Abstract
Interaction of bacteria with mucosal surfaces can modulate the production of proinflammatory cytokines and adhesion molecules produced by epithelial cells. Previously, we showed that expression of interleukin-8 (IL-8) and intercellular adhesion molecule 1 (ICAM-1) by gingival epithelial cells increases following interaction with several putative periodontal pathogens. In contrast, expression of IL-8 and ICAM-1 is reduced after Porphyromonas gingivalis ATCC 33277 challenge. In the present study, we investigated the mechanisms that govern the regulation of these two molecules in bacterially infected gingival epithelial cells. Experimental approaches included bacterial stimulation of gingival epithelial cells by either a brief challenge (1.5 to 2 h) or a continuous coculture throughout the incubation period. The kinetics of IL-8 and ICAM-1 expression following brief challenge were such that (i) secretion of IL-8 by gingival epithelial cells reached its peak 2 h following Fusobacterium nucleatum infection whereas it rapidly decreased within 2 h after P. gingivalis infection and remained decreased up to 30 h and (ii) IL-8 and ICAM-1 mRNA levels were up-regulated rapidly 2 to 4 h postinfection and then decreased to basal levels 8 to 20 h after infection with either Actinobacillus actinomycetemcomitans, F. nucleatum, or P. gingivalis. Attenuation of IL-8 secretion was facilitated by adherent P. gingivalis strains. The IL-8 secreted from epithelial cells after F. nucleatum stimulation could be down-regulated by subsequent infection with P. gingivalis or its culture supernatant. Although these results suggested that IL-8 attenuation at the protein level might be associated with P. gingivalis proteases, the Arg- and Lys-gingipain proteases did not appear to be solely responsible for IL-8 attenuation. In addition, while P. gingivalis up-regulated IL-8 mRNA expression, this effect was overridden when the bacteria were continuously cocultured with the epithelial cells. The IL-8 mRNA levels in epithelial cells following sequential challenge with P. gingivalis and F. nucleatum and vice versa were approximately identical and were lower than those following F. nucleatum challenge alone and higher than control levels or those following P. gingivalis challenge alone. Thus, together with the protease effect, P. gingivalis possesses a powerful strategy to ensure the down-regulation of IL-8 and ICAM-1.
Increasing attention has been drawn to the role of gingival epithelial cells in the innate immune response of local gingival tissues. The epithelial cells express chemokines that attract and activate leukocytes and express adhesion molecules that mediate leukocyte migration. The expression of these molecules that initiate and maintain inflammatory reactions can be regulated by the interaction of bacterial pathogens with epithelial cells. Interleukin-8 (IL-8), a neutrophil chemoattractant and activator (1), is induced in gingival epithelial cells by several periodontal microbes, such as Fusobacterium nucleatum, Actinobacillus actinomycetemcomitans, and Eikenella corrodens (4, 14, 17). Increased IL-8 production is thought to play a role in the transmigration of neutrophils from the submucosa to the sulcular space (38), even though constitutive IL-8 expression in noninflamed gingival epithelium has been reported (9, 18). Intercellular adhesion molecule 1 (ICAM-1) is the ligand for lymphocyte function-associated antigen 1 (LFA-1) or Mac-1 expressed on leukocytes (5, 23). In human gingival epithelium, ICAM-1 expression is restricted to the junctional and sulcular epithelium (3, 13, 18), forming a gradient with the highest ICAM-1 level on epithelial cells facing the tooth surface. This gradient is thought to play a role in directing the migration of leukocytes toward the sulcular space (18, 37, 38).
Our previous report showed that IL-8 and ICAM-1 are up-regulated in gingival epithelial cells following challenge with A. actinomycetemcomitans (17). However, both IL-8 and ICAM-1 are down-regulated by Porphyromonas gingivalis (4, 17, 22). The actual role and outcome of these regulatory processes in the pathogenesis of periodontal diseases in vivo are unknown. However, it is thought that up-regulation of IL-8 and ICAM-1 in gingival epithelial cells by microorganisms such as A. actinomycetemcomitans and F. nucleatum may stimulate the host immune response by recruiting leukocytes to the site of infection. In contrast, P. gingivalis, which attenuates the expression of IL-8 and ICAM-1, may delay the host defense mechanisms and evade the immune system, thus creating more damage to the surrounding tissue (4, 17). The exact mechanism of the attenuated expression of IL-8 and ICAM-1 is not clear, although P. gingivalis invasiveness and proteases have been reported to play a role (4, 25, 43).
The present study was undertaken to investigate the regulation of IL-8 and ICAM-1 at the molecular level in gingival epithelial cells in response to challenge with several periodontal bacteria. Our results revealed the kinetics of the regulation of these two molecules at the protein and mRNA levels. The regulation of IL-8 and ICAM-1 mRNA in epithelial cells challenged with P. gingivalis appears to be governed by antagonistic mechanisms.
MATERIALS AND METHODS
Cell cultures.
HOK-18A and HOK-16B-BaP-T1 cells, obtained from N.-H. Park (University of California Los Angeles, Los Angeles, Calif.) are immortalized oral keratinocyte cell lines derived from primary normal human oral keratinocyte cells (27, 33). The cell culture procedures were performed as described in previous reports with some modifications (17). Briefly, HOK-18A cells were grown in Dulbecco's modified Eagle's medium-F12 (3:1, vol/vol) (GIBCO/BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum, 0.5 ng of human epidermal growth factor per ml, 5 μg of bovine insulin per ml, 0.4 μg of hydrocortisone per ml, 0.1 nM choleratoxin, 0.5 μg of transferrin per ml, 2 nM 3,3′,5-triiodo-l-thyronine, 25 μg of gentamicin per ml, and 250 ng of amphotericin B per ml. HOK-16B-BaP-T1 cells were grown in Dulbecco's modified Eagle's medium containing 4.5 g of d-glucose per liter and supplemented with 10% fetal bovine serum and 0.4 μg of hydrocortisone per ml. Since HOK-16B-BaP-T1 cells normally express low levels of IL-8 and ICAM-1, they were used for bacteria that showed an up-regulating effect on these molecules, i.e., A. actinomycetemcomitans and F. nucleatum. In contrast, HOK-18A cells, which express higher levels of IL-8 and ICAM-1, were mainly used to study the kinetics of IL-8 secretion following P. gingivalis infection, which down-regulated IL-8 and ICAM-1.
Bacterial strains and cytokine.
The following laboratory strains were utilized: A. actinomycetemcomitans Y4 from K. Miyasaki (University of California Los Angeles); F. nucleatum 12230, a clinical isolate from the upper trachea, obtained by S. Finegold (Los Angeles, Calif.); P. gingivalis ATCC 33277 (American Type Culture Collection, Rockville, Md.); P. gingivalis strain W50 from J. Sandros (Gøteborg University, Gøteborg, Sweden); and the P. gingivalis protease mutant strain V2296 (kgp, Lys-gingipain) and its corresponding wild-type strain, W83, from H. M. Fletcher (Loma Linda University, Loma Linda, Calif.) (10). The protease (Arg-gingipain)-defective mutants, MT10 (rgpA) and G-102 (rgpB), were derived from wild-type P. gingivalis strain 381 (35, 36). Escherichia coli HB101 was obtained from Y. Han (Case Western Reserve University, Cleveland, Ohio).
