and remains to be confirmed in clinical trials.
In conclusion, SARS-CoV-2 S protein glycosylation has major influence on virus infection, protein integrity, and immune responses. The S protein from lung epithelial cells contained more sialylated complex-type glycans to facilitate receptor binding, and glycosites N801 and N1194 were shown to be essential for S protein folding and viral infection. The analysis of cell-specific glycoform distribution, sequence conservation, glycan shielding, and their mutual correlations led to the design of SMG vaccine, in which essentially all glycan shields are removed. This made the conserved epitopes better exposed to the immune system so that more effective and broadly protective B cell and T cell responses could be elicited against the virus and variants (30). As illustrated by the broadly neutralizing m31A7 antibody in this study and more to be identified in the future, the conserved epitopes targeted by such antibodies could be used for next-generation vaccine development. In an effort to develop vaccines against the currently circulating and future SARS-CoV-2 variants (31–34), the impact of glycosylation on viral infection, protein integrity, immune response, and vaccine design as demonstrated in this study should be considered together with other parameters.
MATERIALS AND METHODS
Study design
This work aims to analyze the impact of glycosylation of SARS-CoV-2 S protein and investigate the efficiency of a glycoengineered SMG as a vaccine candidate. ACE2 binding ELISA and glycan-engineered pseudovirus neutralization assays were used to understand the impact of glycosylation on receptor binding and viral infectivity. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) was applied to analyze and compare glycan profiles of S protein expressed from different cell lines. Computational modeling and bioinformatics analysis were used to visualize these findings on the three-dimensional (3D) structure in the context of evolutionary conservation. The SMG protein was designed, expressed, purified, characterized for glycan profiling, and injected into BALB/c mice for immune response analyses, including serum titers against SARS-CoV-2 WT and variants by ELISA and neutralization assay, T cell and B cell responses by flow cytometry, and B cell repertoire analysis by single B cell screening assay. The in vivo protection was studied in both hamsters and hACE2 transgenic mice by weight monitoring, virus detection (TCID50 and histopathology and immunostaining analysis), and survival analysis. A broad-spectrum ultrapotent antibody m31A7 was identified from SMG-immunized mice, with its efficiency analyzed by ELISA, flow cytometry, and pseudovirus neutralization, and its protection tested in transgenic mice, binding affinity by BLI, epitope mapping by HDX-MS, and structure determination by cryo-EM and x-ray crystallography. Mouse or hamster group sizes were determined on the basis of animal availability and power analyses using data from our pivotal experiments, and animals were randomly distributed between groups. No blinding was used throughout all studies, and no outliers were skewed to any one group. The selection of end points was made before the start of each study and decided based on the primary objective of investigating the immune responses to SMG vaccination referring to previous studies (12, 19, 20). The number of technical or biological replicates varied between experiments as described in the figure legends. These altogether serve as a foundation for further investigation and development of the SMG.
SARS-CoV-2 S protein production and characterization
The production of S protein was modified from previous studies (10, 35, 36), with details in the Supplementary Materials and Methods. pcDNA3.1 was used for construction and expression in HEK293T cells [American Type Culture Collection (ATCC) CRL-3216] or BEAS-2B cells (Homo sapiens, lung) (ATCC CRL-9609). The pTT vector was used for large-scale expression in HEK293 EBNA (ATCC CRL-10852) or HEK293S GnTI¯ (ATCC CRL-3022) suspension cells for vaccine studies. Purified SHM (from HEK293S GnTI¯ cells) was further treated with Endo H [New England Biolabs (NEB)] overnight at 25°C in a ratio of 50:1 (w/w) to produce SMG. S protein purity was monitored by SDS–polyacrylamide gel electrophoresis, with its binding avidity to hACE2 evaluated by ELISA, its glycan profile analyzed by LC-MS/MS [glycoform categorization followed a previous report (2)], and its structural conformation confirmed by negative staining EM (fig. S10, G and H).
Pseudovirus production with glycan-engineered S protein and glycosite-specific S mutants
The production of glycan-engineered pseudoviruses in (Fig. 1D) followed previous studies (2, 6). For production of glycosite-specific S mutant pseudoviruses (Fig. 1F), HEK293T cells were transiently transfected with pVax-nCoV-SΔ19 construct carrying mutations at each glycosite and luciferase-expressing HIV-1 genome plasmid (pNL4-3.luc.RE). Details of pseudovirus production and infectivity assay can be found in the Supplementary Materials and Methods.