Infection of oral epithelial cells.
Oral epithelial cells were seeded into 24- or 6-well Costar tissue culture plates at a density of 105 cells/ml in volumes of 0.5 ml (24-well) or 2 ml (6-well) per well. Cultures were grown to confluence before being infected with bacteria. The preparation of A. actinomycetemcomitans, P. gingivalis, and F. nucleatum was described in our previous reports (14, 17). Briefly, bacteria were inoculated in 5 ml of appropriate broth medium and grown at 37°C under 80% N2–10% H2–10% CO2 in an anaerobic chamber (Coy Laboratory Production, Ann Arbor, Mich.) overnight. Typically, 1 ml of the bacterial culture was then transferred to 9 ml of fresh broth medium and allowed to grow to an optimal optical density so that the bacteria were in the exponential phase of growth. E. coli HB101 was grown in Luria-Bertani medium at 37°C to the logarithmic growth phase. The bacteria were washed three times with phosphate-buffered saline (pH 7.2) and resuspended at concentrations equivalent to various multiplicities of infection (MOI) in the antibiotic-free medium used to grow the specific cell lines. Bacteria were added in 200-μl volumes to the cell monolayers in 24-well culture plates or in 1-ml volumes to the cell monolayers in 6-well culture plates and were centrifuged onto the monolayers at 900 × g for 5 min at room temperature. The bacteria were cocultured with epithelial cells at 37°C for 1.5 to 2 h to allow interaction between the bacteria and epithelial cells. After incubation, the monolayers were washed three times to remove extracellular bacteria and the cultures were further incubated for 2 to 30 h in fresh medium containing antibiotics (described below) to kill the remaining extracellular bacteria. Alternatively, bacteria were cocultured with the epithelial-cell monolayers for the entire incubation period; i.e., bacteria were continuously cocultured with the epithelial cells throughout the incubation without removal of the bacteria or change of medium. The antibiotics used in the media were as follows: for infection with A. actinomycetemcomitans and F. nucleatum, 0.1 mg of gentamicin per ml; for infection with P. gingivalis, 0.1 mg of metronidazole per ml and 0.5 mg of gentamicin per ml. After incubation, the supernatants were collected for IL-8 detection by enzyme-linked immunosorbent assay (ELISA). Epithelial-cell viability was determined by trypan blue exclusion after the supernatant was collected. Supernatant from epithelial cells in 24-well plates were used for ELISA to measure the amount of secreted IL-8. Epithelial cells in six-well plates were harvested for Northern blot analyses. The epithelial-cell viability was >90% for all the kinetics studies described above when optimal epithelial-cell lines and bacterial doses were used.
Invasion inhibition studies.
The bacterial invasion inhibitor sodium azide (Sigma, St. Louis, Mo.) (50 mM in PBS) was used to block P. gingivalis invasion of gingival epithelial cells (21). Preliminary experiments indicated that the inhibitor had no effect on bacterial viability at the concentrations and under the conditions utilized. The inhibitor was preincubated with P. gingivalis for 4 h and then removed by washing prior to coculture of the epithelial cells with P. gingivalis. The extent of inhibition of invasion was determined by parallel invasion assays using the standard antibiotic protection method (6, 31).
Northern blot analysis.
Cellular RNA was isolated using RNA STAT-60 (Tel-Test “B,” Inc., Friendswood, Tex.). Total RNA (8 to 20 μg) was size fractionated on 1.5% formaldehyde–agarose gels, transferred to nitrocellulose filters, and probed with a 32P-labeled cDNA fragment specific for human ICAM-1 or IL-8. The ICAM-1 probe targeted a 1,400-bp fragment in the coding region. This fragment was released by XhoI digestion from plasmid pICAM-1 (kindly provided by B. Seed, Boston, Mass.), which carries the full-length human ICAM-1 cDNA. The IL-8 probe targeted a 420-bp fragment released by EcoRI digestion from plasmid pBhIL8 (kindly provided by M. Kagnoff, La Jolla, Calif.), which carries the full-length human IL-8 cDNA. A 32P-labeled cDNA fragment of human glyceraldehyde-3-phosphate dehydrogenase was used as the control probe to verify that an equal amount of RNA from each cell line was used in each analysis. The signals were visualized by autoradiography using a PhosphorImager system (Molecular Dynamics, Sunnyvale, Calif.). The images of specific bands were quantitated using an ImageQuant software program (Molecular Dynamics).
ELISA for IL-8.
The procedures for ELISA for IL-8 were described in previous studies (17, 18). Standard ELISA was performed using polyclonal goat anti-human IL-8 antibodies (R&D Systems, Minneapolis, Minn.) as capturing antibodies, polyclonal rabbit anti-human IL-8 antibodies (Endogen Inc., Cambridge, Mass.) as detecting antibodies, and horseradish peroxidase-labeled polyclonal goat anti-rabbit immunoglobulin G (Biosource International, Camarillo, Calif.) as a second-step antibody.
RESULTS
Kinetics of IL-8 secretion by gingival epithelial cells following F. nucleatum or P. gingivalis infection.
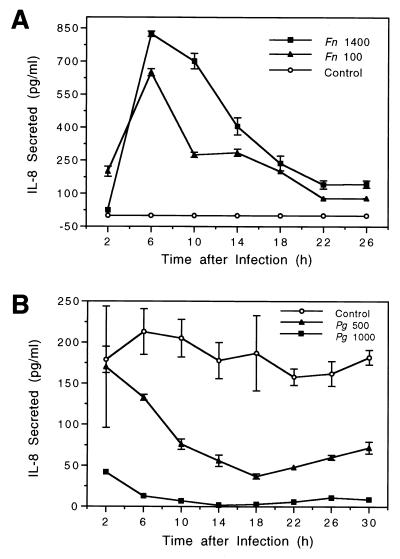
Epithelial cells were cocultured with F. nucleatum or P. gingivalis for 2 h. During the subsequent incubation, extracellular bacteria were removed and fresh medium with antibiotics was added to kill the remaining extracellular bacteria so that any effect of bacterial infection on IL-8 induction or reduction was exerted during the coculturing period due to bacterium–epithelial-cell interactions and/or during the subsequent incubation period due to the activity of invaded or attached bacteria. The production of secreted IL-8 during sequential 4-h intervals after infection was measured by ELISA (Fig. 1). The results showed that IL-8 secretion by gingival epithelial cells increased from 2 to 14 h following F. nucleatum infection and decreased thereafter up to 26 h. In contrast, IL-8 secretion into the supernatant rapidly decreased 2 h after P. gingivalis infection and remained suppressed during the remaining incubation period. As shown in Fig. 1, these effects appeared to be dose dependent, since the higher the bacterial load, the stronger the IL-8-regulatory effect.
FIG. 1.
Kinetics of IL-8 secretion by gingival epithelial cells following bacterial infection. (A) Confluent HOK-16B-BaP-T1 cell monolayers were infected with F. nucleatum 12230 (Fn) at a MOI of 1,400:1 or 100:1. (B) Confluent HOK-18A cell monolayers were infected with P. gingivalis 22377 (Pg) at a MOI of 1:1,000 or 1:500. At 2 h after infection, the cultures were washed and further incubated in the presence of gentamicin (0.5 mg/ml) for F. nucleatum or metronidazole (0.1 mg/ml) plus gentamicin (0.5 mg/ml) for P. gingivalis for up to 26 to 30 h. The MOIs were confirmed retroactively in parallel experiments. IL-8 secretion was determined for each time point by removing the medium and subsequently incubating the cells in freshly added medium containing the same concentration of antibiotics. Data points represent mean and standard error of the mean of the results of triplicate assay determinations from one representative experiment.
Kinetics of IL-8 and ICAM-1 mRNA levels after infection with A. actinomycetemcomitans, F. nucleatum, or P. gingivalis.
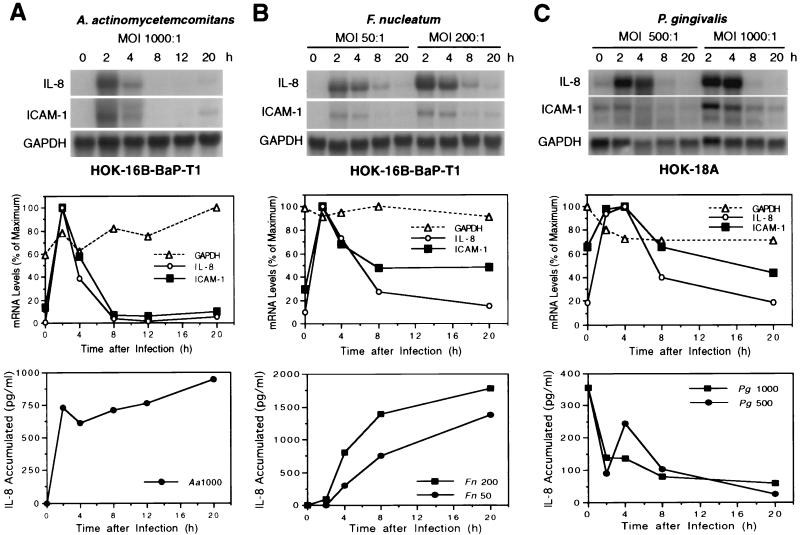
After 2 h of bacterium–epithelial-cell coculture, extracellular bacteria were removed by washing the epithelial-cell cultures and fresh medium containing antibiotics was added for further incubation. At different time points, supernatants were collected for the measurement of accumulated IL-8 and RNA was isolated from the epithelial-cell cultures for analysis. The results presented in Fig. 2 demonstrate that all three bacteria induced IL-8 (4.0- to 91.0-fold) and ICAM-1 (1.2- to 7.2-fold) mRNA production between 2 and 4 h and that this was followed by reduced levels (Fig. 2, top and middle panels). The level of mRNA was proportional to the number of bacteria used in the coculture (Fig. 2B and C). F. nucleatum appeared to be more potent in inducing IL-8 and ICAM-1, as indicated by the same level of induction at a lower MOI compared to the other two microorganisms.
FIG. 2.
Kinetics of IL-8 and ICAM-1 mRNA levels in gingival epithelial cells following bacterial infection. (A and B) Confluent HOK-16B-BaP-T1 cell monolayers were infected with A. actinomycetemcomitans Y4 at a MOI of 1,000:1 (A) or F. nucleatum 12230 at MOIs of 50:1 and 200:1 (B). (C) Confluent HOK-18A cell monolayers were infected with P. gingivalis 22377 at MOIs of 500:1 and 1,000:1. At 2 h after infection, the cultures were washed and further incubated for up to 20 h in the presence of gentamicin (0.5 mg/ml) (A. actinomycetemcomitans and F. nucleatum) or metronidazole (0.1 mg/ml) plus gentamicin (0.5 mg/ml) (P. gingivalis). At various intervals after infection, epithelial cells were harvested and RNA was isolated. A 20-μg sample of total RNA was subjected to Northern blot analysis. The MOIs were confirmed retroactively. The results are shown in the top panels. The levels of mRNA from the analysis are plotted and shown in the middle panels. Data presented in the middle panel for panel B were from a MOI of 50:1, and those in the middle panel for panel C were from a MOI of 1000:1. Supernatants were also collected at each time point for accumulated IL-8 measurement using ELISA (data shown in the bottom panel).
The accumulated IL-8 in the supernatant continued to increase following A. actinomycetemcomitans and F. nucleatum challenge, whereas it decreased in response to P. gingivalis challenge (Fig. 2, bottom panel). The accumulated cell surface ICAM-1 expression could not be tested in this experiment due to the harvesting of cellular RNA. Separate experiments were performed to measure the cell surface ICAM-1 expression following F. nucleatum challenge. The results (data not shown) were similar to those of previous studies in which epithelial cells challenged with A. actinomycetemcomitans showed increased cell surface ICAM-1 expression (17).
The decrease in secreted IL-8 production is facilitated by P. gingivalis attachment.
To determine whether P. gingivalis attachment and invasion plays a role in attenuating IL-8 secretion, we examined the effects of the poorly adherent and poorly invasive P. gingivalis strains W50 and W83 with those of invasive strains 381 and 33277. The data indicate that P. gingivalis W50 and W83 did not attenuate IL-8 production (Table 1), suggesting that attachment and invasion is important for mediating the decrease in secreted IL-8 levels under these experimental conditions. Two protease knockout mutant strains were also used to examine the effect of proteases on IL-8 attenuation. The mutant strains derived from P. gingivalis 381 did not affect IL-8 regulation compared with their wild-type strains. Pretreatment with sodium azide, which inhibited P. gingivalis invasion but not attachment, also did not affect IL-8 attenuation. These results suggest a more important role of attachment than invasion in IL-8 response.
TABLE 1.