Informatics analysis of SARS-CoV-2 S protein
A total of 1,117,474 S protein sequences of all available SARS-CoV-2 strains were extracted from the GISAID (Global Initiative on Sharing Avian Influenza Data) database (version: 18 April 2021) (9). The S protein 3D structure modeling was constructed by CHARMM-GUI (37) and OpenMM (38) based on the Protein Data Bank (PDB) file 6VSB_1_1_1 (39), with the most abundant glycoform of BEAS-2B data as representative glycan profile (fig. S4). O-glycans (T323 and S325) used Neu5Ac (α2,3)Gal (β1,3)GalNAc (α1) as representative (8). The definition of transmembrane region was according to UniProt (P0DTC2), and other parameters in CHARMM-GUI were according to Woo et al. (39). Scripts, parameters, and preoptimized models generated by CHARMM-GUI were used as the input for OpenMM. The protein secondary structure was determined by majority voting in the Dictionary of Secondary Structure of Proteins (DSSP) program (40, 41) and 2Struc web server (42). The relative solvent accessibility (RSA) of S protein was calculated by FreeSASA program (43) and averaged at each site of residues from three chains. The probe radius in FreeSASA was set to 7.2 Å to approximate the average size of hypervariable loops of antibody complementarity-determining regions (CDRs) (44). Residues with RSA above 5% were regarded as exposed, otherwise as buried (45). All 3D structure figures were drawn by UCSF ChimeraX (46).
Animal immunization, virus challenge, and prophylactic study of m31A7
For mice vaccination with a two-dose schedule, female 6- to 8-week-old BALB/c mice (n = 5) were immunized intramuscularly with 10 μg of purified SFG, SHM, or SMG mixed with aluminum hydroxide (50 μg) at days 0 and 14. The serum was collected at day 28 after the first vaccination for evaluation of anti–S IgG abundance, IgG subtype, and neutralizing titers (described in the Supplementary Materials and Methods). The lymph nodes of SFG- or SMG-immunized mice were collected at day 21 after the first vaccination for T cell response analysis (described in the Supplementary Materials and Methods). For B cell repertoire analysis and serum titers against variants, female 6- to 8-week-old BALB/c mice (n = 5) were immunized intramuscularly with 20 μg of purified SFG or SMG mixed with aluminum hydroxide (20 μg) at days 0, 14, and 56; mice were euthanized at day 84 to collect whole blood for anti–S IgG and neutralizing titer evaluation and spleens for sorting of S protein–specific B cells (described in the Supplementary Materials and Methods).
For hamster vaccination and virus challenge study, male 6- to 7-week-old golden Syrian hamsters (n = 5) were immunized intramuscularly with 25 μg of purified SFG or SMG mixed with aluminum hydroxide (250 μg) at days 0 and 14. Four weeks after the second immunization, each hamster was intranasally challenged with 1 × 104 TCID50 of SARS-CoV-2 (hCoV-19/Taiwan/4/2020) in 100 μl of PBS. Body weight was recorded daily after infection. On day 3 after challenge, hamsters were euthanized by carbon dioxide. The superior lobe of the left lung was fixed in 10% paraformaldehyde for histopathological examination, and the rest of the lung was collected for viral load determination (TCID50 assay) as described in the Supplementary Materials and Methods.
For transgenic mouse vaccination and virus challenge study, male 6- to 8-week-old CAG-hACE2 transgenic mice (19) or male 12-week-old K18-hACE2 transgenic mice (20) (purchased from the Jackson Laboratory) were immunized intramuscularly with 10 μg of purified SFG or SMG mixed with aluminum hydroxide (50 μg) at days 0 and 14. CAG-hACE2 transgenic mice were challenged intranasally 4 weeks after the second immunization with 1 × 103 TCID50 of WT SARS-CoV-2. In the first trial (n = 3), all mice were euthanized at 7 dpi for histopathological examination of superior lobe of the left lung; in the second trial (n = 7), three mice were euthanized at 4 dpi for lung virus titer, and four mice were kept until 14 dpi for survival analysis. Serum was collected 1 day before virus challenge.