IL-8 secretion from gingival epithelial cells following P. gingivalis challengea
| Bacterium added | Amt of secreted IL-8 (pg/ml) | % Attachmentb | % Invasionc |
|---|---|---|---|
| None | 950 ± 97 | ||
| P. gingivalis 381 | 128 ± 31 | 3.1563 ± 2.6456 | 1.4231 ± 0.9102 |
| Mutant MT-10 | 151 ± 34 | 2.3943 ± 0.5823 | 1.9674 ± 0.8827 |
| Mutant G-102 | 332 ± 79 | 6.0650 ± 3.2536 | 4.6520 ± 3.6773 |
| P. gingivalis W50 | 944 ± 82 | 0.0038 ± 0.0016 | 0.0083 ± 0.0074 |
| P. gingivalis W83 | 927 ± 63 | 0.0042 ± 0.0025 | 0.0005 ± 0.0003 |
| Mutant V2296 | 1,041 ± 69 | 0.0292 ± 0.0241 | 0.009 ± 0.0085 |
| None | 457 ± 44 | ||
| P. gingivalis ATCC 33277 | |||
| Not treated | 74 ± 74 | 0.807 ± 0.577 | 0.617 ± 0.537 |
| Azide treated | 82 ± 26 | 0.826 ± 0.536 | 0.134 ± 0.051 |
Confluent HOK-18A cell monolayers were infected with P. gingivalis 381, W50, or W83 and their mutants for 2 h at MOIs of 1,200 or with P. gingivalis 33277 and sodium azide treated for 1.5 h at MOIs of 1,500. The cultures were then washed and further incubated for 12 h in the presence of metronidazole (0.1 mg/ml) and gentamicin (0.5 mg/ml). IL-8 secretion was determined by ELISA. The MOIs were confirmed retroactively in parallel experiments by plating bacteria onto appropriate blood agar plates and counting bacteria colonies after incubation. Values represent mean ± standard error of the mean of the results of at least two independent experiments in triplicate (IL-8 measurement) or duplicate (attachment and invasion analyses) assays.
Defined as the percentage of bacteria added that bound to HOK-18A cells. Cultures were subjected to standard attachment assays following the 1.5- to 2-h coculture incubation.
Defined as 100 × number of bacterial CFU recovered following antibiotic treatment/number of bacteria added.
In the studies described above, the bacterium–epithelial-cell coculture time was only 1.5 to 2 h. Therefore, the IL-8 attenuation effect was exerted either during the coculture period or by the attached bacteria after removal of nonattached bacteria or both. To examine these possibilities, we analyzed IL-8 attenuation using two approaches. In one approach, the bacteria were incubated with the epithelial cells for 2 h, and this was followed by washing and the addition of fresh medium containing antibiotics. The supernatants were collected for IL-8 analysis at both the end of the 2-h coculture time and the end the incubation (at 6 h) after the addition of fresh medium. In the other approach, the bacteria were continuously cocultured with the epithelial cells throughout the incubation period (0 to 18 h). The results presented in Table 2 show that when IL-8 production was measured at the end of the 2-h coculture period, all three P. gingivalis strains had attenuated it. After the cultures were washed and fresh medium was added, the level of IL-8 accumulated in the supernatant during the 2- to 6-h incubation period was decreased only for P. gingivalis 381. When these P. gingivalis strains were cocultured with the epithelial cells throughout the entire incubation period (18 h), all the strains attenuated IL-8. The results indicate that the continuous presence of bacteria along with epithelial cells, either by attachment or by their presence in the cocultures, played a key role in this IL-8 attenuation.
TABLE 2.
IL-8 secretion from HOK-18A cells following P. gingivalis challenge
| Bacterium added | Amt of secreted IL-8 (pg/ml)a
|
||
|---|---|---|---|
| 0–2 hb (continuous) (MOI, 1,000:1) | 2–6 hc (washed) (MOI, 1,000:1) | 0–18 hd (continuous) (MOI, 500:1) | |
| None | 292 ± 37 | 1,111 ± 18 | 1,212 ± 111 |
| P. gingivalis 381 | 134 ± 2 | 163 ± 7 | 54 ± 4 |
| P. gingivalis W50 | 24 ± 6 | 1,241 ± 121 | 26 ± 12 |
| P. gingivalis W83 | 54 ± 15 | 1,343 ± 86 | 13 ± 2 |
Values represent mean ± standard error of the mean of the results of duplicate or triplicate assay determinations from one representative experiment. At the end of infection and/or incubation, supernatants were collected for IL-8 determination by ELISA.
Confluent epithelial-cell monolayers infected with P. gingivalis for 2 h.
Epithelial cells infected with P. gingivalis for 2 h followed by washing to remove extracellular bacteria and further incubation to 6 h in the presence of metronidazole (0.1 mg/ml) and gentamicin (0.5 mg/ml).
Epithelial cells infected with P. gingivalis for 18 h continuously without removal of bacteria.
IL-8 secreted from epithelial cells can be attenuated by P. gingivalis supernatant.
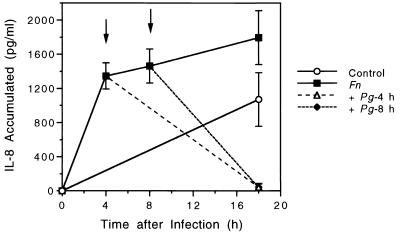
The above data revealed that although P. gingivalis up-regulated the mRNA levels of IL-8 and ICAM-1, both molecules were down-regulated at the protein level. This attenuation was related to the physical association of bacteria with epithelial cells. To determine if soluble proteases released from P. gingivalis into the culture supernatant are capable of degrading IL-8 from HOK-18A cells, we incubated P. gingivalis culture supernatants with IL-8 secreted from epithelial cells. The results showed that supernatants from all P. gingivalis strains used in this study, i.e., both wild-type strains and their protease mutants, were capable of degrading IL-8 secreted from HOK-18A cells (Table 3). In contrast, supernatant from E. coli HB101 or F. nucleatum did not appear to degrade IL-8. The supernatants of mutant strains, MT10 (rgpA) and G-102 (rgpB), appeared less potent in degrading IL-8 than did that of mutant strain V2296 (kgp), suggesting that Arg-gingipain may play a more important role than Lys-gingipain in this degradation. We also incubated IL-8 with P. gingivalis cells directly and found that bacterial cells were much more potent than their supernatant in degrading IL-8 (data not shown). We reasoned that P. gingivalis protease activities should be potent enough to reduce the IL-8 levels secreted in response to a prior infection with F. nucleatum. Therefore, we challenged epithelial cells with P. gingivalis at either 4 or 8 h after infection with F. nucleatum. The results presented in Fig. 3 show that P. gingivalis cells were capable of degrading secreted IL-8 which had been induced by F. nucleatum and had accumulated in the culture supernatant.
TABLE 3.
Degradation of secreted IL-8 by P. gingivalis supernatanta
| Bacterium added | % of IL-8 remaining |
|---|---|
| None | 100 |
| P. gingivalis 381 | 66 ± 3bc |
| Mutant MT-10 | 82 ± 10 |
| Mutant G-102 | 94 ± 11 |
| P. gingivalis W83 | 58 ± 14d |
| Mutant V2296 | 60 ± 11 |
| E. coli HB101 | 106 ± 4 |
| F. nucleatum ATCC 12230 | 99 ± 11 |
Secreted IL-8 from confluent HOK-18A culture medium was collected, and bacterial supernatant was collected from cultures in logarithmic growth. Equal volumes (100 μl) of the HOK-18A culture medium and P. gingivalis, F. nucleatum, or E. coli supernatant were combined in each well of a 24-well plate and incubated for 18 h at 37°C. The remaining IL-8 was determined by ELISA. Values represent mean ± standard error of the mean of the results of at least three independent experiments in triplicate assays, except for F. nucleatum ATCC 12230, for which the values represent results of two independent experiments in a triplicate assay).
P = 0.183 (when result is compared with that for MT-10).
P = 0.043 (significant difference [P < 0.05] by the t test) (when result is compared with that for G-102).
P = 0.939 (when result is compared with that for V2296).
FIG. 3.
IL-8 secretion from HOK-18A following sequential challenge of F. nucleatum and P. gingivalis. (○) Control group without bacteria. Supernatants were collected at 18 h for IL-8 ELISA. (■) F. nucleatum (MOI, 300:1) was added to cell monolayers and supernatants were collected for IL-8 measurement at the 4-, 8-, or 18-h time point. ↓, P. gingivalis 381 (MOI, 500:1) was added to cell monolayers 4 h (▵) or 8 h (⧫) after the addition of F. nucleatum (MOI, 300:1), and the supernatant was collected at 18 h.
Effect of bacterium–epithelial-cell coculture time on IL-8 mRNA expression.
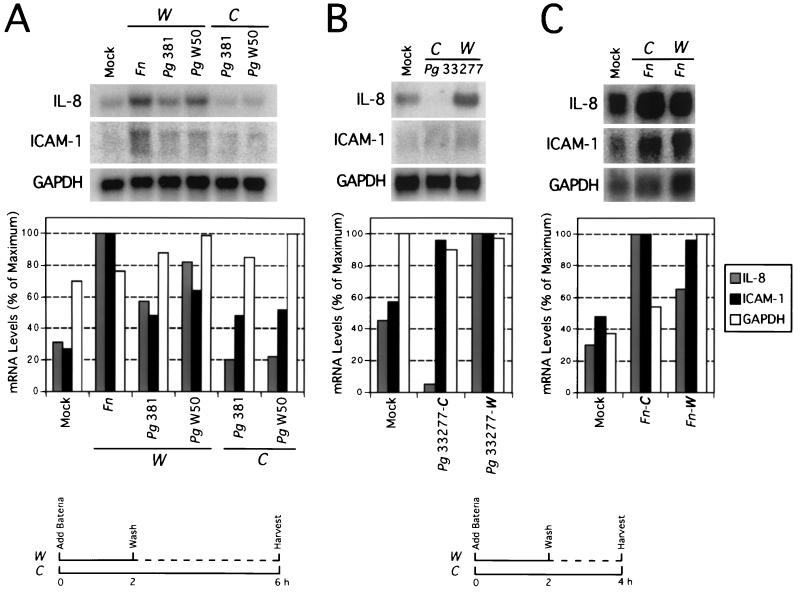
IL-8 and ICAM-1 mRNA levels peaked between 2 and 4 h after a 2-h challenge with P. gingivalis (Fig. 2). We next determined the kinetics of the mRNA levels when bacteria were continuously cultured for 4 to 6 h with the epithelial cells and compared them to the kinetics in the groups using the 2-h challenge followed by washing and addition of fresh medium for further incubation to 4 to 6 h. The results in Fig. 4A and B showed that IL-8 mRNA was detected at lower levels when the continuous challenge with P. gingivalis was used than when the 2-h challenge was used. IL-8 mRNA levels after continuous challenge were even lower than the levels after no infection. Thus, P. gingivalis 381, ATCC 22377, and W50 up-regulated IL-8 and ICAM-1 mRNA when the epithelial-cell cultures were washed at the 2-h time point and down-regulated IL-8 below the baseline level when the bacteria were continuously cocultured with the epithelial cells. However, whereas 381 and ATCC 33277 are adherent and invasive, W50 is not. This suggests that the attachment and invasion phenotype is not required for the IL-8 mRNA regulation. The above phenomenon did not occur when F. nucleatum was used to challenge the epithelial cells (Fig. 4C). In fact, F. nucleatum induced more IL-8 mRNA when continuously cocultured with the epithelial cells.
FIG. 4.
IL-8 and ICAM-1 mRNA levels in gingival epithelial cells following bacterial challenge. Confluent HOK-18A cell monolayers were infected with F. nucleatum or various P. gingivalis strains at a MOI of 500:1. The bacterium–epithelial-cell coculture period was either 2, 4, or 6 h. RNA was isolated at 4 or 6 h after infection, and 8 to 12 μg of total RNA was subjected to Northern blot analysis. For the 2-h coculture group, the culture was washed and fresh medium containing appropriate antibiotics was added at 2 h and the cultures were further incubated for another 2 to 4 h before harvesting. For the 4- or 6-h coculture group, the bacteria were continuously cocultured with the epithelial cells to 4 or 6 h (see diagram in the bottom panel). (A) Epithelial cells were challenged with F. nucleatum 12230, P. gingivalis 381, or P. gingivalis W50 for 2 h (washed) or 6 h (continuous), and RNA was isolated at 6 h. (B and C) Epithelial cells were challenged with P. gingivalis 33277 (B) or F. nucleatum 12230 (C) for 2 h (washed) or 4 h (continuous) and RNA was isolated at 4 h. The levels of mRNA from the Northern blot analysis (shown in the top panel) are plotted and shown in the middle panel. Mock, no bacteria were added to the cultures. Fn, F. nucleatum; Pg, P. gingivalis; W, 2-h coculture group that was washed at the 2-h time point; C, 4- or 6-h coculture group, bacteria continuously cocultured with the epithelial cells.
Regulation of IL-8 mRNA levels following sequential challenge of epithelial cells with P. gingivalis and F. nucleatum.
To determine how IL-8 mRNA regulation is affected by sequential challenge of epithelial cells with P. gingivalis and F. nucleatum, we carried out kinetic studies examining IL-8 mRNA levels at 4 h after stimulation. The results showed that when the cells were challenged first with F. nucleatum and then with P. gingivalis 33277 and when they were challenged first P. gingivalis 33277 and then with F. nucleatum, the IL-8 mRNA levels were similar (Fig. 5). In both cases, the IL-8 mRNA level was still higher than the levels associated with control (mock) and P. gingivalis infection alone but lower than the level associated with F. nucleatum alone.
FIG. 5.