For challenge studies using VOCs, CAG-hACE2 mice were challenged with 1 × 103 TCID50 of the alpha variant (hCoV-19/Taiwan/792/2020) (n = 5) or the gamma variant (hCoV-19/Taiwan/906/2021) of SARS-CoV-2 in 50 μl of PBS per mouse. In addition, K18-hACE2 mice were challenged intranasally 4 weeks after the second immunization with 1 × 104 TCID50 of the delta SARS-CoV-2 (hCoV-19/Taiwan/1144/2021) (n = 4) in 50 μl of PBS per mice. For all SARS-CoV-2 variant challenge models, body weight for each mouse was recorded daily until 14 dpi.
For prophylactic protection test of antibody, male 8-week-old K18-hACE2 transgenic mice (n = 3) were injected intraperitoneally with m31A7 (15 mg/kg) or PBS 1 day before being intranasally challenged with 1 × 103 TCID50 of WT SARS-CoV-2 (hCoV-19/Taiwan/4/2020). Body weight and body temperature were recorded daily until 5 dpi. All animal experiments were evaluated and approved by the Institutional Animal Care and Use Committee of Academia Sinica (approval nos. 21-10-1716, 18-12-1272, and 20-10-1522).
m31A7 isolation by single B cell screening assay and characterization
Primers were designed on the basis of a previous publication (47). Polymerase chain reaction (PCR) was performed at 50°C for 30 min, 95°C for 15 min, followed by 40 cycles of incubation at 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. Seminested second-round PCR was performed using KOD One PCR master mix (TOYOBO) with 1 μl of unpurified first-round PCR product at 98°C for 2 min, followed by 45 cycles of incubation at 98°C for 10 s, 55°C for 10 s, and 68°C for 10 s, with a final extension at 68°C for 1 min. PCR products were then analyzed by electrophoresis and sequencing. The Ig V and L genes were identified on the international ImMunoGeneTics information system (http://imgt.org/IMGT_vquest/input). Genes were then amplified from second-round PCR product with single gene-specific V and L gene primers containing restriction sites for cloning into the vectors containing human IgH or IgL expression backbone. The chimeric IgH and IgL expression constructs were cotransfected into Expi293 for antibody production. After m31A7 was isolated, the antibody was subsequently evaluated for S protein binding by ELISA and fluorescence-activated cell sorting, pseudovirus neutralization potency, binding kinetics, epitope mapping, and structure determination. Details are in the Supplementary Materials and Methods.
Statistical analysis
Raw data can be found in data file S1. All data were expressed as the means ± SEM or SD as mentioned individually. In Fig. 1D, ordinary one-way analysis of variance (ANOVA) test followed by Tukey’s multiple comparisons was used to compare data. In Fig. 5H, multiple t tests with the two-stage step-up method of Benjamini, Krieger, and Yekutieli multiple comparisons were used to compare each time point. For B cell repertoire analysis, a chi-square test was used. For all serum antibody titer analyses, P values were obtained from two-sided Mann-Whitney U tests to compare two experimental groups. Curves were fit by nonlinear regression using GraphPad Prism 9.0, and comparisons were performed by Wilcoxon matched pairs signed rank test (two-tailed). P < 0.05 was considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank the Academia Sinica specific pathogen–free (SPF) Animal Facility for providing animal support and the National RNAi Core Facility at Academia Sinica in Taiwan for providing pseudovirus reagents and related services. We thank the Mass Core Facility of Genomics Research Center, Academia Sinica for glycopeptide LC-MS/MS analysis and the Academia Sinica Proteomics Common Mass Spectrometry Facilities in the Institute of Biological Chemistry for HDX-MS. We thank the technical services provided by the National Synchrotron Radiation Research Center, Taiwan. We thank C.-K. Chang for helping with hamster vaccination, C.-W. Li and H.-C. Huang for providing ACE2-expressing cell lines, K.-Y. A. Huang for providing antibody EY6A, and C.-Y. Chen and K.-C. Lee for glycoform analysis.