IL-8 mRNA levels in gingival epithelial cells following sequential bacterial challenge. Confluent HOK-18A cell monolayers were stimulated with F. nucleatum 12230 (Fn), or P. gingivalis 33277 (Pg) alone or sequentially with P. gingivalis and F. nucleatum at MOIs of 500:1. The bacterium–epithelial-cell coculture period was 4 h. RNA was isolated at 4 h after infection, and 10 μg of total RNA was subjected to Northern blot analysis. For stimulation with one type of bacterium alone, the stimulus and the bacteria were in the culture throughout the 4-h period. For sequential stimulation, the first stimulus or bacterium was added at 0 h and remained in culture for 4 h. The second bacterium was added at 2 h. The levels of mRNA from the Northern blot results (shown in the top panel) are plotted and shown in the bottom panel. Mock, no bacteria were added to the cultures.
DISCUSSION
Our previous studies demonstrated that secreted IL-8 and cell surface ICAM-1 protein expression are increased in gingival epithelial cells challenged with A. actinomycetemcomitans or F. nucleatum whereas both proteins are down-regulated when the cells are challenged with P. gingivalis (14, 17). Similar findings have also been reported by other investigators (4, 22). The present studies further investigated the mechanisms underlying the regulation of IL-8 and ICAM-1 expression. Both A. actinomycetemcomitans and F. nucleatum up-regulated IL-8 at the protein and mRNA levels. Interestingly, whereas P. gingivalis down-regulated IL-8 at the protein level, it could both up- and down-regulate mRNA levels. The up-regulation versus down-regulation was dependent on the duration of the P. gingivalis interaction with epithelial cells. Time course studies demonstrated the kinetic profile of IL-8 and ICAM-1 mRNA expression in gingival epithelial cells in response to a 2-h bacterial challenge followed by washing and incubation with fresh medium. All three bacteria, A. actinomycetemcomitans, F. nucleatum, and P. gingivalis, up-regulated IL-8 and ICAM-1 mRNA with similar kinetics. Although P. gingivalis stimulation of epithelial cells down-regulated IL-8 and ICAM-1 at the protein level, the mRNA induction patterns of these two molecules were almost identical to what was seen with stimulation by A. actinomycetemcomitans and F. nucleatum. This suggests that all three bacteria induce an epithelial-cell response that potentially utilizes identical pathways to activate IL-8 and ICAM-1 transcription. The kinetics of IL-8 secretion in response to F. nucleatum appeared identical to the response to A. actinomycetemcomitans (17). P. gingivalis, on the other hand, rapidly attenuated the production of secreted IL-8 from epithelial cells at 2 h after infection, even as the IL-8 mRNA accumulation reached its peak at 2 h. This suggests that this down-regulation is exerted at the translational and/or posttranslational level.
Based on the potent protease activities possessed by P. gingivalis (2, 4, 11, 12, 20, 25, 43), the most likely cause is the degradation of ICAM-1 and IL-8 proteins by proteases. Consistent with this hypothesis, the greater the bacterial load to the epithelial cells, the stronger this effect. Our findings suggest that P. gingivalis proteases may be responsible for the low levels of IL-8 and ICAM-1 protein expression during the period after the epithelial cells were washed. The results shown in Table 1 suggest a role for P. gingivalis attachment in that after the epithelial cells were washed, the attached P. gingivalis bacteria were able to execute IL-8 degradation. This degradation was not affected by the presence of antibiotics, as shown in our previous study, in which we demonstrated that antibiotic-killed P. gingivalis is still capable of degrading IL-8 (17). The use of nonadherent and noninvasive P. gingivalis strains, W50 and W83 (Table 1), ensured that these bacteria could be easily washed away; i.e., they were not present after washing to degrade IL-8. Furthermore, the deletion of one (rgpA, rgpB, or kgp) protease gene in P. gingivalis did not dramatically affect IL-8 attenuation, indicating that no individual protease was exclusively or dominantly responsible for the degradation. The gingipain proteases released from P. gingivalis have shown potent activity in degrading IL-8 in purified form (25) and may account for the degradation from crude supernatant, as demonstrated by our study (Table 3); however, bacterial cell-associated protease activity appears to be more important in IL-8 protein degradation.
Darveau et al. (4) showed that P. gingivalis added to epithelial-cell cultures halted ongoing IL-8 accumulation induced by F. nucleatum stimulation without the loss of previously secreted IL-8. Their data suggest that P. gingivalis interaction with epithelial cells counteracts the induction of IL-8 by F. nucleatum, but does not affect already secreted IL-8 in the supernatant. The data presented in Fig. 3, however, do not correspond to their finding. It might be that the use of different MOIs of P. gingivalis or the use of primary gingival epithelial cells in their system versus the use of cell lines in our system accounts for this discrepancy. It appears likely that P. gingivalis interacts with epithelial cells with different affinities depending on the strain. For strains that can attach well to the epithelial cells, their secreted or vesicle-associated proteases can establish a high concentration on the cell surface, thus degrading the IL-8 as it is secreted from the epithelial cells and degrading the cell surface ICAM-1. For strains of P. gingivalis that do not attach well (W50 and W83), continuous coculture with the epithelial cells may allow their released soluble protease as well as surface-associated proteases to degrade the accumulated IL-8 in the epithelial cell supernatant (Tables 2 and 3) (4, 43). These data suggest that the proteolytic activity, rather than attachment itself, is likely to be responsible for the IL-8 protein degradation.
It is of interest that P. gingivalis can either up- or down-regulate IL-8 mRNA in epithelial cells (Fig. 4). At 2 h after P. gingivalis challenge, the bacteria were removed, the culture was washed, and fresh medium was added for further incubation until the 4- to 6-h time point. At this time, the IL-8 and ICAM-1 mRNA levels increased. However, when P. gingivalis, regardless of whether the strains were adherent (381 and ATCC 33277) or nonadherent (W50), was cocultured with the cells continuously for 4 to 6 h, the ICAM-1 mRNA levels were not as elevated and the IL-8 mRNA concentrations decreased. These observations suggest that when P. gingivalis interacts with the epithelial cells, a signal is triggered that leads to the up-regulation of IL-8 and ICAM-1 mRNA. At the same time, another signal is triggered that leads to the down-regulation of IL-8 mRNA, and ICAM-1 mRNA to a lesser extent. This latter signal may occur through induced degradation of mRNA. Sequential addition of F. nucleatum and then P. gingivalis to epithelial-cell cultures resulted in the attenuation of IL-8 mRNA levels compared to challenge with F. nucleatum alone (Fig. 5). This appears to contradict the P. gingivalis regulation of IL-8 mRNA kinetics presented above (Fig. 2 and 4). It was expected that stimulation of epithelial cells with P. gingivalis followed by F. nucleatum at 2 h would result in the attenuation of IL-8 mRNA levels compared to challenge with F. nucleatum alone. However, challenge of the epithelial cells with F. nucleatum followed by P. gingivalis at 2 h was expected to induce an even higher level of IL-8 mRNA than challenge with F. nucleatum alone since this would allow P. gingivalis to up-regulate IL-8 mRNA further before the down-regulation took place. A possible explanation is that the intracellular signals in epithelial cells leading to up-regulation of IL-8 mRNA may have been exhausted by F. nucleatum in the first 2 h of infection, so that the P. gingivalis down-regulation activities appeared early.
Bacterial components play important roles in stimulating proinflammatory gene expression (15, 39, 41, 42). Released bacterial surface-associated material from several periodontal microbes, including A. actinomycetemcomitans and P. gingivalis, is capable of inducing gingival fibroblasts and human peripheral blood mononuclear cells to release cytokines (28, 29). A lipid A-associated protein of P. gingivalis is a potent stimulator of IL-6 production (32). Bacterial factors interacting with epithelial cells could initiate intracellular signaling events, leading to activation of genes. For example, Helicobacter pylori induces activation of the transcription factor AP-1 through the ERK/mitogen-activated protein kinase cascade in gastric epithelial cells (24). The bacterial immunodominant antigen CagA from H. pylori enters epithelial cells and becomes wired to the eukaryotic signal transduction pathways (34). Increased NF-κB and AP-1 activities, a result of activation of intracellular signaling events, in intestinal epithelial cells in response to infection with enteroinvasive bacteria play a central role in intestinal innate immunity (8, 16). Studies also showed that P. gingivalis induces phosphotyrosine-dependent intracellular signaling in gingival epithelial cells, which facilitates the invasion of P. gingivalis (30). Although not yet investigated, this activated signal transduction may also lead to activation of genes. P. gingivalis lipopolysaccharide interacts with gingival fibroblasts through toll-like receptor 4 and induces the activation of several intracellular proteins including NF-κB and AP-1 (40). In our experimental setting, it is possible that one of these signal transduction events activate IL-8 and ICAM-1 genes whereas another event induces the degradation of IL-8 mRNA.
The down-regulation of IL-8 may debilitate the recruitment of neutrophils which normally play a critical role in maintaining periodontal health by defending against bacterial infection (4, 18, 22, 25, 26). The data presented here indicate that P. gingivalis is able to regulate the production of IL-8 mRNA at either the transcriptional or posttranscriptional level. The up-regulation of IL-8 in epithelial cells upon interaction with many microbial pathogens is a common feature, as evidenced by many observations (4, 7, 14, 18). Bacterial lipopolysaccharide, in F. nucleatum and E. coli, could be partly responsible for this IL-8 mRNA up-regulation (19). Thus, the ability of P. gingivalis to down-regulate IL-8 mRNA makes it a unique microorganism among the potential periodontal pathogens. The threefold effects of P. gingivalis on IL-8 expression, i.e., increased IL-8 mRNA levels, decreased IL-8 mRNA levels, and decreased IL-8 protein levels by protease-mediated degradation, illustrate its multiple strategies that may ultimately incapacitate local host defenses.
ACKNOWLEDGMENTS
We acknowledge the following individuals for providing epithelial cell lines or bacteria strains required for this study: N.-H. Park, K. Miyasaki, J. Sandros, H. M. Fletcher, and Y. Han. We thank S. Hunt Gerardo for editorial assistance with the manuscript.
This study was supported in part by UCLA Academic Senate Research Grants (G.T.-J.H.), a Faculty Career Development Award (G.T.-J.H.), and UCLA School of Dentistry Research Opportunity Grants (G.T.-J.H. and S.K.H.).
REFERENCES
- 1.Baggiolini M, Walz A, Kunkel S L. Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils. J Clin Investig. 1989;84:1045–1049. doi: 10.1172/JCI114265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calkins C, Platt K, Potempa J, Travis J. Inactivation of TNF-alpha by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis: implications of immune evasion. J Biol Chem. 1998;273:6611–6614. doi: 10.1074/jbc.273.12.6611. [DOI] [PubMed] [Google Scholar]
- 3.Crawford J M. Distribution of ICAM-1, LFA-3 and HLA-DR in healthy and diseased gingival tissues. J Periodontal Res. 1992;27:291–298. doi: 10.1111/j.1600-0765.1992.tb01680.x. [DOI] [PubMed] [Google Scholar]
- 4.Darveau R P, Belton C M, Reife R A, Lamont R J. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond M S, Staunton D E, De Fougerolles A R, Stacker S A, Garcia-Aguillar, Hibbs M L, Springer T A. ICAM (CD54)—a counter receptor for Mac-1 (CD11/CD18) J Cell Biol. 1990;111:3129–3139. doi: 10.1083/jcb.111.6.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan M J, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivalis with epithelial cells. Infect Immun. 1993;61:2260–2265. doi: 10.1128/iai.61.5.2260-2265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckmann L, Kagnoff M F, Fierer J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elewaut D, DiDonato J A, Kim J M, Truong F, Eckmann L, Kagnoff M F. NF-kappa B is a central regulator of the intestinal epithelial cell innate immune response induced by infection with enteroinvasive bacteria. J Immunol. 1999;163:1457–1466. [PubMed] [Google Scholar]
- 9.Fitzgerald J E, Kreutzer D L. Localization of interleukin-8 in human gingival tissues. Oral Microbiol Immunol. 1995;10:297–303. doi: 10.1111/j.1399-302x.1995.tb00158.x. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher H M, Schenkein H A, Morgan R M, Bailey K A, Berry C R, Macrina F L. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher J, Nair S, Poole S, Henderson B, Wilson M. Cytokine degradation by biofilms of Porphyromonas gingivalis. Curr Microbiol. 1998;36:216–219. doi: 10.1007/s002849900297. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher J, Reddi K, Poole S, Nair S, Henderson B, Tabona P, Wilson M. Interactions between periodontopathogenic bacteria and cytokines. J Peridontal Res. 1997;32:200–205. doi: 10.1111/j.1600-0765.1997.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 13.Gemmell E, Walsh L J, Savage N W, Seymour G J. Adhesion molecule expression in chronic inflammatory periodontal disease tissue. J Periodontal Res. 1994;29:46–53. doi: 10.1111/j.1600-0765.1994.tb01090.x. [DOI] [PubMed] [Google Scholar]
- 14.Han Y, Shi W, Huang G T-J, Kinder Haake S, Park N-H, Kuramitsu H, Genco R J. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson B, Wilson M. Cytokine induction by bacteria: beyond lipopolysaccharide. Cytokine. 1996;8:269–282. doi: 10.1006/cyto.1996.0036. [DOI] [PubMed] [Google Scholar]