Funding: The research was funded by Genomics Research Center, Summit Project (AS-SUMMIT-109) and the Translational Medical Research Program (AS-KPQ-109-BioMed), Academia Sinica and Taiwan Bio-Development Foundation (to C.M.). The Academia Sinica SPF Animal Facility, the Academia Sinica Transgenic Core Facility in the Institute of Molecular Biology, the Academia Sinica Biological Electron Microscopy Facility in the Institute of Cellular and Organismic Biology, and the Academia Sinica Proteomics Mass Spectrometry Common Facilities in the Institute of Biological Chemistry were funded by Academia Sinica Core Facility and Innovative Instrument Projects (AS-CFII-111-204, AS-CFII-108–104, AS-CFII-108-119, and AS-CFII-108-107, respectively). The Academia Sinica Cryo-EM Facility (ASCEM) was funded by Academia Sinica Core Facility and Innovative Instrument Projects (AS-CFII-108-110) and Taiwan Protein Project (AS-KPQ-109-TPP2). The Biosafety Level 3 Laboratory of the First Core Laboratory from National Taiwan University College of Medicine was supported by the Ministry of Science and Technology (MOST-110-2740-B-002-006 and MOST-109-2327-B-002-009).
Author contributions: C.M. and C.-H.W. conceived the idea. H.-Y.L., C.-Y.W., H.-Y.H., and Y.-C.C. performed pseudovirus production, infectivity, or neutralization. H.-Y.L. and T.-J.R.C. performed glycan profile analysis. C.-W.C. performed bioinformatics analysis. H.-Y.H., X.C., T.-H.C., and J.M.L. performed S protein vaccine design and preparation. H.-Y.H. performed immunization, serum collection, and vaccine analysis. J.-T.J., H.-H.M., Y.-M.L., S.-Y.C., T.-L.C., H.-C.K., and Y.-M.T. performed virus challenge animal studies and authentic virus neutralization in biosafety level 3. M.S.-A.-M. and Y.-H. Chen performed IHC and IF staining. A.M., X.C., and M.-S.A.-M. purified S protein variants. S.-W.W., Y.-H. Chang, and H.-Y.T. performed T cell analysis, B cell sorting, m31A7 identification, and antibody analysis. Y.-P.H., C.-Y.T., and P.-Y.H. generated and immunized CAG-hACE2 mice. X.C., Y.-M.W., and M.S.-A.-M. performed cryo-EM structure determination. A.M. performed crystal structure determination. C.-H.W., C.M., K.-I.L., H.-Y.H., X.C., S.-W.W., H.-Y.L., and C.-W.C. wrote the paper.
Competing interests: C.M., C.-H.W., and H.-Y.H. are inventors on a patent application (63/190,199) submitted by Academia Sinica that covers improved coronavirus vaccines. K.-I.L., C.M., C.-H.W., S.-W.W., Y.-H. Chang, X.C., and H.-Y.H. are inventors on a patent application (63/251,472) submitted by Academia Sinica entitled “A chimera monoclonal antibody possesses protective activity against various SARS-CoV-2 variants.” All other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. Negative stain volume of SMG was deposited in the Electron Microscopy Data Bank (EMD) under accession number EMD-32825. Cryo-EM data of the m31A7-bound S protein were deposited in EMD under EMD-32832 and in the Protein Data Bank (PDB) under ID 7WUH. Crystal structure of the m31A7-bound RBD was deposited in PDB under ID 7WUE. CAG-hACE2 mice are available from Y.-P.H. and C.-Y.T. lab under a material transfer agreement (MTA) with the Jackson Laboratory and Academia Sinica, Taiwan. SMG protein and m31A7 antibody are available upon request with Academia Sinica for evaluation by Y.-L. Chou (qbowchou@gate.sinica.edu.tw) and completion of an MTA. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using this material.