- 16.Hobbie S, Chen L M, Davis R J, Galan J E. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- 17.Huang G T-J, Kinder Haake S, Kim J W, Park N H. Differential expression of interleukin-8 and intercellular adhesion molecule-1 by human gingival epithelial cells in response to Actinobacillus actinomycetemcomitans or Porphyromonas gingivalis infection. Oral Microbiol Immunol. 1998;13:301–309. doi: 10.1111/j.1399-302x.1998.tb00711.x. [DOI] [PubMed] [Google Scholar]
- 18.Huang G T-J, Zhang X. Immunohistochemical analysis of interleukin-8 and intercellular adhesion molecule-1 in human gingival epithelium. Int J Oral Biol. 1999;24:7–16. [Google Scholar]
- 19.Krisanaprakornkit S, Kimball J R, Weinberg A, Darveau R P, Bainbridge B W, Dale B A. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun. 2000;68:2907–2915. doi: 10.1128/iai.68.5.2907-2915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuramitsu H K. Proteases of Porphyromonas gingivalis: what don't they do? Oral Microbiol Immunol. 1998;13:263–270. doi: 10.1111/j.1399-302x.1998.tb00706.x. [DOI] [PubMed] [Google Scholar]
- 21.Lamont R J, Chan A, Belton C M, Izutsu K T, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madianos P N, Papapanou P N, Sandros J. Porphyromonas gingivalis infection of oral epithelium inhibits neutrophil transepithelial migration. Infect Immun. 1997;65:3983–3990. doi: 10.1128/iai.65.10.3983-3990.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marlin S D, Springer T A. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1) Cell. 1987;51:813–819. doi: 10.1016/0092-8674(87)90104-8. [DOI] [PubMed] [Google Scholar]
- 24.Meyer-ter-Vehn T, Covacci A, Kist M, Pahl H L. Helicobacter pylori activates mitogen-activated protein kinase cascades and induces expression of the proto-oncogenes c-fos and c-jun. J Biol Chem. 2000;275:16064–16072. doi: 10.1074/jbc.M000959200. [DOI] [PubMed] [Google Scholar]
- 25.Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 1998;440:282–286. doi: 10.1016/s0014-5793(98)01461-6. [DOI] [PubMed] [Google Scholar]
- 26.Miyasaki K T. The neutrophil: mechanisms of controlling periodontal bacteria. J Periodontol. 1991;62:761–774. doi: 10.1902/jop.1991.62.12.761. [DOI] [PubMed] [Google Scholar]
- 27.Park N-H, Gujuluva C N, Baek J-H, Cherrick H M, Shin K-H, Min B-M. Combined oral carcinogenicity of HPV-16 and benzo(a)pyrene: an in vitro multistep carcinogenesis model. Oncogene. 1995;10:2145–2153. [PubMed] [Google Scholar]
- 28.Reddi K, Nair S P, White P A, Hodges S, Tabona P, Meghji S, Poole S, Wilson M, Henderson B. Surface-associated material from the bacterium Actinobacillus actinomycetemcomitans contains a peptide which, in contrast to lipopolysaccharide, directly stimulates fibroblast interleukin-6 gene transcription. Eur J Biochem. 1996;236:871–876. doi: 10.1111/j.1432-1033.1996.00871.x. [DOI] [PubMed] [Google Scholar]
- 29.Reddi K, Wilson M, Nair S, Poole S, Henderson B. Comparison of the pro-inflammatory cytokine-stimulating activity of the surface-associated proteins of periodontopathic bacteria. J Periodontal Res. 1996;31:120–130. doi: 10.1111/j.1600-0765.1996.tb00473.x. [DOI] [PubMed] [Google Scholar]
- 30.Sandros J, Madianos P N, Papapanou P N. Cellular events concurrent with Porphyromonas gingivalis invasion of oral epithelium in vitro. Eur J Oral Sci. 1996;104:363–371. doi: 10.1111/j.1600-0722.1996.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 31.Sandros J, Papapanou S J, Dahlen G. Poyphyromonas gingivalis invades oral epithelial cells in vitro. J Periodontal Res. 1993;28:219–226. doi: 10.1111/j.1600-0765.1993.tb01072.x. [DOI] [PubMed] [Google Scholar]
- 32.Sharp L, Poole S, Reddi K, Fletcher J, Nair S, Wilson M, Curtis M, Henderson B, Tabona P. A lipid A-associated protein of Porphyromonas gingivalis, derived from the haemagglutinating domain of the RI protease gene family, is a potent stimulator of interleukin 6 synthesis. Microbiology. 1998;144:3019–3026. doi: 10.1099/00221287-144-11-3019. [DOI] [PubMed] [Google Scholar]
- 33.Shin K H, Min B M, Cherrick H M, Park N-H. Combined effects of human papillomavirus-18 and N-methyl-N′-nitro-N-nitrosoguanidine on the transformation of normal human oral keratinocytes. Mol Carcinogenesis. 1994;9:76–86. doi: 10.1002/mc.2940090205. [DOI] [PubMed] [Google Scholar]
- 34.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokuda M, Duncan M, Cho M I, Kuramitsu H K. Role of Porphyromonas gingivalis protease activity in colonization of oral surfaces. Infect Immun. 1996;64:4067–4073. doi: 10.1128/iai.64.10.4067-4073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tokuda M, Karunakaran T, Duncan M, Hamada N, Kuramitsu H. Role of Arg-gingipain A in virulence of Porphyromonas gingivalis. Infect Immun. 1998;66:1159–1166. doi: 10.1128/iai.66.3.1159-1166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tonetti M S. Molecular factors associated with compartmentalization of gingival immune responses and transepithelial neutrophil migration. J Periodontal Res. 1997;32:104–109. doi: 10.1111/j.1600-0765.1997.tb01389.x. [DOI] [PubMed] [Google Scholar]
- 38.Tonetti M S, Imboden M A, Lang N P. Neutrophil migration into the gingival sulcus is associated with transepithelial gradients of interleukin-8 and ICAM-1. J Periodontol. 1998;69:1139–1147. doi: 10.1902/jop.1998.69.10.1139. [DOI] [PubMed] [Google Scholar]
- 39.Vernier A, Diab M, Soell M, Haan-Archipoff G, Beretz A, Wachsmann D, Klein J P. Cytokine production by human epithelial and endothelial cells following exposure to oral viridans streptococci involves lectin interactions between bacteria and cell surface receptors. Infect Immun. 1996;64:3016–3022. doi: 10.1128/iai.64.8.3016-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P L, Azuma Y, Shinohara M, Ohura K. Toll-like receptor 4-mediated signal pathway induced by Porphyromonas gingivalis lipopolysaccharide in human gingival fibroblasts. Biochem Biophys Res Comm. 2000;273:1161–1167. doi: 10.1006/bbrc.2000.3060. [DOI] [PubMed] [Google Scholar]
- 41.Wilson M, Reddi K, Henderson B. Cytokine-inducing components of periodonto-pathogenic bacteria. J Periodontal Res. 1996;31:393–407. doi: 10.1111/j.1600-0765.1996.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 42.Wilson M, Seymour R, Henderson B. Bacterial perturbation of cytokine networks. Infect Immun. 1998;66:2401–2409. doi: 10.1128/iai.66.6.2401-2409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Dong H, Kashket S, Duncan M J. IL-8 degradation by Porphyromonas gingivalis proteases. Microb Pathog. 1999;26:275–280. doi: 10.1006/mpat.1998.0277. [DOI] [PubMed] [Google Scholar]