Supplementary Materials
This PDF file includes:
Other Supplementary Material for this manuscript includes the following:
Data file S1
MDAR reproducibility checklist
REFERENCES AND NOTES
- 1.Klasse P. J., Nixon D. F., Moore J. P., Immunogenicity of clinically relevant SARS-CoV-2 vaccines in nonhuman primates and humans. Sci. Adv. 7, eabe8065 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watanabe Y., Allen J. D., Wrapp D., McLellan J. S., Crispin M., Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369, 330–333 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao P., Praissman J. L., Grant O. C., Cai Y., Xiao T., Rosenbalm K. E., Aoki K., Kellman B. P., Bridger R., Barouch D. H., Brindley M. A., Lewis N. E., Tiemeyer M., Chen B., Woods R. J., Wells L., Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe 28, 586–601.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen J. D., Chawla H., Samsudin F., Zuzic L., Shivgan A. T., Watanabe Y., He W.-T., Callaghan S., Song G., Yong P., Brouwer P. J. M., Song Y., Cai Y., Duyvesteyn H. M. E., Malinauskas T., Kint J., Pino P., Wurm M. J., Frank M., Chen B., Stuart D. I., Sanders R. W., Andrabi R., Burton D. R., Li S., Bond P. J., Crispin M., Site-specific steric control of SARS-CoV-2 spike glycosylation. Biochemistry 60, 2153–2169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouwman K. M., Tomris I., Turner H. L., van der Woude R., Shamorkina T. M., Bosman G. P., Rockx B., Herfst S., Snijder J., Haagmans B. L., Ward A. B., Boons G.-J., de Vries R. P., Multimerization- and glycosylation-dependent receptor binding of SARS-CoV-2 spike proteins. PLOS Pathog. 17, e1009282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Q., Hughes T. A., Kelkar A., Yu X., Cheng K., Park S., Huang W.-C., Lovell J. F., Neelamegham S., Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. eLife 9, e61552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y., The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 182, 1284–1294.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casalino L., Gaieb Z., Goldsmith J. A., Hjorth C. K., Dommer A. C., Harbison A. M., Fogarty C. A., Barros E. P., Taylor B. C., McLellan J. S., Fadda E., Amaro R. E., Beyond shielding: The roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 6, 1722–1734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbe S., Buckland-Merrett G., Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Chall. 1, 33–46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J.-R., Yu Y.-H., Tseng Y.-C., Chiang W.-L., Chiang M.-F., Ko Y.-A., Chiu Y.-K., Ma H.-H., Wu C.-Y., Jan J.-T., Lin K.-I., Ma C., Wong C.-H., Vaccination of monoglycosylated hemagglutinin induces cross-strain protection against influenza virus infections. Proc. Natl. Acad. Sci. U.S.A. 111, 2476–2481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C.-C., Chen J.-R., Tseng Y.-C., Hsu C.-H., Hung Y.-F., Chen S.-W., Chen C.-M., Khoo K.-H., Cheng T.-J., Cheng Y.-S. E., Jan J.-T., Wu C.-Y., Ma C., Wong C.-H., Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl. Acad. Sci. U.S.A. 106, 18137–18142 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo T.-Y., Lin M.-Y., Coffman R. L., Campbell J. D., Traquina P., Lin Y.-J., Liu L. T.-C., Cheng J., Wu Y.-C., Wu C.-C., Tang W.-H., Huang C.-G., Tsao K.-C., Chen C., Development of CpG-adjuvanted stable prefusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. Sci. Rep. 10, 20085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krause P. R., Fleming T. R., Longini I. M., Peto R., Briand S., Heymann D. L., Beral V., Snape M. D., Rees H., Ropero A.-M., Balicer R. D., Cramer J. P., Muñoz-Fontela C., Gruber M., Gaspar R., Singh J. A., Subbarao K., Van Kerkhove M. D., Swaminathan S., Ryan M. J., Henao-Restrepo A.-M., SARS-CoV-2 variants and vaccines. N. Engl. J. Med. 385, 179–186 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey W. T., Carabelli A. M., Jackson B., Gupta R. K., Thomson E. C., Harrison E. M., Ludden C., Reeve R., Rambaut A., Consortium C.-G. U., Peacock S. J., Robertson D. L., SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19, 409–424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Beltran W. F., Lam E. C., Denis K. S., Nitido A. D., Garcia Z. H., Hauser B. M., Feldman J., Pavlovic M. N., Gregory D. J., Poznansky M. C., Sigal A., Schmidt A. G., Iafrate A. J., Naranbhai V., Balazs A. B., Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2372–2383.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G.-L., Wang Z.-Y., Duan L.-J., Meng Q.-C., Jiang M.-D., Cao J., Yao L., Zhu K.-L., Cao W.-C., Ma M.-J., Susceptibility of circulating SARS-CoV-2 variants to neutralization. N. Engl. J. Med. 384, 2354–2356 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., Myers R., Campbell C. N. J., Amirthalingam G., Edmunds M., Zambon M., Brown K. E., Hopkins S., Chand M., Ramsay M., Effectiveness of covid-19 vaccines against the B.1.617.2 (delta) variant. N. Engl. J. Med. 385, 585–594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corti D., Purcell L. A., Snell G., Veesler D., Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 184, 3086–3108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai C.-Y., Chen C.-Y., Jan J.-T., Chou Y.-C., Chang M.-L., Lu L. A., Huang P.-Y., Chu M. F. C., Hsu T.-T., Hsueh Y.-P., Sex-biased response to and brain cell infection by SARS-CoV-2 in a highly susceptible human ACE2 transgenic model. bioRxiv, 2021.2005.2004.441029 (2021). [Google Scholar]
- 20.Winkler E. S., Bailey A. L., Kafai N. M., Nair S., McCune B. T., Yu J., Fox J. M., Chen R. E., Earnest J. T., Keeler S. P., Ritter J. H., Kang L.-I., Dort S., Robichaud A., Head R., Holtzman M. J., Diamond M. S., SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou D., Duyvesteyn H. M. E., Chen C.-P., Huang C.-G., Chen T.-H., Shih S.-R., Lin Y.-C., Cheng C.-Y., Cheng S.-H., Huang Y.-C., Lin T.-Y., Ma C., Huo J., Carrique L., Malinauskas T., Ruza R. R., Shah P. N. M., Tan T. K., Rijal P., Donat R. F., Godwin K., Buttigieg K. R., Tree J. A., Radecke J., Paterson N. G., Supasa P., Mongkolsapaya J., Screaton G. R., Carroll M. W., Gilbert-Jaramillo J., Knight M. L., James W., Owens R. J., Naismith J. H., Townsend A. R., Fry E. E., Zhao Y., Ren J., Stuart D. I., Huang K.-Y. A., Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 27, 950–958 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Wang L., Zhou T., Zhang Y., Eun S. Y., Chaim A. S., Shi W., Pegu A., Olamide K. O., Amy R. H., Darko S., Sandeep R. N., Hatcher C., David R. M., Tsybovsky Y., Phung E., Olubukola M. A., Antia A., Evan M. C., Lauren A. C., Choe M., Kizzmekia S. C., Rachel L. D., DiPiazza Anthony T., Ingelise J. G., Sabrina H. H., Hermanus T., Kgagudi P., Laboune F., Leung K., Liu T., Rosemarie D. M., Alexandra F. N., Novik L., O’Connell S., O’Dell S., Adam S. O., Stephen D. S., Stephens T., Christopher D. S., Chloe A. T., Teng I. T., Danielle A. W., Alicia T. W., Zhang B., Roederer M., Julie E. L., Tracy J. R., Martin R. G., Penny L. M., Nicole A. D.-R., Ralph S. B., Barney S. G., McDermott Adrian B., Daniel C. D., Peter D. K., John R. M., Nancy J. S., Misasi J., Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science 373, eabh1766 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dejnirattisai W., Zhou D., Ginn H. M., Duyvesteyn H. M. E., Supasa P., Case J. B., Zhao Y., Walter T. S., Mentzer A. J., Liu C., Wang B., Paesen G. C., Slon-Campos J., López-Camacho C., Kafai N. M., Bailey A. L., Chen R. E., Ying B., Thompson C., Bolton J., Fyfe A., Gupta S., Tan T. K., Gilbert-Jaramillo J., James W., Knight M., Carroll M. W., Skelly D., Dold C., Peng Y., Levin R., Dong T., Pollard A. J., Knight J. C., Klenerman P., Temperton N., Hall D. R., Williams M. A., Paterson N. G., Bertram F. K. R., Siebert C. A., Clare D. K., Howe A., Radecke J., Song Y., Townsend A. R., Huang K.-Y. A., Fry E. E., Mongkolsapaya J., Diamond M. S., Ren J., Stuart D. I., Screaton G. R., The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 184, 2183–2200.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Tenchov R., Smoot J., Liu C., Watkins S., Zhou Q., A comprehensive review of the global efforts on COVID-19 vaccine development. ACS Cent. Sci. 7, 512–533 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C.-Y., Cheng C.-W., Kung C.-C., Liao K.-S., Jan J.-T., Ma C., Wong C.-H., Glycosite-deleted mRNA of SARS-CoV-2 spike protein as a broad-spectrum vaccine. Proc. Natl. Acad. Sci. 119, e2119995119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.VanBlargan L. A., Errico J. M., Halfmann P. J., Zost S. J., Crowe J. E., Purcell L. A., Kawaoka Y., Corti D., Fremont D. H., Diamond M. S., An infectious SARS-CoV-2 B.1.1.529 omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 28, 490–495 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stravalaci M., Pagani I., Paraboschi E. M., Pedotti M., Doni A., Scavello F., Mapelli S. N., Sironi M., Perucchini C., Varani L., Matkovic M., Cavalli A., Cesana D., Gallina P., Pedemonte N., Capurro V., Clementi N., Mancini N., Invernizzi P., Bayarri-Olmos R., Garred P., Rappuoli R., Duga S., Bottazzi B., Uguccioni M., Asselta R., Vicenzi E., Mantovani A., Garlanda C., Recognition and inhibition of SARS-CoV-2 by humoral innate immunity pattern recognition molecules. Nat. Immunol. 23, 275–286 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Moss P., The T cell immune response against SARS-CoV-2. Nat. Immunol. 23, 186–193 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Wagh K., Hahn B. H., Korber B., Hitting the sweet spot: Exploiting HIV-1 glycan shield for induction of broadly neutralizing antibodies. Curr. Opin. HIV AIDS 15, 267–274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadarangani M., Marchant A., Kollmann T. R., Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat. Rev. Immunol. 21, 475–484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez D. R., Schäfer A., Leist S. R., De la Cruz G., West A., Atochina-Vasserman E. N., Lindesmith L. C., Pardi N., Parks R., Barr M., Li D., Yount B., Saunders K. O., Weissman D., Haynes B. F., Montgomery S. A., Baric R. S., Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science 373, 991–998 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen A. A., Gnanapragasam P. N. P., Lee Y. E., Hoffman P. R., Ou S., Kakutani L. M., Keeffe J. R., Wu H. J., Howarth M., West A. P., Barnes C. O., Nussenzweig M. C., Bjorkman P. J., Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 371, 735–741 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders K. O., Lee E., Parks R., Martinez D. R., Li D., Chen H., Edwards R. J., Gobeil S., Barr M., Mansouri K., Alam S. M., Sutherland L. L., Cai F., Sanzone A. M., Berry M., Manne K., Bock K. W., Minai M., Nagata B. M., Kapingidza A. B., Azoitei M., Tse L. V., Scobey T. D., Spreng R. L., Rountree R. W., DeMarco C. T., Denny T. N., Woods C. W., Petzold E. W., Tang J., Oguin T. H. 3rd, Sempowski G. D., Gagne M., Douek D. C., Tomai M. A., Fox C. B., Seder R., Wiehe K., Weissman D., Pardi N., Golding H., Khurana S., Acharya P., Andersen H., Lewis M. G., Moore I. N., Montefiori D. C., Baric R. S., Haynes B. F., Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature 594, 553–559 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powell A. E., Zhang K., Sanyal M., Tang S., Weidenbacher P. A., Li S., Pham T. D., Pak J. E., Chiu W., Kim P. S., A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Cent. Sci. 7, 183–199 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papanikolopoulou K., van Raaij M. J., Mitraki A., Creation of hybrid nanorods from sequences of natural trimeric fibrous proteins using the fibritin trimerization motif. Methods Mol. Biol. 474, 15–33 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S.-J., Lee J., Qi Y., Kern N. R., Lee H. S., Jo S., Joung I., Joo K., Lee J., Im W., CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates. Glycobiology 29, 320–331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eastman P., Swails J., Chodera J. D., McGibbon R. T., Zhao Y., Beauchamp K. A., Wang L.-P., Simmonett A. C., Harrigan M. P., Stern C. D., Wiewiora R. P., Brooks B. R., Pande V. S., OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 13, e1005659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woo H., Park S.-J., Choi Y. K., Park T., Tanveer M., Cao Y., Kern N. R., Lee J., Yeom M. S., Croll T. I., Seok C., Im W., Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B 124, 7128–7137 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W., Sander C., Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- 41.Touw W. G., Baakman C., Black J., te Beek T. A. H., Krieger E., Joosten R. P., Vriend G., A series of PDB-related databanks for everyday needs. Nucleic Acids Res. 43, D364–D368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klose D. P., Wallace B. A., Janes R. W., 2Struc: The secondary structure server. Bioinformatics 26, 2624–2625 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitternacht S., FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Res. 5, 189–189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grant O. C., Montgomery D., Ito K., Woods R. J., Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 10, 14991–14991 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kajander T., Kahn P. C., Passila S. H., Cohen D. C., Lehtiö L., Adolfsen W., Warwicker J., Schell U., Goldman A., Buried charged surface in proteins. Structure 8, 1203–1214 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiller T., Busse C. E., Wardemann H., Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods 350, 183–193 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G., iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using x-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1
MDAR reproducibility checklist